Abstract
Background
Gliomasphere cultures are widely utilized for the study of glioblastoma (GBM). However, this model system is not well characterized, and the utility of current classification methods is not clear.
Methods
We used 71 gliomasphere cultures from 68 individuals. Using gene expression-based classification, we performed unsupervised clustering and associated gene expression with gliomasphere phenotypes and patient survival.
Results
Some aspects of the gene expression-based classification method were robust because the gliomasphere cultures retained their classification over many passages, and IDH1 mutant gliomaspheres were all proneural. While gene expression of a subset of gliomasphere cultures was more like the parent tumor than any other tumor, gliomaspheres did not always harbor the same classification as their parent tumor. Classification was not associated with whether a sphere culture was derived from primary or recurrent GBM or associated with the presence of EGFR amplification or rearrangement. Unsupervised clustering of gliomasphere gene expression distinguished 2 general categories (mesenchymal and nonmesenchymal), while multidimensional scaling distinguished 3 main groups and a fourth minor group. Unbiased approaches revealed that PI3Kinase, protein kinase A, mTOR, ERK, Integrin, and beta-catenin pathways were associated with in vitro measures of proliferation and sphere formation. Associating gene expression with gliomasphere phenotypes and patient outcome, we identified genes not previously associated with GBM: PTGR1, which suppresses proliferation, and EFEMP2 and LGALS8, which promote cell proliferation.
Conclusions
This comprehensive assessment reveals advantages and limitations of using gliomaspheres to model GBM biology, and provides a novel strategy for selecting genes for future study.
Keywords: brain tumor stem cell, cancer stem cell, glioma, neurosphere, The Cancer Genome Atlas
Glioblastomas (GBMs) contain cells that have neural stem cell-like properties1 in that they grow as spheroid cultures in the presence of epidermal growth factor (EGF) and basic fibroblast growth factor and are capable of producing the varied cell types found within the parent tumor.2–4 These serum-free sphere cultures are enriched for a set of cells capable of initiating tumors when implanted at very low numbers: the so-called glioma stem or tumor-initiating cells.5–11 The utility of these gliomasphere (sometimes termed neurosphere) cultures as a model has been supported by their ability to retain the major mutations found within the tumor and to produce tumors following xenotransplantation resembling human GBM.4,10,12
Glioma subtypes are defined by histopathological criteria. Several studies have demonstrated that high-grade gliomas, including GBM, can be further classified through gene expression into clinically relevant subgroups.13–15 A study of samples in The Cancer Genome Atlas (TCGA) produced a classification of GBM tumors into 4 subtypes: proneural, neural, classical, and mesenchymal.15 This classification system was based on differential gene expression across a catalog of tumor samples and associates with genetic mutation and other phenotypes such as hypermethylation.16 For the purposes of this paper, we will refer to the Verhaak et al classification as TCGA. The goals of the current study were to determine the relevance of TCGA and other classification schemes to gliomaspheres and how such classifications related to tumor and gliomasphere phenotypes. Furthermore, we sought to determine whether combining gene expression phenotypic and patient survival analysis could be used to discover novel determinants of GBM proliferation. To do this, we established a large bank of gliomaspheres and assayed their phenotypic and gene expression characteristics in relationship to the parent tumor and patient outcome. While our data identify some limitations of the gliomasphere model, we also provide evidence that gliomaspheres preserve important aspects of GBM biology. We demonstrate that the dynamic gliomasphere model can be utilized in a manner distinct from assessment of the original, static tumor sample.
Materials and Methods
A more detailed description of some of the methods can be found in the Supplementary Methods online.
Clinical Data and Tumor Collection
High grade human gliomas from 70 surgical resections were collected under institutional review board-approved protocols and graded by neuropathologists as previously described.17 One widely used18,19 tumor sample was obtained from Duke University after it had been resected and placed as a xenograft. There were 67 distinct patients as four pairs of gliomaspheres were derived from the same patients at different resections.
Gliomasphere Culturing
Gliomaspheres were cultured from GBM tumor samples as previously described.3,17
In Vitro Assays
We studied the percentage of sphere formation for cultures at clonal density (plating 50 cells/100 µL/well of a 96 well plate).
Cell Proliferation Assays
Cells were plated at 5000 cells/100 µL/well of a 96-well plate and grown for 5-7 days. Cell number was assessed using Dojindo Cell Counting Kit-8 (Dojindo Molecular Technologies Inc.).
Ingenuity and TFacts Analyses
Lists of genes associated with outcomes at P < .001 were uploaded to Qiagen's Ingenuity Pathway Analysis (IPA) in order to determine canonical pathways associated with each gene list. Ingenuity pathway analysis is a curated database of associations between molecules and the canonical pathways within which they reside.
Lists of genes associated with outcomes at P < .001 were uploaded to TFacts (http://www.tfacts.org/TFactS-new/TFactS-v2/index1.html). Tfacts generates lists of associated transcription factors based on catalogs of transcription factor signatures in a sign-sensitive manner.
Quantitative Real Time- Polymerase Chain Reaction (qRT-PCR)
qRT-PCR was performed utilizing standard procedures and primers as detailed in supplementary methods. RNA was isolated using TRIzol (Gibco) and 1.5 µg RNA was converted to cDNA by reverse transcription. qRT-PCR was performed after addition of Power SYBR Master Mix (Applied Biosystems) on an ABI PRISM 7700 sequence detection system (Applied Biosystems, Foster City, CA).
Gene Trait Correlations
Gene-trait correlations and p-values were obtained using the standard Pearson correlation coefficient r using the cor() function in R. A P < .001 threshold was used to select the most interesting candidates.
Microarrays
Concentration and quality of RNA samples was examined using the NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies) and the Agilent 2100 Bioanalyzer (Agilent Technologies). RNA samples were reverse transcribed and labeled according to manufacturer's instructions and hybridized to Affymetrix high-density oligonucleotide HG-U133A Plus 2.0 Human Arrays.
GSEA
In order to functionally annotate the expression differences between the two clusters, we performed a gene set enrichment analysis (GSEA) using the GSEA software23.
