Abstract
Background
Diffuse gliomas are poorly immunogenic, fatal brain tumors. The basis for insufficient antitumor immunity in diffuse gliomas is unknown. Gain-of-function mutations in isocitrate dehydrogenases (IDH1 and IDH2) promote diffuse glioma formation through epigenetic reprogramming of a number of genes, including immune-related genes. Here, we identify epigenetic dysregulation of natural killer (NK) cell ligand genes as significant contributors to immune escape in glioma.
Methods
We analyzed the database of The Cancer Genome Atlas for immune gene expression patterns in IDH mutant or wild-type gliomas and identified differentially expressed immune genes. NKG2D ligand expression levels and NK cell–mediated lysis were measured in IDH mutant and wild-type patient-derived glioma stem cells and genetically engineered astrocytes. Finally, we assessed the impact of hypomethylating agent 5-aza-2′deoxycytodine (decitabine) as a potential NK cell sensitizing agent in IDH mutant cells.
Results
IDH mutant glioma stemlike cell lines exhibited significantly lower expression of NKG2D ligands compared with IDH wild-type cells. Consistent with these findings, IDH mutant glioma cells and astrocytes are resistant to NK cell–mediated lysis. Decitabine increases NKG2D ligand expression and restores NK-mediated lysis of IDH mutant cells in an NKG2D-dependent manner.
Conclusions
IDH mutant glioma cells acquire resistance to NK cells through epigenetic silencing of NKG2D ligands ULBP1 and ULBP3. Decitabine-mediated hypomethylation restores ULBP1 and ULBP3 expression in IDH mutant glioma cells and may provide a clinically useful method to sensitize IDH mutant gliomas to NK cell–mediated immune surveillance in patients with IDH mutated diffuse gliomas.
Keywords: IDH mutation, glioma, Natural Killer cells, NKG2D ligands, immune escape, immunotherapy
Mutations in the isocitrate dehydrogenase 1 and 2 (IDH1 and IDH2) genes are present in over 80% of all adult diffuse gliomas and in up to 90% of all secondary glioblastoma (GBM). IDH mutations define a subset of gliomas that progress from histologically low-grade tumors to fatal high-grade gliomas usually over a period of 5–10 years.1 The hallmark of all IDH mutated (IDHmut) cancers is epigenetic dysregulation through hypermethylation of genomic DNA and histones.2 Methylation results in the transcriptional repression of a number of genes, including immune-related genes.3 IDH mutations are gain-of-function events that catalyze the production of an oncometabolite (R)-2-hydroxyglutarate (2-HG). In turn, 2-HG inhibits α-ketoglutarate-dependent dioxygenases, many of which are demethylating enzymes. The net result is extensive genomic methylation and epigenetic remodeling.4–6 These molecular alterations impair differentiation of IDHmut cells.7 A specific temporal pattern of high-grade progression is observed in IDHmut diffuse gliomas: IDH mutations appear to occur earliest, followed by chromosome 1p/19q codeletion or mutation of alpha thalassemia/mental retardation syndrome X-linked and tumor protein 53 loss of heterozygosity. Inactivation of the retinoblastoma or the phosphatase and tensin homolog tumor suppressor pathways is correlate with transformation of diffuse gliomas to glioblastoma.8–10 Early IDHmut glial cells, which lack high-grade molecular alterations, must evade antitumor immune responses prior to acquisition of high-grade features. How and why IDHmut gliomas arise in immunologically competent individuals and whether IDH mutations suppress immunity before high-grade transformation is unknown.
Natural killer (NK) cells are effector lymphocytes that play a central role during mammalian host rejection of tumor cells. They recognize and lyse target cells that display abnormal cell surface expression of stress-induced major histocompatibility complex 1 (MHC I) or MHC I-like proteins. Since activation does not require antigen-specific sensitization, NK cells represent an important first-line defense against neoplasia.11–15
The NK group 2D (NKG2D) receptor is an activating NK and CD8 T-cell receptor that mediates cytotoxicity by ligating stress-inducible ligands on target cells. These ligands, known as NKG2D ligands (NKG2DLs), include MHC class I–related chains A and B (MICA, MICB) and the UL16-binding protein family (ULBP 1–6).16 Oncogenic stress mediated by genomic instability or metabolic derangements induces the expression of NKG2DLs in tumor cells, and expression of these ligands may be required for antitumor immunity during the elimination phase of innate immune tumor surveillance.17,18 Notably, malignant cells that overexpress NKG2DLs undergo robust negative selection in glioma mouse models.19,20 Moreover, studies of NKG2DL expression in human cancer reveal a positive correlation between NKG2DL expression and improved clinical outcomes in various solid tumors.21–23
Malignant tumors utilize a number of tissue- and mutation-specific mechanisms to escape NK-mediated immunity. In acute myelogenous leukemia, the NKG2DL locus is methylated, thus promoting self-tolerance to the cytolytic activity of NK cells.25 Shedding of soluble NKG2DLs to induce premature engagement of NK cells and subsequent NK exhaustion has also been reported in cancer.24,26,27 Lastly, NKG2DL-specific microRNAs may also downregulate NKG2DL expression in glioma.28 Although little is known regarding the mechanisms of immune escape in IDHmut gliomas, the latency between tumor initiation and high-grade transformation suggests a prolonged failure of immune surveillance. Moreover, genomic hypermethylation in IDHmut gliomas provides a plausible mechanism for cell-intrinsic silencing of antitumor immune genes.
Here, we utilize primary glioma stemlike cell lines and genetically modified astrocytes to identify transcriptional silencing of NKG2DL as a mechanism for self-tolerance to NK cells in IDHmut gliomas. Additionally, we characterize a potential therapeutic strategy to promote NK-mediated lysis of IDHmut glioma using the demethylating agent 5-aza-2′deoxycytodine (decitabine [DAC]), a cytosine analogue that induces genomic hypomethylation by inhibiting DNA methyltransferase 1 (DNMT1). Our findings provide novel insight into an immune escape mechanism mediated by a single glioma-inducing mutation and sets the stage for targeted immunotherapy in IDHmut glioma using hypomethylating agents.
