Abstract
Studies have demonstrated that the combination of antimicrobial stewardship programs (ASP) and rapid organism identification improves outcomes in bloodstream infections (BSI) but have not controlled for the incremental contribution of the individual components. Hospitalized adult patients with blood culture pathogens on a rapid, multiplex PCR-based blood culture identification panel (BCID) that included 19 bacterial species, 5 Candida spp., and 4 antimicrobial resistance genes were studied over sequential time periods in a pre-post quasiexperimental study in 3 groups in the following categories: conventional organism identification (controls), conventional organism identification with ASP (AS), and BCID with ASP (BCID). Clinical and economic outcomes were compared between groups. There were 783 patients with positive blood cultures; of those patients, 364 (115 control, 104 AS, and 145 BCID) met inclusion criteria. The time from blood culture collection to organism identification was shorter in the BCID group (17 h; P < 0.001) than in the control group (57 h) or the AS group (54 h). The BCID group had a shorter time to effective therapy (5 h; P < 0.001) than the control group (15 h) or AS group (13 h). The AS (57%) and BCID (52%) groups had higher rates of antimicrobial de-escalation than the control group (34%), with de-escalation occurring sooner in the BCID group (48 h; P = 0.034) than in the AS group (61 h) or the control group (63 h). No difference between the control group, AS group, and BCID group was seen with respect to mortality, 30-day readmission, intensive care unit length of stay (LOS), postculture LOS, or costs. In patients with BSI, ASP alone improved antimicrobial utilization. Addition of BCID to an established ASP shortened the time to effective therapy and further improved antimicrobial use compared to ASP alone, even in a setting of low antimicrobial resistance rates.
INTRODUCTION
Bloodstream infections (BSI) are an important cause of morbidity and mortality (1). The rise of antimicrobial-resistant organisms in recent years warrants empirical use of broad-spectrum antimicrobials for patients with suspicion of serious infections, including BSI, until organism identification and antimicrobial susceptibility data become available (2). Unfortunately, conventional organism identification and susceptibility reporting require 48 to 72 h to produce final results, leading to a substantial delay in the receipt of appropriate antimicrobial therapy, which has been shown to negatively impact patient outcomes, particularly in the setting of multidrug-resistant organisms (3, 4). That the delay in de-escalation of antimicrobial therapy for infections caused by susceptible organisms can result in longer durations of exposure of these patients to broader-spectrum agents, which may lead to development of resistance, Clostridium difficile infections, microbiome disruption, and increased costs, is equally troubling (5, 6).
Several approaches can provide organism identification and detect antimicrobial resistance genes within hours of blood culture positivity, allowing earlier and more effective antimicrobial therapy (7, 8). These include fluorescence in situ hybridization, matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry, and nucleic acid hybridization and amplification assays. Comprehensive, panel-based molecular diagnostic assays that detect all of the major bloodstream pathogens and selected antimicrobial resistance genes are now available for direct testing of positive blood cultures (9, 10, 11). A number of examples in the literature suggest that shortening the time to appropriate therapy due to rapid diagnostic tests may lessen the clinical and economic burden of BSI (12). The greatest impact of rapid diagnostic tests appears to occur when the tests are implemented in combination with antimicrobial stewardship program (ASP) intervention to ensure that the test result is acted on in a timely manner (12). In fact, a recent prospective clinical trial by Banerjee et al. that evaluated a rapid multiplex PCR blood culture identification panel (BCID) found that while BCID reported with templated comments resulted in optimized antibiotic use compared to controls without BCID, the addition of ASP intervention enhanced rates of antimicrobial de-escalation (13).
The purpose of the present study was to evaluate the impact of BCID in combination with ASP intervention on antimicrobial use and the clinical and economic outcomes of patients with bloodstream infections compared to conventional organism identification techniques. To account for the incremental contribution of each individual constituent (ASP or BCID) to the study endpoints, three analysis groups were compared: convention organism identification, conventional organism identification with ASP intervention group, and BCID with ASP intervention.
MATERIALS AND METHODS
Study design.
This single-center, pre-post quasiexperimental study was performed at the Medical University of South Carolina (MUSC), a 709-bed academic medical center in Charleston, SC. The study was approved by the MUSC Institutional Review Board.
All adult (≥18 years of age) patients with a positive blood culture(s) between 1 August and 31 October of three distinct time periods corresponding to the years 2010, 2012, and 2014 were included in the study unless they met any exclusion criterion. Patients who had expired or were placed on hospice care prior to blood culture positivity, those who had been transferred from an outside hospital and had a history of a previously positive blood culture of the same organism, those whose blood cultures were deemed by the treating health care team to contain contaminants, and those who were not admitted to MUSC were excluded from the study. Patients with blood culture pathogens not included on the BCID were excluded from the study. Based on an 82% identification rate of the BCID, the number of patients meeting this criterion was not anticipated to be sizeable (14).
Patients with BSI from 2010, prior to both ASP intervention and the implementation of the BCID technology (controls), were compared to patients with BSI from 2012 after implementation of ASP intervention on all positive blood cultures but without the BCID technology (AS) and to patients with BSI from 2014 with both antimicrobial stewardship intervention and BCID (BCID).
Patients were evaluated for study inclusion by querying the institution's database for positive blood cultures. Only the first positive culture for each patient was included during the study period; any subsequent episode of BSI was excluded. Once patients were identified, information was collected by investigators using REDCap electronic data capture tools (15).
Interventions.
MUSC employed an ASP throughout the study period, with daily activities of positive blood culture review performed by the stewardship pharmacists starting in 2012. Prior to 2012, ASP activities were selected on the basis of drug identification (e.g., audits of restricted and high-cost antimicrobials) rather than on the basis of syndrome- or culture-based interventions. Real-time alerts of positive blood cultures provided by SafetySurveillor (Premier, Inc., Charlotte, NC), a Web-based infection tracking and antimicrobial utilization tool, were available to the ASP beginning in January 2012. The BCID technology was coupled with real-time alerting capabilities of positive blood culture results for ASP activities starting in December 2013. The BCID results were used by the ASP to assess appropriateness of therapy based on the institution's pathogen-specific empirical treatment algorithms according to organism identification, which were posted on the hospital intranet (see Fig. S1 in the supplemental material). The ASP recommended changes to antimicrobial therapy, if needed, to the primary care team (Monday through Friday from h 0800 to h 1700). For results occurring outside these working hours, ASP interventions were made the following working day, if necessary.