Subgroup Classification
The Cancer Genome Atlas (TCGA) unified gene expression dataset for the 173 core tumor samples utilized by Verhaak et al15 was used to create our models.16,21,22,23
Clustering
Hierarchical clustering was obtained using the hclust function in R using standard parameters.
Western Blotting
Samples of protein for Western blots were prepared by collecting equal amounts of cells, counted using the Countess Automated Cell Counter (Life Technologies), and then boiled in Laemmli Sample Buffer (Bio-Rad #161-0737) with 5% beta-mercaptoethanol for 5 min. An equal numbers of cells (125 000 cells/lane) were loaded onto 10% Mini-Protean Pre-cast gels (Bio-Rad #456-1036) and the western blots were performed according to standard procedures. Protein expression of PTEN was determined by western blot (Cell Signaling #9559). Beta Actin was used as a loading control (ABCAM #8277). EGFRvIII mutation in gliomasphere cultures was ascertained by western blots using an EGFR antibody (Millipore #06-847). Samples that showed a 140kD band were considered to be positive.
shRNA
Lentiviral mediated shRNA knockdowns of EFEMP2, LGALS8, and PTGR1 were performed using constructs from the Dharmacon-Harmon library (General Electric: http://dharmacon.gelifesciences.com/shrna/gipz-lentiviral-shrna/?Parent=12884902157).
Pathology Reporting
The readout of clinical diagnosis, the presence of EGFRvIII rearrangement and cytogenetic analysis were obtained from the official pathology report.
Sequencing of IDH1 and IDH2
Genomic DNA was isolated from the cultured glioma-sphere cells by using DNeasy Blood and Tissue Kit (Qiagen). The IDH1 and IDH2 genotypes were determined by Sanger sequencing.
Weighted Gene Co-expression Network Analysis (WGCNA)
WGCNA was conducted using the R package as previously described.20,21 Briefly, correlation coefficients were constructed between expression levels of genes, and a connectivity measure (topological overlap, TO) was calculated for each gene by summing the connection strength with other genes. Genes were then clustered based on their TO, and groups of co-expressed genes (modules) were identified. Each module was assigned a color, and the first principal component (eigengene) of a module was computed and considered to be representative of the gene expression profiles in a module. We then correlated eigengenes for each module with phenotypic traits of interest.
Survival Curves
Time to survival (TTS) for patients in our database was determined by the duration of life from the date of surgery using death certificates and the social security death index.
Statistics
For comparison of small groups we used a cutoff of P < .05 to distinguish significant differences. For analysis of gene correlations to survival, we used a false discovery rate (FDR) P < .05 for greater stringency. Statistics for comparing cell proliferation, sphere formation, sphere diameter, and sphere total volume between groups of control cells and shRNA mediated knockdowns were done in GraphPad Prism software (http://www.graphpad.com/scientific-software/prism/) utilizing the paired T-test.
Results
Classification of Gliomasphere Gene Expression and Associations with Tumor and Patient Characteristics
Patient characteristics, tumor pathological features, EGFRvIII expression, and IDH mutation status of the tumors studied can be found in Supplementary Table S1. Sixty nine of the samples had the diagnosis of GBM. We validated the gliomasphere cultures for EGFRvIII by Western blot. All tumors that were positive for EGFRvIII in pathology were also positive for EGFRvIII in the gliomaspheres (Supplementary Table S1). Only HK361 was EGFRvIII-positive in gliomasphere culture but negative in the tumor pathology report, although this tumor did have EGFR amplification. IDH1 R132H mutation was confirmed in all 6 cultures from IDH1 mutant tumors.
As previously reported about the tumor samples that formed gliomasphere cultures, 6 of 8 formed tumors upon intracranial xenotransplantation into mice.17 Since that publication, we have performed successful transplants on an additional 6 of 6 gliomasphere cultures (data not shown). However, all 6 IDH1 mutant cell cultures failed to produce tumors. From this, we concluded that IDH1 cultures do not readily form tumors, while the vast majority of gliomasphere cultures can.
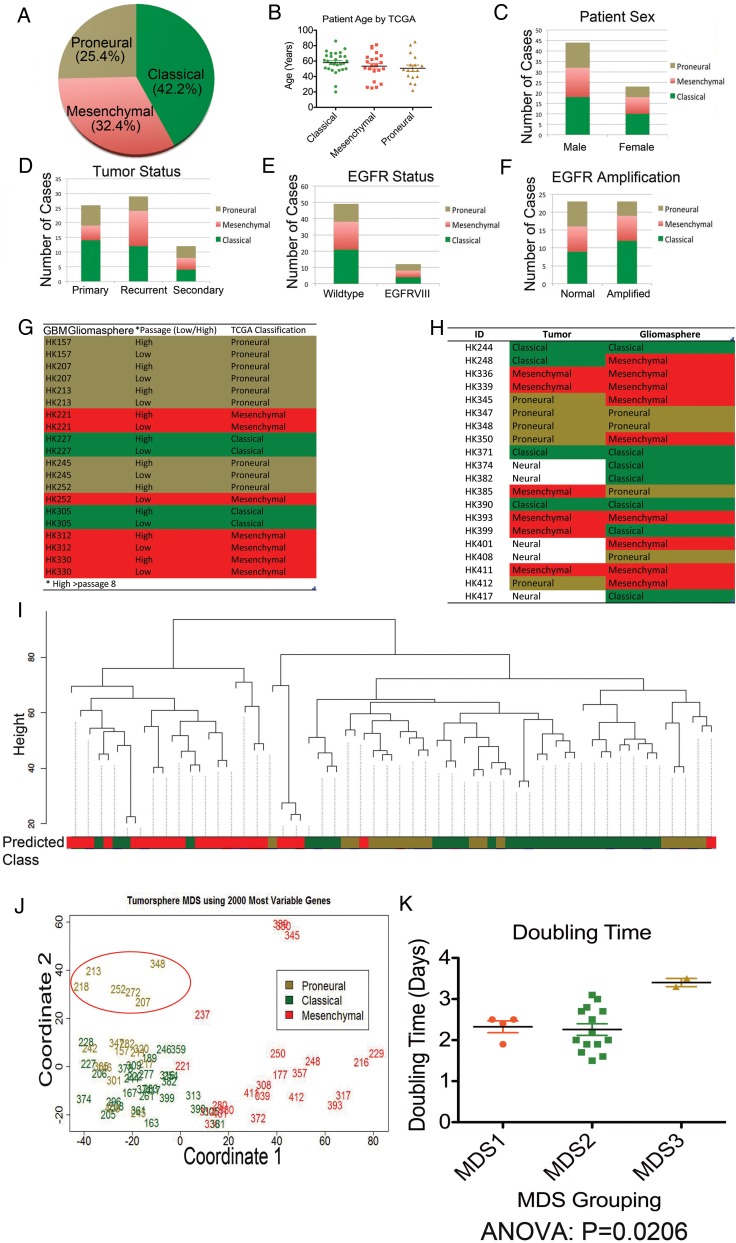
We used all 4 TCGA classifications—neural, classical, mesenchymal, and proneural—initially to classify the expression patterns of the gliomasphere samples However, only 2 of the 71 gliomaspheres were classified as neural. For subsequent analysis, we followed the strategies of others22 and eliminated the neural classification because it may be more representative of normal brain rather than tumor.15 Of the 71 gliomasphere cultures, 30 (42.2%) were categorized as classical, 23 (32.4%) were mesenchymal, and 18 (25.4%) were proneural (Fig. 1A and Supplementary Table S1). We subcategorized patient and tumor phenotypes by TCGA classification (Fig. 1B–F). We found no significant association between TCGA classification and patient age (Fig. 1B), sex (Fig. 1C), or whether the cultures were derived from primary, recurrent, or secondary GBM (Fig. 1D). We found no association between TCGA subclass and EGFRvIII status or EGFR amplification (Fig. 1E and F). As expected, IDH1 mutations were uniformly (6 of 6 cases) associated with the proneural classification (Supplementary Table S1). Of the 7 gliomaspheres derived from gliosarcomas, 6 were classified as mesenchymal subtype (Supplementary Table S1).
