Abstract
A better understanding of the molecules implicated in the growth and survival of glioblastoma (GBM) cells and their response to temozolomide (TMZ), the standard-of-care chemotherapeutic agent, is necessary for the development of new therapies that would improve the outcome of current GBM treatments. In this study, we characterize the role of pericentriolar material 1 (PCM1), a component of centriolar satellites surrounding centrosomes, in GBM cell proliferation and sensitivity to genotoxic agents such as TMZ. We show that PCM1 is expressed around centrioles and ciliary basal bodies in patient GBM biopsies and derived cell lines and that its localization is dynamic throughout the cell cycle. To test whether PCM1 mediates GBM cell proliferation and/or response to TMZ, we used CRISPR/Cas9 genome editing to generate primary GBM cell lines depleted of PCM1. These PCM1-depleted cells displayed reduced AZI1 satellite protein localization and significantly decreased proliferation, which was attributable to increased apoptotic cell death. Furthermore, PCM1-depleted lines were more sensitive to TMZ toxicity than control lines. The increase in TMZ sensitivity may be partly due to the reduced ability of PCM1-depleted cells to form primary cilia, as depletion of KIF3A also ablated GBM cells' ciliogenesis and increased their sensitivity to TMZ while preserving PCM1 localization. In addition, the co-depletion of KIF3A and PCM1 did not have any additive effect on TMZ sensitivity. Together, our data suggest that PCM1 plays multiple roles in GBM pathogenesis and that associated pathways could be targeted to augment current or future anti-GBM therapies.
Introduction
Glioblastoma (GBM) is the most common malignant brain tumor in adults with an extremely poor prognosis, mostly due to primary and acquired resistance to standard-of-care treatments, i.e., chemotherapy and irradiation [1], [2]. A more complete understanding of the cellular and molecular mechanisms that ensure the proliferation and survival of GBM cells despite aggressive therapies is crucial for the development of new treatment modalities that would further inhibit tumor progression.
GBM cells contain a matrix of pericentriolar material that comprises satellite proteins and surrounds centrioles, which form basal bodies during ciliogenesis. Pericentriolar proteins (PCPs) play crucial roles in cell division and survival, particularly in microtubule and actin organization, centrosome stability, centriolar duplication prior to mitosis [3], [4], [5], [6], [7], [8], [9], [10], [11], as well as ciliogenesis [12], [13], [14], [15], [16], [17], [18], [19], [20], in several cell types. Several PCPs are also closely associated with DNA repair proteins in centrosomes [21], [22], [23], [24] and undergo significant subcellular reorganization in response to various cellular stresses (e.g., heat shock and UV radiation [25], [26]), in some cases mediating DNA damage-induced centrosome amplification and tumorigenesis [27], [28]. However, the degree to which various PCPs affect GBM pathogenesis and sensitivity to therapy is poorly understood.
Pericentriolar material 1 (PCM1) is an essential PCP that is required for the cellular processes mentioned above in some normal and malignant cell types [25], [29], [30], [31], [32]. PCM1 has also been shown to bind to several centrosomal proteins, including pericentrin [30], [33] and Cep131/AZI1 [10], ensuring their correct localization [18]. However, the extent of PCM1’s expression in GBM cells or its function in GBM growth and/or stress response has never been studied. We recently reported that PCM1 localizes around the majority of centrioles and basal bodies situated at the base of primary cilia of patient-derived GBM cells in vitro [34]. In the present study, we further examine whether PCM1 is expressed in GBM biopsies and patient-derived GBM cell lines and study its localization during GBM cell division. In addition, we investigate the in vitro consequences of PCM1 depletion on the localization of other PCPs, GBM cell ciliogenesis, cell proliferation, and sensitivity to standard-of-care chemotherapeutic agent temozolomide (TMZ).
Materials and Methods
Generation and Maintenance of PCM1 and KIF3A-Depleted Cell Lines
The primary cell lines used in this study, Line 0 (L0; 43-year-old man) and mCherry-expressing SN186 (S3; 75-year-old man) [34], were isolated from human GBM tumors [35], [36], [37], [38], [39] and cultured as previously described [34]. Briefly, cells were grown as floating spheres and maintained in DMEM/F12 medium supplemented with 2% B27, 1% penicillin-streptomycin, 20 ng/ml human EGF, and 10 ng/ml human bFGF. When the spheres reached approximately 150 μm in diameter, they were enzymatically dissociated by digestion with Accumax (Innovative Cell Technologies, Inc.) for 10 minutes at 37°C. Cells were washed, counted using Trypan blue to exclude dead cells, and replated in fresh medium supplemented with hEGF and bFGF.
To generate PCM1-depleted patient-derived GBM cell lines, we screened and identified CRISPR/Cas9-encoding plasmids containing a GFP reporter gene that could target human PCM1 [Sigma-Aldrich; CRISPR/Cas-GFP vector (pU6-gRNA-CMV-Cas9:2a:GFP); primer pair ID: HS0000328679; PCM1 gRNA target sequence: GACTCCGGAGAAATATCATTGG]. We used the same approach to generate GBM cell lines depleted of KIF3A [Sigma-Aldrich; CRISPR/Cas-GFP vector (pU6-gRNA-CMV-Cas9:2a:GFP); primer pair ID: HS0000342157; KIF3A gRNA target sequence: GGTCATATTGCAAAAGCGGAGG]. Clones depleted of both PCM1 and KIF3A were generated by co-transfection. For transfection experiments, GBM cells were grown on 10-cm2 plates and transfected (Lipofectamine 2000; Life Technologies) at 60% to 70% confluence with 0.5 μg/ml of the CRISPR/Cas9-encoding plasmid DNA. Twenty-four to 48 hours after transfection, GFP-positive cells were sorted as individual clones into 96-well plates containing 250 μl of DMEM/F12 medium supplemented with hEGF and bFGF using a BD FACS Aria II Cell Sorter (BD Biosciences, San Jose, CA), excluding cell debris from the analysis by forward- and side-scatter gating. Stable cell lines from each GFP-positive clone were then expanded and screened for presence or absence of PCM1 or KIF3A proteins by immunostaining and Western blotting (WB). GFP-positive clones with immunopositive PCM1 clusters and detectable PCM1 levels by WB were termed PCM1-positive control clones (L0 clone D5 and S3 clone F11), whereas those with no immunopositive PCM1 clusters and undetectable PCM1 levels by WB were designated PCM1-depleted clones (L0 clone F9 and S3 clone B9). Further, DNA sequencing around the CRISPR/Cas9 target site in exon 4 of PCM1 of S3 clones revealed significant base pair deletions and point mutations in the PCM1-depleted clone B9 compared with controls (data not shown).
For immunostaining, once cells formed spheres greater than 100 μm in diameter in each well, the spheres were mechanically dissociated, replated, and expanded into 24-well plates on glass coverslips in DMEM/F12 medium supplemented with 5% FBS. After 2 to 3 days, cells were fixed with 4% paraformaldehyde in 0.1 M phosphate buffer (4% PFA) for immunohistochemical analysis as described below. For WB, cells were expanded in T25 flasks for 1 week prior to analysis and harvested as described below.
Western Blot
S3 and L0 cells were harvested and lysed in 1× RIPA buffer (Cell Signaling) containing 1× protease inhibitor cocktail (Sigma), phosphatase inhibitor cocktails 1 and 2 (Sigma), and 1× phenylmethanesulfonyl fluoride (Sigma). For WB of PCM1, 20 μg of total protein lysate per lane was separated on a 4% to 12% Bis-Tris gel (NuPage). Proteins were blotted onto PVDF membranes using iBlot (program 3 for 8 minutes; Invitrogen). Blots were blocked in 5% nonfat dry milk (NFDM) in 1× TBS with 0.1% Tween (TBST) for 1 hour and then incubated in primary anti-PCM1, anti-KIF3A, or anti–beta-actin antibodies in 2.5% NFDM in 1× TBST for 24 hours at 4°C. Blots were then rinsed and probed in the appropriate HRP-conjugated secondary antibody (1:10,000; BioRad; cat # 170-6515; lot # 310008703) for 1 hour at room temperature in 2.5% NFDM in 1× TBST. Finally, blots were rinsed in 1× TBS and developed using an ECL-plus chemiluminescence kit (Amersham), and images were captured using an AlphaInnotech Fluorchem Q Imaging System (Protein Simple).