Materials and Methods
TCGA Data Analysis
RNA-seq data from 286 diffuse glioma patient samples were downloaded from The Cancer Genome Atlas (TCGA) (http://cancergenome.nih.gov). We compared IDHmut and IDH wild-type (wt) immune gene expression by evaluating 20 531 reads corresponding to 1639 annotated immune response genes (Gene Ontology [GO] category 0050776). Reads were normalized using log2(rpkm+1) as the input. Paired t-tests were applied for each marker and adjusted P-values generated using the Benjamini–Hochberg method. Thresholds for differential expression were set at P ≤ 1e-7 and absolute fold change >1.5. Average expression values were subject to hierarchical clustering using Cluster 3.0, and visualized using Java TreeView.29 Infinium HM450 mean methylation values for 162 low-grade glioma patients (130 IDHmut; 32 IDHwt) were downloaded from the Memorial Sloan Kettering Cancer Center cBioPortal (http://www.cbioportal.org/public-portal). Mean methylation values were averaged across samples. t-Tests were applied for each marker and adjusted P-values generated using the Benjamini–Hochberg method.
Cell Lines and Cell Culture
Immortalized isogenic human astrocyte cell lines transduced to express endogenous IDHwt (Parental, PA) or the most common IDH mutation occurring in glioma, IDH1mut (R132H), were cultured as previously described and used between passages 25 and 40 for all experiments.2 Peripheral blood mononuclear cells were isolated from healthy donors by Ficoll gradient centrifugation. MACS NK Negative Selection Kit (Miltenyi Biotec) and MidiMACS separation columns were used for NK enrichment. NKs were activated overnight in 500 IU/mL of interleukin-2 (Peprotech). NK92MI cells (ATCC CRL2408) were grown as recommended.
Glioma stemlike cells (GSCs) were derived from freshly resected glioblastoma samples (see Supplementary Table S2). Freshly resected tumors from IDHmut or wt glioma patients were mechanically dissociated to form a single cell suspension and propagated as cell lines in Neurocult basal medium supplemented with Neurocult neuronal supplement, epidermal growth factor, basic fibroblast growth factor, and heparin according to manufacturer's instructions. GSCs were subsequently cultured in Dulbecco's modified Eagle's medium and grown as adherent cells prior to NK cell killing. Tissue was collected in accordance with the University of Pittsburgh institutional review board.
Decitabine and 2-HG Treatment of Astrocytes
For decitabine treatment of astrocytes, DAC (LC Laboratories) dissolved in dimethyl sulfoxide (DMSO) was added to the complete medium at a final concentration of 200 nM for 5 days. The medium was changed every 2 days and split every 3 days. After 5 days, cells were harvested and counted.
For 2-HG treatment, cell-permeable 2-HG octyl ester (SLR Biosciences) was added to the cell medium at a final concentration of 800 nM for 21 days, and cells were split every 3 days. After treatment, cells were either lysed in Trizol or used directly in NK cytotoxicity assays.
Real-Time PCR
We lysed 5 × 105 IDHmut and IDHwt astrocytes or GSCs in Trizol, and RNA was isolated with chloroform extraction.30 Complementary DNA was synthesized using an iScript cDNA synthesis kit (BioRad). Real-time (RT) PCR for NKG2DLs was performed using Taqman primers (Applied Biosystems) on an Applied Biosystems 7900HT Sequence Detection System. Reactions were amplified for 15 s at 95°C and 1 min at 60°C for 40 cycles and run in triplicate. The cycle number at which the reaction crossed an arbitrarily placed threshold (C T) was determined for each gene and the relative amount of each gene to 18S rRNA was described using the equation 2−ΔC T , where ΔC T = (C TtargetRNA − C T18S rRNA).
NK Cytotoxicity
The flow cytometric carboxyfluorescein succinimidyl ester (CFSE)/7-aminoactinomycin (7-AAD) cytotoxicity assay was performed as described.31 Immortalized astrocytes or glioma cells (2.5 × 105) were labeled with 500 nM CFSE in phosphate buffered saline for 15 min at 37°C. CFSE-labeled target cells (25 000 cells) were used at effector:target (E:T) ratios of 1:5, 1:10, and 1:20. After 6 h incubation, cells were stained with 0.25 μg/mL 7-AAD and analyzed immediately by flow cytometry. Cell-free supernatants were harvested from the cocultures and analyzed for tumor necrosis factor-α levels by enzyme-linked immunosorbent assay (ELISA) following manufacturer's instructions (R&D Systems). For interferon (IFN)-γ ELISA (R&D Systems), cultures were maintained for 16 h.
ULBP-3 Surface Staining
Cells were stained using phycoerythrin-conjugated anti-ULBP3 antibody (R&D Systems). After 30 min, cells were washed and fixed in 2% paraformaldehyde. Samples were analyzed using Accuri C6 (BD Biosciences).
Statistical Methods
All independent experiments were repeated at least 3 times. Data are expressed as means ± SEM, unless otherwise specified. Statistical differences were determined by Student's t-test, where P < .05, P < .01, and P < .001.
Results
Identification of Differentially Expressed NKG2DLs in IDHmut Glioma from RNA-seq in TCGA
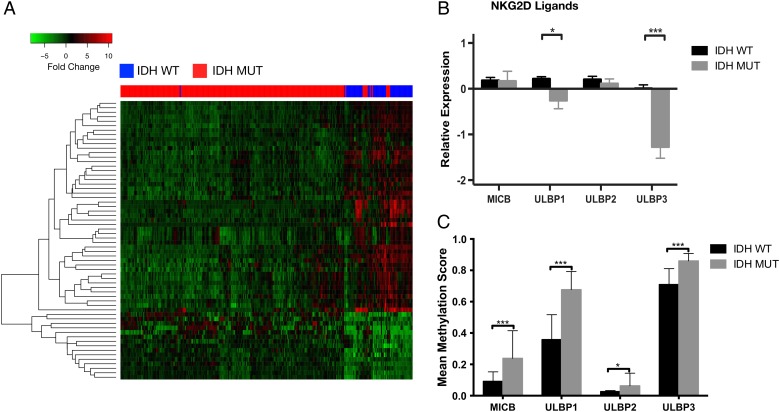
To investigate genotype-dependent differences in the expression of immune-related genes, we compared the expression of 1639 immune-related genes (GO category 0050776) in IDHmut and wt diffuse gliomas using RNA-seq data from TCGA. We identified 62 differentially expressed immune genes between IDHmut and wt gliomas (Supplementary Figures S1 and S3). Hierarchical clustering of the top 20 differentially expressed genes was performed using the Pearson correlation as a distance measure and compared among samples using pairwise complete linkage. We observed a cluster of genes, including NKG2DLs ULBP1 and ULBP3, that were highly expressed in IDHwt tumors but not in IDHmut tumors (Fig. 1A; Supplementary Table S1). We expanded our gene expression analysis to include other NKG2DLs, including ULBP2 and MICB (MICA expression levels were not available from TCGA database) but found that only ULBP1 (P = .03) and ULBP3 (P < .01) were differentially expressed between IDHmut and IDHwt tumors (Fig. 1B). To investigate whether transcriptional repression of NKG2D ligands correlated with promoter methylation, we obtained Infinium HM450 mean methylation values for 130 IDHmut and 32 IDHwt from the low-grade glioma database of TCGA. We identified higher levels of promoter methylation for MICB, ULBP1, and ULBP3 in IDHmut tumors compared with IDHwt tumors (P < .01), suggesting that promoter hypermethylation may lead to reduced NKG2DL expression (Fig. 1C).
