Abstract
Quantitation of the HIV-1 viral load in plasma is the current standard of care for clinical monitoring of HIV-infected individuals undergoing antiretroviral therapy. This study evaluated the analytical and clinical performances of the Aptima HIV-1 Quant Dx assay (Hologic, San Diego, CA) for monitoring viral load by using 277 well-characterized subtype samples, including 171 cultured virus isolates and 106 plasma samples from 35 countries, representing all major HIV subtypes, recombinants, and circulating recombinant forms (CRFs) currently in circulation worldwide. Linearity of the Aptima assay was tested on each of 6 major HIV-1 subtypes (A, B, C, D, CRF01_AE, and CRF02_AG) and demonstrated an R2 value of ≥0.996. The performance of the Aptima assay was also compared to those of the Roche COBAS AmpliPrep/COBAS TaqMan HIV-1 v.2 (CAP/CTM) and Abbott m2000 RealTime HIV-1 (RealTime) assays on all subtype samples. The Aptima assay values averaged 0.21 log higher than the CAP/CTM values and 0.30 log higher than the RealTime values, and the values were >0.4 log higher than CAP/CTM values for subtypes F and G and than RealTime values for subtypes C, F, and G and CRF02_AG. Two samples demonstrated results with >1-log differences from RealTime results. When the data were adjusted by the average difference, 94.9% and 87.0% of Aptima results fell within 0.5 log of the CAP/CTM and RealTime results, respectively. The linearity and accuracy of the Aptima assay in correctly quantitating all major HIV-1 subtypes, coupled with the completely automated format and high throughput of the Panther system, make this system well suited for reliable measurement of viral load in the clinical laboratory.
INTRODUCTION
HIV viral load assays are an essential tool in the management of HIV infections and are used to measure an individual's infectivity, evaluate the risk for disease progression, guide the response to antiretroviral therapy (ART), and monitor failure of virologic control. Current guidelines recommend that ART be provided to all HIV-infected patients regardless of CD4 T-cell count and that viral loads of HIV-infected patients be measured at baseline and at regular (6 to 12 months) intervals after initiation of treatment to monitor treatment effectiveness (1–4). Accurate, sensitive measurement of viral load at regular intervals is required to monitor the effectiveness of therapy. Virologic failure as defined by the World Health Organization (WHO) or the U.S. Department of Health and Human Services (HHS) is an increase in plasma HIV RNA levels to >1,000 copies/ml (1) or ≥200 copies/ml (2) as measured at 3-month intervals on therapy, but even viral loads of <200 copies/ml can be predictive of viral rebound (5, 6) and can be associated with the evolution of drug resistance (7). Initiation of antiretroviral therapy at diagnosis could lead to a significant decrease in HIV transmission rates and, ultimately, to reductions in HIV incidence rates (8).
A potential challenge to the performance of viral load assays is the extensive genetic diversity of HIV subtypes, which can vary by as much as 17% within a single subtype and up to 35% between subtypes, depending on the genomic regions examined (9). HIV diversity affects HIV diagnosis and viral load measurements (10, 11) and may affect the response to antiretroviral treatment and the emergence of drug resistance (12, 13). Subtypes can differ in the rate of disease progression (14–16) and may be transmitted at different rates (17, 18). It is therefore critical that viral load assays deployed worldwide demonstrate high sensitivity and accurate quantification of all HIV-1 subtypes across the analytical range of the assay (19).
The Hologic Aptima HIV-1 Quant Dx assay (Aptima assay; Hologic Inc., San Diego, CA) employs transcription-mediated amplification (TMA) and real-time detection for rapid, high-throughput testing of viral load on the Panther platform (20). This assay is the first fully automated assay to receive CE certification for intended use as an aid in HIV-1 diagnosis as well as for clinical monitoring and to permit physicians to confirm a patient's HIV infection diagnosis, obtain the patient's baseline viral load, and initiate ART immediately. FDA clearance for the Aptima assay with intended use for clinical viral load monitoring of HIV-1 in the United States is pending.
The purpose of this study was to evaluate the analytical and clinical performances of the Aptima assay on a diverse global collection of HIV subtypes. Performance was assessed for accuracy and linearity for each major subtype on well-characterized panels of cultured viruses. The ability of the Aptima assay to accurately quantitate a diverse range of HIV subtypes was evaluated on 277 well-characterized cultured viral isolates and plasma samples from 35 countries and compared to the results of two U.S. FDA-approved HIV-1 quantitative assays: the COBAS AmpliPrep/COBAS TaqMan HIV-1 test, version 2 (CAP/CTM assay; Roche Molecular Systems, Branchburg, NJ), and the m2000 RealTime HIV-1 assay (RealTime assay; Abbott Molecular, Des Plaines, IL). Other assays for viral load measurements include the HIV-1 NucliSens EasyQ v2.0 assay (bioMérieux, Marcy l'Etoile, France) and the Xpert HIV-1 viral load assay (Cepheid, Sunnyvale, CA). These assays are CE marked for plasma viral load testing in Europe but were not included in this evaluation.
MATERIALS AND METHODS
Sources of HIV-1 subtype samples.
A total of 277 well-characterized HIV-1 subtype samples were assembled from various sources to include 171 HIV-1 isolates cultured in peripheral blood mononuclear cells and 106 clinical specimens from 35 countries, representing all the major HIV-1 group M subtypes and HIV-1 group O. Samples from the Walter Reed Army Institute of Research (WRAIR) included 60 isolates, 10 each of the 6 major HIV-1 subtypes (A, B, C, D, CRF01_AE, and CRF02_AG), representing cultured viruses from blood samples collected between 1995 and 2002 (21). An additional group of 46 isolates (Accutype) were obtained from SeraCare (Medford, MA) and included cultures from plasma samples collected by the National Institute of Allergy and Infectious Disease (NIAID), the European Virus Association, and the U.S. Department of Defense (DoD) (NED Collection) from 1992 to 2002 (22), supplemented with more recent worldwide isolates to include four group O viruses and several unique recombinants. An additional 65 worldwide viral isolates, most of which were cultured from samples collected between 2009 and 2014 from individuals with acute HIV infection (23, 24), were obtained from the EQAPOL collection (http://eqapol.dhvi.duke.edu) under a Material Transfer Agreement (MTA) with Duke University (Durham, NC). All cultured viruses included information on the year of collection, country of origin, HIV RNA copies per milliliter, and subtype designation as determined by sequencing of gag, pol, and env and, in most cases, full-genome sequencing. Cultured isolates were obtained at high titers (typically 108 to 109 copies/ml) as quantitated by the respective providers, using the CAP/CTM and RealTime assays. All isolates were diluted at WRAIR to approximately 105 copies/ml by using BaseMatrix, an HIV-1 RNA-negative defibrinated plasma product (SeraCare, Medford, MA), or human plasma.