Antibodies, Immunostaining, and Image Acquisition
The following primary antibodies were used for immunocytochemistry (ICC), immunohistochemistry (IHC), and WB: mouse anti–acetylated alpha-tubulin (aa-tub) [1:3000 (ICC); Sigma (cat # T6793; lot # 088K4829)], mouse anti–gamma-tubulin (gTub) [1:2000 (ICC); Sigma (cat # T6557; lot # 072M4808)], rabbit anti-PCM1 [1:1000 (ICC/IHC/WB); Bethyl Laboratories (cat # A301-150A; lot # A301-150A-1)], rabbit anti-AZI1 [1:1000 (ICC); Abcam (cat # ab84864; lot # GR13395-12)], rabbit anti-Cep290 [1:1000; Abcam (cat # ab84870; lot # GR32927-13)], rabbit anti-Ki67 [1:200 (ICC); Vector (cat # VP-RM04; lot # V0523)], rabbit anti-KIF3A [1:1000 (WB); Abcam (cat # ab11259)], rabbit anti-PHH3 [1:1000 (ICC); Millipore (cat # 06-570; lot # 2066052)], and rat anti-BrdU [1:1000 (ICC); Accurate (cat # OBT0030; lot # H7227)]. For BrdU staining, cells were rinsed with 0.9% NaCl, followed by 2 M HCl for 20 minutes at 37°C to denature DNA prior to immunostaining. Secondary antibodies were species-specific and conjugated with fluorescent tags [1:400 (ICC/IHC); Jackson Immunoresearch]. Stained sections and cells were coverslipped with Prolong Gold antifade media containing DAPI (Invitrogen). For some of our analyses, sections of a tumor formed from an S3 cell intracranial xenograft in a male NOD/SCID mouse were obtained from our previous study [34]. Immunostained cells and sections were examined and imaged using an Olympus IX81-DSU confocal microscope fitted with a 60× water objective, and all images were captured as z-stacks (0.5-μm steps). Nonconfocal images and time-lapse videos were acquired using a Zeiss Axio Observer D1 inverted microscope equipped with an AxioCam MRm camera and analyzed using ZEN software.
Cell Growth, Viability, and Survival Assays
For cell proliferation assay, cells (5 × 104) were seeded in 1 ml of growth media per well in 24-well plates for each experimental group. Cells were enzymatically dissociated and counted after 5 days using a hemocytometer. Cells were stained with 0.04% Trypan blue at the time of dissociation to identify dead (Trypan blue–positive) cells. These cells were counted, and the percentage of dead cells was calculated for each group at the time of each passage.
To assess the effects of in vitro TMZ treatment on cell viability, 10,000 cells per experimental group were plated per well into 96-well cell culture plates and treated daily for 5 days with TMZ (50 or 100 μM, Tocris, Ellisville, MO) in 10% DMSO. Vehicle-control wells received equivalent volumes of DMSO. Cell viability was assessed with the 3-(4, 5-dimethylthiazole-yl)-2, 5-diphenyl tetrazolium bromide (MTT) assay as previously described [34]. The number of viable cells was determined through optical density measurements (by measuring absorbance at 570 nm).
To determine the percentage of cleaved caspase 3–positive cells, cells (5 × 105) were pelleted, resuspended in 1 ml of 4% PFA, and incubated for 15 minutes at room temperature. The suspension was then centrifuged and the pellet washed twice with PBS. Labeling was performed by incubating the cells in rabbit anti–cleaved caspase-3 primary antibody (Asp175) [1:1000; Cell Signaling (cat # 9661S; lot # 42)] overnight at 4°C, followed by incubation in an anti-rabbit, FITC-tagged secondary antibody [1:400 (ICC/IHC); Jackson Immunoresearch] for 1 hour at room temperature. Cells were then washed in PBS, DAPI-stained, and analyzed by flow cytometry. Negative control samples incubated with antirabbit, fluorescent-tagged secondary antibodies were run in parallel.
Data Analysis
Statistical analyses were performed using GraphPad Prizm 5.0 (GraphPad Software, La Jolla, CA). In all analyses, P values less than .05 were considered significant. Comparisons between groups were performed using either a Student's t test or one-way analysis of variance (ANOVA) followed by Tukey's post hoc analysis, as indicated.
Results
Expression of PCM1 in Patient-Derived GBM Cell Lines, Associated Tumors, and Patient Biopsies
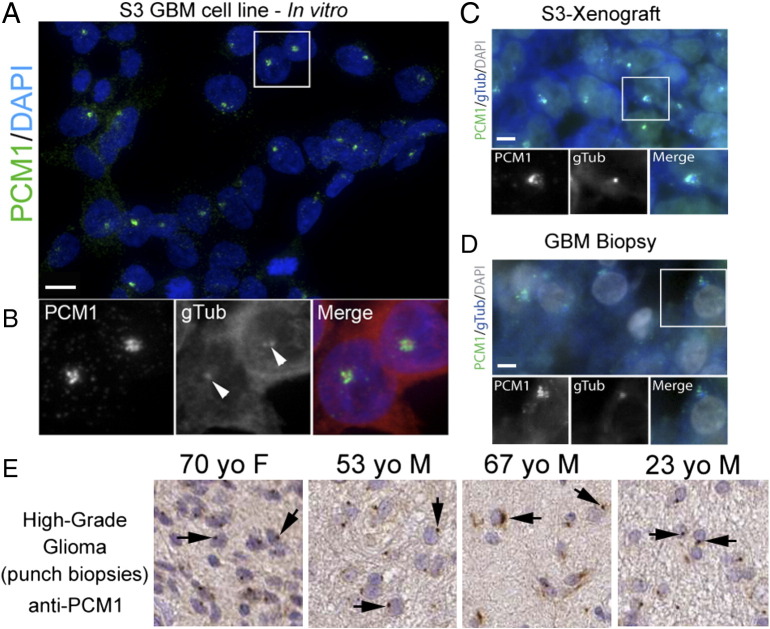
In other cancer cell types (e.g., HeLa, U2OS [3], [7], [25]), PCM1 expression appears in the form of granules or puncta that surround centrioles, structures typically containing gTub. To identify PCM1 in two patient-derived GBM cell lines, L0 and S3, we co-immunostained cells with antibodies raised against PCM1 and gTub. We found small PCM1-positive granules typically concentrated around gTub-positive centrioles in the majority of cells from the GBM cell lines we examined, e.g., S3 (Figure 1, A and B). A similar staining pattern was observed within the tumors that formed following the intracranial xenotransplantation of these S3 cells into mice (Figure 1C) [34]. Furthermore, examination of freshly stained or archived GBM biopsies from patients of various ages [40], [41] also showed clusters of PCM1 granules in every sample (Figure 1, D and E). Thus, PCM1 expression is preserved and ubiquitous in patient-derived GBM cell lines and specimens.
Figure 1.
PCM1 is widely expressed in cells from GBM patient tumor-derived cell lines and biopsies. (A) Example of cells from the S3 cell line immunostained for PCM1 (green). Nuclei are counterstained with DAPI (blue). (B) Magnification of the inset in A. Cells were also immunostained for gTub (red in merged). Note the PCM1-positive cluster of granules surrounding gTub-positive puncta (arrowheads). (C) Immunostaining for PCM1 (green) in a tumor resulting from the xenotransplantation of S3 cells (stained mouse brain section from [34]). (D) Example of a GBM biopsy immunostained for PCM1 (green) with nuclei counterstained with DAPI (blue). The boxed inset shows clustering of PCM1 around gTub + puncta. (E) Examples of archived biopsies from five individuals of various ages with high-grade glioma immunostained for PCM1 (brown). PCM1-positive puncta (arrows) are observed across samples. Images were derived from the Human Protein Atlas (www.proteinatlas.org). Scale bars in A = 5 μm; C, D = 5 μm.
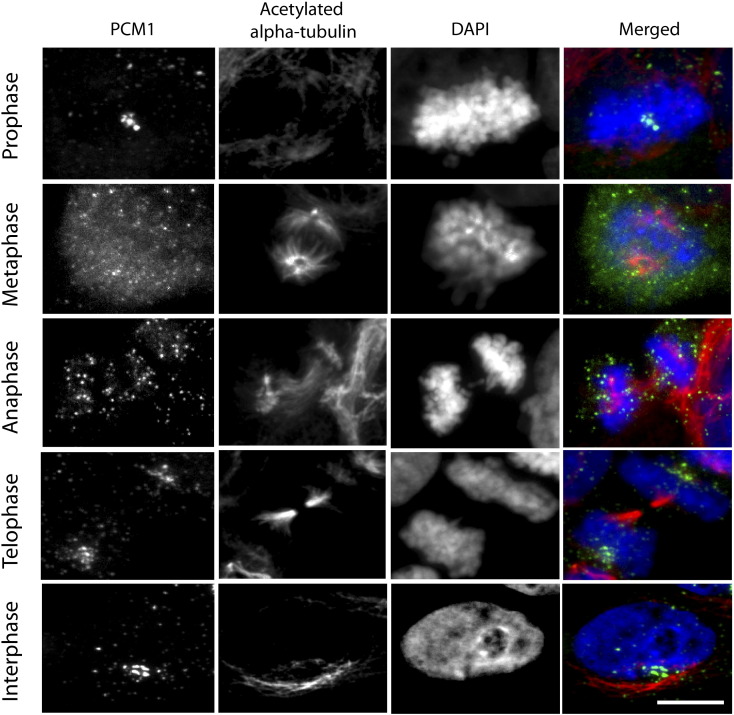
Previous studies have reported that PCM1 subcellular localization is dynamic throughout the cell cycle, with the distribution pattern of PCM1 granules changing as cells proceed from interphase to mitotic telophase [7], [29], [30], [31], [42]. Similarly, in both patient-derived GBM L0 (Figure 2) and S3 (Supplemental Figure S1) cell lines, we found that PCM1 granules were concentrated in clusters during interphase and dispersed around the nucleus during all stages of mitosis. Time-lapse imaging of live GBM cells transfected with a plasmid encoding EGFP-tagged human PCM1 (gift from S. Shi [43]) further illustrates the dynamic nature of these PCM1 granules that are in continuous motion (Supplemental Video 1, Supplemental Video 2), as has been described in Xenopus A6 cells [14]. Together, these data show that PCM1’s localization is dynamic and undergoes subcellular changes during GBM cell division, suggesting that PCM1 may play a role in GBM cell proliferation.