Fig. 1.
NKG2D ligands are differentially expressed in IDHmut gliomas. RNA-seq analysis of 1639 immune-related genes (GO category 0050776) was compared in IDHmut and wt diffuse lower grade gliomas using TCGA data. (A) Unsupervised hierarchical clustering of the resulting 62 differentially expressed genes clustered by expression values using Pearson correlation. (B) Gene expression analysis of other known NKG2DLs from the same cohort as Fig. 1A. ULBP1 and ULBP3 expression was significantly lower in IDHmut compared with wt samples. (C) Comparison of NKGD2L mean methylation values in low-grade glioma patients (130 IDHmut, 32 IDHwt) from TCGA. (*P < .05, ***P < .001).
Expression of NKG2D Ligands Is Reduced in IDH1 Mutant Astrocytes and Primary Glioma Cell Lines
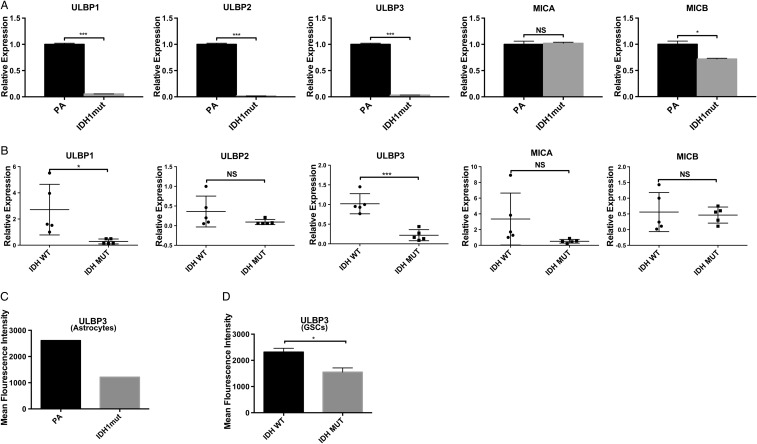
To validate our in silico findings, we quantified gene expression of NKG2DLs in IDHwt and IDHmut immortalized human astrocytes and patient-derived GSCs (Fig. 2A and B, respectively; Supplementary Table S2). We utilized isogenic immortalized human astrocyte lines expressing either endogenous IDH1 (PA) or IDH1mut (R132H).2 This IDHmut cell line effectively reproduces the glioma CpG island hypermethylator phenotype.2,7,32,33
Fig. 2.
Reduced Expression of ULBP1 and ULBP3 in IDHmut gliomas. (A) NKG2DL RT-PCR expression in IDHmut or PA immortalized human astrocytes. Expression levels are relative to 18S RNA (3 independent experiments) (B) Expression of NKGD2Ls in IDHmut or wt primary GSCs by RT-PCR. Each data point represents glioma cells derived from an individual patient. (C, D) Cell surface expression of ULBP3 in astrocytes and GSCs measured by flow cytometry and quantified by mean fluorescence intensity (MFI). For (A) and (B), samples were run in triplicates. Error bars show standard deviation among the replicates. (*P < .05, **P < .01, ***P < .001).
To determine the expression levels of NKG2D ligands in IDHmut or IDHwt astrocytes, we performed RT-PCR for NKG2DLs. We observed a >5-fold reduction in gene expression for ULBP1, ULBP2, and ULBP3 (P < .001) in IDHmut compared with IDHwt astrocytes, and a small but statistically significant reduction in MICB (P < .01) (Fig. 2A). No significant changes were observed for MICA.
To assess whether NKG2DLs were differentially expressed in human glioma specimen, we quantified the expression levels of ULBP1–3, MICA, and MICB in 5 IDHmut and 5 IDHwt patient-derived GSCs using RT-PCR (Fig. 2B). Gene expression was more variable in GSCs than immortalized astrocytes. Nevertheless, all IDHmut GSCs exhibited a significant reduction in the expression of ULBP1 (P = .02) and ULBP3 (P < .01) but not ULBP2, MICA, or MICB (Fig. 2B). To assess concordance of NKG2DL protein expression with our gene expression findings, surface of expression of ULBP1 and ULBP3 was assessed by flow cytometry using GSCs. ULBP3 surface expression was 2-fold higher in IDHwt GSCs and astrocytes than IDHmut specimen (Fig. 2C and D). Unfortunately, no useful signal was obtained with 2 commercially available ULBP1 antibodies tested.
Notably, our analysis of gene expression with 3 data sources (TCGA, immortalized astrocytes, and patient-derived GSCs) shows concordance in NKG2DL expression patterns between IDHmut and wt samples across all 3 sources. These data establish that ULBP1 and ULBP3 are the 2 primary transcriptionally silenced NKG2DLs in IDHmut gliomas.
IDH1 Mutant Astrocytes and Primary Glioma Cell Lines Exhibit Decreased Ability to Activate NK Cells
NKG2DL expression by tumors is proportional to NK-mediated cytotoxicity.34 Therefore, we reasoned that reduced expression of NKG2DLs in IDHmut astrocytes would impede NK-mediated cytotoxicity. To determine whether IDHmut astrocytes are resistant to CD56+/CD3− NK cells, we performed mixed allogeneic cocultures of donor NK cells and IDHmut or IDHwt astrocytes. Astrocytes were then imaged using phase-contrast micrography. After 72 hours of coculture, 97% of initially cultured IDHmut astrocytes remained viable and adherent, compared with 54% of wt astrocytes (Supplementary Fig. 1A and B) (2-tailed Student's t-test, P < .01).