The cultured virus samples were supplemented with an additional 106 well-characterized plasma samples from consenting HIV-infected individuals participating in WRAIR Institutional Review Board (IRB)-approved HIV-1 vaccine or surveillance studies conducted in Africa and Asia. Plasma samples collected between 2010 and 2014 included samples from recent seroconverters in Thailand (WRAIR study 1587) or from infected individuals in Kenya, Nigeria, Tanzania, Uganda, and Thailand (study RV217), Uganda (study RV172), Thailand (study RV91), or Nigeria (study RV230). Characterization of each specimen included the year collected, country of origin, HIV enzyme immunoassay (EIA) result, Western blot result, viral load, and subtype designation based on full-length sequencing or multiregion hybridization assays (MHA) (25). One milliliter of each plasma sample was diluted in BaseMatrix by a minimum of a 1:3 dilution, or to approximately 104 copies/ml in cases where sufficiently high titers were available, to allow testing of one replicate sample in each of the three viral load assays.
The distribution of the 277 subtypes and the 35 countries of origin is shown in Table 1. This extensive collection included representative isolates from North and South America, Europe, and Asia, with a particularly strong focus on African countries (171 specimens), where the epidemic is most developed and subtypes are most diverse. African countries that contributed one or two specimens were grouped under “other” and included Algeria, Ghana, Liberia, Malawi, Rwanda, Zambia, Democratic Republic of Congo, and one unknown. Major subtypes included A, B, C, D, and G as well as the less frequent subtype F and group O. Also represented in the panel were the predominant circulating recombinant forms (CRFs), namely, CRF01_AE and CRF02_AG. “Other recombinants” included G/CRF02_AG (13 isolates), CRF01/B (3 isolates), CRF02/A3, CRF04_CPX (4 isolates), CRF06_cpx, CRF11_cpx, A1B (URF), BF (2 isolates), ACD (4 isolates), CD (2 isolates), AC (3 isolates), AD (7 isolates), and AFGHKU.
TABLE 1.
HIV-1 samples used in the comparative evaluation, by subtype and origina
| Subtype | No. of samples |
||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Americas |
Europe |
Africa |
Asia |
Total | |||||||||||||||||||||||||
| USA | Bolivia | Brazil | Uruguay | Venezuela | France | Germany | Greece | Poland | Romania | Spain | Angola | Cameroon | Djibouti | Ethiopia | Kenya | Nigeria | Senegal | Somalia | South Africa | Tanzania | Uganda | Other | China | India | Indonesia | Kyrgyzstan | Thailand | ||
| A | 1 | 8 | 1 | 2 | 16 | 2 | 1 | 31 | |||||||||||||||||||||
| B | 19 | 1 | 3 | 1 | 1 | 3 | 1 | 3 | 1 | 1 | 3 | 37 | |||||||||||||||||
| C | 1 | 1 | 1 | 1 | 1 | 3 | 1 | 1 | 2 | 2 | 14 | 3 | 2 | 3 | 1 | 37 | |||||||||||||
| D | 2 | 1 | 1 | 1 | 19 | 24 | |||||||||||||||||||||||
| F | 3 | 1 | 1 | 2 | 7 | ||||||||||||||||||||||||
| G | 2 | 1 | 1 | 15 | 19 | ||||||||||||||||||||||||
| O | 1 | 1 | 2 | 4 | |||||||||||||||||||||||||
| CRF01_AE | 3 | 2 | 34 | 39 | |||||||||||||||||||||||||
| CRF02_AG | 1 | 1 | 12 | 3 | 15 | 1 | 33 | ||||||||||||||||||||||
| Other recombinants | 1 | 4 | 4 | 1 | 14 | 4 | 12 | 3 | 3 | 46 | |||||||||||||||||||
| Total | 22 | 1 | 8 | 1 | 1 | 3 | 3 | 4 | 3 | 1 | 11 | 2 | 18 | 4 | 3 | 12 | 46 | 3 | 2 | 14 | 9 | 49 | 9 | 4 | 1 | 2 | 1 | 40 | 277 |
The subtypes include 171 well-characterized cultured virus samples, obtained from WRAIR, SeraCare, and EQAPOL, plus 106 plasma samples from HIV-infected individuals in Africa and Asia. HIV subtype designations were assigned on the basis of sequence analysis or multiregion hybridization assay (MHA), and the country of origin for each specimen/isolate is shown. There were 33, 25, 171, and 48 total samples from the Americas, Europe, Africa, and Asia, respectively.
Viral load assays.
All samples were tested in accordance with the manufacturers' instructions (26) at WRAIR, Silver Spring, MD, by use of the Roche COBAS AmpliPrep/COBAS TaqMan HIV-1 test, v2.0 (CAP/CTM assay), using a COBAS AmpliPrep instrument for automated extraction of RNA and with PCR amplification and detection on a COBAS TaqMan 48 analyzer (Indianapolis, IN); the Abbott RealTime HIV-1 assay (RealTime assay), performed on an automated m2000 system (Chicago, IL); and the Hologic Aptima HIV-1 Quant Dx assay (Aptima assay), performed on a Panther system (San Diego, CA).
(i) Aptima assay.