Figure 2.
PCM1 granules in L0 GBM cells display a dynamic distribution throughout the cell cycle. Co-immunostaining of L0 cells for PCM1 (green) and aa-tub (red) with nuclei counterstained with DAPI (blue). Cells in various stages of the cell cycle (prophase, metaphase, anaphase, telophase, and interphase) show a cell cycle–dependent assembly and disassembly of PCM1 granules and aggregates. Scale bar =5 μm.
Depletion of PCM1 Using CRISPR/Cas9 Genome Editing in GBM Cell Lines and AZI1 Centriolar Satellite Protein Organization
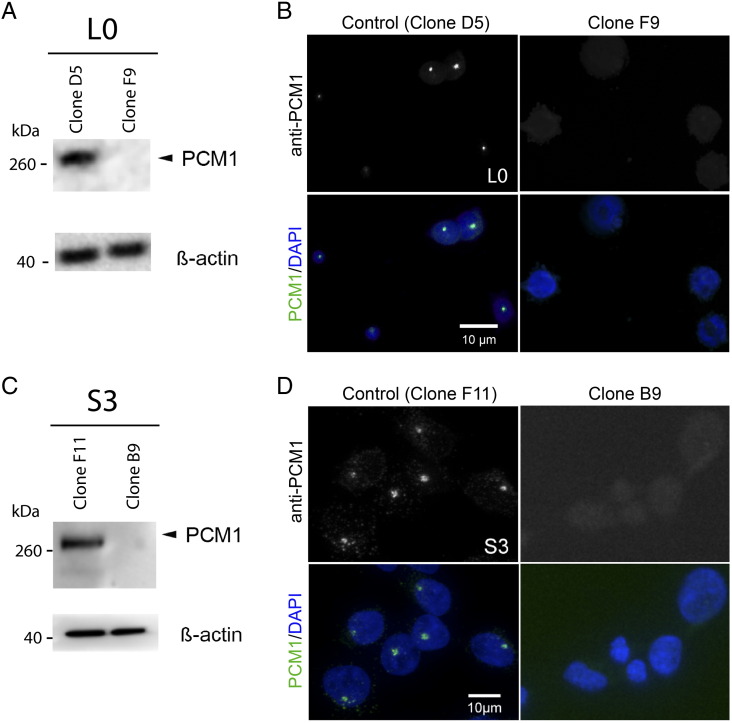
To investigate the functions of PCM1 in GBM cell proliferation, we used the CRISPR/Cas9 genome editing technology to deplete PCM1 expression from our L0 and S3 GBM cell lines. We transfected GBM cells with a plasmid encoding RNA-guided Cas9 endonuclease with a GFP reporter and a trans-activating guide RNA designed against human PCM1 under the control of a U6 promoter. Single GFP-positive clones were isolated using FACS and expanded. To identify PCM1-depleted clones, we screened cell lines derived from those GFP-positive clones using WB and ICC for PCM1. For both L0 and S3 cell lines, we identified clones that were characterized by undetectable PCM1 (e.g., L0 clone F9, S3 clone B9; Figure 3, A and C) as well as clones that expressed PCM1 (“PCM1-positive control” L0 clone D5, S3 clone F11; Figure 3, A and C) using WB. Immunostaining using an antibody against PCM1 revealed PCM1-positive clusters in the majority of cells derived from PCM1-positive control clones but no PCM1-positive clusters in cells originating from PCM1-depleted clones for L0 or S3 cell lines (Figure 3, B and D).
Figure 3.
PCM1-depleted GBM cell lines generated using CRISPR/Cas9. A CRISPR/Cas9 plasmid co-expressing a GFP reporter for Cas9 and gRNA directed against human PCM1 was used to transfect L0 and S3 GBM cells and generate cell clones depleted of PCM1. GFP-positive clones were FAC-sorted and expanded for screening by WB and immunostaining. (A) WB of L0 lysates shows that, compared with clone D5, clone F9–derived cells displayed an absence of a band for PCM1. β-Actin was used as a loading control. (B) Immunostaining of L0 clones shows the presence of PCM1-positive clusters (green) in clone D5 but not clone F9–derived cells (right panels). (C, D) Similarly, S3 cells generated from clone B9 lacked detectable PCM1 by WB (C) and ICC (D) compared with clone F11.
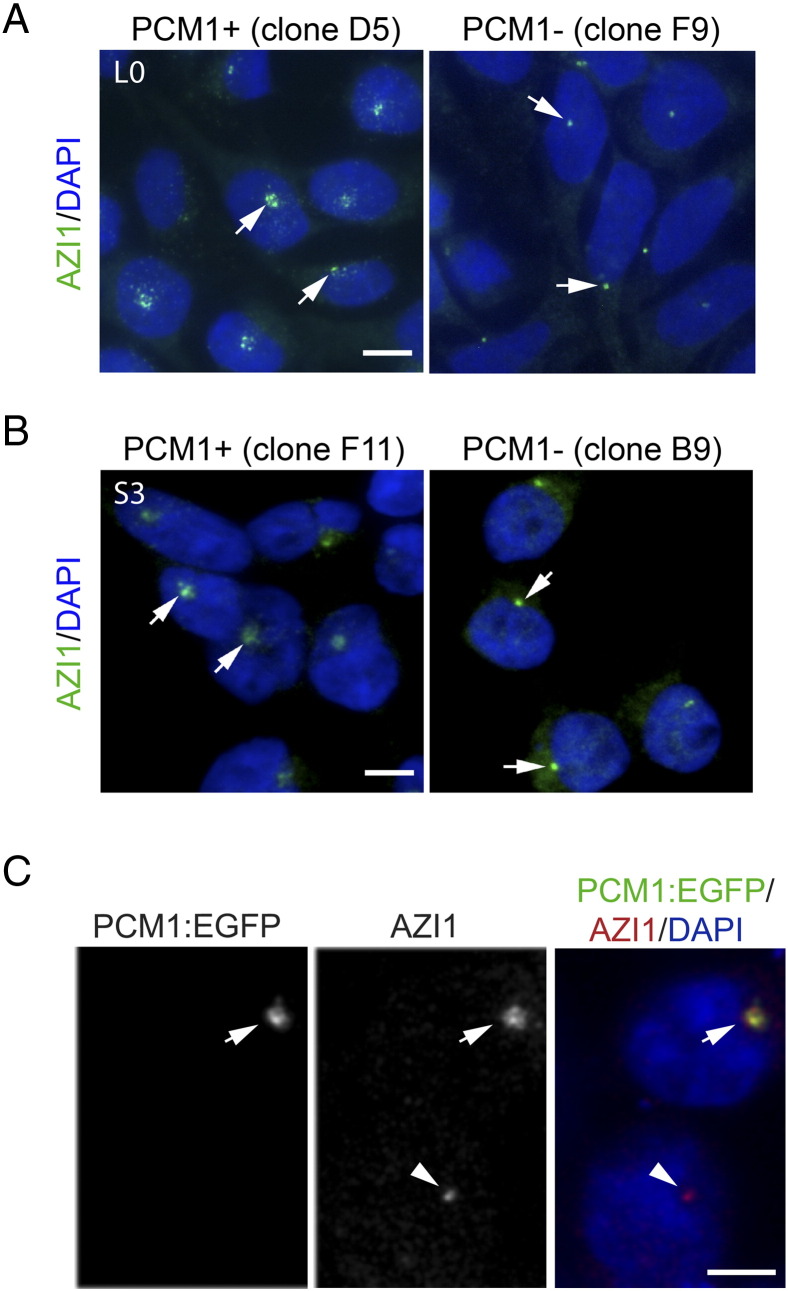
We next examined the localization of known PCM1-associated proteins. We found that L0 and S3 PCM1-depleted lines displayed loss of AZI1 from pericentriolar satellites, whereas its localization at the centriolar core remained unaffected (Figure 4, A and B), observations which are consistent with those in other studies showing that PCM1 knockdown using siRNA leads to the abnormal localization of centrosomal proteins [18]. Surprisingly, not all centrosomal proteins' localization was altered following PCM1 depletion. We observed similar expression patterns for Cep290, a centrosomal protein that interacts with AZI1 and PCM1 [10], in control and PCM1-depleted cells (Supplemental Figure S2). Importantly, transfecting PCM1-depleted GBM cells with a plasmid encoding wild-type, EGFP-tagged human PCM1 restored the AZ1 centriolar satellite localization pattern compared with untransfected cells (Figure 4C). Together, these data support a critical role for PCM1 in organizing and recruiting certain proteins to the centriolar satellites of GBM cells.