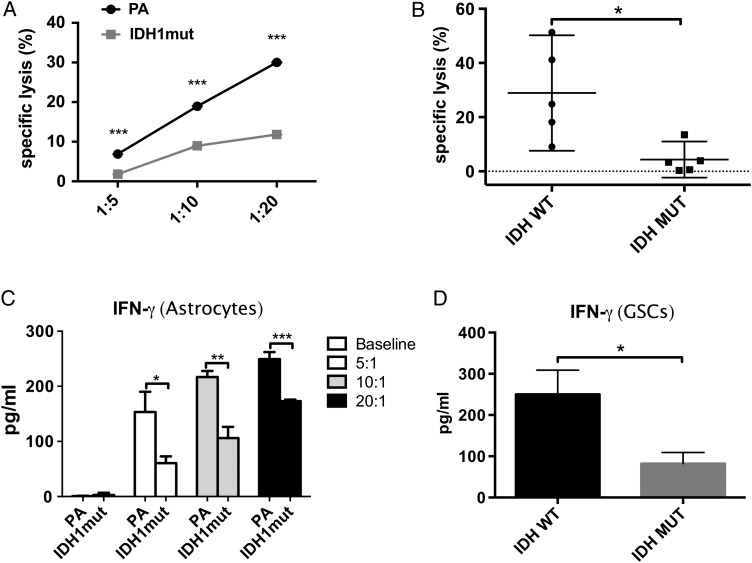
To better quantify the impact of IDH mutations on NK-mediated killing, we cocultured PA and IDHmut astrocytes with NK cells at different E:T ratios and determined specific lysis using multiparametric flow cytometry.31 As shown in Fig. 3A, compared with IDHwt, IDHmut astrocytes were significantly more resistant to NK killing at all E:T ratios tested (P < .05). We then evaluated the relevance of this finding to human glioma by extending our NK cytotoxicity experiments to primary GSCs. Since a 10:1 E:T ratio was sufficient to produce robust killing in IDHwt astrocytes, we used this ratio for cytotoxicity experiments in GSCs and confirmed that human glioma cells carrying IDH mutations are highly resistant to NK killing (P < .01) (Fig. 3B).
Fig. 3.
IDH1mut gliomas cells are resistant to NK-mediated cytotoxicity. NK cell–mediated cytotoxicity measured by 7-AAD-based flow cytometry in astrocytes and GSCs. IFN-γ secretion (ELISA) was measured using supernatant from experiments. (A) NK-mediated specific lysis of IDHmut and parental astrocytes at E:T ratios of 1:5; 1:10, and 1:20. Statistical differences in specific lysis of astrocytes (IDHmut = gray line, squares; PA = black line, circles) were measured using paired Student t-tests. (B) Specific lysis of patient-derived IDHmut (black circles) or wt (black squares) GSCs by NK-92 cells. (C, D) IFN-γ expression measured by ELISA in supernatants from experiments in Fig. 3A and B, respectively. (*P < .05, **P < .01, ***P < .001).
Since classical NK activation is characterized by type 1 cytokine secretion, we investigated whether IDHmut resistance to NK killing was accompanied by reduced IFN-γ secretion. As shown in Fig. 3C and D, respectively, astrocytes and GSCs expressing IDH mutations exhibited significantly lower secretion of IFN-γ, suggesting that tumor-inducing IDH mutations impede NK cell activation.
Natural Killer Cell–Mediated Cytotoxicity in IDHwt Astrocytes and GSCs Is NKG2D Dependent
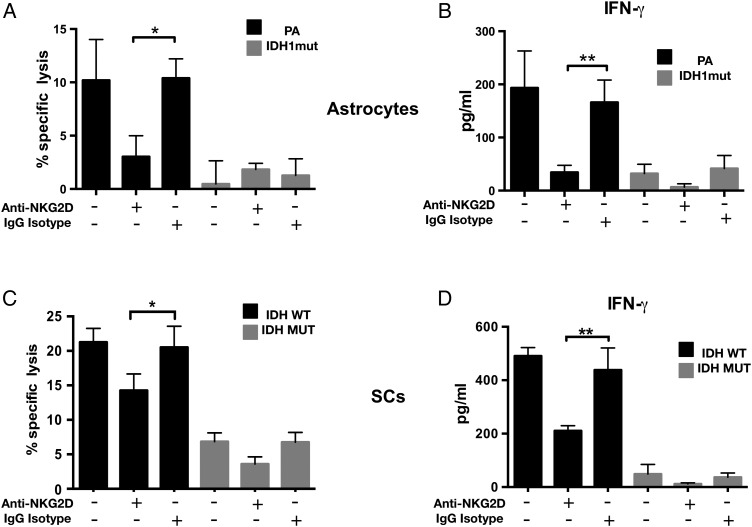
NK cell activity is modulated by the summation of signals from activating and inhibitory surface receptors on NK cells.35 The NKG2D activating receptor promotes both cytokine production and perforin-mediated cytotoxicity after engaging NKG2DLs.15 To understand the degree to which NK activation by astrocytes is dependent on NKG2DL expression, NK cells were pretreated with NKG2D blocking antibody and cocultured with PA or IDHmut astrocytes. As shown in Fig. 4, anti-NKG2D antibodies (but not isotype controls) dramatically reduced NK-mediated cytoxicity (Fig. 4A) and NK activation (Fig. 4B), suggesting that the NKG2D/NKG2DL interaction is the main regulator of NK-mediated astrocyte cytolysis. When similar experiments were performed using GSCs, we also observed significant inhibition of NK-mediated cytoxicity by anti-NKG2D antibodies (Fig. 4B and C). Notably, however, NKG2D blockade did not completely abrogate NK-mediated cytotoxicity in patient-derived GSCs, suggesting that NKG2D/NKG2DL interaction is not the exclusive mechanism of NK lysis in human glioma cells.
Fig. 4.
Natural killer cell–mediated cytotoxicity in IDHwt astrocytes and GSCs is NKG2D dependent. (A) Donor-derived NK-mediated cytolysis of IDHmut or wt astrocytes with or without NKG2D blocking antibodies. Specific lysis was determined by 7-AAD flow cytometry. (B) IFN-γ levels (ELISA) using cell-free supernatants from (A). (C) NK-92 mediated cytolysis of 2 IDHmut and 2 wt patient-derived glioma cell lines in the presence or absence of NKG2D blocking antibodies. (D) IFN-γ levels using supernatants from (C). Data shown are representative of 3 independent experiments. Error bars represent SD between each sample. (*P < .05; **P < .01, ***P < .001).