Fig. 1.
Summary statistics on 71 gliomasphere samples. (A) Gliomasphere distribution for each of 3 The Cancer Genome Atlas (TCGA) classifications. (B–F) Distribution, according to TCGA classification of patients' age in years (B), sex (C), type of parent tumor (primary, recurrent, or secondary) (D), EGFR mutation status, wild-type or EGFRvIII mutant (E), and EGFR-amplified status (F). (G) TCGA classification of gliomaspheres across high and low passages (high > 8 passages). (H) TCGA classification of parent tumors and their derived gliomasphere cultures. (I) Hierarchical clustering of 71 gliomasphere cultures based on gene expression profiles of the top 2000 most variable genes identifies 2 main clusters. Color codes: classical = green, mesenchymal = red, proneural = gold. (J) Multidimensional scaling (MDS) of 71 tumorsphere cultures based on the top 2000 most variable genes distinguishes 2 groups on the first principal component (PC1 = coordinate 1), and 3 groups when incorporating the second principal component (PC2=coordinate 2) with a minor fourth group. The red oval distinguishes the third subgroup of proneural gliomaspheres. Three-digit numbers represent the GBM gliomasphere identification numbers. (K) Cell-doubling time (inverse of proliferation rate) is plotted for the 3 MDS groups from MDS plot (J). Group 1 = right side of plot (all mesenchymal) Group 2 = lower left side of plot (mostly classical with some proneural and mesenchymal), Group 3 = upper left side of plot, delineated by red oval (all proneural). MDS groupings for each gliomasphere sample are reported in Supplementary Table S1. Mean ± sSEM is depicted as bars. Classical = green, mesenchymal = red, and proneural = gold in all figures.
To determine whether gliomasphere cultures maintained expression profiles throughout passaging, we classified a subset of gliomasphere cultures at early (<8) and later (>8) passages. Nine of 10 cultures maintained their classification from low to high passage number (P < .003, chi-square test, Fig. 1G). In 3 of 4 cases, gliomaspheres derived from different resections from the same patient retained their TCGA classification. The classification in one case changed from classical to mesenchymal.
We compared mRNA profiles of 20 brain tumors and gliomaspheres derived from the same piece of tissue (Fig. 1H). Sixty percent of the samples (excluding 5 tumors that fell into the neural subclass) yielded the same TCGA classification in tumor and spheres, but this overlap did not reach statistical significance. For those samples with differing subclassifications, there were no trends in the manner in which they switched categories. Gliomaspheres derived from neural tumor samples did not fall into any one particular classification. In order to identify a measure of similarity that is more sensitive than simple clustering, we identified the transcriptional differences for each of the gliomaspheres and tumors as compared with the remaining catalog, using the 20 paired tumor and gliomasphere samples. After normalizing gliomasphere and tumor samples separately, we obtained the gene changes related to each line by comparing one line versus all remaining lines and using P < .001 as the statistical threshold. We then computed the % overlap between all possible gliomasphere/tumor pairs and plotted these values in Supplementary Fig. S1. For 8 pairs, the overlap of gene expression between tumor and gliomasphere was more closely related than the other comparisons and greater than what would have been expected by chance. Taken together, these data indicate there is a significant relationship between many gliomasphere cultures and their parent tumor, but this relationship is not uniform.
We next determined gene expression differences between tumors and gliomasphere samples as a whole. Pairwise comparisons of gene expression from each tumor and its derived gliomasphere yielded 188 differentially expressed genes using a nominal P value of .001 as our statistical threshold. Ingenuity analysis revealed that the molecular and cellular functions associated with these changes included cellular development, cell growth and proliferation, cell cycle, cell signaling, posttranslational modification, and Clathrin-mediated endocytosis signaling (Table 1). We then used a more conservative threshold (false discovery rate [FDR] adjusted P values of P < .05) and identified a subset of 13 differentially expressed genes between tumor and gliomaspheres (Table 1). Thus, while there are certain changes in expression from tumors to gliomaspheres related to cell proliferation, the vast majority of genes do not undergo consistent changes across gliomasphere cultures.
Table 1.