Samples (0.7 ml; 0.5-ml assay volume) were transferred to specimen aliquot tubes, vortexed, centrifuged at 1,100 × g for 10 min, and loaded directly onto a Panther system. The Panther system performs automated extraction and amplification of the HIV-1 long terminal repeat (LTR) and pol gene target sequences by isothermal transcription-mediated amplification and quantitative analysis. The assay's reported limit of detection (LOD) is 12 copies/ml with the 3rd WHO HIV-1 international standard, and it has a linear range of quantitation of 30 to 10,000,000 copies/ml (20).
(ii) CAP/CTM assay.
Samples (1.05 ml; 0.85-ml assay volume) were transferred into input S tubes and loaded onto a COBAS AmpliPrep instrument for purification of RNA via magnetic silica bead extraction and for PCR setup. A K carrier with K tubes was manually transferred to a COBAS TaqMan 48 analyzer for amplification of the HIV-1 LTR and gag target sequences by reverse transcription-PCR (RT-PCR), detection, and quantification. The assay's reported LOD is 20 copies/ml with the 2nd WHO HIV-1 international standard, and it has a linear range of quantitation of 20 to 10,000,000 copies/ml (26).
(iii) RealTime assay.
Plasma (0.8 ml; 0.6-ml assay volume) was transferred to Simport tubes, vortexed, centrifuged at 2,000 × g for 5 min, and loaded onto an Abbott m2000 sample preparation system for HIV-1 RNA purification. A PCR tray was set up in the m2000sp system and transferred to an Abbott m2000rt PCR instrument for amplification and detection of the HIV-1 integrase gene target sequence. The assay's reported LOD is 40 copies/ml with DAIDS Virology Quality Assurance (VQA) standards, and it has a linear range of quantitation of 40 to 10,000,000 copies/ml (27).
Data analyses.
The linearity of the Aptima assay as measured on serial dilution panels of each of the major subtypes was determined by the correlation coefficient, expressed as the R2 value. HIV-1 RNA quantitative values were compared for each of the subtypes by using linear regression analysis. The fitted regression line was compared to the line of equality (slope = 1, intercept = 0) and a 0.5-log difference on both sides of the line of equality to assess agreement among Aptima, CAP/CTM, and RealTime assay results (log-transformed values) for each subtype. Agreements between the assays were assessed using the Bland-Altman test.
Ethical considerations.
The use of clinical specimens from open protocols to assess HIV-1 viral load assays (studies RV366 and WRAIR 1947) was approved by the WRAIR IRB and local IRB committees in each of the countries from which specimens were obtained. All plasma samples were collected from volunteers of at least 18 years of age who had completed the IRB-approved informed consent process. Preexisting clinical specimens received in this study were deidentified in accordance with IRB-approved study RV372. HIV-1 stocks, HIV-1 panels, and blood products were obtained in accordance with study RV234, an IRB-exempt protocol for use of HIV-1 stocks, HIV-1 panels, and blood products.
RESULTS
Linearity.
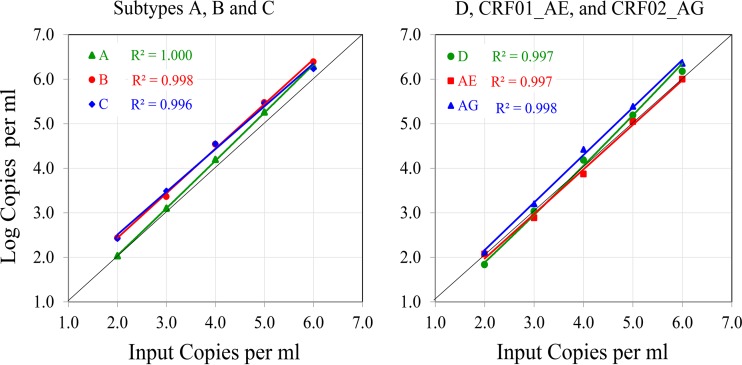
Linearity of the Aptima HIV-1 Quant Dx assay was evaluated on virus isolates from six major HIV-1 subtypes: A, B, C, D, CRF01_AE, and CRF02_AG. Tenfold serial dilutions were prepared for each isolate, targeting concentrations of 100 to 1,000,000 (log 2.0 to 6.0) copies/ml based on quantitation of the respective stocks by the Abbott m2000 RealTime assay. Aptima results were measured in duplicate, and the coefficient of determination (R2) for each subtype was determined (Fig. 1). The Aptima assay demonstrated excellent linearity for each of the subtypes tested, with R2 values of ≥0.996.
FIG 1.
Linearity of Aptima results by HIV subtype. One representative of each subtype (A, B, C, D, CRF01_AE, and CRF02_AG), at dilutions ranging from 100 to 1,000,000 (log 2.0 to 6.0) copies/ml, was tested in duplicate in the Aptima assay. The average viral load for 2 replicates is plotted for each data point. Input copies are based on RealTime results.
Comparison of viral load measurements by Aptima versus CAP/CTM and RealTime assays on major HIV-1 subtypes and groups.