Oncometabolite 2-Hydroxyglutarate Treatment Is Sufficient to Induce NK Resistance in IDHwt Astrocytes and GSCs
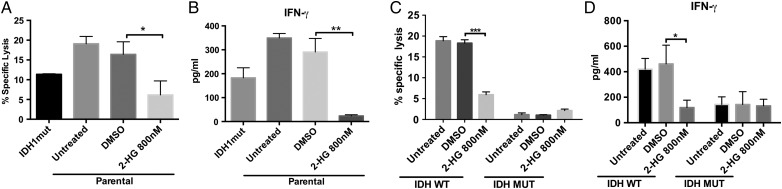
Since oncogenesis in IDHmut cancer is mediated by the 2-HG oncometabolite, we investigated whether cell-permeable 2-HG36 is directly responsible for resistance of astrocytes to NK cells. We treated IDHwt astrocytes and GSCs with 800 nM 2-HG for 21 days and measured NK-mediated cytotoxicity using an immortalized NK cell line (NK-92MI). Reminiscent of IDHmut astrocytes, IDHwt (parental) astrocytes and GSCs acquired resistance to NK cytotoxicity following 2-HG treatment (Fig. 5A and C). Moreover, IFN-γ release from NK cells cocultured with 2-HG-treated astrocytes and GSCs was markedly reduced (Fig. 5B and C), suggesting that 2-HG directly mediates NK cell resistance in IDHmut gliomas.
Fig. 5.
Treatment with 2-HG induces resistance to NK-mediated cytolysis in IDHwt astrocytes and GSCs. IDHwt astrocytes and patient-derived GSCs were treated with 800 nM 2-HG for 21 days. (A, C) NK-mediated cytotoxicity determined by 7-AAD-based flow cytometry after 6 h of NK coculture. (B, D) IFN-γ from corresponding supernatants. (*P < .05, **P < .01, ***P < .001).
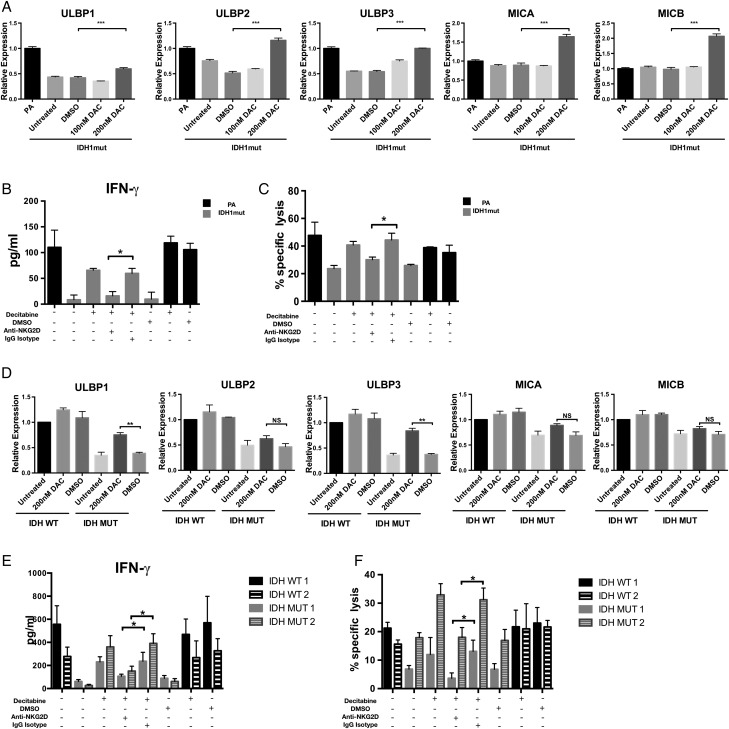
Treatment with DNMT Inhibitor Decitabine Is Sufficient to Restore NKG2D Ligand Expression and Natural Killer Cell Activation by IDH Mutant Astrocytes and GSCs
DNMT inhibitors such as DAC have been shown to effectively reverse hypermethylation in IDH1mut astrocytes and glioma xenografts.5 We investigated whether treating IDH1mut astrocytes and GSCs with DAC would increase NK activation through increased NKG2D ligand expression. We discovered that DAC was able to induce NKG2DLs ULBP1, ULBP2, ULBP3, MICA, and MICB in IDHmut astrocytes (P < .01) compared with the DMSO-treated groups (Fig. 6A). However, this pattern of gene expression is relatively nonspecific, since ULBP2, MICA, and MICB (which are not downregulated in IDHmut gliomas) were also induced by DAC treatment. Interestingly, when GSCs were treated with DAC, we observed specific upregulation of ULBP1 and ULBP3 (P < .01), but not ULBP2, MICA, or MICB (Fig. 6D). This suggests that human patient-derived IDHmut glioma stem cells are uniquely functionally repressed at the ULBP1 and ULBP3 loci.
Fig. 6.
Decitabine restores NKG2DL expression and NK-mediated cytotoxicity. (A, D) IDHmut astrocytes and GSCs (gray bars) were treated for 5 days with DAC or DMSO or left untreated. NKG2D ligand expression levels were assessed by RT-PCR. IDHwt astrocytes or GSCs (black bars) served as controls. Expression levels were normalized to 18S RNA levels. (C, F) IDHmut astrocytes and GSCs (gray bars) were treated with decitabine (200 nM) or DMSO for 5 days and cultured with NK cells pretreated with NKG2D blocking antibody (anti-NKG2D+) or isotype (immunoglobulin G isotype+). Specific lysis was determined by flow cytometry. Percents 7-AAD+ cells are indicated. (B, E) Corresponding IFN-γ levels from supernatants were measured by ELISA. (*P < .05, **P < .01, ***P < .001).
Given the robust induction of NKG2DL expression by DAC, we investigated whether DAC sensitized IDHmut cells to NK-induced cytotoxicity. We treated IDHmut astrocytes and GSCs with DAC and cocultured these cells with NK-92MI NK cells. DAC treatment of IDHmut astrocytes and GSCs nearly doubled NK-mediated cytotoxicity and resulted in a significant increase in IFN-γ secretion by NKs (Fig. 6B and E). To determine whether DAC-mediated increase in NK-induced killing and cytokine production was NKG2D dependent, we exposed DAC-treated cells to anti-NKG2D blocking antibody prior to coculture with NK cells. As shown in Fig. 6C and F, NK killing and IFN- γ release were significantly decreased in the presence of NKG2D blocking antibody, whereas no effect was observed for isotype-treated cells. These results imply that NK resistance by IDHmut cells can be partially reversed by hypomethylating agents.
