Abstract
Engineered in vitro models using human cells, particularly patient-derived induced pluripotent stem cells (iPSCs), offer a potential solution to issues associated with the use of animals for studying disease pathology and drug efficacy. Given the prevalence of muscle diseases in human populations, an engineered tissue model of human skeletal muscle could provide a biologically accurate platform to study basic muscle physiology, disease progression, and drug efficacy and/or toxicity. Such platforms could be used as phenotypic drug screens to identify compounds capable of alleviating or reversing congenital myopathies, such as Duchene muscular dystrophy (DMD). Here, we review current skeletal muscle modeling technologies with a specific focus on efforts to generate biomimetic systems for investigating the pathophysiology of dystrophic muscle.
Introduction
The skeletal musculature in healthy individuals has substantial regenerative capacity. Activation of quiescent muscle stem cells (satellite cells) in response to physical trauma facilitates the production of new myonuclei capable of fusing with damaged muscle fibers to restore structure and contractile function to extant myofibers. If damage to a given fiber is extensive, as in certain disease states, satellite cell-derived myoblasts are capable of proliferating and fusing with each other to generate new fibers to replace those lost to injury. However, genetic defects in sarcolemmal, contractile, or extracellular matrix (ECM) proteins can result in dystrophic phenotypes that are wholly or partially incapable of regenerating damaged muscle tissues. Moreover, concomitant altered expression of other muscle genes can lead to progressive breakdown and scaring of healthy tissue, leading to gradual muscle wasting and eventually death. Often in such cases, the repair mechanisms cannot overcome the high level of fiber necrosis and death that occurs because the structural integrity of the muscle fiber has been lost. To better care for patients with the approximately 30 known muscular dystrophies, improved model systems are required with which to study the mechanisms responsible for disease onset, as well as the pathological progression of the disease and potential therapeutic targets.
Animal models will always have their place in the elucidation and confirmation of disease mechanisms. In terms of muscular dystrophy, there are currently more than 50 animal models of DMD alone, including non-mammalian (Caenorhabditis elegans, zebrafish, among others), mouse, dog, rat, and pig-based systems [1]. The study of these animals, as well as animal models of other dystrophic conditions, over the past 30 years has provided a staggering amount of data on the pathology of dystrophic muscles, and served as successful preclinical platforms for the development of novel therapeutics. However, it has become increasingly clear that animal models cannot fully recapitulate the clinical, physiological, and biochemical manifestations of human disease [2–4]. Furthermore, animal-based screening systems are inherently low throughput, making it expensive, time consuming, and often impossible to delineate tissue-specific responses from compensatory effects. Despite these shortcomings, animal models will be required during preclinical assessment as a means to evaluate off-target responses, behavioral changes, and phenotypic improvement to treatment with novel therapeutics. However, in vitro models using patient-derived cells that are high throughput, fully defined, and biomimetic constitute exciting new technologies to augment current in vivo studies. Such models provide investigators with a more comprehensive understanding of human skeletal muscle physiology and development in dystrophic disease states, and enable collection of more predictive data in terms of the effect of new chemical entities on human tissues. In this way, a combination of in vivo testing and in vitro screening using engineered human muscle models will likely lead to more stringent control of compounds progressing to clinical trial, and thereby ensure better translation of benchtop results to the bed-side.
Here, we discuss a range of skeletal muscle modeling technologies, with a specific focus on efforts to generate biomimetic systems to enhance the future study of dystrophic pathology in vitro.
Skeletal muscle physiology and dystrophic pathophysiology
Skeletal muscle comprises highly oriented, unbranched, contractile fibers arranged to bring about controlled contraction of the tissue [5]. Each muscle fiber is a single multinucleated cell [6] that ranges in length from 1 mm to several cm and can even run the length of the muscle [7] (Fig. 1a). Sarcomere shortening is the basis for muscle contraction and is initiated by a rise in cytosolic Ca2+, upon the arrival of an action potential at the neuromuscular junction and the subsequent depolarization of the postsynaptic fiber [6]. Influx of Ca2+ into the cell triggers the reconfiguration of the actin–tropomyosin structure by the troponin complex, allowing the myosin heads to cyclically bind to previously unavailable sites on the actin molecule and ratcheting the two Z discs together [6, 8]. Rapid sequestration of cytosolic Ca2+ then returns the actin–tropomyosin structure to its original conformation, blocking myosin head binding, and resulting in relaxation [8]. Structural integrity of the muscle cell during contraction is facilitated by costameres; striated muscle-specific focal adhesions that bind sarcomeres to the muscle fiber membrane [9]. These structures laterally transmit contractile forces from active sarcomeres across the sarcolemma to the surrounding ECM, and to nearby muscle cells. Lateral transmission of contractile force ensures the maintenance of uniform sarcomere length between adjacent muscle cells from different motor units (i.e. activated by separate motor neurons) within a skeletal muscle. In doing so, costameres mechanically fortify the tissue to minimize stress imposed on the relatively fragile lipid bilayer, and thereby protect the tissue from undergoing repetitive, shear-induced damage [9].