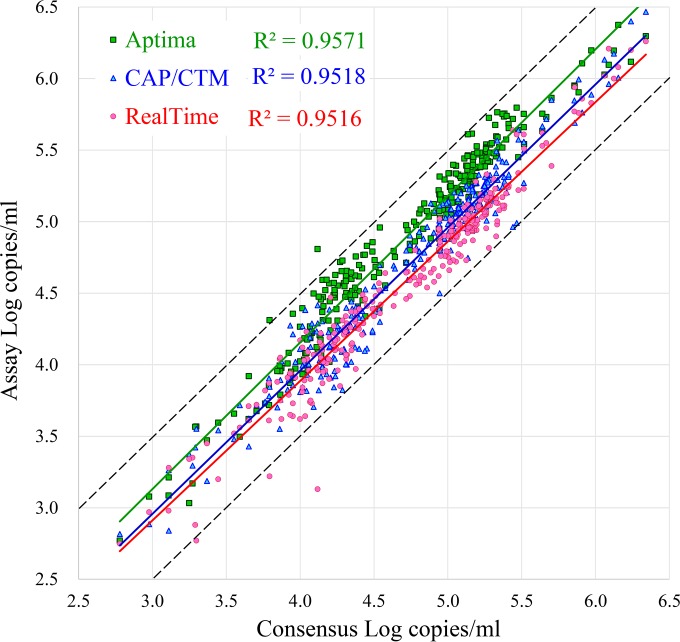
Replicate samples of a total of 277 well-characterized HIV-1 samples (171 cultured HIV-1 isolates from 33 countries and 106 clinical specimens from 5 countries), representing all the major group M subtypes and group O, were tested by the Aptima, CAP/CTM, and RealTime assays. All samples yielded quantitative values in all assays and were included in the data analysis. Figure 2 demonstrates the results for the three assays plotted against consensus (average) values. A plot of Aptima values relative to those of either the CAP/CTM or RealTime assay demonstrates nearly parallel trend lines, with R2 values of 0.95 to 0.96, indicating that all three assays remain proportional throughout the quantitative range tested. On average, the Aptima assay results tended to run 0.17 log higher than the consensus values, the RealTime assay results ran 0.13 log lower, and the CAP/CTM results ran very close to the consensus values (0.04 log lower). All CAP/CTM results were within 0.5 log of the consensus results. When the data were adjusted by the average difference, 94.9% and 87.0% of Aptima results fell within 0.5 log of the CAP/CTM and RealTime results.
FIG 2.
Comparison of three assays on all viral subtypes tested. The results of RealTime, CAP/CTM, and Aptima testing of the 277 subtype samples tested are plotted against consensus values representing the means for the 3 measurements. The black solid and dotted lines represent the consensus ± 0.5 log. The green, blue, and red lines represent trend lines for the Aptima, CAP/CTM, and RealTime assays, respectively.
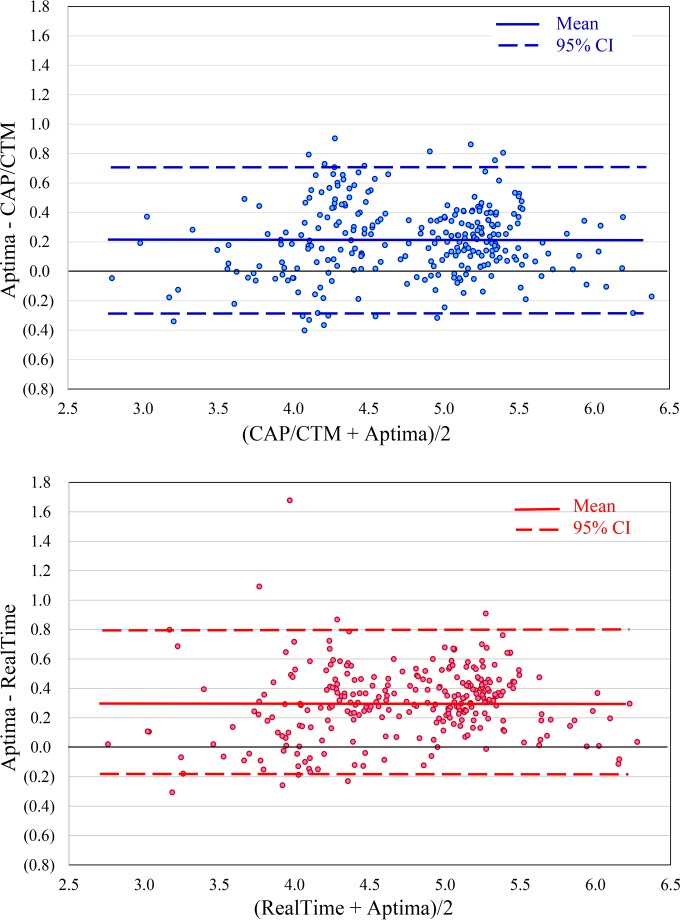
Bland-Altman analyses of the differences in Aptima results versus CAP/CTM and RealTime results relative to the respective average values are shown in Fig. 3. The results show overall biases of 0.214 (95% confidence interval [95% CI], 0.708 to −0.280) and 0.305 (95% CI, 0.790 to −0.180) higher results by the Aptima assay than the CAP/CTM and RealTime assays, respectively, with the 95% confidence intervals falling within 0.5 log of the adjusted value in both cases.
FIG 3.
Bland-Altman analyses of Aptima versus CAP/CTM and RealTime quantitation of the subtype panel. Each point represents a separate sample. The central lines represent the biases of 0.214 (95% CI, 0.708 to −0.280) and 0.305 (95% CI, 0.790 to −0.180) for the CAP/CTM and RealTime assays, respectively.
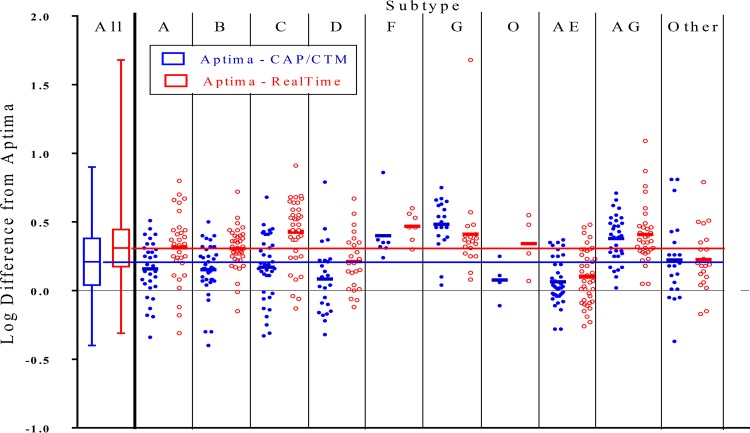
Figure 4 demonstrates a plot of the log difference between Aptima and CAP/CTM results or Aptima and RealTime results for each sample as grouped by major subtype, with other recombinants and other CRFs grouped separately. As seen previously, the Aptima results generally tended to be 0.30 log higher than RealTime results and 0.21 log higher than CAP/CTM results, with the largest differences seen for subtypes C, F, and G and CRF02_AG between Aptima and RealTime results and for subtype G and CRF02_AG between Aptima and CAP/CTM results. Forty-eight of the 277 samples (17.3%) had more than a half-log difference between Aptima and RealTime results. Of these, one subtype G sample tested at 4.81 log in the Aptima assay but at 3.13 log in the RealTime assay, for a 1.68-log difference. Two CRF02_AG samples showed 1.09- and 0.87-log differences between Aptima and RealTime results, a subtype A sample showed a 0.80-log difference, and a subtype C sample showed a 0.91-log difference. The CAP/CTM results tended to run between these two extremes.
