Abstract
The growing number of DNA helicases implicated in hereditary disorders and cancer indicates that this particular class of enzymes plays key roles in genomic stability and cellular homeostasis. Indeed, a large body of work has provided molecular and cellular evidence that helicases act upon a variety of nucleic acid substrates and interact with numerous proteins to enact their functions in replication, DNA repair, recombination, and transcription. Understanding how helicases operate in unique and overlapping pathways is a great challenge to researchers. In this review, we describe a series of experimental approaches and methodologies to identify and characterize DNA helicase inhibitors which collectively provide an alternative and useful strategy to explore their biological significance in cell-based systems. These procedures were used in the discovery of biologically active compounds that inhibited the DNA unwinding function catalyzed by the human WRN helicase-nuclease defective in the premature aging disorder Werner Syndrome. We describe in vitro and in vivo experimental approaches to characterize helicase inhibitors with WRN as the model, anticipating that these approaches may be extrapolated to other DNA helicases, particularly those implicated in DNA repair and/or the replication stress response.
Keywords: helicase, replication, DNA repair, small molecule, inhibitor, Werner syndrome, cancer therapy
Graphical Abstract
1. Introduction
Seminal studies from the Ashworth [1] and Helleday [2] laboratories first published in 2005 described small molecules (<500 Daltons) effective in sensitizing mutant cancer cells, defective in the tumor suppressors BRCA1 or BRCA2, to clinically relevant anti-cancer agents. These studies led to the discovery of poly(ADP)ribose polymerase (PARP) inhibitors that show promise in the clinic for treatment of BRCA1- or BRCA2- negative cancers. Aside from their own success, the PARP inhibitors provide a roadmap for investigation of other DNA repair targets using small molecule screens as the prevailing experimental strategy for initial identification of lead compounds. It is now apparent that DNA repair represents a broad class of proteins which may be valuable for targeting in anti-cancer schemes. While this field is still in its early stages at the translational level, progress continues to be made to identify new targets in DNA repair pathways, particularly homologous recombination (HR) repair where the PARP inhibitor story first got started.
A recent review by Huang and Mazin provides a detailed perspective of small molecule inhibitor screens to identify druggable targets of the HR pathway [3]. This is a very useful review as it discusses proof-of-concept examples for compound library screens adapted to cell-based assays which illustrated the utility of small molecule modulators as research tools and also potential drug candidates to modulate the DNA damaging effects of classical chemotherapy drugs. HR repair is elicited in rapidly dividing cancer cells undergoing prolific DNA synthesis to cope with double-strand breaks due to replication-blocking lesions that occur endogenously or double-strand breaks that are induced directly or indirectly by chemotherapeutic drugs or ionizing radiation; therefore, it is conjectured that HR targets might be a focal area for cancer therapy. Hot on the heels of PARP inhibitors, small molecules that affect the major strand recombinase RAD51 is attracting interest. In addition, the double-strand DNA translocate RAD54, which stimulates DNA strand exchange activity of RAD51 among other activities including Holliday Junction branch-migration and remodeling of protein-DNA complexes, is also a candidate for small molecule modulation.
Our own group has addressed the potential value of targeting molecular motor DNA unwinding enzymes known as helicases for anti-cancer therapies [4-6]. Helicases catalytically disrupt hydrogen bonds between bases in structured nucleic acids, and have important functions in virtually all aspects of nucleic acid metabolism [7]. Because DNA helicases play a unique and early role in a number of DNA damage response and DNA repair pathways especially in dividing cells, we hypothesize that they represent a useful target to exploit synthetic lethal relationships with other DNA damaging agents and/or in specific mutant backgrounds. In a 2013 review, the Frick laboratory provided an overview of the helicase inhibitors described to date with an emphasis on published work that used screens to identify compounds that modulate the human RecQ helicases or the RNA helicase elongation initiation factor 4A [8]. In the current review for this special Methods collection on DNA helicases, we have focused on the actual experimental approaches and assays we employed to perform a small molecule screen for inhibitors of the Werner syndrome helicase implicated in the premature aging disorder Werner syndrome [9-11]. Furthermore, we provide the reader guidance on important experimental approaches that address issues relating to potency, specificity, and reversibility of helicase inhibitors in vitro. In addition, the review places a major emphasis on cell-based assays to characterize the biological effects of WRN helicase inhibitors and synthetic lethal approaches we have used in laboratory experiments. The review is written in plain language so that it may be helpful to many experimental biologists, even those who are relatively new to the helicase field. The review is divided into two over-arching sections: 1) biochemical screen for WRN inhibitors and related in vitro assays; 2) biological assays with the WRN helicases inhibitors and human cells. In keeping with the theme of Methods, we have focused on rapidly developing techniques and strategies to characterize DNA helicases using small molecules as novel tools for basic science investigation and potential development into translational therapies, particularly in the anti-cancer field.
2. Biochemical small molecule helicase inhibitor screens
Screening and characterization of biologically active small molecules that modulate the DNA unwinding function of a target helicase represents a unique approach to studying helicase function in human cells [4,5,8]. We have used this approach to investigate the molecular and cellular functions of the WRN helicase-nuclease defective in the premature aging disorder Werner syndrome. These studies were initially guided by an in vitro radiometric-based helicase assay using the purified recombinant WRN protein in which approximately 500 compounds from the National Cancer Institute Diversity Set were screened [10]. One compound that we identified to inhibit WRN with relatively high potency compared to other compounds in the NCI library was 1-(propoxymethyl)-maleimide, designated NSC 19630 (IC50 ~ 20 μM). Having determined potency for WRN helicase inhibition, the specificity of compounds which tested positively for helicase inhibition in vitro was assessed by evaluating their effects on other DNA helicases. In parallel, DNA binding, ATPase, and WRN exonuclease assays were performed to further characterize compounds which selectively inhibited WRN helicase activity. In addition, selected WRN helicase inhibitory compounds were assayed for displacement of the fluorescently active DNA intercalating compound Thiazole Orange to assess the relative ability of each respective compound from the NCI Diversity Set to bind the DNA substrate used for WRN helicase assays. This effort helped to eliminate those compounds whose effect on WRN helicase activity was mediated by its direct interaction with the DNA helicase substrate and therefore considered to be non-specific in nature. Further testing of structures similar to NSC 19630 led to the identification of a more potent WRN helicase inhibitor designated NSC 617145 [9]. In the following sections, we will describe the procedures for these assays used to identify and characterize the WRN helicase inhibitors NSC 19630 [10] and NSC 617145 [9], and highlight some salient points which are useful to keep in mind when designing experiments and carrying out biochemical assays.
2.1. Semi-high-throughput helicase activity screen
Semi-high-throughput screening of a large number of small molecules for inhibition of helicase activity requires a DNA substrate (either radiolabeled or fluorescently labeled) that is relevant for measuring helicase activity, purified helicase protein devoid of contaminating nuclease activity, reaction salts optimal for helicase activity, a source of energy (typically ATP) for the helicase enzyme, and the library of small molecules in solution (typically dissolved in DMSO). Reactions are typically 20 microliters with 0.5 nM DNA substrate used. A good preliminary experiment prior to screening is to test the effect of DMSO (or whatever solvent is being used to dissolve the small molecules in the screen) on the activity of the helicase, as routinely done for other enzymes [12]. It is recommended to design the experiment so that the solvent is only 1 microliter of the 20 microliter reaction volume (5%) to minimize any non-specific effects of the solvent on enzyme activity. A broad titration of helicase in the presence of the solvent can be used to choose an appropriate enzyme concentration for the screening assay. Ideally, the helicase concentration should result in 50-75% of the DNA substrate being unwound so it is easier to identify inhibitors and even small molecule activators that enhance catalytic DNA unwinding activity.
In a standard Hoefer 400 SE electrophoresis unit with a gel containing 15 wells, up to 11 small molecules can be tested in one experiment. Alternatively, a mixture of 5 selected compounds can be used per helicase reaction mixture, assuming that they do not interact with each other in a manner to modulate the effect of the single compound on the helicase under investigation. This would expedite the testing, essentially resulting in the assessment of 55 compounds per 15-well helicase gel instead of 11 compounds, but also requires an additional step to determine which of the five compounds are responsible for inhibition if there is a positive hit. The controls required are: 1) a DNA substrate alone control; 2) a heat-denatured DNA substrate control; and 3) two helicase alone reactions (in the absence of small molecules but with DMSO). To perform the experiment, a cocktail is prepared containing the reaction salts, water, and DNA substrate. The appropriate volume is dispensed into each tube, followed by the addition of small molecules or solvent with the helicase. Alternatively, the helicase and small molecules can be incubated together for at least 5 minutes followed by the addition of DNA substrate and ATP (or other energy source). We typically use a small molecule concentration of 50 μM for initial screening, which is high enough to ensure potential hits are not missed but low enough to avoid detecting a large number of positive hits. For example, out of 500 tested compounds in our initial screen that identified NSC 19630, we had 7 positive hits [10]. Reaction mixtures are incubated for a specified period of time (e.g., 15 min) and then quenched by the addition of EDTA in the presence of marker dyes (e.g., bromophenol blue, xylene cyanol), glycerol, and unlabeled oligonucleotide of the same sequence to prevent reannealing. The heat-denatured DNA substrate control is boiled for 5-10 min. All samples are then loaded on a non-denaturing PAGE gel with appropriate % acrylamide/bis-acrylamide to resolve the intact DNA substrate from the released radiolabeled unwound strand under standard electrophoresis conditions (e.g., 1.5-2 hr at 200 V). Gels are then exposed to a phosphorimager screen and the screen is scanned on a Typhoon scanner (or other type of scanner that can recognize the labeled substrate and unwound product) and quantitated using ImageQuantTL and Excel software.
