Abstract
Helicases are enzymes involved in nucleic acid metabolism, playing major roles in replication, transcription, and repair. Defining helicases oligomerization state and transient and persistent protein interactions is essential for understanding of their function. In this article we review current methods for the protein-protein interaction analysis, and discuss examples of its application to the study of helicases: Pif1 and DDX3. Proteomics methods are our main focus – affinity pull-downs and chemical cross-linking followed by mass spectrometry. We review advantages and limitations of these methods and provide general guidelines for their implementation in the functional analysis of helicases.
Keywords: Biological networks, helicase, protein oligomerization, protein-protein interactions, mass spectrometry, chemical cross-linking
Graphical Abstract
1. Introduction
1.1 Types of protein-protein interactions
For methodological purposes it is convenient to divide protein-protein interactions (PPIs) into two major classes – persistent and transient. Persistent PPIs occur when the protein of interest functions as a part of a stable multi-subunit protein complex. In contrast, transient PPIs are brief and often weak. Both classes of interactions can be important for enzymatic function – e.g. function of an oligomeric helicase is an example of persistent PPI, while phosphorylation of a protein by a protein kinase is an example of a transient interaction involved in regulation. In this methods article we will be focusing primarily on the persistent PPIs class.
1.2 Protein-protein interaction networks
In a cellular environment, proteins and enzymes rarely act in isolation, but instead rely on a network of interactions with other proteins to perform their functions. Helicases – enzymes responsible for nucleic acid structure remodeling – are a perfect illustration of this principle. Typical helicases function in a variety of quaternary forms – from monomers to double hexamers [1], and their function is often regulated by the oligomeric state. Our group has encountered this type of regulation during analysis of NS3 helicase from the Hepatitis C virus [2]. NS3 is a superfamily II (SF2) helicase, with a 3’-to-5’ polarity of translocation. In prior publications we also have shown that interaction of NS3 with another viral non-structural protein, NS5B, stimulate its function [3]. Similar cases of the role of PPIs in helicase function abound in the literature, including heterohexameric minichromosome maintenance (MCM2–7) helicase, which belongs to AAA+ superfamily of ATPases and is involved in eukaryotic DNA replication, with 3’-to-5’ polarity of translocation [4–7]. The architecture of the complex was determined by in vivo protein-protein crosslinking [7].). In a later publication, the Botchan lab showed that MCM2–7 are associated with CDC45 and the GINS complex by immunoaffinity chromatography [8]. It is this Cdc45/Mcm2–7/GINS (CMG) complex which appears to be the active helicase complex. Recently, the architecture of the entire replisome, including the CMG complex and the leading and lagging strand polymerases was determined by EM and chemical crosslinking [9]. Another example of PPIs in helicase function is the yeast DEAD-box helicase Ded1 and its human homolog DDX3, SF2-type helicases, without a strict polarity of translocation. Interaction of Ded1 with the nuclear exportin complex was found to inhibit its ATPase activity. Similarly, DDX3 has been shown to interact with the CRM1 nuclear pore [10, 11]. In this article we will highlight some of these studies, primarily those that used mass spectrometry and proteomics.
1.3 Protein-protein interaction data repositories and databases
The importance of PPIs for protein and enzyme function is now well appreciated. Hence, in parallel to the tools for sequence analysis – protein sequence database (e.g. Uniprot, NCBI, Blast); structure analysis – structure repositories (e.g. PDB); tools for PPI analysis are becoming just as important at the beginning stages of an exploratory analysis into a given protein’s function. Our group has used the following tools in our work: String database [12], Biogrid and Cytoscape [13], PINA database [14], and iRefIndex database [15, 16]. For advanced users, R and Bioconductor cisPath package [17] may give more creative control over PPI visualization and analysis.
1.4 Network properties and exploratory PPI network analysis
In this section we demonstrate the use of some of the above-mentioned PPI tools and databases for an exploratory analysis of DEAD-Box ATP dependent Dbp2 yeast helicase and its respective human homolog DDX5. These helicase are SF2-type helicases and are members of the DEAD-box RNA helicase family, characterized by nine conserved motifs, and capable of unwinding blunt ended RNA duplexes, without a strict polarity of translocation [18]. In addition to duplex RNA unwinding, DDX5 functions as a transcriptional co-activator for multiple transcription factors including estrogen and androgen receptors and p53 [19]. The variety of functions of DDX5 suggest likely interactions with multiple different protein partners as well as possible posttranslational modifications.
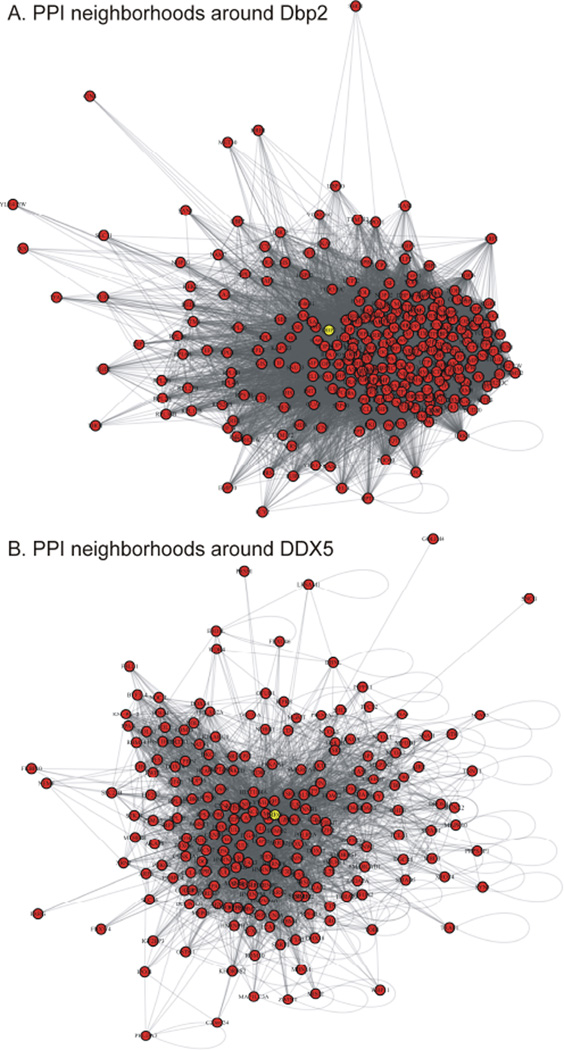
Figures 1A,B shows the initial visualization of yeast and humans PPI networks where Dbp2 and DDX5 were used as the input nodes, using R (version 3.2.2) with igraph package. Specifically, these networks show the first-degree neighborhoods of these proteins in the two organisms. There is no significant utility in these visualizations at this stage as it is not clear how to immediately make sense of the myriad of connections. One approach is to calculate network properties of the nodes in questions, and to see if these changes differ between yeast and human. Table 1 shows such an analysis. At this point we evaluate: betweeness, degree (d), and closeness. Betweeness measures number of paths that go through the node in question, degree measures the number of connections, and closeness measures average pair-wise proximities of nodes in a graph. The high values of betweenness and centrality of a protein in a PPI network were suggested to correlate with the importance of the protein. Both proteins, Dbp2, and DDX5 have high values of betweenness and centrality and are important in both organisms. However, beyond this vague indication of “functional importance” nothing else can be inferred using centrality measures alone. To gain further insight into proteins functional association in PPI networks, one can explore well-established concepts in graph theory. For a graph G(V,E) where V is the set of vertices and E is the set of edges, the first minimum spanning tree (MST) is defined as the acyclic subset T1⊆E that connects all vertices in V and whose total length Σi,j∈T1 d(vi, vj) is minimal. The second MST is defined as the MST of the reduced graph G(V, E-T1). Here the total length is measured as the number of edges connecting all vertices. The union of the first and second MST is denoted by MST2. MST function is available within the igraph package.
