Abstract
Falcipain-2 and falcipain-3, haemoglobin degrading enzymes in Plasmodium falciparum, are validated drug targets for the development of effective inhibitors against malaria. However, no commercial drug targeting falcipains has been developed despite their central role in the life-cycle of the parasites. In this work, in silico approaches are used to identify key structural elements that control the binding and selectivity of a diverse set of non-peptidic compounds onto FP-2, FP-3 and homologs from other Plasmodium species as well as human cathepsins. Hotspot residues and the underlying non-covalent interactions, important for the binding of ligands, are identified by interaction fingerprint analysis between the proteases and 2-cyanopyridine derivatives (best hits). It is observed that the size and chemical type of substituent groups within 2-cyanopyridine derivatives determine the strength of protein-ligand interactions. This research presents novel results that can further be exploited in the structure-based molecular guided design of more potent antimalarial drugs.
Keywords: Malaria, homology modelling, docking, molecular dynamics, Falcipains
1. Introduction
Malaria is an infectious disease caused by a diverse group of erythrocytic protozoan parasites of the genus Plasmodium. It remains an exigent public health problem in the tropical areas of Africa, South America and parts of Asia, and continues to take its toll in morbidity and mortality with half of the world’s population under a permanent risk of infection leading to more than half a million deaths annually (WHO, 2013). Five Plasmodium species namely P. falciparum (Pf), P. vivax (Pv), P. ovale (Po), P. malariae (Pm) and P. knowlesi (Pk) are known to infect humans with Pf responsible for more than 90% of the malarial fatalities reported in sub-Saharan Africa. The predominance of Pf is attributed to its adaptability (Ashley, McGready, Proux, & Nosten, 2006; Prugnolle et al., 2011). Although the high occurrence of the Duffy negative trait among African populations lowers the threat posed by Pv, it is the most frequent and widely causative agent of benign tertian malaria in other parts of the world (Mendis, Sina, Marchesini, & Carter, 2001). In addition to the listed human malarial parasite forms, several other Plasmodium species, which infect non-human laboratory models, have been identified, and are of significant importance in understanding the parasite biology, the host-parasite interactions and in the drug development process (Langhorne et al., 2011).
Malaria still remains a major threat primarily due to the emergence of drug-resistant parasitic mutations, particularly to available frontline antimalarial drugs, a situation aggravated by the absence of an effective vaccine which has remained elusive to date (Hartjes, 2011). In addition, the recent reports indicating the emergence of resistance to artemisinin, which is the cornerstone to the current artemisinin-based combination therapies (ACT), emphasizes the continued need for identification of new drugs with novel scaffolds and mode of action (Mok et al., 2011; Saralamba et al., 2011). A formidable hurdle against successful elimination of malaria is the identification of a broad range of drugs targeting common druggable targets in both the active human forms of Plasmodium as well as the circulating wild species. Several Plasmodium drug targets including but not limited to falcipains (FPs) (Marco & Coteron, 2012), serine/threonine protein phosphatase 5 , serine repeat antigens, apicoplast-targeted tRNA-guanine transglycosylase (Gupta et al., 2015), Pf lactate dehydrogenase (Thillainayagam et al., 2014) and heat shock proteins (Hatherley, Blatch, & Tastan Bishop, 2014) have been identified. In the current study, our focus is on the falcipains which are validated drug targets (Teixeira, Gomes, & Gomes, 2011).
The FPs are a homologous family of four Pf cysteine proteases viz. FP-1, FP-2, FP-2’ and FP-3, which are fairly similar to papain-like human cathepsins but with eccentric features such as longer prodomains and a 14 amino acid (aa) insert between highly conserved residues near the C-terminus (Rosenthal, 2011). FP-1 is distantly related to the other FPs, and its role in the Plasmodium development process remains unclear (Kumar et al., 2007). FP-2 and FP-2’ have a ∼99% sequence identity and differ only in three amino acids; none of which is located in the active site cavity (Mane et al., 2013). Through chemical inhibitory studies against FP-2 and FP-3 as well as FP-2 gene disruption experiments, both enzymes have been shown to play indispensable roles either solely or in concert with other proteases, as they possess a nucleophilic thiolate group in their active site’s cysteine residue which is key for amide bond cleavage (Ehmke et al., 2011). These two proteases of the clan CA family are not only key in the haemoglobin degradation pathway which is the sole source of amino acids for the exponential growth and proliferation of the parasites but also in the erythrocyte egression and rupture process (Marco & Coteron, 2012; Rosenthal, 2011).
Several attempts combining chemical synthesis and in silico screening approaches have been undertaken in the past decade to discover and optimize inhibitors targeting parasite proteases of infectious agents, leading to two broad classes of chemical compounds viz. peptidomimetic and non-peptidic small molecules. Such attempts have successfully led to the discovery of several therapies in other diseases such as those against hepatitis C virus (HCV) (Melnikova, 2008) and human immunodeficiency virus (HIV) (Flexner, Bate, & Kirkpatrick, 2005). As reviewed by Mane et al., scientific efforts to develop FP-2 and FP-3 inhibitors have mainly realised peptide based compounds (Mane et al., 2013) such as α-ketoamides (Lee et al., 2003), peptidyl aldehydes (Rosenthal, Lee, & Smith, 1993), E-64 epoxysuccinyl derivatives (Schulz et al., 2007); but none has ever been commercialized as an antimalarial drug due to inherent poor pharmacological profiles as well as susceptibility to degradation by host enzymes (Ettari et al., 2010). The identification and validation of small non-peptide FP inhibitors is expected to overcome these shortcomings, and so far several compounds belonging to isoquinolenes, chalcones and others (Batra, Sabnis, Rosenthal, & Avery, 2003; Domínguez et al., 2005; Li et al., 1995; Liu, Wilairat, & Go, 2001) have been shown to possess antimalarial potency either on whole Pf parasite cultures or directly on FP-2 protein at low micromolar ranges. Coteron et al., recently reported a new class of compounds which belong to the heteroaryl nitrile class hitherto vouched as the most potent. Through a sequential lead optimization process, several 5-substituted-2-cyanopyrimidines derivatives (CPs) with FP-2 and FP-3 inhibitory potency and anti-plasmodial activity in the picomolar to low nanomolar ranges were obtained (Coteron et al., 2010).
To facilitate the discovery of novel potential antimalarial compounds, our aim was to set up a reliable integrated virtual screening workflow comprising of homology modelling, molecular docking, molecular dynamics (MD) and binding free energy (BFE) calculations, which can be used to identify potential non-peptide compounds against plasmodial cysteine proteases. A diverse set of compounds from the literature with reported potency against FP-2 and/or FP-3 were used to determine their mode of interaction with FP-2 and FP-3 and their plasmodial homologs (targets) and their selectivity towards the human papain-like cathepsins (off-targets). These compounds are CPs (Coteron et al., 2010), chalcones (Domínguez et al., 2005; Li et al., 1995; Liu et al., 2001), isoquinolenes (Batra et al., 2003), and thiosemicarbazones (Chipeleme, Gut, Rosenthal, & Chibale, 2007; Chiyanzu et al., 2003; Greenbaum et al., 2004). After docking, guided by molecular docking energy scores, compounds that showed the best scores were further put through MD simulations, and interaction energy quantified via BFE calculations. The dynamical behaviour and strength of interactions of the protein-compound complexes were analysed to describe the possible binding modes, the interacting structural elements as well as the stability of the complexes, information which may be potentially useful in the drug discovery and development process of novel broad-spectrum anti-malarial agents. The key amino acid residues contributing to different interactions with potential inhibitors were identified. Overall, the following observations were made: (i) there exist amino acid composition differences in the subsites of plasmodial FP-2 and FP-3 homologs and human cathepsins, which could be important for inhibitor design; (ii) out of the four classes of compounds used, the CPs are the best docked inhibitors with very low binding free energies against plasmodial proteases compared to human cathepsins, thus showing a degree of selectivity; (iii) The substituent chemical groups in the main CPs scaffold determine the overall potency of the individual derivatives.
2. Methods and materials
The flow diagram of the methodology followed in this study is presented in Figure 1 and the details are given below.
Figure 1.
Graphical workflow for identification of FP-2/3 homologs (sequence and structure) and analysis of non-peptidic compounds as potential inhibitors via in silico approaches. Bold and in brackets are the key databases, web-servers and tools used.
2.1 Sequence retrieval
FP-2 and FP-3 protein sequences (accession numbers: PF3D7_1115700 and PF3D7_1115400 respectively) were retrieved from PlasmoDB version 9.3 (Aurrecoechea et al., 2009), and used as queries to retrieve homolog sequences of other Plasmodium species and humans using the BLASTP tool in the PlasmoDB and the NCBI database (Sayers et al., 2009) respectively. To confirm if the retrieved hits were indeed the true orthologs reverse BLAST searches were performed. Only sequences with significant query coverage, E-values lower than 1.0 × 10−5 were selected (Text S1). Crystal structures of the human cathepsins (Cat K [PDB ID: 3OVZ], Cat L [PDB ID: 3OF8], Cat S [PDB ID: 1NPZ] together with those of FP-2 [PDB ID: 2OUL] and FP-3 [PDB ID: 3BWK] were retrieved from the Brookhaven Protein Data Bank (PDB) (Bernstein et al., 1977).