Unwound DNA substrate product for each reaction mixture is calculated as being 100 *((ssDNA)/(total DNA)) %. Control unwinding is calculated as the average of the helicase + solvent reactions which is normalized to 100%. All helicase + small molecule reactions are then normalized as a percentage of control unwinding. Small molecules from reactions where % control unwinding is below a chosen threshold (for example ≤25%) are retested to verify them as positive hits and are further analyzed as described in the next section.
2.2. In vitro assessment of small molecule potency and specificity for helicase inhibition
Once a collection of small molecules (using a relatively high concentration of drug (e.g., 50 μM)) are identified that inhibit the helicase under investigation by an arbitrary cut-off (e.g., ≥75% that of control reactions lacking drug), they can be tested for inhibition of DNA unwinding as a function of drug concentration to assess potency. IC50 values for helicase inhibition are determined from compound titrations. These are typically set up in reaction volumes similar to the original screen (for WRN, 20 μl [10]), using the compound dilution solvent DMSO as a control in the reaction mixtures in which the compound is omitted entirely. The contribution of DMSO to the reaction volume is 5%. For the set of 7 compounds identified in the initial WRN helicase screen, the IC50 range was 2-20 μM [10]. The potency screen can be useful, particularly when there are a large number of compounds which test positively for helicase inhibition in the primary screen.
It can be useful to test compounds structurally related to the one that tests positively for helicase inhibition, as naturally occurring or derivative compounds may be better candidates to pursue for subsequent studies based on their potency, specificity (see below), pharmacodynamics behavior, and/or biological activity. In the case of WRN, we identified a close structural analog of NSC 19630, designated NSC 617145, which was reported to inhibit proliferation of certain cancer cell lines in the NCI60 screen [9]. NSC 617145 was tested for in vitro inhibition of WRN helicase activity on a forked duplex substrate and the IC50 value was determined to be 230 nM [9], ~80-fold more potent than NSC 19630. Moreover, NSC 617145 was blocked to thiol reactivity in the two five-membered rings by Cl atoms, which may be useful in a biological context or lead compound development for a potential therapeutic purpose.
It is also very useful to determine if inhibition of the helicase-catalyzed unwinding reaction is reversible or not. Determining the mode of inhibition is an important tool for quantitatively comparing a variety of small molecules that show activity against the helicase or enzyme under study [12]. One method of testing reversibility of the helicase-drug interaction is through dilution. In this case, the helicase (100-fold greater concentration than needed for activity in the standard helicase assay) and drug (10-fold higher concentration than its IC50 value) are incubated together in reaction salts until equilibrium is reached. The reaction is then diluted 100-fold in reactions salts in the presence of ATP (or other energy source) and the DNA substrate. Reactions are quenched by the addition of excess EDTA at specific time-points to determine helicase-catalyzed DNA unwinding kinetics. In this setup, if the enzyme is still inhibited after dilution of the drug from a level 10-fold greater than the IC50 value to a level 10-fold less than the IC50 value, the inhibition may be considered to be irreversible. If normal helicase activity returns after dilution, then the inhibition is considered to be reversible. Partial recovery of helicase activity is an indication that the inhibition is partially reversible.
Upon identification of relatively potent helicase inhibitors, the drugs can be tested for their effects on other purified recombinant helicases available to characterize their specificity. For the WRN helicase inhibitor NSC 19630, a 50 μM concentration of drug inhibited WRN helicase activity by 80% compared to the control DMSO [10]. In similar reactions, little to no inhibition of unwinding by NSC 19630 (50 μM) was observed for two related human RecQ helicases (BLM and RECQ1), the Fanconi Anemia Group J helicase (FANCJ), and three E. coli helicases (UvrD, RecQ, DnaB), suggesting that NSC 19630 acts in a WRN-specific manner. Similarly, NSC 617145 (which is structurally related to NSC 19630) failed to show appreciable inhibition of other DNA helicases tested (human BLM, RECQ1, FANCJ, or ChlR1 (DDX11), E. coli RecQ or UvrD) at a 20-fold greater concentration of drug compared to the IC50 of 230 nM for WRN helicase inhibition [9]. Although in vitro specificity is generally desired for a small molecule compound and its helicase target, this may not always apply. While the BLM helicase inhibitor Ml216 showed very modest effects on helicase activity catalyzed by E. coli UvrD, human RECQ1 or RECQ5, the drug inhibited WRN helicase activity in vitro (IC50 = 5 μM) [13]. However, ML216 acted specifically toward BLM in vivo, as demonstrated by cell-based assays in which the proliferation of WRN mutant and wild-type cells were similarly sensitive to the compound [13] (see below for discussion of helicase inhibitor specificity in vivo).
One perhaps salient point is the choice of DNA substrate for high-throughput helicase activity screens. In both the WRN [10] and BLM [13] helicase inhibitor screens, the DNA substrate was a partial forked duplex substrate with both 5′ and 3′ single-stranded arms. In the case of full-length recombinant BLM, the IC50 for BLM helicase inhibitor ML216 was 3.0 μM [13]. However, inhibition of BLM branch-migration activity on a mobile D-loop substrate or Holliday Junction was significantly more modest, requiring a ML216 concentration of 50 μM. This finding suggests that a small molecule may differentially affect helicase versus branch-migration activity of those helicase proteins that have dual functions; therefore it is important to consider the choice of DNA substrate used for helicase inhibitor screens.
2.3. ATPase assay
More mechanistic information can be acquired for those small molecules believed to bind directly to the helicase protein. A good place to start for these secondary biochemical assays is to assess the compound’s effect on hydrolysis of nucleoside triphosphate (typically ATP) by the helicase because the energy produced from this reaction drives DNA unwinding. ATPase assays are typically performed using a thin layer chromatography (TLC) procedure to analyze reaction mixtures containing a radiolabeled nucleotide (e.g, 32Pγ-ATP) and helicase incubated in the presence of the appropriate reaction salts (e.g., divalent cation such as Mg2+) and DNA effector. Upon quench with an ATPase stop solution containing a large excess of ADP, ATP, and EDTA, the samples are spotted onto a TLC sheet (Baker-flex Cellulose PEI sheet, Philipsburg, NJ (J.T. Baker)) and carefully placed in a tank containing a 1 M formic acid/ 0.8 M LiCl solvent; the solvent front is then allowed to migrate to a position near the top of the TLC sheet (~ 10 cm). This allows for the released 32P to migrate significantly farther than the 32Pγ-ATP so that the spots are readily distinguished on a PhosphorImager and can be quantitated using the ImageQuant software. Details for the WRN ATPase assay using the TLC procedure are described in ref. [14], which can be easily modified to suit the helicase under study. There are also coupled spectrophotometric ATP hydrolysis assays described [15,16].
From a mechanistic view, a nucleotide analog might competitively bind to the ATP binding site of a DNA helicase, resulting in a change in the Michaelis-Menton constant (Km) for ATP binding. Alternatively, a small molecule binding to a helicase may alter its catalytic efficiency, which would be detected by change in Vmax or the kcat for ATP hydrolysis. In the case of the WRN helicase inhibitor NSC 19630, drug concentrations up to 50 μM only modestly inhibited WRN ATPase activity compared to the IC50 of 20 μM for WRN helicase inhibition [10], suggesting that the compound targeted the DNA unwinding function of WRN. The structurally related WRN helicase inhibitor NSC 617145 showed a dose-dependent inhibition of WRN ATPase; however, ATPase inhibition required significantly greater NSC 617145 concentrations compared to helicase inhibition [9]. Therefore, the experimental ATPase data suggest that the mechanism of WRN helicase inhibition by NSC 617145 is similar to that of NSC 19630.
2.4. Thiazole Orange dye displacement assay
As mentioned above, it is generally desirable for a small molecule to be specific in its mechanism of action for helicase inhibition. A compound that directly binds to the DNA substrate may act non-specifically to inhibit DNA unwinding by multiple helicases as well as affect the catalytic functions of other DNA metabolizing enzymes such as polymerases or nucleases. Therefore, it is valuable to biochemically test if the small molecule under investigation directly binds the DNA substrate. An initial indication that a particular small molecule is binding to DNA is the presence of a DNA species migrating more slowly than the intact DNA substrate in a non-denaturing gel during the semi-high throughput helicase assay. A convenient assay for this purpose is the fluorescent intercalator dye displacement assay. The assay measures the displacement of the fluorescent dye that intercalates into the dsDNA by the compound under study. Intercalation of the duplex DNA by the dye enhances the fluorescence of the dye, whereas, subsequent displacement of the intercalator by the DNA binding compound results in the decrease in fluorescence where the percent fluorescent decrease is directly related to the extent of binding. Thiazole Orange (TO) is a preferred dye as an increase in its fluorescent intensity upon intercalation far exceeds that of the other commonly used dye ethidium bromide (3000-fold vs 20-fold); moreover, TO displays less sequence-dependent DNA binding and minimal fluorescence in solution or when bound to ssDNA [17].