Discussion
IDH mutations define a subset of diffuse gliomas that gradually acquire aggressive genetic and histologic features, sometimes well after initial diagnosis.10,37 The progressive nature of this disease presents a difficult treatment challenge, because there are no clearly defined criteria or timelines for initiation of potentially toxic chemotherapy or surgical resection. Consequently, there is a need for antitumor therapy that can be initiated at the time of diagnosis.
Although the mechanisms of tumor initiation and progression in IDHmut gliomas are not fully understood, it is known that overproduction of the 2-HG oncometabolite promotes a program of genomic hypermethylation and reproducibly silences thousands of genes. Since hypermethylation of activating NK ligands is a known mechanism for immune escape in cancer, we theorized that oncogenesis and immune escape in IDHmut gliomas is linked by genomic hypermethylation.
In this study, we discover that transcriptional silencing of NKG2D ligands ULBP1 and ULBP3 in IDHmut glioma cell lines facilitates resistance to NK-mediated cytotoxicity. This finding highlights a previously unknown mechanism of immune escape in IDHmut gliomas and suggests a potential role for NK cell–directed therapies for IDHmut gliomas.
ULBP1 and ULBP3 are MHC class I-like proteins that are membrane-bound through a glycosylphosphatidylinositol anchor.38 Both ligands belong to the UL-16 binding NKG2DLs, of which there are 6 members (ULBP1–ULBP6).39 Membrane-bound ULBP1 and ULBP3 are recognized by NKG2D receptors on NK cells, and binding of these ligands by NKG2D induces NK cell activation and target cell lysis.17 Although ULBP expression by normal glial cells is generally low or absent, stress-induced transcription of these genes occurs following oncogenic transformation.40,41 Therefore, it appears that transcriptional silencing of NKG2DLs in IDHmut gliomas represents a distinct mechanism of innate immune escape that is not shared with IDHwt gliomas.42 As such, mechanisms of NK dysfunction may differ between IDHmut and wt gliomas. For instance, recent studies show that NKG2DL expression is regulated by tumor-derived lactate dehydrogenase, signal transducer and activator of transcription 3 signaling, and microRNA expression IDHwt gliomas.28,43,44
NK cell killing of individual tumor cells is dependent on the balance of activating and inhibitory ligands on the surface of the tumor cells.45 Suppression of NKG2D ligand expression in IDHmut cells would potentially tip the balance of NKG2DL expression in favor of inhibitory ligands and render IDHmut cells resistant to NK cells. Notably, all IDHmut GSCs and astrocytes evaluated in this study exhibited low levels of ULBP1 and ULBP3, and reduced expression of these ligands appear to be synonymous with the glioma hypermethylator phenotype. Since IDH mutations are the earliest known genetic alterations in diffuse gliomas, we speculate that downregulation of NKG2DLs is necessary for immune escape in IDHmut gliomas.
Our data show that DAC robustly sensitizes IDHmut glioma cells to NK cell–mediated cytotoxicity. DAC upregulates the expression of all NKG2D activating ligands tested (ULBP1–3, MICA, and MICB). Importantly, we show that DAC-mediated NK killing of IDHmut astrocytes can be partially reversed by blockade of the NKG2D receptor. These findings strongly imply that hypermethylation of NKG2D ligands constitutes an immune escape mechanism in IDHmut gliomas and that epigenetic therapies might combat immune suppression in these tumors.
As a result of improved understanding of the molecular pathogenesis of IDH-mediated cancer, investigators are currently exploring several targeted therapeutic avenues. For instance, several 2-HG inhibitors have been developed, and some are currently in early-phase clinical trials for glioma and leukemia. Notably, 2-HG production induces both epigenetic and metabolic reprogramming, and both processes may be necessary for glial cell transformation. Whereas its epigenetic effects can become imprinted after multiple cell divisions, metabolic reprogramming by 2-HG is relatively dynamic and potentially reversible. Once the epigenetic phenotype of IDH mutation is established, the reversal of hypermethylation by 2-HG inhibition appears to be exquisitely tissue dependent. In acute myeloid leukemia, 2-HG inhibition induces robust transcription of previously methylated genes, whereas this effect may be more modest in glioma.5,46 Nevertheless, 2-HG inhibition promotes cellular differentiation in IDHmut glioma cells.47 These findings raise the possibility that cellular differentiation and reprogramming of cellular metabolism after inhibition of 2-HG production may be dissociable from its gene transcription effects.
For gliomas, treatment of IDHmut xenografts with AGI-5198 (a specific 2-HG inhibitor) results in very limited induction of pro-inflammatory immune gene expression, a limitation that could impede its potential as monotherapy for IDHmut gliomas. In contrast, DAC appears to produce more robust induction of pro-inflammatory immune genes in IDHmut gliomas.5 These findings suggest that 2-HG inhibition may be insufficient to sensitize IDHmut tumors to immune-mediated cytotoxicity and that hypomethylating agents can effectively initiate innate immune sensitization of IDHmut gliomas.
Immunotherapy for diffuse gliomas has achieved limited success.2,48,49 Although peptide and dendritic cell vaccination strategies are ultimately dependent on successful induction of specific T-cell responses, innate immune activity is required for induction and maintenance of T-cell mediated immunity. We speculate that diminished NK cell licensing secondary to NKG2DL silencing may contribute to the failure of immunotherapy for IDHmut gliomas. Although NK cells are not classical antigen-presenting cells, antigen presentation by dendritic cells is enhanced by NK mediated priming through crosstalk between NK and dendritic cells.50 Our findings may be particularly relevant in light of the recent discovery that the mutant IDH1 protein contains a robustly immunogenic epitope,51 which, nevertheless, is incapable of producing long-term natural antitumor immunity in patients with IDHmut gliomas. Interestingly, Schumacher et al showed that only 4/25 patients with IDHmut gliomas exhibited evidence of spontaneous T-cell responses to this epitope, suggesting the existence of a persistently immunosuppressive tumor microenvironment in these tumors.51
Our study is the first to demonstrate a specific immune escape mechanism caused by the IDH1 mutation in gliomas. We have identified a cell-intrinsic NK escape mechanism characterized by transcriptional repression of NKG2D ligands. It is likely that cell-extrinsic immune escape mechanisms are present in the IDHmut tumor microenvironment. For instance, it is unknown why NK cells and CD8+ T cells appear to be excluded from the tumor microenvironment in World Health Organization grades II/III gliomas52 and whether this phenotype is dependent on 2-HG production. It is also unknown if early NK cells in IDHmut tumors experience exhaustion because of the chronic absence of activating NKG2DLs. Our data provide preclinical data supporting the use of hypomethylating agents as a targeted immune adjuvant in IDHmut gliomas.53 Given the relatively good safety profile of many of the hypomethylating agents, we plan to explore early-phase clinical trials utilizing hypomethylating agents for immune sensitization of recurrent IDHmut gliomas.