2.2 Multiple sequence alignment
Multiple sequence alignment (MSA) was done using web based servers, namely MAFFT (Katoh, Misawa, Kuma, & Miyata, 2002) and PROMALS3D (Pei, Kim, & Grishin, 2008) and alignment outputs compared to determine the alignment accuracy. For MAFFT program, the following sequence alignment parameters were utilized; substitution matrix was set as BLOSUM62 scoring matrix (Eddy, 2004), gap opening and extension penalty of 1.53 and 0.123 respectively. The number of tree building steps was adjusted to 2 with maximum iteration set at 2. For PROMALS3D, structural information from the crystal structures of FP-2 and FP-3 was added to guide the alignment using default parameters with an exception of PSI-BLAST expect value which was adjusted to 0.0001. Alignment parameter for sequences within groups in the first alignment stage was set to the slow but more accurate PROMALS algorithm (Jimin Pei & Grishin, 2007). The initial alignment was done for the full length protein sequences and trimmed using JalView software (Waterhouse, Procter, Martin, Clamp, & Barton, 2009) to obtain the catalytic domain portions of the cysteine proteases which were realigned. From the MSA, sequence identities, similarities and conserved regions were identified. Subsite residues were extracted from the alignment into a Fasta file and submitted to WebLogo (Crooks, Hon, Chandonia, & Brenner, 2004), a web based sequence logo generator, to determine the residue conservation at these specific positions in a WebLogo presentation.
2.3 Homology modelling
For each of the FP-2 and FP-3 plasmodial homologs, the HHpred web server (Söding, Biegert, & Lupas, 2005) was utilized to search for suitable templates for building high quality models to be used in the subsequent docking and molecular dynamics steps. Selection of predicted templates was based on the one with the highest sequence identity, coverage and resolution. Two templates were retrieved from PDB namely [PDB ID: 2OUL] (Wang et al., 2007) and [PDB ID: 3BWK] (Kerr et al., 2009)for FP-2 and FP-3 respectively. Template quality validation was performed using MetaMQAPII (Benkert, Biasini, & Schwede, 2011), ANOLEA (Melo, Devos, Depiereux, & Feytmans, 1997), QMEAN (Benkert, Tosatto, & Schomburg, 2008) and PROCHECK (Laskowski, MacArthur, Moss, & Thornton, 1993). Manual adjustments were made in the template-target alignment files where applicable by comparing the HHpred and PROMALS3D structural alignment output with that of MAFFT. For each homolog, 200 models were calculated using MODELLER version 9.10. Models were ranked using DOPE Z scores (Shen & Sali, 2006). The top three models with the lowest energy scores for each protein were further validated by MetaMQAPII and PROCHECK. Disulphide bond formations in the models were assessed through Protein Interaction Calculator webserver (Tina, Bhadra, & Srinivasan, 2007) in relation to the FP-2 and FP-3 templates. The top best model for each protein was selected for the subsequent steps of molecular docking and molecular dynamics.
2.4 Compounds
All the compounds (CPs, chalcones, isoquinolenes, thiosemicarbazones) and associated activity data used in this study were retrieved from published data. In DS (Discovery Studio version 3.1, Accelrys Software Inc. Discovery Studio Modelling Environment, San Diego: 2011), compound 2D structures were sketched and converted to a 3D format. The 3D geometry of structures was cleaned and optimized to attain stable conformations with minimum energy. Figure 2 shows the chemical 2D structures of all compounds drawn using MarvinSketch 6.1.0 (Marvin Calculator Plugin and Chemical Terms Demo ChemAxon).
Figure 2.
2-D structures of the different sets of non-peptidic compounds used in this study.
2.5 Protein-ligand docking
In order to explore the likely binding modes of the CPs, chalcones, isoquinolenes, thiosemicarbazones onto the substrate pocket of FP-2 and FP-3, the modelled 3D homologs and crystal structures of human cathepsins, molecular docking was performed using AutoDock4.2 (Morris et al., 2009). Prior to docking, identical chain sequence information and co-crystallized water molecules in protein crystal structures were deleted using DS. AutoDock tools (ADT) were used to prepare the ligands, protein structures and calculated models for the actual modelling simulations. Using the Gasteiger-Hückel method in ADT, the partial atomic charges of the ligands were assigned. Cubic grid points were set at 70, 70 and 65 along the x, y and z directions respectively for all the ligands with a grid spacing of 0.3472 Å. A centroid point was generated on the catalytic Cys42 of FP-2 and corresponding positions in all homologs. The grid box spanned an area of residues around a 12 Å radius. Docking simulations were carried out locally on a Linux based cluster with the parameters set as follows; Genetic Algorithm (GA) and the Lamarckian Genetic Algorithm (LGA) variant were used for receptor-ligand conformational search. The population size was set at 150, 100 GA runs, maximum energy evaluations of 450,000 and maximum number of generations set at 27,000. Cluster analysis for docked results was done using a root mean square (RMS) tolerance of 2.0 Å. Ligands in the best predicted poses were visually analysed to identify interactions of specific interest using DS. An energy heat map was created using the gnuplot program (Williams and Kelley, 2011). An ad hoc Perl script utilizing LigPlot+ subroutines (Laskowski & Swindells, 2011) was used to determine the hydrogen bonding and hydrophobic interactions between each ligand and active site residues. From the docking results, protein-compound complexes with low docking energy scores were selected for the molecular dynamics and binding free energy (BFE) calculations.
2.6 Molecular dynamics simulations
To determine the stability and map the intermolecular interactions of CPs with the cysteine proteases, molecular dynamics (MD) simulations of up to 10 nanoseconds (ns) were performed using GROMACS 4.5.5 package (Pronk et al., 2013) with AMBER96 force field (Kollman, 1996). The protonation state of all the protein’s ionisable amino acid groups at a pH of 5.0 were assigned using the DS protein utility module, and protonation was performed accordingly with the pdb2gmx functionality of GROMACS to mimic the acidic environment of the lysosomal vacuole where the proteins reside (Hogg et al., 2006). Ligand topology files were generated using ACPYPE interface (Sousa da Silva & Vranken, 2012) and Open Babel (O’Boyle et al., 2011). The protein-CP complexes were solvated in a triclinic box of dimension 17.5 Å and the flexible SPC water model was used to create an explicit continuous water model. The system was neutralized to a total charge of zero by randomly replacing water molecules with 0.15 M counter ions (Na+ and Cl−). To avoid high energy interactions and steric clashes, the system was subjected to a steepest descent energy minimization process without constraints until a tolerance of 1000 kJ mol−1 nm−1 was obtained. Each energy minimised system was then equilibrated in the canonical ensemble (for 200 ps through the NVT ensemble set at 300 K) and subsequently in the isothermal-isobaric ensemble (for 200 ps). The NPT conditions were set at 1.0 bar of pressure in all directions and a pressure coupling constant (τP) of 2.0 ps using the V-rescale thermostat (Bussi, Donadio, & Parrinello, 2007) and the Parrinello-Rahman barostat (Parrinello, 1981) algorithms respectively. The values of the isothermal compressibility were set at 4.5 × 10−5 bar−1 for water simulations. The pre-equilibrated systems were thereafter subjected to a 10 ns production run with an integration time step of 2 femtoseconds (fs) while maintaining temperature and pressure. All bond lengths during equilibration and production runs were constrained utilizing the LINCS algorithm (Hess, Bekker, Berendsen, & Fraaije, 1997). Long range electrostatic interactions were approximated by the particle-mesh Ewald algorithm (Petersen, 1995) with a 0.16 nm Fourier grid spacing and a fourth order interpolation while the cut-off distances for calculation of Coulomb and van der Waal interactions were set at 1.4 nm. During the sampling process, trajectory snapshots were stored at every 2 ps for structural analysis.
MD trajectories were analysed using the GROMACS in house tools in conjunction with LigPlot +. In house ad hoc Perl and Python scripts were used to automate the analysis process. Xmgrace of Grace 5.1.21 was used to plot MD graphical displays while protein-ligand complex visualizations were performed using PyMOL (The PyMOL Molecular Graphics System, Version 1.6.0.0 Schrödinger, LLC.) software.
2.7 Binding free energy calculations
According to the Molecular Mechanics-Poisson Boltzmann Surface Area (MM-PBSA), the strength of interactions between the different protease-CP complexes (total BFE) was determined using g_mmpbsa tool (Kumari, Kumar, & Lynn, 2014). Using the single trajectory approach, the calculations were performed on 4,000 snapshot structures extracted at 2 ps intervals over the last 8 ns of each of the system’s GROMACS generated trajectories. The following set of equations was used to calculate the BFE:
| (1) |
| (2) |
| (3) |
| (4) |
| (5) |
where Gcomplex, Greceptor and Gligand denote the absolute free energy of the protein-ligand complex, apoprotein, and ligand respectively. Egas, Eint, Evdw and Eele signify gas-phase, internal, van der Waals and electrostatic energies in that order. The solvation free energy (Gsolv) is composed of polar (Gpol) and nonpolar (GSA) terms: the former is estimated from a solution of the linear Poisson-Boltzmann (PB) equation, and the latter from the solvent accessible surface area (SASA) using a water probe of radius 1.4 Å, an offset value (b) of 3.84928 kJ.mol−1 and surface tension proportionality (γ) set at 0.0226778 kJ.mol−1.Å−2. T and S correspond to temperature and solute entropy, respectively. To decipher key determinants involved in the ligand binding process, the BFE energetic contributions by van der Waals forces, electrostatic energy, as well as polar and non-polar solvation energy were decomposed for each the protein-ligand complex systems. A per-residue decomposition analysis of the contribution of each protein residues to the three components of BFE was also calculated.
2.8 System specifications
Unless otherwise stated, all the computational analysis including 3D model building and MSA were performed on a Linux Intel Xeon workstation equipped with four 3.10 GHz parallel E3–1220V2 processors, Quadro K600/PCIe/SSE2 graphics card and 31.1 GB RAM. Molecular dynamics and binding BFE calculations were performed using a local cluster, and the Tsessebe cluster (Sun) high performance computing unit at the CHPC (Centre for high performance computing), Cape Town, South Africa. All webservers and databases used for data set retrieval and analysis are listed in Table S1.