The FilmArray BCID (BioFire Diagnostics, LLC, Salt Lake City, UT) analysis was performed on all index-positive blood cultures beginning on 1 December 2013. The BCID can identify Staphylococcus spp., S. aureus, Streptococcus spp., S. agalactiae, S. pyogenes, S. pneumoniae, Enterococcus spp., Listeria monocytogenes, Enterobacteriaceae, Escherichia coli, Enterobacter cloacae complex, Klebsiella oxytoca, K. pneumoniae, Serratia spp., Proteus spp., Acinetobacter baumannii, Haemophilus influenzae, Neisseria meningitidis, Pseudomonas aeruginosa, Candida albicans, C. glabrata, C. krusei, C. parapsilosis, and C. tropicalis and 4 antibiotic resistance genes, mecA, vanA and vanB (vanA/B), and blaKPC, within 1 h directly from positive blood culture bottles (9). Blood cultures were performed during first two study periods using BacT/Alert standard aerobic, standard anaerobic, Fan Plus aerobic, and Fan Plus anaerobic blood culture bottles (bioMérieux, Durham, NC) and during the last study period using Bactec Plus aerobic/F and Plus anaerobic/F bottles (BD Diagnostic Systems, Sparks, MD). During all study periods, aliquots from bottles that gave positive signals were Gram stained and subcultured for organism identification by conventional methods, and results of the Gram stain were communicated by laboratory personnel to nursing staff via telephone within 1 h of the blood culture being identified as positive. During the BCID procedure, laboratory personnel would wait for both the Gram stain result and the BCID result to be available and would then communicate with the nursing staff via telephone within 2 h of blood culture being identified as positive. The ASP was notified of positive blood cultures only through SafetySurveillor, and the data were updated and monitored in real time.
Results were posted to the electronic medical records once verbal notification was received. No templated comments or clinical interpretations of the results were provided at the time of reporting. Identification and susceptibility testing were performed using conventional phenotypic methods and a MicroScan WalkAway system (Beckman Coulter, Inc., Brea, CA). No other rapid identification techniques were in place except for those used for identification of Staphylococcus aureus (direct tube coagulase testing and plating to methicillin-resistant S. aureus [MRSA] chromogenic medium) at the time of BCID implementation. The microbiology laboratory was staffed 24 h a day, 7 days a week throughout the study.
Outcomes.
The primary outcome was the comparison of times to effective therapy and initial antimicrobial use with the BCID versus conventional methods with and without ASP intervention for BSI. Time to effective therapy was defined as the elapsed time in hours between the index culture collection and receipt of the initial dose of an antimicrobial shown to exhibit activity against the patient-specific organism based on the in vitro susceptibility results with intermediate results considered ineffective. Initial antimicrobial use was defined as the duration of therapy received during the first 96 h after blood culture collection. The hospital's antimicrobial formulary remained largely unchanged over the course of the study, with the exception of a restriction on empirical ciprofloxacin use beginning in 2012. Clinical endpoints were compared between groups and included in-hospital mortality (all cause and infection related), 30-day all-cause readmission, microbiological clearance, hospital length of stay (LOS) following blood culture positivity, and overall patient-specific hospital costs. Microbiological clearance was defined as the time from index culture collection to the time of collection of the first negative blood culture if cultures were repeated and subsequently negative. All-cause mortality was defined as death resulting from any cause at the end of hospitalization, while infection-related mortality was defined as death occurring while the patient was receiving antimicrobials for the bloodstream infection, without any other obvious cause of death. Patients who died during their hospital admission were not included in the LOS stay analysis. Relapse of bloodstream infection was defined as the reoccurrence of the same organism in a blood culture within 30 days after the end of treatment. All costs were adjusted for inflation and converted to 2014 dollars, according to the consumer price index inflation calculator provided by the United States Department of Labor, Bureau of Labor Statistics (http://www.bls.gov/data/inflation_calculator.htm).
Statistical analysis.
Statistical comparisons were performed between study groups with one-way analysis of variance for continuous variables. Dichotomous variables were compared using the chi-square test or Fisher's exact test. All statistical analyses were performed using SigmaPlot version 12.5 (Systat Software, Inc., San Jose, CA). A P value of ≤0.05 (two-tailed test) was considered statistically significant.
RESULTS
Patients.
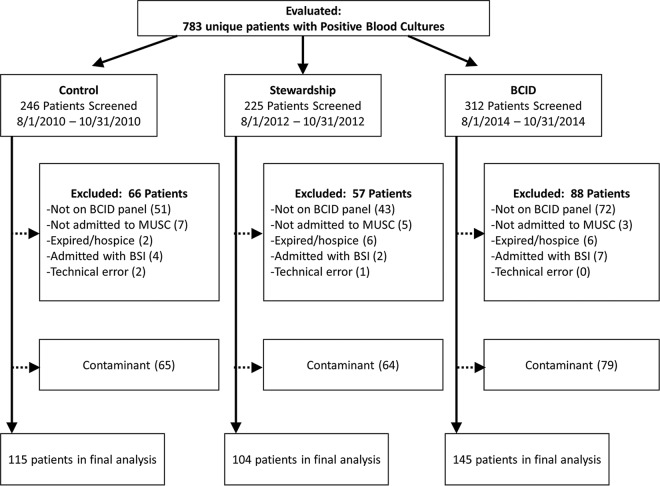
There were 783 unique patients with positive blood cultures identified and screened during the study periods, and 364 (115 control, 104 AS, and 145 BCID) met the inclusion criteria, representing a total of 404 blood culture pathogens. A total of 211 patients were excluded from the study, and another 208 patients had blood culture contaminants (Fig. 1). Patient demographics and baseline characteristics were comparable between groups, with the exception of higher rates of myocardial infarction in the BCID group (Table 1). The BSI was more frequently of intra-abdominal origin in the control group than in the AS and BCID groups.