Molecular expression changes from tumor to gliomasphere
| Molecular Expression Changes from Tumor to Tumorsphere |
|||
|---|---|---|---|
| Significant Molecular and Cellular Functions (Ingenuity Analysis) | |||
| Name | P value | # Molecules | |
| Cellular development | .000657–.0403 | 16 | |
| Cellular growth and proliferation | .000657–.0404 | 14 | |
| Cell cycle | .000981–.0482 | 13 | |
| Cell signaling | .00171–.0313 | 15 | |
| Posttranslational modification | .00171–.0244 | 21 | |
| Significant Canonical Pathways (Ingenuity) Analysis) | |||
| Ingenuity Canonical Pathways | P value | Ratio | Molecules |
| Fcγ receptor-mediated phagocytosis in macrophages and Monocytes | .00724 | 4.3E-02 | ACTR2,ARF6,CBL,FYB |
| Clathrin-mediated endocytosis signaling | .0182 | 2.7E-02 | ACTR2,ARF6,CBL,USP9X,APOC1 |
| Gene changes (FDR P < .05) | |||
| Gene | FDR P value | Mean of differences T-N | |
| ZW10 | .0162 | −0.392 | |
| CLN6 | .00371 | −0.451 | |
| SYNJ2BP | .0245 | −0.713 | |
| SCIN | <.001 | 0.483 | |
| C1QB | .000543 | 0.935 | |
| FCER1G | .0433 | 0.726 | |
| TAF6L | .0289 | −0.324 | |
| ANKRD37 | .00867 | 0.363 | |
| MS4A7 | .0433 | 0.796 | |
| RGS1 | .0433 | 0.865 | |
| PTPRC | .0245 | 0.683 | |
| GIMAP6 | .0268 | 0.681 | |
| ZNF492 | .00197 | 0.463 | |
Ingenuity analysis of gene list comparing significant genes (P < .001) differing between 20 tumors and their derived gliomaspheres. The ratio is the number of genes in our list divided by the number of genes in the pathway. Canonical pathways are shown if they are significant (P < .05) and contain >2 genes in the list. Gene changes depict the list of genes significantly changed between tumors and their derived gliomaspheres with a false discovery rate (FDR) P < .05. Mean of differences T-N depicts the difference of the tumor minus the gliomasphere mean gene expression levels. A negative number represents enrichment of gene expression in the gliomasphere culture as compared with the tumor, while a positive number represents depletion of gene expression in the gliomasphere culture as compared with the tumor.
Next, we used unbiased methods to analyze gene expression in the gliomaspheres. Unsupervised clustering of the gliomasphere cultures revealed 2 general subgroups: one containing most of the mesenchymal cultures and another consisting largely of proneural and classical gliomaspheres (Fig. 1I). Multidimensional scaling (MDS) analysis generally supports the concept of 2 groups when viewed along the first principal component (PC1), which accounts for 48.3% of the variance. In order to functionally annotate the expression differences between the 2 clusters, we performed a gene set enrichment analysis (GSEA) using the GSEA software.23 We identified 261 gene sets overrepresented (n = 174) or depleted (n = 87) at a false discovery rate of 10% in cluster M compared with cluster non-M. These gene sets are listed in Supplementary Table S2. In order to estimate the relationship in terms of functional annotation between cluster M and typical TCGA classification, we then classified our samples using the TCGA classifier and identified transcriptional profiles related to each of the TCGA categories by comparing each TCGA category with the rest of the cohort. We then ran GSEA on each of these transcriptional profiles and assessed the enrichment of the 261 gene sets in each of the TCGA categories using the normalized enrichment score (NES) as a metric of enrichment. Samples classified as mesenchymal were the closest to cluster M in terms of enrichment, followed by classical and proneural categories, which were not enriched for most of these gene sets (Supplementary Fig. S2). These results support our assignment of gliomaspheres into 2 groups: mesenchymal and nonmesenchymal.
When considering the second principal component of the MDS analysis (PC2), which accounts for 9.63% of the variance, a subset of 6 proneural gliomaspheres appeared to segregate separately (red oval in Fig. 1J), as did a minor group of 3 mesenchymal samples (top right quadrant, Fig. 1J). Within this subset of 3 very closely related mesenchymal samples is a pair of gliomaspheres, which were derived from the same patient diagnosed with a secondary GBM in surgeries separated by only 49 days. The third mesenchymal sample from that subgroup was also derived from a secondary GBM.
We examined gene expression differences between gliomaspheres from recurrent and primary GBMs. Four-hundred nine genes were differentially expressed at a P < .005. Supplementary Table S3 shows genes enriched in recurrent samples and genes enriched in primary samples. There were no consistent changes according to gene ontology groups (not shown).
Association of Gliomasphere Gene Expression with Gliomasphere Phenotype
We examined 3 properties of gliomasphere formation: clonal sphere formation (see Supplemental Methods for description of the validation of clonality), sphere total volume, and proliferation rate (the inverse of doubling time). As anticipated, doubling time was inversely correlated with sphere total volume (linear regression, P = .0182, Supplementary Fig. S3). However, doubling time was not significantly correlated with clonal sphere formation, suggesting that clonal sphere formation is not merely a reflection of overall proliferation. None of the 3 gliomasphere characteristics was associated with TCGA classification, EGFR mutational status, or with one of the 2 clusters (mesenchymal and nonmesenchymal) identified by unsupervised clustering. However, if we categorize the gliomaspheres based on the 3 main MDS groups observed (Fig. 1J), then group 3 (the proneural subgroup in the red oval) has the slowest proliferation (largest doubling time) (Fig. 1K).
We next correlated gene expression levels with the 3 gliomasphere phenotypes, as shown in Supplementary Table S4. For example, KLHL9 expression was inversely correlated with sphere formation. Utilizing TFacts (www.TFacts.org), a tool that predicts activation or inhibition of transcription factors through gene expression changes, we identified a significant association between genes correlated to sphere formation and 3 activated transcription factors: HIF1A, TCF7, and CTNNB1 (beta catenin) (Table 2). These results were supported by an ingenuity analysis (www.ingenuity.com), which generated a list of canonical pathways associated with sphere formation including HIF1A, Wnt/beta catenin, and other pathways associated with tumor initiation such as PI3K, EGF, mTOR, and ERK (Supplementary Table S5). PI3Kinase, mTOR, beta catenin, and ERK pathways were associated with both sphere formation and sphere total volume (Supplementary Table S5).
