Abstract
Vancomycin-resistant enterococci (VRE) are an important cause of health care-acquired infections (HAIs). Studies have shown that active surveillance of high-risk patients for VRE colonization can aid in reducing HAIs; however, these screens generate a significant cost to the laboratory and health care system. Digital imaging capable of differentiating negative and “nonnegative” chromogenic agar can reduce the labor cost of these screens and potentially improve patient care. In this study, we evaluated the performance of the WASPLab Chromogenic Detection Module (CDM) (Copan, Brescia, Italy) software to analyze VRE chromogenic agar and compared the results to technologist plate reading. Specimens collected at 3 laboratories were cultured using the WASPLab CDM and plated to each site's standard-of-care chromogenic media, which included Colorex VRE (BioMed Diagnostics, White City, OR) or Oxoid VRE (Oxoid, Basingstoke, United Kingdom). Digital images were scored using the CDM software after 24 or 40 h of growth, and all manual reading was performed using digital images on a high-definition (HD) monitor. In total, 104,730 specimens were enrolled and automation agreed with manual analysis for 90.1% of all specimens tested, with sensitivity and specificity of 100% and 89.5%, respectively. Automation results were discordant for 10,348 specimens, and all discordant images were reviewed by a laboratory supervisor or director. After a second review, 499 specimens were identified as representing missed positive cultures falsely called negative by the technologist, 1,616 were identified as containing borderline color results (negative result but with no package insert color visible), and 8,234 specimens were identified as containing colorimetric pigmentation due to residual matrix from the specimen or yeast (Candida). Overall, the CDM was accurate at identifying negative VRE plates, which comprised 84% (87,973) of the specimens in this study.
INTRODUCTION
Members of the genus Enterococcus are commensal colonizers of the gastrointestinal tract but can cause a variety of serious nosocomial infections, including bacteremia, endocarditis, intra-abdominal and pelvic infections, urinary tract infections, and, in rare cases, central nervous system infections (1–4). Treatment can be difficult, as E. faecalis has been observed to be 10 to 100 times more resistant to β-lactams than other streptococcal species and E. faecium is 4 to 16 times more resistant than E. faecalis (5). Vancomycin had been used for years to successfully treat enterococcal infections; however, in 1988, the first case of a vancomycin-resistant-enterococcus (VRE) infection was reported (6). Resistance is conferred by the van operon, carried in either the chromosome or a plasmid, and is inducible in response to membrane disruption caused by vancomycin (7, 8). Currently, resistance is widespread, with prevalence rates ranging from 1.0% to 35.5% of all Enterococcus specimens isolated, depending on the geographical location, and is more commonly found in E. faecium (4).
The success of Enterococcus as a nosocomial pathogen relies on the organism's ability to colonize the human gut, shed into the environment, and survive on surfaces such as the hands of health care workers for up to 60 min or on surfaces in the rooms of colonized patients for as long as 4 months (9, 10). Transmission from person to person or from surface to person is a serious threat, as infection with VRE has been associated with increased mortality rates (11). Active screening of high-risk patients allows early identification of colonized individuals, which aids in decreasing transmission by implementation of patient cohorting, improving hand hygiene, and increasing the use of barrier protection (12).
The use of chromogenic media is a quick and efficient method to screen for VRE in stool or rectal swab specimens. These media contain an antibiotic cocktail composed of vancomycin along with a specific chromogen to develop colorimetric changes in E. faecalis or E. faecium colonies, with some media using two different reactions to differentiate species. However, depending on the size of the hospital and the screening procedure used, plating and interpreting chromogenic VRE plates can be time-consuming and costly for the laboratory. As chromogenic media use colorimetric reactions to allow technologists to quickly identify VRE, software capable of differentiating colors between images should be able to differentiate chromogenic growth from no growth or growth without a colorimetric reaction.
Previously, we reported on the use of the WASPLab to automatically plate and categorize chromogenic methicillin-resistant Staphylococcus aureus (MRSA) specimens as negative or “nonnegative” (detection of color can be performed after 24 h but requires manual examination to confirm) (13). Using specific algorithms to interpret colony growth and color, the software was highly sensitive and resulted in no false-negative specimen identifications. In this study, we calibrated the software to analyze VRE chromogenic agar. Rectal swab specimens from three sites were plated using the WASPLab, and image analysis reported each plate as positive or negative for VRE at 24 or 40 h postinoculation. The performance of the software was evaluated by comparing each specimen result to manual interpretation as the gold standard.