FIG 1.
Flowchart of study participants. Abbreviations: BCID, blood culture identification panel; BSI, bloodstream infection; MUSC, Medical University of South Carolina.
TABLE 1.
Demographics and baseline characteristics of patients with bloodstream infectionsa
| Parameter | Value(s) |
|||
|---|---|---|---|---|
| Control (n = 115) | Stewardship (n = 104) | BCID (n = 145) | P | |
| Demographics | ||||
| Age, yrs, median (IQR) | 55 (43–69) | 57.5 (47–68) | 58 (44–69) | 0.97 |
| Male | 58 (50.4) | 59 (56.7) | 80 (55.1) | 0.61 |
| White | 67 (58.3) | 53 (51.0) | 77 (53.1) | 0.78 |
| Black or African American | 45 (39.1) | 46 (44.4) | 63 (43.4) | 0.78 |
| Preexisting conditions | ||||
| Immunosuppressed secondary to therapyb | 30 (26.1) | 23 (22.1) | 32 (22.0) | 0.70 |
| Neutropenia (ANC < 500) | 6 (5.2) | 2 (1.9) | 8 (5.5) | 0.34 |
| Myocardial infarction | 2 (1.7) | 2 (1.9) | 11 (7.6) | 0.03 |
| Congestive heart failure | 16 (13.9) | 15 (14.4) | 28 (19.3) | 0.42 |
| HIV/AIDS | 5 (4.3) | 0 | 2 (1.4) | 0.05 |
| Solid-organ malignancy | 23 (20) | 17 (16.3) | 27 (18.6) | 0.78 |
| Peripheral vascular disease(s) | 5 (4.3) | 7 (6.7) | 9 (6.2) | 0.72 |
| Hematologic malignancy | 12 (10.4) | 5 (4.8) | 13 (9.0) | 0.29 |
| Cerebrovascular disease | 10 (8.7) | 12 (11.5) | 22 (15.2) | 0.28 |
| Diabetes mellitus | 29 (25.2) | 33 (31.7) | 52 (35.9) | 0.18 |
| Renal replacement therapy | 12 (10.4) | 12 (11.5) | 15 (10.3) | 0.95 |
| Total parenteral nutrition | 14 (12.2) | 12 (11.5) | 17 (11.7) | 0.99 |
| Liver disease | 16 (13.9) | 10 (9.6) | 16 (11) | 0.59 |
| Presumed source of infection | ||||
| Urine | 19 (16.5) | 27 (26.0) | 36 (24.8) | 0.17 |
| Catheter related | 21 (18.3) | 16 (15.4) | 25 (17.2) | 0.87 |
| Respiratory | 7 (6.1) | 10 (9.6) | 13 (9.0) | 0.59 |
| Intra-abdominal | 38 (33.0) | 12 (11.5) | 30 (20.7) | <0.001 |
| Skin or skin structure related | 12 (10.4) | 16 (15.4) | 13 (9.0) | 0.27 |
| Other | 8 (7.0) | 11 (10.6) | 5 (3.4) | 0.08 |
| Unidentified | 10 (8.7) | 12 (11.5) | 23 (15.6) | 0.21 |
| Clinical features | ||||
| Charlson comorbidity index, median (IQR) | 3 (2–6) | 3 (2–5) | 4 (2–6) | 0.11 |
| Pitt bacteremia score, median (IQR) | 2 (0–3) | 1 (0–3) | 1 (0–3) | 0.30 |
| Mechanical ventilation | 21 (18.2) | 23 (22.1) | 22 (15.1) | 0.37 |
| Hypotension | 46 (40.0) | 33 (31.7) | 54 (37.2) | 0.44 |
| ICU admission ± 48 h of blood culture | 42 (36.5) | 37 (35.6) | 59 (40.1) | 0.67 |
| ICU, LOS, days, median (IQR) | 7 (3–11) | 5 (3–16.5) | 5 (2–11) | 0.45 |
| ID consult | 68 (59.1) | 62 (59.6) | 75 (51.7) | 0.36 |
| Polymicrobial bloodstream infection | 12 (10.4) | 4 (3.8) | 18 (12.4) | 0.06 |
| Community associated (<48-h onset) | 79 (68.7) | 78 (75.0) | 94 (64.8) | 0.23 |
| Hospital associated (>48-h onset) | 36 (31.3) | 26 (25.0) | 51 (35.2) | 0.23 |
Data are presented as number (percent) of patients, unless specified otherwise. Abbreviations: ANC, absolute neutrophil count; BCID, blood culture identification panel; HIV/AIDS, human immunodeficiency virus infection and AIDS; ICU, intensive care unit; ID, infectious diseases; IQR, interquartile range.
Immunosuppressive therapy data represent active systemic chemotherapy, tacrolimus, mycophenolate mofetil, azathioprine, cyclosporine (or equivalent) for more than 7 days, or a systemic steroid for more than 10 days the previous month.
Microbiology.
The blood culture pathogens of the included patients were 41.6% Gram-positive bacteria, 50.5% Gram-negative bacteria, and 7.9% Candida spp. and did not differ in prevalence between study groups. Blood cultures were polymicrobial for 34 (9.3%) patients. Organism distributions did not differ significantly between study groups, with S. aureus, Escherichia coli, and Klebsiella pneumoniae being the most common pathogens in each cohort (Table 2).
TABLE 2.