FIGURE 1.
Skeletal muscle physiology in normal and dystrophic muscles. (a) Schematic representation of skeletal muscle illustrating the hierarchical structure of the tissue. Individual fibers are surrounded by a layer of extracellular matrix (ECM; the endomysium), and are grouped together into fascicles. Multiple fascicles are banded together by a second ECM layer (the perimysium), and the entire tissue is surrounded by a third ECM layer (the epimysium). Breakdown of interactions between muscle cells and the complex network of ECM fibers surrounding them leads to increased levels of cell membrane stress, Ca2+ leakage, cell death, and fibrosis, which in turn manifests as progressive muscle weakness in patients with dystrophy. (b) Cross-sections of normal (i) and Duchenne muscular dystrophy (DMD) (ii) human gastrocnemius skeletal muscle stained with hematoxylin and eosin. The normal muscle biopsy shows relatively uniform fiber diameters, peripherally located nuclei, with no fiber degeneration, inflammation, or endomysial fibrosis. By contrast, the DMD tissue contains smaller and more rounded fibers, reflecting the patient’s younger age, as well as small basophilic regenerating fibers, necrotic fibers, and scattered inflammatory cells. Adapted from [117] (a).
Skeletal muscle dystrophies represent a heterogeneous group of diseases that arise predominantly through mutations in genes that encode structural components of muscle fibers, costameric proteins in the sarcolemma, proteins of the ECM, and some modifying enzymes [10]. DMD is the most prevalent and severe form of muscular dystrophy. This condition, along with a milder form called Becker muscular dystrophy (BMD), arises from mutations in the dystrophin gene, which results in either a complete loss of the protein from muscle fibers, or expression of a truncated, partially functional isoform. Dystrophin links the sarcomere to costameric complexes in the muscle membrane as part of the dystrophin glycoprotein complex (DGC) [10]. Mutations in several genes encoding proteins of the DGC, such as the dystroglycans, dystrobrevin, and the sarcoglycans, have all been linked to various forms of muscular dystrophy. Dystrophin and the DGC proteins collectively act as shock absorbers that reduce the mechanical strains put on the membrane during muscle contraction. In the absence of functional dystrophin and/or components of the DGC, the high tension generated by repetitive cycles of lengthening and contracting produces microtears in the sarcolemma, rendering it permeable to extracellular Ca2+ and leading to Ca2+ overload and mitochondria-dependent necrotic cell death [11–13]. Experiments in transgenic mouse models that have directly manipulated store-operated Ca2+ entry by altering the expression of transient receptor potential (TRPC), stromal interaction molecule (STIM), or calcium release-activated calcium channels (ORAI) have demonstrated that Ca2+ leakage is sufficient to cause a dystrophic disease state [11, 13]. Along with membrane instability, unregulated Ca2+ entry can also occur with a loss of function in genes that encode membrane repair mechanisms, such as dysferlin, which has also been identified as a genetic determinant of limb girdle muscular dystrophy [14].
Despite variation in disease etiologies, one commonality is that dystrophic muscles are constantly undergoing cycles of muscle fiber death and regeneration [15]. Thus, the pathologic hallmarks of the disease include elevated serum creatine kinase, fibrosis, and inflammation from muscle fiber degeneration, as well as fibers with centrally located nuclei and extremely small fiber diameters, indicative of fiber regeneration (Fig. 1b). Moreover, skeletal muscle tissues in patients with dystrophy fail to repair injured fibers adequately, leading to exacerbation of the fibrotic response [16, 17] and the replacement of muscle with adipose tissue [18]. In such patients, this pathology leads to the clinical presentation of muscle weakness and fatigue, pseudohypertrophic muscles, and poor ambulation.