MATERIALS AND METHODS
Specimen processing.
Enrolled specimens were comprised of rectal ESwab (Copan, Brescia, Italy, and Murrieta, CA, USA) specimens sent to the laboratory for VRE screening and to sites 1 and 3 (enrolled from September 2014 through November 2015) and site 2 (enrolled from November 2014 through October 2015). Specimens were plated directly onto chromogenic agar at two sites (sites 1 and 2) following standard-of-care procedures, which included the use of the WASPLab for processing. At site 3, an initial enrichment step was part of the laboratory standard of care. Enrichment consisted of adding 0.5 ml of ESwab liquid to an enrichment broth comprised of brain heart infusion (BHI) broth plus amoxicillin (16 μg/ml) and incubation for 22 h at 37°C. Incubated broths were then loaded onto the WASPLab for processing. The chromogenic agar used in this study was Colorex VRE (BioMed Diagnostics, White City, OR; sites 1 and 2) or Oxoid VRE (Thermo Fisher Scientific, Basingstoke, United Kingdoms; site 3). Inoculated plates were moved by conveyor belt to a digital imager, where an image was obtained time point 0, and then moved into the WASPLab incubator, where the plates were incubated at 35°C without CO2 for 24 h at two sites and for 40 h at one site (site 3). Each site performed testing in accordance with site-specific institutional review board-approved protocols.
Digital analysis of chromogenic media.
The WASPLab contains a digital imager to automatically take images of plates at programmable time points throughout incubation. For this study, an initial image was taken at time point 0 and again at either 24 or 40 h postinoculation in accordance with each site's standard of operations for interpreting chromogenic VRE plates. Using the Chromogenic Detection Module (CDM) image analysis software incorporated into the WASPLab, images were automatically screened by comparing the plate at time point 0 to the plate after incubation. The CDM analysis was previously described by Faron et al. (13). Briefly, the software analyzes the plates to identify differences in growth and colony color and is programmed to correspond specifically to the medium type used by the laboratory (expected colony color per package insert). The software then creates a value score for each plate that is based on the RGB (red, green, and blue) colors found in the image. The software converts the RGB image to a three-dimensional (3D) “bubble” composed of the pixel hue, saturation, and value (HSV) and compares the HSV score to a threshold set for each medium type. If the plate's value falls within the set parameters, the plate was marked as automation positive (AP). When there is no growth observed or growth falls outside the value threshold, the plate is defined as automation negative (AN) for VRE.
Manual scoring of chromogenic plates.
Technologists performed manual screening of plates in a blind manner with respect to the CDM results, and manual analysis was performed following the standard-of-care procedures of the laboratories. Briefly, at 24 or 40 h, images were manually read by a technologist using an imaging workstation (high-definition [HD] monitor). The images were identical to the images scored by the CDM software. Depending on the medium used, the technologist looked for colonies containing pigmentation consistent with the package insert indicating VRE (for Colorex VRE, mauve [VanA or -B] or blue [VanC, -D, or -E]; for Oxoid VRE, light blue [E. faecalis] or indigo [E. faecium]). For this study, as plates containing VanC, -D, or -E can contain chromogenic growth, these colonies were counted as positive. Plates with colonies that contained chromogenic color but appeared to be yeast or non-VRE in morphology were removed from the WASPLab, and a wet mount procedure was performed to differentiate yeast from bacteria. Specimens identified by technologists containing pigmented colonies consistent with the package insert description were reported as manual positive (MP) for VRE. Specimens with a lack of pigmentation or wet mounts containing yeast or negative rods were scored as manual negative (MN).
Discrepancy analysis.
Comparisons of the results of automation and manual analyses were performed after all testing was completed; therefore, plates were not available for discrepancy analysis using biochemical testing methods. To characterize discrepant specimens, digital images of discrepant results were sent back to the corresponding laboratory and reviewed by the laboratory director or supervisor. The images were identical to the images originally read by the technologist and scored by the CDM software. Discrepant plates were categorized into three categories depending on the type of growth that was observed, including plates with results that were automation positive and second manual positive (positive colony upon second review), plates with residual matrix or yeast, and plates with borderline color results.
Statistical analysis.
The results of the manual read were compared to those obtained from the automated image analysis. Sensitivity and specificity data were calculated using standard methods and McNemar's test (14). Ninety-five-percent confidence intervals (CI) were calculated by using the efficiency score method (15).