Table 2.
Transcription factors associated* with genes whose expression correlates with phenotypic outcome of GBM gliomaspheres
| Outcome | Factor | Activated | P value | E value | Q value | FDR control (B-H) | Intersection | Target genes |
|---|---|---|---|---|---|---|---|---|
| % Sphere formation | HIF1A | Activated | <.001 | <0.001 | <0.001 | 0.000455 | 14 | 22 |
| % Sphere formation | TCF7 | Activated | .00015 | 0.0165 | 0.00825 | 0.000909 | 9 | 17 |
| % Sphere formation | CTNNB1 | Activated | .00042 | 0.0462 | 0.0154 | 0.00136 | 62 | 300 |
| Proliferation Rate | TP53 | Activated | <.001 | 0.0108 | 0.0108 | <0.001 | 11 | 97 |
TFacts signed analysis of our list of significant genes (P < .001) associated with the in vitro phenotypic outcomes listed. The E-value takes into account the significance of repeated comparisons and the likelihood of the observed value being different from the expected value. The Q value is the measurement of the false discovery rate. We chose transcription factors as significant if the E value <0.05. The intersection depicts how many genes from our list coincide with the known target genes of the transcription factor. Target genes denote the number of genes that are known targets of the indicated transcription factor.
*Tfacts Signed Analysis (www.Tfacts.org, de Duve Institute).
Ingenuity analysis determined overrepresented pathways from genes associated with proliferation rate (P < .001) that included several proliferation-associated pathways such as HIF1A and p70S6K (Supplementary Table S5). TFacts analysis also indicated that activated TP53 was associated with proliferation rate (Table 2).
Another method of analyzing gene expression is via weighted gene correlation network analysis (WGCNA), which identifies modules of tightly coexpressed genes across multiple conditions or samples. We performed a signed WGCNA analysis of the gene expression for the gliomasphere cultures to determine coregulated modules of genes that were associated with phenotype (Fig. 2A). Sphere formation was most highly associated with the orange-red module (Fig. 2A) comprising 37 genes. Several pathways, as revealed by Ingenuity analysis, were overrepresented within this group of genes including protein kinase A (PRKACA), mTOR, and integrin signaling (Fig. 2B). Both sphere total volume and doubling time were most highly associated with the dark orange module (Fig. 2A). Thus, the dark orange module is inversely correlated to both outcomes of proliferation and may be assumed to be associated with lack of proliferation. Ingenuity analysis of the dark orange module revealed associated canonical pathways including cAMP signaling, and protein kinase A (Fig. 2C). Although the ingenuity analysis of the modules did not signify whether the protein kinase A pathway was activated or inactivated, protein kinase A (PRKACA) itself was directly associated with sphere total volume in the gene-by-gene analysis (Supplementary Table S4). This suggests that the protein kinase A pathway activation is associated with proliferation.
Fig. 2.
Weighted gene co-expression network analysis (WGCNA) of in vitro phenotypes. (A) WGCNA analysis of 71 glioblastoma (GBM) gliomasphere cultures identified 40 modules of coexpressed genes. The module eigengene, representing the expression changes of all the genes in the module, was associated with phenotypic characteristics. The heat-map depicts the Pearson correlation between module eigengens and trait according to the scale on the side P values for each association are in each cell. The orange-red and dark orange modules are outlined in red. (B) Significant (P <.05) canonical pathways associated with the orange-red module, which is associated with sphere formation. The bars represent the –log(P value) from a Fisher exact test. Yellow line:.05 P value threshold. The orange graphing line with the orange box points represents the ratio of the number of genes in the pathway from our list divided by the total number of genes in the pathway. Grey bars represent no activity pattern available for the pathway, and white bars indicate the Z-score = 0, no activation, or inactivation of the pathway. (C) Significant (P < .05) canonical pathways associated with the dark orange module that is associated with both proliferation (sphere doubling time) and sphere total volume.
Aggressive in Vitro Phenotype and Survival Outcome in TCGA
We compiled a list of 89 genes that were significantly associated with aggressive phenotype in all 3 in vitro outcomes (Supplementary Table S6). We tested whether this list was enriched for genes that were also associated with poor patient survival using the TCGA database. We found that 37 of the 89 genes were associated with survival outcome in TCGA, a proportion that is significantly enriched compared with all genes (P < .05). When we used a stringent FDR of P < .05, 5 genes were associated with survival outcome in TCGA: MDK, PLAT, HEATR2, LGALS8, EFEMP2. These 5 genes were all significantly associated with poor survival in the TCGA database as depicted by Kaplan-Meier survival curves (Fig. 3A and B, Supplementary Fig. S4A–C). Since the entire list of 12 042 genes in TCGA only harbored 12 genes significantly associated with patient outcome with a FDR (P < .05), our determination of these 5 genes associated with survival in our database was highly significant (Fisher exact test, P = 1.43 × 10−7). These data indicate that the genes in our list, which is consistently associated with aggressive in vitro phenotypes in gliomaspheres, is enriched for genes associated with poor survival in TCGA data collected on tumor tissue.
Fig. 3.
