Abstract
Background
The filamentous fungus Penicillium oxalicum is a potential alternative to Trichoderma reesei for industrial production of a complete cellulolytic enzyme system for a bio-refinery. Comparative omics approaches can support rational genetic engineering and/or breeding of filamentous fungi with improved cellulase production capacity. In this study, comparative genomic, transcriptomic and secretomic profiling of P. oxalicum HP7-1 and its cellulase and xylanase hyper-producing mutant EU2106 were employed to screen for novel regulators of cellulase and xylanase gene expression.
Results
The 30.62 Mb P. oxalicum HP7-1 genome was sequenced, and 9834 protein-coding genes were annotated. Re-sequencing of the mutant EU2106 genome identified 274 single nucleotide variations and 12 insertion/deletions. Comparative genomic, transcriptomic and secretomic profiling of HP7-1 and EU2106 revealed four candidate regulators of cellulase and xylanase gene expression. Deletion of these candidate genes and measurement of the enzymatic activity of the resultant mutants confirmed the identity of three regulatory genes. POX02484 and POX08522, encoding a putative Zn(II)2Cys6 DNA-binding domain and forkhead protein, respectively, were found to be novel, while PoxClrB is an ortholog of ClrB, a key transcriptional regulator of cellulolytic enzyme gene expression in filamentous fungi. ΔPOX02484 and ΔPOX08522 mutants exhibited significantly reduced β-glucosidase activity, increased carboxymethylcellulose cellulase and xylanase activities, and altered transcription level of cellulase and xylanase genes compared with the parent strain ΔPoxKu70, with Avicel as the sole carbon source.
Conclusions
Two novel genes, POX02484 and POX08522, were found and characterized to regulate the expression of cellulase and xylanase genes in P. oxalicum. These findings are important for engineering filamentous fungi to improve cellulase and xylanase production.
Electronic supplementary material
The online version of this article (doi:10.1186/s13068-016-0616-9) contains supplementary material, which is available to authorized users.
Keywords: Penicillium oxalicum, Genomics, Transcriptomics, Secretomics, Transcription factor, Cellulase, Xylanase, Regulation
Background
Plant cell walls, which primarily consist of cellulose and hemicellulose, are a potential bio-energy source that is an alternative to unsustainable fossil fuels. However, integrated bio-refineries that process lignocellulosic biomass, including agricultural wastes, woody biomass, forestry residues, and grasses, to generate biomaterials such as second-generation biofuels, still face several challenges, including feedstock production and logistics, the development of energy-efficient technologies (pretreatment, enzyme hydrolysis, and microbial fermentation), and societal acceptance [1]. Of these, insufficient amounts and high costs of cellulolytic enzymes hamper the development of bio-refineries. In the biosphere, most organisms do not directly utilize natural cellulose, except for a few cellulose-utilizing microorganisms that are present in soil and the gut of animals [2]. Therefore, to reduce cellulolytic enzyme costs, manipulation of genes regulating the expression of cellulolytic enzymes in genetically amenable microbes has the potential to enhance enzyme production.
Cellulases are produced commercially by filamentous fungi, mainly Trichoderma reesei in recent decades [3]. However, in the native extracellular enzyme system of this organism, secretion of β-glucosidase (BGL, EC 3.2.1.21) is low [4], and cellulase preparations from derivatives of T. reesei must be supplemented with BGL from other sources to improve the efficiency of cellulose hydrolysis [3]. In contrast, the filamentous fungus Penicillium oxalicum secretes a complete cellulase system with a high level of BGL activity [3, 5], and P. oxalicum might be therefore a potential alternative to T. reesei for bioenergy applications [3], although cellulase production must be enhanced if P. oxalicum is to meet the demands of a commercial cellulase source.
Cellulase is a mixture of endo-glucanase (EG, EC 3.2.1.4), cellobiohydrolase (CBH, EC 3.2.1.91), and BGL, that act synergistically with hemicellulases such as endo-β-1,4-xylanases (EC 3.2.1.8) and β-xylosidases (EC 3.2.1.37), along with other enzymes, to hydrolyse cellulose in the plant cell wall into glucose [6]. The expression of genes that encode these plant cell wall-degrading enzymes (CWDEs) is controlled by a complex regulatory system [7].
Several transcription factors involved in cellulase and hemicellulase gene expression have been identified and characterized, including transcriptional repressors CRE1/CreA in T. reesei QM9414 [8] and P. oxalicum 114-2 [9] and Ace1 in T. reesei ALKO2221 [10], as well as activators Clr1 in Neurospora crassa FGSC 2489 [11], Clr2/ClrB in N. crassa FGSC 2489 and P. oxalicum 114-2 [9], Vib1 in N. crassa FGSC 2489 [12], Bgl2 in P. oxalicum 114-2 [13], and XlnR in Aspergillus niger CBS 120.49 [14] and P. oxalicum 114-2 [9]. Of these, Clr2/ClrB, which contains a binuclear zinc cluster, is a key transcriptional activator that is essential for inducing the expression of major cellulases, some major hemicellulases, and mannanolytic enzymes in the presence of plant cell walls (Miscanthus), cellulose, or cellodextrins. Clr2/ClrB is highly conserved in most filamentous ascomycete fungi such as N. crassa, Aspergillus sp., T. reesei and Penicillium sp. [9, 10, 15]. Experimental data showed that manipulating Clr2/ClrB expression in filamentous fungi has great potential for enhancing enzyme production for plant cell wall deconstruction [15]. Very recently, the cellulase yield of a genetically engineered P. oxalicum strain was increased several-fold following induction and/or repression of known transcription factors including ClrB [9, 16]. However, cellulases suitable for use in the industrial-scale bio-refinery of lignocellulosic biomass remain elusive, and the identification and manipulation of additional regulatory genes could be a major step forward in this regard.
In this study, comparative genomic, transcriptomic and secretomic profiling of P. oxalicum HP7-1 and its cellulase and xylanase hyper-producing mutant EU2106 were employed to screen for candidate regulatory genes that regulate cellulase and/or xylanase gene expression. Knockout of candidate transcription factor genes resulted in mutants that were tested for cellulase and xylanase production, and two novel genes regulating the expression of cellulase and/or xylanase genes were identified.
Results
Sequencing of the P. oxalicum HP7-1 genome
Penicillium oxalicum strain HP7-1 was isolated from a decayed forest soil system in China [17]. This strain displayed high cellulase activity [5], particularly towards KOH-pretreated sugarcane bagasse (Fig. 1). The cellulase and xylanase hyper-producing mutant EU2106 was derived from HP7-1 after three rounds of γ-irradiation and two rounds of ethyl methanesulfonate/ultraviolet light mutagenesis [18]. To comprehensively characterize cellulolytic enzymes secreted by EU2106, filter paper cellulase (FPase), Avicelase, KOH-pretreated sugarcane bagasse cellulase (KSBase), carboxymethylcellulose cellulase (CMCase), p-nitrophenyl-β-cellobioside cellulase (pNPCase), p-nitrophenyl-β-glucopyranoside cellulase (pNPGase), and xylanase enzyme activities were measured. The results showed that the secreted FPase activity (2.78 ± 0.16 U/mL) was significantly higher (P ≤ 0.01, Student’s t test) than that of the wild-type HP7-1 (1.79 ± 0.16 U/mL). Similarly, EU2106 possessed higher Avicelase, KSBase, pNPCase and xylanase activities (P ≤ 0.05, Student’s t test; Fig. 1), whereas the CMCase and pNPGase activities of strain EU2106 were similar and lower than those of strain HP7-1, respectively.
Fig. 1.
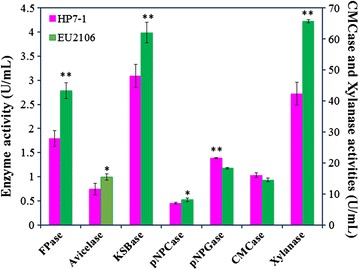
Enzyme activities of FPase, Avicelase, KSBase, CMCase, pNPCase and pNPGase cellulases from P. oxalicum strains HP7-1 and EU2106. The symbols * and ** indicate significant differences (P ≤ 0.05 and P ≤ 0.01, respectively) between the two strains (by Student’s t test)
To further explain the mechanism of enhanced cellulase production in the mutant EU2106, comparative genomics, transcriptomics and secretomics were performed. First, genome sequencing of HP7-1 was conducted using an Illumina HiSeq 2000 system. The genome (accession number JRVD00000000) was assembled using three read libraries, with lengths of 170, 500 and 6000 bp, which were generated by deep sequencing. In total, 30.62 Mbp of nucleotides were generated and covered by 43 scaffolds comprising 263 contigs with 105-fold coverage. The longest contig was 1.18 Mbp, which indicated good assembly continuity. Similar to the genome of P. oxalicum strain 114-2 reported previously [18], 9834 protein-coding genes were annotated in the genome of P. oxalicum strain HP7-1 (Table 1) following curation of genes from four databases (E value <1e−5), namely, the Kyoto Encyclopedia of Genes and Genomes (KEGG), the National Center for Biotechnology Information non-redundant (NCBI NR), UniProt, and Gene Ontology (GO) databases (Additional file 1: Figure S1). In total, 92.3 % of HP7-1 genes shared more than 90 % identity with those in strain 114-2. A comparative analysis of the general features of the P. oxalicum HP7-1 and 114-2 genomes is presented in Table 1.
Table 1.
General genome features of P. oxalicum strains HP7-1 and 114-2
| Genome feature | Value | |
|---|---|---|
| HP7-1 | 114-2 | |
| Size of assembled genome (Mbp) | 30.62 | 30.18 |
| GC content of assembled genome (%) | 50.65 | 50.66 |
| All protein-coding genes | 9834 | 9979 |
| Protein-coding genes ≥60 amino acids | 9602 | 9784 |
| GC content of protein-coding regions (%) | 54.44 | 54.41 |
| Average gene length (bp) | 1621 | 1598 |
| Average number of introns per gene | 1.88 | 1.95 |
| Average intron size (bp) | 103 | 117 |
| Average exon size (bp) | 485 | 464 |
Carbohydrate-active enzymes (CAZymes) and transcription factors may contribute to the ability of strain HP7-1 to hydrolyse plant cell walls [19]. Notably, 477 genes encoding CAZymes and 484 genes encoding predicted transcription factors were annotated in the genome of HP7-1.
The genome of the cellulase and xylanase hyper-producing mutant EU2106 was re-sequenced (accession number SRA399107) and mapped onto the genome of the wild-type strain HP7-1. In total, 1664 Mbp of clean data were generated by constructing a read library with a length of 500 bp using the Illumina HiSeq 2000 system, which covered approximately 30.59 Mbp of the HP7-1 genome with 99.81 % coverage.
A comparative analysis of the HP7-1 and EU2106 genomes revealed 274 single nucleotide variations (SNVs) and 12 insertion/deletions (InDels) (Additional file 2: Table S1) in EU2106, which was far fewer than reported previously for the P. oxalicum mutant JU-A10-T compared with its wild-type strain 114-2 [20]. Of the 274 SNVs, 105, 146 and 23 were located in coding DNA sequences (CDSs), intergenic regions, and introns, respectively (Fig. 2). Of the 12 InDels, 11 are deletions and 1 is an insertion (Fig. 2), but only one deletion is located in a CDS, specifically in the POX01312 gene that encodes a hypothetical protein (according to NCBI BLASTP analysis), resulting in a frameshift mutation.
