Abstract
Background
In glioblastoma (GBM), Id1 serves as a functional marker for self-renewing cancer stem-like cells. We investigated the mechanism by which cyclooxygenase-2 (Cox-2)-derived prostaglandin E2 (PGE2) induces Id1 and increases GBM self-renewal and radiation resistance.
Methods
Mouse and human GBM cells were stimulated with dimethyl-PGE2 (dmPGE2), a stabilized form of PGE2, to test for Id1 induction. To elucidate the signal transduction pathway governing the increase in Id1, a combination of short interfering RNA knockdown and small molecule inhibitors and activators of PGE2 signaling were used. Western blotting, quantitative real-time (qRT)-PCR, and chromatin immunoprecipitation assays were employed. Sphere formation and radiation resistance were measured in cultured primary cells. Immunohistochemical analyses were carried out to evaluate the Cox-2-Id1 axis in experimental GBM.
Results
In GBM cells, dmPGE2 stimulates the EP4 receptor leading to activation of ERK1/2 MAPK. This leads, in turn, to upregulation of the early growth response1 (Egr1) transcription factor and enhanced Id1 expression. Activation of this pathway increases self-renewal capacity and resistance to radiation-induced DNA damage, which are dependent on Id1.
Conclusions
In GBM, Cox-2-derived PGE2 induces Id1 via EP4-dependent activation of MAPK signaling and the Egr1 transcription factor. PGE2-mediated induction of Id1 is required for optimal tumor cell self-renewal and radiation resistance. Collectively, these findings identify Id1 as a key mediator of PGE2-dependent modulation of radiation response and lend insight into the mechanisms underlying radiation resistance in GBM patients.
Keywords: Cox-2, PGE2, Id1, glioblastoma, radioresistance
Cyclooxygenase-2 (Cox-2) is an inducible enzyme that plays a key role in the production of the bioactive lipid prostaglandin E2 (PGE2).1–3 Secreted PGE2 can act in a paracrine manner by binding to ≥1 of a set of 4 G-protein coupled receptors known as EP1-44 that mediate the effects of PGE2 by activating specific downstream effector pathways in a cell-type specific manner.5–8
The Cox-2/PGE2 pathway plays an important role in maintaining tissue-resident stem cell populations.9 PGE2 functions in hematopoietic stem cell (HSC) homeostasis in both embryonic development and in the adult organism.9–11 The cancer stem cell hypothesis argues that tumors contain subpopulations of cells with stem-like qualities that can maintain tumor growth potential and reconstitute the tumor after transplantation.12,13 The Cox-2/PGE2 signaling pathway plays an important role in several human cancers, and Cox-2 is overexpressed in multiple tumor types14,15 including glioblastoma (GBM).16 Cox-2 can be induced by oncogenes, growth factors, and proinflammatory mediators in a variety of cell types including macrophages.14 In the brain, specialized glial cells known as microglia function as resident macrophages and are activated in response to brain injury.17 Microglia have been demonstrated to produce PGE2 and to infiltrate GBM tumors.18
A subpopulation of cells can be purified from human GBMs that express various stem cell markers and show increased self-renewal capacity.19 The GBM stem-like population was also enriched in patients after treatment with ionizing radiation (IR).20 Additionally, therapies targeting the stem-like population have shown therapeutic efficacy when coupled with radiation in mouse xenograft models.21
Id genes (Id1-4) are negative regulators of basic helix-loop-helix factors with important roles in embryonic development, stem cell homeostasis, and tumor progression.22,23 We previously investigated the role of Id1 in a mouse model of GBM24 using an Id1-Venus-YFP knock-in allele that allowed for sorting of Id1-positive cells. These cells showed increased expression of a variety of cancer stem cell markers, displayed strong self-renewal capacity, and generated tumors that recapitulated the histopathology of the original tumor when transplanted into the brains of recipient mice.
The ability of PGE2 to regulate tissue resident stem cell homeostasis, combined with the increased expression of Cox-2 in some human patients, raises the intriguing possibility that PGE2 functions in part to maintain the cancer stem-like cell population and preserve self-renewal capacity within GBM. Furthermore, PGE2 has been shown to induce Id1 transcription in cultured breast cancer cell lines,25 and recent work has demonstrated that forced overexpression of Cox-2 in human GBM cell lines results in increased Id1 expression and cellular invasiveness.26 Here, we sought to investigate the mechanistic basis for Id1 upregulation by Cox-2/PGE2 and to explore the ability of PGE2 signaling to regulate GBM stem cell character. We present evidence that Cox-2-derived PGE2 can induce Id1 in a GBM mouse model through the EP4-ERK1/2 MAPK pathway. Importantly, we demonstrate for the first time that this pathway promotes self-renewal capacity and resistance to radiation in an Id1-dependent manner, a finding with important therapeutic implications for the treatment of human disease.
Materials and Methods
In Vivo Models
Nestin-tva, Arf −/− mice27 and Id1 −/− mice28 have been previously described. Briefly, 4–8 week old mice were anesthetized and administered replication competent avian leukosis virus long terminal repeat with splice-acceptor (RCAS) virus expressing the platelet-derived growth factor B (PDGFB) oncogene via stereotactic intracranial injection at −1 mm/−1 mm from bregma. This was performed in Id1+/+, Id3+/+, Nestin-TVA+, Arf −/− and Id1 −/− , Id3 +/− Nestin-TVA+, Arf −/− animals.24 Mice were monitored until they became symptomatic, at which point they were sacrificed and their tumors harvested. For in vivo experiments utilizing celecoxib, animals were administered 100 mg/kg/day celecoxib (gift from NCI Division of Cancer Prevention) or vehicle via oral gavage over a 4 day time course prior to sacrifice. All mouse experiments were approved by MSKCC's Institutional Animal Care and Use Committee.
Short Interfering RNA Knockdown
Predesigned short interfering (si)RNAs targeting human EP2/4 and scrambled control siRNAs were purchased from Qiagen. Two separate siRNAs targeting different sequences within EP2/4 were used to transfect U87 cells using Lipofectamine 2000 (Invitrogen) versus scrambled control siRNA. Cells were harvested 48 hours post-transfection for analysis of protein and RNA levels.
Immunostaining
Whole brains were fixed in 4% paraformaldehyde and dehydrated in sucrose prior to embedding in optimal cutting temperature compound (Sakura) and sectioning (10 µm) using a Leica Cryostat. Antigen retrieval was performed by boiling slides in 10 mM sodium citrate buffer, followed by blocking/permeabilization in phosphate-buffered saline (PBS), 10% normal goat serum, and 0.5% TritonX-100. Primary antibody incubation was followed by incubation with an appropriate fluorescent secondary antibody (Alexa 488 or Alexa 568 anti-rabbit or anti-mouse [Invitrogen]) for 1 hour and counterstaining with 5 µg/mL DAPI (Invitrogen). As a control for Cox-2 immunostaining, we pre-incubated the Cox-2 primary antibody with a Cox-2 blocking peptide (Cayman Chemical) for 1 hour prior to tissue incubation. The following primary antibodies were used: Id1 (Biocheck), Cox-2 (Cayman Chemical), CD105 (AbCam), cleaved caspase 3 (Cell Signaling Technology), γ-H2AX (Millipore), and Ki67 (Dako). Images were acquired with a Zeiss Axioplan2 Imaging widefield microscope using the AxioVision software (Zeiss).
