Abstract
A potential benefit of digital PCR is a reduction in result variability across assays and platforms. Three sets of PCR reagents were tested on two digital PCR systems (Bio-Rad and RainDance), using three different sets of PCR reagents for quantitation of cytomegalovirus (CMV). Both commercial quantitative viral standards and 16 patient samples (n = 16) were tested. Quantitative accuracy (compared to nominal values) and variability were determined based on viral standard testing results. Quantitative correlation and variability were assessed with pairwise comparisons across all reagent-platform combinations for clinical plasma sample results. The three reagent sets, when used to assay quantitative standards on the Bio-Rad system, all showed a high degree of accuracy, low variability, and close agreement with one another. When used on the RainDance system, one of the three reagent sets appeared to have a much better correlation to nominal values than did the other two. Quantitative results for patient samples showed good correlation in most pairwise comparisons, with some showing poorer correlations when testing samples with low viral loads. Digital PCR is a robust method for measuring CMV viral load. Some degree of result variation may be seen, depending on platform and reagents used; this variation appears to be greater in samples with low viral load values.
INTRODUCTION
Viral load testing has become a routine part of clinical care, particularly for immunocompromised patients (1–3). Such tests are used to diagnose disease, trigger preemptive therapy, and determine treatment responsiveness and endpoints. While load testing is central to viral diagnosis and treatment, many challenges remain in producing uniform results. Numerous studies have demonstrated a high degree of variability among tests for various hematogenous viruses (4–6), which is likely exacerbated by the fact that few commercial tests are approved (in the United States) for in vitro diagnostic use. Both result variability and accuracy have been shown to depend on several factors (7). Some of these owe their impact to the widespread use of real-time PCR as the primary means of viral load determination, typically normalized to quantitative calibrators. In turn, variability in calibrators or in behavior of calibrators (for example, commutability) has been seen as a key factor in the production of disparate results (8, 9). The dependence on rate of amplification also means that any factor affecting amplification efficiency may affect accuracy, agreement, and variability.
Digital PCR (dPCR) has been seen as a potential remedy to these challenges. Based on the principles of limiting dilution or partition, together with endpoint PCR, digital methods remove dependence on rate-based quantitation (10–12). They are therefore potentially less sensitive to the presence of PCR inhibitors or other sources of variation in assay efficiency (13–15), and they no longer require the use of a calibration curve to produce quantitative data. As such, it might be expected that accuracy and interassay agreement will improve over those seen with real-time methods. While some authors have shown that particularly with reverse transcription-based amplification (RNA targets), results may still vary between methods (16), less has been published specifically looking at this question with regard to DNA virus assays. Similarly, while the number of dPCR platforms has begun to increase, interplatform measures of concordance are also lacking. Here, we examine the impact of reagent and platform on dPCR measures of cytomegalovirus (CMV) load in commercially produced quantitative viral standards and in human clinical plasma samples.
MATERIALS AND METHODS
Experimental design.
Four concentrations of AcroMetrix CMVtc panel and 16 human cytomegalovirus (CMV)-positive specimens were tested in four replicates on two droplet digital PCR (ddPCR) systems using each of three CMV analyte-specific reagents (ASRs). A single operator performed all testing. Quantitative agreement was assessed among ASRs in the same digital PCR system and for each ASR between the digital PCR systems.
CMV standard and human plasma specimens.
A five-member AcroMetrix CMVtc panel was purchased from Applied Biosystems, containing human cytomegalovirus (CMV) (strain AD169) in normal human EDTA plasma at concentrations of 2.48, 3.48, 4.48, 5.48, and 6.48 log10 international units (IU)/ml. A total of 16 deidentified human plasma specimens were previously detected as positive for human CMV, using an ASR assay based on MultiCode CMV reagents (Luminex Corporation, Toronto, Canada), at levels ranging from 2.70 to 6.54 log10 copies/ml and had been stored at −80°C for about 3 years prior to use in this study. As the samples were all deidentified, without links to identifiers or other protected health information (PHI), they did not qualify as human subjects and institutional review board (IRB) approval was not required.