Fig. 2.
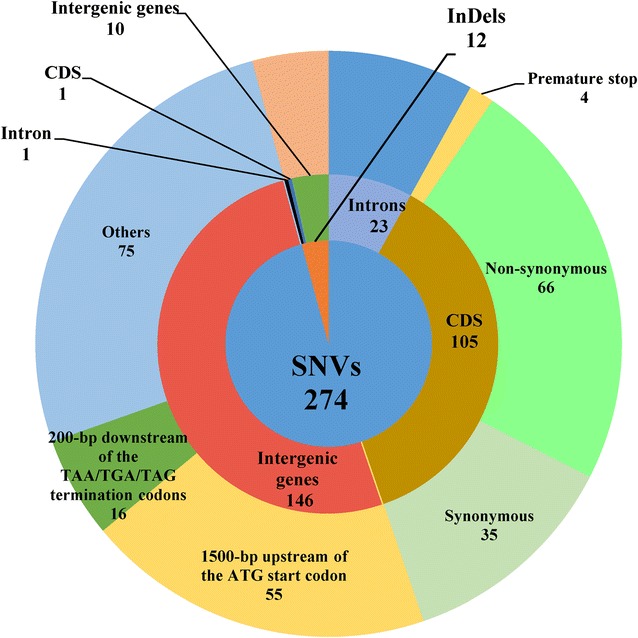
Distribution of single nucleoside variations and InDels in P. oxalicum strain EU2106. CDS coding DNA sequence, InDels insertion/deletions
Comparison of global gene expression in P. oxalicum HP7-1 and EU2106
Penicillium oxalicum HP7-1 and EU2106 were grown in medium containing wheat bran and Avicel as the carbon source (see the “Methods” section for more details) to investigate global gene expression using transcriptome sequencing on an Illumina HiSeq 2000 system. In total, approximately 25–27 million clean reads of 90 bp in length were generated from each sample [accession number GSE80076 in the gene expression omnibus (GEO) database, or SRA399500 for HP7-1 and SRA399511 for EU2106 in the sequence read archive (SRA) database], representing an average 80-fold coverage for the entire HP7-1 genome. Clean reads were mapped to 8441 predicted protein-coding genes in the HP7-1 genome.
Comparative analysis of the available transcriptome data using the R Bioconductor package NOISeq [21] revealed 347 genes that were differentially expressed (|log2 fold change| ≥ 1 and probability ≥ 0.8) in mutant strain EU2106 compared with HP7-1 (grown in medium containing wheat bran and Avicel). Of these, 142 were up-regulated and 205 were down-regulated in EU2106 (Additional file 3: Table S2). After screening the InterPro database [22], approximately 79 % of identified genes were functionally annotated, and the remaining genes were of unknown function and thus could be novel genes involved in cellulase gene expression in P. oxalicum. Among those of unknown function, 17 genes were up-regulated (1.27 ≤ log2 fold change ≤ 5.83), while 56 genes were down-regulated (−12.59 ≤ log2 fold change ≤ −1.28) in EU2106, compared with HP7-1 (Additional file 3: Table S2). In addition, 18 genes (4 up- and 14 down-regulated) included predicted secretion signals, which indirectly indicated that other unknown mechanisms were affected by the mutations.
Transporter genes encoding the major facilitator superfamily (MFS), such as the cellodextrin transporter Cdt-C homolog gene POX06051 [23], together with genes encoding extracellular glycosyl hydrolases, were the most abundant, accounting for 8.1 and 15.9 %, respectively, of all genes that were differentially expressed in EU2106 (Additional file 4: Figure S2A).
Of the up-regulated genes, the transcriptional levels of 10 CAZyme-encoding genes (POX01796, POX01961, POX02490, POX03641, POX05188, POX05260, POX05570, POX05571, POX06079 and POX07971) in EU2106 were 2.38 to 61.93-folds of that in HP7-1. Interestingly, POX05570 and POX05571, POX03641 and POX06079, and POX02490 encode EG, BGL and CBH, respectively (Additional file 4: Figure S2B). In contrast, the expression of POX07535 and POX06835 that encode an EG and a BGL, respectively [19], and which are abundantly secreted by P. oxalicum, were significantly down-regulated in EU2106 (log2 fold change = −3.85 and −1.59, respectively) compared with HP7-1, and the xylanase-encoding gene POX05916 was also down-regulated (log2 fold change = −2.54) in the mutant strain (Additional file 4: Figure S2C). Of note, a reduction in BGL activity in P. oxalicum might stimulate cellulase activity by increasing cellobiose concentration, which is an inducer of the cellulase-induction pathway, which could explain the enhanced cellulase activity in the EU2106 mutant strain.
Also of note, eight major MFS members (POX01812, POX01981, POX05155, POX06051, POX06283, POX06915, POX07416, and POX09632) were up-regulated at least two-fold in EU2106 compared with HP7-1. Among them, POX06051 encodes an ortholog of the cellodextrin transporter Cdt-C (log2 fold change = 1.47). This could also contribute to the enhanced cellulase activity of the mutant strain, since over-expression of cdt-c in P. oxalicum 114-2 enhanced cellobiohydrolase production by stimulating the expression of cellobiohydrolase-encoding gene Cel7A-2 [23]. Expression of two predicted Zn(II)2Cys6 domain-containing transcription factors POX02484 and POX03873 was down-regulated approximately 3.5-fold in EU2106 compared with HP7-1, whereas POX07938, that also encodes a protein with a Zn(II)2Cys6 domain, was up-regulated 14.8-fold (Additional file 3: Table S2).
Comparative analysis of the secretomes of P. oxalicum HP7-1 and EU2106
A total of 240 proteins were identified in the secretomes of HP7-1 and EU2106 (accession number PXD004046) using isobaric tags for relative and absolute protein quantitation, which increases our understanding of the complete enzyme set secreted by P. oxalicum. According to KEGG annotation, these proteins are mainly involved in carbohydrate metabolism (Additional file 5: Figure S3). Approximately 55 % of these proteins lack a predicted secretion signal (Additional file 6: Table S3), which is comparable to the secretome of T. asperellum S4F8 and T. reesei Rut C30 [24]. This could indicate possible non-classic secretory mechanisms or protein leakage due to occasional cell autolysis or autophagy.
Many of the 240 proteins involved in the hydrolysis of plant cell wall polysaccharides were elevated in EU2106 compared with HP7-1. Of particular note, three CBHs (POX02490, POX04786, and POX05587), eight EGs (POX01166, POX01896, POX02740, POX04137, POX05571, POX06147, POX06983, and POX07535), two BGLs (POX003641 and POX06079), and five xylanases (POX00063, POX05916, POX06601, POX08484, and POX08990) were up-regulated (Additional file 6: Table S3), consistent with some of the transcriptome data described above.
Screening of candidate regulators of cellulase and/or xylanase gene expression
To identify novel genes regulating cellulase and/or xylanase gene expression, all 274 SNVs in EU2106 were analyzed in detail. Non-synonymous SNVs in CDSs that result in amino acid substitutions in the translated protein could potentially affect protein function [25], while SNVs that occur in the promoters and terminators of protein-coding genes could influence their transcriptional levels. Among the 70 non-synonymous SNVs, none were located in sequences encoding CWDEs, except for POX00008, which encodes an exo-α-l-1,5-arabinanase. The four SNVs that occurred in POX02087, POX03561, POX03683, and POX07858 would halt translation and possibly affect the functions of their encoded proteins, which are putatively associated with DNA repair and protein modification. Notably, three SNVs were located in the CDSs of PoxClrB (POX01960), POX03199 and POX08522, that encode two predicted transcription factors and a homolog of the ClrB transcription factor that positively regulates cellulase and/or hemicellulase gene expression in P. oxalicum 114-2 in the presence of various carbon sources [9].
In contrast, 53 and 10 SNVs were found within 1500 and 200 bp of ATG start codons and termination codons, respectively. KEGG annotation indicated that most genes containing SNVs were associated with genetic information processing and metabolism (Additional file 2: Table S1).
Transcriptional of all genes containing SNVs in both HP7-1 and the cellulase and xylanase hyper-producing EU2106 mutant was measured in media containing wheat bran and Avicel as the carbon source. In total, eight genes containing SNVs were altered more than twofold in EU2106 compared with HP7-1 (five up- and three down-regulated, respectively; Additional file 3: Table S2). Of these eight genes, five (POX00301, POX06751, POX06820, POX07291, and POX09827) are particularly relevant since the SNVs are located in CDS, promoter, or terminator regions of open reading frames.
Together, these results identified 11 potentially important genes, including five (POX00301, POX06751, POX06820, POX07291, and POX09827) in which SNVs were located in CDSs or within 1500 or 200 bp of ATG start codons or termination codons, respectively, and that were differentially expressed (|log2 fold change| ≥ 1) in the mutant strain. The other six potentially important genes comprised three transcription factor-encoding genes (PoxClrB, POX03199, and POX08522) with SNVs in their CDS regions, and three genes (POX02484, POX03873 and POX07938) with significantly altered expression (|log2 fold change| ≥ 1 and probability ≥ 0.8) in EU2106. Detailed analysis using the NCBI protein basic local alignment search tool (BLASTP) indicated that four proteins (PoxClrB, POX02484, POX07291 and POX08522) were worthy candidates for further characterization of their regulatory functions in cellulase and xylanase gene expression (Additional file 7: Table S4).
Construction and characterization of deletion mutants for four candidate cellulase and xylanase regulators
To comprehensively investigate gene function in filamentous fungi, a highly efficient gene targeting system that allows rapid genetic manipulation is needed, and can be achieved by deleting genes involved in the non-homologous end-joining pathway such as Ku70 and Ku80 [26]. In the present study, the double-stranded break repair complex subunit-encoding gene POX01582, which is homologous to Ku70, was identified and subsequently knocked out in P. oxalicum strain HP7-1 as described previously [26]. The resultant mutant ΔPoxKu70 displayed no significant defects in phenotypes such as vegetative growth and conidiation or cellulase/xylanase activities, and sensitivity to hygromycin B, G418, and ultraviolet light was comparable with HP7-1 (Additional file 8: Figure S4), suggesting ΔPoxKu70 was suitable for the construction of deletion mutants.
