Summary
We identified two SNPs with potentials in risk-stratifying GS 7 prostate cancer patients and predicting reclassification among patients undergoing AS. It may have clinical implication in managing localized patients by optimizing patient selection and personalized monitoring prior to/during AS.
Abstract
Little is known about the genetic predictors of prostate cancer aggressiveness and reclassification in men with localized prostate cancer undergoing active surveillance. The Wnt signaling pathway is important for prostate cancer development and progression. Identifying genetic variants associated with prostate cancer aggressiveness and reclassification may have a potential role in the management of localized patients. In this study, we used a three-phase design. In phases I and II prostate cancer patient cohort, 578 single nucleotide polymorphisms (SNPs) from 45 genes of the Wnt signaling pathway were analyzed in 1762 localized prostate cancer patients. Twelve SNPs from four regions were significantly associated with aggressive disease, among which, three linked SNPs in CSNK1A1 at 5q32 (represented by rs752822) may differentiate GS 4+3 from GS 3+4 patients (OR = 1.44, 95% CI = 1.12–1.87, P = 4.76×10-3). In phase III active surveillance (AS) cohort, genotyping of rs752822 (candidate from phases I and II) and previously identified rs2735839 were determined in 494 GS ≤7 patients. We found a significant association between rs2735839 and prostate cancer reclassification in the AS cohort (AG + AA versus GG, HR = 1.59, 95% CI = 1.11–2.28, P = 0.012) and a suggestive association of rs752822. Jointly, rs752822 and rs2735839 showed good potentials in risk-stratifying GS 7 patients and predicting disease reclassification (OR = 2.71, 95% CI = 1.62–4.51, P = 1×10−4 in phase II; HR = 1.89, 95% CI = 1.13–3.18, P = 0.016 in phase III). In summary, rs752822 and rs2735839 may assist in risk-stratifying GS 7 patients and predict prostate cancer reclassification. The significant associations were independent from GS, T stage and PSA levels at baseline.
Introduction
Central to clinical dilemma in managing localized prostate cancer is the inability to objectively distinguish individual men whose prostate cancer requires and will benefit from treatment from those who do not require or will not benefit from treatment. Active surveillance (AS) strategies permit men with nonaggressive prostate cancer to avoid unnecessary treatment. AS strategies have been applied to narrowly defined low-risk groups and have proven safe in short follow-up studies (1). However, these studies are undermined by early disease reclassification with approximate 20–30% of men undergoing AS to have definitive therapy within a 5-year follow-up (2). Given morbidities associated with prostate biopsies (3), blood-based markers predictive of disease reclassification will have a great clinical impact on managing men with early stage prostate cancer.
Prostate cancer is a genetic disease involving multiyear and multistep processes. Notwithstanding nearly 100 susceptibility loci associated with prostate cancer identified by genome-wide association studies (4,5), among which, only a few SNPs were linked to disease aggressiveness. Recently, two new loci at 5q14.3 and 3q26.31 were identified to be correlated with prostate cancer aggressiveness (6). The study also confirmed a previously reported locus at 19q13.33 (KLK3, gene encodes PSA). In addition, our previous work indicated that this same locus (rs2735839) may be useful in risk-stratifying patients with Gleason score (GS) 7 prostate cancer (7). Furthermore, little is known about the role of genetic susceptibility in determining the clinical course of patients undergoing AS. One study found that rs11568818 on chromosome 11q22 was significantly associated with upgrading (8). In another study, no significant association was observed (9).
Wnt pathway plays a pivotal role in prostate cancer development and progression (10–12). It is well known that the expression of KLK3 is regulated by β-catenin in nucleus, which mediates the cross-talk between the Wnt signaling pathway and androgen receptor (AR) signaling in prostate cancer (13). Thus, it is interesting to investigate whether the genetic variants in the Wnt signaling pathway are associated with prostate cancer aggressiveness and reclassification. In this study, we hypothesize that the genetic variants in the Wnt signaling pathway are associated with prostate cancer aggressiveness and affect the course of disease reclassification in localized patients.
Materials and methods
Study population
The study was approved by the MD Anderson Institutional Review Board. The details of two study populations have been depicted in the previous publications (7,14). In brief, men with previously untreated prostate cancer registered at The University of Texas MD Anderson Cancer Center were recruited. Clinical data were abstracted from medical records, which included diagnosis date, biopsy-proven Gleason score, clinical tumor stage at diagnosis, PSA level at diagnosis, pathologic information and surveillance or treatment. Age at the date when first prostate biopsy was determined to be cancer-positive was used to define the age at diagnosis. All biopsy slides from outside institutions were reviewed by pathologists at MD Anderson. When difference arose, the GS assessment at MD Anderson was used.
Phases I and II MD Anderson prostate cancer patient cohort (MDA-prostate cancer patient cohort).