FIG 4.
Comparison of Aptima quantitative results versus CAP/CTM (red) and RealTime (blue) results by subtype. The difference between Aptima and CAP/CTM results (blue) or Aptima and RealTime results (red) is plotted by subtype, with each dot representing a single sample. The first column shows whisker plots for all the samples, indicating the means (center lines), 25th to 75th percentiles (boxes), and extents of outliers (lines). The solid black horizontal line represents the value for the Aptima assay (no difference), with the blue and red lines representing the average overall differences from the Aptima results of 0.21 log for the CAP/CTM assay and 0.30 log for the RealTime assay.
The means and standard deviations (SD) for differences between the Aptima assay and the CAP/CTM or RealTime assay as grouped by subtype are summarized in Table 2. The Aptima assay yielded higher quantitative values than those of the CAP/CTM assay, by 0.21 log, and those of the RealTime assay, by 0.30 log. Average Aptima values were higher than RealTime values, by 0.43 log for subtype C, 0.47 log for subtype F, and 0.41 log for subtype G and CRF02_AG. Similarly, Aptima results were higher than CAP/CTM results, by 0.40 log for subtype F and 0.48 log for subtype G.
TABLE 2.
Mean differences in Aptima versus CAP/CTM and RealTime assay resultsa
| Subtype | n | Aptima minus CAP/CTM |
Aptima minus RealTime |
||
|---|---|---|---|---|---|
| Mean | SD | Mean | SD | ||
| A | 31 | 0.16 | 0.21 | 0.31 | 0.26 |
| B | 37 | 0.15 | 0.19 | 0.31 | 0.15 |
| C | 37 | 0.17 | 0.24 | 0.43 | 0.24 |
| D | 24 | 0.08 | 0.25 | 0.21 | 0.21 |
| F | 7 | 0.40 | 0.21 | 0.47 | 0.11 |
| G | 19 | 0.48 | 0.19 | 0.41 | 0.33 |
| O | 4 | 0.08 | 0.15 | 0.34 | 0.22 |
| CRF01_A/E | 39 | 0.06 | 0.17 | 0.10 | 0.20 |
| CRF02_A/G | 33 | 0.38 | 0.17 | 0.41 | 0.21 |
| Other recombinants | 46 | 0.28 | 0.27 | 0.27 | 0.22 |
| Total | 277 | 0.21 | 0.25 | 0.30 | 0.24 |
Differences between Aptima and CAP/CTM or RealTime assay results are shown for each of the major subtypes. The number of isolates/plasma samples tested for each subtype is shown, along with the means and standard deviations of the differences between the assays, expressed in log copies per milliliter.
DISCUSSION
Plasma HIV-1 viral load measurements are an important tool for the evaluation of infection status and risk of disease progression, and they provide critical guidance for antiretroviral treatment decisions (13). HIV-1 viral load assays used for clinical monitoring must meet strict performance criteria, including high accuracy and precision, detection sensitivity (low LOD), and linearity across the dynamic range of the assay (1, 2, 7, 28). In view of the genetic diversity of HIV-1 infections worldwide and the impacts of migration and travel on dissemination, it is essential that viral load assays meet these strict performance criteria across a dynamic population of HIV-1 subtypes (23, 24, 29).
According to the product insert, the Aptima assay's LOD (12 copies/ml) is similar to that of the CAP/CTM assay (20 copies/ml) and lower than that of the RealTime assay (40 copies/ml), despite using a smaller sample volume (0.5 ml) than that for the CAP/CTM (0.85 ml) or RealTime (0.6 ml) assay (20, 26, 27). Previous performance evaluations of viral load assays have shown the Aptima assay to be more sensitive than the RealTime assay (30, 31) for detection of HIV-1 RNA in serial dilutions of the WHO 3rd international standard and that the assay is capable of detecting lower viral loads in clinical samples (32, 33). A more recent study showed the Aptima assay to be more sensitive than the CAP/CTM assay on both standards and clinical samples (34). The modest increase (1.5- to 2-fold) in viral loads observed at concentrations above 10,000 copies/ml of HIV-1 by using the Hologic assay relative to the comparator assays should have a minimal impact on treatment decisions because these concentrations are significantly above the 200-copies/ml medical decision point in the NIH treatment guidelines (2). Studies conducted by various investigators showed that although the Aptima assay detected HIV in more samples, the number of quantified results above 50 copies/ml reported by the Aptima assay was lower than those for the CAP/CTM and RealTime assays (30–33). Therefore, the likelihood of a patient categorized as virologically controlled by the CAP/CTM or RealTime assay (below 200 copies/ml) yielding results above the medical decision point (200 copies/ml) in the Aptima assay is low. The higher sensitivity of the Aptima assay may result in more patients receiving an “HIV detected” result than occurs with the CAP/CTM or RealTime assay, although the clinical relevance of persistent low-level HIV-1 RNA near the limit of detection remains undefined.