3. Results and discussion
3.1 Sequence analysis
Besides the protein sequences of FP-2 and FP-3, seven plasmodial and three human homologs were retrieved from PlasmoDB and NCBI databases (Table 1). That the retrieved protein sequences were all FP-2 and FP-3 orthologs was strongly supported by the reverse BLAST results. Interestingly, in most of the reverse BLAST results, the first hit was to FP-3 rather than FP-2, even though the initial BLAST search was performed with FP-2 as query sequence, indicating that some of the homolog proteins are not accurately annotated in the literature.
Table 1.
A summary of FP-2 and FP-3 homologs from different Plasmodium species and Homo sapiens
| Accession number | Common name (Abbreviation) |
Source organism (Abbreviation) |
% SI | Position in whole sequence | Catalytic domain numbering | |
|---|---|---|---|---|---|---|
| FP-2 | FP-3 | |||||
| PF3D7_1115700 | Falcipain-2 (FP-2)§ | P. falciparum (Pf) | 100 | 66 | 244-484 | 1-243 |
| PF3D7_1115400 | Falcipain-3 (FP-3)§ | 66 | 100 | 250-492 | 1-249 | |
| PVX_091415 | Vivapain-2 (VP-2) | P. vivax (Pv) | 62 | 66 | 246-487 | 1-242 |
| PVX_091410 | Vivapain-3 (VP-3) | 57 | 57 | 253-493 | 1-241 | |
| PCHAS_091190 | Chaubapain-2* (CP-2) | P. chabaudi (Pc) | 50 | 48 | 231-471 | 1-241 |
| PKH_091250 | Knowlesipain-2* (KP-2) | P. knowlesi (Pk) | 57 | 57 | 252-495 | 1-244 |
| PVX-091260 | Knowlesipain-3* (KP-3) | 57 | 60 | 240-479 | 1-240 | |
| PBANKA_093240 | Berghepain-2* (BP-2) | P. berghei (Pb) | 51 | 47 | 228-468 | 1-241 |
| PY00783 | Yoelipain-2* (YP-2) | P. yoelii (Py) | 48 | 47 | 232-472 | 1-241 |
| gi | 157830076 | Cathepsin-K (Cat K) | H. sapiens (Hs) | 38 | 41 | 115-329 | 1-215 |
| gi | 313754424 | Cathepsin-L (Cat L) | 37 | 38 | 113-333 | 1-221 | |
| gi | 30749675 | Cathepsin-S (Cat S) | 36 | 37 | 115-331 | 1-217 | |
Marked with indicates the query sequences while
indicates sequences renamed for convenience. % SI (Percentage Sequence Identity) is based on the catalytic domain of FP-2 and FP-3.
MAFFT MSA output after additional minor manual adjustments was considered to be the best for the catalytic domain. Sequence identities (SI) were calculated (Table 1) and conservation of protein sequence features were determined (Figure (3a)). From the primary sequence alignment, 45 aa positions (highlighted in green) inclusive of the clan C1A characteristic catalytic triad residues namely Cys, His and Asn (marked with an asterisk) and the Gly-Cys-X-Gly-Gly motif were fully conserved in all protein sequences. In addition, up to 18 aa positions were only conserved in the plasmodial proteases (highlighted in blue) whereas 23 aa positions were unique in the human cathepsins (highlighted in black). Up to 34 aa positions were conserved exclusively in the rodent plasmodial proteases (highlighted in grey). 4 aa positions were conserved only in the human plasmodial proteases (highlighted in red). Two clearly visible amino acid inserts present only in the plasmodial proteases were observed, one at the N-terminal and the other near the C-terminal (boxed). The first insert of about 17 residues (commonly referred to as nose) has been shown to be important for the correct folding of the catalytic domains of FP-2 and FP-3 before assuming its final active conformation (Pandey & Dixit, 2012). The second insert consisting of approximately 14 aa (commonly referred as arm) structurally forms a highly flexible β-hairpin. Wang et al., associated it with the haemoglobin (substrate) binding process (Wang et al., 2007). Both in FP-2 and FP-3, the arm is fairly conserved while in the rest of the plasmodial orthologues variations were observed. Although it is yet to be determined, it can be assumed that the inserts play similar functions in the other plasmodial homologs as established in FP-2 and FP-3. Interestingly, the rodent plasmodial proteases had a unique aa pattern at the arm region compared to the human plasmodial forms. Besides the catalytic cysteine residue, all plasmodial proteases had a total of eight cysteine residues in the catalytic domain which form a set of four disulphide bonds. In contrast, the human cathepsins had only six extra cysteine residues capable of forming three sets of disulphide bonds.
Figure 3.
Sequence analysis and subsite amino acid composition of FP-2, FP-3 and homologs. (a) MAFFT multiple sequence output of FP-2/3 and homologs showing conservation of functional and structural residues. The Clan CA characteristic catalytic triad residues are indicated with an asterisk. The two unique inserts in all plasmodial proteases are indicated with a black dashed box. (b) Summary of the amino acid composition (table) and conservation (sequence logo) of all four subsites for FP-2/3 and homologs as determined by WebLogo. The relative height of each letter and total height indicates the relative frequency of corresponding aa and information content respectfully per site in all the sequences.
A characteristic feature of cysteine proteases is the presence of an active site pocket constituted by residues surrounding the catalytic triad. It is situated in a cleft between the structurally conserved R and L domains and consists of subsite S1, S2, S3 and S1’ (Kerr et al., 2009). By comparing the active site pockets of human cathepsins and plasmodial cysteine proteases, the structural information that controls the selectivity and functioning of these proteins can be determined, a key feature in designing efficacious and more potent drugs. As shown in Figure (3b), it was observed that S1 and S3 subsite residues are fairly conserved, while S2 as well as a significant number of aa positions in S1’ are highly varied. Although at the sequence level the effect of these observations on the structure and functions of the proteins cannot be determined, it is well known that S2 plays a key role in the specificity of ligand binding (Pandey & Dixit, 2012). Amino acid residues Gln36 (S1), Asn173 (S2) and Asn204 (S1’), which are key in polarizing His174 (S1’) during catalysis, are conserved in all proteases with an exception of the S2 position of Cat L which has Asp residue (numbering as per FP-2 catalytic domain). In S1, the two conserved cysteine residues (Cys39 and Cys80) form one of four disulphide bridges critical in stabilizing the proteins (Hogg et al., 2006). The small polar Gly40 residue is conserved in all homologs except in the rodent associated proteases where it is replaced with a biochemically related nonpolar alanine residue. Even though the fifth position of S1 was highly varied among the proteases, residues occupying this position were all polar suggesting a conserved function. S2, which has been defined as the major pocket that determines ligand specificity in cysteine proteases, was mainly occupied by hydrophobic residues as per previous observation (Pandey & Dixit, 2012). A striking difference between the human plasmodial homologs and the rest including the cathepsins is the presence of a polar charged residue at the S2 pocket’s hollow end of human plasmodial proteases while the rest have a small uncharged residue at the same position. Zhao et al., observed that the S2 of human Cat K was constituted by hydrophobic residues, but from our results this was the case only with Cat L and Cat S (Zhao et al., 1997). S2 pocket volume of FP-2 vis-à vis FP-3 and VP-2 vis-à-vis VP-3 have been shown to vary in size indicating that careful consideration of the size and shape of inhibitor groups targeting this site have to be carefully determined. FP-2 has less bulky Leu84 and Leu172 at the S2 opening groove while in FP-3, these residues are replaced with more bulky Tyr86 and Pro174 resulting in a narrower distal end (Sabnis, Desai, Rosenthal, & Avery, 2003). In VP-3, the area between S1’ and S2 subsites is folded inwards making the S2 narrower than in VP-2 (Desai & Avery, 2004). In the rodent plasmodial proteases, the S2 opening residues are highly conserved with less bulky Ile85 and Ala173 on either side. It was observed that S3 has a rich highly conserved glycine component, a feature that provides additional protease-substrate stability via hydrogen bonding (Desai & Avery, 2004). At the opening of the V-shaped cleft there exists a highly conserved tryptophan residue in all the proteases, which has been found to form hydrophobic interaction with substrates (Sabnis et al., 2003).
3.2 Homology modelling and quality validation
Seven high quality protein models of the catalytic (mature) domains of Plasmodium FP-2 and FP-3 homologs, VP-2 (Vivapain-2), VP-3 (Vivapain-3), BP-2 (Berghepain-2), CP-2 (Chaubapain-2), KP-2 (Knowlesipain-2), KP-3 (Knowlesipain-3) and YP-2 (Yoelipain-2) were calculated using MODELLER version 9.10. The position of the catalytic domain for each of the FP homologs in the whole sequence and the adopted catalytic domain numbering is shown in Table 1. For each of these proteins modelled, two templates (PDB IDs: 2OUL and 3BWK) were consistently identified from HHpred webserver (Table S2). To attain alignment accuracy, the secondary structure prediction in the HHpred alignments were considered, and HHPred, PROMALS3D and MAFFT alignments were compared; if necessary hand adjustments were done (Figure (3a)). The suitability of the obtained templates was scrutinized by assessing the target sequence coverage and resolution. Furthermore, template quality assessment results by MetaMQAPII, ANOLEA, and Ramachandran plot showed that the two selected templates were suitable. Depending on the sequence identity score, KP-2 was modelled using 3BWK while BP-2, CP-2, YP-2 used 2OUL and the rest used both templates. All resulting 3D models were assessed by DOPE-Z score, Global Distant Test-Total Score (GDT-TS) score as well as by the same programs used for template quality check. From Table S2, KP-3 had the best normalized DOPE-Z and GDT-TS scores. Shen & Sali estimated that DOPE Z-score below −0.5 are indicators of a protein structure near to the native form, as such it is evident that all models generated were of high quality (Shen & Sali, 2006). The color code presentation of MetaMQAPII results indicated that they had been accurately modelled, except for some loop regions in some cases (blue for accurate regions and red for problematic regions) as shown in Figure S1. Overall, the obtained evaluation results indicated that the models are of high quality, especially in the active site region, and can be used for inhibitor docking experiments.