In our work, we used the TO dye displacement assay to assess if the WRN helicase inhibitor NSC 19630 binds to the19-base pair forked duplex DNA substrate used for the in vitro helicase activity screen [10]. To evaluate NSC 19630 ability to displace the intercalated fluorescent dye by binding to the DNA substrate, TO (400 nM, Sigma-Aldrich)) was preincubated with the forked duplex DNA substrate (50 nM) in WRN helicase reaction buffer for 10 min followed by addition of a small molecule (0.25 - 50 μM) or Hoechst 33258 (0.05 - 50 μM, Sigma-Aldrich) for 15 min. Hoescht 33258 binds in the minor groove of B-form duplex DNA and has minimal fluorescence in solution or when bound to ssDNA; therefore, it serves as a positive control for DNA binding by a small molecule. As a control, fluorescent intensities of a well with no DNA binding agent (100% fluorescence) and a well with no DNA (0% fluorescence) was measured. Percent fluorescence was reported as the ratio of fluorescence reading obtained from DNA-binding incubations performed in the presence of NSC 19630 or Hoescht 33258 compared with incubations performed in the absence of either compound. It is critical to determine the saturating concentrations of fluorescent dye to be used in the reaction as it may vary for different substrates and hence will affect the final outcomes of the displacement assay. To determine the appropriate amount of TO in the above mentioned assay, each well of the costar black 96-well assay plate was loaded with WRN helicase reaction buffer (50 μl) containing forked duplex substrate (50 nM) and the varying dye concentrations for 10 min. Fluorescence (excitation 485 nm, emission 520 nm) was measured using a FluoSTAR OPTIMA (BMG Labtek) to determine the saturating concentrations. In the case of WRN helicase inhibitor NSC 19630, no change in fluorescence was detected up to a WRN helicase inhibitor concentration of 100 μM, whereas a significant fluorescence decrease by 10 μM Hoescht 33258 was observed, suggesting that inhibition of WRN helicase by NSC 19630 was not attributed to direct binding of the small molecule to the DNA substrate. Consistent with this conclusion, little to no inhibition of WRN exonuclease activity was observed at NSC 19630 concentrations up to 100 μM [10].
3. Cell-based assays to assess biological activity of helicase inhibitors
The most potent and WRN-specific helicase-inhibitors were then assessed for their biological effects in human cells grown in culture using a battery of assays [10] (Fig. 1). Initially, the effects of small molecules were tested on proliferation of the human cervical cancer cell line HeLa 1.2 11 (HeLa) using a conventional WST-1 assay for metabolic activity. Typically, a concentration range of 1.5 – 12 μM NSC 19630 displayed a dose-dependent inhibition of cell proliferation. The specificity of small molecule-mediated inhibition of cell proliferation was assessed in a key set of experiments by evaluating the effect of selected compounds (e.g., NSC 19630) on proliferation of HeLa cells that had been transiently depleted of WRN by transfection with a WRN-specific RNA interference oligonucleotide. These efforts allowed us to determine that the WRN helicase inhibitor exerted a WRN-dependent effect on cell proliferation as well as other biological endpoints (Fig. 2). The simple premise here was that anti-proliferative effect of a compound is likely to be mediated by a direct effect on the helicase in question if the cells depleted of WRN are resistant to the anti-proliferative effect of the WRN helicase inhibitor. Indeed, we found this to be the case for NSC 19630 that was originally identified from the in vitro WRN helicase inhibition screen [10], as well as a structurally related compound NSC 617145 [9].
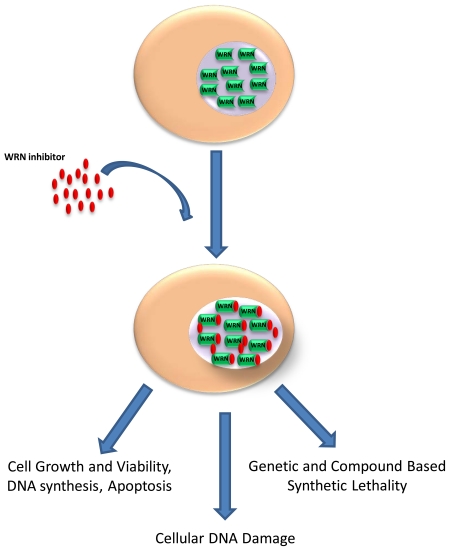
Figure 1. Cancer cells expressing wild-type WRN are sensitive to a WRN helicase inhibitor.
Multiple biological endpoints can be assayed to assess the effect of the WRN helicase inhibitor in cell-based assays. See text for details.
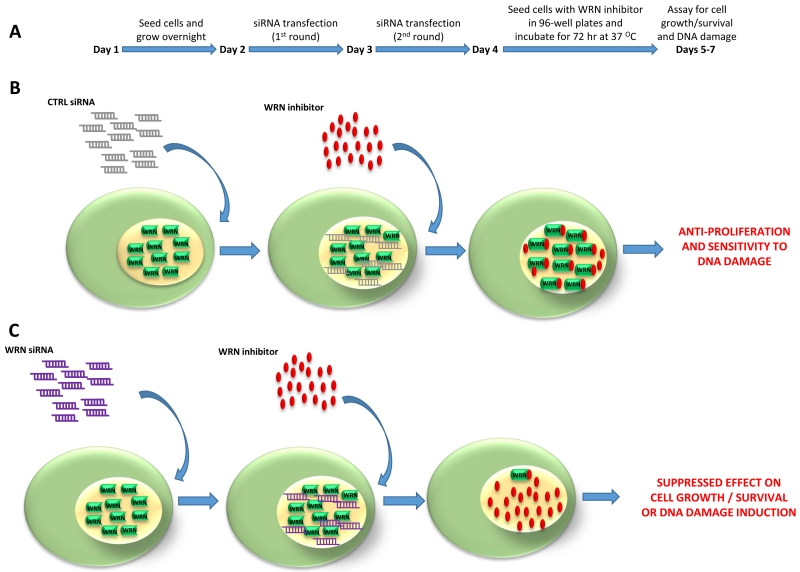
Figure 2. Experimental scheme to assess specificity of a WRN helicase inhibitor in cell-based assays.
(A) Schematic of the RNAi-mediated WRN depletion regime to demonstrate that the effects of the WRN helicase inhibitor on cell proliferation and DNA damage induction are dependent on the expression of the inhibitor target, i.e., WRN helicase. (B) Cells transfected with control siRNA are sensitive to the WRN helicase inhibitor as measured by an impaired DNA damage response, cell growth, and survival. (C) WRN-depleted cells are resistant to the biological effects of the WRN helicase inhibitor.
To further address the specificity of a DNA helicase inhibitor in cells, additional experimental approaches can be taken. One is to assess if the recovery of target helicase expression via siRNA dilution by cell division renders the cells once again sensitive to the helicase inhibitor. To do this for the WRN helicase inhibitor NSC 19630, HeLa cells that had been transfected with the WRN siRNA were allowed to grow for an additional two weeks [10]. At that time, Western blot analysis was used to confirm that WRN protein expression had been restored. The cells were then treated with DMSO or the WRN helicase inhibitor NSC 19630 (3 μM), and cell proliferation was determined for the following three days. These experiments demonstrated that the HeLa cells were sensitive to NSC 19630. A second experimental approach used to address the specificity of the WRN helicase inhibitor was to test its effect on an isogenic pair of Bloom’s syndrome-null and Bloom’s syndrome-corrected cells that are defective in the RecQ helicase family member BLM related in helicase core domain sequence to WRN [10]. Demonstration that the cells were similarly sensitive to NSC 19630 irrespective of BLM status indicates that BLM is unlikely to play a role in the anti-proliferative effects of the WRN helicase inhibitor. Analogous to this set of experiments, the BLM helicase inhibitor ML216 was found to similarly inhibit proliferation of WRN mutant and wild-type cells, as well as sensitize both cell lines comparably to the replication inhibitor aphidicolin [13].
Having identified a WRN helicase inhibitor that specifically inhibited cell proliferation in a WRN-dependent manner, a number of cell-based assays were used to determine the effects of the compound on other biological endpoints including cell growth (colony formation), mitogenic efficiency (DNA synthesis as measured by EdU staining), apoptosis (accumulation of histone-associated DNA fragments), or DNA damage induction (immunofluorescent detection of γ-H2AX foci as a marker of double-strand breaks) [10]. The specificity of the WRN helicase inhibitor on apoptosis and γ-H2AX induction was assessed using HeLa cells depleted of WRN by RNA interference, analogous to the cell proliferation assays. The WRN helicase inhibitor NSC 19630 which tested positively for inhibition of cell proliferation in a WRN-dependent manner also induced apoptosis and caused double-strand breaks in a WRN-dependent manner as well. Consistent with accumulation of double strand breaks, there was elevated auto-phosphorylated ataxia telangiectasia (ATM) in NSC 19630-treated cells.
Because WRN is implicated in DNA replication, we were prompted to measure effects of the WRN helicase inhibitor on cell cycle progression by flow cytometry [10]. This analysis demonstrated that HeLa cells exposed to NSC 19630 were enriched in the S-phase population, which occurred in a WRN-dependent manner. Consistent with a model in which NSC 19630 caused replication fork stalling by inhibiting WRN helicase function, the number of immunofluorescent proliferating cell nuclear antigen (PCNA) foci increased dramatically in HeLa cells exposed to the WRN helicase inhibitor, and this effect was also WRN-dependent as shown by its suppression in WRN-depleted cells.