Cell Culture
Tumors were digested using papain (Worthington Biochemical), triturated, and neutralized with ovomucoid (Worthington Biochemical) according to the manufacturer's instructions. Resulting cells were seeded into tissue culture plates precoated with 10 µg/mL Laminin (Sigma) in NeuroCult stem cell basal medium with proliferation supplements, 20 ng/mL EGF, 10 ng/mL FGF, and 2 µg/mL heparin (Stem Cell Technologies). U87, LN-229, and U118 cells were acquired from ATCC and maintained in Eagle's minimum essential medium (ATCC) supplemented with 10% fetal bovine serum (FBS; ATCC), Dulbecco's modified Eagle medium (DMEM, ATCC) supplemented with 5% FBS (ATCC), or DMEM (ATCC) supplemented with 10% FBS (ATCC), respectively.
Western Blotting
Fifteen micrograms of protein were separated on 10% acrylamide/bisacrylamide gels and transferred to PVDF membranes. The following primary antibodies were used: β-actin (Sigma), Id1 (Biocheck), Cox-2 (Cayman Chemical), phospho-ERK1/2 (Cell Signaling Technology), ERK1/2 (Cell Signaling Technology), Egr1 (Cell Signaling Technology), cleaved caspase 3 (Cell Signaling Technology), cleaved Parp-1 (Cell Signaling Technology), and phospho-ATM (Cell Signaling Technology).
ELISA Assay
ELISA assays were performed on conditioned medium using a PGE2 EIA kit (Cayman Chemical) according to the manufacturer's instructions.
Chromatin Immunoprecipitation
Cells were fixed with 1% formaldehyde at room temperature for 10 minutes. Fixation was stopped with 0.125 M glycine, and cells were lysed in a solution containing 1% SDS, 0.5% Empigen BB, 10 mM EDTA, and 50 mM Tris/HCL pH 7.4. Soluble chromatin was generated by sonication, and individual immunoprecipitations were performed overnight at 4°C using 1.5 µg of either Egr1 antibody (Cell Signaling Technology) or normal rabbit IgG (Santa Cruz Biotechnology). From each sample, 10% total volume was retained as input. Antibody-chromatin complexes were isolated using Protein-A Sepharose beads (Sigma), washed, and eluted in buffer containing 1% SDS and 0.1 M NaHCO3. ChIP DNA and 10% input DNA were purified via phenol-chloroform extraction followed by ethanol precipitation. The following PCR primers were used to amplify the purified DNA via quantitative PCR: Egr1 binding site primers – Forward: 5′- AGAATGCTCCAGCCCAGTTT – 3′, Reverse: 5′ – ACCCCCTCCCTTTCCTTT – 3′, Upstream control primers – Forward: 5′ – AGGATCCCTGCCAAGCTAAT – 3′, Reverse: 5′ – TGCCTGGATTTTGCTACACA – 3′.
Quantitative Real-time PCR
Total RNA was extracted from cells in culture using an RNAEasy Kit according to the manufacturer's instructions (Qiagen), and cDNA synthesis was performed using SuperScript II (Invitrogen). Assays were performed using SYBR Green PCR Master Mix (Applied Biosystems) with a 7900HT Fast Real Time PCR System. Predesigned primers were purchased from Qiagen (Id1, EP1-4, β-Actin). Gene transcript levels were calculated using the ΔΔCt method.
Tumorsphere Assays
Tumors were digested using papain (Worthington Biochemical), triturated, neutralized with ovomucoid and seeded into 24-well ultra-low adhesion tissue culture plates at clonal density29 (0.5 cells/µL, 500 cells/1 mL/well) in NeuroCult stem cell basal medium with proliferation supplements, 20 ng/mL EGF, 10 ng/mL FGF, and 2 µg/mL heparin (Stem Cell Technologies). Sphere formation was quantitated after 4–7 days depending on passage number. The total number of spheres was manually counted for each well and divided by the number of cells initially plated to determine the percentage of sphere-forming cells in the starting population.
Mass Spectrometry
Frozen tissue samples were quickly homogenized in the mortar of a clean Tenbroeck homogenization apparatus along with PGE2-d4 and 4 mL 1:1 methanol:PBS. A portion of the homogenate was dried under nitrogen and diluted and acidified such that the pH < 3.5 and the organic component of the final solution was approximately 10% by volume.
For solid phase extraction, the dilute sample was loaded onto Phenomenex C18 SPE cartridges (6 cc, 200 mg) that were conditioned with 4 mL methanol followed by 4 mL 0.5% acetic acid (aq). The SPE cartridges were rinsed with 4 mL 0.5% acetic acid (aq), then 12% methanol in 0.5% acetic acid (aq), dried at full vacuum for 1-2 minutes, and then eluted with 2 mL methanol. Samples were then dried and stored at −20°C.
For liquid-liquid extraction, 4.5 mL of ethyl acetate with 10% cyclohexane was added to the acidified, dilute sample. This was mixed well and centrifuged for 10 minutes at 4C and 3000 rpm. The, upper layer was removed, dried, and stored at −20°C. Immediately prior to analysis, samples were reconstituted in 100 µL methanol and 80 µL H2O and analyzed via LC-MS. All solvents were HPLC grade and purchased from Thermo Fisher Scientific.
PGE2 was chromatographed on a Supelco C18 column (50 × 2 cm, with a C18 guard cartridge) using the following gradient: 20%B to 65%B over 4 minutes followed by a 1.0 minute hold at 95%B. The column was NeuroCult stem cell basal medium with proliferation supplements re-equilibrated at 20%B for at least 2 minutes prior to each injection. Component A was water, and component B was acetonitrile, with each component containing 0.025% formic acid. C18 SPE cartridges (6 cc, 200 mg) were obtained from Phenomenex Corp.
The analytes were detected on a Thermo Vantage triple quadrupole mass spectrometer via selected reaction monitoring. The following SRM transitions were employed: PGE2 (m/z 351Ù271); PGE2-d4 (m/z 355Ù275). Unknown samples were quantitated via stable isotope dilution against the tetra-deuterated internal standard.
Statistics
Unpaired t test and 1-way ANOVA were performed on relevant datasets using GraphPad Prism 6 software. Where appropriate, Tukey's test was used for post hoc analysis of ANOVA. For all analyses, P values <.05 were considered significant. Data are shown as mean ± SEM.