Two criteria were used to define aggressiveness (phase I): GS ≥8 or high D’Amico risk was defined as more aggressive, while GS ≤6 or low D’Amico risk was defined as less aggressive. The definition of D’Amico risk stratification is defined according to modified NCCN (http://www.nccn.org/professionals/physician_gls/f_guidelines.asp): (1) low-risk, T1–T2a and GS ≤6 and PSA ≤10ng/ml; (2) intermediate-risk, T2b or GS =7 or PSA >10–20ng/ml; (3) high-risk, ≥T2c or GS 8–10 or PSA >20ng/ml. Less aggressive cancer was defined as disease with GS 3+4 and more aggressive as GS 4+3 in phase II.
Phase III AS cohort.
The trial started in February 2006 and is currently ongoing (registered with clinical.trials.gov: NCT00490763). Briefly, after study enrollment the patients are stratified to groups I (favorable risk), II (patient’s choice) or III (therapy prevented by comorbidities) (14). Inclusion criteria for group I are diagnosis within 6 months before enrollment, a biopsy of ≥10 cores showing either a 3 + 3 Gleason score (GS) in one core (tumor focus <3.0mm) or a 3 + 4 GS in one core (tumor focus <2.0mm), and baseline PSA<4ng/ml (adjusted for volume). Patients with GS ≤7 prostate cancer but not meeting the criteria for group I were stratified to group II. Patients in group III had comorbidities which prevent local therapy were determined by the managing physician. Since June 2007 patients were required to have a confirmatory biopsy at study enrollment unless their diagnostic biopsy was performed at MDACC. A total of 494 localized patients with GS ≤7 prostate cancer and at least 1 repeated biopsy during the follow-up were included in the current analyses. Patients were evaluated at baseline and every 6 months by clinical examination (DRE) and laboratory studies (serum PSA, testosterone). Prostate biopsies were repeated every 1–2 years; if the biopsy was negative, then the following year’s biopsy was omitted, unless requested by the patient. An event of disease reclassification was defined as an increase in number of positive core or tumor length outside of the study entry criteria, or increase in GS on repeated biopsy. The upgrading was defined as an increase in GS only. Due to small number of upgrading events, we only focused on disease reclassification. Patients without event were censored in March 31, 2015 when the dataset was prepared for analysis. The time to event was defined as the time from the date of diagnosis to the date of reclassification/last follow-up/March 31, 2015.
Genes of interest in Wnt signaling pathway
We generated a cancer-related gene list for the Wnt signaling pathway using the Gene Ontology (GO) database (http://www.geneontology.org). Extensive literature review of genes from GO database was done using HUGO names and common aliases. A total of 45 genes were selected based on their relevance and significance to prostate cancer. The chromosome positions of start and end of the gene were obtained from USCS Genome Browser (http://genome.ucsc.edu/, build version: GRCh37/hg19).
Genotyping and quality control
All DNA samples were extracted from peripheral whole blood using the QIAamp DNA extraction Kit (QIAGEN).
Phases I and II.
A total of 1823 DNA samples were genotyped for this project. Custom Infinium Oncoarray-500K Beadchip was used for genotyping. The assay was run on the iScan system (illumina). Genotyping data were analyzed and exported using the GenomeStudio software (illumina). All subjects had a call rate >95%. The following exclusion criteria were applied to all samples: gender disparity which was identified by checking X chromosome (n = 2), disease not localized (N1/M1, n = 3), ethnicity other than non-Hispanic white (n = 21), histology other than adenocarcinoma (n = 12) and duplicated samples (n = 23). After all exclusions, there remained an analytic set of 1762 participants. The mean concordance rate of duplicated samples was 99.2%. SNPs with minor allele frequency (MAF) <0.01 (n = 83 738) and call rate <0.90 (n = 2945) were excluded. A total of 412 487 SNPs remained after quality control. Finally, genotyping data of 578 SNPs within 10kb flanking regions upstream and downstream of each gene of interest were extracted from Oncoarray dataset for this study.
Phase III.
rs752822 (a candidate SNP at 5q32 identified from phase I study) and rs2735839 (a previously-identified prostate cancer risk- and aggressiveness-associated SNP at 19q13.13) (7) were genotyped using TaqMan Pre-Designed SNP Genotyping Assays (Applied Biosystems, Foster City, CA) in the AS cohort.
Statistical analysis
Phases I and II.
About 578 SNPs were compared in the additive genetic model between more aggressive phenotype to less aggressive phenotype for two criteria of disease aggressiveness, i.e. patients with GS ≥8 versus patients with GS ≤6, and patients classified as D’Amico high-risk versus low-risk. To ensure the consistency, SNPs nominally associated with both aggressiveness phenotypes (P < 0.05) were considered good candidates for phase II analysis. The candidate SNPs were tested for their ability to stratify patients with GS 7 disease by comparing GS 4+3 to GS 3+4 patients (P < 0.05) in phase II analysis.