The four gene candidates (PoxClrB, POX02484, POX07291, and POX08522) were individually knocked out in the ΔPoxKu70 strain by homologous recombination, and their roles in regulating cellulase and xylanase activities were investigated by measuring FPase, CMCase, pNPCase, pNPGase, and xylanase enzyme activity. All deletion mutants were verified by PCR with gene-specific primers and by Southern hybridization analysis (Additional file 9: Figure S5). A crude enzyme solution from each strain was collected after 6 days of cultivation in medium containing Avicel as the sole carbon source. As expected, all enzyme activities were significantly decreased (by 62.18 ± 4.60 to 94.51 ± 1.36 %; P ≤ 0.01, Student’s t test) in the ΔPoxClrB mutant, compared with the ΔPoxKu70 parent strain, except for pNPGase activity, which was increased by 134.55 ± 26.39 %. This result is consistent with previously findings [9], and confirmed that PoxClrB is a global transcription factor that positively regulates cellulolytic enzyme production. Similarly, FPase, CMCase, pNPCase, and pNPGase enzyme activities were decreased by 36.42 ± 5.58, 17.74 ± 2.96, 35.91 ± 6.61 and 76.64 ± 5.23 %, respectively, (P ≤ 0.01, Student’s t test) in the ΔPOX02484 mutant (Additional file 10: Figure S6). The ΔPOX08522 mutant lost 85.85 ± 1.22 % (P ≤ 0.01, Student’s t test) of its pNPGase activity, whereas xylanase activity was increased by 17.38 ± 1.93 % (P ≤ 0.01, Student’s t test) in this mutant strain. In contrast, there were no significant differences in enzyme activity between the ΔPOX07291 mutant and ΔPoxKu70 (data not shown). In addition, the yield of secreted protein was decreased by 33.50 ± 4.95 and 62.18 ± 4.60 % respectively in the ΔPOX02484 and ΔPoxClrB mutants (P ≤ 0.01, Student’s t test), compared with ΔPoxKu70 (Additional file 10: Figure S6).
Together, these preliminary results indicated that POX02484 and POX08522 are novel transcriptional factors that are potentially involved in cellulase and/or xylanase production in P. oxalicum. ΔPOX02484 and ΔPOX08522 mutants, and the ΔPoxClrB strain in which the known regulator PoxClrB was deleted, were therefore selected for further investigation.
Cell growth and enzyme production in ΔPoxClrB, ΔPOX02484 and ΔPOX08522 under Avicel induction
To determine whether the candidate regulators were involved in cellulase and xylanase production under Avicel induction conditions, we investigated the growth of ΔPoxClrB, ΔPOX02484 and ΔPOX08522 mutants, along with the ΔPoxKu70 parent strain, in glucose and Avicel. As shown in Fig. 3a, the growth of all three mutants showed no significant differences from the parent strain in medium containing glucose as the sole carbon source, suggesting these genes are not involved in fungal basic metabolism. In contrast, ΔPOX02484 and ΔPOX08522 grew significantly faster before 12 h with Avicel as the sole carbon source than did mutant ΔPoxClrB or the ΔPoxKu70 parent strain. Furthermore, after 24 h of cultivation, the growth of ΔPoxKu70 was fastest, followed by ΔPOX08522 and ΔPOX02484, and the ΔPoxClrB grew at slowest rate (Fig. 3b).
Fig. 3.
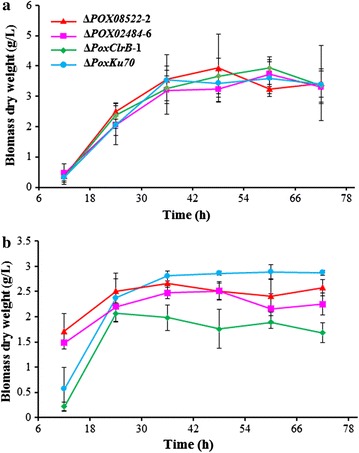
Growth profiles of P. oxalicum grown on glucose (a) and Avicel (b). The biomass dry weight per litre was directly measured gravimetrically for glucose or by calculating from the intracellular protein content for Avicel on the basis of 0.28 g intracellular proteins per gram dry biomass. Data are presented as the mean of three biological replicates
Enzyme production was then tested after 3 and 5 days of induction by Avicel. The activity of all tested enzymes was reduced to below 14 % that of ΔPoxKu70 in mutant ΔPoxClrB after 3 days, except for pNPGase activity, which was 526 % higher than that of ΔPoxKu70 (P ≤ 0.01, Student’s t test; Fig. 4), consistent with enzyme production in Avicel after 6 days as described above. Interestingly, in the ΔPOX08522 mutant, only pNPGase activity was significantly decreased, by 71.82 ± 3.62 % (P ≤ 0.01, Student’s t test), whereas CMCase and xylanase activities were increased by 17.31 ± 2.98 % (P ≤ 0.05, Student’s t test) and 80.76 ± 10.88 % (P ≤ 0.01, Student’s t test), compared with ΔPoxKu70. Similar to the ΔPOX08522 mutant, ΔPOX02484 also showed significantly decreased pNPGase activity, while CMCase, pNPCase and xylanase activities were increased to varying extents, and xylanase activity was particularly increased, by 94.63 ± 16.83 % (Fig. 4).
Fig. 4.
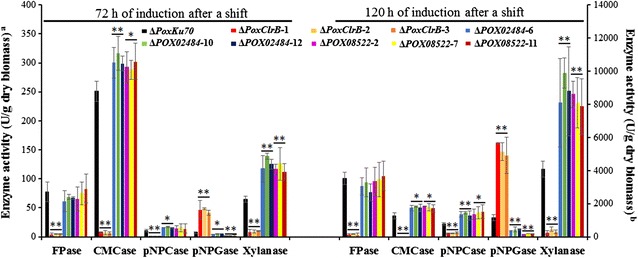
Activities of crude enzymes from PoxClrB, POX02484 and POX08522 deletion mutants following a shift from glucose to Avicel. Crude enzymes were produced by fungal strains grown in 2.0 % Avicel as the sole carbon source. Enzymatic activity was measured at 3 and 5 days after the shift. The symbols * and ** indicate significant differences (P ≤ 0.05 and P ≤ 0.01, respectively) between the candidate mutants and the parent ΔPoxKu70 strain, as assessed by Student’s t test. a FPase, pNPCase, pNPGase activities and CMCase activity from 72 h of induction; b Xylanase activity and CMCase activity from 120 h of induction
After 5 days of Avicel induction, the trends in enzyme activities in all three mutant strains were in agreement with those observed after 3 days, although the variation was larger (Fig. 4).
POX02484 and POX08522 regulate the expression of cellulase and xylanase genes
To directly elucidate transcriptional regulatory roles of these candidate transcription factors, real-time quantitative reverse transcription-PCR (qRT-PCR) was used to measure the expression of their target genes at 4, 12 and 24 h after a shift from glucose to Avicel. Target genes were chosen based on the secretome data described above, and included three cellobiohydrolase genes (cbh), eight endo-glucanase genes (eg), three β-glucosidase genes (bgl), and four xylanase genes (xyn) (Fig. 5). The results showed that in the ΔPOX02484 mutant, expression of a number of cellulase and xylanase genes was down-regulated at 4 h. Specifically, two major cbh genes (POX05587/Cel7A-2 and POX04786/Cel6A), five eg genes (POX06147/Cel5A, POX01166/Cel5B, POX01896/Cel5C, POX05571/Cel7B, and POX02740, P ≤ 0.01, Student’s t test), three bgl genes (POX07963, POX08882, P ≤ 0.01, Student’s t test; POX06835/Bgl3A, P ≤ 0.05, Student’s t test), and one xyn gene (POX05916, P ≤ 0.01, Student’s t test) were significantly down-regulated by 34.30–87.29 %, compared with ΔPoxKu70. Conversely, cbh gene POX02490/Cel7A-1 (P ≤ 0.01, Student’s t test), three eg genes (POX07535/Cel12A, POX04137, P ≤ 0.01, Student’s t test; POX06983, P ≤ 0.05, Student’s t test), and three xyn genes (POX00063/xyn10A, POX06783/xyn11A and POX08484/xyn11B, P ≤ 0.01, Student’s t test) were significantly up-regulated from 34.01 to 236.31 % in the ΔPOX02484 mutant (Fig. 5a). Notably, the expression levels of cellulase and xylanase genes at 12 h were significantly different from those at 4 h. For example, two cbh genes (POX05587/Cel7A-2 and POX04786/Cel6A), four eg genes (POX06147/Cel5A, POX01896/Cel5C, POX07535/Cel12A and POX04137), and bgl gene POX08882 in ΔPOX02484 were comparable to ΔPoxKu70, whereas three eg genes (POX01166/Cel5B, POX05571/Cel7B, P ≤ 0.01, Student’s t test) and POX02740 (P ≤ 0.05, Student’s t test), two bgl genes (POX06835/Bgl3A, P ≤ 0.05, Student’s t test; POX07963, P ≤ 0.01, Student’s t test), and xyn gene POX05916 (P ≤ 0.01, Student’s t test) were significantly up-regulated compared with ΔPoxKu70. At 24 h, most cellulase and xylanase genes were expressed at levels comparable with those measured at 12 h (Fig. 5a).
Fig. 5.
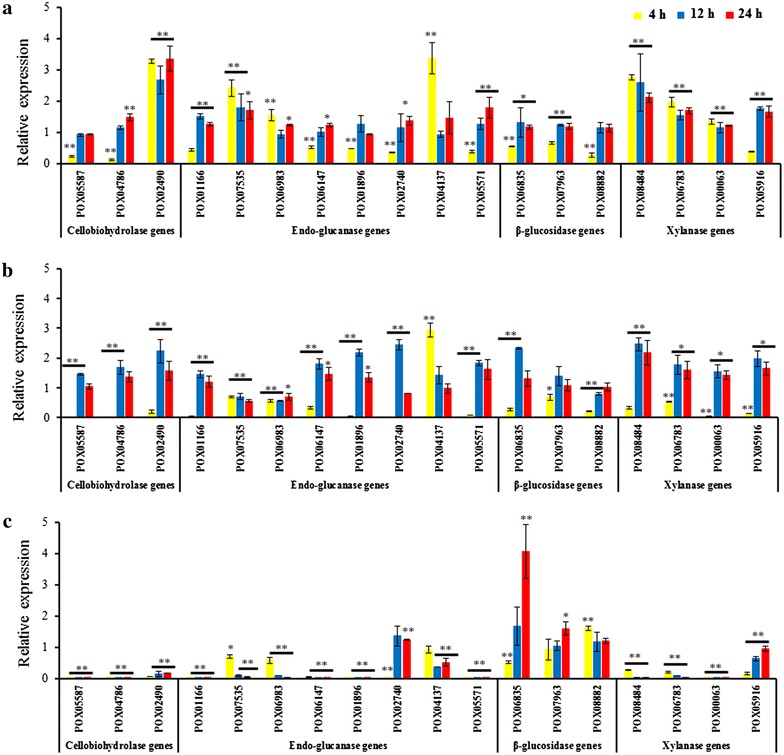
Regulation of gene expression by PoxClrB, POX02484, and POX08522. Under induction by the same carbon source, the expression levels of genes encoding three cellobiohydrolases, eight endo-glucanases, three β-glucosidases and four xylanases were determined in the ΔPOX02484-6 (a), ΔPOX08522-2 (b) and ΔPoxClrB-1 (c) mutants at three different time points after the shift (4, 12 and 24 h) by qRT-PCR. Expression levels were normalized against the ΔPoxKu70 parent strain. The symbols * and ** indicate significant differences (P ≤ 0.05 and P ≤ 0.01, respectively) between the tested samples and the ΔPoxKu70 strain, as assessed by Student’s t test
Like POX02484, another regulator POX08522 also positively regulated all tested cellulase and xylanase genes at 4 h (by 31.31–98.43 %), except for eg gene POX04137 that was increased by 193.67 ± 29.75 % (P ≤ 0.01, Student’s t test) compared with ΔPoxKu70 at 4 h (Fig. 5b). In contrast, most cellulase and xylanase genes were significantly up-regulated at 12 h, including three cbh genes (POX05587/Cel7A-2, POX02490/Cel7A-1 and POX04786/Cel6A), five eg genes (POX06147/Cel5A, POX01166/Cel5B, POX01896/Cel5C, POX05571/Cel7B, and POX02740), one bgl gene (POX06835/Bgl3A), and four xyn genes (POX00063/Xyn10A, POX06783/Xyn11A, POX08484/Xyn11B and POX05916; Fig. 5b) compared with ΔPoxKu70 at 12 h. Again, expression levels of most of cellulase and xylanase genes at 24 h were comparable to those observed at 12 h (Fig. 5b).