Results
Cox-2-derived PGE2 Induces Id1 in Glioblastoma
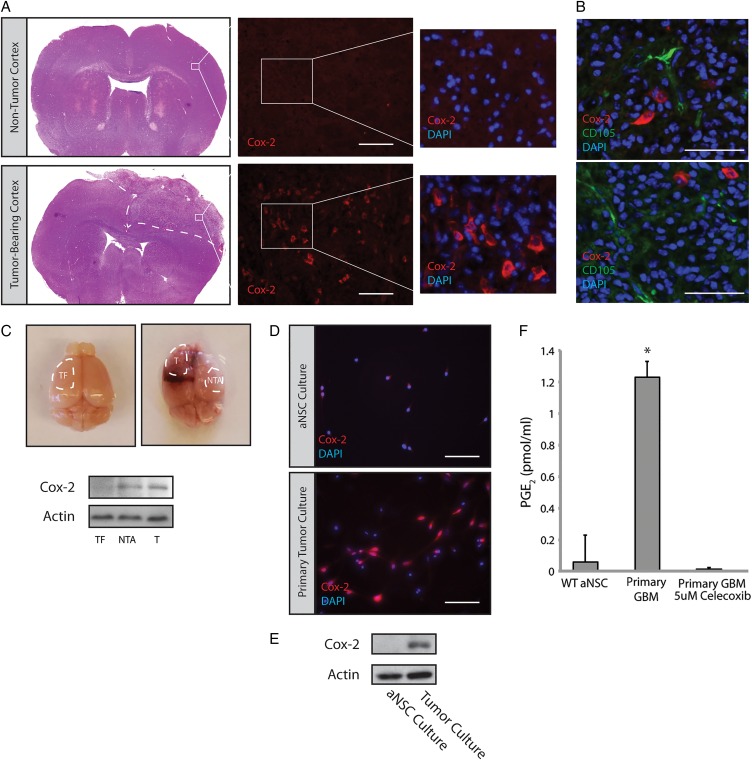
To investigate the role of Cox-2 in GBM, we utilized the RCAS-Tva system to generate mouse brain tumors. Previous studies have demonstrated that these tumors are very similar to human GBM in regards to histopathology and gene expression profiling.27 Tumors generated in Nestin-Tva, Arf −/− mice were sectioned and stained using a Cox-2 antibody. While age-matched, nontumor-bearing mouse brains displayed minimal Cox-2 immunoreactivity in the dorsolateral cortex, a significant number of cells displayed cytoplasmic Cox-2 immunoreactivity within the tumor mass (Fig. 1A, Supplementary Material, Fig. S1). Cox-2 did not overlap with CD105, a marker of endothelium30,31 (Fig. 1B). We also observed Cox-2 immunoreactivity outside of the main tumor mass (data not shown), consistent with previous reports of Cox-2 induction in the tumor periphery and nontumor tissue.18,32 Western blot analysis of tumor versus adjacent nontumor tissue confirmed elevated Cox-2 levels in both regions relative to a tumor-free control (Fig. 1C). Tumors were harvested and used to generate primary adherent cultures. Cells derived from PDGFB-expressing tumors displayed significant Cox-2 immunoreactivity, while Cox-2 was not detected in adherent cultures of adult subventricular zone neural stem cells (aNSC) generated from nontumor-bearing mice (Fig. 1D). Western blot analysis of these primary cultures confirmed overexpression of Cox-2 in GBM cells (Fig. 1E). Consistent with the immunoblot findings, elevated levels of PGE2 were found in the conditioned media of GBM cultures compared with that from aNSC (Fig. 1F). Treatment of GBM cultures with celecoxib, a selective Cox-2 inhibitor, led to a dramatic reduction in PGE2 levels (Fig. 1F). Collectively, these results suggest that analogous to what has been observed in a subset of human GBMs,16 tumors arising in our mouse model display increased levels of active Cox-2. Cox-2 expression and PGE2 production by primary mouse tumor cultures further suggests that Cox-2 is expressed by tumor cells themselves and is not exclusively expressed by activated microglia that can account for extratumoral Cox-2 expression.
Fig. 1.
Cox-2 is overexpressed in glioblastoma (GBM)-bearing mouse brains. (A) GBMs generated via injection of PDGFB-expressing virus into the lateral ventricle of Nestin-tva, Arf −/− mice showed significant expression of Cox-2 compared with a corresponding cortical region in a nontumor-bearing mouse. Scale bar = 0.2 mm. (B) Double-labeling with Cox-2 and the tumor vasculature marker CD105 showed minimal overlap. Scale bar = 0.1 mm. (C) Tumor (T) and Nontumor adjacent (NTA) tissue were isolated from tumor-bearing brains (n = 3) and analyzed by Western blot. Cox-2 was consistently elevated in T and NTA tissue relative to tumor-free (TF) tissue. (D) Dissociated GBMs were used to generate adherent primary tumor cultures. These cultures continued to express Cox-2 by immunofluorescence staining. Scale bar = 0.1 mm. (E) Cox-2 protein levels were determined by Western blot using lysates from primary cultures of GBM versus adult neural stem cells (aNSC). (F) PGE2 levels were analyzed by ELISA on conditioned media from primary aNSC and GBM cultures treated with 5 µM celecoxib or vehicle for 24 hours. Error bars represent mean ± SEM (n = 3). *, P < 0.05.
To investigate the possibility that Cox-2-derived PGE2 is associated with Id1 expression in GBM as has been observed in other cell types,25 we examined mRNA levels in 136 patient samples with RNA-seq data from the The Cancer Genome Atlas database using the cBioPortal for Cancer Genomics.33 Although Id1 and Cox-2 were only upregulated in a minority of samples, a strong tendency toward overall co-occurrence was observed (log odds ratio >3 for Id1 and Cox-2, P = .005).