The association between SNP and aggressive prostate cancer defined by GS (i.e. GS ≥8 versus GS ≤6 or GS 4+3 versus GS 3+4) was evaluated using unconditional multivariable logistic regression with adjustment of age at diagnosis (continuous), PSA levels at diagnosis (categorical) and clinical T stage (T1 versus T2 versus T3–4). Only age and PSA levels were adjusted for comparisons between D’Amico high-risk and low-risk groups using logistic regression.
Phase III.
Univariable Cox regression model was used to determine covariates associated with disease reclassification. Associations of rs752822 and rs2735839 with reclassification were analyzed using multivariable Cox regression model with adjustment of age at diagnosis, ethnicity, GS, T stage and PSA levels at baseline. The proportional hazard assumption was examined by plotting and testing the Schoenfeld Residuals and including time varying covariates in the model (interaction terms for GS, T stage and natural logarithms of time).
The associations of T stage and log-transformed PSA were further assessed separately for rs752822 and rs2735839 in two study populations using ordinal logistic regression or linear regression with adjustment of age, GS, clinical T stage and PSA levels where appropriate. The assumptions of linear regression were assessed by plotting residuals against the predicted PSA levels and Q–Q plot. To assess the joint effect, simple genetic risk score (GRS) was generated by adding unfavorable genotypes of rs752822 and rs2735839 and tested in the two phases. Additive genetic model was assumed unless otherwise stated. All P values were two-sided, with values less than 0.05 considered statistically significant. All statistical analyses were performed using Stata version 13.
Results
Clinicopathologic characteristics of the study populations at diagnosis are shown in Table 1. The mean age at diagnosis was 61.6 and 63.7 years for subjects enrolled in phases I + II and III, respectively. The enrolled subjects were primarily non-Hispanic whites (100 and >80% for phases I + II and III, respectively). For patients in phases I and II, 657 and 218 had less aggressive (GS ≤6) and more aggressive prostate cancer (GS ≥8), respectively. A 3:1 ratio of GS 3+4 versus GS 4+3 was observed among GS 7 patients. The majority of patients had T1–T2 tumors (>90%) and low PSA levels at diagnosis (<10ng/ml, 88%). Based on D’Amico classification system, 598, 829 and 330 patients were grouped into low-, intermediate-, high-risk categories, respectively. Radical prostatectomy and radiotherapy were two primary treatments received by three quarters of the patients (n = 1296). Patients in phase III AS cohort were mainly classified as low-risk (GS ≤6, 82%; T1, 89%; PSA levels <10ng/ml, 96%). After a median follow-up of 41.9 months (range: 9.3–110.8 months), approximately one-third of patients were reclassified (n = 159) and 20% of patients had upgrading of GS (n = 98).
Table 1.
Clinical characteristics of the study patients with localized prostate cancer
| Characteristics | MDA-PCa patient cohort patient cohort | MDA-AS cohort |
|---|---|---|
| N (%) | N (%) | |
| Total | 1762 | 494 |
| Age at diagnosis, years | ||
| Mean(SD) | 61.6 (7.9) | 63.8 (8.3) |
| Ethnicity | ||
| Non-Hispanic White | 1762 (100) | 403 (81.58) |
| Others | 0 | 91 (18.42) |
| Biopsy-proven GS | ||
| ≤6 | 657 (37.29) | 406 (82.19) |
| 3+4 | 647 (36.72) | 77 (15.59) |
| 4+3 | 240 (13.62) | 11 (2.23) |
| ≥8 | 218 (12.37) | 0 |
| Clinical tumor stage, n (%) | ||
| T1 | 1109 (62.94) | 441 (89.27) |
| T2 | 575 (32.63) | 52 (10.53) |
| T3–4 | 69 (3.92) | 0 |
| Unknown | 9 (0.51) | 1 (0.20) |
| PSA level at diagnosis, ng/mla | ||
| <4 or <2.5 | 442 (25.11) | 114 (23.08) |
| 4–9.9 or 2.5–3.9 | 1108 (62.95) | 128 (25.91) |
| 10–19.9 or 4–9.9 | 145 (8.24) | 234 (47.37) |
| ≥20 or ≥10 | 65 (3.69) | 18 (3.64) |
| D’Amico risk group | ||
| Low | 598 (33.94) | 391 (79.15) |
| Intermediate | 829 (47.05) | 95 (19.23) |
| High | 330 (18.73) | 7 (1.42) |
| Not grouped | 5 (0.28) | 1 (0.20) |
| Disease reclassification | ||
| Yes | — | 159 (32.19) |
| No | — | 334 (67.81) |
| Disease upgrading | ||
| Yes | — | 98 (19.84) |
| No | — | 396 (80.16) |
| Primary treatment | ||
| Radical prostatectomy | 918 (52.10) | — |
| Radiotherapy | 378 (21.45) | — |
| Surveillance or unknownb | 429 (24.35) | — |
| Other treatmentc | 37 (2.10) | — |
aDifferent criteria were used for PSA levels categorizing in two study populations. Retrospective case series: <4 versus 4–9.9 versus 10–19.9 versus ≥20ng/ml; AS cohort: <2.5 versus 2.5–3.9 versus 4–9.9 versus ≥10ng/ml.
bPatients undergoing active surveillance/watchful waiting or whose initial treatment information was unavailable.
cCryoablation, high−intensity focused ultrasound, transurethral resection of prostate or androgen deprivation therapy.