Microbiology, treatment, and clinical outcomes for all patientsa
| Parameter | Value(s) |
|||
|---|---|---|---|---|
| Control (n = 115) | Stewardship (n = 104) | BCID (n = 145) | P | |
| Microbiology-related outcomes | ||||
| Time to blood culture positivity, h, median (IQR) | 17.3 (14.6–22.0) | 14.2 (12.9–18.2)b | 13.9 (11.9–18.5)b | <0.001 |
| Time to organism identification, h, median (IQR) | 57.4 (42.5–68.5) | 53.9 (40.8–62.5) | 17.2 (13.3–24.8)b,c | <0.001 |
| Time to in vitro susceptibility results, h, median (IQR) | 64.4 (57.4–70.6) | 61.5 (55.6–67.8) | 64.4 (58.6–78.1)c | 0.03 |
| Blood culture pathogens | ||||
| Gram-positive bacteria (n = 168) | 51 (40.2) | 48 (44.4) | 59 (37.1) | 0.58 |
| Enterococcus faecalis (VRE) | 0 | 0 | 1 | |
| Enterococcus faecalis (not VRE) | 8 | 5 | 13 | |
| Enterococcus faecium (VRE) | 0 | 0 | 1 | |
| Enterococcus faecium (not VRE) | 1 | 0 | 0 | |
| Enterococcus avium | 0 | 0 | 2 | |
| Methicillin-resistant Staphylococcus aureus | 14 | 14 | 13 | |
| Methicillin-sensitive Staphylococcus aureus | 18 | 15 | 18 | |
| Staphylococcus spp. (coagulase negative) | 2 | 2 | 4 | |
| Streptococcus agalactiae | 3 | 3 | 2 | |
| Streptococcus pneumoniae | 2 | 1 | 2 | |
| Streptococcus pyogenes | 0 | 1 | 1 | |
| Streptococcus anginosus group | 2 | 0 | 5 | |
| Streptococcus group C | 0 | 1 | 0 | |
| Streptococcus group G | 0 | 1 | 0 | |
| Streptococcus mitis | 4 | 5 | 4 | |
| Gram-negative bacteria (n = 204) | 63 (49.6) | 54 (50) | 87 (54.7) | 0.58 |
| Acinetobacter baumannii | 2 | 2 | 4 | |
| Enterobacter cloacae | 6 | 6 | 7 | |
| Enterobacter aerogenes | 2 | 1 | 4 | |
| Escherichia coli | 20 | 27 | 34 | |
| Klebsiella oxytoca | 1 | 0 | 1 | |
| Klebsiella pneumoniae | 19 | 13 | 21 | |
| Proteus mirabilis | 3 | 2 | 4 | |
| Pseudomonas aeruginosa | 5 | 3 | 6 | |
| Serratia marcescens | 4 | 0 | 4 | |
| Salmonella spp. | 1 | 0 | 2 | |
| Candida spp. (n = 32) | 13 (10.2) | 6 (5.6) | 13 (8.2) | 0.58 |
| Candida albicans | 6 | 5 | 2 | |
| Candida glabrata | 4 | 1 | 9 | |
| Candida krusei | 1 | 0 | 0 | |
| Candida parapsilosis | 1 | 0 | 1 | |
| Candida tropicalis | 1 | 0 | 1 | |
| Treatment-related outcomes | ||||
| Time to effective therapy, h, median (IQR) | 15.0 (4.3–25.7) | 13.0 (4.3–23.1) | 4.9 (1.0–19.6)b,c | <0.001 |
| Repeat positive blood cultures | 21/118 (17.8) | 11/103 (10.7) | 25/155 (16.1) | 0.31 |
| Microbiologic clearance within 3 days after blood culture draw | 100/127 (78.7) | 90/108 (83.3) | 128/159 (80.5) | 0.67 |
| Clinical outcomes | ||||
| Length of hospitalization (LOS), no. of days, median (IQR) (n = 319) | 12 (7–20) | 9 (6–18) | 11 (7–24) | 0.33 |
| LOS, no. of days, following index blood culture collection, median (IQR) (n = 319) | 8.4 (5.5–15.6) | 7.8 (5.1–14.8) | 8.2 (5.2–15.9) | 0.88 |
| In-hospital mortality (all cause) | 13 (11.3) | 12 (11.5) | 20 (13.8) | 0.80 |
| Infection-related mortality | 7 (6.1) | 8 (7.7) | 13 (9.0) | 0.69 |
| 30-day all-cause hospital readmission (n = 319) | 26 (25.5) | 24 (26.1) | 23 (18.4) | 0.31 |
| Relapse of bacteremia | 6 (5.2) | 4 (3.8) | 4 (2.7) | 0.59 |
| Total hospital costs (per patient), $, mean (median) | 57,442 (27,564) | 58,306 (21,222) | 47,992 (23,840) | 0.26 |
Data are presented as number (percent) of patients, unless specified otherwise. Abbreviations: BCID, blood culture identification panel; IQR, interquartile range; VRE, vancomycin-resistant enterococci.
Statistically significant compared to control group.
Statistically significant comparison between the 2 intervention groups.
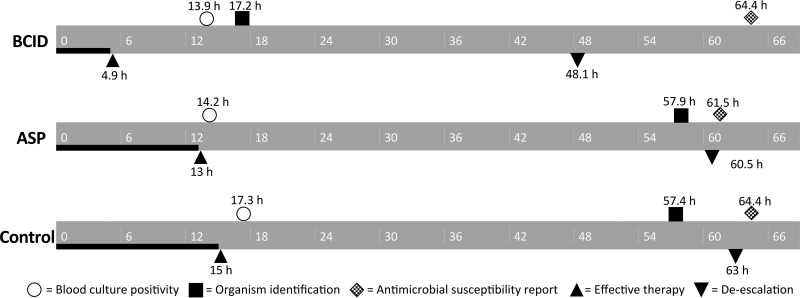
The median time to blood culture positivity from the time of blood culture collection was significantly longer in the control group (17.3 h) than in the AS (14.2 h) and BCID (13.9 h) groups. This was probably due to increased use of blood culture bottles containing antimicrobial neutralizing substances. The median time to organism identification from the time of blood culture collection was significantly shorter in the BCID group (17.2 h) than in the AS (53.9 h) and control (57.4 h) groups even when the shorter time to blood culture positivity was taken into account. Although the median time to conventional antimicrobial susceptibility test results was significantly longer (2.9 h) in the BCID group than in the AS group, the test methods remained the same throughout all three study periods (Fig. 2 and Table 2).