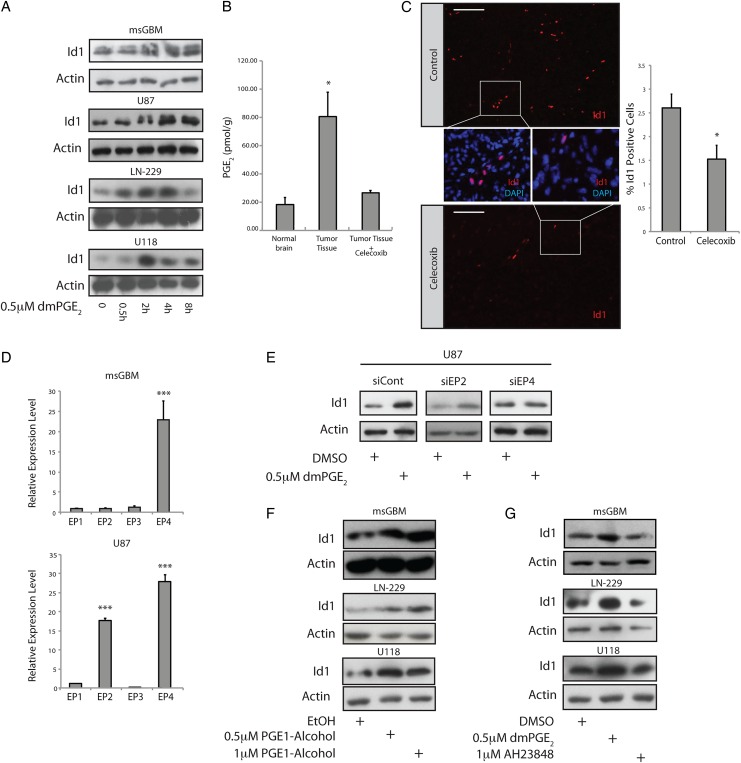
In mouse GBM cells, we observed basal levels of Id1 that were comparable to those in aNSC cultures (Supplementary Material, Fig. S2A). Treatment of either mouse GBM cells or a panel of 3 human GBM cell lines (U87, LN-229, U118) with the stable PGE2 analogue dimethyl-PGE2 (dmPGE2)11,34 induced Id1 protein (Fig. 2A). To determine the importance of PGE2 for regulating Id1 in vivo, tumor-bearing mice were treated with celecoxib (100 mg/kg/day) or vehicle for 4 consecutive days prior to sacrifice and tumor analysis. Control tumor tissue showed elevated PGE2 levels relative to nontumor-bearing cortex (Fig. 2B). Importantly, treatment with celecoxib led to a significant reduction in both intratumor PGE2 and Id1 levels (Fig. 2B and C). Id1 immunostaining in control tumors demonstrated a specific nuclear signal that localized primarily to vascular and perivascular regions and did not localize to regions of high Cox-2 immunoreactivity (Supplementary Material, Fig. S2B–E), suggesting a paracrine regulatory mechanism.
Fig. 2.
The EP4 receptor is responsible for PGE2-mediated induction of Id1. (A) Treatment of primary mouse glioblastoma (GBM) cultures or human U87, LN-229, or U118 glioblastoma cells with 0.5 µM dmPGE2 led to time-dependent induction of Id1 protein. (B) GBMs were initiated in Nestin-tva, Arf −/− mice at day zero via stereotactic injection RCAS-PDGFB virus. At day 23, tumor-bearing mice were treated with 100 mg/kg/day celecoxib or vehicle control (n = 4 per group) by gavage for 4 days, followed by sacrifice and tumor harvest. Harvested tissue was subjected to mass spectrometry to quantify PGE2 levels in normal brain versus GBM with and without celecoxib treatment. *, P < .05, Error bars represent mean ± SEM. (n = 4). (C) Immunohistological analysis of Id1 in tumors derived from mice treated with vehicle (n = 4) or celecoxib (n = 3). Three non–overlapping fields were quantitated for each tumor. Scale bar = 0.1 mm. Error bars represent mean ± SEM. (n = 3 or 4). *, P < .05. (D) To identify the relevant PGE2 receptor in GBM cells, we performed quantitative real-time (qRT) PCR for the 4 EP receptors in mouse GBM cells (upper panel) and human U87 cells (bottom panel). The level of EP4 was notably higher than EP1-3, when EP1 was used as the point of comparison in both cell types, while U87 cells also displayed elevated EP2. Error bars represent mean ± SEM. (n = 3). ***, P < .001. (E) EP2/4 receptor expression was knocked down in human U87 GBM cells via transient transfection with specific siRNAs. Id1 upregulation in response to dmPGE2 treatment was inhibited specifically with EP4 knockdown. (F) Treatment of primary mouse GBM cells or human LN-229 or U118 glioblastoma cell lines with the EP4 receptor agonist PGE1-alcohol resulted in a dose-dependent increase in Id1 protein levels. (G) Treatment with an EP4 antagonist (AH23848) effectively blocked dmPGE2-mediated induction of Id1.
Next, we attempted to identify the EP receptor that was responsible for PGE2-mediated induction of Id1. qRT-PCR was performed using cDNA from primary mouse GBM cells and human U87 cells to assess the relative expression level of EP1-4. In both cell types, EP4 was the most highly expressed receptor, although U87 cells also showed significant levels of EP2, raising the possibility that these GPCRs are important for PGE2–mediated induction of Id1 (Fig. 2D). siRNA knockdown of EP4, but not EP2, blocked Id1 upregulation in U87 cells in response to dmPGE2 treatment (Fig. 2E, Supplementary Material, Fig. S3). Receptor knockdown was confirmed by qRT-PCR (Supplementary Material, Fig. S3). In mouse primary GBM cells, human LN-299, and human U118 cells, pharmacological activation of EP4 using the receptor agonist PGE1-alcohol35 caused a dose-dependent induction of Id1 protein (Fig. 2F). Conversely, treatment with AH23848, an EP4 receptor antagonist,36 blocked dmPGE2-mediated induction of Id1 in these cells (Fig. 2G). Thus, mouse and human GBM cells respond to treatment with exogenous PGE2 by upregulating Id1 in an EP4-dependent manner.
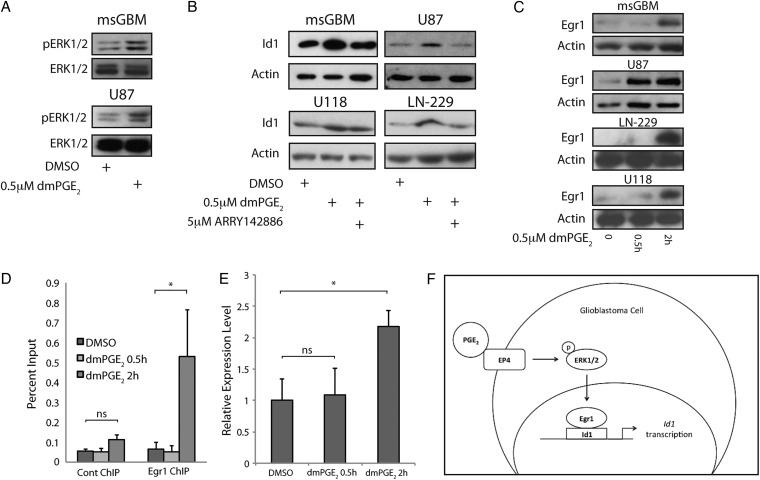
Ligand binding to EP4 can activate the MAPK cascade.5–7 Here we demonstrated that dmPGE2 stimulated ERK1/2 phosphorylation in both mouse GBM cells and human U87 cells (Fig. 3A). To determine whether MAPK signaling contributed to dmPGE2 –mediated induction of Id1, we utilized ARRY1428886,37,38 a potent noncompetitive MEK inhibitor. As shown in Fig. 3B, co-treatment with ARRY142886 blocked dmPGE2-mediated induction of Id1 in both mouse and human cells, although only a partial block was observed in the U118 cell line (Fig. 3B). The transcription factor Early growth response 1 (Egr1) is a downstream target of PGE2-EP4 signaling25 that directly activates Id1 transcription.25,39,40 Treatment with dmPGE2 led to rapid Egr1 induction (Fig. 3C). To determine whether Egr1 mediated the induction of Id1 in mouse GBM cells, chromatin IP analysis was performed. As shown in Fig. 3D, Egr1 was recruited to a conserved binding site in the mouse Id1 promoter 2 hours post-treatment with dmPGE2. This time point corresponded to the observed increase in Egr1 expression in these cells after dmPGE2 treatment. Consistent with the timing of Egr1 recruitment to the Id1 promoter, qRT-PCR demonstrated an increase in Id1 mRNA expression at 2 hours post-treatment with dmPGE2 (Fig. 3E). Collectively, these data suggest that PGE2 stimulates the EP4-ERK1/2 MAPK-Egr1 pathway leading to induction of the stem cell factor Id1 (Fig. 3F).