DNA extraction of the four panel members (2.48, 3.48, 4.48, and 5.48 log10 IU/ml) and human plasma specimens was performed on the Qiagen EZ1 advanced XL using the Qiagen EZ1 DSP virus kit (Qiagen, Inc., Valencia, CA). Internal controls specific to each assay were added to the samples prior to the extraction. Four aliquots of 200 μl from each panel member and plasma specimen were processed, and DNA was eluted in 90 μl. Extracts were pooled, aliquoted, and stored at −20°C until molecular analysis.
ddPCR.
Two droplet digital PCR (ddPCR) systems were used: the QX200 droplet digital PCR system with an automated droplet generator (Bio-Rad, Pleasanton, CA) and the RainDrop digital PCR system (RainDance Technologies, Billerica, MA). The latter consists of two parts, the RainDrop Source (droplet generator) and Sense instrument (reader/counter). Both systems were used with RealStar (RS) CMV ASR (altona Diagnostics) reagents (hydrolysis probes), CMV set one real-time primer/probe ASR (Abbott Laboratories, Des Plaines, IL) (single-stranded, linear probes), and CMV primer pair ASR (Focus Diagnostics, Inc., Cypress, CA) (Scorpion primers). As noted below in the individual assay methodology descriptions (and in Discussion), the different assays were each run in different reaction volumes. This was a necessary consequence of the manufacturers providing reagents packaged for different volumes of use. In particular, Focus reagents are sold based on the presumption of a lower reaction volume, and increasing that volume to match that of other manufacturers would have been cost-prohibitive. All standards were tested in a single run for each reagent on Bio-Rad dPCR, while they were tested in multiple independent runs on RainDance dPCR. This was of necessity, based on the low number of samples (8) which can be processed on a single run of the RainDance instrument.
Bio-Rad system. (i) altona CMV reagents.
The ddPCR mixture consisted of 5 μl of 4× dPCR Supermix for Probe (Bio-Rad), 0.5 μl each of RS-ASR CMV-Prm and RS-ASR CMV-Prb (altona), 0.5 μl of RS-internal control (IC) primer/probe mix (altona), 0.5 μl of RS-IC DNA template (altona), 3 units of restriction enzyme HindIII (New England BioLabs, Inc., Ipswich MA), and 10 μl of nucleic acid solution in a final volume of 20 μl. The use of restriction endonuclease has been recommended by the manufacturer (Droplet Digital PCR Applications Guide, bulletin 6407 [Bio-Rad]) to improve template accessibility for droplet generation. HindIII was demonstrated to be a noncutter in all amplicons of three ASRs used in this study (data not shown).
(ii) Abbott reagents.
The ddPCR mixture consisted of 5 μl of 4× dPCR Supermix for Probe (Bio-Rad), 0.2 μl each of CMV set one forward primer, reverse primer, and probe (Abbott), 0.2 μl of each of internal control (IC) forward primer, IC reverse primer, and IC probe, (Abbott), 2 μl of IC DNA template DNA (Abbott), 3 units of restriction enzyme HindIII (New England BioLabs, Inc., Ipswich MA), and 10 μl of nucleic acid solution in a final volume of 20 μl.
(iii) Focus reagents.
The ddPCR mixture consisted of 5 μl of 4× dPCR Supermix for Probe (Bio-Rad), 0.4 μl of CMV primer pair (Focus), 0.2 μl of 25× CMV TM IC (Focus), 0.5 μl of Simplexa CMV molecular control DNA template DNA (Focus), 3 units of restriction enzyme HindIII (New England BioLabs, Inc., Ipswich MA), and 10 μl of nucleic acid solution in a final volume of 20 μl.
Each reaction mix was used to produce droplets on the automated droplet generator (Bio-Rad). A 96-well PCR plate (Eppendorf, Germany) containing the droplets was amplified on a T100 thermal cycler (Bio-Rad) for 40 cycles. The thermal protocol for altona and Abbott CMV reagents began with a denaturation at 95°C for 10 min, followed by 40 cycles of 94°C for 30 s and 58°C for 60 s and 1 cycle of 98°C for 10 min (omitted for Focus reagents, as this step would eliminate binding of the Scorpion primers, upon which that assay depends) and ending at 12°C. The plate was read on the QX200 droplet reader (Bio-Rad) at a rate of 32 wells per hour. dPCR data were analyzed with QuantaSoft software version 1.7.4 (Bio-Rad), and results were expressed as copies per μl of PCR mixture.