Supplementary material
Funding
This work was supported by a Copeland Foundation Award (N.A., C.D., X.Z.), Brain Awareness 5K Award (N.A.), NIH 5R01CA175052 (P.G.); University of Pittsburgh Clinical Scientist Training Program (CSTP) and Clinical and Translational Science Institute (UL1 TL1TR000005) (X.Z.).
Supplementary Material
Acknowledgments
The authors would like to acknowledge Robert Ferris and Eric Holland for their critical reading of the manuscript. Additionally, we would like to thank our glioma patients and their families for supporting our research.
Conflict of interest statement. None declared.
References
- 1. Wu JS, Lever JE. Developmentally regulated 75-kilodalton protein expressed in LLC-PK1 cultures is a component of the renal Na+/glucose cotransport system. J Cell Biochem. 1989;40 (1):83–89. [DOI] [PubMed] [Google Scholar]
- 2. Turcan S, Rohle D, Goenka A et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483 (7390):479–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Verhaak RG, Hoadley KA, Purdom E et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cells. 2010;17 (1):98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Losman JA, Looper R, Koivunen P et al. (R)-2-Hydroxyglutarate is sufficient to promote leukemogenesis and its effects are reversible. Science. 2013;339 6127:1621–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Turcan S, Fabius AW, Borodovsky A et al. Efficient induction of differentiation and growth inhibition in IDH1 mutant glioma cells by the DNMT Inhibitor Decitabine. Oncotarget. 2013;4 (10):1729–1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Xu W, Yang H, Liu Y et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer Cells. 2011;19 (1):17–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lu C, Ward PS, Kapoor GS et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483 (7390):474–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Phillips HS, Kharbanda S, Chen R et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cells. 2006;9 (3):157–173. [DOI] [PubMed] [Google Scholar]
- 9. Ohgaki H, Kleihues P. Population-based studies on incidence, survival rates, and genetic alterations in astrocytic and oligodendroglial gliomas. J Neuropathol Exp Neurol. 2005;64 (6):479–489. [DOI] [PubMed] [Google Scholar]
- 10. Cancer Genome Atlas Research Brat DJ, Verhaak RG et al. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med. 2015;372 (26):2481–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Herberman RB, Nunn ME, Holden HT et al. Natural cytotoxic reactivity of mouse lymphoid cells against syngeneic and allogeneic tumors. II. Characterization of effector cells. Int J Cancer. 1975;16 (2):230–239. [DOI] [PubMed] [Google Scholar]
- 12. Ljunggren HG, Karre K. Host resistance directed selectively against H-2-deficient lymphoma variants. Analysis of the mechanism. J Exp Med. 1985;162 (6):1745–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Algarra I, Cabrera T, Garrido F. The HLA crossroad in tumor immunology. Hum Immunol. 2000;61 (1):65–73. [DOI] [PubMed] [Google Scholar]
- 14. French AR, Yokoyama WM. Natural killer cells and viral infections. Curr Opin Immunol. 2003;15 (1):45–51. [DOI] [PubMed] [Google Scholar]
- 15. Bauer S, Groh V, Wu J et al. Activation of NK cells and T cells by NKG2D, a receptor for stress-inducible MICA. Science. 1999;285 (5428):727–729. [DOI] [PubMed] [Google Scholar]
- 16. Eagle RA, Trowsdale J. Promiscuity and the single receptor: NKG2D. Nat Rev Immunol. 2007;7 (9):737–744. [DOI] [PubMed] [Google Scholar]
- 17. Gonzalez S, Groh V, Spies T. Immunobiology of human NKG2D and its ligands. Curr Top Microbiol Immunol. 2006;298:121–138. [DOI] [PubMed] [Google Scholar]
- 18. Nausch N, Cerwenka A. NKG2D ligands in tumor immunity. Oncogene. 2008;27 (45):5944–5958. [DOI] [PubMed] [Google Scholar]
- 19. Friese MA, Platten M, Lutz SZ et al. MICA/NKG2D-mediated immunogene therapy of experimental gliomas. Cancer Res. 2003;63 (24):8996–9006. [PubMed] [Google Scholar]
- 20. Eisele G, Wischhusen J, Mittelbronn M et al. TGF-beta and metalloproteinases differentially suppress NKG2D ligand surface expression on malignant glioma cells. Brain. 2006;129 (Pt 9):2416–2425. [DOI] [PubMed] [Google Scholar]
- 21. McGilvray RW, Eagle RA, Watson NF et al. NKG2D ligand expression in human colorectal cancer reveals associations with prognosis and evidence for immunoediting. Clin Cancer Res. 2009;15 (22):6993–7002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Duan X, Deng L, Chen X et al. Clinical significance of the immunostimulatory MHC class I chain-related molecule A and NKG2D receptor on NK cells in pancreatic cancer. Med Oncol. 2011;28 (2):466–474. [DOI] [PubMed] [Google Scholar]
- 23. de Kruijf EM, Sajet A, van Nes JG et al. NKG2D ligand tumor expression and association with clinical outcome in early breast cancer patients: an observational study. BMC Cancer. 2012;12:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Salih HR, Antropius H, Gieseke F et al. Functional expression and release of ligands for the activating immunoreceptor NKG2D in leukemia. Blood. 2003;102 (4):1389–1396. [DOI] [PubMed] [Google Scholar]
- 25. Baragano Raneros A, Martin-Palanco V, Fernandez AF et al. Methylation of NKG2D ligands contributes to immune system evasion in acute myeloid leukemia. Genes Immun. 2015;16 (1):71–82. [DOI] [PubMed] [Google Scholar]
- 26. Mamessier E, Sylvain A, Bertucci F et al. Human breast tumor cells induce self-tolerance mechanisms to avoid NKG2D-mediated and DNAM-mediated NK cell recognition. Cancer Res. 2011;71 (21):6621–6632. [DOI] [PubMed] [Google Scholar]
- 27. Song H, Kim J, Cosman D et al. Soluble ULBP suppresses natural killer cell activity via down-regulating NKG2D expression. Cell Immunol. 2006;239 (1):22–30. [DOI] [PubMed] [Google Scholar]