One of the main challenges for implementation of viral load assays in an era of increasing global mobility is ensuring accurate quantitation of all major HIV-1 subtypes, groups, and recombinants. Previous studies on the performance of quantitative assays carried out on limited numbers of HIV-1 subtypes showed significant variability between viral load assays on different subtypes. Underquantitation of subtype A1 samples by the NucliSens v2.0 and RealTime assays, of subtype C by the RealTime assay, and of subtypes G and CRF02_AG by the NucliSens EasyQ v2.0 test was observed relative to the Xpert and Aptima assays, particularly for samples in the lower viremic range (35). Similarly, subtype CRF02_AG was underestimated by the NucliSens EasyQ v2.0 test (36), while the Aptima assay showed greater sensitivity than the CAP/CTM or RealTime assay on viral isolates and clinical samples but equivalent quantification on clinical samples and isolates belonging to HIV groups M, N, O, and P (34). An additional concern is the ability to test low-volume specimens and alternative specimen matrices. The Aptima assay product insert describes dilution (1:3) testing of low-volume specimens by use of Aptima specimen diluent (Hologic). Alternative sample types successfully run on the Panther system using the Aptima assay include cervicovaginal lavage fluid (33), serum (34), and dried blood spots (37).
In this study, we examined the performance of the Hologic Aptima HIV-1 Quant Dx assay in accurately quantitating plasma viral loads of HIV-1 subtypes from diverse geographic origins, including those currently in widespread circulation as well as various common and unique recombinants. The HIV RNA concentrations measured in the Aptima assay for the subtype samples studied were higher than those measured by the CAP/CTM and RealTime assays, by 0.21 and 0.30 log, respectively, on average. Aptima results were higher than CAP/CTM results for subtypes F and G and higher than RealTime results for subtypes C, F, and G and CRF02_AG, by >0.4 log. Two outliers (one subtype G and one CRF01_AG specimen) showed more than a log difference between Aptima and RealTime results, while all remaining CAP/CTM and RealTime results were within 1 log of the Aptima results. Differences observed among assay values could be accounted for by calibration against the WHO standard. According to the manufacturers' package inserts, the Aptima assay is calibrated at 0.35 copy per WHO international unit, the CAP/CTM assay at 0.59, and the RealTime assay at 0.58, which would transform to Aptima values of 0.23 and 0.22 log higher. Thus, differences in quantitation between the three assays can be normalized to nearly equivalent levels if the data are expressed in international units, although quantitation of subtypes C, F, and G and CRF02_AG would still be 0.20 to 0.25 log lower by RealTime assay than by Aptima assay. Overall differences in quantitation among the three assays are relatively small and are comparable across diverse group M subtypes, recombinant circulating forms, and group O samples, and the results are consistent with previous studies (30–36). Our study demonstrates the excellent linearity and accuracy of the Aptima assay in quantifying viral load measurements on diverse HIV-1 subtypes, groups, and circulating recombinant forms, even at very low RNA levels. The Panther instrument features random access of samples to permit simultaneous testing of different pathogens within the same test run, has easy-to-use workflows, requires less stringent technical skills, and can generate up to 320 results in an 8-h period. The linearity and accuracy of the Aptima assay in the quantitation of HIV-1 on a panel of diverse HIV-1 isolates and clinical specimens collected worldwide, along with full automation on the Panther system, make the Aptima HIV-1 Quant Dx assay well suited for routine use in the clinical laboratory.
ACKNOWLEDGMENTS
This work was supported by a cooperative agreement (agreement W81XWH-11-2-0174) between the Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc., and the U.S. Department of Defense (DoD) and by a Cooperative Research and Development Agreement with Hologic, Inc.
We thank Florence Paillard for her assistance in drafting the manuscript. We also thank Merlin Robb and Ogbonnaya Njoku and other WRAIR investigators for kindly providing access to the clinical samples used in this evaluation.
The views expressed are those of the authors and should not be construed to represent the positions of the U.S. Army or the Department of Defense.
S.V.N. and A.W. are employees of Hologic, Inc.
REFERENCES
- 1.World Health Organization (WHO). 2014. March 2014 supplement to the 2013 consolidated guidelines on the use of antiretroviral drugs for treating and preventing HIV infection: recommendations for a public health approach. World Health Organization, Geneva, Switzerland: http://www.who.int/hiv/pub/guidelines/arv2013/arvs2013upplement_march2014/en/ Accessed 20 February 2016. [Google Scholar]
- 2.Department of Health and Human Services (DHHS) Panel on Antiretroviral Guidelines for Adults and Adolescents. 28 January 2016. Guidelines for the use of antiretroviral agents in HIV-1-infected adults and adolescents. http://www.aidsinfo.nih.gov/ContentFiles/AdultandAdolescentGL.pdf. Accessed 16 February 2016. [Google Scholar]
- 3.Williams I, Churchill D, Anderson J, Boffito M, Bower M, Cairns G, Cwynarski K, Edwards S, Fidler S, Fisher M, Freedman A, Geretti AM, Gilleece Y, Horne R, Johnson M, Khoo S, Leen C, Marshall N, Nelson M, Orkin C, Paton N, Phillips A, Post F, Pozniak A, Sabin C, Trevelion R, Ustianowski A, Walsh J, Waters L, Wilkins E, Winston A, Youle M. 2014. British HIV Association guidelines for the treatment of HIV-1-positive adults with antiretroviral therapy 2012. (Updated November 2013. All changed text is cast in yellow highlight.) HIV Med 15(Suppl 1):S1–S85. doi: 10.1111/hiv.12119. [DOI] [PubMed] [Google Scholar]
- 4.European AIDS Clinical Society. October 2015. EACS guidelines, version 8.0. http://www.eacsociety.org/guidelines/eacs-guidelines/eacsguidelines.html. Accessed 26 February 2016.