3.1. WST-1 cell proliferation assay
The WST-1 assay is a nonradioactive method to measure metabolic activity of cells which can be used to assay cytotoxic or cytostatic effects of compounds such as DNA damaging drugs used for chemotherapy or other types of potential pharmaceutical molecules. The principle of the assay is that a stable form of the tetrazolium salt WST-1 is cleaved to a soluble form at the cell surface in a process that is largely dependent on glycolytic synthesis of the reduced form of nicotinamide adenine dinucleotide phosphate (NADPH), which is a form of chemical energy produced by the pentose phosphate pathway. The WST-1 assay provides a facile technique to spectrophotometrically quantitate the proliferative capacity of cells and is readily adapted to a 96-well format. If a number of positive small molecule hits are obtained from a primary screen such as the in vitro helicase assay, then these can be easily screened for biological activity using the WST-1 assay. Moreover, the WST-1 assay requires less time than alternative approaches such as measurement of DNA synthesis by BrdU incorporation. WST-1 assays are widely used to perform screens for compounds that have an effect on cell viability [18-20].
After the initial radiometric strand displacement screen of the compounds in the library to determine which small molecules exerted an inhibitory effect on WRN helicase activity in a relatively potent and specific manner, we performed cell proliferation assays using WST-1 (Sigma-Aldrich Cellpro-Ro Roche) to test the effect of these compounds on the viability of HeLa cells [10]. Selected small molecules from the NCI Diversity Set that tested positive for WRN helicase inhibition (IC50 ≤ 20 μM) were diluted in DMSO to yield the desired concentration of each compound at 1% DMSO in 1 ml of trypsinized and resuspended HeLa cells (50,000 cells/ml). 100 μl of this cell suspension with the compound was seeded (in triplicate) in each of the wells of a 96 well plate. Relevant controls with only DMSO or media containing the compounds but without cells were kept for background correction. The plates were then incubated at 37 °C and 5% CO2 and cell viability measured over 0-3 days by the WST-1 assay. Briefly, 10 μl of WST-1 reagent was added to each well at specified time points and incubated for 2 hr. Optical density readings were measured in a microplate reader at OD450. For best results, care should be taken that WST-1 addition is done in the absence of fluorescent light. The raw data generated from the microplate reader is then processed by first subtracting for background followed by determining the fold increase or decrease in viable cells compared to the vehicle (DMSO) control well. Results are graphically interpreted as fold increase or decrease in viable cells as a function of drug dose over time.
3.2. Colony survival assay
Having narrowed down the number of helicase inhibitors that affect cell proliferation, it is valuable to perform colony survival assays with a small subset of compounds. The colony survival assay is different from cell proliferation or viability assays because the latter are difficult to interpret whether the effect of the compound is cytostatic or cytotoxic. Cytotoxic refers to cell killing, whereas cytostatic refers to stopping cells from dividing. The colony formation assay is useful to asses if a helicase inhibitor prevents cells from dividing because it relies on the potential of a single cell to multiply numerous times to generate a colony. Colony formation assays are time-consuming, but the results can be informative.
To assess the effect of the WRN helicase inhibitor NSC 19630 on colony formation, 250-300 HeLa cells were seeded in each well of a 6-well plate and allowed to grow overnight [10]. The cells were then treated with different doses of the compound or DMSO vehicle control and allowed to incubate for 72 hr at 37 °C and 5% CO2. Following the incubation with the drug, the small molecule containing media was discarded and the cells were washed twice in 1× PBS. The cells were then replenished with DMEM media containing 10% FBS and 1% Pen-Step cocktail and incubated an additional 7-10 days to allow colony formation with a change of media every 3 days. Finally the cells were fixed in methanol for 2 min, followed by staining with 2% Methylene Blue in 50% methanol for 2-5 min. The dye was then discarded and plates washed under tap water. Care should be taken while washing under tap water to ensure that colonies do not get washed away under the flow of water. The plates were then dried and the colonies counted and compared with vehicle control. Results are graphically represented as number of colonies formed as a function of dose of the compound. The assay is done in triplicate and at least three independent times to determine statistical significance and reproducibility.
3.3. DNA synthesis measured by EdU staining
A growing number of DNA helicases are implicated in the replication stress response by either dealing directly with aberrant DNA structures that arise at the fork or facilitating DNA damage response pathways that involve checkpoint signaling. Therefore, helicase deficiency may give rise to replication stalling, failure to restart replication forks, and/or DNA damage at the fork such as single-stranded or double-stranded breaks. This issue is particularly relevant in cancer cells, in which the rapidly dividing cells may accumulate replicative lesions. Therefore, it is conjectured that helicases may be good targets for inhibition in chemotherapy strategies. To assess the effects of helicase inhibitors on mitogenic capacity, DNA synthesis in proliferating cells can be detected by measuring the incorporation of labeled DNA precursors into cellular DNA during S-phase of the cell cycle. Several methods are available to measure DNA synthesis based on the incorporation of [3H] Thymidine, bromo deoxyuridine (BrdU), or 5-ethynyl-2′-deoxyuridine (EdU) into genomic DNA. In our experience, we prefer the EdU staining method as the small size of the fluorescent azides has much higher rates of diffusion, and the method does not require sample DNA denaturation or fixation, thereby allowing high-resolution microscopic analysis in the context of well-preserved chromatin structure.
We evaluated if exposure of HeLa cells to the WRN helicase inhibitor NSC 19630 impaired DNA synthesis as measured by EdU incorporation (4). In principle, a thymidine analogue (EdU) characterized by the replacement of a methyl group with a terminal alkyne group at the 5th position of the base moiety is first incorporated into cellular DNA. The incorporated terminal alkyne group is then detected in a Cu(I)-catalyzed [3 + 2] cycloaddition (“click” chemistry) reaction [21]. HeLa cells (25,000 cells per well) were treated with NSC 19630 (2 μM) for 72 hr, followed by the addition of EdU (10 μM) (Invitrogen) for 6 hr. After cell fixation (3.7% paraformaldehyde, 15 min) and cell permeabilization (0.5 % Triton X-100, 20 min), 0.5 ml Click-iT reaction mixture was added for 30 min in the dark. This was followed by coating with Prolong Gold Anti-Fade reagent containing DAPI (Invitrogen) and cells were imaged using a Zeiss LSM 510 META inverted Axiovert 200 M laser scan microscope with a Plan-Apochromat 100× 1.4-numerical-apperture oil immersion differential interference contrast objective lens. Images were analyzed using the LSM Browser software package. The fluorescent analysis demonstrated that the WRN helicase inhibitor NSC 19630 impaired the ability of HeLa cells to synthesize DNA by nearly 3-fold compared to the DMSO-treated cells.
3.4. Cellular apoptosis assay
Biologically active small molecule helicase inhibitors that inhibit cell proliferation may cause programmed cell death (apoptosis). To address this possibility for a WRN helicase inhibitor, HeLa cells (5,000 cells per well of 96-well plates) were exposed to NSC 19630 (3 μM) for 72 hr, and the Cell Death Detection ELISA PLUS photometric enzyme immunoassay (Roche) was used for quantitative determination of cytoplasmic histone-associated DNA fragments indicative of ongoing apoptosis [10]. The highly sensitive and nonradioactive sandwich enzyme immunoassay utilizes mouse monoclonal antibodies directed against both DNA and histones. After NSC 19639 exposure, cells are chemically lysed and samples of cell lysate solution (20 μL) were placed into wells of a streptavidin-coated microplate followed by incubation for 2 hr with immunoreagent containing a mixture of anti-histone-biotin and anti-DNA-POD (80 μL). In principle, the anti-histone antibody binds to the histone component of the nucleosome and will capture the immunocomplex to the streptavidin-coated microplate via its biotinylation, whereas the anti-DNA-POD antibody will bind to the DNA component of the nucleosomes. The extent of apoptosis corresponds to the amount of nucleosomes retained by POD in the immunocomplex which is then measured spectrophotometrically with 2,2′-azinobis-3-ethyl-benzothiazoline-6-sulfonic acid (ABTS) substrate using a microplate reader at a wavelength of 405 nm and reference wavelength of 490 nm. In the case of the WRN helicase inhibitor NSC 19630, the exposed HeLa cells displayed a significant increase in apoptosis (4). Of note, WRN knockdown HeLa cells were resistant to the anti-apoptotic effects of NSC 19630, indicating that induction of apoptosis by the compound is WRN-dependent.