RainDrop digital PCR system. (i) altona reagents.
The ddPCR mixture consisted of 20 μl of 2× Universal master mix (Life Technologies, Inc.), 1 μl each of RS-ASR CMV-Prm and RS-ASR CMV-Prb (altona), 1 μl of RS-internal control (IC) primer/probe mix (altona), 1 μl of RS-IC DNA template (Focus), 5 units of restriction enzyme HindIII (New England BioLabs, Inc., Ipswich MA), and 10 μl of nucleic acid solution in a final volume of 40 μl.
(ii) Abbott reagents.
The ddPCR mixture consisted of 15 μl of 2× Universal master mix (Life Technologies), 0.25 μl each of CMV set one forward primer, reverse primer, and probe (Abbott), 0.25 μl of each of internal control (IC) forward primer, IC reverse primer, and IC probe, (Abbott), 2 μl of IC DNA template DNA (Abbott), 5 units of restriction enzyme HindIII (New England BioLabs, Inc., Ipswich MA), and 10 μl of nucleic acid solution in a final volume of 30 μl.
(iii) Focus reagents.
The ddPCR mixture consisted of 12.5 μl of 2× Universal master mix (Life Technologies), 0.5 μl of CMV primer pair (Focus), 0.25 μl of 25× CMV TM IC (Focus), 0.5 μl of Simplexa CMV molecular control DNA template DNA (Focus), 5 units of restriction enzyme HindIII (New England BioLabs, Inc., Ipswich MA), and 10 μl of nucleic acid solution in a final volume of 25 μl.
Each reaction mixture was transferred to one of the 8 wells on a Source Chip (RainDance Technologies). The loaded Source Chip and an 8- by 0.2-ml tube strip were inserted into the RainDance Source instrument for droplet generation. After processing, droplets in the tube strip were amplified on a C1000 thermal cycler (Bio-Rad): 1 cycle at 95°C for 10 min, followed by 40 cycles of 95°C for 30 s and 58°C for 60 s and 1 cycle of 98°C for 10 min (omitted for Focus reagent) and ending at 12°C. After amplification, the 8-tube strip and a Sense Chip (RainDance Technologies) were inserted into the Sense instrument. This instrument identifies and counts droplets at a rate of 8 samples (50 μl) per 5 h. Run data were analyzed with RainDrop Analyst software and the result generated in copies per PCR.
Statistical analysis.
Nominal concentration and nonzero digital PCR measurements were log10 transformed. The limit of detection (LOD) was defined as the lowest concentration at which all tested replicates were positive. A simple linear regression model was used to examine the quantitative correlations of digital PCR measurements at or above the LOD against nominal concentrations.
Clinical samples were tested in one run in quadruplicate, and the mean log10-transformed measurement was computed for each instrument and assay. Linear regressions and Bland-Altman plots were applied to assess the quantitative agreement among assays in the same instrument as well as between instruments with the same assay. Levene's test (17) was used to compare variability across different assays or concentrations; a small P value from Levene's test indicates that there is significant evidence that the compared groups have unequal variability.
Statistical analyses were performed using SAS (SAS Institute, Cary, NC), Windows version 9.3. No adjustment for multiple comparisons was undertaken; a P value of 0.05 or less was considered statistically significant.