Figure 1.
Respective PPI neighborhoods around Dbp2 yeast helicase (A) and its human homolog DDX5 (B) has been visualized using information retrieved from PINA and String databases of interactions. The input Dbp2 and DDX5 nodes are highlighted in yellow. Additional analysis is clearly needed to make sense of these interactions – compare to the derivative MST2 networks in Fig. 2.Figure 2
Table I.
Comparison of network properties of yeast Dbp2 helicase and its human homolog DDX5.
| Network Property | Yeast DBP2 | Human DDX5 |
|---|---|---|
| betweeness | 3282.609 | 46090.11 |
| degree | 284 | 248 |
| closeness | 1.11e-05 | 6.17e-07 |
Betweeness centrality gives the number of paths going through a protein node Degree centrality is the number of immediate interactors (those that are shown in Figure 1), Closeness centrality measures average distance of paths going through the input node, the higher it is, the closer nodes are to each other. Higher centrality measures may indicate higher functional importance.
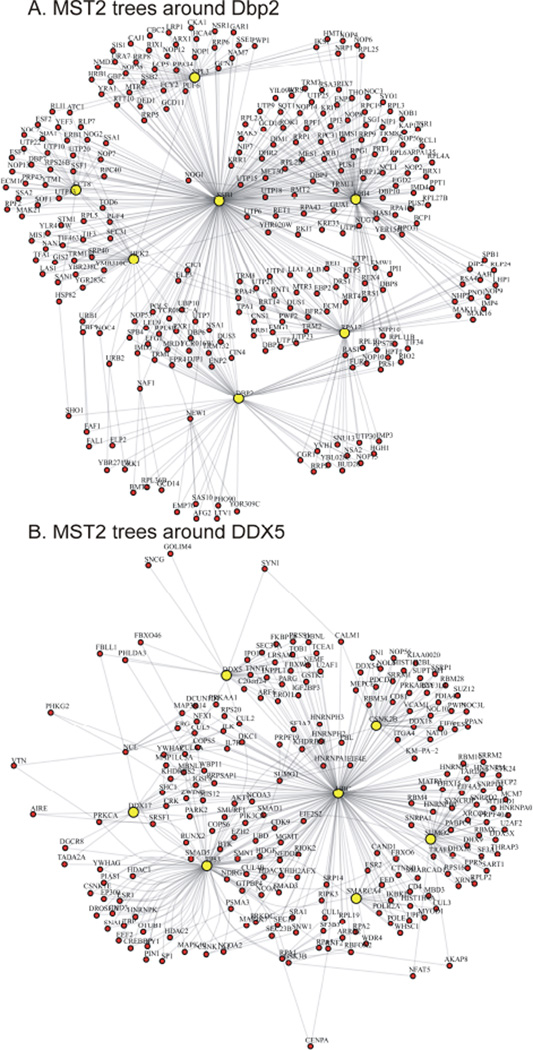
In gene expression studies MST analysis was used for expression data clustering [20, 21] and module inference [22]. Rahmatallah et al recently suggested to employ MST2 to visualize gene coexpression networks with all correlations present [23]. Similarly, the MST2 approach can also be informative in deciphering characteristic properties of PPI networks. Figure 2 presents MST2 constructed from the first-degree neighborhood of Dbp2 that highlights the minimal set of essential interactions among proteins in the neighborhood. A highly connected protein tends to occupy a central position and has a relatively high degree in the MST2 because the shortest paths connecting the vertices of the first and second MSTs tend to pass through this protein [23]. In contrast, a protein with low degree most likely occupies a non-central position in the MST2 and has a degree of 2. Using MST2 representation one can see the structure of the first-degree neighborhood of Dbp2 more clearly (Figure 2, compare to the clutter of connections in Figure 1). There are several highly connected proteins (HEK2 (d=33), SSB1 (d=224), NPL3 (d=44), CCT8 (d=32), UBI4 (d=110), RPA12 (d=59), DBP2 (d=61)) that form a backbone structure of the neighborhood, while most of the proteins have degree of 2. Similarly, MST2 representation of the structure of the first degree neighborhood of DDX5 highlights the presence of seven hubs (TP53 (d=99), UBC (d=176), SMARCA4 (d=18), CSNK2B (d=32), SUMO2 (d=39), DDX17 (d=14), DDX5 (d=27)). As a result, more biological insights can be gained from studying MST2 trees compared to the original PPIs graphs. In the example of Dbp2/DDX5 one could see functions of DDX5 that are specific to humans – interactions with p53 and chromatin modifiers. At the same time, interactions with RNA-binding proteins persist in both organisms.
Figure 2.
MST2 trees around PPIs of Dbp2 yeast helicase (A) and its human homolog DDX5 (B), derived from the respective networks shown in Fig. 1 (using R with iGraph package). Fine network structure becomes visible, with highly connected molecular chaperons, protein modifiers and regulators occupying central positions (UBI4, SSB1 in yeast, and Ubiquitin and p53 in human). Figure 4
2. Methods of protein-protein interaction discovery
2.1 Affinity pull-down approach
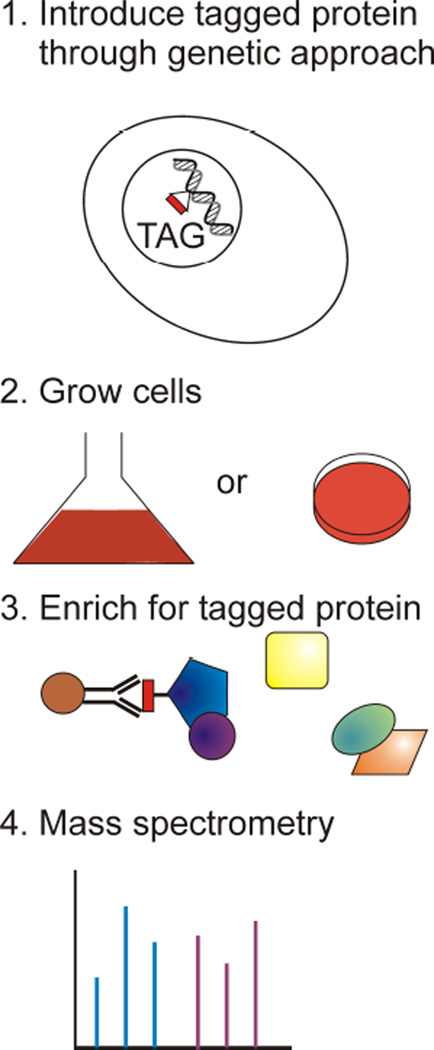
Affinity pull-down exploits specific interactions between proteins and antibodies or between proteins and other protein-specific molecules, immobilized in a column, magnetic beads, or in the form of an array. For example, to isolate phosphorylated proteins from the rest of the proteome, one can use an immuno-affinity column prepared with antibodies specific to the phosphorylated amino acids. Similarly, to isolate proteins tagged with a His6 tag, one can use Ni2+ or Co2+ resins. Epitope tagging is used in large-scale analyses of protein–protein interactions and protein complexes (Figure 3). In the epitope tagging strategy, proteins to be isolated are fused with a motif recognizable by a specific antibody. This fusion is done by incorporating the epitope sequence into the gene that encodes the protein of interest. Next, the tagged protein is expressed and purified along with its interaction partners by immuno-affinity chromatography or magnetic beads using antibodies specific to the particular epitope. Combinations of different tags, or multiples of the same tags are often used to increase specificity. If a particular protein is of low abundance, it can be overexpressed to increase the overall recovery.