Similar to previously reported data [9], transcription of all tested cbh, eg and xyn genes at 4, 12 and 24 h was down-regulated in the ΔPoxClrB mutant compared with ΔPoxKu70 (P ≤ 0.01, Student’s t test; Fig. 5c), except for eg genes POX02740 at 12 and 24 h and POX04137 at 4 h, and xyn gene POX05916 at 24 h. Interestingly, transcription of all three bgl genes varied significantly with induction time. POX06835/Bgl3A expression in the ΔPoxClrB mutant was decreased by 48.11 ± 3.59 % at 4 h, but increased to levels comparable with ΔPoxKu70 at 12 h, and continued to increase to levels that were 406.41 ± 86.43 % higher than those of the ΔPoxKu70 strain at 24 h. In comparison, expression levels of POX07963 and POX08882 in the ΔPoxClrB mutant were 159.38 ± 20.87 and 161.45 ± 67.32 % higher than in ΔPoxKu70 at 24 and 4 h, respectively (Fig. 5c).
Sequence analysis of POX02484 and POX08522
The CDSs of POX02484 and POX08522 (accession numbers KU597419 and KU597417) were found to be 2067 and 2151 bp, respectively, encoding proteins of 688 and 716 aa, respectively. The POX02484 and POX08522 protein sequences were used as queries in a BLASTP search of the NCBI database (http://www.ncbi.nlm.nih.gov/), and phylogenetically analyzed.
POX02484 was found to contain a GAL4-like Zn2Cys6 binuclear cluster DNA-binding domain (cd00067, E value = 5.80e−11) and a conserved fungi-specific transcription factor domain (pfam11951, 1.14e−77). POX02484 shares 69, 60, 45 and 43 % identity with the C6 transcription factor AFUA_4G09710 from A. fumigatus AF293 (XP_751855.2), PDE_05453 from P. oxalicum 114-2 (EPS30502.1), transcriptional regulatory protein NCU07392 from N. crassa (EAA29240.3), and TRIREDRAFT_76590 from T. reesei QM6a (XP_006963971.1), respectively. Phylogenetic tree analysis clustered POX02484 homologs in Penicillium with homologs in Aspergillus (Fig. 6a).
Fig. 6.
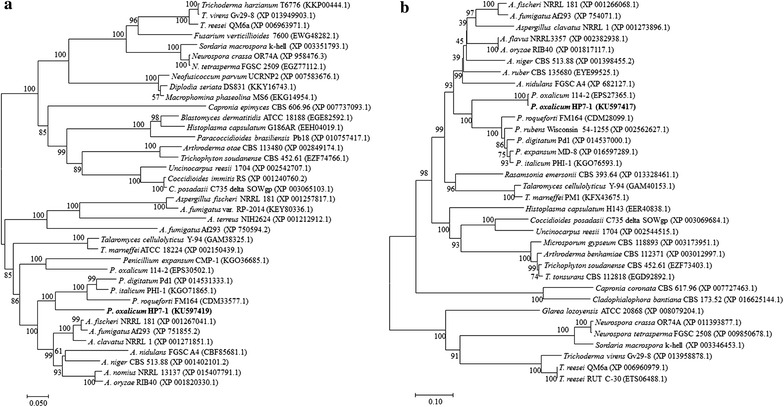
Unrooted phylogenetic analysis of POX02484 (a) and POX08522 (b) and putative homologs. The dendrogram was constructed using MEGA 7.0 software with the neighbour-joining method and Poisson model. Bootstrap values shown at the nodes are derived from 1000 replicates, and branch lengths corresponding to the divergence of sequences are indicated by the scale bar
POX08522 contains a forkhead box (cd00059, E value 1.15e−44) between aa 195 to 271. This protein shares 99 % identity with PDE_02308 (EPS27365.1) from P. oxalicum 114-2, and 69 and 61 % identity with Pc20g00650 from P. rubens Wisconsin 54-1255 (XP_002562627.1) and AOR_1_1186184 of A. oryzae RIB40 (XP_001817117), respectively, and a somewhat lower identity (37.35 and 43 %) with NCU06173 of N. crassa (ESA43188.1) and M419DRAFT_67752 of T. reesei Rut C-30 (ETS06488.1). As with POX02484, phylogenetic tree analysis clustered POX08522 homologs in Penicillium with their counterparts in Aspergillus (Fig. 6b).
Discussion
In this study, we performed a comparative omics analysis combining genomics, transcriptomics and secretomics of P. oxalicum strain HP7-1 and its cellulase and xylanase hyper-producing mutant EU2106. This microorganism has potential for application in the bio-refinery of lignocellulosic biomass [18, 27, 28]. Unsurprisingly, there were no mutations in the catalytic residues of the major cellulase and hemicellulase enzymes in EU2016. However, expression of these enzymes was significantly up-regulated in the mutant strain, as previously observed in P. oxalicum JU-A10-T and T. reesei cellulase over-producing mutants [20, 29], although fewer mutations in genes encoding transcription factors were found in P. oxalicum EU2106 than in JU-A10-T [20]. Functional studies on P. oxalicum in this work demonstrated that some of the mutated transcription factors were involved in cellulase gene expression. In particular, the master transcription factor PoxClrB included a SNV in its CDS, which changed S725 to N, and transcription factor POX08522 underwent a SNV in its CDS that changed H612 to Y. However, comparative transcriptomics showed that these two transcription factors were expressed equally in the EU2106 mutant and wild-type strain HP7-1. Therefore, whether these alterations affect protein function requires further investigation.
In addition, the transcription factor POX02484 was found to be down-regulated (log2 fold change = −1.82) in mutant EU2106, which resulted in an increase in the expression of major cellulase and xylanase genes at 12 and 24 h after carbon source induction, which could partially explain the enhanced cellulase activity in the mutant strain. However, the detailed molecular mechanism underlying the cellulase hyper-producing activity of EU2106 remains unknown.
Phylogenetic analysis indicated that homologs of POX02484 and POX08522 exist in a wide variety of filamentous fungi, including Neosartorya spp., Penicillium spp., Aspergillus spp. and N. crassa. Interestingly, POX02484 contains a GAL4-like binuclear Zn2Cys6 cluster DNA-binding domain, which is found in many transcription factors and plays a role in transcriptional regulation in a wide range of processes, including carbon and nitrogen metabolism, amino acid and vitamin synthesis, and morphogenesis, in combination with a regulation domain such as Fungal_trans_2 (pfam11951) and Fungal_TF_MHR (cd12148) [28]. POX08522 harbours a forkhead domain, which can bind nucleosomal DNA and participate in a wide variety of cellular processes, including cell cycle regulation, control of cell death, pre-mRNA processing, and morphogenesis, in both yeast and filamentous fungi, as well as mammals [30].
Multiple alignment of homologs revealed that POX02484 shares 45–69 % identity with Pro1 in N. crassa OR74A (NCU07392; XP_958476.3) and Sordaria macrospora K-hell (SMAC_00338; XP_003351793.1), and NosA (number of sexual spores) in A. fumigatus AF293 (AFUA_4G09710; XP_751855.2) and A. nidulans FGSC A4 (ANIA_01848; CBF85681.1). Transcription factor Pro1 from S. macrospora is a developmental regulator that is required for fruiting body development and preventing the transition of primordia into mature fruiting bodies [31]. NosA from Aspergillus spp., shares ~45 % identity with S. macrospora Pro1, and is involved in the control of sexual development. Deletion of NosA impaired the development of sexual primordia, drastically reduced sexual spore production during self-mating, and increased vegetative growth and germination rates by regulating the expression of several genes associated with sexual development, including the glucose transporter hxtA and the catalase peroxidase cpeA [32, 33]. Furthermore, NosA function is conserved in other Aspergillus species, and when constitutively expressed during the life cycle of A. nidulans, it was up-regulated during late asexual development following carbon starvation [33].
In addition, multiple alignment of homologs also revealed that POX08522 shares 49 % identity with transcription factor Sep1 in Schizosaccharomyces pombe 972 h (SPBC4C3.12; NP_596301.1). Sep1 as a transcription activator that controls mitosis in S. pombe through the binding and activation of a small subset of mitotic genes involved in septation [34].
Although there are mechanistic differences between Pro1/NosA and Sep1, deletion of either results in an increase in vegetative growth [32, 35]. Here, deletion of POX02484 and POX08522 in P. oxalicum HP7-1 increased growth during the early stages of development under carbon starvation, but growth was slightly slower than the wide-type during the later stages. Notably, vegetative growth was similar between mutants ΔPOX02484 and ΔPOX08522 and the parent ΔPoxKu70 in the present study when grown on glucose, suggesting POX02484 and POX08522 act through a mechanism that is different from their homologs in other species.
This is the first study to report the involvement of POX02484 and POX08522 or their homologs in cellulase and xylanase production under cellulosic induction, and the first to conclude that these proteins act as regulators of cellulase and xylanase gene expression in filamentous fungi. Enzymatic activity assays under insoluble cellulose induction indicated that both POX02484 and POX08522 regulated BGL activity, as did PoxClrB. BGL activity is generally considered rate-limiting and can act as a bottleneck that limits the efficient enzymatic degradation of cellulose [4]. Deletion of either POX02484 or POX08522 increased the activity of all cellulases tested except for pNPGase, which is a measure of the proportion of BGL activity responsible for the degradation of cellobiose, a major inducer of cellulase gene expression. In the presence of a strong inducer, cellulase gene expression is up-regulated resulting in very high levels of cellulase activity. This is reminiscent of intracellular β-glucosidase Bgl2 in P. oxalicum 114-2, since production of all extracellular cellulolytic enzymes including CMCase, pNPCase, pNPGase and xylanases were up-regulated in the ΔBgl2 strain when grown on insoluble cellulose [13]. Conversely, all cellulase activities in the ΔPoxClrB mutant were reduced except for pNPGase, indicating that a low level of intracellular cellobiose could not stimulate cellulase gene expression in this mutant. To our knowledge, this is the first report on the involvement of PoxClrB on BGL activity. As expected, all three regulators regulated BGL gene expression under induction conditions, to varying degrees. Cellobiose is reported to act as an adaptive BGL inducer rather than a cellulase inducer [36], and may therefore perform a more specific regulatory role in P. oxalicum than the other transcription factors identified in this study.