RESULTS
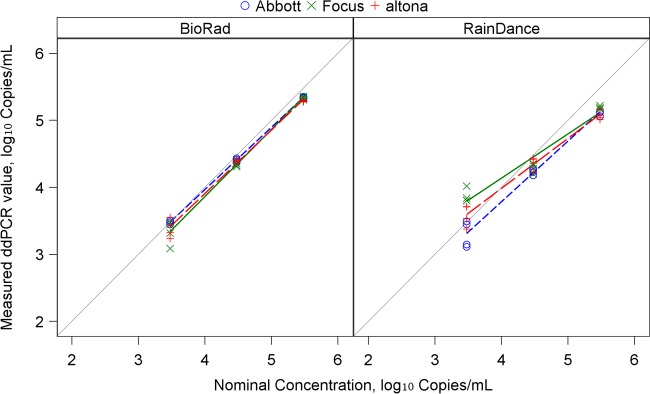
Descriptive statistics for the standards are shown in Table 1. Each assay had the same LOD when using Bio-Rad (3.48 log10 copies/ml). When RainDance was used, the LOD was 2.48 log10 copies/ml for altona and Focus and 3.48 log10 copies/ml for Abbott.
TABLE 1.
Descriptive statistics for standards
| Assay | Nominal concn, log10 copies/ml | Bio-Rad |
RainDance |
||||
|---|---|---|---|---|---|---|---|
| No. of positive replicates/total | Viral load, log10 copies/ml |
No. of positive replicates/total | Viral load, log10 copies/ml |
||||
| Mean | SD | Mean | SD | ||||
| altona | 2.48 | 2/4 | 2.45 | 0 | 4/4 | 3.5 | 0.15 |
| 3.48 | 4/4 | 3.39 | 0.14 | 4/4 | 3.58 | 0.16 | |
| 4.48 | 4/4 | 4.39 | 0.02 | 4/4 | 4.36 | 0.09 | |
| 5.48 | 4/4 | 5.29 | 0.01 | 4/4 | 5.08 | 0.07 | |
| Abbott | 2.48 | 2/4 | 2.38 | 0.34 | 3/4 | 2.94 | 0.82 |
| 3.48 | 4/4 | 3.48 | 0.03 | 4/4 | 3.3 | 0.2 | |
| 4.48 | 4/4 | 4.4 | 0.03 | 4/4 | 4.24 | 0.04 | |
| 5.48 | 4/4 | 5.34 | 0.01 | 4/4 | 5.11 | 0.03 | |
| Focus | 2.48 | 3/4 | 2.34 | 0.19 | 4/4 | 3.59 | 0.3 |
| 3.48 | 4/4 | 3.33 | 0.18 | 4/4 | 3.87 | 0.1 | |
| 4.48 | 4/4 | 4.35 | 0.04 | 4/4 | 4.3 | 0.07 | |
| 5.48 | 4/4 | 5.33 | 0.02 | 4/4 | 5.19 | 0.02 | |
All three assays with the use of Bio-Rad showed excellent linearity above the LOD. The estimated intercepts and slopes were close to 0 (−0.14 to 0.25) and 1 (0.93 to 1.00), respectively (Fig. 1; Table 2). The r2 values were very close to 1 (≥0.99). Abbott based on RainDance had more markedly reduced linearity (intercept, 0.16; slope, 0.91; r2, 0.98). Linearity and correlation were further reduced for altona and Focus in RainDance (Table 2 and Fig. 1). Variability was similar for the two platforms, although RainDance showed higher variability than Bio-Rad using Abbott at a low nominal concentration (standard deviation [SD], 0.20 versus 0.03; P < 0.001) (Table 3). SD values were not significantly different among the three reagents when results were compared across each platform, irrespective of the concentration of standard used (Table 3).
FIG 1.
Regression analysis of measured values of ddPCR against nominal values.
TABLE 2.
Regression analysis of measured values in ddPCR against nominal values
| Instrument | Assay | n | Intercept (95% CIa) | Slope (95% CI) | r2 |
|---|---|---|---|---|---|
| Bio-Rad | altona | 12 | 0.11 (−0.18, 0.40) | 0.95 (0.88, 1.01) | 0.99 |
| Abbott | 12 | 0.25 (0.17, 0.33) | 0.93 (0.91, 0.95) | >0.99 | |
| Focus | 12 | −0.14 (−0.51, 0.22) | 1.00 (0.92, 1.08) | 0.99 | |
| RainDance | altona | 12 | 1.00 (0.61, 1.39) | 0.75 (0.66, 0.83) | 0.97 |
| Abbott | 12 | 0.16 (−0.25, 0.56) | 0.91 (0.82, 1.00) | 0.98 | |
| Focus | 12 | 1.49 (1.00, 1.99) | 0.66 (0.55, 0.77) | 0.95 |
CI, confidence interval.