3.5. γ-H2AX and PCNA foci detection
Detection of DNA damage or markers of stalled forks or the DNA damage response is a valuable and facile approach to assess the consequences of cellular exposure to a compound that modifies genomic DNA and interferes with replication fork progression and causes fork collapse (e.g., alkylating agents such as methylmethanesulfonate) or when a DNA damaging agent such as ionizing radiation or a compound (e.g., bleomycin) directly introduces strand breaks. Alternatively, inhibitors of replication or DNA repair may also lead to accumulation of double strand breaks. Based on the anti-proliferative and apoptotic effects of NSC 19630, we reasoned that the WRN helicase inhibitor would likely induce double strand breaks by interfering with the replication stress response that cells use to cope even with endogenous DNA damage. This led us to experimentally assess DNA damage induction by assaying for phosphorylation of the histone H2A, member X (H2AX). H2AX phosphorylation on serine-139 (γ-H2AX) is enacted by cellular PI3-family kinases (Ataxia-telangiectasia mutated (ATM), ATR, and DNA-PKcs. γ-H2AX is generally considered a good marker of double strand breaks that can be detected by immunofluorescent foci. Depending on the intensity and number of foci within each cell it is also possible to interpret the severity of DNA damage and how well the cell can cope with the stress. In addition to DNA damage, a marker of stalled replication foci is the accumulation of proliferating cell nuclear antigen (PCNA), a replication processivity and DNA repair factor.
The cell-based immunostaining technique to detect γ-H2AX or PCNA involves primary antibody recognition of one of these markers and detection using a fluorescently labelled secondary antibody. About 20,000-30,000 HeLa cells are seeded in each well of a four chambered slide (Nalge Nunc International, Rochester, NY) and allowed to grow overnight as a monolayer [10]. The cells are then treated with the small molecule NSC 19630 or vehicle control (DMSO) and incubated for 72 hr at 37 °C and 5% CO2. The media containing the compound was then discarded and the cells washed in 1× PSB twice. The cells were next fixed in 4% formaldehyde at room temperature for 10 min. To avoid over-fixation of cells, the duration of the incubation period should be carefully monitored. Following fixation, the cells are next washed in 1× PBS twice and then treated with 0.5% Triton X-100 (Sigma) for 5 min at room temperature. The cells are again washed in 1× PBS containing 0.5% Tween-20 and blocked with goat serum (Sigma) overnight at 4 °C. The blocking serum will depend on the host in which the secondary antibody has been raised. The cells are then washed in 1× PBS with 0.1% Tween-20 and incubated with anti γ-H2AX monoclonal antibody (1:300)(Upstate) or mouse proliferating cell nuclear antigen (PCNA) F-2 monoclonal antibody (1:500) (Santa Cruz) overnight at 4 °C. Cells are washed in 1× PBS with 0.1% Tween-20 and incubated with Alexa Fluor 488-conjugated goat anti-mouse IgG (1:400) (Invitrogen) at room temperature for 2 h. Cells are washed four times with 1× PBS with 0.1% Tween-20 and coated with Prolong Gold Anti-Fade reagent (Invitrogen) containing DAPI. Coverslips are placed on the chamber slides, and cells are cured at room temperature in the dark for 24 hr. The slides are observed under a fluorescent microscope under 60× oil magnification and the number of foci for γ-H2AX and PCNA quantified in each sample and compared to vehicle control. Each experiment is done in duplicate and at least three independent experiments are performed to determine statistical significance. The experimental data can be expressed quantitatively as number of γ-H2AX (or PCNA) foci per cell. Alternatively, an arbitrary threshold of γ-H2AX foci per cell can be assigned, and percent cells with greater than or less than that number of foci (e.g., 15) is determined from microscopic inspection of fixed cells, typically 50-100.
From our studies with WRN helicase inhibitors, we observed a dramatic 17-fold increase in γ-H2AX foci and 20-fold increase in PCNA foci in HeLa cells exposed to NSC 19630 (2 μM) for 3 days compared to the DMSO control [10]. Moreover, γ-H2AX and PCNA induction was WRN-specific as evidenced by the low level of foci in WRN-depleted cells. NSC 617145 exerted a similar inducing effect on γ-H2AX and PCNA foci in HeLa cells that was WRN-dependent, only the effect of NSC 617145 was more potent than that of NSC 19630 as a lower concentration of compound (0.25 μM) was required to elicit the effect [9]. Moreover, significantly elevated number of foci corresponding to 53-binding protein 1 (53BP1), a signaling factor that rapidly recruits to chromatin surrounding the site of a double strand break [22], were also observed in HeLa cells exposed to NSC 617145 (0.25 μM), as detected using mouse anti-53BP1 monoclonal antibody (1:500, Millipore) [9]. Together, these results confirmed that the WRN helicase inhibitor NSC 617145 exerted a DNA damage inducing effect.
3.6. Helicase chromatin binding, degradation, and extract-derived DNA unwinding
The impairment of cell proliferation and strong DNA damage induction upon cellular exposure to the WRN helicase inhibitor in a WRN-dependent manner suggested that a toxic ternary complex of WRN helicase-drug-genomic DNA may form in vivo. Presumably, the small molecule traps helicase-active WRN on a key DNA replication/repair intermediate resulting in a toxic chromatin DNA-drug complex. This scenario led us to the hypothesis that poisoned WRN would be enriched in the chromatin fraction in WRN inhibitor-treated cells. To address this for the WRN inhibitor, HeLa cells were exposed to increasing concentrations of NSC 617145 (0.75-2.0 μM) or the small molecule solvent DMSO (1%) for 4 hr, followed by cell freezing at −80 °C and subsequent preparation of various subcellular fractions according to the manufacturer’s recommendations (Subcellular protein fractionation kit, Thermo Scientific) [9]. This procedure involves simple sequential steps to lyse cultured mammalian cells in the presence of a protease inhibitor cocktail so that nuclear soluble and chromatin-bound fractions can be isolated within 2-3 hr, exploiting the inclusion of a stabilized micrococcal nuclease to free chromatin-bound proteins from other nuclear proteins. The solubilized proteins from the respective fractions are then in a compatible form to be readily resolved on denaturing gradient (4-12%) polyacrylamide-SDS gels and probed for WRN protein by immunoblotting using a WRN mouse monoclonal antibody (1:1000; Spring Valley Laboratories). To control for loading, blots were also probed for the nuclear proteins Topoisomerase I (BD Biosciences) and Histone H3 (Abcam). This approach demonstrated that HeLa cells exposed to NSC 617145 displayed a greater percentage of endogenous WRN in the chromatin fraction compared to control DMSO-treated cells.
The toxic WRN protein-DNA complexes that result from cellular exposure to NSC 617145 may elicit cellular degradation of the inhibited WRN helicase to remove it from genomic DNA so that normal DNA transactions (e.g., replication, transcription, DNA repair) can be at least partially restored. In our studies, we observed that a 6 hr incubation of HeLa cells with NSC 617145 (0.75 μM) led to a significantly diminished amount of WRN in the total cell extract, as detected by immunoblotting [9]. Cellular WRN protein could be restored to that of DMSO-treated cells by the inclusion of 10 μM MG132 (Sigma-Aldrich), a proteasome inhibitor that inhibits degradation of ubiquitin-conjugated proteins in mammalian cells, in the cell culture media. Based on Western blot analysis of extracts from HeLa cells exposed to NSC 617145 or DMSO in the presence or absence of MG132, the results suggested that cellular exposure to the WRN helicase inhibitor caused its degradation by the proteasome in a ubiquitin-mediated pathway.
The proteasomal degradation of WRN in NSC 617145-treated cells suggested that these cells would have lower WRN helicase activity compared to DMSO-treated cells. To test this, we immunoprecipitated endogenous WRN from extracts of HeLa cells (5 × 107) that were exposed to 0.75 μM NSC 617145 or DMSO for 4 hrs [9]. Rabbit anti-WRN polyclonal antibody (H300, Santa Cruz) was used to precipitate WRN from the precleared soluble fraction after detergent solubilization of cells. A normalized amount of total extract protein from NSC 617145 versus DMSO treated HeLa cells was then tested for ATP-dependent unwinding of a simple radiolabeled 19-base pair forked duplex in a conventional strand displacement helicase assay in which products are resolved on a native 12% polyacrylamide gel [23]. Consistent with a lower amount of WRN in the NSC 617145-treated cells, the helicase activity of these extracts was 4-fold reduced compared to those extracts prepared from DMSO-treated cells [9].
Endogenous helicase activity isolated from extracts of cells exposed to the inhibitor or DMSO control can be measured by a radiometric helicase assay. One approach is to immunoprecipitate the helicase targeted for small molecule inhibition with a well-characterized antibody from extracts of HeLa cells exposed to drug or DMSO; the immunoprecipitated helicase is tested for DNA unwinding with a radiolabeled DNA substrate in standard reaction conditions. This experiment may require a large amount of cells and inhibitor. In experiments with the WRN inhibitor NSC 617145 we used 5 × 107 cells per experiment to have enough activity to measure DNA unwinding by the immunoprecipitated WRN in the helicase assay [9]. Based on a previously published protocol [24], HeLa cells were treated with either the inhibitor or DMSO and harvested after sufficient time had elapsed for the inhibitor to enter cells and elicit an effect. Protein extracts were isolated from the cell pellets using dounce homogenization (20 strokes, 30 min on ice) in extraction buffer (20 mM Tris-HCl (pH 8.0), 0.5 M NaCl, 1 mM EDTA, 0.5 mM DTT, 0.5% NP-40, 25% glycerol, 0.2 mM PMSF, and 10 μg/ml each of aprotinin, pepstatin A and leupeptin). Protein samples were taken to determine protein concentration using the Bio-Rad Protein Assay. The clarified lysate was pre-cleared using Pansorbin (EMD Chemicals) and then incubated with the helicase-specific antibody or IgG control. The helicase:antibody complexes were precipitated using Pansorbin, washed and resuspended in 15 ul extraction buffer for each mg of protein as measured after the lysis step. Immunoprecipitated WRN protein from an equivalent amount of total extract protein from NSC 617145-treated cells compared to DMSO-treated cells was then incubated in a standard helicase reaction with a radiolabeled forked duplex DNA substrate. At a minimum, the set of helicase assays for a typical experiment should include: 1) no enzyme control; 2) immunoprecipitated helicase:antibody + ATP; 3) immunoprecipitated helicase:antibody + ATPγS; 4) IgG antibody control + ATP; 5) IgG antibody control + ATPγS; 6) recombinant helicase + ATP; 7) heat-denatured DNA substrate control. A representative result from this type of experiment is shown in Fig 2C of ref. [9].