Figure 3.
Epitope tagging approach in affinity pull-down mass spectrometry experiments. Proteins constructs with a fused tag are prepared in a model organism, cells are lysed, and the tag-specific antibodies are used to enrich for protein complexes, followed by the mass spectrometry analysis.
After initial capture of the tagged protein, the recovered mixture is washed to remove weakly bound, or non-specific proteins. Notably, affinity pull-down differs from affinity purification only in the stringency of washes. Typically, higher and higher salt concentration in buffer is used in the successive washes, so that only the most stable of the interactions remain. Next, the protein bait along with its interactors is eluted in denaturing conditions and submitted to a protein analysis. Proper control accounting for non-specific interactions is very important, especially in the context of pull-down from whole cells, where highly abundant proteins – e.g. chaperones, house-keeping proteins, ribosomal proteins - will typically show up as hits. Such a control can be achieved for example, by using a non-specific antibody or an untagged construct. The majority of the interactions shown in Figure 1 were obtained through affinity pull-down methods.
2.1.1 Identification of proteins obtained by affinity pull-down using mass spectrometry based proteomics
Due to the development of soft ionization techniques and methods of predictable fragmentations of peptides, mass-spectrometry became a primary tool for identification of proteins in complex protein mixtures [24]. In a typical proteomics experiment, protein mixture (e.g. obtained by an affinity pull-down) is resolved by a denaturing polyacrylamide gel. The individual gel lanes are then cut into 20–30 small bands followed by proteolytic digestion of the proteins in individual gel slices, and the subsequent analysis of the resulting peptide mixtures by liquid-chromatography coupled to a mass spectrometer. Peptide sequences are derived from the mass spectra of peptide fragments, usually by automated algorithms, followed by protein inference and assembly from multiple peptide sequences.
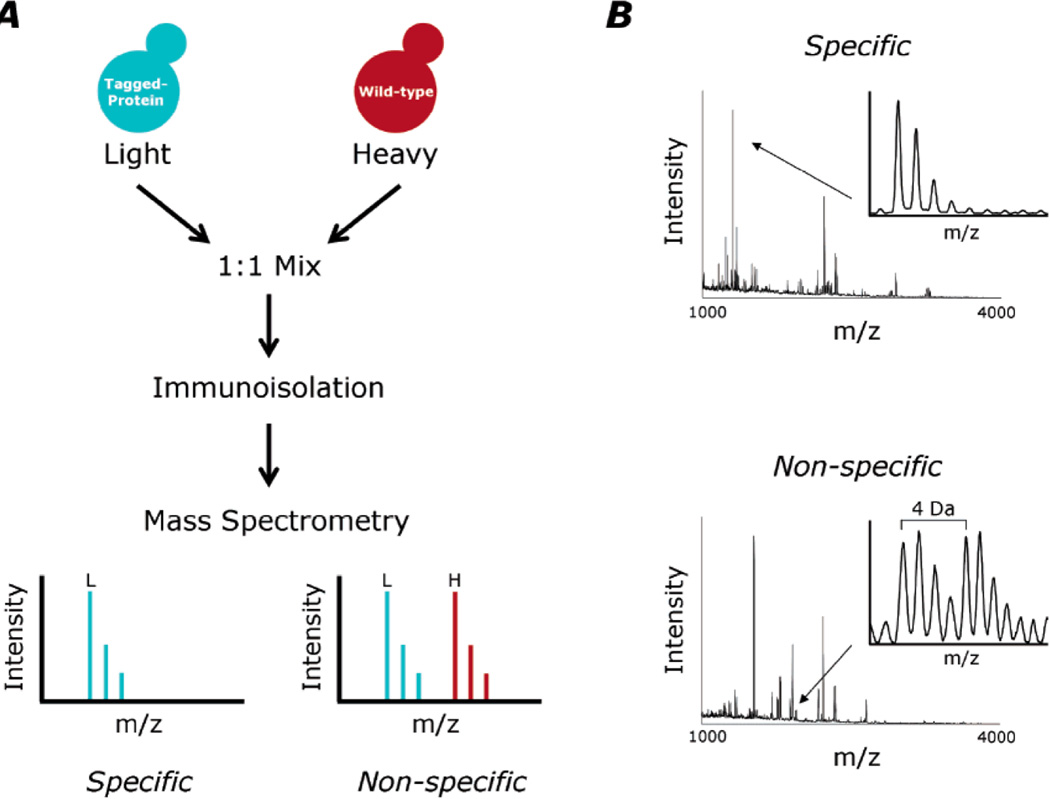
Notably, an affinity pull-down is a necessarily quantitative experiment – it is important to know quantities protein hits relative to a bait, and also, relative to the non-specific binding control. The most precise quantification in a mass spectrometry analysis is achieved by using stable isotopes – peptide mixtures from one of the conditions are labeled with heavy isotopes (e.g. 15N, 13C, 2H, or 18O) and compared to the other condition labeled with the light isotopes (14N, 12C, 1H, or 16O). In such an experiment, total protein concentration is determined in each of the conditions, and 1:1 mixture is prepared. The protein ratios are determined from the heavy:light ratios of the representative peptide ions in mass spectra. In application to the affinity pull-downs this technic is known as Isotopic Differentiation of Interactions as Random or Targeted (I-DIRT), developed by Tackett et. al. [25] – shown in Figure 4.
Figure 4.
I-DIRT method for determining specific interactions in a protein complex. Figure is reproduced from [21], by permission. A) General scheme of the I-DIRT – cellular lysates from cells expressing tagged proteins are labeled with light isotopic label, and un-tagged cells are labeled with the heavy version of the label. The lysates are mixed, and the affinity isolation is performed. Non-specific interactions correspond to 1:1 ratio of light vs. heavy while specific interactions result in an increased ratio of light to heavy peptides. B) Examples of mass spectra from a tryptic peptide indicative of specific protein interaction (top), and non-specific (bottom). The bottom spectrum shows a 4 Da shift corresponding to a lysine containing 4 deuterium atoms used as a heavy label for the untagged cell lysate.
Label-free approaches can be used when isotopic labeling is not available. One of commonly used label-free protein quantitation approaches is spectral counting – e.g. counting of number of mass spectra matched to a given protein in a tandem mass spectrometry experiment. Typically, the number of these counts is proportional to the proteins abundance in a complex mixtures [26–29]. Powerful statistical methods have been developed, that rely on the label-free quantitation using normalized spectral counts to infer PPI networks from pull-down experiments using multiple baits [30].