3.3 Docking studies
A total of 20 non-peptide inhibitors of the chemical classes of isoquinolines, chalcones, thiosemicarbazones and CPs were docked to twelve cysteine proteases. Out of these twelve, three were human cysteine proteases, namely Cat L, Cat S and Cat K. They were used to assess protein-ligand interaction characteristics which could provide significant information with regard to selectivity. Two were crystal structures of FP-2 and FP-3 (2OUL and 3BWK respectively) and the remaining seven proteins were the 3D models of FP-2 and FP-3 Plasmodium homologs. The results are presented in Figure 4 and Table 2. The CP series of compounds exhibited the lowest docking free energy of binding (strong affinity) and lowest inhibition constants against almost all proteins compared to the other classes of compounds used, and thus were selected for the subsequent steps of MD simulations and BFE calculations.
Figure 4.
A heatmap showing the interaction free energies of all compounds when docked against plasmodial cysteine proteases and human cathepsins using AutoDock4.2. The energy code ranges from high (yellow) to low (black).
Table 2.
AutoDock inhibition constants (μM) of the CPs onto FP-2 and FP-3 and homologs. Enclosed in brackets are the IC50 from wet lab assays. The references for the assays were given in the main text.
| Protein | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Plasmodial | Human homologs | |||||||||||
| CMPD | FP-2 | FP-3 | VP-2 | VP-3 | KP-2 | KP-3 | BP-2 | CP-2 | YP-2 | Cat S | Cat L | Cat K |
| CPA | 0.80 (0.0005) | 0.33 (0.006) | 0.14 | 3.32 | 2.51 | 0.45 | 0.38 | 0.54 | 1.02 | 1.68 | 2.09 | 0.24 |
| CPB | 0.78 (<0.0005) | 0.11 (0.011) | 0.52 | 3.14 | 2.71 | 0.18 | 0.39 | 0.85 | 3.42 | 3.19 | 2.27 | 0.43 |
| CPC | 0.53 (0.0005) | 0.12 (0.006) | 0.12 | 1.61 | 1.22 | 0.21 | 0.34 | 0.36 | 0.50 | 0.71 | 0.79 | 0.13 |
| CPD | 1.23 | 0.35 | 0.46 | 2.63 | 1.06 | 0.36 | 0.58 | 0.32 | 0.13 | 6.80 | 0.32 | 1.08 |
| CPE | 1.62 | 0.27 | 0.72 | 1.91 | 1.35 | 0.48 | 0.38 | 0.47 | 0.82 | 11.16 | 0.36 | 1.14 |
| CPF | 1.58 | 0.29 | 0.36 | 2.34 | 1.91 | 0.20 | 0.94 | 0.81 | 1.68 | 12.19 | 0.31 | 0.94 |
| CPG | 0.16 | 0.03 | 0.02 | 0.81 | 0.11 | 0.04 | 0.05 | 0.16 | 0.04 | 0.27 | 0.14 | 0.10 |
| CPH | 0.08 | 0.01 | 0.01 | 0.25 | 0.06 | 0.02 | 0.01 | 0.04 | 0.04 | 0.16 | 0.06 | 0.13 |
| CPI | 0.08 | 0.01 | 0.01 | 0.06 | 0.02 | 0.02 | 0.03 | 0.03 | 0.02 | 0.22 | 0.03 | 0.15 |
| CL-A | 1.63 | 3.50 | 9.03 | 2.98 | 2.25 | 1.60 | 4.10 | 2.82 | 6.10 | 14.95 | 5.02 | 2.36 |
| CL-B | 15.64 | 22.94 | 4.18 | 28.42 | 8.58 | 18.86 | 12.27 | 6.60 | 10.26 | 7.20 | 21.99 | 12.36 |
| CL-C | 3.57 (1.80) | 2.08 | 1.53 | 8.62 | 4.60 | 7.85 | 2.45 | 2.94 | 1.52 | 7.18 | 6.31 | 0.92 |
| IQ-A | 0.34 (3.00) | 0.58 | 0.34 | 0.63 | 0.85 | 0.60 | 0.17 | 0.08 | 0.37 | 0.33 | 0.54 | 0.24 |
| IQ-B | 3.41 (3.00) | 5.41 | 3.55 | 5.75 | 4.02 | 5.84 | 2.50 | 3.73 | 4.45 | 4.66 | 5.88 | 1.67 |
| TSC-A | 24.99 | 40.27 | 15.17 | 42.72 | 26.92 | 18.64 | 14.00 | 12.67 | 14.11 | 33.75 | 26.48 | 22.12 |
| TSC-B | 5.25 (4.40) | 10.79 | 11.14 | 10.51 | 14.58 | 6.45 | 23.34 | 19.48 | 15.47 | 12.58 | 9.35 | 19.20 |
| TSC-C | 20.28 (10.00) | 35.73 | 14.57 | 87.47 | 33.82 | 35.30 | 22.57 | 27.24 | 31.24 | 36.38 | 14.69 | 30.51 |
| TSC-D | 12.72 (5.80) | 45.65 | 13.09 | 48.25 | 30.33 | 14.39 | 13.83 | 42.55 | 24.61 | 42.84 | 5.88 | 62.04 |
| TSC-E | 9.84 (3.80) | 40.69 | 15.66 | 43.92 | 10.84 | 8.54 | 19.89 | 12.99 | 16.43 | 52.21 | 13.66 | 37.77 |
| TSC-F | 2.13 (2.25) | 2.13 | 1.87 | 1.85 | 2.29 | 3.75 | 0.21 | 0.29 | 0.87 | 3.97 | 1.72 | 3.60 |
CMPD=Compound, CPX=2-cyanopyrimidine, CL=Chalcone, IQ=Isoquinolene, TSC=Thiosemicarbazone
In Table 2, thiosemicarbazones had high free energy of binding which implied lower estimated binding affinities and consequently, higher inhibition constants. AutoDock inhibition constants of these compounds onto FP-2 were comparable with results obtained from experimental enzyme assays (Chiyanzu et al., 2003; Greenbaum et al., 2004; Liu et al., 2001). In both approaches, inhibition constants were at μM levels, although in some cases the experimental values were half or even a third of the docking values. Docking results were varied between 2.13 to 24.99 μM. Chalcones showed the same trend as thiosemicarbazones both in in silico results and experimental data (Li et al., 1995). The two docked isoquinoline derivatives showed varied binding affinity. Isoquinoline-A had considerably good docking free energies against all proteins compared to thiosemicarbazones and chalcones. Interestingly, while assay results were indicating inhibition at 3 μM against FP-2 for both compounds (Batra et al., 2003), the in silico result for isoquinoline-B agreed, but the isoquinoline-A was 10 fold less. In fact, isoquinoline-A was good in the context of Plasmodium homologs, with estimated inhibition constants in the nanomolar range. Even though its uniform docking free energy among Plasmodium proteins and the human cathepsins raises concerns of selectivity, it could be used as a starting scaffold for further studies involving chemical modifications leading to derivatives with increased potency and safety profiles.