RESULTS
Specimen characteristics and prevalence.
In total, 104,730 rectal swab specimens were collected and enrolled into the study from the three sites. Two sites (site 1 and site 2) used Colorex VRE agar and performed reading at 24 h postinoculation, whereas testing was performed using Oxoid VRE agar after 40 h at site 3. The overall prevalence of VRE determined by manual reading was 6.1%. The prevalence rates at sites 1, 2, and 3 were 12.9%, 3.7%, and 11.8%, respectively.
Comparison of the automatic imaging to manual detection of VRE-positive chromogenic agar.
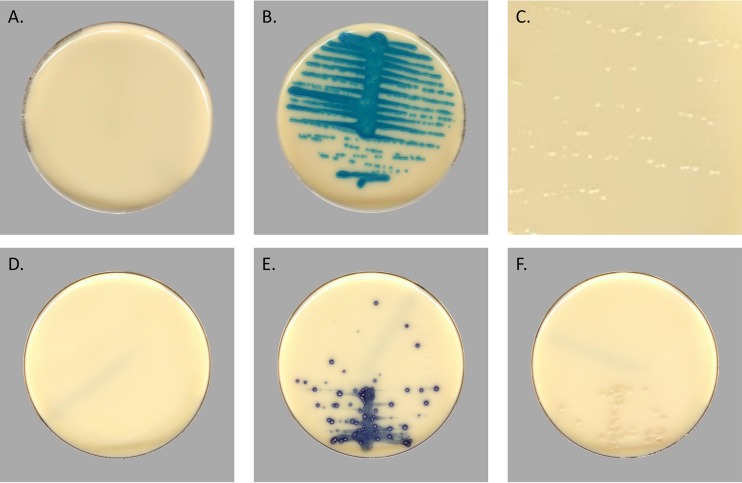
Comparisons between manual and automatic reading were performed using identical images taken at either 24 or 40 h postinoculation. Representative images with positive and negative results and breakthrough growth on the Colorex VRE and Oxoid VRE plates are shown in Fig. 1. Images viewed on HD monitors are observed at the dimensions of the monitor, so individual colonies are easily distinguishable.
FIG 1.
Representative digital images of Colorex VRE chromogenic media and Oxoid VRE captured by the WASPLab. (A) Negative Colorex plate with no growth. (B) Positive Colorex plate containing VRE. (C) A zoomed-in image of clear breakthrough growth with no colorimetric reaction occurring on Colorex media. (D) Negative Oxoid agar. (E) Positive Oxoid agar. (F) A negative plate with breakthrough growth on Oxoid agar.
Automation analysis was concordant with manual testing for 90.1% (94,382/104,730) of all specimens tested. Concordant specimens consisted of 6,403 (6.1%) MP/AP (true positive) and 87,979 (84.0%) MN/AN (true negative) specimens (Table 1). The CDM software reported an additional 10,348 (9.9%) specimens as positive that were MN. No specimens were reported as manual positive and automation negative (false negative). Together, these data demonstrate that the overall sensitivity and specificity for all sites were 100% (99% to 100%; 95% CI) and 89.5% (89% to 90%; 95% CI), respectively. Sites 1 and 3 had statistically similar specificities of 91.6% and 91.4% based on confidence intervals. Site 2 had a specificity of 88.8%, which may have been a consequence of the high volume tested and the prevalence being lower than that seen with the other sites.
TABLE 1.
Performance of WASPLab digital imaging of VRE plates compared to manual reading
| Clinical test site | No. of specimens tested | Resulta |
% sensitivity (95% CI)b | % specificity (95% CI)b | PPVc (%) | NPVd (%) | |||
|---|---|---|---|---|---|---|---|---|---|
| MP/AP | MN/AN | MN/AP | MP/AN | ||||||
| 1 | 11,438 | 1,474 | 9,129 | 835 | 0 | 100 (99–100) | 91.6 (91–92) | 64 | 100 |
| 2 | 75,518 | 2,822 | 64,535 | 8,161 | 0 | 100 (99–100) | 88.8 (88–89) | 26 | 100 |
| 3 | 17,774 | 2,107 | 14,315 | 1,352 | 0 | 100 (99–100) | 91.4 (91–92) | 61 | 100 |
| Total | 104,730 | 6,403 | 87,979 | 10,348 | 0 | 100 (99–100) | 89.5 (89–90) | 38 | 100 |
MP/AP, number of specimens with positive manual results and positive automation results; MN/AN, number of specimens with negative manual results and negative automation results; MN/AP, number of specimens with negative manual results and positive automation results; MP/AN, number of specimens with positive manual results and negative automation results.