Once a protein has been identified as being part of the interaction network with the bait protein, confirmation of the interaction can achieved by a reciprocal pull-down in which the newly identified interactor becomes the bait. In a multiple-bait approach, each of the candidate subunits of a protein complex serve as baits in multiple pull-down, and the interaction network emerges through a statistical consensus, after the removal of non-specific interactions.
2.1.2. Validation of protein-protein interactions discovered by affinity pull-down
The gold standard of protein detection is a western blot, and this is how mass spectrometry-derived protein identifications are often validated. The discovered PPIs can be further explored by co-localization techniques using antibodies for proteins in a pair-wise interaction. Notably, affinity pull-downs obtain a subset of the whole interaction network, and the detected PPIs are not necessarily direct or pair-wise. To validate pair-wise interactions, one could use the yeast-two-hybrid method [31] or mammalian-two-hybrid method [32], where proteins of interest activate transcription of a reporter, if the interaction takes place. Finally, the prospective interacting partners can be purified and their interaction can be studied in detail by using biophysical methods. Typical approaches for evaluating the strength of specific protein interactions include isothermal calorimetry [33], surface plasmon resonance [34], and analytical ultracentrifugation [35]. Recombinant protein can be fluorescently labeled so that a variety of fluorescence techniques can be used to examine binding. Qualitative confirmation of specific protein-protein interactions can be obtained from size-exclusion chromatography, native polyacrylamide gel electrophoresis, and simple co-precipitation experiments.
2.2. Chemical Crosslinking
Affinity methods of PPIs discovery and analysis primarily yield information about identities of the interacting partners, and at most, about stoichiometry of stable protein complexes. Additional structural information about protein-protein interfaces and topology of interactions can be gained when chemical cross-linking is employed for PPIs capture. By definition, chemical cross-linking introduces covalent bonds between interacting partners. Typically, chemical cross-links survive protein denaturation and digestion, and cross-linked peptide pairs, along with the cross-linking sites can be identified in a mass spectrometer. We recently reviewed this topic extensively in [36]. We also recommend earlier review by Trakselis et al, which discusses chemical cross-linking for structural analysis of protein complexes [37].
The following experimental difficulties need to be considered when implementing a cross-linking method: 1) in a cross-linked protein mixture concentration of the linear peptides is always higher compared to the cross-linked ones; 2) cross-links within the same molecule (intra-protein) are typically several fold higher than cross-links between different molecules (inter-protein); 3) data analysis and interpretation is not as straightforward for cross-linked species and requires specialized software; 4) multiple types of cross-linkers and experimental conditions need to be screen for optimal specificity and sensitivity.
Long-length cross-linkers tend to capture interactions away from the interaction interface, while shorter cross-linkers tend to capture interactions close to the interface. Also, if one uses a chemoselective cross-linker, for example zero-length cross-linker 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide (EDC), which catalyzes condensation of primary amines with carboxylic acids [38]; only those interactions, that have these residues at right positions will be obtained. Other protein-protein contacts that do not involve carboxy-to-amino contacts will be lost. At the same time, when a carboxylic acid/amine pair is buried deep within interaction interface, the cross-linker will not get to it, thereby ignoring most stable of interactions.
In contrast, if a very long, flexible cross-linker is chosen that couples a specific amino acid pair, then there is an improved chance that the majority of interactions will have at least one such pair at the right position within the cross-linker’s reach to be captured. Another thing to consider is that long-length cross-linker may detect spurious, non-specific interactions and alter the native protein structure more so, compared to the short-length cross-linkers.
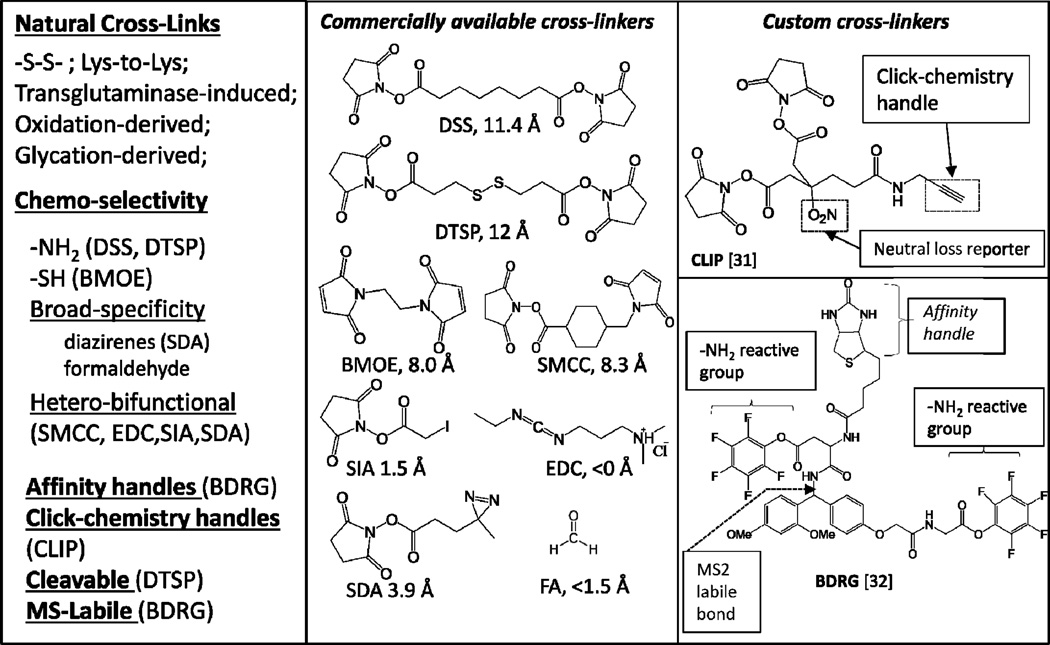
As a consequence, in [36] we suggested that long-length cross-linkers, selective for frequently occurring amino acids, are useful for establishing identities of interacting partners; while shorter, broadly selective cross-linkers are better suited for analyzing structure of PPI interfaaces. An affinity handle, such as biotinyl, or alkynyl for conjugation using click-chemistry; and MS2-labile reporter groups can be introduced into a long-length cross-linker [39, 40]. These custom multi-functional cross-linkers allows both for selective detection and specific enrichment of cross-linked species. Figure 5 summarizes cross-linkers frequently used in PPI analysis; examples of the multifunctional cross-linkers are given in the right-most panel.
Figure 5.
Cross-linking agents for the capture and analysis of protein protein interactions. Reproduced from [29], an open access publication.