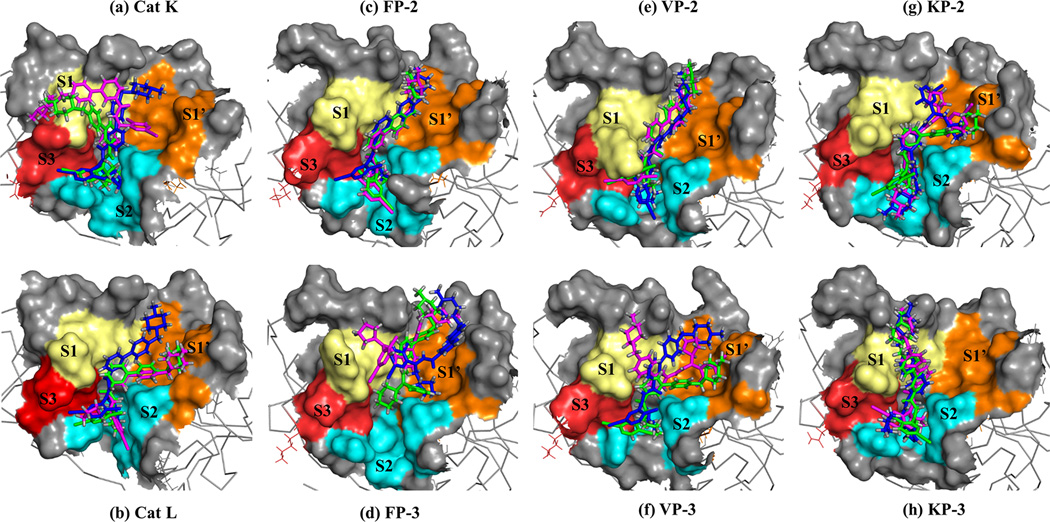
Among the CPs, compound CPG, CPH and CPI had the lowest inhibition constants as determined by AutoDock (See Table 2) and had interaction energies ranging between, −8 and −10 kcal.mol−1 across all homologs (Figure 4). According to docking results these inhibitors had high binding affinity for FP-3 than FP-2. The estimated inhibition constants were all in the nanomolar range. Interestingly, experimental inhibition constants for CPA-CPC for FP-2 and FP-3 were much lower than the in silico results (Coteron et al., 2010). Compounds with a cyclohexyl or cyclopentyl at the P2 position (corresponds to S2 in protease) and pyridinyl-phenyl combinations yielded anti parasitic activity at sub nanomolar and nanomolar levels against FP-2 and FP-3 respectively (Coteron et al., 2010). According to Table 2, CPs are better inhibitors against FP-3, VP-2, and KP-3 as compared to FP-2, VP-3 and KP-2 respectively. In the case of vivapains, Desai et al.,(Desai & Avery, 2004) observed that VP-2 was more sensitive to inhibitors than VP-3 as its S2 opening was wider. In contrast, the other classes of inhibitors seemed to inhibit FP-2 more than FP-3. The activity of CPs seemed to be largely influenced by the chemical composition of R1 and R2 (Figure 2). Compound CPG, CPH and CPI were the strongest binders against all proteases (See Figure 4 and Table 2). Interestingly, R1 group which was the same in CPA-CPC, CPD-CPF and CPG-CPI seemed to affect the overall docking energy score in the human cathepsins and falcipains. CPs had a cyclopentyl or cyclohexyl at R2 which bound at or near the S2 position. Compounds CPC, CPF and CPI which had a cyclohexyl group at R2 exhibited lower docking energies and inhibition constants when in complex with most proteins. This was dependent on whether the opening of S2 was wide enough to allow the entry of the R2 group. Rosenthal et al., (Rosenthal, Wollish, Palmer, & Rasnick, 1991) previously established that the S2 of FP-2 preferred the phenyl group of Phenylalanine, a group that is chemically similar to the cyclohexyl. Additionally, the binding poses of CPs varied in different proteins as seen in Figure 5, for example for CPG, CPH and CPI, thus explaining the differential docking energies and inhibition constants in Figure 4 and Table 2. In Cat K (Figure (5a)), CPG fits in well interacting with most subsite residues as compared to CPH and CPI, hence the resulting low docking energy and inhibition constant. In Cat L, the three compounds have similar pose resulting in almost similar inhibition constants (Figure (5b)). For FP-2 and FP-3, the gate to S2 seems to play a key role in determining what goes in. In FP-2, the compounds can access and thus interact with S2 subsite residues (Figure (5c)), while in FP-3 the narrower gate hinders their entry (Figure (5d)). Despite this observation, FP-3 has lower docking energies and inhibition constants. A similar observation is made in the case of VP-2 and VP-3 (Figure (5e) and (5f)). In the case of knowlesipains, the compounds interacted mainly with S2 residues (Figure (5g) and (5h)). A comparison to a recent study using novel artemisinin derivatives as FP-2 inhibitors by Liu et al., shows that the aa interactions pattern between CPG, CPH and CPI and FP-2 and FP-3 were similar (Liu et al., 2012).
Figure 5.
Binding modes of CPG, CPH and CPI compounds in the binding pocket of human cathepsins and plasmodial homologs. Surface view of the orientation of CPG (blue), CPH (magenta) and CPI (green) on the S1 (pale yellow), S2 (cyan), S3 (brick-red) and S1’ (orange) of (a) Cat K, (b) Cat L, (c) FP-2, (d) FP-3), (e) VP-2, (f) VP-3, (g) KP-2) and (h) KP-3.
3.4 Stability of protein-ligand (CP) complexes
10 ns all-atom MD simulation of each of the proteases in complex with the CP derivatives was performed to obtain a dynamical picture of the conformational changes as a function of time. For quality assurance, the convergence of thermodynamic parameters namely temperature, total and potential energies was performed beforehand (data not shown). To determine if the simulated protein-ligand system formed stable complexes, the root mean square deviation (RMSD), a global measure of conformational diversity, the radius of gyration (Rg), a measure of compactness and the root mean square fluctuation (RMSF), a local measure of conformational diversity relative to the initial structures, were monitored. Using an in house Perl script, the mean and standard deviations of RMSD for the apo structures, CPs and protein-CP complexes as well as the Rg during the last 6 ns of MD simulations were determined. In the first 2 ns, the RMSD values increased after which they converged.
From Figure (6a), resulting RMSD values of all apo structures used in this study indicated that they were quite stable based on the error bars. In Figure (6b), the human cathepsins-CPs attained RMSD values ranging from 0.15-0.19 nm with standard deviations of up to 0.02 nm while the plasmodial-CPs complexes had RMSD values ranging from 0.18 up to 0.29 nm. The little fluctuations as depicted by the standard deviations suggested that the complexes were stable and snapshots of structures at different times could be collected for further analysis. The effect of a ligand is either to stabilize or destabilize a receptor upon binding, and thus considering the small size of CPs compared to the respective proteins used in this study and the resultant RMSD values (Figure (6c)), all CPs compounds seemed to stabilize the systems. As seen from Figure (6d), during the simulation, all structures were compact. From the RMSF plots (Figure S2) the plasmodial proteases (falcipains and vivapains) exhibited huge local conformational diversity within the β-hairpin (∼aa 175 – aa 200), an inimitable structure associated with the binding of haemoglobin (Hb) (Hogg et al., 2006). This observation was consistent with the other plasmodial proteases (Data not shown). In contrast, the human cathepsins (Cat K and Cat L) exhibited minimal RMSF fluctuations, a fact linked to the much shorter arm region. All other residues showing huge fluctuations were located on loop regions. As these fluctuations were not occurring in the active site, it can be concluded that the protein-ligand complex system remained stable during the simulation period. Structure visualization using PyMOL showed the ligands fitted well onto the binding pockets and their stabilization was facilitated mostly by a network of hydrophobic and hydrogen bonding between the ligand’s atoms and residues lining the S1, S2 S3 and S1’ subsites.
Figure 6.
Stability of Protein-CPs complexes as determined by GROMACS tools during the last 6 ns of MD simulations. Graphs showing the mean values of RMSD for apo structures (a), protein-ligand complexes (b), ligands only (c) and the radius of gyration for protein-ligand complexes (d). Error bars show the standard deviations.
3.5 Structural chemical features of binding
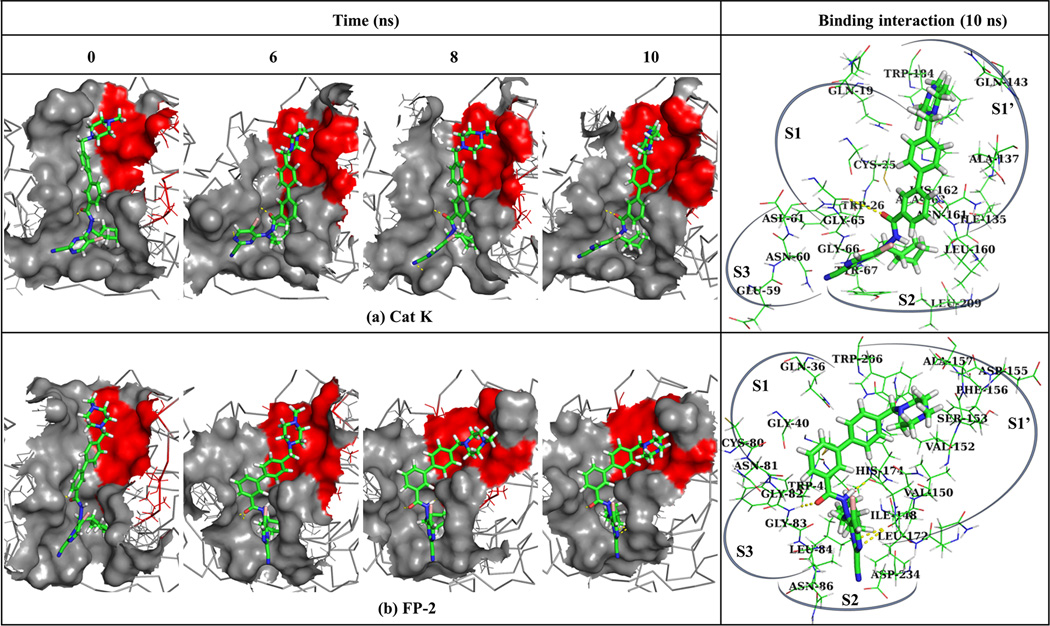
Several interactions between the residues lining the active pocket subsites and the ligand play key role in stabilizing it depending on the chemical group and nature of the surrounding environment. To extract the aa-ligand binding footprint along the MD simulations for all generated 3D structures, a Perl script with LigPlot + batch processing functionality was used. Table S4 shows the number and individual aa involved in van der Waals interactions and hydrogen bonds respectively. Notably, from these interaction results, the number of van der Waals and hydrogen interactions between CPD, CPE and CPF and protein residues were drastically fewer compared to when the proteins were interacting with the rest of CPs, an observation that could be explained by their observed binding modes and the short length of R1. In contrast, CPG, CPH and CPI maintained van der Waals interactions with most subsite aa residues mainly because of the extended chemical nature of R1. Figure 7 illustrates the conformational changes of compound CPG in complex with Cat K (Figure (7a)) and FP-2 (Figure (7b)) during MD simulations.
Figure 7.
Conformational changes of CPG within active site pockets (a) Cat K and (b) FP-2 during MD simulations. The brick red surface shows the S1’ position. The corresponding panels on the right show the residues interacting with CPG in Cat K and FP-2 at 10 ns respectively. The selected regions show the different subsites that constitute the binding pocket. Yellow dashes represent hydrogen bonds between heavy atoms.
From these results, CPG attains a stable conformation all through from 6 ns with the R1 chemical group stably interacting with S1’. These results were observed in the other proteins when in complex with CPG-CPI. Shown also in Figure 7 are the zoomed view of the key subsite residues in Cat K and FP-2 interacting with CPG through van der Waals and hydrogen interactions. In both cases, it is evident that CPG interacted with most subsite S1’ residues.
