Abstract
Background
Cell culture plays a pivotal role in cancer research. However, culture-induced changes in biological properties of tumor cells profoundly affect research reproducibility and translational potential. Establishing culture conditions tailored to the cancer cell of origin could resolve this problem. For glioma research, it has been previously shown that replacing serum with defined growth factors for neural stem cells (NSCs) greatly improved the retention of gene expression profile and tumorigenicity. However, among all molecular subtypes of glioma, our laboratory and others have previously shown that the oligodendrocyte precursor cell (OPC) rather than the NSC serves as the cell of origin for the proneural subtype, raising questions regarding the suitability of NSC-tailored media for culturing proneural glioma cells.
Methods
OPC-originated mouse glioma cells were cultured in conditions for normal OPCs or NSCs, respectively, for multiple passages. Gene expression profiles, morphologies, tumorigenicity, and drug responsiveness of cultured cells were examined in comparison with freshly isolated tumor cells.
Results
OPC media-cultured glioma cells maintained tumorigenicity, gene expression profiles, and morphologies similar to freshly isolated tumor cells. In contrast, NSC-media cultured glioma cells gradually lost their OPC features and most tumor-initiating ability and acquired heightened sensitivity to temozolomide.
Conclusions
To improve experimental reproducibility and translational potential of glioma research, it is important to identify the cell of origin, and subsequently apply this knowledge to establish culture conditions that allow the retention of native properties of tumor cells.
Keywords: cell culture, glioma, oligodendrocyte precursor cell (OPC), neural stem cell (NSC), research reproducibility
Cell culture is a cornerstone technique in biomedical research. For cancer research, tumor cell lines for almost all types of human cancers have been invaluable for the discovery of novel drug targets through mechanistic studies and high-throughput screening efforts.1 However, cultured tumor cells often have phenotypic and genetic alterations that could confound research reproducibility2–4 and mislead therapeutic development.5 Therefore, it is imperative to improve cell culture practices based on concrete scientific rationales.
For glioma research, efforts have been devoted to keeping cultured cell lines as similar to their parental tumors as possible.6 About a decade ago, cancer stem-like cells (CSLCs) were first described in glioblastoma (GBM),7,8 which behave similarly to neural stem cells (NSCs).9,10 This revelation cast doubt on the usage of serum in glioma cell culture as serum is known to induce NSC differentiation.11 Furthermore, recent studies showed that serum induces differentiation of glioma CSLCs, alters gene expression patterns,6 and even impairs tumorigenicity.12 On the contrary, culturing glioma cells in serum-free media supplemented with NSC-supporting growth factors epidermal growth factor (EGF) and basic fibroblast growth factor (FGF-2) preserved salient features of glioma CSLCs such as multilineage differentiation and tumor-initiating capacity.6,9 Together, these findings suggested that the NSC culture condition should be adopted to maintain glioma CSLCs.
One recent advance in tumor pathology is the use of molecular profiling to define subtypes in histopathologically indistinguishable cancers, which led to the subdivision of GBMs into 4 subtypes: classical, mesenchymal, proneural, and neural.13,14 While NSCs may serve as the cell of origin for some subtypes, our lab and others have shown that the oligodendrocyte precursor cell (OPC), a glial cell progenitor, is the cell of origin for at least the proneural subtype.15–18 Since culture conditions differ between OPCs and NSCs, it raises an important question on how to properly maintain OPC-originated proneural glioma cells, which is the main line of investigation in this paper.
Materials and Methods
This is a brief description. Please see supplemental files for detailed information such as vendors, catalog numbers for reagents, etc.
Mouse Lines and Genotyping
Mouse genotype was MADM (TG11, GT11), hGFAP-Cre, p53KO, NF1 flox or MADM (TG11, GT11), NG2-Cre, p53KO, NF1 null. p53KO, NF1 flox, p53 flox, hGFAP-Cre mice were used to purify p53/NF1 double-null OPCs and p53/NF1 double-null NSCs.1 Wild-type (WT) GFP OPCs were purified from NG2-eGFP (WT) mice.1 Genotyping was performed as described.1 All animal procedures were in compliance with animal care guidelines and approval by the IACUC of University of Virginia (approval #3955).
Immunopanning Procedure
OPCs and OPC-originated tumor cells were dissociated and purified through immunopanning with PDGFRα as a primary antibody.
Cell Culture
Cells were maintained in Neurobasal (NB) media supplemented with either EGF/FGF-2 for NmA cells and eNSCs, or PDGF-AA for WT OPCs and OmA cells. Glioma cell lines between had 12–17 passages unless otherwise stated. Human glioma samples collected by the University of Virginia Hospital were approved by the institutional review board under protocol IRB-HSR#17626. Tumor tissue was digested and dissociated as described.17 Primary tumor cells were maintained in indicated media.
Sphere Formation Assay
Cells were dissociated from mouse brains (E15.5 for NSCs or P10 for OPCs), cell number was adjusted, and cells were cultured in different media in plates coated with Poly (2-hydroxyethyl methacrylate) (Sigma 192066-1G) to inhibit cell adhesion.
Lentivirus Production and Cell Infection
Lentivirus production was performed with a third-generation packaging system cotransfected in HEK293T-cell line through a calcium phosphate method. Supernatant was collected, filtered, and used to infect target cells.
Tumor-cell Grafting
For grafting, either MADMmodel-derived mouse glioma cells or human patient-derived glioma cells were grafted into the striatum of NOD-SCID mouse brains with the following coordinates, measured according to bregma: 1 mm posterior, 1 mm lateral, and 2.5 mm deep under the pia surface.
qGRATIs Analysis
The q-GRATIs system is a set of lentiviral vectors with unique DNA tags that can be quantitatively detected with quantitative (q)PCR) (Other data (C.L.), unpublished data, 2016). After 2 cell lines, which are separately infected with viral particles and contain distinct DNA tags, are mixed together, genomic DNA of the cell mixture is extracted, and qPCR is performed on those tags to indicate the relative abundance of each cell line initially (I). The remaining cell mixture is used for grafting. After tumor formation, genomic DNA is extracted, and qPCR is performed on those tags to indicate the relative abundance of each cell line in the end (E). The end-to-initial (E/I) ratio reflects the difference in propagation rate of each cell line (Fig. 5E). Mouse or human glioma cells infected by qGRATIs vectors were orthografted as described.17 Genomic DNA was extracted from tumors formed as a template for qPCR reactions to measure abundance of individual barcoded tags.