CI, confidence interval.
PPV, positive predictive value.
NPV, negative predictive value.
Analysis of discrepant results.
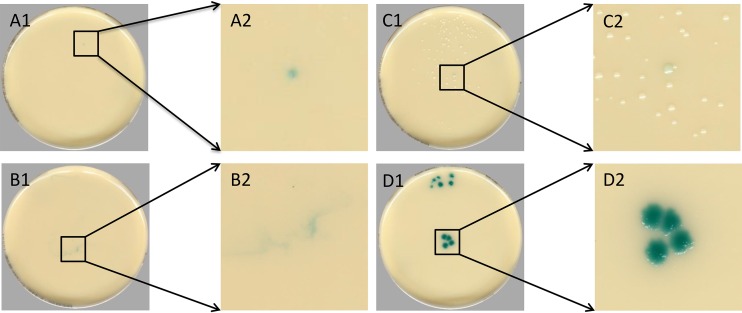
Discordant specimen images were sent to the clinical laboratory supervisor or director for further review. Upon review, discrepant images were characterized as containing positive colonies that were undetected by technologists but positive after a second review (automation positive and second manual read positive), pigmentation of the agar without microbial growth (residual matrix), colonies with coloration that would not be classified as representing a positive result based on the package insert instructions (borderline color), or pigmented breakthrough yeast; representative images are shown in Fig. 2. Wet mount results were not collected for the study, and digital imaging alone could not always differentiate yeast from residual matrix, and so these false positives (FPs) were combined and classified as representing residual matrix or yeast. In total, 10,348 images were sent back for review. The most common discrepant results were categorized as having residual matrix or yeast growth, consisting of 8,234 (79.6%) discrepant specimens (Table 2), followed by 1,616 (15.6%) specimens with borderline colors and 499 (4.8%) discordant specimens that were automation positive and second manual positive (Table 2).
FIG 2.
Examples of discrepant categories determined by a second manual reading. (A1) Automation-positive second manual positive showing a positive single colony that was identified as manual positive after a second observation. (A2) Magnification of inset in panel A1. (B1) Residual matrix creating a colorimetric reaction with the agar but containing growth. (B2) Magnification of inset in panel B1. (C1) A single colony containing colorimetric reaction but not falling within package insert definition of positive VRE. (C2) Magnification of inset in panel C1. (D1) An example of yeast breakthrough developing colors similar to those seen with positive VRE specimens. (D2) Magnification of inset in panel D1.
TABLE 2.
Discrepancy analysis of plates with negative manual results and positive automation results
| Plate category | No. of plates |
|||
|---|---|---|---|---|
| MN/APa | Positive automation result/2nd manual result positive | Residual matrix or yeast | Borderline colors | |
| Total no. of plates | 10,348 | 499 | 8,234 | 1,616 |
| Colorex VRE | 8,996 | 432 | 7,684 | 881 |
| Oxiod VRE | 1,352 | 67 | 550 | 735 |
MN/AP, negative manual result and positive automation result.
Comparison of results determined using the CDM software to detect VRE from various chromogenic agars.
Two sites (sites 1 and 2) in the study performed all testing using Colorex VRE, while the third site used Oxoid VRE. Comparisons were made between the two medium types to determine if performance of the CDM was affected by the medium type used (Table 3). A total of 86,956 Colorex VRE plates and 17,774 Oxoid VRE plates were tested, and both agars had sensitivity of 100% compared to manual reading of the corresponding chromogenic agar. The specificity of Colorex VRE was 89.1%, and the specificity of Oxoid VRE was 91.4%.
TABLE 3.
Comparison of 2 chromogenic agars for the detection of VRE using automated scoring
| Chromogenic medium | No. of specimens tested | Resulta |
% sensitivity (95% CI)b | % specificity (95% CI)b | PPVc | NPVdc | |||
|---|---|---|---|---|---|---|---|---|---|
| MP/AP | MN/AN | MN/AP | MP/AN | ||||||
| Colorex VRE | 86,956 | 4,296 | 73,664 | 8,996 | 0 | 100 (99–100) | 89.1 (89–89) | 32 | 100 |
| Oxoid VRE | 17,774 | 2,107 | 14,315 | 1,352 | 0 | 100 (99–100) | 91.4 (91–92) | 61 | 100 |
MP/AP, number of specimens with positive manual results and positive automation results; MN/AN, number of specimens with negative manual results and negative automation results; MN/AP, number of specimens with negative manual results and positive automation results; MP/AN, number of specimens with positive manual results and negative automation results.