EFEMP2 and LGALS8 play functional roles in promoting glioblastoma (GBM) proliferation and are associated with poor survival in the Cancer Genome Atlas (TCGA). (A and B) TCGA Kaplan-Meier curves for EFEMP2 (A) and LGALS8 (B) FDR P values are shown. (C) Confirmation of knockdown of EFEMP2 and LGALS8 by qRT-PCR in HK296 GBM cells. Relative expression of EFEMP2 and LGALS8 as indicated on the Y-axis are each normalized to GAPDH. shEFEMP2 depicts standard error bars for 2 biological replicates (mean = 0.238, ±0.0276 SD.). shLGALS8 depicts results for one biological replicate (mean = 0.653). (D) shEFEMP2 inhibits cell proliferation. Estimation of cell numbers of HK296 GBM cell cultures after 6 days of proliferation with control cells or shRNA-mediated knockdown of EFEMP2 or LGALS8. Mean values, normalized to Ctrl are depicted ± SEM. Each of the 3 biological replicates represents 16 technical replicates. shEFEMP2 is significantly lower than control (paired t test, P = 00166, N = 3). shLGALS8 is not significantly lower than control (paired t test, P = .0611 N = 3). (E) shEFEMP2 and shLGALS8 inhibit clonal sphere formation. Graph of HK296 GBM cell cultures after 15 days of clonal sphere formation with control cells, or knockdown of EFEMP2, or LGALS8. Mean values of % sphere formation are depicted ± SEM. Each of the 3 biological replicates represents the mean of 20 technical replicates. Sphere formation for shEFEMP2 (mean = 12.7 ± 11) is significantly lower than control (paired t test, P = .0069, N = 3). shLGALS8 is significantly lower than control (paired t test, P = .0043, N = 3). F. shEFEMP2 and shLGALS8 inhibit clonal sphere total volume. Graph of HK296 GBM cell cultures after 15 days of clonal sphere formation with control cells or knockdown of EFEMP,2 or LGALS8. Mean values of clonal sphere total volume in uM3 are depicted ± SEM. shEFEMP2 is significantly lower than control (paired t test, P = .0029, N = 3). shLGALS8 is significantly lower by than control (paired t test, P = .0037, N = 3).
Two of the 5 genes associated with both aggressive in vitro phenotype and tumor malignancy have already been shown to regulate GBM proliferation: MDK and PLAT.24,25 The remaining 3 genes (HEATR2, EFEMP2, and LGALS8) are novel candidates of potential interest. EFEMP2 is interesting because it is associated with both MDK and PLAT in an association matrix (STRING-db.org), indicating that these 3 genes may interact (Supplementary Fig. S4D).
We chose to further study EFEMP2 and LGALS8. Lentiviral shRNA infection resulted in mRNA depletion of either EFEMP2 or LGALS8 (Fig. 3C). EFEMP2 depletion resulted in a 66% decrease in cell number after 6 days in culture (Fig. 3D). Depletion of LGALS8 reduced cell proliferation by 53.2% but did not reach statistical significance over this brief period of time. Depletion of EFEMP2 and LGALS8 in HK296 GBM cells resulted in 85% and 58% reduction, respectively, in clonal sphere formation (Fig. 3E). The change in sphere formation resulted in an 89% decrease in sphere total volume after EFEMP2 knockdown and an 86% decrease after LGALS8 knockdown (Fig. 3F). These results demonstrate that EFEMP2 and LGALS8 play functional roles in promoting GBM tumor cell proliferation and clonal sphere formation. Similar results were obtained in the HK308 GBM cell culture (Supplementary Fig. S5).
Gliomasphere Genes Associated with Patient Survival
Next, we focused our analysis on factors associated with the survival of patients included in our own dataset. Neither the gliomasphere TCGA classification (Supplementary Table S7 and Supplementary Fig. S6A) nor the mesenchymal versus nonmesenchymal unsupervised clustering classification of gliomaspheres was significantly associated with patient survival, although we observed a nonsignificant trend for poorer survival in the mesenchymal cluster (Supplementary Fig. S6B). EGFR or PTEN mutation status of gliomaspheres was also not significantly associated with patient outcome in our cohort (Supplementary Table S7). Factors that were associated with poorer survival in our study were recurrent tumors, male sex, and age (Supplementary Fig. S7A,B and Supplementary Table S7).
We analyzed various outcomes for associations with patient survival in our gliomasphere database using Cox regression and Kaplan-Meier analysis. We found 22 genes that were significantly associated with patient outcome (FDR P < .05, Supplementary Table S8). To validate these findings, we searched the Repository for Molecular Brain Neoplasia Data (REMBRANDT) dataset. Ten of the 22 genes identified in our study were also significantly associated (P < .05) with patient outcome in REMBRANDT glioma cases. However, only 7 of these 10 genes had consistent (sign-sensitive) associations in both databases (Supplementary Table S8). Of the genes with consistent associations, 57% (4 of 7) were also significantly (P < .05) associated with outcome in the REMBRANDT database of tumor expression within the restricted subpopulation of glioblastoma (Supplementary Table S8). Thus, we have identified 4 genes that are significantly associated with patient outcome in a robust manner, both in GBM-derived gliomaspheres (Fig. 4A–D) and in GBM tumors themselves (Supplementary Fig. S8).
Fig. 4.
PTGR1 plays a functional role in impairing glioblastoma (GBM) proliferation. (A–D) Kaplan-Meier survival curves from our gliomasphere dataset of gene expression in gliomasphere cultures and patient survival for 4 genes associated with survival in both our gliomasphere dataset and in the REMBRANDT dataset for glioblastoma (P values shown are for the FDR of the Mantel-Haenszel test). (E) Western blot confirmation of knockdown. (F) Normalized sphere formation percentage displays a significant increase in sphere formation upon knockdown of PTGR1 in HK248 GBM cell culture (paired t test, P = .0062, N = 4). Cells were grown at clonal density (50 cells/well) for 21 days.
A gene's association with patient outcome was not necessarily reflected in its association with in vitro gliomasphere phenotype. For example, higher expression of MAP2K5 is associated with shorter survival, but MAP2K5 levels are associated with a slower proliferation rate of the gliomasphere cultures. VTI1A, whose expression is associated with better patient survival, is also significantly associated with a faster proliferation rate of the gliomasphere cultures. Thus, a complex system exists wherein gene expression associated with patient outcome is not strictly related to gliomasphere proliferation in a simple, predictive manner.
PTGR1 has a complex phenotypic characterization. Underexpression of PTGR1, as compared with the mean, is associated with poor survival in our gliomasphere dataset (Fig. 4B). However, PTGR1 expression in gliomaspheres is consistently related to the aggressive phenotype: a faster rate of proliferation, greater sphere total volume, and increased sphere formation. We knocked down PTGR1 in GBM cell culture HK248 (Fig. 4E) and assayed sphere formation under clonal conditions. After 21 days of growth, PTGR1 depletion resulted in a 4.8-fold increase in mean percentage sphere formation (Ctrl = 4.76%, shPTGR1 = 22.3%) (Fig. 4F). These significant results demonstrate that PTGR1 plays a functional role in impeding GBM tumor cell proliferation and sphere formation.