Figure 4.
AZI1 recruitment to GBM centriolar satellites is dependent on PCM1. Immunostaining for AZI1 (green; arrows) with nuclei counterstained with DAPI (blue) for (A) L0 cells derived from PCM1-positive control clone D5 and PCM1-depleted clone F9 and (B) S3 cells derived from PCM1-positive control clone F11 and PCM1-depleted clone B9. In both lines, note the reduced localization of AZI1 granules at pericentriolar satellites in PCM1-depleted compared with wild-type clones. (C) S3 cells derived from PCM1-depleted clone B9 were transfected with a plasmid encoding human wild-type EGFP-tagged PCM1 and immunostained for AZI1. Arrows show a transfected cell expressing EGFP-tagged PCM1 (green) co-localizing with AZI1 (red). The arrowhead points to AZI1 expression in an adjacent nontransfected cell. Scale bars in A, B, C = 5 μm.
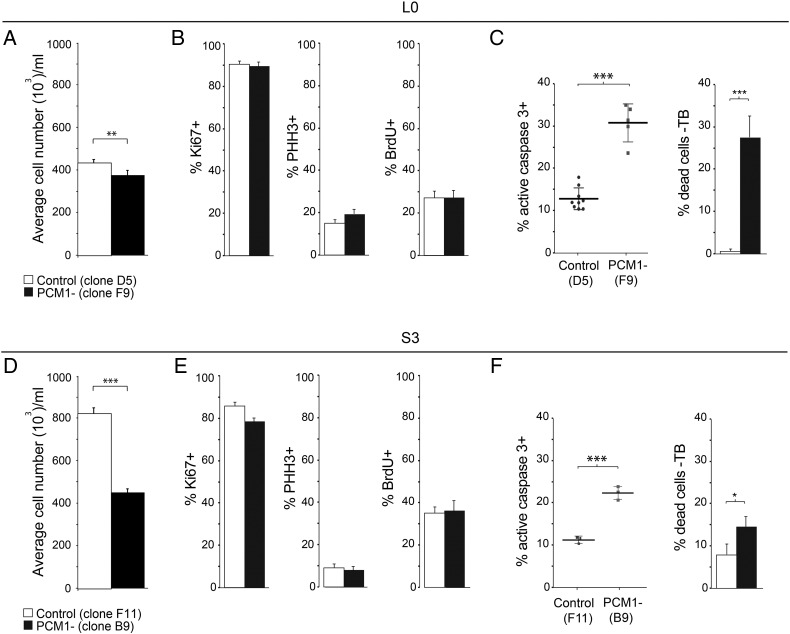
Decreased Cell Proliferation Through Increased Apoptotic Cell Death Following PCM1 Depletion
Considering the reported influence of PCM1 on microtubule organization in other cell types [30], its dynamic localization around the centrosome during cell cycle progression (Figure 2, Supplemental Figure S1), and the centriolar satellite changes observed following its depletion in GBM cells (Figure 4) [18], we next examined whether the depletion of PCM1 from L0 and S3 GBM cells affected these cells' baseline proliferation. We observed a significant decrease in cell proliferation for PCM1-depleted L0 (Figure 5A) and S3 (Figure 5D) cell lines compared with cells derived from PCM1-positive control clones. To assess whether this decrease was due to aberrant cell division, we examined the distribution of these cells among the various phases of the cell cycle using immunostaining for markers of actively proliferating cells (Ki67+), as well as cells in the G2/M (phospho-histone H3 + (PHH3+)) and S (BrdU+) phases (Supplemental Figure S3A). We found no significant differences in cell cycle distribution between cell populations derived from PCM1-positive control clones and those derived from PCM1-depleted clones for both L0 (Figure 5B) and S3 (Figure 5E) cell lines.
Figure 5.
PCM1 depletion results in reduced GBM cell proliferation due to increased apoptotic cell death. (A, D) Average cell numbers per well 5 days after plating for L0 (A) and S3 (D) cell lines derived from PCM1-positive control clones (open-filled bars) or PCM1-depleted clones (black-filled bars). (B, E) Percentages of cells from control or PCM1-depleted clones that were Ki67+, PHH3+, or BrdU+ for L0 (B) and S3 (E) cell lines 5 days after plating. (C, F) Percentages of cleaved caspase 3–positive immunolabeled cells detected by FACS and percentages of Trypan blue–labeled cells 5 days after passaging for L0 (C) and S3 (F) cell lines. *P < .05, **P < .01, ***P < .001, Student's t test.
We next assessed apoptotic cell death by flow cytometry using active caspase 3 immunostaining and observed an increased percentage of apoptotic cells in PCM1-depleted clones compared with PCM1-positive control clones for both L0 (Figure 5C; Supplemental Figure S3B) and S3 (Figure 5F) cell lines. The percentage of dead cells assessed by Trypan blue staining was also increased in PCM1-depleted clones compared with control clones (Figure 5, C and F). Notably, the percentages of active caspase 3–positive and Trypan blue–positive dead cells were higher in L0 than in S3 PCM1-depleted cell line, despite a smaller decrease in cell proliferation for L0 than for S3 cell line after PCM1 depletion. This discordance could be due to a loss of dead/fragmented S3 cells during the dissociation and staining process. Together, these results suggest that the decrease in cell proliferation following PCM1 depletion is due to an increase in apoptotic cell death rather than a change in cell cycle phase distribution, a result that has been observed in multiple other cancer cell lines following siRNA-mediated knockdown of PCM1 [7], [30].
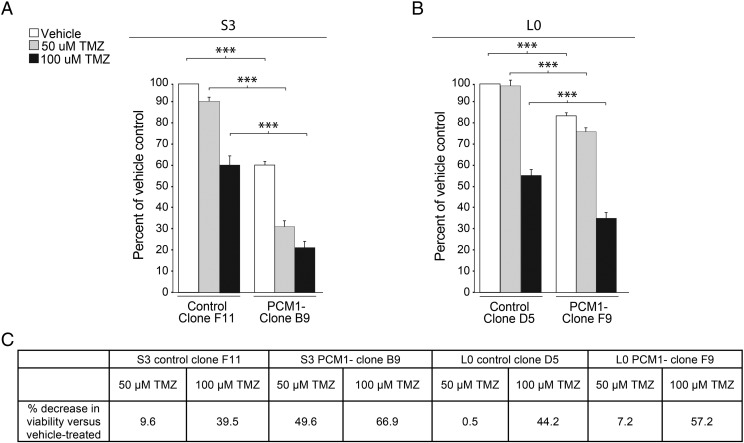
Sensitivity of GBM Cells to TMZ After PCM1 Depletion
Because the loss of PCM1 induced GBM cell apoptosis, we hypothesized that PCM1 depletion prevents GBM cells from engaging in PCM1-associated survival pathways, thus rendering those cells more sensitive to TMZ toxicity. To test this hypothesis, we examined whether TMZ treatment differentially affected the viability of control and PCM1-depleted cells. Consistent with our cell proliferation data shown above, our MTT assay results show that baseline (vehicle-treated) cell viability is significantly decreased in PCM1-depleted S3 (by ~40%; Figure 6A) and L0 (~15%; Figure 6B) cells compared with their respective controls. Notably, we found that daily treatment with 50 μM TMZ for 5 days significantly decreased the viability of S3 control cells by ~10% and the viability of S3 PCM1-depleted cells by ~50%. The same 50-μM TMZ treatment course also decreased the viability of L0 control cells by ~44% as well as the viability of L0 PCM1-depleted cells by ~57% (Figure 6C). Furthermore, using an increased TMZ concentration of 100 μM, PCM1-depleted cells' viability was reduced by 67% for S3 cells and by 57% for L0 cells compared with 40% and 44%, respectively, for the PCM1-positive control cells. Thus, these data suggest that PCM1-depleted cells are more sensitive to TMZ exposure than control cells. Together, the induced apoptosis and increased sensitivity to TMZ resulting from PCM1 depletion have significant additive effects in inhibiting GBM cell proliferation.