CI, confidence interval.
PPV, positive predictive value.
NPV. negative predictive value.
Discrepant results were also classified by the chromogenic agar used to determine if there were issues that were more prevalent with either of the two media used in testing. The proportions of automation false positives (FP) observed with each chromogenic agar were 10.3% with Colorex VRE and 7.6% with Oxoid VRE, giving positive predictive values (PPV) of 32.3% (32% to 33%; 95% CI) and 60.9% (59% to 62%; 95% CI). False-positive specimens on both plates had similar occurrences of automation-positive, second-manual-read-positive plates at 4.8% and 4.7% FP specimens (Table 2). Differences were observed between residual matrix or yeast and borderline color false-positive results. The Colorex VRE had 85.4% FP resulting from residual matrix or breakthrough yeast and only 9.8% FP having colonies containing borderline colors. In contrast, the Oxoid VRE agar contained only 40.7% FP due to residual matrix and breakthrough yeast and 54.4% were due to borderline color colony growth. As the Oxoid VRE was read at 40 h of incubation, additional breakthrough may have resulted in a higher number of specimens containing borderline colors.
DISCUSSION
The prevalence of VRE has continued to rise, with 35.5% of all Enterococcus health care-acquired infections (HAIs) in the United States being resistant to vancomycin and the proportion ranging between <1% and >30% in European countries (16, 17). In response to VRE outbreaks, the CDC has recommended that patients identified as colonized or infected should be isolated or quarantined with other colonized patients along with mandatory hand washing by health care workers and cleaning of the surrounding environment. When implemented, these recommendations decreased the presence of the organism from 2.2% to 0.5% in the Siouxland region of Iowa, Nebraska, and South Dakota (18). Currently, it is recommended by the CDC that institutions that contain moderate to high rates of VRE perform active surveillance using either stool or rectal swab specimens (19). At these institutions, the cost of screening can vary, with one study estimating a cost of $29,570 per year, with approximately 30% of the cost associated with labor (20).
In this study, 84% of specimens plated were negative by both methods of analysis. With the current software, a technologist is required to confirm negatives; however, the software allows the technologist to view these negatives in batches of 40 images per screen. From this batched screen, the technologist can quickly view these plates and report them as negative all at once; if such an approach had been used over the course of this study, the 87,979 screens viewed for each image would have been reduced to 2,200 (87,979/40) screens viewed. Our data suggest that automatic reporting of negatives would be accurate, as no images that were manual read positive and automation negative were observed after 104,730 specimens were tested. Automatic reporting of negative plates would have several benefits, such as decreasing turnaround time, especially in laboratories that are not open 24 hours a day and 7 days a week; support of antimicrobial stewardship by allowing pharmacists and physicians to guide practices based on both patient health and laboratory results; and reduction of laboratory labor costs, as technologists would not have to view any plates with negative results, which in this study represented the vast majority of specimens tested.