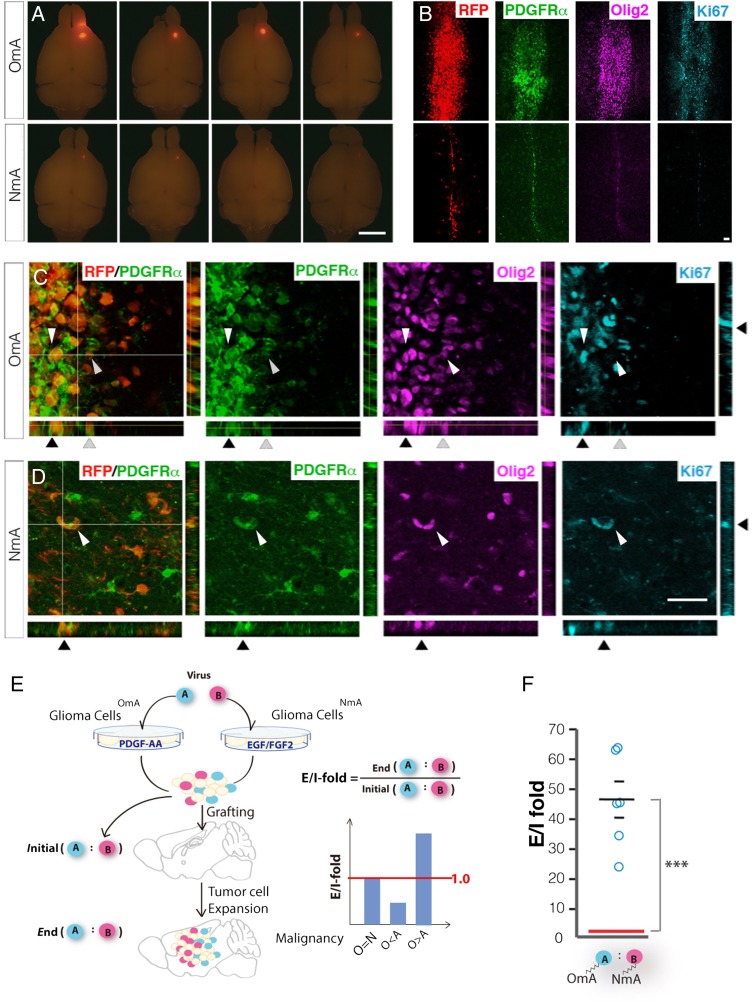
Fig. 5.
Oligodendrocyte precursor cell (OPC) media maintained tumorigenic potential of OPC-originated glioma cells much better than neural stem cell media. (A) Gross images of tumors generated by grafting with 100,000 cells of either gliomaOmA or gliomaNmA cells. Tumor cell RFP expression shows bigger tumors from gliomaOmA cells than from gliomaNmA cells. Images are representative of 3 grafting experiments. Scale bar: 2 mm. (B) Low-magnification images of sections transversing the injection tract of tumor-bearing brains shown in (A). Scale bar: 200 μm. (C) and (D). High-magnification images showing the co-localization of PDGFRα and Olig2 in Ki67+ proliferating tumor cells from both gliomaOmA and gliomaNmA-derived tumor cells. Arrows point to a few cells in all imaging channels, showing that tumor cells (RFP+) positive for OPC markers (PDGFR+, Olig2+) are dividing (Ki67+). Scale bar: 100 μm. (E). Schematic demonstration on how to use q-GRATIs to compare the relative in vivo propagating capacity between gliomaOmA and gliomaNmA cells. (F). q-GRATIs analysis revealed that gliomaOmA cells propagated 40-fold more than gliomaNmA cells in vivo. Circles represent individual animals; N = 6 mice; 100,000 cells were grafted per mouse. ***P ≤ .001.
Quantitative Real-time PCR
qRT-PCR was performed with an Applied Biosystems StepOnePlus Real-Time PCR System by using the KAPA SYBR FAST ABI Prism qPCR kit. Beta-actin (ActB) and/or glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were used to normalize the expression of target genes. For details on RNA extraction, cDNA synthesis and DNA extraction please refer to Supplementary material.
MTT Assay and Temozolomide Treatment
GliomaNmA and gliomaOmA cells were plated at a density of 2 × 105 cells/well in 96-well plates and treated with various concentrations of temozolomide (Temodar, Merck) as indicated. MTT assay was used to assess cell viability 48 hours after treatment.
Immunofluorescence Staining
Immunofluorescent staining on cell culture or frozen tissue sections was performed as described.17 For antibodies, refer to Supplementary material.
Western Blotting
Cells were lysed in cold lysis buffer 17 (R&D, 895943), supplemented with protease inhibitor cocktail tablets (Roche, 11836153001) and Halt phosphatase inhibitor (Thermo Scientific, 1862495). Total protein was adjusted according to concentration measured by Pierce BCA protein assay kit (Thermo Scientific, cat. # 23227). Protein samples were subjected to SDS-PAGE and transferred to polyvinylidene difluoride membranes. For antibodies, see Supplementary material.
Tissue Microarray Analysis
Tissue microarrays (TMAs) containing 238 glioma samples were prepared as described.19 Five characteristic microscopic fields were randomly selected for quantifying data points. Images were analyzed using Image-Pro Plus 6.0 software (Media Cybernetics). To ensure unbiased scoring, 2 individuals without knowledge of clinical data blindly evaluated immunohistochemical results of TMA. This study was approved by the ethics committee of the Xiangya Hospital of Centre-South University (IRB# 201012047).
Clustering Analysis
Gene expression values from qPCR were normalized to values between −1 and +1 for each gene. Values were used for unsupervised hierarchical clustering with R software,20 (http://www.R-project.org/; version 3.0.2, by gplots package,21 version 2.13.0). Euclidean distance was the distance metric employed.
Single-cell Profile Analysis
We used single-cell RNASeq data available at GEO provided by Patel et al (2014)22 (GSE57872). Tumor identification names were retained from the original dataset. The cell-cycle meta-signature score was reproduced as described and based on the gene list from Fig. 2B of the paper.22 Cells with high cell-cycle (CC) meta-signature scores were divided according to PDGFRA and GFAP. Expression levels were normalized (−5 and 5) in order to compare expression of both genes.
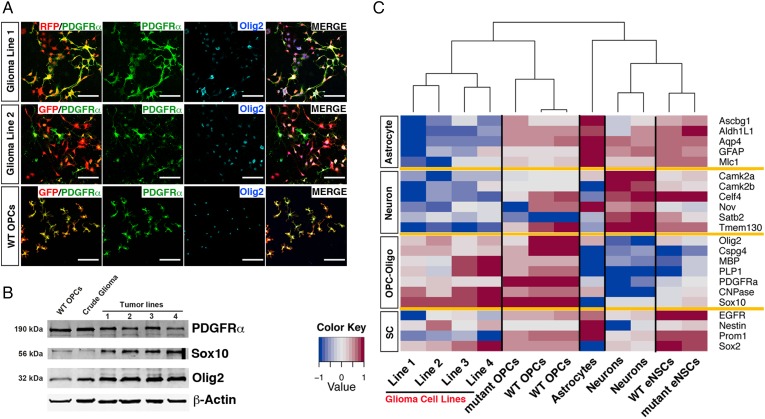
Fig. 2.