3. Example of PPI analysis: DEAD-box RNA helicases
Human ATP-dependent DEAD-box RNA helicases have been implicated in a variety of cellular processes important for carcinogenesis such as cell survival, cell-cycle control, and apoptosis. DDX3 is a prime example of this class of helicases, which has been demonstrated to have both oncogenic and tumor-suppressor properties and as a consequence, suggested as a potential target for cancer treatment [41, 42]. DDX3 is a DEAD-box RNA helicase that can also unwind DNA with a 5’-single-stranded overhang in vitro [43]). It has multiple functions in RNA processing including transcription, mRNA maturation, export, and translation [44]). For example, when nucleases are found to interact with helicases, nucleic acid degradation is implied. If helicases bind to polymerases, then nucleic acid synthesis is likely to be a functional outcome. Regulation of inherent helicase properties such as processivity and rate of unwinding can be impacted through self-interactions. DDX3 directly interacts with DDX5 as shown by the yeast two hybrid assay and co-localization studies [45], and therefore is present in the networks that we discussed in introduction – Figures 1B and 2B. Other RNA helicase are often in the vicinity of the PPI network, directly interacting, or being in close proximity – DDX17, DHX36, DDX6. Here we will summarize some of the experiments that identified interactome of the ATP-dependent DEAD/H-box RNA helicases.
The experimental papers that use a given helicase (e.g. DDX3) as a bait, or find it as a hit, as well the papers that present strong biochemical evidence for pair-wise interactions can be easily retrieved from String database of protein interactions [12]. Using this approach for DDX3 and DDX6, the following study become visible: studies by Shih et al. and Bish et al [46, 47], both implicating DDX3 and DDX6 in stress granule assembly.
Shih et. al. report DDX3 suppressing translation by directly inhibiting eIF4E (eukaryotic initiation factor 4E) and establish interaction of DDX3 with the stress granule marker PABP1 (poly(A)-binding protein 1). Bish et. al. performed the large-scale affinity pull-down using another ATP-dependent RNA helicase DDX6, which is a known component of P-bodies, along with DDX3. Multiple proteins involved in RNA metabolism and ribosomal proteins has been identified. The authors performed impressive validation of the found hits, confirming 81% of interactions. Eleven new splicing factors has been confirmed, implicating DDX6 into in this previously unknown role. The authors further used native gel electrophoresis and separated different protein complexes involving DDX6. In summary, this report is an excellent illustration of PPI methodologies, which separates multiple types of interactions and performs their extensive validation: direct pairwise interactions are sorted out from complex interactions and from RNA-dependent interactions.
In a recent study Jain et al. focused on the biology of stress granules formation and dynamics [48]. Affinity pull-down using known stress granule markers as baits in yeast and mammalian cells was the key methodology allowing the authors to isolate stress granules and characterize the stress granule proteome. Pull-downs from formaldehyde cross-linked cells allowed characterization of less stable PPIs within the stress granules. Based on co-localization microscopy experiments and proteomics data the authors suggested existence of stable stress granule cores and dynamic outer shell. A number of ATP-dependent RNA and DNA helicases has been identified as stress granule components in both yeast and mammals (e.g. DDX3X, DDX6, EIF4A). DEAD-box proteins specific to mammalian cells included DDX1, DDX5, DDX17, and others. Interestingly, minichromosome maintenance complex, consisting of MCM helicases, and RuvBI helicase complex has been also found with stress granules of both organisms.
4. Example of PPI analysis: the case of mitochondrial yeast proteins – Pif1 helicase and Rim1 single-strand binding protein
Pif1 is a highly conserved eukaryotic helicase, which has nuclear and mitochondrial forms [49], and we were motivated to study mechanism of its action and its PPIs because of reports of its involvement in the resolution of non-canonical DNA structures and mitochondrial DNA maintenance during conditions of DNA damage [50]. In an initial pull-down experiment using the I-DIRT method we found only one strongly interacting partner – single strand binding protein Rim1 [51], using MALDI-TOF and LTQ mass spectrometry. In the same publication we characterized this interaction in detail, using recombinant proteins, demonstrating that Rim1 enhances Pif1 activity.
Recently, we revisited the Pif1 interactome, as a more sensitive and accurate LTQ-Orbitrap-Velos mass spectrometer became available. Using 60,000 resolving power, allowed routine measurements of peptide masses within 3 ppm mass error; and employing online reverse phase separation and nano-electrospray ionization further increased sequence coverage of identified proteins. In addition, we were interested in capturing the Rim1-Pif1 interaction using chemical cross-linker, aiming to define the structure of the protein-protein interaction interface [52]. The more sensitive affinity pull-down using TAP-tagged Pif1 produced more interacting partners, with Rim1 still being among the top hits (Table II). Notably, we used label-free spectral counting method for this analysis, as opposed to the I-DIRT technique used earlier. The label-free analysis became possible due to higher sensitivity of the new mass spectrometer and the subsequent hundred-fold increase in the number of spectra. Other mitochondrial proteins were identified in addition to Rim1 (bold face, Table II).
Table II. The most significant Pif1 interactors as determined by the TAP-Pif1 pull down and LTQ-Orbitrap mass spectrometry.
(Originally published in Zybailov et al., 2015, Journal of Proteomics and Bioinformatics, and open access journal, reference [52]).
| accession a | descriptionb | Replicate 1 | Replicate 2 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| S | I | F | P-val | S | I | F | P-val | ||
| P01870 | RABBIT Ig gamma chain C region | 4 | 4247 | 3639 | 0 | 3 | 4333 | 5006 | 0 |
| P07271 |
DNA repair and recombination protein PIF1 |
0.1 | 1595 | 54663 | 0 | 0.1 | 755 | 2616 8 |
0 |
| P07259 | Protein URA2 | 17 | 133 | 27 | 5.1E-67 | 14 | 76 | 19 | 5.1E-36 |
| P35732 | RNA polymerase II degradation factor 1 |
44 | 53 | 4 | 8.4E-12 | 12 | 50 | 14 | 2.0E-22 |
| P07342 |
Acetolactate synthase catalytic subunit, mitochondrial |
0.1 | 58 | 1988 | 3.8E-39 | 0.1 | 25 | 866 | 9.6E-18 |
| P53297 | PAB1-binding protein 1 | 0.1 | 54 | 1851 | 1.5E-36 | 0.1 | 25 | 866 | 9.6E-18 |
| P08566 | Pentafunctional AROM polypeptide | 4 | 47 | 40 | 1.3E-26 | 4 | 30 | 26 | 2.4E-16 |
| P32445 |
Single-stranded DNA-binding protein RIM1 |
5 | 58 | 40 | 2.2E-32 | 5 | 29 | 20 | 5.8E-15 |
| P25367 | [PIN+] prion protein RNQ1 | 3 | 61 | 70 | 1.9E-36 | 3 | 26 | 30 | 9.1E-15 |
| Q04119 | Endopolyphosphatase | 0.1 | 10 | 343 | 8.5E-08 | 0.1 | 17 | 589 | 1.8E-12 |
| P02407 | 40S ribosomal protein S17-A | 19 | 30 | 5 | 6.8E-09 | 23 | 38 | 6 | 2.4E-11 |
| P0CX41 | 60S ribosomal protein L23-A | 28 | 56 | 7 | 7.3E-18 | 33 | 44 | 5 | 4.7E-11 |
| P20606 | Small COPII coat GTPase SAR1 | 15 | 20 | 5 | 1.1E-05 | 7 | 21 | 10 | 3.4E-09 |
| P12612 | T-complex protein 1 subunit alpha | 1 | 25 | 86 | 3.6E-16 | 0.1 | 11 | 381 | 1.7E-08 |
| P07244 | Bifunctional purine biosynthetic protein ADE5,7 |
10 | 33 | 11 | 5.1E-14 | 6 | 15 | 9 | 1.8E-06 |
| P28007 | H/ACA ribonucleoprotein complex subunit 1 |
1 | 9 | 31 | 5.1E-06 | 1 | 9 | 31 | 4.7E-06 |
| P47079 | T-complex protein 1 subunit theta | 4 | 35 | 30 | 3.0E-19 | 4 | 12 | 10 | 7.9E-06 |
| P53123 | Ribosomal RNA methyltransferase MRM2 |
0.1 | 7 | 240 | 8.5E-06 | 0.1 | 7 | 243 | 7.9E-06 |
| P53305 |
Mitochondrial 37S ribosomal protein S27 |
1 | 6 | 21 | 3.8E-04 | 0.1 | 6 | 208 | 3.8E-05 |
| P00958 | Methionine--tRNA ligase, cytoplasmic |
0.1 | 19 | 651 | 1.0E-13 | 1 | 7 | 24 | 8.6E-05 |
| P25626 | 54S ribosomal protein IMG1 | 3 | 9 | 10 | 1.2E-04 | 0.1 | 5 | 173 | 1.8E-04 |
Columns named S and I show spectral counts in the whole-cell lysate and the pull-down, respectively. Column named F shows fold enrichment as calculated using adjusted spectral counts [27]. P-values were determined by G-test of independence for each replicate, as in [27]. Mitochondrial proteins are highlighted in bold. Bait related proteins are shown in italics.