Fig. 3.
MAPK signaling is required for PGE2-mediated induction of Id1 in glioblastoma (GBM) cells. (A) ERK1/2 phosphorylation was induced in mouse primary GBM cells (upper panel) and human U87 cells (bottom panel) in response to 0.5 hour treatment with 0.5 µM dmPGE2. (B) Cotreatment with 5 µM ARRY-142886, a MEK inhibitor, effectively blocked dmPGE2-mediated induction of Id1 at 24 hours in mouse primary GBM cells and U87 cells, and at 4 hours in LN-229 and U118 cells. (C) Expression of the transcription factor Egr1 increased in murine GBM cells and human U87, LN-229, or U118 cells in response to 0.5 µM dmPGE2 treatment. (D) Chromatin IP was performed on mouse GBM primary cultures, revealing specific recruitment of Egr1 to a conserved Egr1 binding site within the Id1 promoter 2 hours after 0.5 µM dmPGE2 treatment. Quantitative PCR was used to calculate the percent input for each chromatin immunoprecipitation (ChIP) sample. Error bars represent mean ± SEM (n = 3). *, P < .05. (E) Egr1 binding coincided with an increase in Id1 mRNA following treatment with dmPGE2 for 2 hours, as measured by qRT-PCR. Error bars represent mean ± SEM (n = 3). *, P < .05. (F) A general schematic of the signal transduction pathway by which PGE2 induces Id1 transcription.
Cox-2-derived PGE2 Acts via Id1 to Promote Glioblastoma Self-renewal and Radioresistance
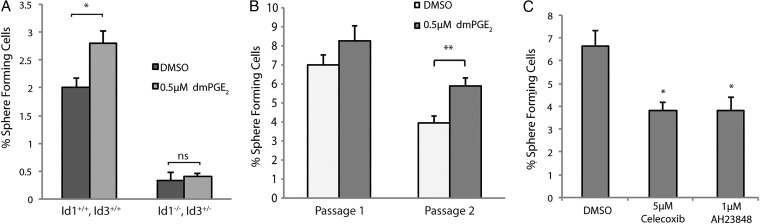
Id1 has been previously characterized as a marker of GBM stem-like cells.24 Two features of these cells are self-renewal capacity and resistance to radiation. To test the possibility that PGE2 signaling promotes self-renewal in GBM cells via the induction of Id1, we used an established tumor sphere-forming assay. Briefly, adherent monolayer cultures of early passage mouse GBM cells were subjected to 24 hours treatment with relevant factors and then replated under nonadherent conditions at clonal density29 (0.5 cell/µL, 1 cell/well). Treatment with dmPGE2 led to a significant increase in the percentage of sphere-forming cells (Fig. 4A, Supplementary Material, Fig. S4), which was maintained after sphere passage (Fig. 4B). To evaluate whether this effect of dmPGE2 was mediated via Id proteins, we employed a genetic strategy. Because previous studies have shown a significant functional redundancy between Id1 and Id3,28 we generated GBM cells from Id1 −/− , Id3 +/− mice. Consistent with what had been previously reported,24 the basal sphere-forming potential of Id-deficient GBM cells was greatly reduced (Fig. 4A). Importantly, dmPGE2 treatment did not enhance self-renewal capacity when Id1 −/− , Id3 +/− cells were used.
Fig. 4.
PGE2 signaling regulates self-renewal in mouse glioblastoma (GBM) primary cultures in an Id1-dependent manner. (A) Primary mouse GBM cells treated with 0.5 µM dmPGE2 were replated at 0.5 cell/µL under nonadherent conditions. The percentage of spheres formed was significantly higher in the dmPGE2-treated group than for control cells. This stimulatory effect was not seen in Id1 −/− , Id3 +/− tumor cells, which also displayed notably lower sphere-forming potential. Error bars represent mean ± SEM (n = 3). *, P < .05. (B) Mouse GBM cells were cultured as spheres at clonal density (0.5 cell/uL) under DMSO or 0.5 uM dmPGE2 treatment and subsequently harvested, dissociated into single cells, and replated under identical conditions. Percentage of sphere-forming cells was quantified at passage 1 and passage 2. Error bars represent mean ± SEM (n = 6). **, P < .01. (C) A significant decrease in sphere-forming potential occurred in GBM cells treated with either 5 µM celecoxib or 1 µM AH23848. Error bars represent mean ± SEM (n = 3). *, P < .05.
To further test the dependence of sphere-forming capacity on PGE2, GBM cells were treated with either celecoxib or AH23848. Notably, 24 hours pretreatment with either compound resulted in a significant reduction in the percentage of sphere-forming cells (Fig. 4C).