4. Research strategies to investigate helicase inhibitors and chemically induced synthetic lethality
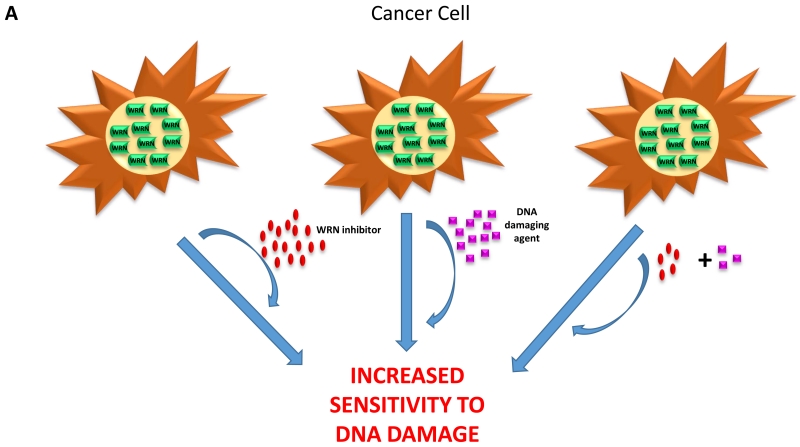
Genetic-based or chemically induced synthetic lethality is an emerging approach to target tumors with novel DNA repair inhibitors. Because rapidly dividing cancer cells accumulate replicative lesions at a significantly greater rate than normally dividing cells, cancer cells may be hyper-sensitive to compounds that target the DNA repair machinery. We have been interested in this concept because helicases play critical roles in distinct steps of multiple DNA repair pathways and are integral for the DNA damage response [7]. Chemically induced synthetic lethality may occur between a helicase inhibitor and an agent that induces DNA damage when cells cannot tolerate even lower doses of the DNA damaging agent when the helicase-dependent pathway is pharmacologically compromised (Figure 3A). Although this outcome may be observed when the helicase operates in a DNA repair pathway directly responsible for the correction of the lesion introduced by cellular exposure to the DNA damaging agent, this may not always be the case. For example, small molecule inhibition of a helicase involved in the replication stress response (e.g., WRN, RECQ1) may behave synergistically with an alkylating agent that introduces a bulky lesion which blocks fork progression. Another mechanism for chemically induced synthetic lethality can occur when cells are exposed to a helicase inhibitor and a compound that inhibits a DNA repair enzyme. The following two sections describe cell-based assays used to show synthetic lethality of a WRN helicase inhibitor with a DNA damaging agent or DNA repair inhibitor.
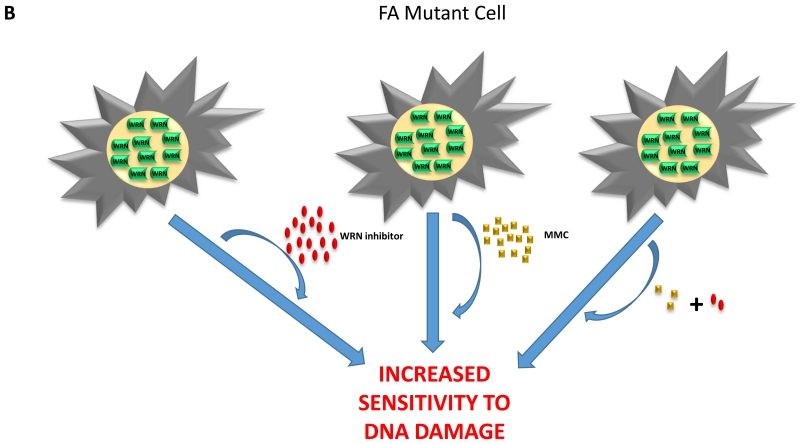
Figure 3. Chemical or genetic synthetic lethality of the WRN helicase inhibitor.
(A) Cancer cells are sensitive to the synergistic effects induced by an agent that causes replication stress and a WRN helicase inhibitor. (B) Fanconi anemia mutant cells are hypersensitive to the synergistic effects induced by low doses of the DNA cross-linking agent mitomycin C and WRN helicase inhibitor.
4.1. Cell-based experiments with helicase inhibitor and DNA damaging agents
Our studies with newly discovered WRN-specific helicase inhibitors have provided proof-of-principle evidence for how these compounds can be used in synthetic lethal approaches with other pharmacological agents or in defined genetic mutant backgrounds. In our discovery of the first WRN helicase inhibitor (NSC 19630), we evaluated several DNA-interacting or damaging compounds as well as a DNA repair inhibitor for chemically induced synthetic lethality with NSC 19630 [10]. The first of these was telomestatin (TMS), a drug known to disrupt normal telomere capping and bind G-quadruplex (G4) DNA [25-27]. Previous work had implicated a role of WRN helicase to unwind DNA structure arising from the G-rich strand of the telomeric tail at the chromosome end [28]. Given WRN’s ability to unwind G4 DNA in vitro [29,30], we tested if the osteosarcoma U2OS cell line was sensitive to co-treatment with a low dose of the G4-interactive ligand telomestatin (TMS) (0.6 μM) combined with a concentration of NSC 19630 (1 μM) that would otherwise display low toxicity. WST-1 assays showed that TMS with NSC 19630 behave synergistically to inhibit cell proliferation, suggesting that loss of WRN G4 unwinding rendered cells hyper-sensitive to the G4-binding ligand [10]. In a set of experiments similar to those of TMS, we also found that HeLa cell proliferation was markedly reduced and γ-H2AX induction elevated by co-exposure to NSC 19630 and the topoisomerase inhibitor and chemotherapy drug topotecan (TPT) (0.1 μM) [10].
4.2. Cell-based experiments with helicase inhibitor and DNA repair inhibitors
As mentioned in the Introduction, perhaps the most high profile example of synthetic lethality in the field of DNA repair that may have promising clinical application is that of poly(ADP ribose) polymerase (PARP) inhibitors [31,32]. PARP inhibitors prevent efficient single-strand break repair, and are lethal to cancer cells defective in homologous recombination (HR) repair. Indeed, a number of PARP inhibitors are in clinical trials for breast cancer 1 (BRCA1) and breast cancer 2 (BRCA2) negative patients. We found that NSC 19630 with the PARP inhibitor KU0058948 acted synergistically to inhibit proliferation of HeLa cells, supporting the hypothesis that impairment of WRN helicase activity compromised HR repair in cancer cells, rendering them sensitive to even a very low KU0058948 dose (1 nM) [10]. Overall, the results demonstrated that cancer cells exposed to low doses of pharmacological agents that induce replication stress or faulty DNA repair are selectively killed by the presence of a relatively low dose of WRN helicase inhibitor NSC 19630 (1 μM).
5. Cell-based studies to assess genetic based synthetic lethality of helicase inhibitors
Tumors that are defective in a DNA repair pathway due to mutational inactivation may be more sensitive to DNA damaging chemotherapy drugs and/or DNA repair inhibitors. Compensatory mechanisms, such as those mediated by WRN helicase, may provide cellular resistance to DNA damage by enabling redundant repair or signal transduction pathways to be elicited. This is likely to be highly relevant in the emerging field of personalized medicine in which the genetic mutations of a tumor or blood cancer are known. For example, a number of cancers are known to show low expression or defects in genes encoding proteins of a specialized interstrand cross-link DNA repair pathway that are implicated in Fanconi Anemia (FA) [33-36]. Because WRN helps cells cope with replication stress and WRN-deficient cells show moderate sensitivity to DNA cross-linking agents, we hypothesized that cancer cells defective in the FA DNA cross-link repair pathway may be further sensitized to the DNA cross-linker and chemotherapy drug mitomycin C (MMC) (Figure 3B). This concept was tested in a series of experiments in which isogenic cancer cell lines either deficient or proficient in the FA pathway were exposed to a very low MMC concentration (9 nM) and low dose (0.125 μM) of NSC 617145, a compound structurally related to NSC 19630 that was found to inhibit WRN helicase in vitro in a more potent manner (IC50 ~ 230 nM) [9]. The results demonstrated that the WRN helicase inhibitor NSC 617145 operated synergistically with MMC in a FA genetic mutant background (FANCD2 or FANCA), resulting in reduced cell proliferation and significant induction of DNA damage and chromosomal abnormalities, the latter evidenced by metaphase spreads. Importantly, the effect of NSC 617145 was WRN-dependent, as experimentally addressed with WRN-depleted FA-D2−/− mutant and corrected cells. Interestingly, the synergistic inhibition of cell proliferation by NSC 617145 and MMC was not observed when FA-D2 mutant cells were co-treated with NSC 617145 and hydroxyurea [9], suggesting that pharmacological inhibition of WRN helicase activity impairs residual interstrand cross-link repair in the absence of a functional FA pathway but does not interfere with resistance to replication stress imposed by hydroxyurea-induced nucleotide pool exhaustion. Furthermore, drug-induced WRN helicase inhibition exacerbated MMC-induced DNA damage when both the FA pathway and nonhomologous end-joining (NHEJ) (a branch of double strand break repair) were defective in human cells mutated in both FANCD2 and the DNA protein kinase catalytic subunit (DNA-PKcs) [11]. We interpret this result to mean that the FA pathway and NHEJ together relieve stress of interstrand cross-links which accumulate in cells when the FA pathway is defective and the WRN helicase-dependent pathway of HR repair is compromised by a WRN inhibitor.