Oligodendrocyte precursor cell (OPC) culture condition maintained OPC features of glioma cells. (A) Immunofluorescence staining shows tumor cell lines derived from OPC-originated glioma cells expressed OPC markers PDGFRα and Olig2 under OPC culture conditions. Wild-type (WT) OPCs were used as positive control. RFP in glioma line 1 and GFP in glioma line 2 permanently express in glioma cells due to MADM model labeling of mutant cells.17 GFP channel was pseudo-colored red in the middle row for simplification. Scale bars, 100 μm. (B) Western blot revealed that crude glioma tissue (acutely dissociated from mouse brains without immunopanning) and OPC-like glioma lines at passage 12 (P12) express OPC markers. Lines 1–2 were originated from NG2-Cre initiated tumors, and lines 3–4 were from hGFAP-Cre initiated tumors. For full genotype, see M&M. (C). Heat map of gene expression patterns of different brain cells and OPC-originated glioma lines by quantitative real-time PCR. Representative markers for neurons, astrocytes, OPC-oligodendrocyte lineage, and stem cells were used. Samples were grouped based on unsupervised hierarchical clustering analysis. Lines 1–4 are the same as those used in panel B. WT OPCs were enriched to >95% purity (based on PDGFRα staining) from WT mice at P10 via immunopanning. Mutant OPCs were purified via immunopanning from MADM-p53KO,NF1flox mice at P 10, prior to tumor formation. WT embryonic neural stem cells (eNSCs) were purified from E15.5 WT embryonic brains. Mutant eNSCs were purified from E15.5 hGFAP p53/NF1 double-null brains.
Results
OPC-like Tumor Cells From a Mouse Glioma Model Function as Cancer Stem-like Cells
Using a genetic mouse model in which p53 and neurofibromatosis 1 (NF1) were inactivated, we have shown that OPCs can function as the cell of origin and directly transform into high-grade glioma.17,23 After transformation, tumor cells maintained prominent OPC features including stellate morphology and expression of OPC-specific markers such as Olig2, PDGFRα, Sox10, NG2/Cspg4, etc.17,23 Transcriptome analysis of tumor cells further revealed a gene expression profile closely resembling OPCs.17 To determine whether these OPC-like glioma cells function as CSLCs, we enriched them to ≥95% purity through an immunopanning procedure that captures cells expressing OPC-specific surface marker PDGFRα.17,24
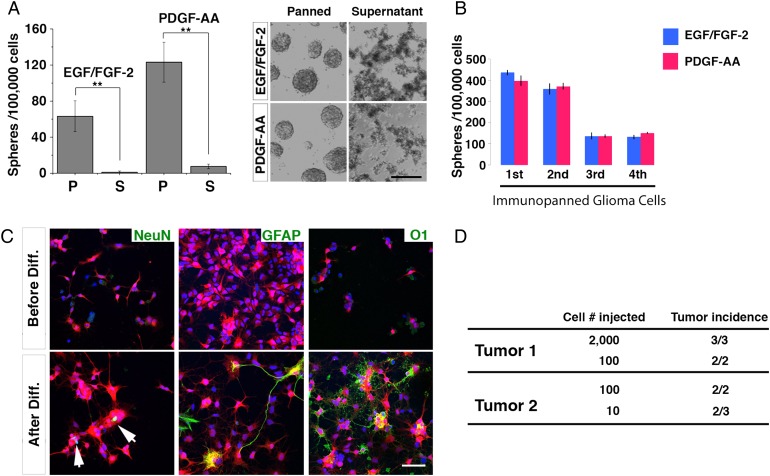
First, we examined the sphere-forming capacity of immunopanned OPC-like tumor cells. We found that OPC-like tumor cells (but not cells in the supernatant fraction where tumor cells were largely depleted) efficiently formed spheres in serum-free culture media (Fig. 1A). While the initial assays were performed in media supplemented with EGF and FGF-2 (known growth factors for NSCs), we wondered if similar results could be obtained if PDGF-AA25,26 (the growth factor essential for normal OPC proliferation) was used. We found that spheres could form in PDGF-supplemented as well as in EGF/FGF-2 supplemented media (Fig. 1A). Furthermore, spheres formed in both EGF/FGF-2 and PDGF-AA conditions could be further passaged for multiple generations, indicating their self-renewal capacity (Fig. 1B). We then determined whether OPC-like glioma cells could differentiate into multiple neuroglial cell types, a hallmark of glioma CSLCs.7 As shown in Fig. 1C, OPC-like glioma cells differentiated into cells expressing markers for mature neurons, astrocytes, and oligodendrocytes in the presence of serum, demonstrating their multipotentiality. Finally, we confirmed that as few as 10 OPC-like cells were sufficient to initiate secondary tumors in immune-compromised mouse brains (Fig. 1D), suggesting that most OPC-like tumor cells—if not all—carry the ability to initiate tumors. Taken together, our data support that OPC-like cells fulfill the criteria of glioma CSLCs.
Fig. 1.
Tumor cells enriched with oligodendrocyte precursor cell (OPC)-specific cell surface marker PDGFRα functioned as cancer stem-like cells. (A) Tumor cells enriched by anti-PDGFRα immunopanning, but not those from supernatant, were capable of forming tumor spheres in serum-free media supplemented with either EGF/FGF-2 or PDGF-AA. Left panel: quantification of sphere-forming capacity; right panel: images of tumor spheres cultured in indicated conditions. Scale bar: 1 mm. P: immunopanned fraction; S: supernatant fraction. At least 4 wells were quantified for each column. Error bars represent SEM. ** P < 0.01. (B) Self-renewability of tumor spheres cultured in either EGF/FGF-2 or PDGF-AA was demonstrated for 3 continuous passages. (C) Multilineage differentiation of OPC-like glioma cells induced by 1% fetal bovine serum and 1% NT-3. NeuN, GFAP, and O1 were used to stain mature neurons, astrocytes, and oligodendrocytes, respectively. RFP is a Cre reporter that labels all tumor cells.17 Scale bar: 100 μm. (D) In vivo tumor-initiating capacity of OPC-like glioma cells was examined in 2 different primary tumors after grafting into brains of 8-week old NOD/SCID mice.
OPC-like Glioma Cells Retained Salient Features of Their Primary Tumors When Maintained in the Media Condition for Normal OPC Culture
Considering the OPC origin, we first expanded glioma cells in serum-free media containing PDGF-AA.25,26 At the time this manuscript was being prepared, we had cultured them successfully for at least 28 passages without detectable changes in proliferative rate, morphology, and the expression of OPC markers PDGFRα, Olig2, and Sox10 (Fig. 2A and B). To further investigate whether these cell lines resemble OPCs, we applied quantitative RT-PCR (qRT-PCR) to evaluate the expression of genes enriched in OPCs, neurons, astrocytes, or oligodendrocytes.27 We also included genes that are considered highly expressed in NSCs and CSLCs, although many of them are also expressed in normal OPCs (see below and discussion).28,29 All established tumor cell lines strongly expressed OPC-oligodendrocyte lineage markers—but not astrocyte or neuron markers—and were grouped closer to WT OPCs by unsupervised clustering (Fig. 2C), suggesting that these cell lines share a gene expression pattern more similar to OPCs than to other cell types. Taken together, these findings demonstrate that OPC-originated glioma cells cultured in PDGF-containing media for multiple passages could maintain salient molecular and cellular features of OPCs.