Discussion
In determining the relevance of the TCGA groupings to gliomasphere cultures, we obtained mixed results. We found that some features of the TCGA categorization were robust and retained in cultures. For example, TCGA classification was retained over long-term passaging in culture, all our IDH mutant cultures were categorized as being proneural, and 6 out of 7 gliosarcoma cultures were categorized as mesenchymal. These classifications are consistent with what is expected for these tumors.15,26 Interestingly, we did not find association with EGFR amplification or EGFRvIII rearrangement with any of the categorization schemes of the cultures. In the original Verhaak et al study, EGFR amplification was found predominantly but was not restricted to the classical group.15
We found that there is a limited relationship between gliomaspheres and their parent tumors, especially as it relates to TCGA subgroups. There was a better correlation between a subset of gliomaspheres and their tumors when we asked what genes were significantly expressed in these pairs compared with all the other samples. However, the majority of samples did not reveal a high level of correlation. A possible explanation for these differences in gene expression may be that brain tumor samples contain many other cells besides GBM cells, but we are selecting for GBM cells when culturing tumor cells. Another important factor lies in which particular cells are being propagated in our cultures. Single brain tumors contain a variety of cells with different gene expression patterns and exhibiting heterogeneity in TCGA classifications.27 A subset enriched by culturing tumor cells may represent tumor-initiating cancer stem cells. However, the cells selected may also not tell the entire story of the tumor. Our cultures are nonclonal, avoiding the bias of clonal selection and providing a gene expression pattern more consistent with the heterogeneity of the tumor. It is also important to consider the possibility that gene expression differences between the tumor and the cultures are based simply on the act of culturing itself. However, our analysis did not reveal many genes that were consistently different between gliomaspheres and parent tumors, suggesting that tissue culture artifact is not the principle driving force for their gene expression differences.
In addition to utilizing TCGA subgroupings, we also utilized unbiased means to categorize cultures according to their gene expression. Similar to another study,26 our hierarchical clustering revealed 2 groups. However, Bhat et al characterized the groups as being mesenchymal and proneural, while our findings indicate that the groups are divided into generally mesenchymal and nonmesenchymal. The third group revealed by MDS, containing proneural samples, had significantly slower in vitro proliferation suggesting that unbiased the MDS classification may distinguish biologically relevant subgroups that the other classification systems (TCGA and Clustering) missed. These results are consistent with a recent study of 20 gliomasphere cultures that distinguished 2 clusters including a predominately proneural subtype (ie, the stem-like group) associated with slower sphere formation rates and longer survival upon xenograft transplantation.28
We employed 2 methods of gene expression-phenotype correlation: gene-by-gene correlation analysis and WGCNA analysis. While not identical in their findings, the 2 methods did appear to reinforce each other. We distinguished associations between proliferation and canonical pathways that bind in vitro phenotypes with established pathways that play essential roles in tumor biology and malignancy. The data, from both WGCNA and gene-by-gene correlation, support the hypothesis that protein kinase A signaling is directly associated with proliferation, which was consistent with a previous study.29 KLHL9 expression was inversely correlated with sphere formation, consistent with a previous study reporting that KLHL9 expression reduced tumor viability in glioblastoma.30
Finally, we utilized a multistep approach to determine whether we could discover novel, functionally relevant targets for future research and possible therapy. As gliomasphere cultures contain a pure set of glioma cells, we reasoned that genes associated with phenotypes of these cultures and with patient survival would have a high degree of functional significance. Both EFEMP2 and LGALS8 facilitated proliferation and clonal sphere formation of GBM gliomaspheres. Our data are consistent with a previous report that EFEMP2 (also known as MBP1) promotes proliferation and transformation in rat embryonic fibroblasts.31 While this manuscript was under review for publication, a new study was published that demonstrated a role for EFEMP2 in proliferation of glioma.32 Galectin 8, the product of LGALS8, has been shown to have a role in the proliferation of tumor cells, although its role is inconsistent (either inducing or inhibiting proliferation in different tumor lines).33 Interestingly, both EFEMP2, an extracellular matrix protein, and LGALS8 (a galectin) are secreted proteins. Therefore, they may serve as suitable biomarkers for GBM tumor proliferation and may be amenable to pharmacologic intervention.
PTGR1, or prostaglandin reductase 1, is involved in the inactivation of some prostaglandins as well as maintaining redox balance in the cell,34 although a role in brain tumors has not been described. PTGR1 expression was associated with all 3 outcomes of aggressive in vitro phenotype. Yet, in the gliomasphere dataset, underexpression of PTGR1 was associated with poor survival, consistent with a role for PTGR1 as an inhibitor of proliferation. Indeed, our experimental data indicate that PTGR1 has a functional role in suppressing GBM sphere formation. Our data support the need for more study of PTGR1 and indicate that correlations between genes and in vitro phenotypes do not always predict their functional roles.
In summary, our data validate the gliomasphere system as a suitable model of fundamental molecular pathways involved in GBM biology. Gliomaspheres demonstrate in vitro phenotypes, which are associated with the canonical pathways involved in GBM tumor biology. Moreover, the gliomasphere model provides a unique platform for the discovery of novel genes involved in proliferation and may be informative for distinguishing important molecular determinants of malignancy.
Supplementary Material
Funding
National Institutes of Neurological Disorders and Stroke (NINDS) grant (NS052563), The Dr. Miriam and Sheldon G. Adelson Medical Research Foundation, California Institute of Regenerative Medicine (CIRM) Training Grant (TG2-01169), and a Whitcome Fellowship.
Supplementary Material
Acknowledgments
We acknowledge the support of the NINDS Informatics Center for Neurogenetics and Neurogenomics (P30 NS062691). Support was provided by the UCLA Jonsson Comprehensive Cancer Center Molecular Screening Shared Resource.