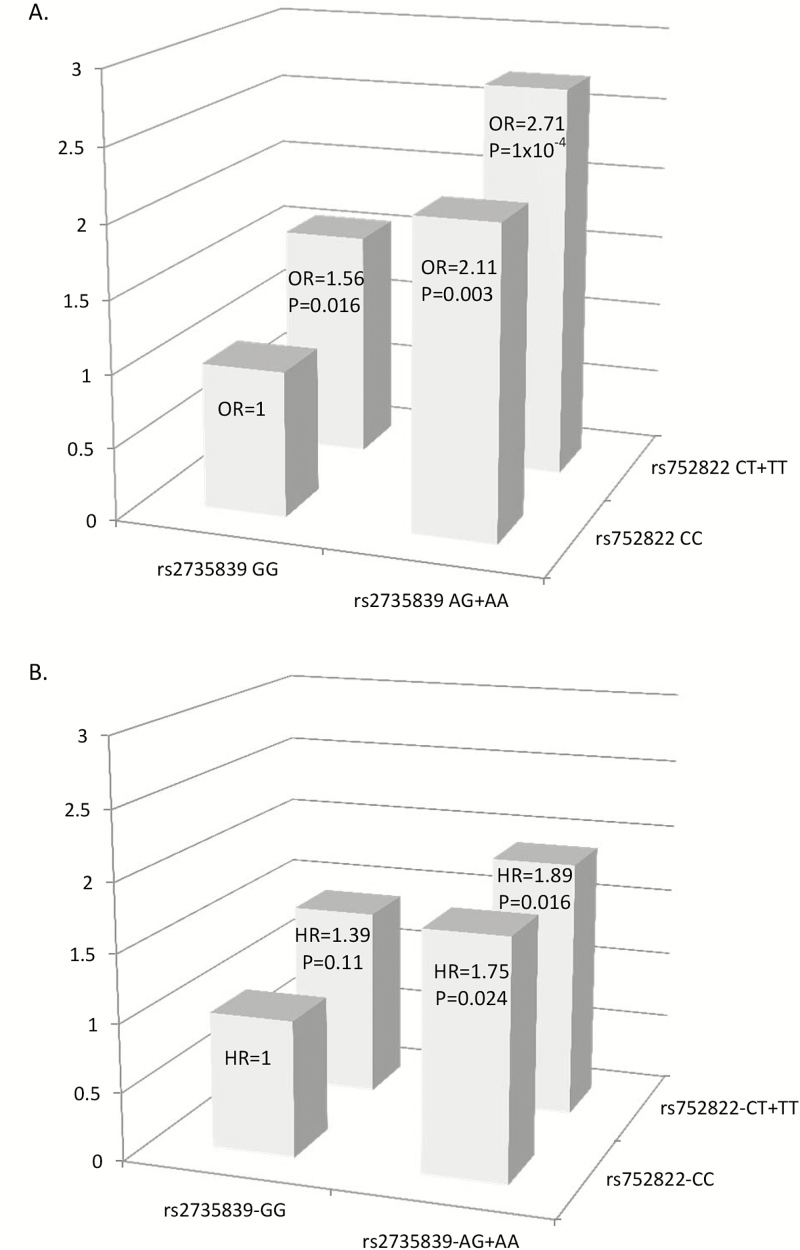
The analyses we conducted were summarized by Supplementary Figure 1, available at Carcinogenesis Online. Among 578 SNPs, 12 were nominally associated with risks of aggressive prostate cancer in phase I analysis (P < 0.05, Table 2). Due to moderate to strong LD, these SNPs represent four different loci where casein kinase 1, alpha 1 (CSNK1A1), catenin, beta 1 (CTNNB1), transcription factor 7-like 1 (TCF7L1) and transcription factor 7-like 2 (TCF7L2) are located. All minor alleles of the 12 SNPs conferred a higher risk of aggressive prostate cancer. In phase II analysis, one locus on CSNK1A1 at 5q32 represented by rs752822 (most statistically significant) was able to risk-stratify GS 4+3 and GS 3+4 prostate cancer patients (OR = 1.44, 95% CI = 1.12–1.87, P = 0.005, Table 3). We further evaluated the association of rs752822 with clinical T stage and log-transformed PSA levels separately in the entire study population in phases I and II. We found that the association between rs752822 and prostate cancer aggressiveness was not driven by T stage or PSA levels (Table 4). Joint analysis of rs752822 and rs2735839 showed a substantial increased risk of primary Gleason 4 pattern when comparing patients with the highest risk genotypes (rs752822 CT+TT and rs2735839 AG+AA genotypes) to the reference (rs752822 CC and rs2735839 GG genotypes) among GS 7 patients (OR = 2.71, 95% CI = 1.62–4.51, P = 1×10−4, Figure 1A).