FIG 2.
Timeline for microbiology and antimicrobial therapy among study groups. Symbols represent the median values for the study population. Abbreviations: ASP, antimicrobial stewardship program; BCID, blood culture identification panel.
BCID provided a result in 76.9% of the blood cultures in the study group. Discrepant results between the BCID and conventional test occurred with 10 blood cultures (Table 3). Seven discrepancies occurred in polymicrobial cultures, with BCID detecting additional organisms in two and conventional culture detecting additional organisms in five. Of the 10 discrepant results, antimicrobial therapy was affected by the BCID result in only 3 cases. Patient 32 was empirically treated with ceftriaxone for K. pneumoniae by BCID and was then transitioned to cefepime when Enterobacter aerogenes was identified. Patient 337 received fluconazole and micafungin for the entire treatment course in order to treat both yeast species identified by BCID, although only C. glabrata was recovered in culture. Patient 342 received 1 dose of daptomycin for treatment of presumed VRE by BCID, and then the patient's treatment was de-escalated back to vancomycin.
TABLE 3.
Discrepancies between BCID and conventional culture and phenotypic antimicrobial susceptibility test resultsa
| Patient | BCID result(s) | Conventional culture and phenotypic susceptibility testing result(s) |
|---|---|---|
| Polymicrobic cultures | ||
| 342 | Enterococcus spp. (vanA/B positive), Candida glabrata, Staphylococcus spp. | Vancomycin-susceptible Enterococcus faecium |
| 13 | Enterococcus spp. (vanA/B negative) | Vancomycin-susceptible Enterococcus avium, Serratia marcescens |
| 28 | Enterobacter cloacae | Enterobacter cloacae, Pseudomonas aeruginosa |
| 41 | Klebsiella pneumoniae, Enterobacter cloacae | Klebsiella pneumoniae, Enterobacter cloacae, Acinetobacter baumannii |
| 337 | Staphylococcus aureus (mecA positive), Candida glabrata, Candida parapsilosis | Methicillin-resistant Staphylococcus aureus, Candida glabrata |
| 51 | Staphylococcus aureus (mecA negative), Klebsiella pneumoniae | Methicillin-susceptible Staphylococcus aureus, Klebsiella pneumoniae, Enterobacter aerogenes |
| 2 | Enterobacter cloacae | Enterobacter cloacae, Klebsiella pneumoniae |
| Discrepancy in organism identification | ||
| 32 | Klebsiella pneumoniae | Enterobacter aerogenes |
| 285 | Enterococcus spp. (vanA/B negative) | Not recovered in culture |
| 5 | Klebsiella pneumoniae | Enterobacter aerogenes |
Two subjects included in the BCID group did not have a BCID panel performed due to operator error. Abbreviation: BCID, blood culture identification panel.
Antimicrobial resistance levels remained low and did not differ across the study periods, with a total of 2 VRE species, 5 extended-spectrum-β-lactamase (ESBL)-producing species, and 1 meropenem-resistant species (Acinetobacter baumannii) encountered throughout the study. There were no Klebsiella pneumoniae carbapenemase (KPC)-producing organisms detected by PCR during the BCID study period. No discrepancies were found between the molecular and phenotypic susceptibility tests for mecA in Staphylococcus spp. One patient with Enterococcus faecium bacteremia had vanA/B detected by BCID, but the phenotypic susceptibility testing revealed the organism to be vancomycin susceptible even when the bottle was subcultured to VRE chromogenic medium.
Antimicrobial utilization.
The BCID group had a shorter median time to effective therapy (4.9 h; P < 0.001) than the control group (15.0 h) or AS group (13.0 h). Between 19% (BCID) and 29% (control) of patients failed to receive effective therapy within 24 h of blood culture collection, and by 48 h less than 10% of patients in each group failed to receive effective therapy. These rates were numerically lower in the BCID group than in the ASP and control groups but did not reach statistical significance. The AS and BCID groups had significantly higher rates of antimicrobial de-escalation (56.7% and 52.4%, respectively) in the first 96 h than the control group (33.9%), with the first antimicrobial de-escalation occurring approximately 12 h sooner in the BCID group (48.1 h; P = 0.034) than in the AS group (60.5 h) or control group (63.0 h) (Table 4).
TABLE 4.
Antimicrobial use during the first 96 h after blood culture collectiong
| Parameter | Value(s) |
|||
|---|---|---|---|---|
| Control (n = 115) | Stewardship (n = 104) | BCID (n = 145) | P | |
| Antibiotic therapy modification in first 96 h | ||||
| De-escalation of any antimicrobial therapyc | 39 (33.9) | 59 (56.7)a | 76 (52.4)a | 0.001 |
| Time to first antimicrobial de-escalation, h, median (IQR) (n = 174) | 63.0 (43.6–73.6) | 60.5 (44.8–70.4) | 48.1 (24.3–69.3)a,b | 0.03 |
| Antibiotic therapy in the first 96 h | ||||
| Receipt of an anti-MRSA agentd | 74 (64.3) | 66 (63.4) | 79 (54.5) | 0.20 |
| Duration of anti-MRSA therapy, h, median (IQR) (n = 219) | 62.7 (45.6–83.0) | 56.4 (32.3–82.0) | 60 (30.7–84.0) | 0.62 |
| Receipt of an antipseudomonal β-lactam agente | 71 (61.7) | 70 (67.3) | 106 (73.1) | 0.15 |
| Duration of anti-pseudomonal β-lactam, h, median (IQR) (n = 247) | 60.6 (39.9–84.7) | 51.5 (32.4–76.3) | 63.1 (29.7–90.1) | 0.26 |
| Receipt of narrow-spectrum β-lactam agentf | 28 (24.3) | 48 (46.2)a | 60 (41.3)a | 0.002 |
| Duration of narrow-spectrum β-lactam, h, median (IQR) (n = 136) | 26.9 (23.2–56.6) | 35.0 (24.3–70.2) | 52.6 (25.4–75.4)a | 0.02 |
| Receipt of fluoroquinolone | 39 (33.9) | 5 (4.8)a | 11 (7.5)a | <0.001 |
| Antimicrobial resistance (no. of documented nonsusceptible strains) | ||||
| Ceftriaxone | 5 | 3 | 4 | |
| Fluoroquinolone (ciprofloxacin) | 9 | 8 | 11 | |
| Cefepime | 3 | 1 | 5 | |
| Meropenem | 0 | 0 | 1 | |
| Piperacillin-tazobactam | 5 | 1 | 8 | |
Statistically significant compared to control group.