TABLE 3.
Results for standards
| Nominal concn, log10 copies/ml | Assay | Bio-Rad |
RainDance |
P value (platform)a | ||
|---|---|---|---|---|---|---|
| No. positive (n = 4 replicates) | Mean (SD) viral load, log10 copies/ml | No. positive (n = 4 replicates) | Mean (SD) viral load, log10 copies/ml | |||
| 3.48 | Abbott | 4 | 3.48 (0.03) | 4 | 3.30 (0.20) | <.001 |
| altona | 4 | 3.39 (0.14) | 4 | 3.58 (0.16) | 0.655 | |
| Focus | 4 | 3.33 (0.18) | 4 | 3.87 (0.10) | 0.366 | |
| P value (reagents)b | 0.092 | 0.069 | ||||
| 4.48 | Abbott | 4 | 4.40 (0.03) | 4 | 4.24 (0.04) | 0.548 |
| altona | 4 | 4.39 (0.02) | 4 | 4.36 (0.09) | 0.104 | |
| Focus | 4 | 4.35 (0.04) | 4 | 4.30 (0.07) | 0.278 | |
| P value (reagents) | 0.526 | 0.402 | ||||
| 5.48 | Abbott | 4 | 5.34 (0.01) | 4 | 5.11 (0.03) | 0.137 |
| altona | 4 | 5.29 (0.01) | 4 | 5.08 (0.07) | 0.071 | |
| Focus | 4 | 5.33 (0.02) | 4 | 5.19 (0.02) | 0.895 | |
| P value (reagents) | 0.218 | 0.223 | ||||
P value from Levene's test comparing the variability between Bio-Rad and RainDance for each reagent at each nominal concentration.
P value from Levene's test comparing the variability of three reagents in each platform at each nominal concentration.
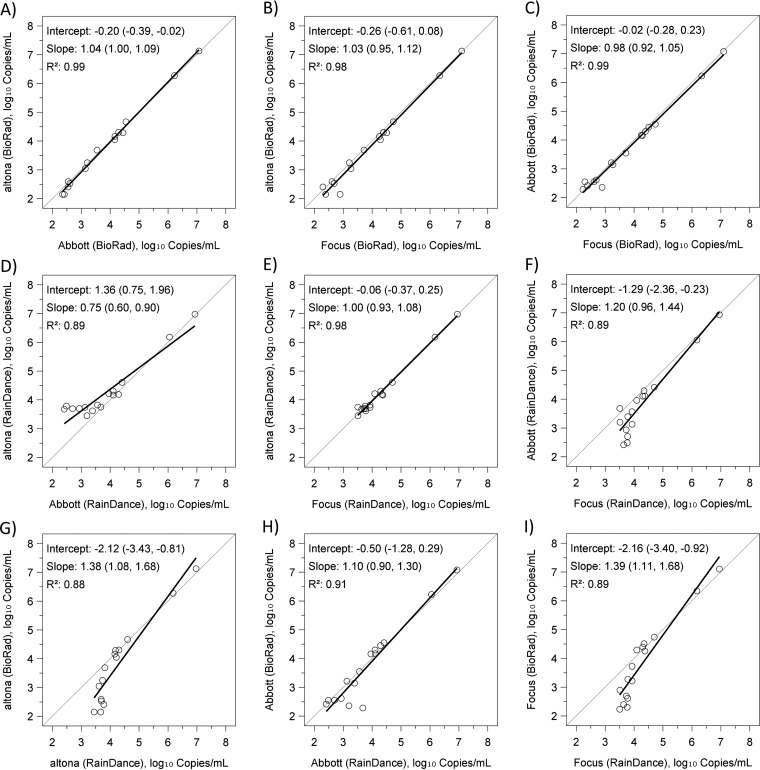
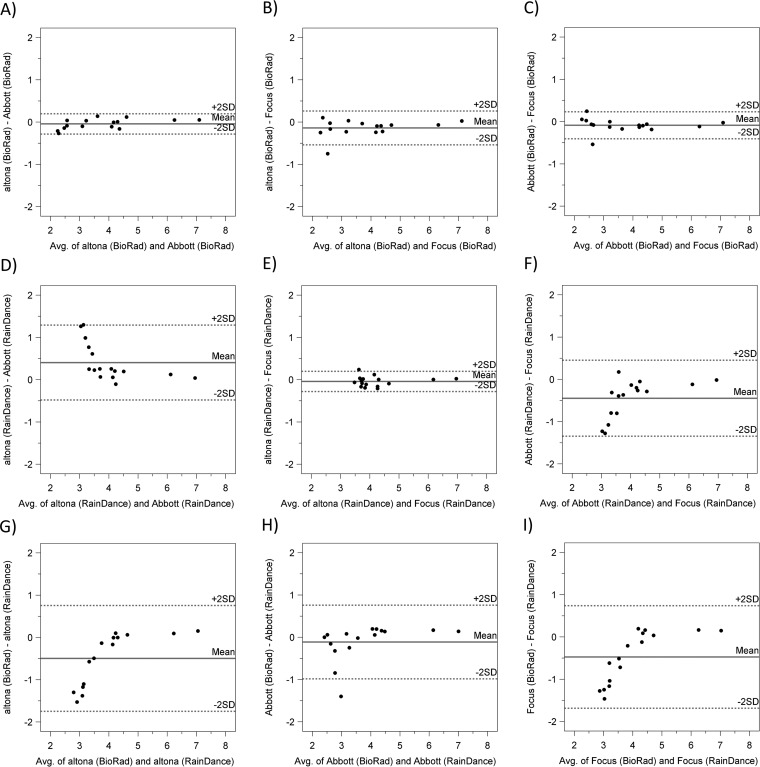
Figures 2 and 3 show pairwise linear regression and Bland-Altman (difference) plots, comparing results from all three assays in both instruments when clinical samples were tested. All three assays with Bio-Rad showed close agreement with each other (Fig. 2A to C and 3A to C). When RainDance was used, quantitative results from altona also agreed well with those from Focus (Fig. 2E and 3E), but the agreement of altona and Focus with Abbott was reduced (Fig. 2D and F and 3D and F). Further, clinical sample results were not significantly different between Bio-Rad and RainDance when Abbott was used (Fig. 2H and 3H). Using altona (Fig. 2G and 3G) or Focus (Fig. 2I and 3I), however, measured results from RainDance were greater than those from Bio-Rad at low viral load concentrations. Quantitative agreement improved in all pairwise comparisons with higher viral loads (above approximately 4 log10 copies/ml). Patient sample results showed differences in variability between the two instruments. RainDance was more likely than Bio-Rad to have high SD values. SD values were not much different among reagents using Bio-Rad, but when using RainDance, altona tended to have higher SD values than Abbott or Focus (see Table S1 in the supplemental material).