Table 2.
Selected SNPs significantly associated with prostate cancer aggressiveness (GS ≥ 8 versus GS ≤ 6 or D’Amico high versus low) in Wnt signaling pathway
| GS ≥8 versus GS ≤6 | D’Amico high versus low | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| SNP | Chr | BP | Risk allele | Gene | Annotation | OR (95% CI) | P value | OR (95% CI) | P value |
| rs752822 | 5 | 148889018 | T | CSNK1A1 | Intron | 1.58 (1.15–2.18) | 4.53×10−3 | 1.54 (1.20–1.99) | 7.88×10−4 |
| rs6886243a | 5 | 148883634 | C | CSNK1A1 | Intron | 1.58 (1.15–2.17) | 4.98×10−3 | 1.53 (1.19–1.97) | 9.54×10−4 |
| rs13170358a | 5 | 148887713 | C | CSNK1A1 | Intron | 1.58 (1.15–2.18) | 4.78×10−3 | 1.53 (1.19–1.97) | 9.30×10−4 |
| rs9883073 | 3 | 41284536 | A | CTNNB1 | Downstream | 2.15 (1.55–2.98) | 4.11×10−6 | 1.39 (1.10–1.76) | 6.43×10−3 |
| rs62258388b | 3 | 41286419 | A | CTNNB1 | Downstream | 2.14 (1.55–2.97) | 4.43×10−6 | 1.38 (1.09–1.75) | 7.34×10−3 |
| rs2953b | 3 | 41281388 | G | CTNNB1 | 3’ UTR | 2.13 (1.54–2.95) | 4.70×10−6 | 1.37 (1.08–1.74) | 8.35×10−3 |
| rs3864004b | 3 | 41240177 | A | CTNNB1 | Upstream | 2.17 (1.54–3.05) | 9.23×10−6 | 1.36 (1.06–1.73) | 1.44×10−2 |
| rs3774371b | 3 | 41276166 | A | CTNNB1 | Intron | 2.05 (1.48–2.84) | 1.44×10−5 | 1.40 (1.11–1.78) | 5.27×10−3 |
| rs55868746 | 2 | 85442740 | A | TCF7L1 | Intron | 1.84 (1.03–3.27) | 3.84×10−2 | 1.75 (1.09–2.82) | 2.09×10−2 |
| rs72840151c | 2 | 85441851 | A | TCF7L1 | Intron | 1.91 (1.01–3.59) | 4.53×10−2 | 2.12 (1.20–3.73) | 9.16×10−3 |
| rs72840119d | 2 | 85402814 | C | TCF7L1 | Intron | 1.73 (1.00–3.00) | 4.96×10−2 | 1.78 (1.12–2.82) | 1.14×10−2 |
| rs10885398 | 10 | 114715930 | G | TCF7L2 | Intron | 1.43 (1.03–1.99) | 3.03×10−2 | 1.33 (1.04–1.72) | 2.53×10−2 |
The models were adjusted for age, clinical tumor stage and PSA levels at diagnosis for GS comparisons; age and PSA levels at diagnosis were adjusted for D’Amico comparisons. Additive genetic model was tested for all SNPs.
aSNPs in high LD (R2 > 0.8) with rs752822.
bSNPs in high LD (R2 > 0.8) with rs9883073.
cSNPs in high LD (R2 > 0.8) with rs55868746.
dSNPs in moderate LD (R2 = 0.6–0.8) with rs55868746.
Table 3.
The associations between identified SNPs and risk of GS 4+3 disease
| GS 4+3 versus GS 3+4 | |||
|---|---|---|---|
| SNP | Risk allele | OR (95% CI) | P value |
| rs752822 | T | 1.44 (1.12–1.87) | 4.76×10−3 |
| rs9883073 | A | 0.98 (0.79–1.23) | 0.884 |
| rs55868746 | A | 1.04 (0.62–1.74) | 0.875 |
| rs72840119a | C | 1.02 (0.62–1.67) | 0.934 |
| rs10885398 | G | 1.09 (0.85–1.40) | 0.483 |
The models were adjusted for age, clinical tumor stage and PSA levels at diagnosis. Additive genetic model was tested for all SNPs.
aaSNP in moderate LD (R2 = 0.6–0.8) with rs55868746.
Table 4.