Figure 6.
PCM1-depleted cells are more sensitive to TMZ-associated toxicity than control cells. MTT assays were used to measure cell viabilities after five daily exposures to TMZ (50 or 100 μM) or vehicle (DMSO) for S3 (A) and L0 (B) cell lines. Bar graphs show the percentages of viable cells derived from control clones (S3-F11 and L0-D5) or PCM1-depleted clones (S3-B9 and L0-F9) after exposure to vehicle or TMZ. Data were normalized to control-vehicle groups for each cell line. (C) Table shows the percent decrease in cell viability that was calculated relative to the vehicle groups for control and PCM1-depleted cell lines. ***P < .001, ANOVA with Tukey's post hoc.
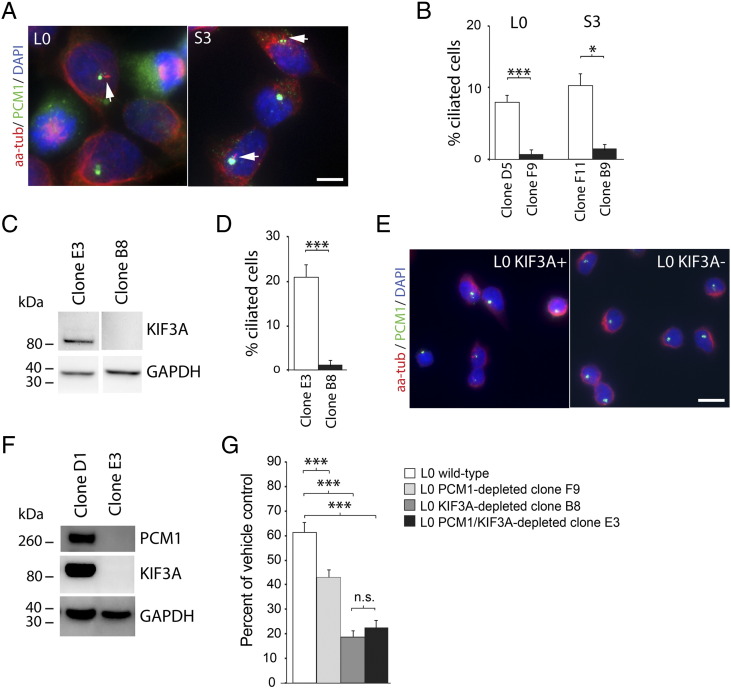
We next investigated possible mechanisms that might underlie the increased sensitivity of GBM cells to TMZ following PCM1 depletion. In multiple cell types, PCM1 has been reported to promote the biogenesis of cilia [18], [25], [32], [44], [45], [46], [47], [48], [49], cellular “signaling antennae” structures typically formed during the G0/G1 phases of the cell cycle. Notably, in U2OS osteosarcoma cells, various cellular stresses including UV radiation, transcriptional inhibitors, and heat shock have been reported to stimulate ciliogenesis via PCM1 [25]. We visualized the primary cilia of S3 and L0 cells using an antibody against aa-tub, a molecule that is enriched in the ciliary axonemes of GBM cells [34], [35] (Figure 7A), and found a significant decrease in the percentage of ciliated cells in L0 and S3 PCM1-depleted cell lines compared with control cell lines (Figure 7B). We then examined whether the increased sensitivity of PCM1-depleted cells to TMZ might be associated to the reduced ability of these cells to form cilia. We compared the TMZ sensitivity of L0 control and PCM1-depleted clones to that of another line of L0 GBM cells in which we used CRISPR/Cas9 genome editing technique to deplete KIF3A, a kinesin motor protein essential for GBM ciliogenesis [34] (Figure 7C). L0 GBM cells depleted of KIF3A were rarely ciliated (Figure 7D), while retaining a normal PCM1 expression pattern, compared with L0 KIF3A-positive control cells (Figure 7E). We found that five daily 100-μM TMZ treatments significantly reduced the viability of both PCM1 and KIF3A-depleted L0 cells compared with vehicle-control cells, and KIF3A-depleted cells were more sensitive than PCM1-depleted cells to TMZ (Figure 7G). To determine whether there were additive effects of KIF3A and PCM1 co-depletion, we generated L0 cells depleted of both PCM1 and KIF3A using CRISP/Cas9 (Figure 7F), and although these cells were also significantly more sensitive to TMZ compared with control cells, this increase in TMZ sensitivity was not significantly different from that of KIF3A-depleted cells (Figure 7G). Thus, there were no additive effects of PCM1 and KIF3A co-depletion on TMZ sensitivity, and the increased TMZ sensitivity resulting from the double depletion was largely due to the loss of KIF3A/cilia and not to extraciliary effects resulting from PCM1 depletion. These results, and the fact that KIF3A/cilia-depleted cells retained a normal PCM1 expression pattern, suggest that the loss of cilia in PCM1-depleted cells is associated to the increased TMZ sensitivity of those cells. These data also suggest that the loss of genes critical for ciliogenesis may lead to a heightened sensitivity of certain GBM cells to TMZ.
Figure 7.
Loss of cilia in PCM1- and KIF3A-depleted GBM cells is associated with increased TMZ sensitivity. (A) Examples of L0 and S3 cell cilia (arrows) co-immunostained for aa-tub (red), enriched in the ciliary axoneme, and PCM1 granules (green), enriched in the region surrounding the basal bodies of cilia. Nuclei were counterstained with DAPI (blue). (B) Percentages of aa-tub–positive cilia on L0 cells, derived from PCM1-positive control clone D5 or PCM1-depleted clone F9, and S3 cells, derived from PCM1-positive control clone F11 and PCM1-depleted clone B9. (C) A CRISPR/Cas9 plasmid co-expressing a GFP reporter for Cas9 and gRNA directed against human KIF3A was used to transfect L0 GBM cells and generate cell clones depleted of KIF3A. GFP-positive clones were FAC-sorted and expanded for screening by WB and immunostaining. WBs of L0 lysates show that, compared with clone E3, clone B8–derived cells displayed an absence of a band for KIF3A. GAPDH was used as a loading control. (D) Percentages of aa-tub–positive cilia on L0 cells derived from KIF3A-positive control clone E3 or KIF3A-depleted clone B8. (E) Examples of L0 KIF3A-positive control and KIF3A-depleted cells generated using CRISPR/Cas9 and co-immunostained for aa-tubulin (red) and PCM1 granules (green). (F) CRISPR/Cas9 plasmids co-expressing a GFP reporter for Cas9 and gRNA directed against human KIF3A or human PCM1 were used to co-transfect L0 GBM cells and generate cell clones depleted of both PCM1 and KIF3A. GFP-positive clones were FAC-sorted and expanded for screening by WB and immunostaining. WBs of L0 lysates show that, compared with clone D1, clone E3–derived cells displayed an absence of bands for both PCM1 and KIF3A. GAPDH was used as a loading control. (G) Bar graphs show the percentages of L0 viable cells derived from control (wild type), PCM1-depleted (clone F9), KIF3A-depleted (clone B8), and PCM1/KIF3A-depleted (clone E3) cell lines after five daily exposures to 100 μM TMZ or DMSO vehicle. *P < .05, ***P < .001, n.s. = nonsignificant; (B) and (D) Student's t test; (G) ANOVA with Tukey's post hoc. Scale bars in A = 5 μm; E = 10 μm.
Discussion
Very little is known about the role of PCM1 in brain tumors. Here, we characterize the expression of PCM1 in GBM cells and the effects of depleting PCM1 on GBM centriolar satellite protein expression, ciliogenesis, cell proliferation, and sensitivity to TMZ. PCM1 is widely expressed around the centrioles in GBM cell lines and biopsies. As others have reported in normal and malignant cell types (e.g., HeLa, osteosarcoma cells, myoblasts) [29], [30], we show that PCM1 distribution is dynamic throughout the cell cycle in GBM cells, with PCM1 granules being particularly concentrated around the centrosomes during interphase and dispersed around the cytoplasm during mitosis. PCM1’s role in recruiting other proteins to centriolar satellites appears to be preserved in GBM cells. Indeed, PCM1 has been shown to bind and recruit AZ1 to centriolar satellites [10], [50]. We found that PCM1-depleted cells displayed a loss of AZI1 from pericentriolar satellites, whereas AZI1’s localization at the centriolar core remained unaffected, similarly to previous reports in Hela and U2OS osteosarcoma cells [10]. The presence of small concentrated clusters of AZI1 observed following PCM1 depletion suggests that the localization of AZI1 is not entirely dependent on PCM1 but also other centrosomal proteins. For instance, Cep290 interacts with AZI1 and promotes its localization in other cell types [10]. We found that Cep290’s expression pattern was unaltered following PCM1 depletion; thus, Cep290 could help promote the localization of AZI1 to the centriolar core in PCM1-depleted GBM cells. Interestingly, Kim et al. [32] showed that hTERT-RPE cells depleted of PCM1 using small interfering RNA displayed disrupted Cep290 localization at centriolar satellites, suggesting that PCM1’s function may differ between GBM cells and other cell types. We also observed that exogenously expressing wild-type PCM1 in PCM1-depleted cells restored the pattern of AZI1 localization around the centriolar satellites. This observation is consistent with recent findings in human RPE1 cells showing that the amino-terminal domain of PCM1 is required for AZI1 centriolar satellite localization [18]. Together, our data show that PCM1, a ubiquitous centrosomal protein in GBM cells, is dynamically localized throughout cell division and regulates the recruitment of other centriolar satellite proteins such as AZI1.