OPC-like Glioma Cells Cultured in NSC Media Lost the Expression of OPC Markers but Upregulated NSC Markers
Given that OPC-like glioma cells can form spheres in EGF/FGF-2 media (Fig. 1A), we wondered whether OPC-like glioma cells could be effectively maintained in NSC culture conditions. Therefore, we split primary glioma cells from the same tumors into 2 fractions and then maintained them in either PDGF-AA- or EGF/FGF-2-containing media to establish cell lines. Similar to cell lines maintained in PDGF-AA described in Fig. 2, those maintained in EGF/FGF-2 could be readily established and maintained for at least 28 passages. Hereafter, we refer to cell lines established in PDGF-AA media as “OPC-media-accustomed glioma” (gliomaOmA) and those in EGF/FGF-2 media as “NSC-media-accustomed glioma” (gliomaNmA).
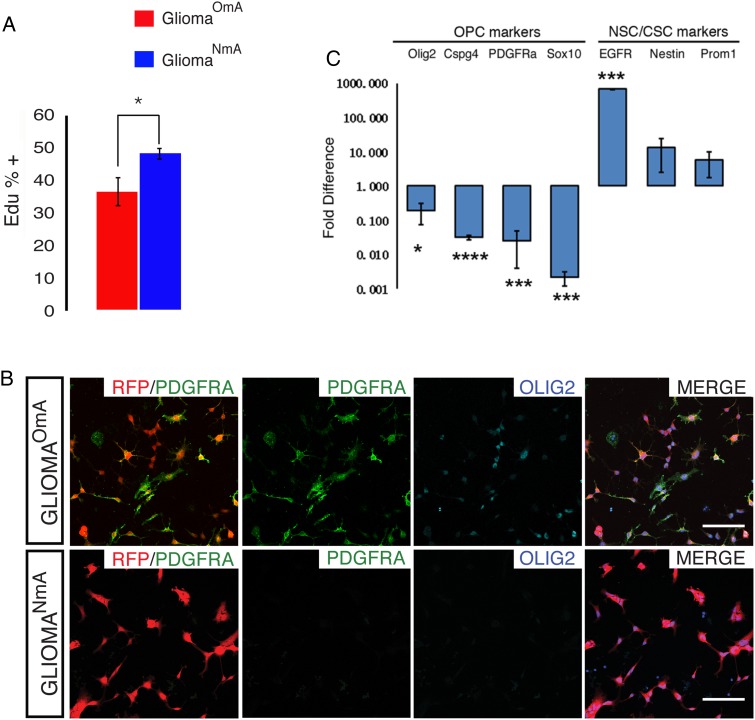
GliomaNmA cells proliferated significantly faster than their gliomaOmA counterparts (Fig. 3A), had less elaborate processes and larger cell bodies compared with gliomaOmA cells and normal OPCs (Fig. 3B, see also Fig. 4A). Furthermore, gliomaNmA cells dramatically downregulated the expression of many OPC-specific genes (Fig. 3B and C) while upregulating NSC genes such as Nestin and Prom1 (CD133) (Fig. 3C). It should be noted that EGFR, reported to be expressed in active NSCs and human GBM CSLCs,30,31 was expressed in the gliomaNmA line but not in the gliomaOmA line or the primary OPC-like tumor cells (Fig. 3C). While it was reported that WT NSCs could contaminate glioma culture and contribute to EGFR expression,32 we confirmed that these NmA cells were derived from glioma cells rather than from contaminating normal NSCs based on their prominent RFP expression (Fig. 3B), the fluorescent marker genetically and permanently introduced into the primary tumor cells from which the cell line was established.17 Finally, while gliomaNmA and gliomaOmA cells are different; it would be important to determine which one better matches acutely purified parental tumor cells. Unsupervised clustering based on gene expression patterns of a matched set of acute tumor-gliomaNmA-gliomaOmA showed that gliomaOmA cells clustered with parental tumor cells WT OPCs, while gliomaNmA cells clustered with NSCs (Supplementary Fig. S1). In summary, our data suggest that long-term exposure to EGF/FGF-2 induces mouse glioma cells to deviate from OPC-like gene expression patterns.
Fig. 3.
Neural stem cell (NSC) culture condition altered proliferative rate and marker gene expression of glioma cells (A) EdU incorporation assay revealed that gliomaNmA cells grow significantly faster than their gliomaOmA counterpart. Graph shows quantification of 3 independent assays with one pair of cell lines. Bars represent mean +/− SEM. *P ≤ .05. A similar trend was confirmed with a second pair of cell lines. (B) Tumor cell lines maintained oligodendrocyte precursor cell (OPC) markers PDGFRα and Olig2 when cultured in OPC media (gliomaOmA) but lost expression of these markers when cultured in NSC media (gliomaNmA). gliomaOmA and gliomaNmA were established from immunopanned OPC-like tumor cells derived from the same primary tumor, established from TG11/GT11, p53, NF1; hGFAP-Cre mice. Scale bars, 100 μm. Images are representative of 3 independent experiments. (C) Quantitative real-time -PCR shows that glioma cells cultured in NSC media had diminished expression of OPC markers PDGFRα, Olig2, Sox10, and NG2 but elevated expression of EGFR and Nestin when compared with their gliomaOmA counterpart. Results represent an average of 3 independent experiments with the same lines as in 3B. Bars represent mean +/- SEM. *P ≤ .05; ***P ≤ 0.001. Experiments in this figure were done with glioma line 1 from Fig. 2 and were confirmed with glioma line 2 to ensure reproducibility.
Fig. 4.