In the comparative secretome analysis, more proteins were found to be associated with secondary metabolism pathways in the secretome of strain HP7-1, particularly carbohydrate metabolism, amino acid metabolism, xenobiotics biodegradation and metabolism, and energy metabolism, compared with the 105 proteins in the secretomes of both P. oxalicum 114-2 and JA-10-T [20]. Specifically, 42 additional CAZymes and one extra MFS were detected, including EGs POX04137 and POX06983, polysaccharide monooxygenases POX05968 and POX08897, expansin-like protein POX01524, and cellodextrin transporter POX06051, indicating a complex synergism between enzymes involved in plant cell wall degradation. Surprisingly, POX03641 and POX06079, previously defined as intracellular BGLs [13], were found in the secretomes of P. oxalicum strains HP7-1 and EU2106, which may merit further investigation.
In P. oxalicum strain 114-2, Li et al. [9] recently reported that 20 transcription factors were involved in the regulation of cellulase and/or hemicellulase gene expression by constructing a single-gene disruptive mutant library of its putative transcription factors. Hydrolysis zones on plates containing cellulose produced by the constructed mutants were used for screening. In this study, we directly tested the activities of five enzymes (FPase, CMCase, pNPCase, pNPGase and xylanase) in crude enzyme preparations from each mutant cultivated in the presence of Avicel as the sole carbon source. Through this approach, we identified two novel regulatory genes (POX02484 and POX08522) that were not listed among the regulatory genes reported by Li et al. [9]. The novel transcription factors POX02484 and POX08522 extend the list of fungal gene regulators known to regulate cellulase and xylanase gene expression. These findings may be useful for the genetic engineering of filamentous fungi to improve cellulase and xylanase activities for use in bio-refinery applications.
Conclusions
In this study, comparative genomic, transcriptomic and secretomic profiling of Penicillium oxalicum HP7-1 and its cellulase and xylanase hyper-producing mutant EU2106 were employed to screen for novel regulators of cellulase and xylanase gene expression. Two novel genes (POX02484 and POX08522) were discovered and characterized to regulate the expression of cellulase and xylanase genes. These findings may prove important for engineering filamentous fungi to improve cellulase and xylanase production.
Methods
Strains and growth conditions
P. oxalicum strain HP7-1 (China General Microbiological Culture Collection (CGMCC) 10781) was isolated from a subtropical forest soil system in Guangxi Zhuang Autonomous Region, China [17], and its cellulase hyper-producing mutant, EU2106, (CGMCC 6471) was obtained by multiple rounds of mutagenesis [18]. The strains were maintained on potato-dextrose-agar (PDA) plates at 4 °C. In this study, the constructed deletion mutants ΔPoxKu70, ΔPoxClrB, ΔPOX02484 and ΔPOX08522 were respectively deposited in CGMCC with numbers of 3.15650, 3.15649, 3.15647 and 3.15648, respectively. Penicillium oxalicum strains were cultivated on PDA plates at 28 °C for 6 days to complete sporulation.
For gene expression assays for comparative transcriptomics, a spore suspension of 1 mL containing approximately 1.0 × 108 spores per mL was inoculated into 100 mL of modified minimal medium (MMM) containing (g/L) KH2PO4 (4.0), (NH4)2SO4 (4.0), MgSO4·7H2O (0.60), CaCl2 (0.6), FeSO4·7H2O (0.0050), MnSO4 (0.001600), ZnCl2 (0.0017), CoCl2 (0.002), 1 mL of Tween, and Avicel and wheat bran that were added to a final concentration of 1 % (w/v) and 4 % (w/v), respectively, as the carbon source. Cultures were incubated at 28 °C with shaking at 180 rpm for 72 h. Mycelia were separated from the culture by an eight-layer filter fabric and washed three times using diethyl pyrocarbonate-treated water prior to RNA extraction.
For gene deletion mutants, Avicel at a final concentration of 2 % (w/v) in 100 mL of MMM was used as sole carbon source. The inoculated medium was incubated at 28 °C with shaking at 180 rpm for 6 days. The supernatant was collected after centrifugation at 11,300g for 10 min, and filtered through a Whatman No. 1 filter paper for enzyme activity and secretome analyses, while the mycelia were collected for DNA extraction.
A shift experiment was performed for qRT-PCR analysis of gene expression and measurement of enzymatic activity in P. oxalicum. Cultures containing approximately 1.0 × 108 spores per mL were pre-grown for 20 h in MMM containing 1 % (w/v) glucose at 28 °C with shaking at 180 rpm. Then, equal portions of the harvested and washed mycelial sample were aseptically re-placed into MMM containing 2 % Avicel as the sole carbon source. After cultivation at 28 °C with shaking at 180 rpm for 4, 12 and 24 h, mycelia were harvested for qRT-PCR assay, while supernatants were collected at 3 and 5 days for enzymatic activity assays.
DNA and RNA extraction
Total DNA was extracted from mycelia using the modified phenol-chloroform method [37]. Briefly, after washing with sterile water, mycelia were immediately ground in liquid nitrogen, and 1 mL of lysate reagent [40 mM Tris-HCl, 20 mM sodium acetate, 10 mM ethylenediaminetetraacetic acid (EDTA), and 1 % sodium dodecyl sulfate, pH 8.0] was added per 100 mg of powder. Whole genomic DNA was collected after precipitation via centrifugation at 11,300g for 10 min.
Total RNA was extracted from mycelia using the TRIzol RNA Kit (Life Technologies, Carlsbad, CA, USA) according to the manufacturer’s instructions. The concentration and integrity of the extracted RNA were determined by measuring the absorbance at 260 and 280 nm (the A260/A280 ratio) by spectrophotometer and electrophoresis on 1 % agarose gels, respectively.
Genome sequencing, assembly, and gene prediction and annotation
The genome of P. oxalicum strain HP7-1 was sequenced using an Illumina HiSeq 2000 system (Illumina, San Diego, CA, USA) at the Beijing Genomics Institute (Shenzhen, China). Genomic DNA was sheared randomly to construct three read libraries with lengths of 170, 500 and 6000 bp by a Bioruptor ultrasonicator (Diagenode, Denville, NJ, USA) and physico-chemical methods. The paired-end fragment libraries were sequenced according to the Illumina HiSeq 2000 system’s protocol. The sequenced reads were assembled using SOAPdenovo v1.05 software (http://soap.genomics.org.cn/soapdenovo.html), while Augustus v2.6.1 (http://bioinf.uni-greifswald.de/augustus/), GeneMark v2.3e (http://exon.gatech.edu/) and GeneWise v2.20 (http://www.sanger.ac.uk/Software/Wise2/) were used for gene prediction. The predicted genes were functionally annotated by homologous alignment using the GO, KEGG, UniProt, Cluster of Orthologous Groups of proteins (COG), and NCBI NR databases, using the BLAST algorithm (http://www.ncbi.nlm.nih.gov/blast/executables/blast+/LATEST/). Transfer RNA and rRNA were detected using RNAmmer v1.2 (http://www.cbs.dtu.dk/services/RNAmmer/) and tRNAscan-SE v1.23 (http://gtrnadb.ucsc.edu/) software, respectively. Secreted proteins were predicted by both SignalP v3.0 (http://www.cbs.dtu.dk/services/SignalP/) and TargetP v1.1 (http://www.cbs.dtu.dk/services/TargetP/). Transcription factors were annotated according to their InterPro IDs in the Fungal Transcription Factor Database [38]. Transporters were identified and classified by BLASTP searches of the TransportDB database (http://www.membranetransport.org) with an E value threshold of 1e−5. CAZymes were predicted by searching the dbCAN database (http://csbl.bmb.uga.edu/dbCAN/), with an E value threshold of 1e−7, F3 = 1e−10, F2 = 1e−7, and F1 = 0.001.
Transcription profiling data assays
Samples for transcriptional profiling were collected after 72 h of induction in the presence of wheat bran and Avicel. Three biological replicates of each sample were analyzed. A cDNA library was constructed, tested by an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) and an ABI StepOnePlus Real-Time PCR System (Applied Biosystems, Foster City, CA, USA), and subsequently sequenced using the Illumina HiSeq 2000 system. The generated clean reads were mapped to the P. oxalicum HP7-1 genome using BWA v0.7.10-r789 (http://sourceforge.net/projects/bio-bwa/files/) and Bowtie2 v2.1.0 software [39]. The gene expression level (fragments per kilobase of exon per million mapped reads) was analyzed with RSEM v1.2.12 software [40]. Differentially expressed genes were detected using the NOISeq tool (http://www.bioconductor.org/packages/release/bioc/html/NOISeq.html) (log2 fold change| ≥ 1, P ≤ 0.01 and probability ≥ 0.8 as thresholds). Blast2GO v2.3.5 (https://www.blast2go.com), and BLAST v2.2.26 (http://blast.ncbi.nlm.nih.gov/Blast.cgi) was used for gene homology and function annotation.
Comparative secretome assays
Culture supernatants from P. oxalicum strains HP7-1 and EU2106 were filtered through a 0.22 μm membrane filter (Pall Corp., Ann Arbor, MI, USA) after adding two cOmplete, EDTA-free tablets (cOmplete EASYpack, Roche, Basel, Switzerland) per 100 mL of medium, and subsequently concentrated by ultrafiltration with a 10 kDa centrifugal concentrator (Millipore, Darmstadt, Germany). Secreted proteins were precipitated with acetone at −20 °C overnight. The collected proteins were denatured, digested, and subsequently labelled using the isobaric tags for relative and absolute protein quantitation Reagent 8 Plex One Assay Kit (AB Sciex, Framingham, MA, USA) according to the manufacturer’s protocol. The labelled peptides were separated on an XBridge C18 chromatographic column (250 mm × 4.6 mm, 3.5 μm, Waters, Milford, MA, USA). The collected peptide mixtures were then desalted with a ZipTip C18 column (Sigma-Aldrich, St. Louis, MO, USA) and dissolved in 10 μL of 2 % (v/v) acetonitrile in 0.1 % (v/v) formic acid for the liquid chromatography coupled with tandem mass spectrometry (LC–MS/MS) assay. After screening using the Proteome Discovery with SEQUEST search engine software (Thermo Fisher Scientific, Waltham, MA, USA), LC–MS/MS data were mapped to the P. oxalicum protein database and analyzed.