3.6 Binding free energy analysis
To obtain the predicted binding free energy (BFE) and the energetic contributions of van der Waals (vdW), electrostatic (ele) interactions, polar solvation (PB) and entropy (SASA), MM-PBSA method was used. Table 3 shows a summary of the overall BFE underlying the binding of CPs to FP-2 and FP-3 as well as their homologs. From the results, it was noted that in most cases, CPG, CPH and CPI had the lowest binding energies, an indication of stronger interactions compared to the other CPs. This was in agreement with the docking results as the same ligands had the lowest docking energies. From the different energetic contributions (see Table S5), it is evident that the binding process was principally favoured by van der Waals and electrostatic terms while the polar solvation impaired it. Surprisingly, the nonpolar solvation energies which correspond to the burial of SASA upon binding contributed slightly in equal order to the binding process in all complexes. To determine the key amino acid residues determining the strength of interactions in each of the protein-CP complexes, the final binding free energy was further decomposed into individual residue contributions. For compounds CPG, CPH and CPI, the sum total energy contribution of residues lining the four individual subsites was calculated. As seen from Figure 8, Figure S1’ subsite residues contributed to negative energy scores for the complexes of the three ligands and all proteases used in the study. However, S1-S3 had significant varying net energies (positive or negative) for the different ligands. For the binding of CPG, CPH and CPI to FP-2, S1 and S3 residues entirely contributed to a net positive energy as opposed to S2 and S1’… S2 and S3 residues contributed to negative energies for all non-human plasmodial proteases in complex with the three ligands (Figure (8b) and (8c)). This explains why Cat L had lower BFE value for the three ligands compared to Cat K. In the case of the plasmodial proteases binding hot spots were mainly found within the binding cleft subsites. As seen in Figure (8c), S3 aa impaired binding of the ligands onto the plasmodial cysteine as opposed to the human cathepsins. This information is quite important in understanding the residues giving attractive contributions to binding and should be considered during the rational drug design process of novel plasmodial cysteine protease inhibitors with increased selectivity towards the human proteases.
Table 3.
Protein-CP complexes overall binding free energy (ΔGbind) in kJ.mol-1 as determined by g_mmpbsa tool.
| Compound | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Protein | CPA | CPB | CPC | CPD | CPE | CPF | CPG | CPH | CPI |
| FP-2 | −84.9±0.2 | −71.8±0.3 | −80.8±0.2 | −91.1±2.7 | −93.1±0.2 | −80.5±0.2 | −131.5±0.2 | −103.4±0.2 | −99.6±0.2 |
| FP-3 | −77.0±0.2 | −67.1±0.2 | −66.7±0.2 | −74.2±0.2 | −74.7±0.2 | −102.8±0.4 | −87.6±0.2 | −111.7±0.2 | −105.7±0.3 |
| VP-2 | −95.0±0.2 | −78.7±0.2 | −60.6±0.2 | −85.8±0.2 | −72.4±0.6 | −77.7±0.3 | −116.2±0.3 | −81.8±0.2 | −93.0±0.2 |
| VP-3 | −112.4±0.2 | −72.6±0.2 | −59.0±0.5 | −98.6±0.3 | −55.0±0.2 | −62.1±0.3 | −104.8±0.2 | −93.7±0.2 | −85.7±0.2 |
| PK-2 | −115.5±0.3 | −68.8±0.3 | −68.8±0.4 | −92.8±0.3 | −99.0±0.2 | −85.7±0.3 | −129.2±0.2 | −61.9±0.3 | −86.8±0.2 |
| PK-3 | −80.3±0.4 | −63.0±0.2 | −81.7±0.3 | −82.1±0.2 | −80.4±0.2 | −80.9±0.2 | −68.5±0.3 | −76.2±0.3 | −80.9±0.3 |
| BP-2 | −92.9±0.2 | −92.1±0.4 | −82.8±0.4 | −133.3±0.5 | −80.5±0.5 | −85.0±0.3 | −135.4±0.4 | −131.7±0.3 | −94.9±0.3 |
| CP-2 | −73.0±0.2 | −85.4±0.2 | −71.9±0.1 | −108.3±0.3 | −122.6±0.3 | −83.7±0.3 | −103.9±0.4 | −103.4±0.3 | −98.1±0.3 |
| YP-2 | −91.6±0.3 | −67.0±0.3 | −82.1±0.2 | −103.0±0.2 | −114.6±0.3 | −89.1±0.3 | −92.3±0.2 | −104.8±0.2 | −97.8±0.3 |
| Cat S | −97.7±0.2 | −72.2±0.2 | −83.9±0.3 | −85.8±0.3 | −85.6±0.2 | −84.9±0.3 | −93.8±0.3 | −98.2±0.3 | −81.8±0.3 |
| Cat K | −86.4±0.3 | −91.3±0.2 | −93.6±0.2 | −90.8±0.3 | −104.1±0.2 | −76.6±0.3 | −96.3±0.3 | −80.0±0.2 | −81.8±0.2 |
| Cat L | −132.4±0.3 | −99.2±0.2 | −44.7±0.5 | −129.2±0.3 | −117.5±0.2 | −111.8±0.3 | −117.3±0.3 | −149.7±0.3 | −147.6±0.2 |
Figure 8.
Subsite amino acid contributions to BFE for compound CPG, CPH and CPI.
4. Conclusion
We report a comprehensive in silico approach encompassing several computational methods namely homology modelling, molecular docking, MD simulations and BFE calculations to determine the broad spectrum inhibitory activity and selectivity of non-peptidic compounds against FPs and their homologs from different Plasmodium species and human cathepsins. Although the compounds used in this study have been tested for activity mainly against FP-2 and FP-3 via wet lab assays (Batra et al., 2003; Chipeleme et al., 2007; Chiyanzu et al., 2003; Coteron et al., 2010; Domínguez et al., 2005; Greenbaum et al., 2004; Iain D Kerr et al., 2009; Li et al., 1995; M. Liu et al., 2001; Verissimo et al., 2008), to our knowledge this was the first attempt to report the broad spectrum inhibitory activity of these compounds using in silico approaches against FPs and their homologs. Through molecular docking, MD simulations and BFE calculations, key amino acids within the receptors’ active pocket responsible for the strong binding of the CPs ligands were deciphered. In all cases, van der Waals forces and polar solvation energies were proposed as the main BFE terms favouring and inhibiting the binding of CPs to the receptors respectfully. CPs represents a class of non-peptidic compounds with a broad spectrum activity against plasmodial FP homologs. Although, there was no clear cut selectivity profiles of these compounds against human cathepsins, previous pre-clinical studies involving cysteine protease inhibitors with little or no selectivity to target trypanosomal and plasmodial parasites were tolerated in animal models. This can be linked to the fact that the human-cathepsins are present in elevated concentrations compared to that of the parasite origin and the redundant nature of the mammalian cysteine proteases family (Ang et al., 2011). However, besides the beneficial intracellular housekeeping roles of human cathepsins, they can also be attractive targets for drug discovery as they have been found to play specific roles in antigen presentation, bone resorption and pro-hormone activation, processes critical for the progression of a variety of disease states such as rheumatoid arthritis, osteoporosis and autoimmune maladies. Hence this protocol may also be valuable to identification of potential inhibitors against the human cathepsins.
A 3D–QSAR study by Cátia et al., on peptidyl vinyl sulfone derivatives as FP-2 inhibitors showed the major structural requirements necessary for optimal activity (Teixeira, Gomes, Couesnon, & Gomes, 2011). A similar approach on non-peptidic heteroarynitrile derivatives by Wang et al., led to comparable conclusions where different subsite pockets preferred groups with certain chemical properties (Wang et al., 2013). A comparison of these results to our docking, MD and BFE shows great consistency. For example, from the per-residue energy decomposition, key aa contributing to the BFE of F2-CPI, a total of 10 residue including Asp35, Cys42, Leu84, Val152, Ser153, Asp154, Asp155, Asn173, His174 and Ala175 were identified in both cases as important in the binding process. The consideration of the important players (energetics and residues) in binding of non-peptidic compounds to proteases of key human Plasmodium species as well as the human cathepsins presents unique results that can be exploited in the structure-based molecular guided design of more potent antimalarial drugs.
Supplementary Material
Figure 9.
Per-residue decomposition fingerprint of compound CPG, CPH and CPI in complex with (a) Cat K (b) Cat L (c) FP-2 (d) FP-3 (e) VP-2 and (f) VP-3.
Acknowledgments
The authors greatly acknowledge CHPC for providing access to the cluster computational facility for MD simulations and BFE calculations. Special thanks to Dane Kennedy (Bioinformatics Application Scientist, CHPC) for his technical assistance in compiling the required software onto the cluster system. The original LigPlot + batch processing script was written by Ricardo Ferreira of University of Lisbon.
Funding:
TMM and AMK thank Rhodes University for postgraduate scholarships. This work is partially supported by the National Institutes of Health Common Fund under grant number U41HG006941 to H3ABioNet. The content of this publication is solely the responsibility of the authors and does not necessarily represent the official views of the funders.