FIG 2.
Pairwise linear regressions for clinical samples.
FIG 3.
Quantitative differences between assays. Values are log10 copies/ml with differences between assays on the y axis and the average on the x axis. The mean difference between assays is represented by the solid line, and ±2 SDs is represented by the dotted lines.
DISCUSSION
The increasing use of digital PCR for viral load quantitation, together with increasing availability of different reagents and platforms, raises the question of comparative performance. The lack of reliance on quantitative standards suggests that the use of this methodology should improve concordance among methods, while reducing result variability. Indeed, the level of agreement seen in this study appears to be increased over that seen in studies with real-time methods. Nonetheless, the data here also show that results vary between reagents and platforms studied; accuracy and agreement cannot be assumed with the use of dPCR methods, even when quantifying DNA targets.
These findings support previous work demonstrating various potential sources of inaccuracy and result variation using digital methods (16, 18, 19). Differing results have been attributed to varying reverse transcriptase efficiency, molecular “dropout” (20), nonspecific amplification, partition volume, and pipetting variation, among other potential causes. We have previously shown variability, particularly in samples with low viral loads, which approaches or exceeds that of real-time PCR (21). Similarly, much of the variability seen in the present study was present at lower target concentrations. Others, however, have supported the advantages of dPCR in reducing susceptibility to PCR inhibitors (13–15). Here and elsewhere, when using a common platform, results have been similar or identical irrespective of the reagents used. As seen here, it might prove that susceptibility to changes in reagents are platform and target concentration dependent.