Protoplast preparation and transformation
Penicillium oxalicum protoplasts were prepared as described by Churchill et al. [41], with some modifications. Fresh conidia were inoculated into 200 mL of CM medium containing 50 ml of 20× nitrate [(g/L) NaNO3 (120.0), KCl (10.40), MgSO4·7H2O (10.40), KH2PO4 (30.40)], d-glucose (10.0), peptone (2.0), yeast extract (1.0), and acid hydrolysed casein (1.0) at pH 6.5, and cultivated at 28 °C with shaking at 180 rpm for 8 h. Mycelia were then harvested, washed three times with 0.6 M MgSO4·7H2O, and lysed in OM solution (1.2 M MgSO4·7H2O, 10 mM NaH2PO4, 6 g/L snailase, 4 g/L lysozyme, and 6 g/L lysing enzymes from T. harzianum (Sigma-Aldrich) at pH 5.8 for 2.5 h. Protoplasts were separated by adding trapping buffer (0.4 M sorbitol and 0.1 M Tris-HCl at pH 7.0), and subsequently collected in 50 mL centrifuge tubes. The collected protoplasts were precipitated using 1 M sorbitol and washed at least three times with 30 mL of STC (1 M sorbitol, 0.1 M Tris-HCl, and 0.1 M CaCl2 at pH 8.0). Finally, the protoplasts were resuspended in 0.5 mL of 4× STC and 1× PTC (40 % PEG3350, 0.1 M Tris-HCl, and 0.1 M CaCl2 at pH 8.0), adjusted to a concentration of 2 × 107 protoplasts per mL, and stored at −80 °C for further study.
For transformations, approximately 5 μg of DNA fragments were dissolved in 10 μL of 0.1 M spermidine and added to 100 μL of the protoplast suspension, followed by incubation on ice for 30 min. The mixture of DNA fragments and protoplasts was cultured for 25 min at room temperature after adding 1 mL of PTC. The cultivated mixture was added to 50 mL of OCM medium at 50 °C containing (g/L) casein enzymatic hydrolysate (1.0), yeast extract (1.0), sucrose (273.6), and agar (10.0), mixed briefly, and poured into Petri plates. After 30 min, PDA medium containing 250 μg/mL hygromycin B and 500 μg/mL G418 was added to the OCM medium. Transformants became visible after 5 days of cultivation at 28 °C.
Construction of gene deletion mutants
The ΔPoxKu70 strain derived from P. oxalicum strain HP7-1 was constructed to improve the efficiency of homologous recombination, as described by Li et al. [26], with some modifications. Briefly, approximately 2 kb of upstream and downstream flanking sequences of the PoxKu70 gene and 2.1 kb of the hygromycin phosphotransferase (hph) resistance gene were amplified from the genomic DNA of P. oxalicum strain HP7-1 and plasmid pCPXHY2GFP, respectively, which were maintained in our laboratory, and fused to construct the knockout cassette by double-joint PCR. The knockout cassette was introduced into wild-type P. oxalicum HP7-1 protoplasts as described above to generate the ΔPoxKu70 strain, in which the Poxku70 gene was replaced by the hph gene.
Subsequently, candidate transcription factor genes were knocked out in the ΔPoxKu70 strain using the method for deleting the PoxKu70 gene in P. oxalicum strain HP7-1. In this instance, the G418 resistance gene, which was amplified from the plasmid pCPXG418 and maintained in our laboratory, was used to replace the target gene. Three transformants for each gene knockout were randomly chosen for further study.
Southern hybridization analysis
Probes used for southern hybridization were amplified with the primers shown in Additional file 11: Table S5. Genomic DNA of the five mutants (ΔPoxKu70, ΔPoxClrB, ΔPOX02484, ΔPOX07291 and ΔPOX08522) was digested with ApaI, SacI, EcoRV and/or BglI (TaKaRa, Shiga, Japan). Enzyme-digested products were separated by 0.8 % agarose gel and transferred to Hybond-N+ nylon membranes (GE Healthcare Limited, Buckinghamshire, UK). A DIG-High prime DNA labelling and detection starter kit (Life Technologies, Carlsbad, CA, USA) was used to label and detect DNA according to the manufacturer’s protocol.
Measurement of biomass dry weight
Biomass formation of P. oxalicum strains in medium containing glucose as sole carbon source was determined gravimetrically as previously described [42]. Biomass concentration in medium containing Avicel as sole carbon source was indirectly measured by calculating the amount of essential intracellular proteins [42]. Briefly, the solid fraction was collected from the culture after centrifugation and immediately ground in liquid nitrogen, and 1 mL of protein extraction reagent consisting of PBS buffer containing 5 mM EDTA, 5 mM Phenylmethanesulfonyl fluoride (PMSF), and one complete ULTRA protease inhibitor cocktail tablet (Life Technologies, Carlsbad, CA, USA) at pH 7.4 was added per 100 mg of powder. Total protein was collected after precipitation via centrifugation at 11,300g for 10 min. Protein concentration was determined by the Bradford method (TIANQEN, Beijing, China). The final protein content was corrected using a set of substrate controls in which no inoculum was added to the Avicel medium. The biomass dry weight was calculated assuming an average content of 0.28 g intracellular protein per gram of dry cell mass. All experiments were repeated three times and the mean of the three experiments is reported.
Enzyme activity and protein concentration assays
Enzyme activities, including FPase, CMCase (EG), xylanase, Avicelase, KSBase, pNPCase (CBH), and pNPGase (BGL) activities, were determined as previously reported [5, 9]. FPase activity was investigated by incubating Whatman No. 1 filter paper (50 mg, 1.0 × 6.0 cm) in a 1.5 mL reaction system containing 1.0 mL of citrate buffer (100 mM, pH 5.0) and 0.5 mL of suitably diluted crude cellulase for 1 h at 50 °C. CMCase activity was measured in a 0.5 mL reaction system containing 0.45 mL of 1.0 % CMC-Na solution in citrate buffer (100 mM, pH 5.0) and 0.05 mL of crude cellulase. This mixture was incubated at 50 °C for 30 min. Xylanase, Avicelase, and KSBase activities were determined under similar conditions, except that 1.0 % xylan, 1.0 % Avicel, or 1.0 % KSB were used as substrates in place of CMC-Na, and the incubation time was 10, 60, or 30 min, respectively. The concentration of reducing sugars produced, in terms of glucose or xylose equivalents, was measured at 540 nm after adding two volumes of 3,5-dinitrosalicylic acid. One unit of enzymatic activity was defined as the amount of enzyme capable of producing 1 μmol of glucose or xylose from the appropriate substrates per min.
Measurement of pNPCase and pNPGase activity was conducted using p-nitrophenyl-β-cellobioside (pNPC) and p-nitrophenyl-β-glucopyranoside (pNPG), respectively, as substrates. A 140 μL mixture comprising 116 μL of citrate buffer (100 mM, pH 5.0), 14 μL of 25 mM pNPC or pNPG solution, and 10 μL of diluted crude cellulase was incubated at 50 °C for 15 min. Liberated p-nitrophenol was measured at 410 nm after adding 70 μL of sodium carbonate (0.4 M). One unit of enzyme activity was defined as the amount of enzyme capable of producing 1 μmol of p-nitrophenol from the appropriate substrates per min. Triplicate experiments were analyzed independently for each sample.
Determination of total protein concentration was performed using a Bradford assay kit (Pierce Biotechnology, Rockford, IL, USA) according to the manufacturer’s instructions.
qRT-PCR
The expression levels of cellulase and hemicellulase genes in mutant and wild-type strains were measured by qRT-PCR on a LightCycle480 instrument with version 4.0 software (Life Technologies, Carlsbad, CA, USA). First-strand cDNA was synthesized using the PrimeScript RT Reagent Kit with gDNA Eraser (TaKaRa, Shiga, Japan) according to the manufacturer’s instructions. Each qRT-PCR was conducted in a final volume of 20 μL that contained 6.4 μL of sterile water, 0.8 μL of 10 μM of the corresponding primers, 2.0 μL of suitable cDNA as the template, and 10 μL of SYBR Premix ExTaq II (TaKaRa, Shiga, Japan). All reactions were run for 40 cycles, comprising 3 s at 95 °C and 30 s at 60 °C. The melting curve program was set for 5 s at 95 °C, 1 min at 60 °C, and 15 s at 95 °C. The fluorescence signal was measured at the end of each extension step at 80 °C. Gene expression levels for cellulase and xylanase genes were measured using actin gene (POX09428) as a control and normalized to the parental strain ΔPoxKu70. Primers used for qRT-PCR analysis are shown in Additional file 11: Table S5. All qRT-PCRs were independently repeated in triplicate.
Phylogenetic analysis
POX02484 and POX08522 homologs were identified from NCBI (http://blast.ncbi.nlm.nih.gov/) using BLASTP, and phylogenetic tree analysis was performed using MEGA software version 7.0 [43] with the neighbour-joining method and a Poisson correction model. In this process, 1000 replicates were used for bootstrap values and gaps, and missing data treatments.
Statistics analysis
The Student’s t test (two-tailed) was performed using Microsoft Excel (Office 2013) (Microsoft, Redmond, WA, USA).
Sequence accession numbers
The genome sequence of P. oxalicum strain HP7-1 reported in this study was deposited in the GenBank database under accession number JRVD00000000. The re-sequenced clean data of P. oxalicum mutant EU2106 was submitted to SRA (http://www.ncbi.nlm.nih.gov/sra) under accession number SRA399107. The transcriptome of P. oxalicum HP7-1 and EU2106 grown on wheat bran and Avicel as the carbon source were respectively deposited at the GEO (http://www.ncbi.nlm.nih.gov/geo/) and the SRA databases under accession numbers GSE80076 and SRA399500 for HP7-1 and SRA399511 for EU2106. The secretomes of P. oxalicum HP7-1 and EU2106 grown on wheat bran and Avicel as the carbon source were submitted to the proteomics identifications (PRIDE) database (http://www.ebi.ac.uk/pride/archive/) under the accession numbers PXD004046. Sequences of PoxClrB, POX02484 and POX08522 from P. oxalicum HP7-1 and EU2106 were deposited in the GenBank database under accession numbers KU597415 to KU597419.
Authors’ contributions
JXF designed and supervised the research, involved in data analysis, and prepared the manuscript. SZ prepared the manuscript and involved in data analysis and secreted proteins extraction. YSY constructed deletion mutants of candidate genes and subsequently tested enzyme activities, performed qRT-PCR, and involved in secretome analysis. QPH, LSL, XY and YRX performed the construction of deletion mutants, enzyme activity assays of mutants and biomass weight measurement. CXL involved in bioinformatic analysis of genome and transcriptome. LY, JQH and SBX performed genome and transcriptome sequencing and the generated data analysis. LCM constructed the mutant ΔPoxKu70. LJ tested enzyme activities from P. oxalicum HP7-1 and EU2106. ZZ isolated and screened microbial strains to obtain P. oxalicum strain HP7-1. QDN obtained the mutant EU2106 by multiple mutagenesis of strain HP7-1. CJD and JLL involved in data analysis. All authors read and approved the final manuscript.
Acknowledgements
Wen-Jian Cen at Guangxi University, China, is greatly appreciated for her assistance with secretome assays.
Competing interests
The authors declare that they have no competing interests.