5.1. Double strand break response in FA mutant cells exposed to a WRN helicase inhibitor and DNA cross-linker
An impaired replication stress response in a genetic mutant background that is already compromised in DNA repair may result in replication fork blockage and double strand breaks. Because a number of DNA helicases are critical responders to replication stress, they may play instrumental roles in this capacity. Following this logic, we investigated if small molecule inhibition of WRN helicase activity in FA mutant cells would exasperate the number of double strand breaks induced by the DNA cross-linker MMC [9]. Using the γ-H2AX immunofluorescence foci detection procedure described above, we determined that co-exposure of FA-D2−/− cells to NSC 617145 (0.125 mM) and MMC (9.4 nM) for 3 hr induced 80% of the population to display greater than 15 foci per cell, whereas only 30% of the isogenic FA-D2 +/+ cells behaved in this manner [9].
Consistent with the DSB induction in FA mutant cells caused by synergistic effect of NSC 617145 and MMC, the double strand break sensor kinase ATM was activated, as detected by immunoblotting [9]. Briefly, FA-D2−/− and FA-D2+/+ cells were co-treated with 0.125 μM NSC 617145 and 9.4 nM MMC for 3 hr. Cell lysates were prepared by resuspension in a RIPA buffer containing 1% Nonidet P-40 and 0.1% SDS and proteins resolved by electrophoresis on a 4% polyacrylamide SDS gel. After transfer to a PVDF membrane, the blots were probed using anti-pATM Ser1981 antibody (1:500, Santa Cruz) followed by a peroxidase-labeled anti-mouse IgG (1:1000, Vector). As a control, blots were reprobed with anti-ATM antibody (1:500, Millipore). The ECL Plus Western Blot Detection Kit (Amersham) was used according to the manufacturer’s recommendations to detect ATM and pATM-Ser1981.
A hallmark feature of FA is a marked sensitivity of the cells to the interstrand cross-linking agents, indicating a crucial role for FA proteins in resolving double strand breaks created at cross-links [37]. Double strand breaks can be repaired by NHEJ or homologous recombination (HR). Because NHEJ proteins have a high affinity for DNA ends [38], we reasoned that the treatment of FA mutant cells with a cross-linking agent in the presence of the WRN helicase inhibitor NSC 617145 might cause the phosphorylation of the DNA-dependent protein kinase complex (DNA-PKcs) at serine-2056, an event associated with the accumulation of the NHEJ factor at double strand breaks. Indeed, exposure of FA-D2−/− cells to 0.125 μM NSC 617145 and 9.4 nM MMC resulted in a significantly greater percentage of cells with more than 15 DNA-PKcs pS2056 foci compared to similar treatment of NSC 617145+/+ cells, as detected by immunofluorescent foci using a specific antibody against DNA-PKcs p2056 (1:500, Abcam) [9]. Thus, the experimental data altogether suggest that WRN helicase helps cells deal with double-strand breaks arising from MMC-induced cross-links when the FA pathway is non-operational.
5.2. Analyses of chromosomal instability by metaphase spreads in NSC 617145-treated FA mutant cells
The results described in the previous section suggested that the residual DNA cross-links which accumulated in FA mutant cells exposed to a low dose of MMC led to double-strand breaks in the pharmacologically compromised WRN helicase inhibitor treated cells. This result coupled with the ATM activation and accumulation of DNA-PKcs pS2056 foci in FA-D2 mutant cells exposed to MMC and NSC 617145 led us to evaluate chromosomal instability that might be attributed to aberrant DNA repair by NHEJ [9]. From an experimental standpoint, chromosomal instability is readily detected by microscopic visualization. Metaphase chromosomes are fundamental substrates for cytogenetic studies and metaphase spreads are usually analyzed to quantitatively evaluate chromosomal abnormalities. To perform this metaphase spread analysis, FA-D2−/− or FA-D2+/+ cells were co-treated with WRN helicase inhibitor NSC 617145 (0.125 μM) and MMC) (9.4 nM) or with either agent alone for 2 days followed by Colcemid (0.1 mg/ml) for 1 h. For metaphase spreads, cells were swollen in KCL (0.075 M) for 12 min, fixed with methanol/acetic acid (3:1), dropped on microscope slides and stained with Giemsa (5%) [39]. The slides were imaged using a Zeiss LSM 510 META inverted Axiovert 200 M laser scan microscope with a Plan-Apochromat 100× 1.4-numerical-apperture oil immersion differential interference contrast objective lens. Images were analyzed using the LSM Browser software package. In the case of FA-D2−/− cells, co-treatment with WRN helicase inhibitor NSC 617145 and MMC resulted in a significant increase in abnormal chromosomal structures including chromatid breaks, chromatid loss and radials compared to either agent alone. Of note, FA-D2+/+ cells co-treated with NSC 617145 and MMC showed no significant increase in chromosomal aberrations, thus indicating that chromosomal instability was dependent on the mutational status of FANCD2.
6. Concluding remarks
In this Methods article, we have described experimental approaches and strategies we have used to identify and characterize small molecule helicase inhibitors, with an emphasis placed on the WRN helicase-nuclease which is defective in a monogenic accelerated aging disorder known as Werner syndrome. The experimental strategy begins with a simple in vitro screen to measure the effects of compounds on helicase-catalyzed DNA unwinding using a radiometric strand displacement assay, and is followed by cell-based assays using a small subset of compounds identified from the in vitro screen. This tactic was successful for the discovery of the first human helicase inhibitor described in the literature. One advantage to this approach is that a library can be chosen in which it is known that the compounds are biologically active. However, there are advantages to performing more high-throughput screens (typically fluorometric) on vastly larger libraries, even ones that are composed of a large number of compounds whose biological activity was not previously assessed. In this case, lead compound development of positive hits with the appropriate chemistry scaffold may be undertaken to optimize the compound for potency, specificity, and pharmacological parameters.
Although this review is modeled after our experimental studies with the WRN helicase inhibitor, we believe it provides some guiding concepts and experimental strategies that can be extrapolated to many helicases, particularly those implicated in the DNA damage response or DNA repair. Helicase inhibitors provide an alternative strategy for investigating the molecular and cellular functions of their targets, and in a broader scope, the sophisticated orchestration of overlapping and intersecting DNA metabolic pathways. In addition, we visualize helicases as suitable small molecule targets that might enhance existing anti-cancer strategies or emerge as novel therapeutic treatments not yet envisioned.
Highlights.