These potential caveats to dPCR reliability can only be suggested by the present study, which was inherently limited by the dynamic range of available quantitative standards and patient samples. While the RainDance system appeared to show somewhat more result variability and reduced linearity for some of the studied reagents, this platform might have advantages for samples with higher viral loads (not represented here), due to the much larger number of partitions it utilizes (107) compared to the Bio-Rad system (2 × 104). Comparability of the tests may also have been confounded by the fact that different assays were run in different reaction volumes. This was a necessary consequence of the manufacturers providing reagents packaged for differing volumes of use (based on component volumes). To assemble altona reagents, a minimum volume of 40 μl is required, while Focus requires at least 25 μl. As Focus reagents are sold based on the presumption of a lower reaction volume, increasing that volume to match that of other manufacturers would be cost-prohibitive. Furthermore, cost constraints and limitations in throughput and sample availability prevented testing a higher number of runs or replicates per sample; this may have enabled improved evaluation of result variability.
The results demonstrate a high degree of concordance among results achieved using different reagents and platforms, particularly at higher target concentrations. While the use of dPCR as a reference standard or for routine clinical testing continues to require thorough validation for any given assay and instrument, data continue to support its value for viral load determination. As instrumentation and reagents continue to improve and become better characterized, this methodology may prove advantageous in settings where real-time PCR provides insufficient reliability.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by ALSAC.
A.M.C. and R.T.H. serve on Roche Molecular advisory boards, and A.M.C. serves on a Cepheid advisory board. None of the authors have any other conflicts to disclose.
Reagents for the study were kindly provided by Abbott Molecular, Inc.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.01474-16.
REFERENCES
- 1.Gartner B, Preiksaitis JK. 2010. EBV viral load detection in clinical virology. J Clin Virol 48:82–90. doi: 10.1016/j.jcv.2010.03.016. [DOI] [PubMed] [Google Scholar]
- 2.Ljungman P. 2010. Molecular monitoring of viral infections after hematopoietic stem cell transplantation. Int J Hematol 91:596–601. doi: 10.1007/s12185-010-0570-4. [DOI] [PubMed] [Google Scholar]
- 3.Razonable RR, Hayden RT. 2013. Clinical utility of viral load in management of cytomegalovirus infection after solid organ transplantation. Clin Microbiol Rev 26:703–727. doi: 10.1128/CMR.00015-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hayden RT, Hokanson KM, Pounds SB, Bankowski MJ, Belzer SW, Carr J, Diorio D, Forman MS, Joshi Y, Hillyard D, Hodinka RL, Nikiforova MN, Romain CA, Stevenson J, Valsamakis A, Balfour HH Jr, U.S. EBV Working Group. 2008. Multicenter comparison of different real-time PCR assays for quantitative detection of Epstein-Barr virus. J Clin Microbiol 46:157–163. doi: 10.1128/JCM.01252-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoffman NG, Cook L, Atienza EE, Limaye AP, Jerome KR. 2008. Marked variability of BK virus load measurement using quantitative real-time PCR among commonly used assays. J Clin Microbiol 46:2671–2680. doi: 10.1128/JCM.00258-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pang XL, Fox JD, Fenton JM, Miller GG, Caliendo AM, Preiksaitis JK. 2009. Interlaboratory comparison of cytomegalovirus viral load assays. Am J Transplant 9:258–268. doi: 10.1111/j.1600-6143.2008.02513.x. [DOI] [PubMed] [Google Scholar]