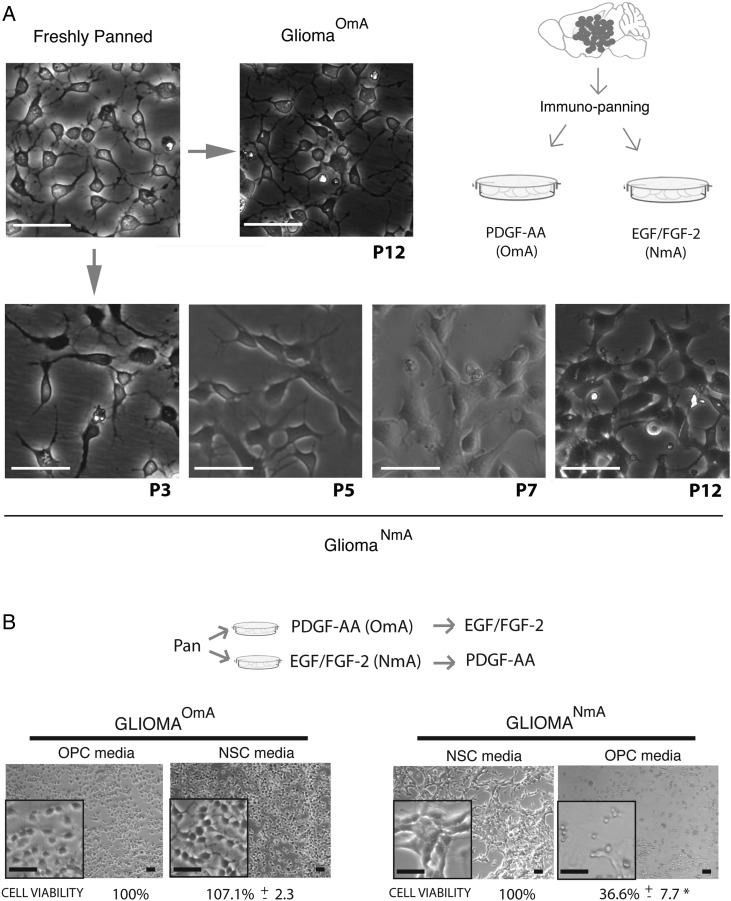
Neural stem cell (NSC) media-induced phenotypic changes happened gradually at the population level and were mostly irreversible. (A) GliomaOmA cells cultured in oligodendrocyte precursor cell (OPC) media maintain morphologies for up to 12 passages (top panel). In contrast, after being cultured in NSC media for multiple passages, glioma cells gradually changed morphologies at the population level (bottom panel). Images are representative of 3 different tumors that were freshly purified and cultured in the respective media. Scale bars, 100 μm. (B) Switching gliomaOmA cells into NSC media did not affect cell viability, in stark contrast to the <40% cell viability after switching gliomaNmA cells into OPC media, suggesting that adaptation of glioma cells to NSC media is mostly an irreversible process. Images are representative of 3 different freshly purified tumors that were cultured in the respective media. Cell viability was assessed by MTT assay. Percentages are at the bottom of each picture, and represent an average of 3 experiments ± SEM. *P ≤ 0.05. Scale bars, 100 μm.
GliomaNmA Cells Were Gradually Converted From OPC-like Tumor Cells When Cultured in NSC Media, a Process That is Mostly Irreversible
There are 2 possible explanations for how gliomaNmA cell lines were derived in the presence of EGF/FGF-2: gradual conversion at population level or outgrowth from a few rare, pre-existing NSC-like glioma cells. To tease apart these possibilities, we carefully monitored glioma cells cultured in NSC media for >10 passages. We found that tumor cells exhibited neither detectable morphological change nor decreased cell viability during early passages (Fig. 4A). After 4–5 passages in NSC media, morphological alterations such as enlargement of cell body and disappearance of fine processes became noticeable in all cells (Fig. 4A). After 7 passages, cells became morphologically indistinguishable from established gliomaNmA cells (shown at P12 on Fig. 4A). Therefore, we concluded that gliomaNmA cells were converted from gliomaOmA cells through gradual phenotypic alterations at the population level rather than selected from rare, pre-existing cells.
To determine whether the phenotypes of gliomaNmA and gliomaOmA cell lines are reversible, we performed a cross-culture experiment. While gliomaOmA cells grew well after being switched into NSC media, gliomaNmA cells went through massive detachment and reduction of cell viability when switched into OPC media (Fig. 4B). These observations suggest that the phenotypic plasticity is largely unidirectional; while gliomaOmA cells could readily adapt to the new culture condition, the majority of gliomaNmA cells could not readapt to OPC media.
The Tumorigenic Potential of OPC-like Glioma Cells Was Significantly Reduced After Long-term Maintenance in NSC Culture Condition
To compare the tumorigenic potential of glioima cells cultured in each condition, we grafted the same number of gliomaNmA and gliomaOmA cell lines established from the same primary tumor into NOD-SCID mouse brains, respectively. GliomaOmA cells efficiently initiated secondary tumors in vivo even after long-term culture (Fig. 5A, top row), and tumor cells in grafted glioma exhibited prominent expression of OPC markers (Fig. 5B, top row). Of note, tumor latency of long-term cultured gliomaOmA cells (>18 passages) was comparable to that of freshly purified ones (∼4 weeks when 100 000 cells are grafted), indicating that OPC culture condition not only retained OPC properties but also their tumorigenic potential. In stark contrast, gliomaNmA cells did not generate sizable tumors (Fig. 5A, bottom row). Surviving gliomaNmA cells along the needle track of the graft exhibited low proliferative activity (Fig. 5B, bottom row) and restored expression of OPC markers PDGFRα and Olig2 (Fig. 5D, in comparison with 5C), further indicating that OPC-features are critical for in vivo propagation of tumor cells.
To quantitatively compare tumorigenicity of grafted gliomaNmA and gliomaOmA cells within the same mouse, we employed a method termed “quantitative- Grafting assay based on Ratio Alteration between Targeted and Internal control cells (q-GRATIs), which was recently developed in our laboratory (Fig. 5E) (Other data (C.L.), unpublished data, 2016). GliomaNmA and gliomaOmA cells were permanently labeled with unique DNA sequences (q-GRATIs tags named A and B tags, respectively) via lentiviral-mediated genome integration mixed together and then grafted into NOD-SCID mouse brains. The relative in vivo propagation potential between these cell lines was subsequently determined by the E/I ratio, which is defined by the change in relative abundance between the 2 GRATIs tag-labeled cell lines before and after in vivo tumor expansion (Fig. 5E). By design, a 1.0 E/I-ratio would suggest that gliomaOmA and gliomaNmA cells propagate equally during tumor progression, while an E/I-ratio >1.0 would indicate that the former propagates more than the latter in vivo. Using this method, we determined that gliomaOmA outgrew the gliomaNmA cell line in vivo by more than 40-fold (Fig. 5F). This finding is in stark contrast to the relatively similar in vitro proliferative rate of both cell lines (Fig. 3A), cautioning us not to solely base on in vitro growth to choose optimal culture condition for glioma cells.