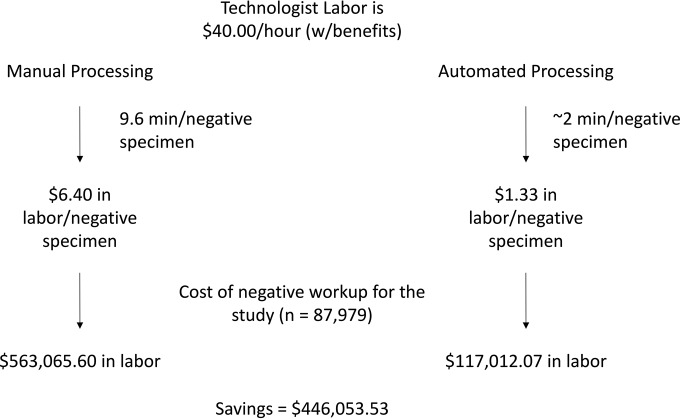
A cost analysis was not performed, but the potential savings can be modeled by predicting the changes in workflow with incorporation of the CDM software and estimating the reduction in labor with its implementation. The process of reading plates by technologists is often batched, and so it is difficult to determine precisely the time that it takes for an individual specimen to be called up, reviewed, and reported. To estimate this time, technologists were observed while reading plates; in general, the workflow was found to require approximately 15 s per negative plate. During the course of this study, technologists reviewed and discarded 87,979 negative plates using manual reading; however, if the technologists had viewed these negatives in batches of 40 plates per screen, this would have reduced the number of screens required to be reviewed from 87,979 to 2,200 (87,979/40). To analyze and view the extra 85,779 specimens (total of 87,979 negative screens minus 2,200 negative screens viewed by batching) at 15 s per negative specimen, it would require 357 h of a technologist's time and cost approximately $14,280 (estimation at an average of $40.00 an hour with benefits). In addition, the technologist would have more time to perform other tasks, further reducing labor costs. The potential savings for the laboratory may be larger for laboratories that switch from manual processing to automation processing of specimens. In one study performing a cost analysis of VRE screening, the authors estimated that it took 9.6 min of a technologist's time to process, read, and report each negative VRE specimen sent to the laboratory (20). Using $40.00 an hour as an average wage, the cost per specimen at 9.6 min of technologist time is $6.40. Roughly estimating that automated processing would require 2 min of hands-on time, the labor cost per negative specimen would be reduced from $6.40 to $1.33 with automation. A summary of the predicted cost savings from this study is modeled in Fig. 3.
FIG 3.
Cost analysis model of incorporation of CDM automation. The average technologist makes approximately $40.00 an hour with benefits included. In a previous study measuring the labor cost of VRE screening, a negative culture result required 9.6 min of hands-on time, equating to $6.40 of labor cost per negative specimen. Using the WASPLab, the hands-on time required for a negative specimen result is reduced to approximately 2 min, which reduces the cost to $1.33 in labor. In this study, 87,979 negative plates were analyzed. Comparing the two costs, using automation to process these specimens would result in a total savings of $446,053.53 and reductions in costs for the three sites of $46,284.03 (site 1, 14 months), $327,192.45 (site 2, 9 months), and $72,577.05 (site 3, 14 months). Adding the cost savings from batching 40 specimens per screen would increase the total savings to $460,333.53.
This is the second study reporting on the performance characteristics of the CDM software. In the first study, analysis using the CDM software was compared to manual reading of MRSA chromogenic plates (13). In both studies, the software was 100% sensitive in differentiating negative from “nonnegative” plates. These data demonstrated that the specificity determined using chromogenic plate screening for VRE (89.5%) was lower than that determined using the MRSA agar (90.7%) (13). However, chromogenic VRE agar is often associated with breakthrough growth of yeast and other enteric bacteria that can contain pigmentation (21, 22). In addition, VRE prevalence tends to be lower than MRSA prevalence at clinical laboratories, as many chromogenic plates can cause false chromogen coloring; the lower prevalence likely affected the specificity differences between the studies. The effect of the prevalence differences between sites on the specificities and positive predictive values (PPV) was demonstrated in this study. Sites 1 and 3 had prevalence rates of around 12% and specificity and PPV values of 91.6% and 64% (site 1) and 91.4% and 61% (site 3), respectively, whereas site 2, which had a prevalence rate of 3.7%, showed specificity of 88.8% and a PPV of 26%. The percentages of discrepant specimens in the two studies were similar, with 9.9% seen using VRE agar and 9.0% using MRSA agar. Use of the software showed that the prevalence rates of categorical FP specimens did differ between medium types. The most common FP specimen results were caused by the presence of residual matrix or yeast using VRE chromogenic media, whereas MRSA media contained more borderline colors.
Specimens were unavailable for further workup, which limited the capabilities of the studies to characterize the discrepant results. The software identified 499 specimens that, upon second review, appeared to be positive for VRE, indicating that the software may be more sensitive than the manual reading of chromogenic plates. The addition of 499 positive specimens could have large downstream effects on patient care; appropriate infection control procedures could be implemented, as unidentified reservoirs can lead to clonal outbreaks (23). However, further studies measuring the impact on patient care of implementation of the software are required.