a, b, protein accession and description are as in Swiss-Prot database (www.uniprot.org). Only proteins that had P-values below 0.001, fold enrichment above 4, and spectral counts in the pull down above 5, in each of the replicates are shown. The fractional spectral count of 0.1 indicates absence of a protein.
The proteins in Table II are sorted by descending total counts within pull-down summed across the two replicates. P-values as well as fold enrichment relative to control are also used to judge the significance of interactions. The top two hits in table 2 when considering both P-value and fold change in the pull-down relative to the lysate (column F) are Pif1 and rabbit IgG. This is to be expected – in a pull-down experiments bait proteins (Pif1 and IgG) are typically the most visible. Rim1 is considered one of the top hits because it is highly enriched in the Pif1-pulldown and the P-value for this enrichment is quite small. However, in this experiment, it was not the most highly enriched protein. For example, mitochondrial acetolactate synthase catalytic subunit, if using fold enrichment as the criteria, or URA2 if using the P-value as the criteria showed higher degree of enrichment. In order to make the most out of this type of experiment, additional experiments are needed. In the case of Rim1, the protein was cloned, over-expressed and purified. The recombinant protein was examined for interactions with Pif1 in vitro, leading to identification of potential sites of interaction (see below).
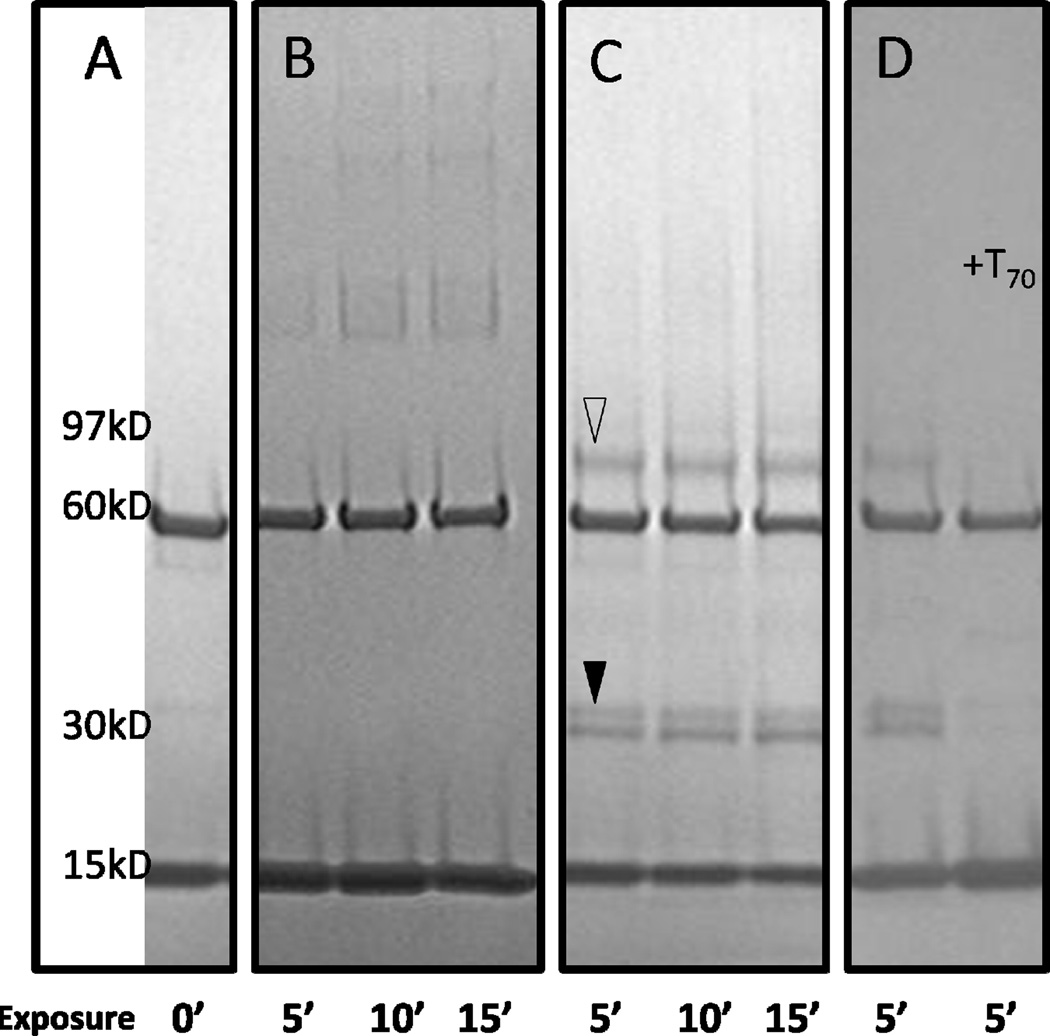
Next, we used a combination of chemical cross-linkers to capture the Rim1-Pif1 interaction on a gel between the recombinant proteins, to find that only one short cross-linker, SDA, produced sufficient quantities of the Rim1-Pif1 heterodimer for mass-spectrometry analysis (Figure 6):
Figure 6.
Rim1-Pif1 heterodimer detected with the SDA cross-linker (reproduced from [41], an open access publication). Rim1 and Pif1 proteins were cross-linked with SDA using a 2-step procedure. The reaction products were resolved on 5–15% gradient SDS-PAGE. Open triangle marks the position of Rim1-Pif1 heterodimer. Filled triangle marks the position of Rim1-Rim1 homodimer. Arrow points to the position of Pif1 bound by two molecules of Rim1. A, Rim1 and Pif1 without cross-linker. The Pif1 and Rim1 isoforms used in this work localize at 60 kD and 15 kD regions, respectively. B, lanes 1–3, Pif1 was reacted with SDA first, followed by addition of Rim1 and UV irradiations. C, lanes 1–3, Rim1 was reacted with SDA first, followed by the addition of Pif1 and UV irradiation. D, lane 1 – same reaction conditions as in C, lane 2 – ssDNA (T70) was added prior to cross-linking with SDA. UV exposure times at the second step of cross-linking are indicated, in minutes.