- 7.Hayden RT, Yan X, Wick MT, Rodriguez AB, Xiong X, Ginocchio CC, Mitchell MJ, Caliendo AM. 2012. Factors contributing to variability of quantitative viral PCR results in proficiency testing samples: a multivariate analysis. J Clin Microbiol 50:337–345. doi: 10.1128/JCM.01287-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holden MJ, Madej RM, Minor P, Kalman LV. 2011. Molecular diagnostics: harmonization through reference materials, documentary standards and proficiency testing. Expert Rev Mol Diagn 11:741–755. doi: 10.1586/erm.11.50. [DOI] [PubMed] [Google Scholar]
- 9.Madej RM, Davis J, Holden MJ, Kwang S, Labourier E, Schneider GJ. 2010. International standards and reference materials for quantitative molecular infectious disease testing. J Mol Diagn 12:133–143. doi: 10.2353/jmoldx.2010.090067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hindson BJ, Ness KD, Masquelier DA, Belgrader P, Heredia NJ, Makarewicz AJ, Bright IJ, Lucero MY, Hiddessen AL, Legler TC, Kitano TK, Hodel MR, Petersen JF, Wyatt PW, Steenblock ER, Shah PH, Bousse LJ, Troup CB, Mellen JC, Wittmann DK, Erndt NG, Cauley TH, Koehler RT, So AP, Dube S, Rose KA, Montesclaros L, Wang S, Stumbo DP, Hodges SP, Romine S, Milanovich FP, White HE, Regan JF, Karlin-Neumann GA, Hindson CM, Saxonov S, Colston BW. 2011. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem 83:8604–8610. doi: 10.1021/ac202028g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sykes PJ, Neoh SH, Brisco MJ, Hughes E, Condon J, Morley AA. 1992. Quantitation of targets for PCR by use of limiting dilution. Biotechniques 13:444–449. [PubMed] [Google Scholar]
- 12.Vogelstein B, Kinzler KW. 1999. Digital PCR. Proc Natl Acad Sci U S A 96:9236–9241. doi: 10.1073/pnas.96.16.9236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dingle TC, Sedlak RH, Cook L, Jerome KR. 2013. Tolerance of droplet-digital PCR vs real-time quantitative PCR to inhibitory substances. Clin Chem 59:1670–1672. doi: 10.1373/clinchem.2013.211045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Racki N, Dreo T, Gutierrez-Aguirre I, Blejec A, Ravnikar M. 2014. Reverse transcriptase droplet digital PCR shows high resilience to PCR inhibitors from plant, soil and water samples. Plant Methods 10:42. doi: 10.1186/s13007-014-0042-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sedlak RH, Kuypers J, Jerome KR. 2014. A multiplexed droplet digital PCR assay performs better than qPCR on inhibition prone samples. Diagn Microbiol Infect Dis 80:285–286. doi: 10.1016/j.diagmicrobio.2014.09.004. [DOI] [PubMed] [Google Scholar]
- 16.Sanders R, Huggett JF, Bushell CA, Cowen S, Scott DJ, Foy CA. 2011. Evaluation of digital PCR for absolute DNA quantification. Anal Chem 83:6474–6484. doi: 10.1021/ac103230c. [DOI] [PubMed] [Google Scholar]
- 17.Levene H. 1960. Robust tests for equality of variances, p 278–292. In Olkin I. (ed), Contributions to probability and statistics: essays in honor of Harold Hotelling. Stanford University Press, Palo Alto, CA. [Google Scholar]
- 18.Huggett JF, Cowen S, Foy CA. 2015. Considerations for digital PCR as an accurate molecular diagnostic tool. Clin Chem 61:79–88. doi: 10.1373/clinchem.2014.221366. [DOI] [PubMed] [Google Scholar]
- 19.Jacobs BK, Goetghebeur E, Clement L. 2014. Impact of variance components on reliability of absolute quantification using digital PCR. BMC Bioinformatics 15:283. doi: 10.1186/1471-2105-15-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Whale AS, Cowen S, Foy CA, Huggett JF. 2013. Methods for applying accurate digital PCR analysis on low copy DNA samples. PLoS One 8:e58177. doi: 10.1371/journal.pone.0058177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hayden RT, Gu Z, Ingersoll J, Abdul-Ali D, Shi L, Pounds S, Caliendo AM. 2013. Comparison of droplet digital PCR to real-time PCR for quantitative detection of cytomegalovirus. J Clin Microbiol 51:540–546. doi: 10.1128/JCM.02620-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.