Surprisingly, depletion of PCM1 did not lead to changes in GBM cell cycle phase distribution, despite the dynamic distribution of PCM1 observed throughout the cell cycle. Rather, we observed a decrease in net cell proliferation due to induced apoptotic cell death, which is consistent with results obtained following siRNA-mediated knockdown of PCM1 in other cell types [7]. Interestingly, the apoptotic cell death caused by reducing PCM1 levels using small interfering RNA in other cancer cell lines was minimal compared with that caused by knocking down other centrosomal proteins (e.g., ninein, aurora-A, TOG, and TACC3) [7]. These findings suggest that targeted loss or inhibition of other GBM centrosomal proteins in addition to PCM1 could result in further cell death. While our data indicate a role for PCM1 in GBM cell proliferation, the exact mechanisms require further investigation.
We observed that PCM1 depletion resulted in reduced ciliogenesis in both L0 and S3 cells, result which is consistent with the role of PCM1 in cilia biogenesis that has been found in other cell types [18], [20], [25], [32], [44], [45], [46], [47], [48], [49]. Although some studies have reported that primary cilia are either rarely or poorly formed in many of the widely used GBM lines [51], [52], examination of more recently derived primary lines and biopsies has revealed the presence of structurally intact cilia with PCM1 around their basal bodies [34], [35]. PCM1, through its interaction with other centriolar proteins, can promote various critical ciliogenesis processes such as ciliary vesicle docking to the mother centriole (e.g., Rab8-positive vesicles), displacement of cilia inhibitory proteins, and recruitment of other essential ciliogenic proteins [18], [25], [32], [44], [45], [46], [47], [48], [49]. Similarly, loss of AZI1 has also been found to inhibit ciliogenesis [12], [15], [16], and the abnormal localization of AZI1 could have contributed to the decreased ciliation of GBM cells following PCM1 depletion. Our finding that PCM1 is required for GBM ciliogenesis may be significant if GBM cilia serve as venues for promoting tumorigenesis, as has been observed in other cancer types (e.g., medulloblastoma, basal cell carcinoma, choroid plexus tumors) [53], [54], [55]. Our previous findings show that GBM cells are capable of forming ciliated progeny and that, in some GBM cell lines (e.g., L0), primary cilia transduce signals, such as Sonic hedgehog, that stimulate GBM cell proliferation [34].
For both L0 and S3 GBM cell lines, PCM1-depleted cells showed enhanced TMZ sensitivity compared with the respective PCM1-positive control cells. Given the ubiquity of PCM1 across lines and biopsies, it will be important to verify whether all GBM cell lines display higher TMZ sensitivity following PCM1 suppression in future studies. Our findings raise the possibility that PCM1 may play a role in survival-associated pathways in GBM cells. It is noteworthy that under starvation/stressed extracellular conditions, hedgehog signaling through mouse astrocyte primary cilia promoted the survival of astrocytes, whereas disrupting the formation of astrocyte cilia increased apoptosis under the same conditions [56]. Recent studies also suggest that stress/DNA damage response pathways engage mechanisms that lead to ciliogenesis [57]. For example, it was demonstrated that PCM1 subcellular localization is significantly altered in response to genotoxic stresses in U2OS cells [25]. That study reported that PCM1 granules undergo rapid displacement from centriolar satellites and centrosomes following exposure to heat shock, UV light, or transcriptional inhibitors such as DRB and Actinomycin D, which resulted in the formation of new cilia [25]. Thus, the reduced capacity of PCM1-depleted and KIF3A-depleted GBM cells to form cilia may render these cells less capable of responding to DNA-damaging reagents such as TMZ. Considering that KIF3A/cilia-depleted cells did not show a change in PCM1 expression pattern compared with control cells and PCM1 depletion did not have an additive effect on TMZ sensitivity with KIF3A depletion, the increased TMZ sensitivity of PCM1-depleted GBM cells may be caused by their reduced ability to form or sustain ciliogenesis; however, the mechanisms underlying this process still need to be elucidated.
In conclusion, the results of our studies highlight functional relationships between PCM1, ciliogenesis, and cell proliferation and sensitivity to TMZ in GBM. Future studies will need to examine the downstream molecular targets of PCM1 and whether disrupting PCM1 and other centrosomal proteins/associated pathways could further sensitize GBM cells to TMZ and/or other conventional therapies such as irradiation.
The following are the supplementary data related to this article.
Figure S1. PCM1 granules in S3 GBM cells display a dynamic distribution during the cell cycle. Co-immunostaining for PCM1 (green) and acetylated alpha-acetylated tubulin (red), with nuclei counterstained with DAPI (blue). Examples of staining of prophase, metaphase, anaphase, telophase and interphase of the cell cycle are shown. Scale bar = 5 μm.
Figure S2. Similar Cep290 subcellular distribution in PCM1-positive control and PCM1-depleted clones. Examples of S3 cells derived from PCM1-positive control clone F11 or PCM1-depleted clone B9 and immunostained for Cep290 (green), with nuclei counterstained with DAPI (blue). Scale bar = 5 μm.
Figure S3. Examples of Ki67, BrdU, and PHH3 immunostaining in L0 GBM cells and detection of active caspase 3-positive cells by flow cytometry. (A) Example of immunostaining of L0 cells derived from PCM1-positive control clone D5 and PCM1-depleted clone F9 for markers of cell cycle active phases (Ki67), G2/M (phospho-histone H3 (PHH3)), and S phase (BrdU). (B) Apoptotic cell death assessed by flow cytometry of L0 cells derived from PCM1-positive control clone D5 or PCM1-depleted clone F9 and labeled with rabbit active caspase 3 antibody followed by Alexa Fluor 488–conjugated anti-rabbit secondary antibody. Scale bar = 5 μm.
PCM1 granules in GBM cells display constant motion. Example of time-lapse recording of L0 cells transfected with GFP-tagged human PCM1 (green). Images were collected every 10 seconds for 4 minutes and 49 sececonds. Movie is shown at 8 frames per sec.
PCM1 granules in GBM cells display constant motion. Another example of time-lapse recording of L0 cells transfected with GFP-tagged human PCM1. Images were collected every 2 seconds for 1 minute and 59 seconds. Movie is shown at 8 frames per sec.
Acknowledgements
We would like to thank N. Benson in the UF COM FACS facility, D. Smith in the UF CTAC facility, J. Harrison, D. Luo, D. Mitchell, D. Tran, as well as members of the Mitchell, Tran, and Huang laboratories for technical assistance and helpful comments. The pPCM1-EGFP-N1 expression vector was a kind gift from S. Shi (Memorial Sloan Kettering Cancer Center). This work was supported by startup funds from the McKnight Brain Research Foundation and the University of Florida (to M.R.S. and B.A.R.), an American Cancer Society Research Scholar Grant (#RSG-13-031-01-DDC) (to M.R.S.), and an American Brain Tumor Association Basic Science Research Fellowship Grant (to L.H.M.).
Footnotes
This work was supported by startup funds from the McKnight Brain Research Foundation and the University of Florida (to M.R.S. and B.A.R.), an American Cancer Society Research Scholar Grant (#RSG-13-031-01-DDC) (to M.R.S.), and an American Brain Tumor Association Basic Science Research Fellowship Grant (to L.H.M.).