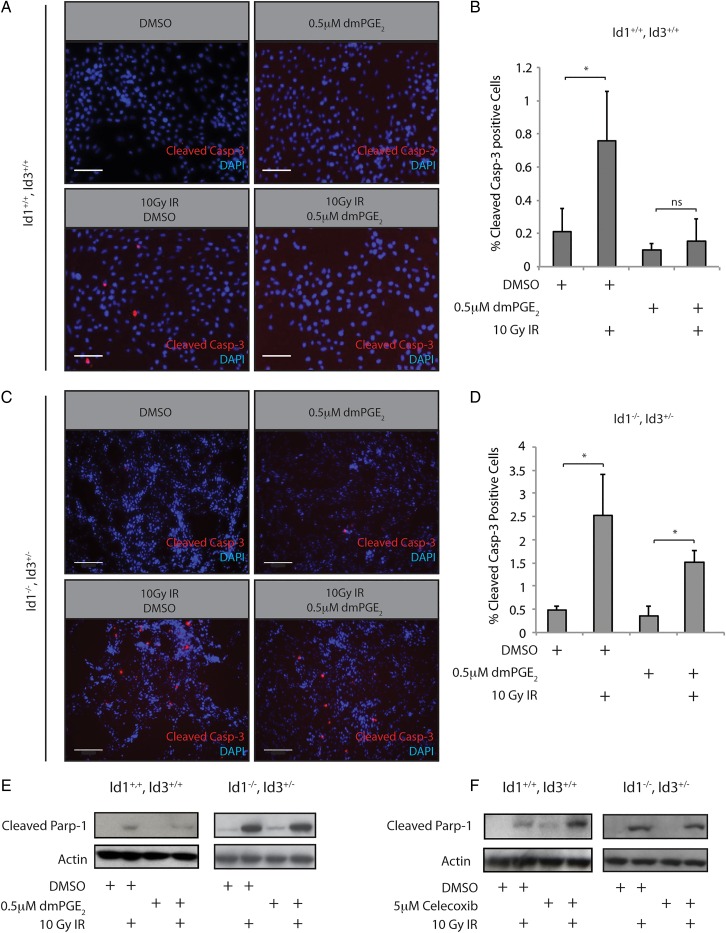
We next sought to determine whether the PGE2-Id1 signaling axis could impact the apoptotic response to radiation-induced DNA damage. Cleavage of caspase-3 and its downstream target Parp-1 are commitment steps for cellular apoptosis. Significantly, Id1+/+, Id3+/+ cells treated with 0.5 µM dmPGE2 24 hours prior to treatment with 10 Gy ionizing radiation (IR) showed markedly reduced caspase-3 cleavage (Fig. 5A and B); this protective effect of dmPGE2 was attenuated in Id1 −/− , Id3 +/− cells (Fig. 5C and D). The attenuation is likely incomplete due to the presence of 1 copy of Id3 in these cells. Western blot analysis demonstrated that Id1+/+, Id3+/+ cells treated with dmPGE2 prior to irradiation also showed reduced Parp-1 cleavage post-IR. Importantly, we did not observe a similar reduction in Parp-1 cleavage after dmPGE2 treatment in Id1 −/− , Id3 +/− cells (Fig. 5E), indicating that Id proteins play an important role in mediating increased radiation resistance imparted by PGE2 signaling. Of note, Id1 −/− , Id3 +/− cells displayed a higher apoptotic response to IR, which coincided with reduced phosphorylation of the DNA damage response (DDR) kinase ATM (Supplementary Material, Fig. S5A) and its substrate, H2AX (Supplementary Material, Fig. S5B). These results are consistent with previous reports demonstrating increased activation of the DDR in GBM stem-like cells.20 Since PGE2 signaling promoted resistance to radiation-induced apoptosis, we speculated that inhibiting this pathway with celecoxib might promote radiosensitivity. Indeed, pretreatment of mouse GBM cells with celecoxib resulted in increased Parp-1 cleavage relative to vehicle-treated cells (Fig. 5F). As with dmPGE2 pretreatment, this effect was not observed in an Id1 −/− , Id3 +/− genetic background, further implicating Id1 in the cellular radiation response.
Fig. 5.
PGE2 signaling regulates radiation resistance in mouse glioblastoma (GBM) primary cultures in an Id1-dependent manner. (A) Treatment of mouse Id1+/+, Id3+/+ GBM cultures with 10 Gy ionizing radiation (IR) induced significant cleavage of caspase-3 after 4 hours. This increase was attenuated by pretreatment with 0.5 µM dmPGE2. Scale bar = 0.01 mm. (B) Quantification of immunostaining from (A). Error bars represent mean ± SEM (n = 3). *, P < .05. C, Id1 −/− , Id3 +/− GBM cultures were treated as described in panel A. In comparison with the Id1+/+, Id3+/+ cells, the radioprotective effects of dmPGE2 were blunted. Scale bar = 0.2 mm. (D) Quantification of immunostaining from (C). Error bars represent mean ± SEM (n = 3). *, P < .05. (E) Treatment of mouse Id1+/+, Id3+/+ GBM cultures with 0.5 µM dmPGE2 for 24 hours was associated with decreased Parp1 cleavage. This protective effect was not observed in Id1 −/− , Id3 +/− GBM cells. F, Pretreatment with 5 µM celecoxib for 24 hours increased Parp1 cleavage in Id1/3 wild type cells, but not in Id1 −/− , Id3 +/− cells.
Significantly, irradiation of GBM primary cultures established from human patients induced a marked increase in the percentage of Id1+ cells while simultaneously decreasing the Ki67+, Id1- population (Supplementary Material, Fig. S6A–C). We note that in unirradiated cultures, the majority of Ki67+ cells are Id1- (approximately 75%) and that 85% of the Id1+ cells are Ki67- (data not shown), suggesting that the Id1+ glioma stem-like cells are of relatively low proliferative potential consistent with the observation in mouse models.24 As seen in primary mouse GBM cultures, the human cells responded to dmPGE2 treatment by inducing Id1 (Supplementary Material, Fig. S6D). Thus, in both mouse and human GBM cells, radiation resistance is a feature of the slowly proliferating, Id1+ stem-like population.
Discussion
In this study, we investigated the functional role of Cox-2-dependent PGE2 signaling in GBM. Using mouse and human GBM cells, we demonstrated that Cox-2-derived PGE2 signals through the EP4 receptor to activate the MAPK signaling cascade, ultimately resulting in upregulation of Id1 via the Egr1 transcription factor. Previous studies have shown that PGE2 can stimulate cross talk between EP4 and the epidermal growth factor receptor leading to activation of MAPK signaling, which could explain the observed increase in Egr1 levels.41 Functionally, the activation of this pathway increases self-renewal capacity and radiation resistance of cultured tumor cells in an Id1-dependent manner. The signal transduction pathway detailed in our studies is similar to that described in cultured breast cancer cell lines.25 Thus, it may be that the Cox-2-PGE2-Id1 signaling pathway is conserved in multiple solid tumor types, in which it functions to maintain expression of Id1 and homeostasis of cancer stem-like cells.
Multiple pathways have been implicated in maintenance of Id1 expression and stem cell character in GBM.42,43 For example, TGF-β signaling has been demonstrated to activate Id1 in a panel of GBM cell lines established from human patients.44 While disparate pathways most likely contribute to GBM cancer stem cell maintenance, our data suggest that Cox-2-derived PGE2 may also play an important role in promoting stemness in human tumors. Importantly, we demonstrated a direct biochemical signaling pathway that acts to induce transcriptional activation of the stem cell factor Id1. Since GBM is a highly heterogeneous tumor type, it will also be important to assess the conservation of the Cox-2-PGE-Id1 signaling pathway in a wider range of patient-derived primary GBM cell lines in future studies.