Finally, we used high resolution mass spectrometry to identify sites of the interactions –the C-terminus of Rim1 with a beta-hairpin of Pif1 rich in acidic residues, Rim1’s Lysine 29 residue in the OB-fold with an acidic patch within the second RecA-like domain of Pif1 (domain 2A). The interactions were validated by performing cross-linking with 15N-labeled Rim1 and performing site directed mutagenesis at the K29. We also demonstrated that the Rim1-Pif1 interaction is disrupted by a single-stranded DNA.
5. Conclusions and perspectives
The advent of powerful mass spectrometric approaches for identifying and quantifying proteins has greatly expanded the approaches one can apply to identify protein interaction networks. When coupled with protein crosslinking, the confidence in identifying such protein networks is further enhanced. In fact, these high-throughput experiments often produce so much data that identification of the most meaningful interactions becomes problematic. Current approaches to define functional significance of protein interactions lag somewhat behind methods to identify these interactions. Determining key biological endpoints that allow one to rapidly screen for function can greatly increase the power of the mass spectrometric/proteomics approach.
Highlights.
Reviews current methods of protein-protein interaction (PPI) analysis.
Affinity pull-down followed by mass spectrometry is a method of PPI discovery.
Minimum spanning trees help to visualize functional features in PPI networks.
Chemical cross-linking provides information on PPI topology.
ATP-depended RNA helicases participate in a diverse set of PPI networks.
Abbreviations
- PPI
protein-protein interaction
- MST
minimum spanning tree
- MS
mass spectrometry
- SF
superfamily
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Singleton MR, Dillingham MS, Wigley DB. Structure and mechanism of helicases and nucleic acid translocases. Annu Rev Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- 2.Jennings TA, et al. NS3 helicase from the hepatitis C virus can function as a monomer or oligomer depending on enzyme and substrate concentrations. J Biol Chem. 2009;284(8):4806–4814. doi: 10.1074/jbc.M805540200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jennings TA, et al. RNA unwinding activity of the hepatitis C virus NS3 helicase is modulated by the NS5B polymerase. Biochemistry. 2008;47(4):1126–1135. doi: 10.1021/bi701048a. [DOI] [PubMed] [Google Scholar]
- 4.Douglas ME, Diffley JF. Recruitment of Mcm10 to Sites of Replication Initiation Requires Direct Binding to the MCM Complex. J Biol Chem. 2015 doi: 10.1074/jbc.M115.707802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Drissi R, et al. Quantitative Proteomics Reveals Dynamic Interactions of the Minichromosome Maintenance Complex (MCM) in the Cellular Response to Etoposide Induced DNA Damage. Mol Cell Proteomics. 2015;14(7):2002–2013. doi: 10.1074/mcp.M115.048991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li N, et al. Structure of the eukaryotic MCM complex at 3.8 Å. Nature. 2015;524(7564):186–191. doi: 10.1038/nature14685. [DOI] [PubMed] [Google Scholar]
- 7.Yu Z, Feng D, Liang C. Pairwise interactions of the six human MCM protein subunits. J Mol Biol. 2004;340(5):1197–1206. doi: 10.1016/j.jmb.2004.05.024. [DOI] [PubMed] [Google Scholar]
- 8.Moyer SE, Lewis PW, Botchan MR. Isolation of the Cdc45/Mcm2–7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc Natl Acad Sci U S A. 2006;103(27):10236–10241. doi: 10.1073/pnas.0602400103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun J, et al. The architecture of a eukaryotic replisome. Nat Struct Mol Biol. 2015;22(12):976–982. doi: 10.1038/nsmb.3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mahboobi SH, Javanpour AA, Mofrad MR. The interaction of RNA helicase DDX3 with HIV-1 Rev-CRM1-RanGTP complex during the HIV replication cycle. PLoS One. 2015;10(2):e0112969. doi: 10.1371/journal.pone.0112969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hauk G, Bowman GD. Formation of a Trimeric Xpo1-Ran[GTP]-Ded1 Exportin Complex Modulates ATPase and Helicase Activities of Ded1. PLoS One. 2015;10(6):e0131690. doi: 10.1371/journal.pone.0131690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Szklarczyk D, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43(Database issue):D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lopes CT, et al. Cytoscape Web: an interactive web-based network browser. Bioinformatics. 2010;26(18):2347–2348. doi: 10.1093/bioinformatics/btq430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cowley MJ, et al. PINA v2.0: mining interactome modules. Nucleic Acids Res. 2012;40(Database issue):D862–D865. doi: 10.1093/nar/gkr967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Razick S, et al. iRefScape. A Cytoscape plug-in for visualization and data mining of protein interaction data from iRefIndex. BMC Bioinformatics. 2011;12:388. doi: 10.1186/1471-2105-12-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Razick S, Magklaras G, Donaldson IM. iRefIndex: a consolidated protein interaction database with provenance. BMC Bioinformatics. 2008;9:405. doi: 10.1186/1471-2105-9-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang L, et al. cisPath: an R/Bioconductor package for cloud users for visualization and management of functional protein interaction networks. BMC Syst Biol. 2015;9(Suppl 1):S1. doi: 10.1186/1752-0509-9-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang Y, Liu ZR. The ATPase, RNA unwinding, and RNA binding activities of recombinant p68 RNA helicase. J Biol Chem. 2002;277(15):12810–12815. doi: 10.1074/jbc.M200182200. [DOI] [PubMed] [Google Scholar]
- 19.Fuller-Pace FV. The DEAD box proteins DDX5 (p68) and DDX17 (p72): multi-tasking transcriptional regulators. Biochim Biophys Acta. 2013;1829(8):756–763. doi: 10.1016/j.bbagrm.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 20.Xu Y, Olman V, Xu D. Minimum spanning trees for gene expression data clustering. Genome Inform. 2001;12:24–33. [PubMed] [Google Scholar]
- 21.Xu Y, Olman V, Xu D. Clustering gene expression data using a graph-theoretic approach: an application of minimum spanning trees. Bioinformatics. 2002;18(4):536–545. doi: 10.1093/bioinformatics/18.4.536. [DOI] [PubMed] [Google Scholar]
- 22.Prom-On S, et al. Enhancing biological relevance of a weighted gene co-expression network for functional module identification. J Bioinform Comput Biol. 2011;9(1):111–129. doi: 10.1142/s0219720011005252. [DOI] [PubMed] [Google Scholar]
- 23.Rahmatallah Y, Emmert-Streib F, Glazko G. Gene Sets Net Correlations Analysis (GSNCA): a multivariate differential coexpression test for gene sets. Bioinformatics. 2014;30(3):360–368. doi: 10.1093/bioinformatics/btt687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jaiswal M, Washburn M, Zybailov B. Mass Spectrometry-Based Methods of Proteome Analysis. In: Meyers RA, editor. Reviews in Cell Biology and Molecular Medicine. Wiley-VCH Verlag: GmbH & Co. KGaA; 2015. pp. 51–103. [Google Scholar]