Availability of supporting data
The genome sequence of P. oxalicum strain HP7-1 reported in this study was deposited in the GenBank database (https://www.ncbi.nlm.nih.gov/genbank/) under accession number JRVD00000000. The re-sequenced clean data of P. oxalicum mutant EU2106 was submitted to Sequence Read Archive (SRA) (http://www.ncbi.nlm.nih.gov/sra) under accession number SRA399107. The transcriptome of P. oxalicum HP7-1 and EU2106 grown on wheat bran and Avicel as the carbon source were respectively deposited at the GEO (http://www.ncbi.nlm.nih.gov/geo/) and the SRA databases under accession numbers GSE80076 and SRA399500 for HP7-1 and SRA399511 for EU2106. The secretomes of P. oxalicum HP7-1 and EU2106 grown on wheat bran and Avicel as the carbon source were submitted to the proteomics identifications (PRIDE) database (http://www.ebi.ac.uk/pride/archive/) under the accession numbers PXD004046. Sequences of PoxClrB, POX02484 and POX08522 from P. oxalicum HP7-1 and EU2106 were deposited in the GenBank database under accession numbers KU597415 to KU597419.
Consent for publication
All authors consent for publication.
Funding
This work was financially supported by grants from the National Natural Science Foundation of China (Grant No. 31260017), the Guangxi BaGui Scholars Program Foundation (Grant No. 2011A001), the Guangxi Natural Science Foundation (Grant No. 2012GXNSFGA060005) and “One Hundred Person” Project of Guangxi.
Abbreviations
- BGL
β-glucosidase
- BLASTP
protein basic local alignment search tool
- CBH
cellobiohydrolase
- CAZymes
carbohydrate-active enzymes
- CDSs
coding DNA sequences
- CMCase
carboxymethylcellulose cellulase
- CWDEs
plant cell wall-degrading enzymes
- EDTA
ethylenediaminetetraacetic acid
- EG
endo-glucanase
- FPase
filter paper cellulase
- GEO
gene expression omnibus
- GO
gene ontology
- Inter
intergenic gene
- hph
the hygromycin phosphotransferase
- InDels
insertion/deletions
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- KSBase
KOH-pretreated sugarcane bagasse
- LC-MS/MS
the liquid chromatography coupled with tandem mass spectrometry
- MFS
major facilitator family
- MMM
modified minimal medium
- NCBI NR
National Center for Biotechnology Information non-redundant
- PMSF
phenylmethanesulfonyl fluoride
- pNPGase
p-nitrophenyl-β-glucopyranoside cellulase
- PCR
polymerase chain reaction
- PDA
potato-dextrose-agar
- pNPCase
p-nitrophenyl-β-cellobioside cellulase
- PRIDE
the proteomics identifications
- qRT-PCR
real-time quantitative reverse transcription-PCR
- SB
sugarcane bagasse
- SNVs
single nucleoside variations
- SRA
sequence read archive
Additional files
10.1186/s13068-016-0616-9 Venn diagram showing unique and shared proteins in P. oxalicum strain HP7-1 annotated using the non-redundant (NR), UniProt, Kyoto Encyclopedia of Genes and Genomes (KEGG), and Gene Ontology (GO) databases.
10.1186/s13068-016-0616-9 List of 274 single nucleotide variations and 12 Insertion/deletions (InDels) that occurred in P. oxalicum mutant strain EU2106 compared with the wild-type strain HP7-1.
10.1186/s13068-016-0616-9 List of 347 genes displaying altered expression in the cellulase and xylanase hyper-producing mutant P. oxalicum strain EU2106 compared with the wild-type strain HP7-1 when cultivated in medium containing wheat bran and Avicel as the carbon source.
10.1186/s13068-016-0616-9 Gene expression profiles of P. oxalicum strains HP7-1 and EU2106 during growth in the presence of wheat bran and Avicel as the carbon source. (A) Gene expression profile (|log2 fold change| ≥ 1, P ≤ 0.01 and probability ≥ 0.8 were used as thresholds). (B) Up-regulated genes encoding plant cell wall degrading enzymes (CWDEs) and major facilitator superfamily (MFS) members. (C) Down-regulated genes encoding CWDEs and MFS members. The expression scale is represented as the Log2 fold-change in B and C.
10.1186/s13068-016-0616-9 Functional annotation of secreted proteins as determined using the Kyoto Encyclopedia of Genes and Genomes database, and comparative analysis of the proteins identified in P. oxalicum strains HP7-1 and 114-2.
10.1186/s13068-016-0616-9 List of 240 proteins secreted by P. oxalicum strains HP7-1 and EU2106 when grown in medium containing wheat bran and Avicel as the carbon source.
10.1186/s13068-016-0616-9 Eleven candidate transcription factors identified from the initial screen.
10.1186/s13068-016-0616-9 Phenotypic comparison of wild-type P. oxalicum strain HP7-1 and the ΔPoxKu70 mutant. (A) Comparison of growth on potato-dextrose-agar plates between HP7-1 and ΔPoxKu70. (B–E) Comparison of the sensitivity to different concentrations of hygromycin B, G418, and ultraviolet light, as well as the cellulase/xylanase activities, between HP7-1 and ΔPoxKu70.
10.1186/s13068-016-0616-9 PCR and Southern hybridization analysis of deletion mutants of four candidate genes derived from the ΔPoxKu70 parent strain. (A) Mutant ΔPoxClrB, M, 1 kb marker; 1, ΔPoxKu70; 2, ddH2O; 3, ΔPoxClrB-1; 4, ΔPoxClrB-2; 5, ΔPoxClrB-3; (B) Mutant ΔPOX02484, M, 1 kb marker; 1, ΔPoxKu70; 2, ddH2O; 3, ΔPOX02484-6; 4, ΔPOX02484-10; 5, ΔPOX02484-17; (C) Mutant ΔPOX07921, M, 1 kb marker; 1, ΔPoxKu70; 2, ddH2O; 3, ΔPOX07291-1; 4, ΔPOX07291-2; 5, ΔPOX07291-3; (D) Mutant ΔPOX08522, M, 1 kb marker; 1, ΔPoxKu70; 2, ddH2O; 3, ΔPOX08522-2; 4, ΔPOX08522-11; 5, ΔPOX08522-13; (E) Southern hybridization of ΔPoxClrB, M, 1 kb marker; 1, ΔPoxKu70; 2, ΔPoxClrB-1; 3, ΔPoxClrB-2; 4, ΔPoxClrB-3; (F) Southern hybridization of ΔPOX02484, M, 1 kb marker; 1, ΔPoxKu70; 2, ΔPOX02484-6; 3, ΔPOX02484-10; 4, ΔPOX02484-17; (G) Southern hybridization of ΔPOX07291, M, 1 kb marker; 1, ΔPoxKu70; 2, ΔPOX07291-1; 3, ΔPOX07291-2; 4, ΔPOX07291-3; (H) Southern hybridization of ΔPOX08522, M, 1 kb marker; 1, ΔPoxKu70; 2, ΔPOX08522-2; 3, ΔPOX08522-11; 4, ΔPOX08522-13. In A–D, the top figure shows the production of each target gene, the middle figure shows the production of the fragment on the left of the target gene, and the bottom figure shows the production of the fragment on the right of the target gene.
10.1186/s13068-016-0616-9 Activities of crude enzymes from PoxClrB, POX02484, and POX08522 deletion mutants following direct inoculation in Avicel. Crude enzymes were produced by fungal strains grown in 1.0 % Avicel as the sole carbon source. The symbols * and ** indicate significant differences (P ≤ 0.05 and P ≤ 0.01, respectively) between candidate mutants and the ΔPoxKu70 parent strain, as assessed by Student’s t test.
10.1186/s13068-016-0616-9 Primers used in this study.
Footnotes
Shuai Zhao, Yu-Si Yan, Qi-Peng He, and Lin Yang contributed equally to this work
Contributor Information
Shuai Zhao, Email: shuaizhao0227@163.com.
Yu-Si Yan, Email: cherryyanyusi@qq.com.
Qi-Peng He, Email: 917407759@qq.com.
Lin Yang, Email: yanglin1@genomics.cn.
Xin Yin, Email: 317621503@qq.com.
Cheng-Xi Li, Email: 904375941@qq.com.
Li-Chun Mao, Email: 1439080483@qq.com.
Lu-Sheng Liao, Email: 1528746151@qq.com.
Jin-Qun Huang, Email: huangjinqun@genomics.cn.
Shang-Bo Xie, Email: xieshangbo@genomics.cn.
Qing-Dong Nong, Email: 574294151@qq.com.
Zheng Zhang, Email: zhangz_05@163.com.
Lei Jing, Email: jinglei@live.cn.
Ya-Ru Xiong, Email: 524926636@qq.com.
Cheng-Jie Duan, Email: duancj2004@163.com.
Jun-Liang Liu, Email: junliang.liu@163.com.
Jia-Xun Feng, Email: jiaxunfeng@sohu.com.