Statistically significant comparison between the 2 intervention groups.
De-escalation was defined as cessation of treatment with 1 or more antibiotics and/or switching to an antibiotic with a narrower spectrum of activity.
Anti-MRSA therapy included vancomycin, daptomycin, or linezolid therapy.
Antipseudomonal β-lactams included cefepime, ceftazidime, doripenem, imipenem-cilastatin, meropenem, and piperacillin-tazobactam.
Narrow-spectrum β-lactams were defined as β-lactams devoid of antipseudomonal activity.
Data are presented as number (percent) of patients, unless specified otherwise. Abbreviations: BCID, blood culture identification panel; IQR, interquartile range; MRSA, methicillin-resistant S. aureus.
The durations of administration of anti-MRSA therapy and antipseudomonal β-lactams among all patients did not differ between study groups (Table 4). However, among patients with monomicrobial Gram-negative bacteremia (n = 68), the duration of anti-MRSA therapy was significantly shorter in the AS (26.4 h) and BCID (12 h) groups than in the control group (60.8 h, P = 0.02) (data not shown). More patients received a narrow-spectrum β-lactam during the initial 96 h in the AS (46.2%) and BCID (41.3%) groups than in the control group (24.3%, P = 0.002). However, it was only in the BCID (52.6 h) group that the duration of narrow-spectrum β-lactam therapy was greater than in the control group (35.0 and 26.9 h, P = 0.019). Consistent with the institutional formulary change, ciprofloxacin use was greatly reduced in the AS and BCID groups compared with the control group.
Clinical and economic outcomes.
There were no significant differences in all-cause or infection-related LOS, in mortality, or in 30-day readmission between cohorts (Table 2). Clinical outcomes were similar between time periods in subgroup analyses of monomicrobial Gram-positive or Gram-negative bacteremia and in analyses controlling for the presumed source of infection. The median total hospital costs were lower in both the AS and BCID groups than in the control group, but the differences did not reach statistical significance.
DISCUSSION
This retrospective study revealed that real-time ASP intervention in patients with bloodstream infections improved antimicrobial utilization in the early course of illness. Moreover, when ASP intervention was supplemented with rapid organism identification and antimicrobial resistance results, effective therapy was prescribed more rapidly and antimicrobials were used in a more judicious manner.
The preponderance of the literature published to date evaluating the impact of rapid molecular diagnostics in BSI has demonstrated that the maximal benefit occurs when molecular diagnostics is implemented in conjunction with real-time ASP intervention compared to reporting results alone (12, 13). Recently, Banerjee et al. validated the beneficial impact of BCID on antibiotic use in a prospective randomized controlled trial of all patients with positive blood cultures at their institution between August 2013 and March 2014 (13). Patients were randomized into one of three groups: conventional blood culture processing (control), BCID with templated comments, or BCID with templated comments and real-time antimicrobial stewardship. The authors found that, while BCID results reported with template comments resulted in optimized antibiotic use, the addition of ASP intervention enhanced antimicrobial de-escalation, further supporting the hypothesis of the advantage of an integrated approach incorporating ASP for implementing rapid diagnostics. There were no differences in clinical or microbiologic outcomes among the groups.
However, none of the previous reports accounted for the contribution of ASP intervention alone to outcomes or assessed whether the addition of rapid molecular results to an established ASP further improves outcomes. To our knowledge, this study was the first to evaluate these endpoints in a sequential manner (no intervention, ASP intervention alone, and ASP intervention and BCID).
Due to the establishment of ASP intervention in all positive blood cultures prior to the implementation of the BCID at our institution, we were in an ideal situation to account for the individual contribution of ASP intervention. Compared with the control group results, ASP intervention did not reduce the time to effective antimicrobial therapy but was associated with higher rates of antimicrobial de-escalation during the first 4 days of BSI. Adding the BCID to our ASP intervention was clearly beneficial, as effective therapy and the first antimicrobial de-escalation occurred more rapidly and utilization of narrow-spectrum therapies significantly increased, with no harmful effects on patient care. While not statistically significant, the average hospital cost was approximately $10,000 less per patient in the BCID group than in the control and AS groups. Applying this to each patient in the study, the BCID group was associated with a nearly $1,500,000 difference in cost. As with the BCID study by Banerjee et al., our institution's active ASP, minimal antimicrobial resistance rates, and high rates of infectious disease consultation (56%) in all study groups likely contributed to the initiation of effective therapy early in the course of BSI, which may have limited the differences in clinical outcomes (13).
Unlike Banerjee et al., we excluded patients with organisms not included on the BCID, as the BCID was not anticipated to have a considerable impact on their clinical outcomes. Additionally, we excluded patients with blood cultures that were considered to contain contaminants (e.g., a single culture of coagulase-negative staphylococcus), which comprised nearly 30% of the population in the study by Banerjee et al., in order to minimize confounding of the impact in patients with active BSI. Even though the studied patient populations differed, the findings were strikingly similar with regard to antimicrobial use and clinical and economic outcomes. As the vast majority of patients in each of these studies were receiving effective therapies at the time of BCID results, it is likely that the greatest individual benefit of BCID lies in reducing unnecessary antimicrobial use. At institutions with higher rates of drug-resistant (VRE and KPC) bloodstream infections, the clinical benefit of the BCID may be greater, and further studies in these settings are needed to realize the full potential of this technology.