List of abbreviations
- 3D–QSAR
Three dimensional quantitative structure-activity relationship
- ACPYPE
AnteChamber PYthon Parser interfacE
- BFE
Binding free energy
- BP-2
Bergheipain-2
- Cat L
Cathepsin L
- Cat K
Cathepsin K
- Cat S
Cathepsin S
- CP-2
Chabaudipain-2
- CPs
5-substituted-2-cyanopyrimidines
- FP
Falcipain
- GROMACS
GROningen MAchine for Chemical Simulations
- KP
Knowlesipain
- MD
Molecular dynamics
- MM-PBSA
Molecular mechanics – Poisson-Boltzmann surface area
- RMSD
Root Mean Square Deviation
- RMSF
Root Mean Square Fluctuation
- VP
Vivapain
- WHO
World Health Organization
- YP
Yoeliipain
References
- Ang KKH, Ratnam J, Gut J, Legac J, Hansell E, Mackey ZB, Renslo AR. Mining a cathepsin inhibitor library for new antiparasitic drug leads. PLoS Neglected Tropical Diseases. 2011;5(5):e1023. doi: 10.1371/journal.pntd.0001023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashley E, McGready R, Proux S, Nosten F. Malaria. Travel Medicine and Infectious Disease. 2006;4(3-4):159–173. doi: 10.1016/j.tmaid.2005.06.009. [DOI] [PubMed] [Google Scholar]
- Aurrecoechea C, Brestelli J, Brunk BP, Dommer J, Fischer S, Gajria B, Wang H. PlasmoDB: a functional genomic database for malaria parasites. Nucleic Acids Research. 2009;37(Database issue):D539–D543. doi: 10.1093/nar/gkn814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batra S, Sabnis YA, Rosenthal PJ, Avery MA. Structure-based approach to falcipain-2 inhibitors: synthesis and biological evaluation of 1,6,7-trisubstituted dihydroisoquinolines and isoquinolines. Bioorganic & Medicinal Chemistry. 2003;11(10):2293–2299. doi: 10.1016/s0968-0896(03)00117-2. [DOI] [PubMed] [Google Scholar]
- Benkert P, Biasini M, Schwede T. Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics (Oxford, England) 2011;27(3):343–350. doi: 10.1093/bioinformatics/btq662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benkert P, Tosatto SCE, Schomburg D. QMEAN: A comprehensive scoring function for model quality assessment. Proteins. 2008;71(1):261–277. doi: 10.1002/prot.21715. [DOI] [PubMed] [Google Scholar]
- Bernstein FC, Koetzle TF, Williams GJ, Meyer EF, Brice MD, Rodgers JR, Tasumi M. The Protein Data Bank: a computer-based archival file for macromolecular structures. Journal of Molecular Biology. 1977;112(3):535–542. doi: 10.1016/s0022-2836(77)80200-3. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/875032. [DOI] [PubMed] [Google Scholar]
- Bussi G, Donadio D, Parrinello M. Canonical sampling through velocity rescaling. The Journal of Chemical Physics. 2007;126(1):014101. doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- Centre for high performance computing. (n.d.) Retrieved October 22, 2014, from http://www.chpc.ac.za/
- Chipeleme A, Gut J, Rosenthal PJ, Chibale K. Synthesis and biological evaluation of phenolic Mannich bases of benzaldehyde and (thio)semicarbazone derivatives against the cysteine protease falcipain-2 and a chloroquine resistant strain of Plasmodium falciparum. Bioorganic & Medicinal Chemistry. 2007;15(1):273–282. doi: 10.1016/j.bmc.2006.09.055. [DOI] [PubMed] [Google Scholar]
- Chiyanzu I, Hansell E, Gut J, Rosenthal PJ, McKerrow JH, Chibale K. Synthesis and evaluation of isatins and thiosemicarbazone derivatives against cruzain, falcipain-2 and rhodesain. Bioorganic & Medicinal Chemistry Letters. 2003;13(20):3527–3530. doi: 10.1016/s0960-894x(03)00756-x. [DOI] [PubMed] [Google Scholar]
- Coteron JM, Catterick D, Castro J, Chaparro MJ, Diaz B, Fernandez E, Fiandor JM. Falcipain inhibitors: optimization studies of the 2-pyrimidinecarbonitrile lead series. Journal of Medicinal Chemistry. 2010;53(16):6129–6152. doi: 10.1021/jm100556b. [DOI] [PubMed] [Google Scholar]
- Crooks G, Hon G, Chandonia J, Brenner S. NCBI GenBank FTP Site\nWebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai PV, Avery MA. Structural characterization of vivapain-2 and vivapain-3, cysteine proteases from Plasmodium vivax: comparative protein modeling and docking studies. Journal of Biomolecular Structure & Dynamics. 2004;21(6):781–790. doi: 10.1080/07391102.2004.10506968. [DOI] [PubMed] [Google Scholar]
- Domínguez JN, León C, Rodrigues J, Gamboa de Domínguez N, Gut J, Rosenthal PJ. Synthesis and evaluation of new antimalarial phenylurenyl chalcone derivatives. Journal of Medicinal Chemistry. 2005;48(10):3654–3658. doi: 10.1021/jm058208o. [DOI] [PubMed] [Google Scholar]
- Eddy SR. Where did the BLOSUM62 alignment score matrix come from? Nature Biotechnology. 2004;22(8):1035–1036. doi: 10.1038/nbt0804-1035. [DOI] [PubMed] [Google Scholar]
- Ehmke V, Rottman M, Brun R, Schirmeister T, Diederich FF, Heindl C, Stich A. Potent and selective inhibition of cysteine proteases from Plasmodium falciparum and Trypanosoma brucei. Abstracts of Papers of the American Chemical Society. 2011;242(2):273–278. doi: 10.1002/cmdc.201000449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettari R, Bova F, Zappala M, Grasso S, Micale N, Zappalà M. Falcipain-2 inhibitors. Medicinal Research Reviews. 2010;30(1):136–167. doi: 10.1002/med.20163. [DOI] [PubMed] [Google Scholar]
- Flexner C, Bate G, Kirkpatrick P. Tipranavir. Nature Reviews. Drug Discovery. 2005;4(12):955–956. doi: 10.1038/nrd1907. [DOI] [PubMed] [Google Scholar]
- Greenbaum DC, Mackey Z, Hansell E, Doyle P, Gut J, Caffrey CR, Chibale K. Synthesis and Structure - Activity Relationships of Parasiticidal Thiosemicarbazone Cysteine Protease Inhibitors against Plasmodium falciparum, Trypanosoma brucei , and Trypanosoma cruzi. Journal of Medicinal Chemistry. 2004;47(12):3212–3219. doi: 10.1021/jm030549j. [DOI] [PubMed] [Google Scholar]
- Gupta S, Jadaun A, Kumar H, Raj U, Varadwaj PK, Rao AR. Exploration of new drug-like inhibitors for serine/threonine protein phosphatase 5 of Plasmodium falciparum: a docking and simulation study. Journal of Biomolecular Structure & Dynamics. 2015;33(11):2421–2441. doi: 10.1080/07391102.2015.1051114. [DOI] [PubMed] [Google Scholar]
- Hartjes LB. Preventing and detecting malaria infections. The Nurse Practitioner. 2011;36(6):45–53. doi: 10.1097/01.NPR.0000397912.05693.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatherley R, Blatch GL, Tastan Bishop O. Plasmodium falciparum Hsp70-x: a heat shock protein at the host-parasite interface. Journal of Biomolecular Structure & Dynamics. 2014;32(11):1766–1779. doi: 10.1080/07391102.2013.834849. [DOI] [PubMed] [Google Scholar]
- Hess B, Bekker H, Berendsen HJC, Fraaije JGEM. LINCS: A linear constraint solver for molecular simulations. Journal of Computational Chemistry. 1997;18(12):1463–1472. [Google Scholar]
- Hogg T, Nagarajan K, Herzberg S, Chen L, Shen X, Jiang H, Schmidt CL. Structural and functional characterization of Falcipain-2, a hemoglobinase from the malarial parasite Plasmodium falciparum. The Journal of Biological Chemistry. 2006;281(35):25425–25437. doi: 10.1074/jbc.M603776200. [DOI] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Research. 2002;30(14):3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr ID, Lee JH, Farady CJ, Marion R, Rickert M, Sajid M, Brinen LS. Vinyl sulfones as antiparasitic agents and a structural basis for drug design. The Journal of Biological Chemistry. 2009;284(38):25697–25703. doi: 10.1074/jbc.M109.014340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr ID, Lee JH, Pandey KC, Harrison A, Sajid M, Rosenthal PJ, Brinen LS. Structures of falcipain-2 and falcipain-3 bound to small molecule inhibitors: implications for substrate specificity. Journal of Medicinal Chemistry. 2009;52(3):852–857. doi: 10.1021/jm8013663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Kumar K, Korde R, Puri SK, Malhotra P, Singh Chauhan V. Falcipain-1, a Plasmodium falciparum cysteine protease with vaccine potential. Infection and Immunity. 2007;75(4):2026–2034. doi: 10.1128/IAI.01533-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari R, Kumar R, Lynn A. g_mmpbsa-A GROMACS Tool for High-Throughput MM-PBSA Calculations. Journal of Chemical Information and Modeling. 2014;54(7):1951–1962. doi: 10.1021/ci500020m. [DOI] [PubMed] [Google Scholar]
- Langhorne J, Buffet P, Galinski M, Good M, Harty J, Leroy D, Duffy P. The relevance of non-human primate and rodent malaria models for humans. Malaria Journal. 2011;10(1):23. doi: 10.1186/1475-2875-10-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. Journal of Applied Crystallography. 1993;26(2):283–291. [Google Scholar]
- Laskowski RA, Swindells MB. LigPlot +: Multiple Ligand-Protein Interaction Diagrams for Drug Discovery. Journal of Chemical Information and Modeling. 2011;51(10):2778–2786. doi: 10.1021/ci200227u. [DOI] [PubMed] [Google Scholar]
- Lee BJ, Singh A, Chiang P, Kemp SJ, Goldman EA, Weinhouse MI, Rosenthal PJ. Antimalarial activities of novel synthetic cysteine protease inhibitors. Antimicrobial Agents and Chemotherapy. 2003;47(12):3810–3814. doi: 10.1128/AAC.47.12.3810-3814.2003. Retrieved from http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=296233&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Kenyon GL, Cohen FE, Chen X, Gong B, Dominguez JN, McKerrow JH. In Vitro Antimalarial Activity of Chalcones and Their Derivatives. Journal of Medicinal Chemistry. 1995;38(26):5031–5037. doi: 10.1021/jm00026a010. [DOI] [PubMed] [Google Scholar]