The associations of T stage and PSA levels for rs752822 and rs2735839
| T stage | Log-transformed PSA | ||||
|---|---|---|---|---|---|
| SNPa | Risk allele | OR (95% CI) | P value | β (95% CI) | P value |
| Phases I and II: PCa patient cohort (N = 1762) | |||||
| rs752822 | T | 0.88 (0.74–1.05) | 0.17 | 0.03 (−0.02 to 0.08) | 0.28 |
| rs2735839 | A | 1.10 (0.88–1.37) | 0.41 | −0.15 (−0.22 to −0.09) | 2.19×10−6 |
| Phase III: AS cohort (N = 494) | |||||
| rs752822 | T | 0.96 (0.51–1.79) | 0.89 | 0.015 (−0.10 to 0.13) | 0.80 |
| rs2735839 | A | 0.68 (0.32–1.45) | 0.32 | −0.31 (−0.44 to −0.17) | 1.1×10−5 |
The models were adjusted for age, ethnicity, Gleason score, T stage and PSA levels at diagnosis where appropriate.
aDominant genetic model was tested for all SNPs due to small sample size in subcategories.
Figure 1.
Joint effect of rs752822 and rs2735839 on prostate cancer aggressiveness and reclassification. (A) Joint effect of rs752822 and rs2735839 on risk-stratification of localized GS 7 cancers in phase II. Comparing GS 7 patients with rs752822 CC and rs2735839 GG genotypes, the risks of primary Gleason 4 pattern were significantly increased for GS 7 patients with rs752822 CC+TT and rs2735839 GG genotypes (OR = 1.56, 95% CI = 1.09–2.24, P = 0.016); rs752822 CC and rs2735839 AG+AA (OR = 2.11, 95% CI = 1.30–3.46, P = 0.003); rs752822 CT+TT and rs2735839 AG+AA (OR = 2.71, 95% CI = 1.62–4.51, P = 1.31×10-4). The model was adjusted for age, T stage and PSA levels. (B) Joint effect of rs752822 and rs275839 on association with prostate cancer reclassification among localized prostate cancer patients with GS ≤7 in the AS cohort. Comparing to GS 7 patients with rs752822 CC and rs2735839 GG, the risks for prostate cancer reclassification were marginally or significantly increased for GS ≤7 patients with rs752822 CC+TT and rs2735839 GG (HR = 1.39, 95% CI = 0.93–2.08, P = 0.108); rs752822 CC and rs2735839 AG+AA (HR = 1.75, 95% CI = 1.08–2.83, P = 0.024); rs752822 CT+TT and rs2735839 AG+AA (HR = 1.90, 95% CI = 1.13–3.18, P = 0.016). The model was adjusted for age, ethnicity, GS, T stage and PSA levels.
Results of univariable Cox regression showed that only GS at baseline was significantly associated with reclassification in the AS cohort (Supplementary Table 1, available at Carcinogenesis Online). The association of rs2735839 was independent from GS, T stage and PSA levels at baseline (AG + AA versus GG, HR = 1.59, 95% CI = 1.11–2.28, P = 0.012, Table 5). Suggestive association was found for rs752822 (CT + TT versus CC, HR = 1.29, 95% CI = 0.93–1.80, P = 0.124, Table 5). Mutual adjustment for two SNPs did not drastically alter the estimates (data not shown). Inclusion of time varying covariates in the models subtly changed the estimates (data not shown). In the analysis of joint effect of rs2735839 and rs752822, up to 1.9-fold increased risk of reclassification was observed in the AS cohort when compared patients with the highest risk genotypes (rs752822 CT+TT and rs2735839 AG+AA genotypes) to the reference (rs752822 CC and rs2735839 GG genotypes, HR = 1.89, 95% CI = 1.13–3.18, P = 0.016, Figure 1B). Highly significant association was found for rs2735839 allele A with log-transformed PSA levels (phases I and II: β = −0.15, 95% CI = −0.22 to −0.09, P = 2.19×10−5; phase III: β = −0.31, 95% CI = −0.44 to −0.17, P = 1.1×10−5, Table 4). In agreement with findings in phases I and II, rs752822 was not associated with T stage and PSA levels in the AS cohort (Table 4).
Table 5.
Associations between SNPs and prostate cancer reclassification in AS cohort
| SNPs | Reclassified, N (%) | Stable, N (%) | Univariable | Multivariable | ||
|---|---|---|---|---|---|---|
| HR (95% CI) | P value | HR (95% CI) | P value | |||
| rs752822, CC | 82 (29.7) | 194 (70.3) | Ref. | Ref. | ||
| CT + TT | 72 (36.5) | 125 (63.5) | 1.29 (0.93–1.78) | 0.124 | 1.29 (0.93–1.80) | 0.124 |
| rs2735839, GG | 104 (28.7) | 258 (71.3) | Ref. | Ref. | ||
| AG + AA | 51 (42.9) | 68 (57.1) | 1.50 (1.06–2.12) | 0.021 | 1.59 (1.11–2.28) | 0.012 |
aThe model was adjusted for age at diagnosis, ethnicity, Gleason score, T stage and PSA level at diagnosis. Dominant genetic model was tested for all SNPs due to small sample size in subcategories.