Despite the more rapid species identification of blood culture pathogens in the BCID group, the utilization and duration of anti-MRSA therapy and antipseudomonal β-lactam antibiotics were unchanged compared to those in the groups with conventional organism identification. Possible explanations for this observation might include but are not limited to the following: a preference for fourth-generation cephalosporins or carbapenems for use against AmpC-producing organisms, a policy of restriction of the empirical use of ciprofloxacin, suspicion of a concomitant infection warranting empirical broad-spectrum coverage (e.g., in cases of immunocompromised patients), lack of an ESBL resistance determinant on the BCID, and/or potential hesitancy of providers to narrow the spectrum of antimicrobial activity based on the PCR result alone, prior to antimicrobial susceptibility test results. In addition, by excluding patients with blood culture contaminants, it is possible that the current study results underrepresent the beneficial impact of BCID on antimicrobial utilization, which may explain why the duration of anti-MRSA therapy was not significantly reduced in the BCID group. Nonetheless, the availability of BCID results significantly reduced the time to effective therapy while also enabling clinicians and ASP to streamline the spectrum of antimicrobial therapy approximately 12 h earlier than in the groups without the BCID technology. Note that the clinical implications of early antimicrobial de-escalation, such as limiting selective pressure, are likely of societal benefit but cannot easily be quantified and that such assessments were beyond the scope of this study design.
Several previous studies documented the excellent sensitivity and specificity of the BCID (9, 16, 17). The multiplexed BCID provided results in 76.9% of positive blood cultures, and those results were concordant with results of conventional methods in 93.1% of positive cultures. There were only three organism misidentifications by BCID. Enterobacter aerogenes was misidentified as K. pneumoniae in two blood cultures, and Enterococcus spp. were not recovered by conventional subculture in one blood culture. The BCID K. pneumoniae assay is known to cross-react with E. aerogenes and Raoultella ornithinolytica. The misidentification of Enterococcus spp. may have been due to contamination of the blood culture medium with enterococcal DNA or to the presence of nonviable organisms in the patient's blood (18). The results of the BCID resistance gene detection were concordant with the phenotypic results, with the exception of one patient with Enterococcus faecium bacteremia; that patient had vanA/B detected by BCID, but susceptibility testing revealed the organism to be vancomycin susceptible. This result was due to either a false-positive vanA/B test or the presence of a vancomycin-resistant subpopulation that was not detected by the conventional methods or of an organism that did not express vancomycin resistance.
BCID results were available on average 1.7 days sooner than the results determined by the conventional methods. BCID can be used whether the blood culture contains Gram-positive or Gram-negative bacteria or yeast and, because of its ease of use, can be performed in many clinical laboratory settings with minimal hands-on time. The known limitations of the BCID include failure to detect all organisms in mixed cultures, cross-reactions with K. pneumoniae assay, and the limited susceptibility information provided for Gram-negative bacteria. BCID supplements but does not replace conventional methods for identification and antimicrobial susceptibility testing and, as such, adds significant expense to the laboratory budget.
Our study was not without limitations. The retrospective nature and lack of randomization in the study may have failed to capture potential changes in the standard of care over time, although we are not aware of any such changes in the management of BSI. While the randomized controlled trial undoubtedly represents a superior study design, it is our belief that it would have been unethical to randomize patients to slower methods of organism identification (conventional methods) when a rapid FDA-approved method (BCID) was available and was our standard of care prior to initiation of this study. Additionally, our study time periods allowed comparisons of individual components of antimicrobial stewardship intervention and BCID, an assessment not performed in prior investigations. Another potential limitation of the study was that the stewardship intervention did not occur 24 h a day. Looking at the time of day of blood culture positivity, approximately 50% of the positive blood cultures were obtained between h 0800 and h 1700 in each group (P = 0.95) when an ASP team member was monitoring in real time, suggesting no difference between study groups in the time of day of blood culture positivity. It is possible that having ASP available for intervention around the clock may have further improved the time to effective therapy and antimicrobial utilization. However, it is our opinion that since most interventions are related to nonurgent antimicrobial de-escalation, overnight staffing of the ASP may not be cost-effective. Further studies should be performed to determine the cost-benefit of the availability of ASP intervention on a 24 h/day, 7 days/week basis.
There were some differences in baseline characteristics between the cohorts; most notably, the source of bacteremia was more frequently intra-abdominal in the control group than in the ASP group or the BCID group. This may be concerning, as patients with intra-abdominal infections do tend to have more-complicated hospital stays than patients with urinary tract infections. However, accounting for the source of bacteremia, there were no differences in secondary clinical outcomes such as infection-related length of stay, mortality, and readmission rates. Importantly, rates of antimicrobial resistance and severity of illness (as measured by Pitt bacteremia score and intensive care unit [ICU] admission) appeared to be similar between groups. Despite these limitations, our study findings were largely congruent with those of the randomized controlled study performed by Banerjee et al. using the same rapid blood culture identification system.
In conclusion, in patients with bloodstream infections, antimicrobial stewardship intervention was associated with improved antimicrobial utilization. The addition of rapid organism identification and antimicrobial resistance marker detection to an established ASP shortened the time to effective therapy and further improved antimicrobial use compared to ASP alone, even in a setting of low rates of antimicrobial resistance.
Supplementary Material
ACKNOWLEDGMENTS
We thank the staff of the Medical University of South Carolina Clinical Microbiology Laboratory for their assistance. We also thank the South Carolina Clinical and Translational Research Institute Nexus Research Coordination and Management core for their assistance with data collection.
F.S.N. reports having received grant support from BioFire Diagnostics and GenMark Diagnostics and served on scientific advisory boards for BioFire Diagnostics. S.H.M. reports no conflicts of interest.
This work was supported by a grant from BioFire Diagnostics.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.00996-16.
For a commentary on this article, see doi:10.1128/JCM.01484-16.