The Prevalence of Highly Proliferative OPC-like Cells in Human High-grade Glioma
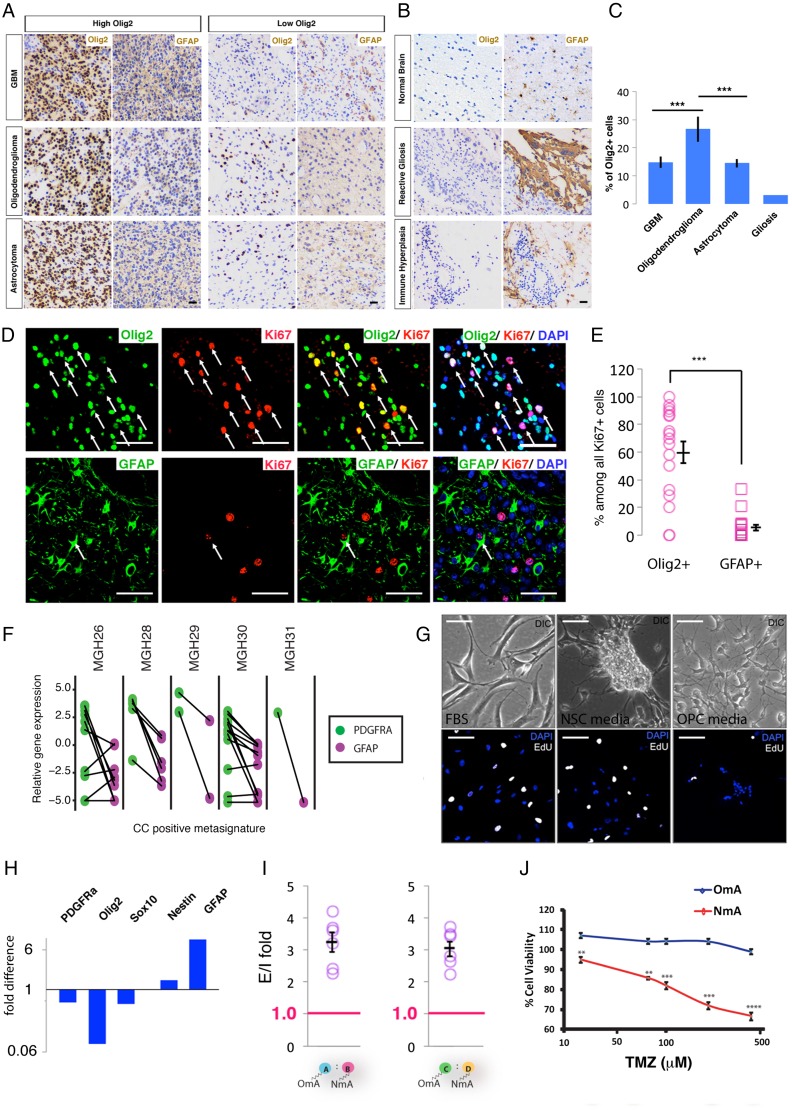
To examine the prevalence of OPC-like glioma cells in human high-grade glioma, we performed immunohistochemical analysis of Olig2 (as an OPC marker) or GFAP (as a non-OPC marker) on a set of tissue microarrays that contained 236 patient samples (see Supplementary Table S1 for a summary of patient characteristics). Consistent with previous findings, Olig2+ cells existed in almost all glioma samples to various extents33 (Fig. 6A and C). In addition to oligodendroglioma, GBMs or even pathologically well-defined astrocytomas also harbor a significant portion of tumor cells with prominent Olig2 staining (Fig. 6A, high Olig2 groups). This finding concurs with a recent study in which well-differentiated astrocytoma and oligodendroglioma were shown to contain OPC-like tumor cells.34 In contrast, there was no significant enrichment of GFAP-positive cells in any group of high-grade glioma. As a negative control, only marginal or no Olig2 staining was found in normal brain tissue or pathological tissues irrelevant to glioma such as reactive gliosis and immune hyperplasia, despite detection of prominent GFAP-staining signals (Fig. 6B).
Fig. 6.
Oligodendrocyte precursor cell (OPC)-like glioma cells are prevalent in human gliomas, and the choice of culture condition is critical to maintaining them in vitro. (A) and (B) Representative immunohistochemical images of tissue microarrays showing Olig2+ staining in glioblastomas (GBMs), astrocytomas, and oligodendrogliomas with varied abundance but not as significant a portion as in normal brains and pathological situations other than glioma. Scale bar: 100 μm. (C) Quantification of the percentage of Olig2+ cells in GBMs (N = 82), astrocytomas (N = 109), and oligodendrogliomas (N = 29). Gliosis tissue was used as control. (D) Representative images of immunostaining of human GBM samples co-stained with Ki67 and Olig2 (top row) or Ki67 and GFAP (bottom row). Arrows point to cells stained for both markers. Scale bar: 100 μm. (E) Quantification of Olig2+ or GFAP+ cells among all Ki67+ proliferating tumor cells (from 17 human GBM samples, manually counted). The percentage of Olig2+ or GFAP+ cells was calculated among the Ki67+ population. Each percentage was plotted in the graph, and the average ± SEM is shown. ***P ≤ 0.001. (F) Analysis of Patel et al single-cell sequencing data from human GBM cells. Cells with a positive cell-cycle meta-signature had their PDGFRα or GFAP expression analyzed. Solid lines connect the same cells. (G) Primary human glioma cells cultured in OPC, neural stem cell (NSC), or serum-containing media exhibited distinct morphologies (top row), and proliferation rates revealed by EdU labeling (bottom row). Scale bar: 100 μm. (H) NSC-media downregulated OPC-specific marker genes but upregulated NSC/astrocyte genes. Expression level was normalized to that of the cells cultured in OPC media. (I) q-GRATIs assay revealed that human gliomaOmA cells propagated significantly more than their NmA counterparts in vivo. N = 7 mice. Circles represent fold difference of individual animals; SEM is also represented. ***P ≤ .001. (J) Human gliomaOmA cells are more resistant to temozolomide than gliomaNmA cells. TMZ doses used were 25, 75, 100, 200, and 400 μM. **P < .01, ***P ≤ .001, ****P ≤ .0001.
To assess the proliferative rate of Olig2+ and GFAP+ cells in the tumor mass, we further co-stained Olig2 with Ki67 or GFAP with Ki67 on 17 GBM samples. Intriguingly, despite varied percentages of Olig2+ cells in these samples, a large proportion of them were undergoing active proliferation based on Ki67 staining (Fig. 6D, top row, and Fig. 6E), consistent with previous reports.33 In contrast, GFAP+ cells were less frequently Ki67+ (Fig. 6D, bottom row, and Fig. 6E). While our attempt to co-stain Ki67 with a second OPC marker (PDGFRα) failed due to unreliable staining on formalin-fixed paraffin-embedded samples, we took advantage of a recently published single cell RNASeq dataset in which 430 cells from 5 human GBMs were sequenced.22 We examined the expression of PDGFRα and GFAP in cells that were considered actively cycling, according to a cell-cycle meta-signature score (Material and Methods). Data in Fig. 6F show that among proliferating cells, most of them exhibited prominent PDGFRα expression, but few had GFAP expression. Notably, despite MGH28 and MGH30 being classified as mesenchymal and classical subtypes, respectively,13 the majority of proliferating cells in these GBMs were PDGFRα+. Taken together, our findings indicate that tumor cells resembling OPCs could contribute more prevalently to human high-grade glioma than previously thought and consist of the main proliferating pool in the tumor mass.
Culture Conditions Significantly Affected Cellular Properties and Tumorigenic Potential of Human Glioma Cells
Our mouse model studies thus far have demonstrated that culture condition could have a dramatic impact on cellular behaviors and tumor-propagating capacities. To evaluate the human relevance of such a notion, we directly cultured surgically resected and dissociated human GBM cells in NSC and OPC media, respectively. Two of 13 tumor samples have been successfully established into cell lines. Notably, despite a generally suboptimal success rate, both gliomaNmA and gliomaOmA sublines could be established from these 2 GBM samples. Although the small amount of tumor tissues precluded us from determining their subtype via gene expression profiling, we retrospectively confirmed the existence of Olig2+ tumor cells in the primary tumors used to establish cell lines (Supplementary Fig. S2).