Decreased Id1 expression was seen to increase the sensitivity of GBM cells to IR. This is potentially consistent with studies in human and mouse GBM cell lines that demonstrate a clear increase in radiosensitivity in response to celecoxib treatment.45 Depletion of cells that are inherently resistant to radiation therapy through loss of signaling networks that maintain cancer stem cell character could result in an overall tumor that is sensitized to radiation. However, since Id1-positive stem-like cells constitute a small percentage of the overall tumor mass, the immediate effect of decreased Id1 expression on apoptotic response to radiation therapy could be difficult to discern in vivo. Indeed, we did not observe a significant increase in apoptosis within celecoxib-treated mouse tumors subjected to radiation therapy (data not shown).
Our laboratory previously reported that genetic loss of function for Id1 had only modest effects on overall survival, despite strongly impacting the self-renewal capacity of GBM stem-like cells.24 This result suggested that self-renewing cells were not the only cells responsible for driving proliferation in the primary tumor. The failure of celecoxib to be of significant benefit in the treatment of GBM46,47 may reflect the fact that targeting stem-like cells alone is insufficient to effect complete remissions. Therapeutic targeting of stem-like cells would need to be combined with near-complete ablation of nonstem progenitor cells for a robust effect on overall survival. Targeting the Cox-2-Id1 pathway in combination with improved therapies that effectively target and rapidly eliminate progenitor cells is predicted to improve the outcome of patients with GBM.
Supplementary Material
Funding
American Cancer Society Postdoctoral Fellowship, Grant Number: PF-12-056-01-DDC (P.J.C.). National Institutes of Health/National Cancer Institute grant P30 CA008748.
Supplementary Material
Acknowledgments
We wish to thank Lindy Barrett for technical assistance and advice, Courtney Coker and Riddhi Shah for animal husbandry expertise, and Andrea Ventura for help in editing this manuscript.
Conflicts of interest statement. Robert Benezra wishes to disclose his position as CSO and chairman of the Scientific Advisory Board, as well as ownership of shares in AngioGenex, a company devoted to antagonizing Id protein activity in patients. The remaining authors have no conflicts of interest to declare.
References
- 1. Jones DA, Carlton DP, McIntyre TM, Zimmerman GA, Prescott SM. Molecular cloning of human prostaglandin endoperoxide synthase type II and demonstration of expression in response to cytokines. J Biol Chem. 1993;268 (12):9049–9054. [PubMed] [Google Scholar]
- 2. Kujubu DA, Fletcher BS, Varnum BC, Lim RW, Herschman HR. TIS10, a phorbol ester tumor promoter-inducible mRNA from Swiss 3T3 cells, encodes a novel prostaglandin synthase/cyclooxygenase homologue. J Biol Chem. 1991;266 (20):12866–12872. [PubMed] [Google Scholar]
- 3. Inoue H, Yokoyama C, Hara S, Tone Y, Tanabe T. Transcriptional regulation of human prostaglandin-endoperoxide synthase-2 gene by lipopolysaccharide and phorbol ester in vascular endothelial cells. Involvement of both nuclear factor for interleukin-6 expression site and cAMP response element. J Biol Chem. 1995;270 (42):24965–24971. [DOI] [PubMed] [Google Scholar]
- 4. Narumiya S, Sugimoto Y, Ushikubi F. Prostanoid receptors: structures, properties, and functions. Physiol Rev. 1999;79 (4):1193–1226. [DOI] [PubMed] [Google Scholar]
- 5. Mendez M, LaPointe MC. PGE2-induced hypertrophy of cardiac myocytes involves EP4 receptor-dependent activation of p42/44 MAPK and EGFR transactivation. Am J Phys Heart Circ Physiol. 2005;288 (5):H2111–7. [DOI] [PubMed] [Google Scholar]
- 6. Qian JY, Leung A, Harding P, LaPointe MC. PGE2 stimulates human brain natriuretic peptide expression via EP4 and p42/44 MAPK. Am J Phys Heart Circ Phys. 2006;290 (5):H1740–6. [DOI] [PubMed] [Google Scholar]
- 7. Fujino H, Xu W, Regan JW. Prostaglandin E2 induced functional expression of early growth response factor-1 by EP4, but not EP2, prostanoid receptors via the phosphatidylinositol 3-kinase and extracellular signal-regulated kinases. J Biol Chem. 2003;278 (14):12151–6. [DOI] [PubMed] [Google Scholar]
- 8. Krysan K, Reckamp KL, Dalwadi H et al. Prostaglandin E2 activates mitogen-activated protein kinase/Erk pathway signaling and cell proliferation in non-small cell lung cancer cells in an epidermal growth factor receptor-independent manner. Cancer Res. 2005;65 (14):6275–6281. [DOI] [PubMed] [Google Scholar]
- 9. Durand EM, Zon LI. Newly emerging roles for prostaglandin E2 regulation of hematopoiesis and hematopoietic stem cell engraftment. Curr Opin Hematol. 2010;17 (4):308–312. [DOI] [PubMed] [Google Scholar]
- 10. Goessling W, North TE, Loewer S et al. Genetic interaction of PGE2 and Wnt signaling regulates developmental specification of stem cells and regeneration. Cell. 2009;136 (6):1136–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. North TE, Goessling W, Walkley CR et al. Prostaglandin E2 regulates vertebrate haematopoietic stem cell homeostasis. Nature. 2007;447 (7147):1007–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414 (6859):105–111. [DOI] [PubMed] [Google Scholar]
- 13. Magee JA, Piskounova E, Morrison SJ. Cancer stem cells: impact, heterogeneity, and uncertainty. Cancer Cell. 2012;21 (3):283–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dannenberg AJ, Subbaramaiah K. Targeting cyclooxygenase-2 in human neoplasia: rationale and promise. Cancer Cell. 2003;4 (6):431–436. [DOI] [PubMed] [Google Scholar]
- 15. Wang D, Dubois RN. The role of COX-2 in intestinal inflammation and colorectal cancer. Oncogene. 2010;29 (6):781–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shono T, Tofilon PJ, Bruner JM, Owolabi O, Lang FF. Cyclooxygenase-2 expression in human gliomas: prognostic significance and molecular correlations. Cancer Res. 2001;61 (11):4375–4381. [PubMed] [Google Scholar]
- 17. Hanisch UK, Kettenmann H. Microglia: active sensor and versatile effector cells in the normal and pathologic brain. Nat Neurosci. 2007;10 (11):1387–1394. [DOI] [PubMed] [Google Scholar]
- 18. Charles NA, Holland EC, Gilbertson R, Glass R, Kettenmann H. The brain tumor microenvironment. Glia. 2011;59 (8):1169–1180. [DOI] [PubMed] [Google Scholar]