References
- 1.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 2.Stupp R, Weber DC. The role of radio- and chemotherapy in glioblastoma. Onkologie. 2005;28(6–7):315–317. doi: 10.1159/000085575. [DOI] [PubMed] [Google Scholar]
- 3.Dammermann A, Müller-Reichert T, Pelletier L, Habermann B, Desai A, Oegema K. Centriole assembly requires both centriolar and pericentriolar material proteins. Dev Cell. 2004;7(6):815–829. doi: 10.1016/j.devcel.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 4.Bärenz F, Mayilo D, Gruss OJ. Centriolar satellites: busy orbits around the centrosome. Eur J Cell Biol. 2011;90(12):983–989. doi: 10.1016/j.ejcb.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 5.Gomez-Ferreria MA, Rath U, Buster DW, Chanda SK, Caldwell JS, Rines DR, Sharp DJ. Human Cep192 is required for mitotic centrosome and spindle assembly. Curr Biol. 2007;17(22):1960–1966. doi: 10.1016/j.cub.2007.10.019. [DOI] [PubMed] [Google Scholar]
- 6.Kim S, Rhee K. NEK7 is essential for centriole duplication and centrosomal accumulation of pericentriolar material proteins in interphase cells. J Cell Sci. 2011;124(Pt 22):3760–3770. doi: 10.1242/jcs.078089. [DOI] [PubMed] [Google Scholar]
- 7.Kimura M, Yoshioka T, Saio M, Banno Y, Nagaoka H, Okano Y. Mitotic catastrophe and cell death induced by depletion of centrosomal proteins. Cell Death Dis. 2013;4 doi: 10.1038/cddis.2013.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kumar A, Rajendran V, Sethumadhavan R, Purohit R. CEP proteins: the knights of centrosome dynasty. Protoplasma. 2013;250(5):965–983. doi: 10.1007/s00709-013-0488-9. [DOI] [PubMed] [Google Scholar]
- 9.Lee S, Rhee K. CEP215 is involved in the dynein-dependent accumulation of pericentriolar matrix proteins for spindle pole formation. Cell Cycle. 2010;9(4):774–783. [PubMed] [Google Scholar]
- 10.Staples CJ, Myers KN, Beveridge RD, Patil AA, Lee AJ, Swanton C, Howell M, Boulton SJ, Collis SJ. The centriolar satellite protein Cep131 is important for genome stability. J Cell Sci. 2012;125(Pt 20):4770–4779. doi: 10.1242/jcs.104059. [DOI] [PubMed] [Google Scholar]
- 11.Farina F, Gaillard J, Guérin C, Couté Y, Sillibourne J, Blanchoin L, Théry M. The centrosome is an actin-organizing centre. Nat Cell Biol. 2015;18(1):65–75. doi: 10.1038/ncb3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hall EA, Keighren M, Ford MJ, Davey T, Jarman AP, Smith LB, Jackson IJ, Mill P. Acute versus chronic loss of Mammalian azi1/cep131 results in distinct ciliary phenotypes. PLoS Genet. 2013;9(12) doi: 10.1371/journal.pgen.1003928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huong PT, Soung NK, Jang JH, Cha-Molstad HJ, Sakchaisri K, Kim SO, Jang JM, Kim KE, Lee KS, Kwon YT. Regulation of CEP131 gene expression by SP1. Gene. 2013;513(1):75–81. doi: 10.1016/j.gene.2012.10.074. [DOI] [PubMed] [Google Scholar]
- 14.Kubo A, Sasaki H, Yuba-Kubo A, Tsukita S, Shiina N. Centriolar satellites: molecular characterization, ATP-dependent movement toward centrioles and possible involvement in ciliogenesis. J Cell Biol. 1999;147(5):969–980. doi: 10.1083/jcb.147.5.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma L, Jarman AP. Dilatory is a Drosophila protein related to AZI1 (CEP131) that is located at the ciliary base and required for cilium formation. J Cell Sci. 2011;124(Pt 15):2622–2630. doi: 10.1242/jcs.084798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilkinson CJ, Carl M, Harris WA. Cep70 and Cep131 contribute to ciliogenesis in zebrafish embryos. BMC Cell Biol. 2009;10:17. doi: 10.1186/1471-2121-10-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang G, Chen Q, Zhang X, Zhang B, Zhuo X, Liu J, Jiang Q, Zhang C. PCM1 recruits Plk1 to the pericentriolar matrix to promote primary cilia disassembly before mitotic entry. J Cell Sci. 2013;126(Pt 6):1355–1365. doi: 10.1242/jcs.114918. [DOI] [PubMed] [Google Scholar]
- 18.Wang L, Lee K, Malonis R, Sanchez I, Dynlacht BD. Tethering of an E3 ligase by PCM1 regulates the abundance of centrosomal KIAA0586/Talpid3 and promotes ciliogenesis. Elife. 2016;5 doi: 10.7554/eLife.12950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hori A, Barnouin K, Snijders AP, Toda T. A non-canonical function of Plk4 in centriolar satellite integrity and ciliogenesis through PCM1 phosphorylation. EMBO Rep. 2016;17(3):326–337. doi: 10.15252/embr.201541432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hori A, Toda T. Regulation of centriolar satellite integrity and its physiology [published online ahead of print August 2, 2016]. Cell Mol Life Sci. http://dx.doi.org/10.1007/s00018-016-2315-x. [DOI] [PMC free article] [PubMed]
- 21.Shimada M, Komatsu K. Emerging connection between centrosome and DNA repair machinery. J Radiat Res. 2009;50(4):295–301. doi: 10.1269/jrr.09039. [DOI] [PubMed] [Google Scholar]
- 22.Zhang S, Hemmerich P, Grosse F. Centrosomal localization of DNA damage checkpoint proteins. J Cell Biochem. 2007;101(2):451–465. doi: 10.1002/jcb.21195. [DOI] [PubMed] [Google Scholar]
- 23.Griffith E, Walker S, Martin CA, Vagnarelli P, Stiff T, Vernay B, Sanna Al, Saggar A, Hamel B, Earnshaw WC. Mutations in pericentrin cause Seckel syndrome with defective ATR-dependent DNA damage signaling. Nat Genet. 2008;40(2):232–236. doi: 10.1038/ng.2007.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y, Dantas TJ, Lalor P, Dockery P, Morrison CG. Promoter hijack reveals pericentrin functions in mitosis and the DNA damage response. Cell Cycle. 2013;12(4):635–646. doi: 10.4161/cc.23516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Villumsen BH, Danielsen JR, Povlsen L, Sylvestersen KB, Merdes A, Beli P, Yang YG, Choudhary C, Nielsen ML, Mailand N. A new cellular stress response that triggers centriolar satellite reorganization and ciliogenesis. EMBO J. 2013;32(23):3029–3040. doi: 10.1038/emboj.2013.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tollenaere MA, Villumsen BH, Blasius M, Nielsen JC, Wagner SA, Bartek J, Beli P, Mailand N, Bekker-Jensen S. p38- and MK2-dependent signalling promotes stress-induced centriolar satellite remodelling via 14-3-3–dependent sequestration of CEP131/AZI1. Nat Commun. 2015;6:10075. doi: 10.1038/ncomms10075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Löffler H, Fechter A, Liu FY, Poppelreuther S, Krämer A. DNA damage-induced centrosome amplification occurs via excessive formation of centriolar satellites. Oncogene. 2013;32(24):2963–2972. doi: 10.1038/onc.2012.310. [DOI] [PubMed] [Google Scholar]
- 28.Douthwright S, Sluder G. Link between DNA damage and centriole disengagement/reduplication in untransformed human cells. J Cell Physiol. 2014;229(10):1427–1436. doi: 10.1002/jcp.24579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Balczon R, Bao L, Zimmer WE. PCM-1, A 228-kD centrosome autoantigen with a distinct cell cycle distribution. J Cell Biol. 1994;124(5):783–793. doi: 10.1083/jcb.124.5.783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dammermann A, Merdes A. Assembly of centrosomal proteins and microtubule organization depends on PCM-1. J Cell Biol. 2002;159(2):255–266. doi: 10.1083/jcb.200204023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kubo A, Tsukita S. Non-membranous granular organelle consisting of PCM-1: subcellular distribution and cell-cycle–dependent assembly/disassembly. J Cell Sci. 2003;116(Pt 5):919–928. doi: 10.1242/jcs.00282. [DOI] [PubMed] [Google Scholar]
- 32.Kim J, Krishnaswami SR, Gleeson JG. CEP290 interacts with the centriolar satellite component PCM-1 and is required for Rab8 localization to the primary cilium. Hum Mol Genet. 2008;17(23):3796–3805. doi: 10.1093/hmg/ddn277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Q, Hansen D, Killilea A, Joshi HC, Palazzo RE, Balczon R. Kendrin/pericentrin-B, a centrosome protein with homology to pericentrin that complexes with PCM-1. J Cell Sci. 2001;114(Pt 4):797–809. doi: 10.1242/jcs.114.4.797. [DOI] [PubMed] [Google Scholar]