Intriguingly, human GBM cell lines clearly recapitulated behaviors of mouse glioma cells. First, human gliomaNmA and gliomaOmA sublines exhibited distinct morphologies, with gliomaNmA frequently growing into tumor spheres and gliomaOmA having spindle morphology resembling OPCs (Fig. 6G, top row). Second, gliomaNmA cells are more proliferative in vitro when compared with gliomaOmA cells based on EdU incorporation (Fig. 6G bottom row). Third, qRT-PCR revealed that the gliomaOmA subline expressed OPC markers such as PDGFRα, Olig2, Sox10, NG2, and ERBB3 at a higher level than its gliomaNmA counterpart. (Fig. 6H). In contrast, NSC marker Nestin and astrocyte marker GFAP were upregulated in the gliomaNmA subline (Fig. 6H). Finally, intracranial xenografting assay using the q-GRATIs approach (Other data (C.L.), unpublished data, 2016) showed that the human gliomaOmA subline possessed significantly higher tumor-propagating activity than its gliomaNmA counterpart (Fig. 6I) despite an apparently slower proliferation in culture (Fig. 6G bottom row). Because temozolomide (TMZ) is the most commonly used chemotherapy drug in clinics, we also tested drug responsiveness of human glioma cells cultured in each condition, and found that gliomaNmA cells were significantly more sensitive to TMZ than gliomaOmA cells (Fig. 6J), suggesting that culture conditions could skew therapeutic predictions.
Discussion
Cell-of-origin Properties Are Critical for Sustaining Tumor-propagating Activity of Transformed Cells
Using genetic mouse models, we and others have previously shown that OPC can function as the cell of origin for high-grade glioma.15–18 The fact that OPCs can be transformed by certain oncogenic mutations while other brain cells fail to do so suggests that the signaling context within OPC is particularly susceptible to those mutations. However, given the plasticity of tumor cells, it is unclear whether OPC properties remain important for sustaining glioma after the initial transformation. Our current study demonstrated that tumor cells retaining OPC features can function as CSLCs, possessing the capacity to self-renew and differentiate into multiple neuroglial lineages in vitro as well as initiating secondary tumors when implanted into the brains of immune-compromised mice. However, after being cultured in the NSC culture condition, tumor cells lost OPC properties and had compromised capacity for forming tumors in vivo. Therefore, the properties of cell of origin are not only important for tumor initiation but also critical for tumor progression.
The Importance of OPC-like Tumor Cells in Human Glioma
Previously, based on an 840-gene signature, pathologically indistinguishable GBMs were stratified into 4 distinctive subtypes.13 Our comparative transcriptome analysis assigned OPC-originated glioma mouse models to human proneural subtypes,17,23 implicating the important role of OPCs in this subtype of human GBMs. Our current work further demonstrates that OPC-like glioma cells are critical for sustaining tumor progression. Therefore, understanding OPC biology in both its normal and malignant states should provide crucial insights for developing effective therapeutic strategies for the proneural subtype of glioma.
The proneural subtype was previously estimated to con stitute ∼25% of all human GBMs;13 however, recent studies suggest that prevalence of the proneural signature may be underestimated. It was found that glioma tissues from different locations within the same tumor could be classified into different subtypes, which frequently include proneural.35 Intriguingly, single-cell RNA sequencing techniques have revealed that even the same GBM can possess cells with signatures of both proneural and other subtypes.22 Moreover, altering a single gene was often sufficient to convert the proneural subtype of GBMs into other subtypes such as mesenchymal.36,37 Consistent with some other studies,38,39 the histological analyses in our work revealed that tumor cells with OPC features have been detected to varying extents in almost all gliomas including astrocytomas. Finally, regardless of their pathological classification, a significant portion of proliferative tumor cells in lower- and higher-grade gliomas that we have examined exhibited OPC features, suggesting that OPC-like glioma cells may be a promising therapeutic target for a large subset of human glioma patients.
The Importance of Culturing Tumor Cells in Media Conditions Tailored for the Cell of Origin
The importance of OPC properties led us to reflect on the conditions used for primary cell culture. Previously, it was elegantly demonstrated that serum-free media, commonly used for NSC culture, maintained features of parental glioma cells better than serum-containing media.6 Since OPC-like glioma cells exhibit some stem cell properties, we wondered whether NSC media, OPC media, or both could be used to maintain these cells. To our surprise, while NSC media could support tumor cell growth and sphere formation in vitro, it significantly impaired—rather than augmented—the tumorigenic potential of OPC-originated glioma cells in vivo. On the contrary, OPC media preserved OPC features of glioma cells in vitro and their tumorigenicity in vivo. Together, these observations highlight the fact that stem cell features may not always indicate tumorigenic potential, in line with a previous study showing that self-renewal does not correlate with tumorigenicity.40 Instead, the features of cell of origin need to be retained with corresponding culture conditions for the maintenance of tumorigenicity.
In the context of human glioma, how might our findings impact the practice of culturing CSLCs from patient samples, in which the cell of origin cannot be identified prior to cell culture? We propose that multiple types of media should be used, including those tailored for NSCs, OPCs, and astrocytes, to cover all possibilities. Once sublines are established, gene expression profiles of each cell line and the original tumor should be examined to see which one(s) recapitulate parental tumor profile. Such practices should greatly increase the chance of obtaining in vitro readouts that closely reflect clinical outcomes in drug discovery processes. In the case of highly heterogeneous cellular composition in the tumor mass, compounds should be tested in all sublines to identify the most broadly efficacious ones as the optimal therapeutic strategy against tumor heterogeneity or plasticity.
Funding
National Institutes of Health /National Cancer Institute (R01-CA136495 to H.Z.); Thousand Talent Program for Young Outstanding Scientists, China (to C.L.); National Natural Science Foundation of China (81302187 to H.H.); Fundação de Amparo a Pesquisa do Estado do Rio Grande do Sul (11/2072-2 to P.L. and G.L.). P.L. received a Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (Brazil) scholarship.
List of unpublished papers cited
Liu, C., Goldfarb, J., Ledur, P., Minussi, D and Zong, H. (2016) q-GRATIs: A convenient and cost-effective assay to precisely determine tumor functional genes both in vitro and in vivo. (In preparation).
Supplementary material
Supplementary data are available online at http://ndt.oxfordjournals.org.
Acknowledgments
We thank Ben Barres, B.J. Purow, Roger Abounader, John Lazo, Shwetal Mehta, Woo-Ping Ge, Kwon-Sik Park, David Kashatus, and Yuan Zhu for their critical comments on the manuscript.
Conflict of interest statement. None declared.