- 19. Singh SK, Hawkins C, Clarke ID et al. Identification of human brain tumour initiating cells. Nature. 2004;432 (7015):396–401. [DOI] [PubMed] [Google Scholar]
- 20. Bao S, Wu Q, McLendon RE et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444 (7120):756–760. [DOI] [PubMed] [Google Scholar]
- 21. Lee JK, Chang N, Yoon Y et al. USP1 targeting impedes GBM growth by inhibiting stem cell maintenance and radioresistance. Neuro Oncol. 2015;18 1:37–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lasorella A, Benezra R, Iavarone A. The ID proteins: master regulators of cancer stem cells and tumor aggressiveness. Nat Rev Cancer. 2014;14 2:77–91. [DOI] [PubMed] [Google Scholar]
- 23. Ling F, Kang B, Sun XH. Id proteins: Small molecules, mighty regulators. Curr Top Dev Biol. 2014;110:189–216. [DOI] [PubMed] [Google Scholar]
- 24. Barrett LE, Granot Z, Coker C et al. Self-renewal does not predict tumor growth potential in mouse models of high-grade glioma. Cancer Cell. 2012;21 (1):11–24. [DOI] [PubMed] [Google Scholar]
- 25. Subbaramaiah K, Benezra R, Hudis C, Dannenberg AJ. Cyclooxygenase-2-derived prostaglandin E2 stimulates Id-1 transcription. J Biol Chem. 2008;283 (49):33955–33968. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26. Xu K, Wang L, Shu HK. COX-2 overexpression increases malignant potential of human glioma cells through Id1. Oncotarget. 2014;5 (5):1241–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hambardzumyan D, Amankulor NM, Helmy KY, Becher OJ, Holland EC. Modeling adult gliomas using RCAS/t-va technology. Transl Oncol. 2009;2 (2):89–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lyden D, Young AZ, Zagzag D et al. Id1 and Id3 are required for neurogenesis, angiogenesis and vascularization of tumour xenografts. Nature. 1999;401 (6754):670–677. [DOI] [PubMed] [Google Scholar]
- 29. Coles-Takabe BL, Brain I, Purpura KA et al. Don't look: growing clonal versus nonclonal neural stem cell colonies. Stem Cells. 2008;26 (11):2938–2944. [DOI] [PubMed] [Google Scholar]
- 30. Dallas NA, Samuel S, Xia L et al. Endoglin (CD105): a marker of tumor vasculature and potential target for therapy. Clin Cancer Res. 2008;14 (7):1931–1937. [DOI] [PubMed] [Google Scholar]
- 31. Wang R, Chadalavada K, Wilshire J et al. Glioblastoma stem-like cells give rise to tumour endothelium. Nature. 2010;468 (7325):829–833. [DOI] [PubMed] [Google Scholar]
- 32. Tafani M, Di Vito M, Frati A et al. Pro-inflammatory gene expression in solid glioblastoma microenvironment and in hypoxic stem cells from human glioblastoma. J Neuroinflammation. 2011;8:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cerami E, Gao J, Dogrusoz U et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2 (5):401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Robert A, Schultz JR, Nezamis JE, Lancaster C. Gastric antisecretory and antiulcer properties of PGE2, 15-methyl PGE2, and 16, 16-dimethyl PGE2. Intravenous, oral and intrajejunal administration. Gastroenterology. 1976;70 (3):359–370. [PubMed] [Google Scholar]
- 35. Gardiner PJ, Copas JL, Schneider C, Collier HO. 2-decarboxy-2-hydroxymethyl prostaglandin E1 (TR4161), a prostaglandin bronchodilator of low tracheobronchial irritancy. Prostaglandins. 1980;19 (3):349–370. [DOI] [PubMed] [Google Scholar]
- 36. Abramovitz M, Adam M, Boie Y et al. The utilization of recombinant prostanoid receptors to determine the affinities and selectivities of prostaglandins and related analogs. Biochim Biophys Acta. 2000;1483 (2):285–293. [DOI] [PubMed] [Google Scholar]
- 37. Davies BR, Logie A, McKay JS et al. AZD6244 (ARRY-142886), a potent inhibitor of mitogen-activated protein kinase/extracellular signal-regulated kinase kinase 1/2 kinases: mechanism of action in vivo, pharmacokinetic/pharmacodynamic relationship, and potential for combination in preclinical models. Mol Cancer Ther. 2007;6 (8):2209–2219. [DOI] [PubMed] [Google Scholar]
- 38. Duncia JV, Santella JB III, Higley CA et al. MEK inhibitors: the chemistry and biological activity of U0126, its analogs, and cyclization products. Bioorg Med Chem Lett. 1998;8 (20):2839–2844. [DOI] [PubMed] [Google Scholar]
- 39. Tournay O, Benezra R. Transcription of the dominant-negative helix-loop-helix protein Id1 is regulated by a protein complex containing the immediate-early response gene Egr-1. Mol Cell Biol. 1996;16 (5):2418–2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Passiatore G, Gentilella A, Rom S, Pacifici M, Bergonzini V, Peruzzi F. Induction of Id-1 by FGF-2 involves activity of EGR-1 and sensitizes neuroblastoma cells to cell death. J Cell Physiol. 2011;226 (7):1763–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dannenberg AJ, Lippman SM, Mann JR, Subbaramaiah K, DuBois RN. Cyclooxygenase-2 and epidermal growth factor receptor: pharmacologic targets for chemoprevention. J Clin Oncol. 2005;23 (2):254–266. [DOI] [PubMed] [Google Scholar]
- 42. Charles N, Ozawa T, Squatrito M et al. Perivascular nitric oxide activates notch signaling and promotes stem-like character in PDGF-induced glioma cells. Cell Stem Cell. 2010;6 (2):141–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Li Z, Bao S, Wu Q et al. Hypoxia-inducible factors regulate tumorigenic capacity of glioma stem cells. Cancer Cell. 2009;15 (6):501–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Anido J, Saez-Borderias A, Gonzalez-Junca A et al. TGF-beta receptor inhibitors target the CD44(high)/Id1(high) glioma-Initiating cell population in human glioblastoma. Cancer Cell. 2010;18 (6):655–668. [DOI] [PubMed] [Google Scholar]
- 45. Suzuki K, Gerelchuluun A, Hong Z et al. Celecoxib enhances radiosensitivity of hypoxic glioblastoma cells through endoplasmic reticulum stress. Neuro Oncol. 2013;15 (9):1186–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kesari S, Schiff D, Henson JW et al. Phase II study of temozolomide, thalidomide, and celecoxib for newly diagnosed glioblastoma in adults. Neuro Oncol. 2008;10 (3):300–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Levin VA, Giglio P, Puduvalli VK et al. Combination chemotherapy with 13-cis-retinoic acid and celecoxib in the treatment of glioblastoma multiforme. J Neurooncol. 2006;78 (1):85–90. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.