- 5.Eron JJ, Cooper DA, Steigbigel RT, Clotet B, Gatell JM, Kumar PN, Rockstroh JK, Schechter M, Markowitz M, Yeni P, Loutfy MR, Lazzarin A, Lennox JL, Strohmaier KM, Wan H, Barnard RJ, Nguyen BY, Teppler H, BENCHMRK Study Teams . 2013. Efficacy and safety of raltegravir for treatment of HIV for 5 years in the BENCHMRK studies: final results of two randomised, placebo-controlled trials. Lancet Infect Dis 13:587–596. doi: 10.1016/S1473-3099(13)70093-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Laprise C, de Pokomandy A, Baril JG, Dufresne S, Trottier H. 2013. Virologic failure following persistent low-level viremia in a cohort of HIV-positive patients: results from 12 years of observation. Clin Infect Dis 57:1489–1496. doi: 10.1093/cid/cit529. [DOI] [PubMed] [Google Scholar]
- 7.Taiwo B, Gallien S, Aga E, Ribaudo H, Haubrich R, Kuritzkes DR, Eron JJ Jr. 2011. Antiretroviral drug resistance in HIV-1-infected patients experiencing persistent low-level viremia during first-line therapy. J Infect Dis 204:515–520. doi: 10.1093/infdis/jir353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bennett B, Hardy B, Fordan S, Haddock-Morilla L, Rowlinson MC, Crowe S. 2013. HIV single staging algorithm: integration and maximization of resources by reducing time between HIV diagnosis and treatment. J Clin Virol 58(Suppl 1):e34–e37. doi: 10.1016/j.jcv.2013.07.020. [DOI] [PubMed] [Google Scholar]
- 9.Korber B, Gaschen B, Yusim K, Thakallapally R, Kesmir C, Detours V. 2001. Evolutionary and immunological implications of contemporary HIV-1 variation. Br Med Bull 58:19–42. doi: 10.1093/bmb/58.1.19. [DOI] [PubMed] [Google Scholar]
- 10.Plantier JC, Djemai M, Lemée V, Reggiani A, Leoz M, Burc L, Vessière A, Rousset D, Poveda JD, Henquell C, Gautheret-Dejean A, Barin F. 2009. Census and analysis of persistent false-negative results in serological diagnosis of human immunodeficiency virus type 1 group O infections. J Clin Microbiol 47:2906–2911. doi: 10.1128/JCM.00602-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim JE, Beckthold B, Chen Z, Mihowich J, Malloch L, Gill MJ. 2007. Short communication: identification of a novel HIV type 1 subtype H/J recombinant in Canada with discordant HIV viral load (RNA) values in three different commercial assays. AIDS Res Hum Retroviruses 23:1309–1313. doi: 10.1089/aid.2007.0080. [DOI] [PubMed] [Google Scholar]
- 12.Geretti AM, Harrison L, Green H, Sabin C, Hill T, Fearnhill E, Pillay D, Dunn D, UK Collaborative Group on HIV Drug Resistance. 2009. Effect of HIV-1 subtype on virologic and immunologic response to starting highly active antiretroviral therapy. Clin Infect Dis 48:1296–1305. doi: 10.1086/598502. [DOI] [PubMed] [Google Scholar]
- 13.Martinez-Cajas JL, Pai NP, Klein MB, Wainberg MA. 2009. Differences in resistance mutations among HIV-1 non-subtype B infections: a systematic review of evidence (1996–2008). J Int AIDS Soc 12:11. doi: 10.1186/1758-2652-12-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kiwanuka N, Laeyendecker O, Robb M, Kigozi G, Arroyo M, McCutchan F, Eller LA, Eller M, Makumbi F, Birx D, Wabwire-Mangen F, Serwadda D, Sewankambo NK, Quinn TC, Wawer M, Gray R. 2008. Effect of human immunodeficiency virus type 1 (HIV-1) subtype on disease progression in persons from Rakai, Uganda, with incident HIV-1 infection. J Infect Dis 197:707–713. doi: 10.1086/527416. [DOI] [PubMed] [Google Scholar]
- 15.Baeten JM, Chohan B, Lavreys L, Chohan V, McClelland RS, Certain L, Mandaliya K, Jaoko W, Overbaugh J. 2007. HIV-1 subtype D infection is associated with faster disease progression than subtype A in spite of similar plasma HIV-1 loads. J Infect Dis 195:1177–1180. doi: 10.1086/512682. [DOI] [PubMed] [Google Scholar]
- 16.Vasan A, Renjifo B, Hertzmark E, Chaplin B, Msamanga G, Essex M, Fawzi W, Hunter D. 2006. Different rates of disease progression of HIV type 1 infection in Tanzania based on infecting subtype. Clin Infect Dis 42:843–852. doi: 10.1086/499952. [DOI] [PubMed] [Google Scholar]
- 17.Kiwanuka N, Laeyendecker O, Quinn TC, Wawer MJ, Shepherd J, Robb M, Kigozi G, Kagaayi J, Serwadda D, Makumbi FE, Reynolds SJ, Gray RH. 2009. HIV-1 subtypes and differences in heterosexual HIV transmission among HIV-discordant couples in Rakai, Uganda. AIDS 23:2479–2484. doi: 10.1097/QAD.0b013e328330cc08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Renjifo B, Gilbert P, Chaplin B, Msamanga G, Mwakagile D, Fawzi W, Essex M, Tanzanian Vitamin and HIV Study Group. 2004. Preferential in-utero transmission of HIV-1 subtype C as compared to HIV-1 subtype A or D. AIDS 18:1629–1636. doi: 10.1097/01.aids.0000131392.68597.34. [DOI] [PubMed] [Google Scholar]
- 19.Hemelaar J, Gouws E, Ghys PD, Osmanov S, WHO-UNAIDS Network for HIV Isolation and Characterisation . 2011. Global trends in molecular epidemiology of HIV-1 during 2000–2007. AIDS 25:679–689. doi: 10.1097/QAD.0b013e328342ff93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hologic, Inc. November 2014. Aptima HIV-1 Quant Dx assay product information. Hologic, Inc, San Diego, CA: http://stage.hologic.com/products/clinical-diagnostics-and-bloodscreening/assays-and-tests/Aptima-hiv-1-quant-dx-assay. [Google Scholar]