Strategies to identify small molecule inhibitors of DNA helicases
Biochemical assays for high-throughput screening of small molecule libraries
Cell-based assays to assess effects of helicase inhibitors on DNA damage and viability
Determination of chemical- and genetic-based synthetic lethality
Acknowledgments
This research was supported in full by the Intramural Research Program of the NIH, National Institute on Aging.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, Martin NM, Jackson SP, Smith GC, Ashworth A. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- [2].Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, Helleday T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- [3].Huang F, Mazin AV. Targeting the homologous recombination pathway by small molecule modulators. Bioorg. Med. Chem. Lett. 2014;24:3006–3013. doi: 10.1016/j.bmcl.2014.04.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Aggarwal M, Brosh RM., Jr. Hitting the bull’s eye: novel directed cancer therapy through helicase-targeted synthetic lethality. J. Cell Biochem. 2009;106:758–763. doi: 10.1002/jcb.22048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Gupta R, Brosh RM., Jr. DNA repair helicases as targets for anti-cancer therapy. Curr. Med. Chem. 2007;14:503–517. doi: 10.2174/092986707780059706. [DOI] [PubMed] [Google Scholar]
- [6].Sharma S, Doherty KM, Brosh RM., Jr. DNA helicases as targets for anti-cancer drugs. Curr. Med. Chem. Anti. -Canc. Agents. 2005;5:183–199. doi: 10.2174/1568011053765985. [DOI] [PubMed] [Google Scholar]
- [7].Brosh RM., Jr. DNA helicases involved in DNA repair and their roles in cancer. Nature Reviews Cancer. 2013;13:542–558. doi: 10.1038/nrc3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Shadrick WR, Ndjomou J, Kolli R, Mukherjee S, Hanson AM, Frick DN. Discovering new medicines targeting helicases: challenges and recent progress. J. Biomol. Screen. 2013;18:761–781. doi: 10.1177/1087057113482586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Aggarwal M, Banerjee T, Sommers JA, Iannascoli C, Pichierri P, Shoemaker RH, Brosh RM., Jr. Werner syndrome helicase has a critical role in DNA damage responses in the absence of a functional Fanconi Anemia pathway. Cancer Research. 2013;73:5497–5507. doi: 10.1158/0008-5472.CAN-12-2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Aggarwal M, Sommers JA, Shoemaker RH, Brosh RM., Jr. Inhibition of helicase activity by a small molecule impairs Werner syndrome helicase (WRN) function in the cellular response to DNA damage or replication stress. Proc. Natl. Acad. Sci. U. S. A. 2011;108:1525–1530. doi: 10.1073/pnas.1006423108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Aggarwal M, Banerjee T, Sommers JA, Brosh RM., Jr. Targeting an Achilles’ heel of cancer with a WRN helicase inhibitor. Cell Cycle. 2013;12:3329–3335. doi: 10.4161/cc.26320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Copeland RA. Assay considerations for compound library screening, Evaluaiton of enzyme inhibitors in drug discovery: a guide for medicinal chemists and pharmacologists. John Wiley & Sons, Inc.; Hoboken, NJ: 2013. [PubMed] [Google Scholar]
- [13].Nguyen GH, Dexheimer TS, Rosenthal AS, Chu WK, Singh DK, Mosedale G, Bachrati CZ, Schultz L, Sakurai M, Savitsky P, Abu M, McHugh PJ, Bohr VA, Harris CC, Jadhav A, Gileadi O, Maloney DJ, Simeonov A, Hickson ID. A small molecule inhibitor of the BLM helicase modulates chromosome stability in human cells. Chem. Biol. 2013;20:55–62. doi: 10.1016/j.chembiol.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Brosh RM, Jr., Opresko PL, Bohr VA. Enzymatic mechanism of the WRN helicase/nuclease. Methods Enzymol. 2006;409:52–85. doi: 10.1016/S0076-6879(05)09004-X. [DOI] [PubMed] [Google Scholar]
- [15].Abd WS, Choi M, Bianco PR. Characterization of the ATPase activity of RecG and RuvAB proteins on model fork structures reveals insight into stalled DNA replication fork repair. J. Biol. Chem. 2013;288:26397–26409. doi: 10.1074/jbc.M113.500223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Shadrick WR, Mukherjee S, Hanson AM, Sweeney NL, Frick DN. Aurintricarboxylic acid modulates the affinity of hepatitis C virus NS3 helicase for both nucleic acid and ATP. Biochemistry. 2013;52:6151–6159. doi: 10.1021/bi4006495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Boger DL, Tse WC. Thiazole orange as the fluorescent intercalator in a high resolution fid assay for determining DNA binding affinity and sequence selectivity of small molecules. Bioorg. Med. Chem. 2001;9:2511–2518. doi: 10.1016/s0968-0896(01)00243-7. [DOI] [PubMed] [Google Scholar]
- [18].Bialkowska AB, Du Y, Fu H, Yang VW. Identification of novel small-molecule compounds that inhibit the proproliferative Kruppel-like factor 5 in colorectal cancer cells by high-throughput screening. Mol. Cancer Ther. 2009;8:563–570. doi: 10.1158/1535-7163.MCT-08-0767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Herrero MD, Boro A, Schafer BW. Cell-based small-molecule compound screen identifies fenretinide as potential therapeutic for translocation-positive rhabdomyosarcoma. PLoS. ONE. 2013;8:e55072. doi: 10.1371/journal.pone.0055072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Rzuczek SG, Southern MR, Disney MD. Studying a drug-like, RNA-focused small molecule library Identifies compounds that inhibit RNA toxicity in Myotonic Dystrophy. ACS Chem. Biol. 2015;10:2706–2715. doi: 10.1021/acschembio.5b00430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Salic A, Mitchison TJ. A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc. Natl. Acad. Sci. U. S. A. 2008;105:2415–2420. doi: 10.1073/pnas.0712168105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Panier S, Boulton SJ. Double-strand break repair: 53BP1 comes into focus. Nat. Rev. Mol. Cell Biol. 2014;15:7–18. doi: 10.1038/nrm3719. [DOI] [PubMed] [Google Scholar]
- [23].Brosh RM, Jr., Waheed J, Sommers JA. Biochemical characterization of the DNA substrate specificity of Werner syndrome helicase. J. Biol. Chem. 2002;277:23236–23245. doi: 10.1074/jbc.M111446200. [DOI] [PubMed] [Google Scholar]
- [24].Moser MJ, Kamath-Loeb AS, Jacob JE, Bennett SE, Oshima J, Monnat RJ., Jr. WRN helicase expression in Werner syndrome cell lines. Nucleic Acids Res. 2000;28:648–654. doi: 10.1093/nar/28.2.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Gomez D, O’Donohue MF, Wenner T, Douarre C, Macadre J, Koebel P, Giraud-Panis MJ, Kaplan H, Kolkes A, Shin-Ya K, Riou JF. The G-quadruplex ligand telomestatin inhibits POT1 binding to telomeric sequences in vitro and induces GFP-POT1 dissociation from telomeres in human cells. Cancer Res. 2006;66:6908–6912. doi: 10.1158/0008-5472.CAN-06-1581. [DOI] [PubMed] [Google Scholar]
- [26].Tahara H, Shin-Ya K, Seimiya H, Yamada H, Tsuruo T, Ide T. G-Quadruplex stabilization by telomestatin induces TRF2 protein dissociation from telomeres and anaphase bridge formation accompanied by loss of the 3′ telomeric overhang in cancer cells. Oncogene. 2006;25:1955–1966. doi: 10.1038/sj.onc.1209217. [DOI] [PubMed] [Google Scholar]
- [27].Temime-Smaali N, Guittat L, Sidibe A, Shin-Ya K, Trentesaux C, Riou JF. The G-quadruplex ligand telomestatin impairs binding of topoisomerase IIIalpha to G-quadruplex-forming oligonucleotides and uncaps telomeres in ALT cells. PLoS. ONE. 2009;4:e6919. doi: 10.1371/journal.pone.0006919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Crabbe L, Verdun RE, Haggblom CI, Karlseder J. Defective telomere lagging strand synthesis in cells lacking WRN helicase activity. Science. 2004;306:1951–1953. doi: 10.1126/science.1103619. [DOI] [PubMed] [Google Scholar]
- [29].Fry M, Loeb LA. Human werner syndrome DNA helicase unwinds tetrahelical structures of the fragile X syndrome repeat sequence d(CGG)n. J. Biol. Chem. 1999;274:12797–12802. doi: 10.1074/jbc.274.18.12797. [DOI] [PubMed] [Google Scholar]
- [30].Mohaghegh P, Karow JK, Brosh RM, Jr., Bohr VA, Hickson ID. The Bloom’s and Werner’s syndrome proteins are DNA structure-specific helicases. Nucleic. Acids. Res. 2001;29:2843–2849. doi: 10.1093/nar/29.13.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Helleday T. Putting poly (ADP-ribose) polymerase and other DNA repair inhibitors into clinical practice. Curr. Opin. Oncol. 2013;25:609–614. doi: 10.1097/CCO.0000000000000016. [DOI] [PubMed] [Google Scholar]
- [32].Lord CJ, Tutt AN, Ashworth A. Synthetic lethality and cancer therapy: lessons learned from the development of PARP inhibitors. Annu. Rev. Med. 2015;66:455–470. doi: 10.1146/annurev-med-050913-022545. [DOI] [PubMed] [Google Scholar]
- [33].Chen CC, Kennedy RD, Sidi S, Look AT, D’Andrea A. CHK1 inhibition as a strategy for targeting Fanconi Anemia (FA) DNA repair pathway deficient tumors. Mol. Cancer. 2009;8:24. doi: 10.1186/1476-4598-8-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Hucl T, Gallmeier E. DNA repair: exploiting the Fanconi anemia pathway as a potential therapeutic target. Physiol Res. 2011;60:453–465. doi: 10.33549/physiolres.932115. [DOI] [PubMed] [Google Scholar]
- [35].Kennedy RD, Chen CC, Stuckert P, Archila EM, l. de, V, Moreau LA, Shimamura A, D’Andrea AD. Fanconi anemia pathway-deficient tumor cells are hypersensitive to inhibition of ataxia telangiectasia mutated. J. Clin. Invest. 2007;117:1440–1449. doi: 10.1172/JCI31245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Taniguchi T, D’Andrea AD. Molecular pathogenesis of Fanconi anemia: recent progress. Blood. 2006;107:4223–4233. doi: 10.1182/blood-2005-10-4240. [DOI] [PubMed] [Google Scholar]
- [37].Niedzwiedz W, Mosedale G, Johnson M, Ong CY, Pace P, Patel KJ. The Fanconi anaemia gene FANCC promotes homologous recombination and error-prone DNA repair. Mol. Cell. 2004;15:607–620. doi: 10.1016/j.molcel.2004.08.009. [DOI] [PubMed] [Google Scholar]
- [38].Bunting SF, Nussenzweig A. Dangerous liaisons: Fanconi anemia and toxic nonhomologous end joining in DNA crosslink repair. Mol. Cell. 2010;39:164–166. doi: 10.1016/j.molcel.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Schlacher K, Wu H, Jasin M. A distinct replication fork protection pathway connects Fanconi Anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell. 2012;22:106–116. doi: 10.1016/j.ccr.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]