Discussion
In this study, we found that rs752822 and rs2735839 can be jointly used as a risk-stratification tool for prostate cancer aggressiveness and a prediction tool for prostate cancer reclassification among localized patients. Identifying novel blood-borne biomarkers for distinguishing indolent from aggressive prostate cancer and predicting reclassification among low-/intermediate-risk patients undergoing AS may have a substantial impact on patient selection and management. Therefore, our findings have potentials to be implemented in the clinic. For example, localized prostate cancer patients with favorable genetic variants might benefit from delayed curative therapy therefore should be encouraged for AS enrollment. Serial biopsies with a longer interval which makes AS less invasive could be an option for these patients. On the other hand, more intensive disease progression monitoring and earlier decision to switch to curative therapy might be adopted for AS participants who carry unfavorable genotypes which increase their likelihood of disease reclassification. Currently, most of the criteria for AS subject selection are on the basis of clinical stage, serum PSA level and GS (15). We further suggest that patients’ genetic makeup should be taken into account for the patient selection and management in the future AS.
The genetic determinants of prostate cancer aggressiveness and reclassification are not well understood. Wnt pathway plays an important role in prostate cancer development and progression (10–12). Studies have shown that the crosstalk between Wnt pathway and other signaling pathways including AR, IGF-1, PI3K/Akt pathways can promote prostate cancer cell survival and growth (12). Somatic genetic alterations in the top 6 mutated Wnt pathway genes were observed in 30% of prostate cancer cases (FZD3: 11%, DVL2: 8%, PPP2CB: 8%, FZD2: 7%, FZD6: 7%, APC: 5%; cBioportal) (16), which further support the importance of Wnt pathway. We previously found that chromosome 19q13.13 (represented by rs2735839) was associated with prostate cancer aggressiveness, where the locus harbors PSA-coding gene KLK3 (7). Importantly, the locus was repeatedly reported to be associated with prostate cancer risk and aggressiveness (6,17,18). However, the interpretation of the associations of rs2735839 with prostate cancer risk and aggressiveness remain to be explored. It is not clear that whether the locus directly affects the predisposition to aggressive prostate cancer or the observed association is mainly mediated through the strong effect of rs2735839 (or other SNPs in the same LD block) on PSA levels and is therefore driven by PSA screening based early detection. Our findings corroborate with previous studies that the minor allele (A) of rs2735839 was strongly associated with low PSA levels (17,19), although the exact biological mechanisms involved are not fully understood. It has been shown that the frequency of allele A was higher among aggressive prostate cancer patients (7). Since the majority of patients were diagnosed on the basis of PSA screening, major allele G of rs2735839 are likely to be of higher frequency particularly in low-grade, less aggressive screen-detected patients. One study also found that the association between rs2735839 and prostate cancer risk was confined to the group of cases diagnosed in 1992 or later, when PSA screening was introduced and started to gain its acceptance (20). However, previous studies with retrospective case series designs cannot provide strong evidence that there is a direct effect of rs2735839 on prostate cancer aggressiveness. A more convincing argument can be made from the results of a prospective study such as our AS cohort. Only a few studies have assessed prostate cancer risk-associated SNPs for their associations with prostate cancer upgrading in a prospective AS cohort (8,9). No significant association was found in one study (9) and rs2735839 was not included in another (8). The discrepancy may stem from a different clinical endpoint used in the earlier study (9).
Unlike locus at 19q13.13, the association of prostate cancer aggressiveness with rs752822 at 5q32 might be mainly driven by GS, indicating that the PSA-related early prostate cancer detection may not play a role in this association. rs752822 is located in the intron of gene Casein Kinase 1, Alpha 1 (CSNK1A1). Together with adenomatous polyposis coli (APC), Axin, glycogen synthase kinase 3 (GSK-3) and protein phosphatase 2A (PP2A), casein kinase 1 (CK1) forms the β-catenin destruction complex (21). With Wnt signals, it results in an accumulation of β-catenin in nucleus, which subsequently increases expressions of KLK3 and other target genes that are essential for prostate cancer development and progression (22–24). Thus, our findings provided evidence that genetic variants in the CSNK1A1 may affect the predisposition to aggressive prostate cancer and the effect is independent from T stage and PSA levels.
Although our study is strengthened by its prospective nature of design, a relatively large sample size and adequate follow-up time, we acknowledge several limitations. In our AS cohort, we did not have sufficient samples within the group consisted of homozygous minor allele carriers. Thus, we only evaluated dominant model for each SNP. Another limitation is that our findings may not be generalized to other ethnicities as the majority of our populations are non-Hispanic whites. Due to small sample size limitation, we could not evaluate its associations in other racial groups. Further investigations are required to determine the risk-stratifying and prediction value of our genetic tool in African Americans and other ethnicities. Finally, further validations of our results on predictors of prostate cancer aggressiveness in large consortiums and predictors of reclassification in clinical trials like independent AS cohorts are warranted before potential clinical implementation.
In summary, we found a new SNP in the Wnt signaling pathway that is involved in the development of more aggressive prostate cancer. We added a new layer of evidence that rs2735839 may directly affect prostate cancer aggressiveness and reclassification in men with localized prostate cancer undergoing AS. Jointly, rs752822 and rs2735839 may have a potential role in the management of localized prostate cancer patients by optimizing patient selection and personalized monitoring process prior to/during AS.