There were limitations to this study. Due to the complexity of data analysis, the volume of testing, and the differences in the geographical locations of the test sites, discrepant results were not identified prior to the discarding of the plates by the laboratory personnel. This limitation required discrepancy analysis to be performed as a second viewing of saved images, which, given the subjective nature of interpretations based on the use of chromogenic agar, might lead to misinterpretation. In addition, secondary reviewers were sent only discrepant results. They were unaware of whether these were MN/AP results or MP/AN results, but additional bias may have resulted from knowledge that images were discrepant. In an ideal experiment, all discrepancy analysis would have been performed while the plates were still maintained, allowing biochemical characterization of potential VRE colonies. Biochemical analysis of borderline colors and potential positives would have confirmed or disallowed the identification of the presence of VRE in these specimens. Finally, there were limitations to our cost analysis. Laboratory technician time per specimen viewing had to be estimated, and each site did not directly report individual material costs for screens. Future studies to improve the cost estimates could add time stamps per specimen, such as the time of the inoculation in WASPLab, the time that the software finished the analysis, the time that image was opened, and the time that the technologist reported the result, to more accurately address the time difference between manual screening and automated screening.
Overall, these data demonstrate that the CDM software is highly sensitive, ensuring that plates with negative results can be accurately sorted to reduce the time and cost required for laboratories to perform large-volume screens. In addition, this was the second study demonstrating the robustness of the software, as 5 different chromogenic media evaluating both VRE and MRSA have been used without loss of performance, suggesting that any chromogenic media can be calibrated to the software, allowing laboratories to perform analyses without the need to change reagents in incorporating the software into the laboratory workflow.
ACKNOWLEDGMENTS
We thank Copan for supporting data analytics and manuscript review.
N.A.L. has served as a consultant to Copan.
Footnotes
For a commentary on this article, see doi:10.1128/JCM.01279-16.
REFERENCES
- 1.Bouza E, Kestler M, Beca T, Mariscal G, Rodríguez-Créixems M, Bermejo J, Fernández-Cruz A, Fernández-Avilés F, Muñoz P; Grupo de Apoyo al Manejo de la Endocarditis. 2015. The NOVA score: a proposal to reduce the need for transesophageal echocardiography in patients with enterococcal bacteremia. Clin Infect Dis 60:528–535. doi: 10.1093/cid/ciu872. [DOI] [PubMed] [Google Scholar]
- 2.Garrison RN, Fry DE, Berberich S, Polk HC Jr. 1982. Enterococcal bacteremia: clinical implications and determinants of death. Ann Surg 196:43–47. doi: 10.1097/00000658-198207000-00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang QY, Li RH, Shang XH. 2015. Urinary tract infection caused by Enterococcus isolates: aetiology and antimicrobial resistance patterns. J Chemother 27:117–119. doi: 10.1179/1973947814Y.0000000192. [DOI] [PubMed] [Google Scholar]
- 4.O'Driscoll T, Crank CW. 2015. Vancomycin-resistant enterococcal infections: epidemiology, clinical manifestations, and optimal management. Infection and drug resistance 8:217–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murray BE. 1990. The life and times of the Enterococcus. Clin Microbiol Rev 3:46–65. doi: 10.1128/CMR.3.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Uttley AH, Collins CH, Naidoo J, George RC. 1988. Vancomycin-resistant enterococci. Lancet i:57–58. [DOI] [PubMed] [Google Scholar]
- 7.Arthur M, Depardieu F, Gerbaud G, Galimand M, Leclercq R, Courvalin P. 1997. The VanS sensor negatively controls VanR-mediated transcriptional activation of glycopeptide resistance genes of Tn1546 and related elements in the absence of induction. J Bacteriol 179:97–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arthur M, Molinas C, Depardieu F, Courvalin P. 1993. Characterization of Tn1546, a Tn3-related transposon conferring glycopeptide resistance by synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J Bacteriol 175:117–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kramer A, Schwebke I, Kampf G. 2006. How long do nosocomial pathogens persist on inanimate surfaces? A systematic review. BMC Infect Dis 6:130. doi: 10.1186/1471-2334-6-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Noskin GA, Siddiqui F, Stosor V, Kruzynski J, Peterson LR. 1999. Successful treatment of persistent vancomycin-resistant Enterococcus faecium bacteremia with linezolid and gentamicin. Clin Infect Dis 28:689–690. doi: 10.1086/517221. [DOI] [PubMed] [Google Scholar]