REFERENCES
- 1.Bearman GM, Wenzel RP. 2005. Bacteremias: a leading cause of death. Arch Med Res 36:646–659. doi: 10.1016/j.arcmed.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 2.Klevens RM, Edwards JR, Gaynes RP. 2008. The impact of antimicrobial-resistant, healthcare-associated infections on mortality in the United States. Clin Infect Dis 47:927–930. doi: 10.1086/591698. [DOI] [PubMed] [Google Scholar]
- 3.Cosgrove SE. 2006. The relationship between antimicrobial resistance and patient outcomes: mortality, length of hospital stay, and health care costs. Clin Infect Dis 42(Suppl 2):S82–S89. doi: 10.1086/499406. [DOI] [PubMed] [Google Scholar]
- 4.Tumbarello M, Sanguinetti M, Montuori E, Trecarichi EM, Posteraro B, Fiori B, Citton R, D'Inzeo T, Fadda G, Cauda R, Spanu T. 2007. Predictors of mortality in patients with bloodstream infections caused by extended spectrum-β-lactamase-producing Enterobacteriaceae: importance of inadequate initial antimicrobial treatment. Antimicrob Agents Chemother 51:1987–1994. doi: 10.1128/AAC.01509-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vernaz N, Hill K, Leggeat S, Nathwani D, Philips G, Bonnabry P, Davey P. 2009. Temporal effects of antibiotic use and Clostridium difficile infections. J Antimicrob Chemother 63:1272–1275. doi: 10.1093/jac/dkp128. [DOI] [PubMed] [Google Scholar]
- 6.Dellit TH, Owens RC, McGowan JE Jr, Gerding DN, Weinstein RA, Burke JP, Huskins WC, Paterson DL, Fishman NO, Carpenter CF, Brennan PJ, Billeter M, Hooton TM. 2007. Infectious Diseases Society of America and the Society for Healthcare Epidemiology of America guidelines for developing an institutional program to enhance antimicrobial stewardship. Clin Infect Dis 44:159–177. doi: 10.1086/510393. [DOI] [PubMed] [Google Scholar]
- 7.Buehler SS, Madison B, Snyder SR, Derzon JH, Cornish NE, Saubolle MA, Weissfeld AS, Weinstein MP, Liebow EB, Wolk DM. 2016. Effectiveness of practices to increase the timeliness of providing targeted therapy for inpatients with bloodstream infections: a laboratory medicine best practices systematic review and meta-analysis. Clin Microbiol Rev 29:59–103. doi: 10.1128/CMR.00053-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pence MA, McElvania TeKippe E, Burnham CA. 2013. Diagnostic assays for identification of microorganisms and antimicrobial resistance determinants directly from positive blood culture broth. Clin Lab Med 33:651–684. doi: 10.1016/j.cll.2013.03.010. [DOI] [PubMed] [Google Scholar]
- 9.Blaschke AJ, Heyrend C, Byington CL, Fisher MA, Barker E, Garrone NF, Thatcher SA, Pavia AT, Barney T, Alger GD, Daly JA, Ririe KM, Ota I, Poritz MA. 2012. Rapid identification of pathogens from positive blood cultures by multiplex polymerase chain reaction using the FilmArray system. Diagn Microbiol Infect Dis 74:349–355. doi: 10.1016/j.diagmicrobio.2012.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buchan BW, Ginocchio CC, Manii R, Cavagnolo R, Pancholi P, Swyers L, Thomson RB Jr, Anderson C, Kaul K, Ledeboer NA. 2013. Multiplex identification of Gram-positive bacteria and resistance determinants directly from positive blood culture broths: evaluation of an automated microarray-based nucleic acid test. PLoS Med 10:e1001478. doi: 10.1371/journal.pmed.1001478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ledeboer NA, Lopranski BK, Dhiman N, Cavagnolo R, Carroll KC, Granato P, Thomson R Jr, Butler-Wu SM, Berger H, Samuel L, Pancholi P, Swyers L, Hansen GT, Tran NK, Polage CR, Thomson KS, Hanson ND, Winegar R, Buchan BW. 2015. Identification of Gram-negative bacteria and genetic resistance determinants from positive blood culture broths by use of the Verigene Gram-negative blood culture multiplex microarray-based molecular assay. J Clin Microbiol 53:2460–2472. doi: 10.1128/JCM.00581-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bauer KA, Perez KK, Forrest GN, Goff DA. 2014. Review of rapid diagnostic tests used by antimicrobial stewardship programs. Clin Infect Dis 59(Suppl 3):S134–S145. doi: 10.1093/cid/ciu547. [DOI] [PubMed] [Google Scholar]
- 13.Banerjee R, Teng CB, Cunningham SA, Ihde SM, Steckelberg JM, Moriarty JP, Shah ND, Mandrekar JN, Patel R. 2015. Randomized trial of rapid multiplex polymerase chain reaction–based blood culture identification and susceptibility testing. Clin Infect Dis 61:1071–1080. doi: 10.1093/cid/civ447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nolte FS, Gullett JC, Youngberg LA, Steed LL. 2014. Clinical evaluation of a rapid multiplex polymerase chain reaction blood culture identification panel. Abstr 54th Intersci Conf Antimicrob Agents Chemother, abstr D-914. [Google Scholar]
- 15.Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. 2009. Research electronic data capture (REDCap) - a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform 42:377–381. doi: 10.1016/j.jbi.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Altun O, Almuhayawi M, Ullberg Ozenci V. 2013. Clinical evaluation of the FilmArray blood culture identification panel in identification of bacteria and yeasts from positive blood culture bottles. J Clin Microbiol 51:4130–4136. doi: 10.1128/JCM.01835-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salimnia H, Fairfax MR, Lephart PR, Schreckenberger P, DesJarlais SM, Johnson JK, Robinson G, Carroll KC, Greer A, Morgan M, Chan R, Loeffelholz M, Valencia-Shelton F, Jenkins S, Schuetz AN, Daly JA, Barney T, Hemmert A, Kanack KJ. 2016. Evaluation of the Filmarray blood culture identification panel: results of a multicenter controlled trial. J Clin Microbiol 54:687–698. doi: 10.1128/JCM.01679-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fredricks DN, Relman DA. 1998. Improved amplification of microbial DNA from blood cultures by removal of the PCR inhibitor sodium polyanetholesulfonate. J Clin Microbiol 36:2810–2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.