References
- 1. Reynolds BA, Weiss S. Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science. 1992;255 (5052):1707–1710. [DOI] [PubMed] [Google Scholar]
- 2. Ignatova TN, Kukekov VG, Laywell ED, Suslov ON, Vrionis FD, Steindler DA. Human cortical glial tumors contain neural stem-like cells expressing astroglial and neuronal markers in vitro. Glia. 2002;39 (3):193–206. [DOI] [PubMed] [Google Scholar]
- 3. Hemmati HD, Nakano I, Lazareff JA et al. Cancerous stem cells can arise from pediatric brain tumors. Proc Natl Acad Sci USA. 2003;100 (25):15178–15183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Singh SK, Clarke ID, Terasaki M et al. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63 (18):5821–5828. [PubMed] [Google Scholar]
- 5. Laks DR, Visnyei K, Kornblum HI. Brain tumor stem cells as therapeutic targets in models of glioma. Yonsei Med J. 2010;51 (5):633–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414 (6859):105–111. [DOI] [PubMed] [Google Scholar]
- 7. Pardal R, Clarke MF, Morrison SJ. Applying the principles of stem-cell biology to cancer. Nat Rev Cancer. 2003;3 (12):895–902. [DOI] [PubMed] [Google Scholar]
- 8. Jordan CT, Guzman ML, Noble M. Cancer stem cells. N Engl J Med. 2006;355 (12):1253–1261. [DOI] [PubMed] [Google Scholar]
- 9. Nakano I, Kornblum HI. Brain tumor stem cells. Pediatr Res. 2006;59 (4 Pt 2):54R–58R. [DOI] [PubMed] [Google Scholar]
- 10. Galli R, Binda E, Orfanelli U et al. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res. 2004;64 (19):7011–7021. [DOI] [PubMed] [Google Scholar]
- 11. Singh SK, Hawkins C, Clarke ID et al. Identification of human brain tumour initiating cells. Nature. 2004;432 (7015):396–401. [DOI] [PubMed] [Google Scholar]
- 12. Lee J, Kotliarova S, Kotliarov Y et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9 (5):391–403. [DOI] [PubMed] [Google Scholar]
- 13. Phillips HS, Kharbanda S, Chen R et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9 (3):157–173. [DOI] [PubMed] [Google Scholar]
- 14. TCGA. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455 (7216):1061–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Verhaak RG, Hoadley KA, Purdom E et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17 (1):98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Noushmehr H, Weisenberger DJ, Diefes K et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17 (5):510–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Laks DR, Masterman-Smith M, Visnyei K et al. Neurosphere formation is an independent predictor of clinical outcome in malignant glioma. Stem Cells. 2009;27 (4):980–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sarkaria JN, Yang L, Grogan PT et al. Identification of molecular characteristics correlated with glioblastoma sensitivity to EGFR kinase inhibition through use of an intracranial xenograft test panel. Mol Cancer Ther. 2007;6 (3):1167–1174. [DOI] [PubMed] [Google Scholar]
- 19. Nathanson DA, Gini B, Mottahedeh J et al. Targeted therapy resistance mediated by dynamic regulation of extrachromosomal mutant EGFR DNA. Science. 2014;343 (6166):72–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4:Article17. [DOI] [PubMed] [Google Scholar]
- 21. Langfelder P, Zhang B, Horvath S. Defining clusters from a hierarchical cluster tree: the Dynamic Tree Cut package for R. Bioinformatics. 2008;24 (5):719–720. [DOI] [PubMed] [Google Scholar]
- 22. Meyer M, Reimand J, Lan X et al. Single cell-derived clonal analysis of human glioblastoma links functional and genomic heterogeneity. Proc Natl Acad Sci USA. 2015;112(3):851–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Subramanian A, Tamayo P, Mootha VK et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102 (43):15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Luo J, Wang X, Xia Z et al. Transcriptional factor specificity protein 1 (SP1) promotes the proliferation of glioma cells by up-regulating midkine (MDK). Mol Biol Cell. 2015;26 (3):430–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yamashita D, Kondo T, Ohue S et al. miR340 suppresses the stem-like cell function of glioma-initiating cells by targeting tissue plasminogen activator. Cancer Res. 2015;75 (6):1123–1133. [DOI] [PubMed] [Google Scholar]
- 26. Bhat KP, Balasubramaniyan V, Vaillant B et al. Mesenchymal differentiation mediated by NF-kB promotes radiation resistance in glioblastoma. Cancer Cell. 2013;24 (3):331–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Patel AP, Tirosh I, Trombetta JJ et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344 (6190):1396–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cusulin C, Chesnelong C, Bose P et al. Precursor states of brain tumor initiating cell lines are predictive of survival in xenografts and associated with glioblastoma subtypes. Stem Cell Reports. 2015;5 1:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Feng H, Hu B, Vuori K et al. EGFRvIII stimulates glioma growth and invasion through PKA-dependent serine phosphorylation of Dock180. Oncogene. 2014;33 (19):2504–2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chen JC, Alvarez MJ, Talos F et al. Identification of causal genetic drivers of human disease through systems-level analysis of regulatory networks. Cell. 2014;159 (2):402–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gallagher WM, Argentini M, Sierra V, Bracco L, Debussche L, Conseiller E. MBP1: a novel mutant p53-specific protein partner with oncogenic properties. Oncogene. 1999;18 (24):3608–3616. [DOI] [PubMed] [Google Scholar]
- 32. Wang L, Chen Q, Chen Z et al. EFEMP2 is upregulated in gliomas and promotes glioma cell proliferation and invasion. Int J Clin Exp Pathol. 2015;8 (9):10385–10393. [PMC free article] [PubMed] [Google Scholar]
- 33. Ruiz FM, Scholz BA, Buzamet E et al. Natural single amino acid polymorphism (F19Y) in human galectin-8: detection of structural alterations and increased growth-regulatory activity on tumor cells. FEBS J. 2014;281 (5):1446–1464. [DOI] [PubMed] [Google Scholar]
- 34. Sanchez-Rodriguez R, Torres-Mena JE, De-la-Luz-Cruz M et al. Increased expression of prostaglandin reductase 1 in hepatocellular carcinomas from clinical cases and experimental tumors in rats. Int J Biochem Cell Biol. 2014;53:186–194. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.