- 11.DiazGranados CA, Zimmer SM, Klein M, Jernigan JA. 2005. Comparison of mortality associated with vancomycin-resistant and vancomycin-susceptible enterococcal bloodstream infections: a meta-analysis. Clin Infect Dis 41:327–333. doi: 10.1086/430909. [DOI] [PubMed] [Google Scholar]
- 12.Perencevich EN, Fisman DN, Lipsitch M, Harris AD, Morris JG Jr, Smith DL. 2004. Projected benefits of active surveillance for vancomycin-resistant enterococci in intensive care units. Clin Infect Dis 38:1108–1115. doi: 10.1086/382886. [DOI] [PubMed] [Google Scholar]
- 13.Faron ML, Buchan BW, Vismara C, Lacchini C, Bielli A, Gesu G, Liebregts T, van Bree A, Jansz A, Soucy G, Korver J, Ledeboer NA. 30 December 2015. Automated scoring of chromogenic media for the detection of MRSA using the WASPLab image analysis software. J Clin Microbiol doi: 10.1128/JCM.02778-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Julious SA. 2005. Two-sided confidence intervals for the single proportion: comparison of seven methods by Robert G. Newcombe, Statistics in Medicine 1998; 17:857–872. Stat Med 24:3383–3384. [DOI] [PubMed] [Google Scholar]
- 15.McNemar Q. 1947. Note on the sampling error of the difference between correlated proportions or percentages. Psychometrika 12:153–157. doi: 10.1007/BF02295996. [DOI] [PubMed] [Google Scholar]
- 16.Sievert DM, Ricks P, Edwards JR, Schneider A, Patel J, Srinivasan A, Kallen A, Limbago B, Fridkin S; National Healthcare Safety Network (NHSN) Team and Participating NHSN Facilities. 2013. Antimicrobial-resistant pathogens associated with healthcare-associated infections: summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2009–2010. Infect Control Hosp Epidemiol 34:1–14. doi: 10.1086/668770. [DOI] [PubMed] [Google Scholar]
- 17.Werner G, Coque TM, Hammerum AM, Hope R, Hryniewicz W, Johnson A, Klare I, Kristinsson KG, Leclercq R, Lester CH, Lillie M, Novais C, Olsson-Liljequist B, Peixe LV, Sadowy E, Simonsen GS, Top J, Vuopio-Varkila J, Willems RJ, Witte W, Woodford N. 2008. Emergence and spread of vancomycin resistance among enterococci in Europe. Euro Surveill 13(47):pii=19046 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19046. [PubMed] [Google Scholar]
- 18.Ostrowsky BE, Trick WE, Sohn AH, Quirk SB, Holt S, Carson LA, Hill BC, Arduino MJ, Kuehnert MJ, Jarvis WR. 2001. Control of vancomycin-resistant enterococcus in health care facilities in a region. N Engl J Med 344:1427–1433. doi: 10.1056/NEJM200105103441903. [DOI] [PubMed] [Google Scholar]
- 19.Siegel JD, Rhinehart E, Jackson M, Chiarello L; Healthcare Infection Control Practices Advisory Committee. 2007. Management of multidrug-resistant organisms in health care settings, 2006. Am J Infect Control 35(Suppl 2):S165–S193. doi: 10.1016/j.ajic.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 20.Shadel BN, Puzniak LA, Gillespie KN, Lawrence SJ, Kollef M, Mundy LM. 2006. Surveillance for vancomycin-resistant enterococci: type, rates, costs, and implications. Infect Control Hosp Epidemiol 27:1068–1075. doi: 10.1086/507960. [DOI] [PubMed] [Google Scholar]
- 21.Jenkins SG, Raskoshina L, Schuetz AN. 2011. Comparison of performance of the novel chromogenic spectra VRE agar to that of bile esculin azide and Campylobacter agars for detection of vancomycin-resistant enterococci in fecal samples. J Clin Microbiol 49:3947–3949. doi: 10.1128/JCM.00180-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suwantarat N, Roberts A, Prestridge J, Seeley R, Speser S, Harmon C, Zhang C, Henciak S, Stamper PD, Ross T, Carroll KC. 2014. Comparison of five chromogenic media for recovery of vancomycin-resistant enterococci from fecal samples. J Clin Microbiol 52:4039–4042. doi: 10.1128/JCM.00151-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Valdezate S, Miranda C, Navarro A, Freitas AR, Cabrera JJ, Carrasco G, Coque TM, Jimenez-Romano E, Saez-Nieto JA. 2012. Clonal outbreak of ST17 multidrug-resistant Enterococcus faecium harbouring an Inc18-like::Tn1546 plasmid in a haemo-oncology ward of a Spanish hospital. J Antimicrob Chemother 67:832–836. doi: 10.1093/jac/dkr545. [DOI] [PubMed] [Google Scholar]