- 25.Tackett AJ, et al. I-DIRT, a general method for distinguishing between specific and nonspecific protein interactions. J Proteome Res. 2005;4(5):1752–1756. doi: 10.1021/pr050225e. [DOI] [PubMed] [Google Scholar]
- 26.Mosley AL, et al. Highly reproducible label free quantitative proteomic analysis of RNA polymerase complexes. Mol Cell Proteomics. 2011;10(2):M110.000687. doi: 10.1074/mcp.M110.000687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zybailov B, et al. Large scale comparative proteomics of a chloroplast Clp protease mutant reveals folding stress, altered protein homeostasis, and feedback regulation of metabolism. Mol Cell Proteomics. 2009;8(8):1789–1810. doi: 10.1074/mcp.M900104-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zybailov BL, Florens L, Washburn MP. Quantitative shotgun proteomics using a protease with broad specificity and normalized spectral abundance factors. Mol Biosyst. 2007;3(5):354–360. doi: 10.1039/b701483j. [DOI] [PubMed] [Google Scholar]
- 29.Zybailov B, Florens L, Washburn M. Quantitative shotgun proteomics using a protease with broad specificity and normalized spectral abundance factors. Mol Biosyst. 2007;3(5):354–360. doi: 10.1039/b701483j. [DOI] [PubMed] [Google Scholar]
- 30.Sardiu ME, et al. Probabilistic assembly of human protein interaction networks from label-free quantitative proteomics. Proc Natl Acad Sci U S A. 2008;105(5):1454–1459. doi: 10.1073/pnas.0706983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stasi M, De Luca M, Bucci C. Two-hybrid-based systems: powerful tools for investigation of membrane traffic machineries. J Biotechnol. 2015;202:105–117. doi: 10.1016/j.jbiotec.2014.12.007. [DOI] [PubMed] [Google Scholar]
- 32.Lievens S, Lemmens I, Tavernier J. Mammalian two-hybrids come of age. Trends Biochem Sci. 2009;34(11):579–588. doi: 10.1016/j.tibs.2009.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liang Y. Applications of isothermal titration calorimetry in protein science. Acta Biochim Biophys Sin (Shanghai) 2008;40(7):565–576. doi: 10.1111/j.1745-7270.2008.00437.x. [DOI] [PubMed] [Google Scholar]
- 34.Lalonde S, et al. Molecular and cellular approaches for the detection of protein-protein interactions: latest techniques and current limitations. Plant J. 2008;53(4):610–635. doi: 10.1111/j.1365-313X.2007.03332.x. [DOI] [PubMed] [Google Scholar]
- 35.Harding SE, Rowe AJ. Insight into protein-protein interactions from analytical ultracentrifugation. Biochem Soc Trans. 2010;38(4):901–907. doi: 10.1042/BST0380901. [DOI] [PubMed] [Google Scholar]
- 36.Zybailov BL, et al. Large Scale Chemical Cross-linking Mass Spectrometry Perspectives. J Proteomics Bioinform. 2013;6(Suppl 2):001. doi: 10.4172/jpb.S2-001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trakselis MA, Alley SC, Ishmael FT. Identification and mapping of protein-protein interactions by a combination of cross-linking, cleavage, and proteomics. Bioconjug Chem. 2005;16(4):741–750. doi: 10.1021/bc050043a. [DOI] [PubMed] [Google Scholar]
- 38.Leszyk J, et al. Characterization of zero-length cross-links between rabbit skeletal muscle troponin C and troponin I: evidence for direct interaction between the inhibitory region of troponin I and the NH2-terminal, regulatory domain of troponin C. Biochemistry. 1990;29(1):299–304. doi: 10.1021/bi00453a041. [DOI] [PubMed] [Google Scholar]
- 39.Chowdhury SM, et al. Identification of cross-linked peptides after click-based enrichment using sequential collision-induced dissociation and electron transfer dissociation tandem mass spectrometry. Anal Chem. 2009;81(13):5524–5532. doi: 10.1021/ac900853k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luo J, et al. An integrated chemical cross-linking and mass spectrometry approach to study protein complex architecture and function. Mol Cell Proteomics. 2012;11(2):M111.008318. doi: 10.1074/mcp.M111.008318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bol GM, Xie M, Raman V. DDX3, a potential target for cancer treatment. Mol Cancer. 2015;14(1):188. doi: 10.1186/s12943-015-0461-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heerma van Voss MR, et al. Identification of the DEAD box RNA helicase DDX3 as a therapeutic target in colorectal cancer. Oncotarget. 2015;6(29):28312–28326. doi: 10.18632/oncotarget.4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Garbelli A, et al. A motif unique to the human DEAD-box protein DDX3 is important for nucleic acid binding, ATP hydrolysis, RNA/DNA unwinding and HIV-1 replication. PLoS One. 2011;6(5):e19810. doi: 10.1371/journal.pone.0019810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sharma D, Jankowsky E. The Ded1/DDX3 subfamily of DEAD-box RNA helicases. Crit Rev Biochem Mol Biol. 2014;49(4):343–360. doi: 10.3109/10409238.2014.931339. [DOI] [PubMed] [Google Scholar]
- 45.Choi YJ, Lee SG. The DEAD-box RNA helicase DDX3 interacts with DDX5, co-localizes with it in the cytoplasm during the G2/M phase of the cycle, and affects its shuttling during mRNP export. J Cell Biochem. 2012;113(3):985–996. doi: 10.1002/jcb.23428. [DOI] [PubMed] [Google Scholar]
- 46.Bish R, et al. Comprehensive Protein Interactome Analysis of a Key RNA Helicase: Detection of Novel Stress Granule Proteins. Biomolecules. 2015;5(3):1441–1466. doi: 10.3390/biom5031441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shih JW, et al. Critical roles of RNA helicase DDX3 and its interactions with eIF4E/PABP1 in stress granule assembly and stress response. Biochem J. 2012;441(1):119–129. doi: 10.1042/BJ20110739. [DOI] [PubMed] [Google Scholar]
- 48.Jain S, et al. ATPase-Modulated Stress Granules Contain a Diverse Proteome and Substructure. Cell. 2016;164(3):487–498. doi: 10.1016/j.cell.2015.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Futami K, Shimamoto A, Furuichi Y. Mitochondrial and nuclear localization of human Pif1 helicase. Biol Pharm Bull. 2007;30(9):1685–1692. doi: 10.1248/bpb.30.1685. [DOI] [PubMed] [Google Scholar]
- 50.Paeschke K, et al. Pif1 family helicases suppress genome instability at G-quadruplex motifs. Nature. 2013;497(7450):458–462. doi: 10.1038/nature12149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ramanagoudr-Bhojappa R, et al. Physical and functional interaction between yeast Pif1 helicase and Rim1 single-stranded DNA binding protein. Nucleic Acids Res. 2013;41(2):1029–1046. doi: 10.1093/nar/gks1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zybailov B, et al. Analysis of Protein-protein Interaction Interface between Yeast Mitochondrial Proteins Rim1 and Pif1 Using Chemical Cross-linking Mass Spectrometry. J Proteomics Bioinform. 2015;8(11):243–252. doi: 10.4172/jpb.1000376. [DOI] [PMC free article] [PubMed] [Google Scholar]