- 34.Hoang-Minh LB, Deleyrolle LP, Siebzehnrubl D, Ugartemendia G, Futch H, Griffith B. Disruption of KIF3A in patient-derived glioblastoma cells: effects on ciliogenesis, hedgehog sensitivity, and tumorigenesis. Oncotarget. 2016;7(6):7029–7043. doi: 10.18632/oncotarget.6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sarkisian MR, Siebzehnrubl D, Hoang-Minh L, Deleyrolle L, Silver DJ, Siebzehnrubl FA, Breunig JJ, De Leon G, Mitchell DA, Semple-Rowland S. Detection of primary cilia in human glioblastoma. J Neuro-Oncol. 2014;117(1):15–24. doi: 10.1007/s11060-013-1340-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deleyrolle LP, Harding A, Cato K, Siebzehnrubl FA, Rahman M, Azari H, Olson S, Gabrielli B, Osborne G. Evidence for label-retaining tumour-initiating cells in human glioblastoma. Brain. 2011;134(Pt 5):1331–1343. doi: 10.1093/brain/awr081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hothi P, Martins TJ, Chen L, Deleyrolle L, Yoon JG, Reynolds B, Foltz G. High-throughput chemical screens identify disulfiram as an inhibitor of human glioblastoma stem cells. Oncotarget. 2012;3(10):1124–1136. doi: 10.18632/oncotarget.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Siebzehnrubl FA, Silver DJ, Tugertimur B, Deleyrolle LP, Siebzehnrubl D, Sarkisian MR, Sarkisian MR, Devers KG, Yachnis AT, Kupper MD, Neal D. The ZEB1 pathway links glioblastoma initiation, invasion and chemoresistance. EMBO Mol Med. 2013;5(8):1196–1212. doi: 10.1002/emmm.201302827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Piccirillo SG, Vescovi AL. Bone morphogenetic proteins regulate tumorigenicity in human glioblastoma stem cells. Ernst Schering Found Symp Proc. 2006;5:59–81. doi: 10.1007/2789_2007_044. [DOI] [PubMed] [Google Scholar]
- 40.Hiddingh L, Raktoe RS, Jeuken J, Hulleman E, Noske DP, Kaspers GJ, Vandertop WP, Wesseling P, Wurdinger T. Identification of temozolomide resistance factors in glioblastoma via integrative miRNA/mRNA regulatory network analysis. Sci Rep. 2014;4:5260. doi: 10.1038/srep05260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Uhlen M, Oksvold P, Fagerberg L, Lundberg E, Jonasson K, Forsberg M, Zwahlen M, Kampf C, Wester K, Hober S. Towards a knowledge-based Human Protein Atlas. Nat Biotechnol. 2010;28(12):1248–1250. doi: 10.1038/nbt1210-1248. [DOI] [PubMed] [Google Scholar]
- 42.Sedjaï F, Acquaviva C, Chevrier V, Chauvin JP, Coppin E, Aouane A, Coulier F, Tolun A, Pierres M, Birnbaum D. Control of ciliogenesis by FOR20, a novel centrosome and pericentriolar satellite protein. J Cell Sci. 2010;123(Pt 14):2391–2401. doi: 10.1242/jcs.065045. [DOI] [PubMed] [Google Scholar]
- 43.Insolera R, Shao W, Airik R, Hildebrandt F, Shi SH. SDCCAG8 regulates pericentriolar material recruitment and neuronal migration in the developing cortex. Neuron. 2014;83(4):805–822. doi: 10.1016/j.neuron.2014.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kurtulmus B, Wang W, Ruppert T, Neuner A, Cerikan B, Viol L, Dueñas-Sánchez R, Gruss OJ, Pereira G. WDR8 is a centriolar satellite and centriole-associate protein that promotes ciliary vesicle docking during ciliogenesis. J Cell Sci. 2015;129(3):621–636. doi: 10.1242/jcs.179713. [DOI] [PubMed] [Google Scholar]
- 45.Staples CJ, Myers KN, Beveridge RD, Patil AA, Howard AE, Barone G. Ccdc13 is a novel human centriolar satellite protein required for ciliogenesis and genome stability. J Cell Sci. 2014;127(Pt 13):2910–2919. doi: 10.1242/jcs.147785. [DOI] [PubMed] [Google Scholar]
- 46.Ye X, Zeng H, Ning G, Reiter JF, Liu A. C2cd3 is critical for centriolar distal appendage assembly and ciliary vesicle docking in mammals. Proc Natl Acad Sci U S A. 2014;111(6):2164–2169. doi: 10.1073/pnas.1318737111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee JY, Stearns T. FOP is a centriolar satellite protein involved in ciliogenesis. PLoS One. 2013;8(3) doi: 10.1371/journal.pone.0058589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stowe TR, Wilkinson CJ, Iqbal A, Stearns T. The centriolar satellite proteins Cep72 and Cep290 interact and are required for recruitment of BBS proteins to the cilium. Mol Biol Cell. 2012;23(17):3322–3335. doi: 10.1091/mbc.E12-02-0134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Keryer G, Pineda JR, Liot G, Kim J, Dietrich P, Benstaali C, Smith K, Cordelières FP, Spassky N, Ferrante RJ. Ciliogenesis is regulated by a huntingtin-HAP1-PCM1 pathway and is altered in Huntington disease. J Clin Invest. 2011;121(11):4372–4382. doi: 10.1172/JCI57552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Akimov V, Rigbolt KT, Nielsen MM, Blagoev B. Characterization of ubiquitination dependent dynamics in growth factor receptor signaling by quantitative proteomics. Mol BioSyst. 2011;7(12):3223–3233. doi: 10.1039/c1mb05185g. [DOI] [PubMed] [Google Scholar]
- 51.Moser JJ, Fritzler MJ, Rattner JB. Primary ciliogenesis defects are associated with human astrocytoma/glioblastoma cells. BMC Cancer. 2009;9:448. doi: 10.1186/1471-2407-9-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moser JJ, Fritzler MJ, Rattner JB. Ultrastructural characterization of primary cilia in pathologically characterized human glioblastoma multiforme (GBM) tumors. BMC Clin Pathol. 2014;14:40. doi: 10.1186/1472-6890-14-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wong SY, Seol AD, So PL, Ermilov AN, Bichakjian CK, Epstein EH, Dlugosz AA, Reiter JF. Primary cilia can both mediate and suppress Hedgehog pathway-dependent tumorigenesis. Nat Med. 2009;15(9):1055–1061. doi: 10.1038/nm.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li L, Grausam KB, Wang J, Lun MP, Ohli J, Lidov HG, Calicchio ML, Zeng E, Salisbury JL, Wechsler-Reya RJ. Sonic Hedgehog promotes proliferation of Notch-dependent monociliated choroid plexus tumour cells. Nat Cell Biol. 2016;18(4):418–430. doi: 10.1038/ncb3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Han YG, Kim HJ, Dlugosz AA, Ellison DW, Gilbertson RJ, Alvarez-Buylla A. Dual and opposing roles of primary cilia in medulloblastoma development. Nat Med. 2009;15(9):1062–1065. doi: 10.1038/nm.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yoshimura K, Kawate T, Takeda S. Signaling through the primary cilium affects glial cell survival under a stressed environment. GLIA. 2011;59(2):333–344. doi: 10.1002/glia.21105. [DOI] [PubMed] [Google Scholar]
- 57.Johnson CA, Collis SJ. Ciliogenesis and the DNA damage response: a stressful relationship. Cilia. 2016;5:19. doi: 10.1186/s13630-016-0040-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. PCM1 granules in S3 GBM cells display a dynamic distribution during the cell cycle. Co-immunostaining for PCM1 (green) and acetylated alpha-acetylated tubulin (red), with nuclei counterstained with DAPI (blue). Examples of staining of prophase, metaphase, anaphase, telophase and interphase of the cell cycle are shown. Scale bar = 5 μm.
Figure S2. Similar Cep290 subcellular distribution in PCM1-positive control and PCM1-depleted clones. Examples of S3 cells derived from PCM1-positive control clone F11 or PCM1-depleted clone B9 and immunostained for Cep290 (green), with nuclei counterstained with DAPI (blue). Scale bar = 5 μm.
Figure S3. Examples of Ki67, BrdU, and PHH3 immunostaining in L0 GBM cells and detection of active caspase 3-positive cells by flow cytometry. (A) Example of immunostaining of L0 cells derived from PCM1-positive control clone D5 and PCM1-depleted clone F9 for markers of cell cycle active phases (Ki67), G2/M (phospho-histone H3 (PHH3)), and S phase (BrdU). (B) Apoptotic cell death assessed by flow cytometry of L0 cells derived from PCM1-positive control clone D5 or PCM1-depleted clone F9 and labeled with rabbit active caspase 3 antibody followed by Alexa Fluor 488–conjugated anti-rabbit secondary antibody. Scale bar = 5 μm.
PCM1 granules in GBM cells display constant motion. Example of time-lapse recording of L0 cells transfected with GFP-tagged human PCM1 (green). Images were collected every 10 seconds for 4 minutes and 49 sececonds. Movie is shown at 8 frames per sec.
PCM1 granules in GBM cells display constant motion. Another example of time-lapse recording of L0 cells transfected with GFP-tagged human PCM1. Images were collected every 2 seconds for 1 minute and 59 seconds. Movie is shown at 8 frames per sec.