References
- 1. Ocana A, Pandiella A, Siu LL et al. Preclinical development of molecular-targeted agents for cancer. Nat Rev Clin Oncol. 2011;8 (4):200–209. [DOI] [PubMed] [Google Scholar]
- 2. Freedman LP, Gibson MC, Ethier SP et al. Reproducibility: changing the policies and culture of cell line authentication. Nat Methods. 2015;12 (6):493–497. [DOI] [PubMed] [Google Scholar]
- 3. Boquest AC, Shahdadfar A, Frønsdal K. Isolation and transcription profiling of purified uncultured human stromal stem cells: alteration of gene expression after in vitro cell culture. Mol Biol Cell. 2005;16:1131–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Maitra A, Arking DE, Shivapurkar N et al. Genomic alterations in cultured human embryonic stem cells. Nat Genet. 2005;37 (10):1099–1103. [DOI] [PubMed] [Google Scholar]
- 5. Deschavanne PJ, Fertil B. A review of human cell radiosensitivity in vitro. Int J Radiat Oncol Biol Phys. 1996;34 (1):251–266. [DOI] [PubMed] [Google Scholar]
- 6. Lee J, Kotliarova S, Kotliarov Y et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9 (5):391–403. [DOI] [PubMed] [Google Scholar]
- 7. Singh SK, Clarke ID, Terasaki M et al. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63 (18):5821–5828. [PubMed] [Google Scholar]
- 8. Magee JA, Piskounova E, Morrison SJ. Cancer stem cells: impact, heterogeneity, and uncertainty. Cancer Cell. 2012;21 (3):283–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yuan X, Curtin J, Xiong Y et al. Isolation of cancer stem cells from adult glioblastoma multiforme. Oncogene. 2004;23 (58):9392–9400. [DOI] [PubMed] [Google Scholar]
- 10. Sanai N, Alvarez-Buylla A, Berger MS. Neural stem cells and the origin of gliomas. N Engl J Med. 2005;353 8:811–822. [DOI] [PubMed] [Google Scholar]
- 11. Reynolds BA, Tetzlaff W, Weiss S. A multipotent EGF-responsive striatal embryonic progenitor cell produces neurons and astrocytes. J Neurosci. 1992;12 (11):4565–4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Suvà ML, Rheinbay E, Gillespie SM et al. Reconstructing and reprogramming the tumor-propagating potential of glioblastoma stem-like cells. Cell. 2014;157 (3):580–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Verhaak RGW, Hoadley KA, Purdom E et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17 (1):98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Phillips HS, Kharbanda S, Chen R et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9 (3):157–173. [DOI] [PubMed] [Google Scholar]
- 15. Lindberg N, Kastemar M, Olofsson T et al. Oligodendrocyte progenitor cells can act as cell of origin for experimental glioma. Oncogene. 2009;28 (23):2266–2275. [DOI] [PubMed] [Google Scholar]
- 16. Persson AI, Petritsch C, Swartling FJ et al. Non-stem cell origin for oligodendroglioma. Cancer Cell. 2010;18 (6):669–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liu C, Sage JC, Miller MR et al. Mosaic analysis with double markers reveals tumor cell of origin in glioma. Cell. 2011;146 (2):209–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Assanah M, Lochhead R, Ogden A et al. Glial progenitors in adult white matter are driven to form malignant gliomas by platelet-derived growth factor-expressing retroviruses. J Neurosci. 2006;26 (25):6781–6790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Simon R, Mirlacher M, Sauter G. Tissue microarrays. Bio t echniques . 2004;36 (1):98–105. [DOI] [PubMed] [Google Scholar]
- 20. R Development Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2011. [Google Scholar]
- 21. Warnes GR. Gplots: Various R Programming Tools for Plotting Data. 2013. Available at https://cran.r-project.org/web/packages/gplots/index.html. [Google Scholar]
- 22. Patel AP, Tirosh I, Trombetta JJ et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344 (6190):1396–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Galvao RP, Kasina A, McNeill RS et al. Transformation of quiescent adult oligodendrocyte precursor cells into malignant glioma through a multistep reactivation process. Proc Natl Acad Sci USA. 2014;111 (40):E4214–E4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dugas JC, Tai YC, Speed TP et al. Functional genomic analysis of oligodendrocyte differentiation. J Neurosci. 2006;26 (43):10967–10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Richardson WD, Pringle N, Mosley MJ et al. A role for platelet-derived growth factor in normal gliogenesis in the central nervous system. Cell. 1988;53 (2):309–319. [DOI] [PubMed] [Google Scholar]
- 26. Raff MC, Lillien LE, Richardson WD et al. Platelet-derived growth factor from astrocytes drives the clock that times oligodendrocyte development in culture. Nature. 1988;333 (6173):562–565. [DOI] [PubMed] [Google Scholar]
- 27. Cahoy JD, Emery B, Kaushal A et al. A Transcriptome database for astrocytes, eurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci. 2008;28 (1):264–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Obayashi S, Tabunoki H, Kim SU et al. Gene expression profiling of human neural progenitor cells following the serum-induced astrocyte differentiation. Cell Mol Neurobiol. 2009;29 (3):423–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Galli R, Binda E, Orfanelli U et al. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma . Cancer Res. 2004;64 (19):7011–7021. [DOI] [PubMed] [Google Scholar]
- 30. Codega P, Silva-Vargas V, Paul A et al. Prospective identification and purification of quiescent adult neural stem cells from their in vivo niche. Neuron. 2014;82 (3):545–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Emlet DR, Gupta P, Holgado-Madruga M et al. Targeting a glioblastoma cancer stem-cell population defined by EGF receptor variant III. Cancer Res. 2014;74 (4):1238–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jiang Y, Boije M, Westermark B et al. PDGF-B Can sustain self-renewal and tumorigenicity of experimental glioma-derived cancer-initiating cells by preventing oligodendrocyte differentiation. Neoplasia. 2011;13 (6):492–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ligon KL, Alberta JA, Kho AT et al. The oligodendroglial lineage marker OLIG2 is universally expressed in diffuse gliomas. J Neuropathol Exp Neurol. 2004;63 (5):499–509. [DOI] [PubMed] [Google Scholar]
- 34. Lindberg N, Jiang Y, Xie Y et al. Oncogenic signaling is dominant to cell of origin and dictates astrocytic or oligodendroglial tumor development from oligodendrocyte precursor cells. J Neurosci. 2014;34 (44):14644–14651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sottoriva A, Spiteri I, Piccirillo SGM et al. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc Natl Acad Sci USA. 2013;110 (10):4009–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Carro MS, Lim WK, Alvarez MJ et al. The transcriptional network for mesenchymal transformation of brain tumours. Nature. 2010;463 (7279):318–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ozawa T, Riester M, Cheng Y-K et al. Most human non-GCIMP glioblastoma subtypes evolve from a common proneural-like precursor glioma. Cancer Cell. 2014;26 (2):288–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bhat KPL, Balasubramaniyan V, Vaillant B et al. Mesenchymal differentiation mediated by NF-κB promotes radiation resistance in glioblastoma. Cancer Cell. 2013;24 (3):331–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ligon KL, Huillard E, Mehta S et al. Olig2-regulated lineage-restricted pathway controls replication competence in neural stem cells and malignant glioma. Neuron. 2007;53 (4):503–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Barrett LE, Granot Z, Coker C et al. Self-renewal does not predict tumor growth potential in mouse models of high-grade glioma. Cancer Cell. 2012;21 (1):11–24. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.