- 28. Codo P, Weller M, Meister G et al. MicroRNA-mediated down-regulation of NKG2D ligands contributes to glioma immune escape. Oncotarget. 2014;5 (17):7651–7662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. de Hoon MJ, Imoto S, Nolan J et al. Open source clustering software. Bioinformatics. 2004;20 (9):1453–1454. [DOI] [PubMed] [Google Scholar]
- 30. Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162 (1):156–159. [DOI] [PubMed] [Google Scholar]
- 31. Kim GG, Donnenberg VS, Donnenberg AD et al. A novel multiparametric flow cytometry-based cytotoxicity assay simultaneously immunophenotypes effector cells: comparisons to a 4 h 51Cr-release assay. J Immunol Methods. 2007;325 (1–2):51–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sonoda Y, Ozawa T, Aldape KD et al. Akt pathway activation converts anaplastic astrocytoma to glioblastoma multiforme in a human astrocyte model of glioma. Cancer Res. 2001;61 (18):6674–6678. [PubMed] [Google Scholar]
- 33. Vivanco I, Palaskas N, Tran C et al. Identification of the JNK signaling pathway as a functional target of the tumor suppressor PTEN. Cancer Cells. 2007;11 (6):555–569. [DOI] [PubMed] [Google Scholar]
- 34. Pende D, Rivera P, Marcenaro S et al. Major histocompatibility complex class I-related chain A and UL16-binding protein expression on tumor cell lines of different histotypes: analysis of tumor susceptibility to NKG2D-dependent natural killer cell cytotoxicity. Cancer Res. 2002;62 (21):6178–6186. [PubMed] [Google Scholar]
- 35. Moretta A, Bottino C, Vitale M et al. Activating receptors and coreceptors involved in human natural killer cell-mediated cytolysis. Annu Rev Immunol. 2001;19:197–223. [DOI] [PubMed] [Google Scholar]
- 36. Losman JA, Looper RE, Koivunen P et al. (R)-2-hydroxyglutarate is sufficient to promote leukemogenesis and its effects are reversible. Science. 2013;339 (6127):1621–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Watanabe T, Nobusawa S, Kleihues P et al. IDH1 mutations are early events in the development of astrocytomas and oligodendrogliomas. Am J Pathol. 2009;174 (4):1149–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Paulick MG, Bertozzi CR. The glycosylphosphatidylinositol anchor: a complex membrane-anchoring structure for proteins. Biochemistry. 2008;47 (27):6991–7000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cosman D, Mullberg J, Sutherland CL et al. ULBPs, novel MHC class I-related molecules, bind to CMV glycoprotein UL16 and stimulate NK cytotoxicity through the NKG2D receptor. Immunity. 2001;14 (2):123–133. [DOI] [PubMed] [Google Scholar]
- 40. Gasser S, Raulet DH. Activation and self-tolerance of natural killer cells. Immunol Rev. 2006;214:130–142. [DOI] [PubMed] [Google Scholar]
- 41. Unni AM, Bondar T, Medzhitov R. Intrinsic sensor of oncogenic transformation induces a signal for innate immunosurveillance. Proc Natl Acad Sci U S A. 2008;105 (5):1686–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Di Tomaso T, Mazzoleni S, Wang E et al. Immunobiological characterization of cancer stem cells isolated from glioblastoma patients. Clin Cancer Res. 2010;16 (3):800–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Crane CA, Austgen K, Haberthur K et al. Immune evasion mediated by tumor-derived lactate dehydrogenase induction of NKG2D ligands on myeloid cells in glioblastoma patients. Proc Natl Acad Sci U S A. 2014;111 (35):12823–12828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ciaglia E, Torelli G, Pisanti S et al. Cannabinoid receptor CB1 regulates STAT3 activity and its expression dictates the responsiveness to SR141716 treatment in human glioma patients’ cells. Oncotarget. 2015;6 (17):15464–15481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Moretta L, Moretta A. Unravelling natural killer cell function: triggering and inhibitory human NK receptors. EMBO J. 2004;23 (2):255–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kernytsky A, Wang F, Hansen E et al. IDH2 mutation-induced histone and DNA hypermethylation is progressively reversed by small-molecule inhibition. Blood. 2015;125 (2):296–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rohle D, Popovici-Muller J, Palaskas N et al. An inhibitor of mutant IDH1 delays growth and promotes differentiation of glioma cells. Science. 2013;340 (6132):626–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Silver DJ, Sinyuk M, Vogelbaum MA et al. The intersection of cancer, cancer stem cells, and the immune system: therapeutic opportunities. Neuro Oncol. 2016;18 2:153–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Okada H, Butterfield LH, Hamilton RL et al. Induction of robust type-I CD8+ T-cell responses in WHO grade 2 low-grade glioma patients receiving peptide-based vaccines in combination with poly-ICLC. Clin Cancer Res. 2015;21 (2):286–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Walzer T, Dalod M, Vivier E et al. Natural killer cell-dendritic cell crosstalk in the initiation of immune responses. Expert Opin Biol Ther. 2005;5 (Suppl 1):S49–S59. [DOI] [PubMed] [Google Scholar]
- 51. Schumacher T, Bunse L, Pusch S et al. A vaccine targeting mutant IDH1 induces antitumour immunity. Nature. 2014;512 (7514):324–327. [DOI] [PubMed] [Google Scholar]
- 52. Farmer JP, Antel JP, Freedman M et al. Characterization of lymphoid cells isolated from human gliomas. J Neurosurg. 1989;71 (4):528–533. [DOI] [PubMed] [Google Scholar]
- 53. Pellegatta S, Valletta L, Corbetta C et al. Effective immuno-targeting of the IDH1 mutation R132H in a murine model of intracranial glioma. Acta Neuropathol Commun. 2015;3:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.