References
- 1.Meng X, Ragauskas AJ. Recent advances in understanding the role of cellulose accessibility in enzymatic hydrolysis of lignocellulosic substrates. Curr Opin Biotechnol. 2014;27:150–158. doi: 10.1016/j.copbio.2014.01.014. [DOI] [PubMed] [Google Scholar]
- 2.Himmel ME, Ding SY, Johnson DK, Adney WS, Nimlos MR, Brady JW, Foust TD. Biomass recalcitrance: engineering plants and enzymes for biofuels production. Science. 2007;315:804–807. doi: 10.1126/science.1137016. [DOI] [PubMed] [Google Scholar]
- 3.Gusakov AV. Alternatives to Trichoderma reesei in biofuel production. Trends Biotechnol. 2011;29:419–425. doi: 10.1016/j.tibtech.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 4.Singhania RR, Patel AK, Sukumaran RK, Larroche C, Pandey A. Role and significance of β-glucosidases in the hydrolysis of cellulose for bioethanol production. Bioresour Technol. 2013;127:500–507. doi: 10.1016/j.biortech.2012.09.012. [DOI] [PubMed] [Google Scholar]
- 5.Jing L, Zhao S, Xue JL, Zhang Z, Yang Q, Xian L, Feng JX. Isolation and characterization of a novel Penicillium oxalicum strain Z1-3 with enhanced cellobiohydrolase production using cellulase-hydrolyzed sugarcane bagasse as carbon source. Ind Crops Prod. 2015;77:666–675. doi: 10.1016/j.indcrop.2015.09.052. [DOI] [Google Scholar]
- 6.Amore A, Giacobbe S, Faraco V. Regulation of cellulase and hemicellulase gene expression in fungi. Curr Genom. 2013;14:230–249. doi: 10.2174/1389202911314040002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tani S, Kawaguchi T, Kobayashi T. Complex regulation of hydrolytic enzyme genes for cellulosic biomass degradation in filamentous fungi. Appl Microbiol Biotechnol. 2014;98:4829–4837. doi: 10.1007/s00253-014-5707-6. [DOI] [PubMed] [Google Scholar]
- 8.Ilmen M, Thrane C, Penttila M. The glucose repressor gene cre1 of Trichoderma: isolation and expression of a full-length and a truncated mutant form. Mol Gen Genet. 1996;251(4):451–460. doi: 10.1007/BF02172374. [DOI] [PubMed] [Google Scholar]
- 9.Li ZH, Yao G, Wu R, Gao L, Kan Q, Liu M, Yang P, Liu G, Qin Y, Song X, Zhong Y, Fang X, Qu Y. Synergistic and dose-controlled regulation of cellulase gene expression in Penicillium oxalicum. PLoS Genet. 2015;11:e1005509. doi: 10.1371/journal.pgen.1005509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aro N, Ilmen M, Saloheimo A, Penttila M. ACEI of Trichoderma reesei is a repressor of cellulase and xylanase expression. Appl Environ Microbiol. 2003;69:56–65. doi: 10.1128/AEM.69.1.56-65.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coradetti ST, Craig JP, Xiong Y, Shock T, Tian C, Glass NL. Conserved and essential transcription factors for cellulase gene expression in ascomycete fungi. Proc Natl Acad Sci USA. 2012;109:7397–7402. doi: 10.1073/pnas.1200785109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiong Y, Sun J, Glass NL. VIB1, a link between glucose signaling and carbon catabolite repression, is essential for plant cell wall degradation by Neurospora crassa. PLoS Genet. 2014;10:e1004500. doi: 10.1371/journal.pgen.1004500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen M, Qin YQ, Cao Q, Liu GD, Li J, Li ZH, Zhao J, Qu YB. Promotion of extracellular lignocellulolytic enzymes production by restraining the intracellular β-glucosidase in Penicillium decumbens. Bioresour Technol. 2013;137:33–40. doi: 10.1016/j.biortech.2013.03.099. [DOI] [PubMed] [Google Scholar]
- 14.van Peij NN, Visser J, de Graaff LH. Isolation and analysis of xlnR, encoding a transcriptional activator coordinating xylanolytic expression in Aspergillus niger. Mol Microbiol. 1998;27:131–142. doi: 10.1046/j.1365-2958.1998.00666.x. [DOI] [PubMed] [Google Scholar]
- 15.Coradetti ST, Xiong Y, Glass NL. Analysis of a conserved cellulase transcriptional regulator reveals inducer-independent production of cellulolytic enzymes in Neurospora crassa. Microbiologyopen. 2013;2:595–609. doi: 10.1002/mbo3.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yao G, Li Z, Gao L, Wu R, Kan Q, Liu G, Qu Y. Redesigning the regulatory pathway to enhance cellulase production in Penicillium oxalicum. Biotechnol Biofuels. 2015;8:71. doi: 10.1186/s13068-015-0253-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Z, Liu JL, Lan JY, Duan CJ, Ma QS, Feng JX. Predominance of Trichoderma and Penicillium in cellulolytic aerobic filamentous fungi from subtropical and tropical forests in China, and their use in finding highly efficient β-glucosidase. Biotechnol Biofuels. 2014;7:107. doi: 10.1186/1754-6834-7-107. [DOI] [Google Scholar]
- 18.Huang Y, Qin XL, Luo XM, Nong QD, Yang Q, Zhang Z, Gao Y, Lv FX, Chen Y, Yu ZW, Liu JL, Feng JX. Efficient enzymatic hydrolysis and simultaneous saccharification and fermentation of sugarcane bagasse pulp for ethanol production by cellulase from Penicillium oxalicum EU2106 and thermotolerant Saccharomyces cerevisiae ZM1-5. Biomass Bioenergy. 2015;77:53–63. doi: 10.1016/j.biombioe.2015.03.020. [DOI] [Google Scholar]
- 19.Liu G, Zhang L, Wei X, Zou G, Qin Y, Ma L, Li J, Zheng H, Wang S, Wang C, Xun L, Zhao GP, Zhou Z, Qu Y. Genomic and secretomic analyses reveal unique features of the lignocellulolytic enzyme system of Penicillium decumbens. PLoS One. 2013;8:e55185. doi: 10.1371/journal.pone.0055185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu G, Zhang L, Qin Y, Zou G, Li Z, Yan X, Wei X, Chen M, Chen L, Zheng K, Zhang J, Ma L, Li J, Liu R, Xu H, Bao X, Fang X, Wang L, Zhong Y, Liu W, Zheng H, Wang S, Wang C, Xun L, Zhao GP, Wang T, Zhou Z, Qu Y. Long-term strain improvements accumulate mutations in regulatory elements responsible for hyper-production of cellulolytic enzymes. Sci Rep. 2013;3:1569. doi: 10.1038/srep01569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huber W, Carey VJ, Gentleman R, Anders S, Carlson M, Carvalho BS, Bravo HC, Davis S, Gatto L, Girke T, Gottardo R, Hahne F, Hansen KD, Irizarry RA, Lawrence M, Love MI, MacDonald J, Obenchain V, Oleś AK, Pagès H, Reyes A, Shannon P, Smyth GK, Tenenbaum D, Waldron L, Morgan M. Orchestrating high-throughput genomic analysis with bioconductor. Nat Methods. 2015;12:115–121. doi: 10.1038/nmeth.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mitchell A, Chang HY, Daugherty L, Fraser M, Hunter S, Lopez R, McAnulla C, McMenamin C, Nuka G, Pesseat S, Sangrador-Vegas A, Scheremetjew M, Rato C, Yong SY, Bateman A, Punta M, Attwood TK, Sigrist CJ, Redaschi N, Rivoire C, Xenarios I, Kahn D, Guyot D, Bork P, Letunic I, Gough J, Oates M, Haft D, Huang H, Natale DA, Wu CH, Orengo C, Sillitoe I, Mi H, Thomas PD, Finn RD. The InterPro protein families database: the classification resource after 15 years. Nucleic Acids Res. 2015;43:D213–D221. doi: 10.1093/nar/gku1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li J, Liu G, Chen M, Li Z, Qin Y, Qu Y. Cellodextrin transporters play important roles in cellulase induction in the cellulolytic fungus Penicillium oxalicum. Appl Microbiol Biotechnol. 2013;24:10479–10488. doi: 10.1007/s00253-013-5301-3. [DOI] [PubMed] [Google Scholar]
- 24.Marx IJ, van Wyk N, Smit S, Jacobson D, Viljoen-Bloom M, Volschenk H. Comparative secretome analysis of Trichoderma asperellum S4F8 and Trichoderma reesei Rut C30 during solid-state fermentation on sugarcane bagasse. Biotechnol Biofuel. 2013;6:172. doi: 10.1186/1754-6834-6-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–1082. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 26.Li Z, Du C, Zhong Y, Wang T. Development of a highly efficient gene targeting system allowing rapid genetic manipulations in Penicillium decumbens. Appl Microbiol Biotechnol. 2010;87:1065–1076. doi: 10.1007/s00253-010-2566-7. [DOI] [PubMed] [Google Scholar]
- 27.Liao HP, Fan XT, Mei XL, Wei Z, Raza W, Shen QR, Xu YC. Production and characterization of cellulolytic enzyme from Penicillium oxalicum GZ-2 and its application in lignocellulose saccharification. Biomass Bioenergy. 2015;74:122–134. doi: 10.1016/j.biombioe.2015.01.016. [DOI] [Google Scholar]
- 28.Xue JL, Zhao S, Liang RM, Yin X, Jiang SX, Su LH, Yang Q, Duan CJ, Liu JL, Feng JX. A biotechnological process efficiently co-produces two high value-added products, glucose and xylooligosaccharides, from sugarcane bagasse. Bioresour Technol. 2016;204:130–138. doi: 10.1016/j.biortech.2015.12.082. [DOI] [PubMed] [Google Scholar]
- 29.Vitikainen M, Arvas M, Pakula T, Oja M, Penttiläet M, Saloheimo M. Array comparative genomic hybridization analysis of Trichoderma reesei strains with enhanced cellulase production properties. BMC Genom. 2010;11:441. doi: 10.1186/1471-2164-11-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murakami H, Aiba H, Nakanishi M, Murakami-Tonami Y. Regulation of yeast forkhead transcription factors and FoxM1 by cyclin-dependent and polo-like kinases. Cell Cycle. 2010;9:3233–3242. doi: 10.4161/cc.9.16.12599. [DOI] [PubMed] [Google Scholar]
- 31.Masloff S, Pöggeler S, Kück U. The pro1+ gene from Sordaria macrospora encodes a C6 zinc finger transcription factor required for fruiting body development. Genetics. 1999;152:191–199. doi: 10.1093/genetics/152.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Soukup AA, Farnoodian M, Berthier E, Keller NP. NosA, a transcription factor important in Aspergillus fumigatus stress and developmental response, rescues the germination defect of a laeA deletion. Fungal Genet Biol. 2012;49:857–865. doi: 10.1016/j.fgb.2012.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vienken K, Fischer R. The Zn(II)2Cys6 putative transcription factor NosA controls fruiting body formation in Aspergillus nidulans. Mol Microbiol. 2006;61(2):544–554. doi: 10.1111/j.1365-2958.2006.05257.x. [DOI] [PubMed] [Google Scholar]
- 34.Garg A, Futcher B, Leatherwood J. A new transcription factor for mitosis: in Schizosaccharomyces pombe, the RFX transcription factor Sak1 works with forkhead factors to regulate mitotic expression. Nucleic Acids Res. 2015;43:6874–6888. doi: 10.1093/nar/gkv274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ribár R, Grallert A, Oláh É, Szállási Z. Deletion of the sep1+ forkhead transcription factor homologue is not lethal but causes hyphal growth in Schizosaccharomyces pombe. Biochem Biophys Res Commun. 1999;263:465–474. doi: 10.1006/bbrc.1999.1333. [DOI] [PubMed] [Google Scholar]
- 36.Nitta M, Furukawa T, Shida Y, Mori K, Kuhara S, Morikawa Y, Ogasawara W. A new Zn(II)2Cys6-type transcription factor BglR regulates β-glucosidase expression in Trichoderma reesei. Fungal Genet Biol. 2012;49:388–397. doi: 10.1016/j.fgb.2012.02.009. [DOI] [PubMed] [Google Scholar]
- 37.Sun X, Liu Z, Qu Y, Li X. The effects of wheat bran composition on the production of biomass-hydrolyzing enzymes by Penicillium decumbens. Appl Biochem Biotechnol. 2008;146:119–128. doi: 10.1007/s12010-007-8049-3. [DOI] [PubMed] [Google Scholar]
- 38.Wilson D, Charoensawan V, Kummerfeld SK, Teichmann SA. DBD—taxonomically broad transcription factor predictions: new content and functionality. Nucleic Acids Res. 2008;36:D88–D92. doi: 10.1093/nar/gkm964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Churchill ACL, Ciuffetti LM, Hansen DR, Van Etten HD, Van Alfen NK. Transformation of the fungal pathogen Cryphonectria parasitica with a variety of heterologous plasmids. Curr Genet. 1990;17:25–31. doi: 10.1007/BF00313245. [DOI] [Google Scholar]
- 42.Bischof R, Fourtis L, Limbeck A, Gamauf C, Seiboth B, Kubicek CP. Comparative analysis of the Trichoderma reesei transcriptome during growth on the cellulase inducing substrates wheat straw and lactose. Biotechnol Biofuel. 2013;6:127. doi: 10.1186/1754-6834-6-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016 doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]