- Liu M, Wilairat P, Go M-L. Antimalarial Alkoxylated and Hydroxylated Chalones: Structure−Activity Relationship Analysis. Journal of Medicinal Chemistry. 2001;44(25):4443–4452. doi: 10.1021/jm0101747. [DOI] [PubMed] [Google Scholar]
- Liu Y, Lu W-Q, Cui K-Q, Luo W, Wang J, Guo C. Synthesis and biological activities of novel artemisinin derivatives as cysteine protease falcipain-2 inhibitors. Archives of Pharmacal Research. 2012;35(9):1525–1531. doi: 10.1007/s12272-012-0902-4. [DOI] [PubMed] [Google Scholar]
- Mane UR, Gupta RC, Nadkarni SS, Giridhar RR, Naik PP, Yadav MR. Falcipain inhibitors as potential therapeutics for resistant strains of malaria: a patent review. Expert Opinion on Therapeutic Patents. 2013;23(2):165–187. doi: 10.1517/13543776.2013.743992. [DOI] [PubMed] [Google Scholar]
- Marco M, Coteron JM. Falcipain inhibition as a promising antimalarial target. Current Topics in Medicinal Chemistry. 2012;12(5):408–444. doi: 10.2174/156802612799362913. [DOI] [PubMed] [Google Scholar]
- Marvin Calculator Plugin and Chemical Terms Demo « ChemAxon – cheminformatics platforms and desktop applications. (n.d.) Retrieved October 22, 2014, from http://www.chemaxon.com/marvin/sketch/index.php. [Google Scholar]
- Melnikova I. Hepatitis C therapies. Nature Reviews Drug Discovery. 2008;7(10):799–800. [Google Scholar]
- Melo F, Devos D, Depiereux E, Feytmans E. ANOLEA: a www server to assess protein structures. Proceedings / … International Conference on Intelligent Systems for Molecular Biology; ISMB. International Conference on Intelligent Systems for Molecular Biology. 1997;5:187–190. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/9322034. [PubMed] [Google Scholar]
- Mendis K, Sina BJ, Marchesini P, Carter R. The neglected burden of Plasmodium vivax malaria. The American Journal of Tropical Medicine and Hygiene. 2001;64(1–2 Suppl):97–106. doi: 10.4269/ajtmh.2001.64.97. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/11425182. [DOI] [PubMed] [Google Scholar]
- Models MM, Philosophy A, Kollman PA. Advances and Continuing Challenges in Achieving Realistic and Predictive Simulations of the Properties of Organic and Biological Molecules. Accounts of Chemical Research. 1996;29(10):461–469. [Google Scholar]
- Mok S, Imwong M, Mackinnon MJ, Sim J, Ramadoss R, Yi P, Bozdech Z. Artemisinin resistance in Plasmodium falciparum is associated with an altered temporal pattern of transcription. BMC Genomics. 2011;12:391. doi: 10.1186/1471-2164-12-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris GM, Ruth H, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. Software news and updates AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. Journal of Computational Chemistry. 2009;30(16):2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR. Open Babel: An open chemical toolbox. Journal of Cheminformatics. 2011;3(1):33. doi: 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey KC, Dixit R. Structure-function of falcipains: malarial cysteine proteases. Journal of Tropical Medicine. 2012;2012:345195. doi: 10.1155/2012/345195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrinello M. Polymorphic transitions in single crystals: A new molecular dynamics method. Journal of Applied Physics. 1981;52(12):7182. [Google Scholar]
- Pei J, Grishin NV. PROMALS: towards accurate multiple sequence alignments of distantly related proteins. Bioinformatics (Oxford, England) 2007;23(7):802–808. doi: 10.1093/bioinformatics/btm017. [DOI] [PubMed] [Google Scholar]
- Pei J, Kim BH, Grishin NV. PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucleic Acids Research. 2008;36(7):2295–2300. doi: 10.1093/nar/gkn072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen HG. Accuracy and efficiency of the particle mesh Ewald method. The Journal of Chemical Physics. 1995;103(9):3668. [Google Scholar]
- Pronk S, Páll S, Schulz R, Larsson P, Bjelkmar P, Apostolov R, Lindahl E. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics (Oxford, England) 2013;29(7):845–854. doi: 10.1093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prugnolle F, Durand P, Ollomo B, Duval L, Ariey F, Arnathau C, Renaud F. A fresh look at the origin of Plasmodium falciparum, the most malignant malaria agent. PLoS Pathogens. 2011;7(2):e1001283. doi: 10.1371/journal.ppat.1001283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal PJ. Falcipains and other cysteine proteases of malaria parasites. Advances in Experimental Medicine and Biology. 2011;712:30–48. doi: 10.1007/978-1-4419-8414-2_3. [DOI] [PubMed] [Google Scholar]
- Rosenthal PJ, Lee GK, Smith RE. Inhibition of a Plasmodium vinckei cysteine proteinase cures murine malaria. The Journal of Clinical Investigation. 1993;91(3):1052–1056. doi: 10.1172/JCI116262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal PJ, Wollish WS, Palmer JT, Rasnick D. Antimalarial effects of peptide inhibitors of a Plasmodium falciparum cysteine proteinase. The Journal of Clinical Investigation. 1991;88(5):1467–1472. doi: 10.1172/JCI115456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabnis YA, Desai PV, Rosenthal PJ, Avery MA. Probing the structure of falcipain-3, a cysteine protease from Plasmodium falciparum: comparative protein modeling and docking studies. Protein Science: A Publication of the Protein Society. 2003;12(3):501–509. doi: 10.1110/ps.0228103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saralamba S, Pan-Ngum W, Maude RJ, Lee SJ, Tarning J, Lindegardh N, White LJ. Intrahost modeling of artemisinin resistance in Plasmodium falciparum. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(1):397–402. doi: 10.1073/pnas.1006113108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayers EW, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Ye J. Database resources of the National Center for Biotechnology Information. Nucleic Acids Research. 2009;37(Database issue):D5–D15. doi: 10.1093/nar/gkn741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz F, Gelhaus C, Degel B, Vicik R, Heppner S, Breuning A, Schirmeister T. Screening of protease inhibitors as antiplasmodial agents. Part I: Aziridines and epoxides. ChemMedChem. 2007;2(8):1214–1224. doi: 10.1002/cmdc.200700070. [DOI] [PubMed] [Google Scholar]
- Shen M-Y, Sali A. Statistical potential for assessment and prediction of protein structures. Protein Science: A Publication of the Protein Society. 2006;15(11):2507–2524. doi: 10.1110/ps.062416606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Research. 2005;33(Web Server issue):W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa da Silva AW, Vranken WF. ACPYPE - AnteChamber PYthon Parser interfacE. BMC Research Notes. 2012;5:367. doi: 10.1186/1756-0500-5-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira C, Gomes JRB, Couesnon T, Gomes P. Molecular docking and 3D–quantitative structure activity relationship analyses of peptidyl vinyl sulfones: Plasmodium Falciparum cysteine proteases inhibitors. Journal of Computer-Aided Molecular Design. 2011;25:763–775. doi: 10.1007/s10822-011-9459-4. [DOI] [PubMed] [Google Scholar]
- Teixeira C, Gomes JRB, Gomes P. Falcipains, Plasmodium falciparum cysteine proteases as key drug targets against malaria. Current Medicinal Chemistry. 2011;18:1555–1572. doi: 10.2174/092986711795328328. [DOI] [PubMed] [Google Scholar]
- Thillainayagam M, Pandian L, Murugan KK, Vijayaparthasarathi V, Sundaramoorthy S, Anbarasu A, Ramaiah S. In silico analysis reveals the anti-malarial potential of quinolinyl chalcone derivatives. Journal of Biomolecular Structure and Dynamics. 2014;33(5):961–977. doi: 10.1080/07391102.2014.920277. [DOI] [PubMed] [Google Scholar]
- Tina KG, Bhadra R, Srinivasan N. PIC: Protein Interactions Calculator. Nucleic Acids Research. 2007;35(Web Server issue):W473–W476. doi: 10.1093/nar/gkm423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verissimo E, Berry N, Gibbons P, Cristiano MLS, Rosenthal PJ, Gut J, O’Neill PM. Design and synthesis of novel 2-pyridone peptidomimetic falcipain 2/3 inhibitors. Bioorganic & Medicinal Chemistry Letters. 2008;18(14):4210–4214. doi: 10.1016/j.bmcl.2008.05.068. [DOI] [PubMed] [Google Scholar]
- Wang J, Li F, Li Y, Yang Y, Zhang S, Yang L. Structural features of falcipain-3 inhibitors: an in silico study. Molecular bioSystems. 2013 doi: 10.1039/c3mb70105k. [DOI] [PubMed] [Google Scholar]
- Wang SX, Pandey KC, Scharfstein J, Whisstock J, Huang RK, Jacobelli J, McKerrow JH. The structure of chagasin in complex with a cysteine protease clarifies the binding mode and evolution of an inhibitor family. Structure (London, England: 1993) 2007;15(5):535–543. doi: 10.1016/j.str.2007.03.012. [DOI] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics (Oxford, England) 2009;25(9):1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO. (n.d.). Retrieved June 22, 2014, from http://www.who.int/malaria/publications/world_malaria_report_2013/wmr2013_no_profiles.pdf?ua=1.
- Williams T. (n.d.) No Title. Retrieved September 10, 2012, from http://gnuplot.info/
- Zhao B, Janson Ca, Amegadzie BY, D’Alessio K, Griffin C, Hanning CR, Abdel-Meguid SS. Crystal structure of human osteoclast cathepsin K complex with E-64. Nature Structural Biology. 1997;4(2):109–111. doi: 10.1038/nsb0297-109. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.