Supplementary material
Supplementary Table 1 and Figure 1 can be found at http://carcin.oxfordjournals.org/
Funding
This study was financially supported by the NCI (grant CA140388) and the MD Anderson Cancer Center institutional support for the Center for Translational and Public Health Genomics.
Conflict of Interest Statement: None declared.
Supplementary Material
Glossary
Abbreviations
- AS
active surveillance
- SNP
single nucleotide polymorphism
References
- 1. Cooperberg M.R., et al. (2011) Active surveillance for prostate cancer: progress and promise. J. Clin. Oncol., 29, 3669–3676. [DOI] [PubMed] [Google Scholar]
- 2. Thomsen F.B., et al. (2014) Active surveillance for clinically localized prostate cancer–a systematic review. J. Surg. Oncol., 109, 830–835. [DOI] [PubMed] [Google Scholar]
- 3. Loeb S., et al. (2013) Systematic review of complications of prostate biopsy. Eur. Urol., 64, 876–892. [DOI] [PubMed] [Google Scholar]
- 4. Eeles R., et al. (2014) The genetic epidemiology of prostate cancer and its clinical implications. Nat. Rev. Urol., 11, 18–31. [DOI] [PubMed] [Google Scholar]
- 5. Al Olama A.A., et al. (2014) A meta-analysis of 87,040 individuals identifies 23 new susceptibility loci for prostate cancer. Nat. Genet., 46, 1103–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Berndt S.I., et al. (2015) Two susceptibility loci identified for prostate cancer aggressiveness. Nat. Commun., 6, 6889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. He Y., et al. (2014) The prostate cancer susceptibility variant rs2735839 near KLK3 gene is associated with aggressive prostate cancer and can stratify gleason score 7 patients. Clin. Cancer Res., 20, 5133–5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kearns J.T., et al. (2016) Associations Between iCOGS Single Nucleotide Polymorphisms and Upgrading in Both Surgical and Active Surveillance Cohorts of Men with Prostate Cancer. Eur. Urol., 69, 223–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Goh C.L., et al. (2013) Clinical implications of family history of prostate cancer and genetic risk single nucleotide polymorphism (SNP) profiles in an active surveillance cohort. BJU Int., 112, 666–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Giles R.H., et al. (2003) Caught up in a Wnt storm: Wnt signaling in cancer. Biochim. Biophys. Acta, 1653, 1–24. [DOI] [PubMed] [Google Scholar]
- 11. MacDonald B.T., et al. (2009) Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev. Cell, 17, 9–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Verras M., et al. (2006) Roles and regulation of Wnt signaling and beta-catenin in prostate cancer. Cancer Lett., 237, 22–32. [DOI] [PubMed] [Google Scholar]
- 13. Kypta R.M., et al. (2012) Wnt/beta-catenin signalling in prostate cancer. Nat. Rev. Urol., 9, 418–428. [DOI] [PubMed] [Google Scholar]
- 14. Davis J.W., et al. (2016) Disease reclassification risk with stringent criteria and frequent monitoring in men with favourable-risk prostate cancer undergoing active surveillance. BJU Int., 118, 68–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dall’Era M.A., et al. (2012) Active surveillance for prostate cancer: a systematic review of the literature. Eur. Urol., 62, 976–983. [DOI] [PubMed] [Google Scholar]
- 16. Gao J.J., et al. (2013) Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal., 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Eeles R.A., et al. (2008) Multiple newly identified loci associated with prostate cancer susceptibility. Nat. Genet., 40, 316–321. [DOI] [PubMed] [Google Scholar]
- 18. Kader A.K., et al. (2009) Individual and cumulative effect of prostate cancer risk-associated variants on clinicopathologic variables in 5,895 prostate cancer patients. Prostate, 69, 1195–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lindstrom S., et al. (2011) Characterizing associations and SNP-environment interactions for GWAS-identified prostate cancer risk markers–results from BPC3. PLoS One, 6, e17142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gudmundsson J., et al. (2010) Genetic correction of PSA values using sequence variants associated with PSA levels. Sci. Transl. Med., 2, 62ra92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Stamos J.L., et al. (2013) The beta-catenin destruction complex. Cold Spring Harb. Perspect. Biol., 5, a007898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mulholland D.J., et al. (2003) Functional localization and competition between the androgen receptor and T-cell factor for nuclear beta-catenin: a means for inhibition of the Tcf signaling axis. Oncogene, 22, 5602–5613. [DOI] [PubMed] [Google Scholar]
- 23. Yang X., et al. (2006) Complex regulation of human androgen receptor expression by Wnt signaling in prostate cancer cells. Oncogene, 25, 3436–3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li Y., et al. (2009) LEF1 in androgen-independent prostate cancer: regulation of androgen receptor expression, prostate cancer growth and invasion. Cancer Res., 69, 3332–3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.