- 21.Brown B, Darden J, Tovanabutra S, Oblander T, Frost J, Sanders-Buell E, de Souza M, Birx D, McCutchan F, Polonis V. 2005. Biologic and genetic characterization of a panel of 60 human immunodeficiency virus type 1 isolates, representing clades A, B, C, D, CRF01_AE, and CRF02_AG, for the development and assessment of candidate vaccines. J Virol 79:6089–6101. doi: 10.1128/JVI.79.10.6089-6101.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang D, Giesler T, Bremer J. 2003. Sequence characterization of the protease and partial reverse transcriptase proteins of the NED panel, an international HIV type 1 subtype reference and standards panel. AIDS Res Hum Retroviruses 19:321–328. doi: 10.1089/088922203764969528. [DOI] [PubMed] [Google Scholar]
- 23.Manak M, Sina S, Anekella B, Sanders-Buell E, Garrett P, Ragupathy V, Hewlett I, Kim J, Vermeulen M, Stramer S, Sabino E, Michael N, Peel S, Tovanabutra S, Busch M, Schito M. 2012. Pilot studies for development of an HIV subtype panel for surveillance of global diversity. AIDS Res Hum Retroviruses 28:594–606. doi: 10.1089/aid.2011.0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanchez AM, DeMarco CT, Hora B, Keinonen S, Chen Y, Brinkley C, Stone M, Tobler L, Keating S, Schito M, Busch MP, Gao F, Denny TN. 2014. Development of a contemporary globally diverse HIV viral panel by the EQAPOL program. J Immunol Methods 409:117–130. doi: 10.1016/j.jim.2014.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kijak GH, Tovanabutra S, Rerks-Ngarm S, Nitayaphan S, Eamsila C, Kunasol P, Khamboonruang C, Thongcharoen P, Namwat C, Premsri N, Benenson M, Morgan P, Bose M, Sanders-Buell E, Paris R, Robb M, Birx D, De Souza M, McCutchan F, Michael N, Kim J. 2013. Molecular evolution of the HIV-1 Thai epidemic between the time of RV144 immunogen selection to the execution of the vaccine efficacy trial. J Virol 87:7265–7281. doi: 10.1128/JVI.03070-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roche. 2015. COBAS AmpliPrep/COBAS TaqMan HIV-1 test, version 2.0, product insert 05212308 190. Roche Diagnostics, Indianapolis, IN. [Google Scholar]
- 27.Abbott. 2012. RealTime HIV-1 product insert 6L18 51-602146/R7. Abbott Molecular, Des Plaines, IL. [Google Scholar]
- 28.Clinical and Laboratory Standards Institute (CLSI). 2012. Evaluation of detection capability for clinical laboratory measurement procedures; approved guideline—2nd ed. CLSI document EP17-A2. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 29.Luft LM, Gill MJ, Church DL. 2011. HIV-1 viral diversity and its implications for viral load testing: review of current platforms. Int J Infect Dis 15:e661–e670. doi: 10.1016/j.ijid.2011.05.013. [DOI] [PubMed] [Google Scholar]
- 30.Amendola A, Pisciotta M, Aleo L, Ferraioli V, Angeletti C, Capobianchi MR. 2016. Evaluation of the Aptima® HIV-1 Quant Dx assay for HIV-1 RNA viral load detection and quantitation in plasma of HIV-1 infected individuals: a comparison with Abbott RealTime HIV-1 assay. J Med Virol 88:1535–1544. doi: 10.1002/jmv.24493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schalasta G, Börner A, Speicher A, Enders M. 2016. Comparative evaluation of the Aptima HIV-1 Quant Dx assay and COBAS TaqMan HIV-1 v2.0 assay using the Roche High Pure system for the quantification of HIV-1 RNA in plasma. Clin Chem Lab Med 54:493–499. doi: 10.1515/cclm-2015-0522. [DOI] [PubMed] [Google Scholar]
- 32.Hopkins M, Hau S, Tiernan C, Papadimitropoulos A, Chawla A, Beloukas A, Geretti AM. 2015. Comparative performance of the new Aptima HIV-1 Quant Dx assay with three commercial PCR-based HIV-1 RNA quantitation assays. J Clin Virol 69:56–62. doi: 10.1016/j.jcv.2015.05.020. [DOI] [PubMed] [Google Scholar]
- 33.Sam SS, Kurpewski JR, Cu-Uvin S, Caliendo AM. 2016. Evaluation of performance characteristics of the Aptima HIV-1 Quant Dx assay for the detection and quantitation of human immunodeficiency virus type 1 in plasma and cervicovaginal lavage samples. J Clin Microbiol 54:1036–1041. doi: 10.1128/JCM.03289-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nair SV, Kim HC, Fortunko J, Foote T, Peling T, Tran C, Nugent CT, Joo S, Kang Y, Wilkins B, Lednovich K, Worlock A. 2016. Aptima HIV-1 Quant Dx-A fully automated assay for both diagnosis and quantification of HIV-1. J Clin Virol 77:46–54. doi: 10.1016/j.jcv.2016.02.002. [DOI] [PubMed] [Google Scholar]
- 35.Mor O, Gozlan Y, Wax M, Mileguir F, Rokovsky A, Noy B, Mendelson E, Levy I. 2015. Evaluation of the RealTime HIV-1, Xpert HIV-1, and Aptima HIV-1 Quant Dx assays in comparison to the NucliSens EasyQ HIV-1 v2.0 assay for quantification of HIV-1 viral load. J Clin Microbiol 53:3458–3465. doi: 10.1128/JCM.01806-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ndiaye O, Diop-Ndiaye H, Ouedraogo AS, Fall-Malick FZ, Sow-Sall A, Thiam M, Diouara AA, Ndour CT, Gaye-Diallo A, Mboup S, Toure-Kane C, Study Group. 2015. Comparison of four commercial viral load techniques in an area of non-B HIV-1 subtypes circulation. J Virol Methods 222:122–131. doi: 10.1016/j.jviromet.2015.06.002. [DOI] [PubMed] [Google Scholar]
- 37.Cunningham P, Carrera A, McNally L, Catlett B, Sherring J. 2015. Performance of the Aptima HIV-1 Quant Dx assay for quantitation of HIV-1 RNA in patients' plasma samples and in dried blood spots, poster 15-05. Abstr World STI HIV Congr, Brisbane, Australia. [Google Scholar]