Several informative animal models for muscular dystrophy have been studied for years. Such models have been successful in providing proof of concept and preclinical validation for several therapeutic strategies, including targeted gene therapy to promote exon skipping and the formation of truncated dystrophin proteins [1]. As a result of inefficiencies with current chemistries, such therapies have yet to transition effectively to humans. Regardless, such data serve to highlight the potential utility of animal models in developing treatments for muscular dystrophy. Despite the numerous successes of animal studies, inherent biochemical and physiological disparities between species, including differences in absorption, distribution, metabolism, and excretion (ADME) of xenobiotics, can cause substantial differences in the efficacy and toxicity of novel compounds between animals and humans [19]. This, coupled with the inherently low-throughput nature of animal studies, limits the utility of such systems for the effective and reliable preclinical evaluation of novel therapeutics. Therefore, humanized in vitro models offer an attractive opportunity to augment current preclinical disease modeling and drug screening studies and improve their ability to predict compound action in humans [20]. However, given the need for aggressive mechanical manipulations and a high degree of cellular maturation to elicit clinically relevant phenotypes in dystrophic muscle, traditional culture systems can be ineffective for studying the cellular and molecular underpinnings of dystrophic pathology. Therefore, there is a need to develop more accurate representations of skeletal muscle in vitro, to better model the physiology of the adult tissue and its breakdown during disease initiation and progression.
Tissue engineering approaches for generating skeletal muscle models
The ability to predict adult human tissue responses accurately during preclinical assessment is predicated based on the capacity of the culture system to generate myogenic cells that are sufficiently mature to mimic the physiology of their counterparts in the intact organism. To complicate matters, several muscular dystrophies, such as DMD, are developmentally late-onset conditions, suggesting that engineered skeletal muscle comprising immature cells will be incapable of accurately recapitulating the disease phenotype seen in the adult patient. Therefore, to gain widespread acceptance, engineered skeletal muscle will need to be created using methodologies that recapitulate embryonic development as much as possible and promote maturation of cultured skeletal muscle cells so that they closely resemble native architecture and function [20].
The anisotropic functional architecture of skeletal muscle dictates several key factors that any in vitro model must emulate, namely the ability to promote differentiation of muscle precursor cells (myoblasts) into densely packed myotubes (myofibers), oriented into fascicles capable of performing uniaxial contraction [5, 6] (Fig. 1a). While such platforms are relatively widespread [21–23], each has deficiencies that cause the functional and physiological maturation of cultured muscle constructs to fall short of the characteristics of intact muscle.
Methods for maturing cells in vitro typically center on the ability to more precisely mimic the in vivo niche for a given cell type [24]. Consequentially, engineered skeletal muscle tissues require the creation of an in vitro microenvironment that provides anisotropic guidance cues [25], biomimetic substrate elasticities [26], 3D matrices (including anchoring moieties) [27, 28], incoming mechanical and electrical stimuli [29], and interaction with support cells and tissue types (including cartilage [30], endothelial [21], and peripheral nervous system elements [31]). Successful integration of such a comprehensive battery of maturation signals will lead to the generation of more biologically accurate engineered muscle tissues for both in vitro and in vivo applications. However, exactly which cues are necessary and sufficient to promote the generation of constructs exhibiting adult phenotypes indistinguishable from native tissues are yet to be elucidated. The major methods so far used to promote physiologically relevant skeletal muscle development and maturation in vitro, and the implications of the work to date in the context of effective dystrophic disease modeling, are discussed below.
Aging
Although most sources of cultured human myoblasts reach a degree of functionality within 2–3 weeks, such cells typically take months or even years to attain an adult phenotype in vivo, indicating a time-dependent mechanism underlying the regulation of tissue development [32]. Given the importance of time in promoting maturation in living systems, it is not surprising that the most well-established and reliable method for improving the maturation state of cells in vitro is to increase the length of time they are maintained in culture before analysis.
Studies of the contractile properties of human and rodent skeletal myotubes in vitro highlight that myotube contraction generates significantly more force after 3 and 4 weeks in culture than is observed following 2 weeks [21, 33]. Improvements in sarcomeric development [34], myosin heavy chain (MyHC) expression patterns [35], and myotube hypertrophy [21] have also been reported, indicating that improved functional outputs correlate with physiological and structural markers of myogenic maturation over this time course. Gene expression analysis of long-term skeletal muscle cultures has likewise demonstrated an upregulation of developmentally more mature MyHC isoforms [35], suggesting that these cells are able to modulate their transcriptome with age.
Although long-term culture is a well-established means to promote cellular maturation, reliance on such extended developmental timelines limits enthusiasm for this method of generating tissues for widespread preclinical and clinical applications. Given the obvious issues with throughput and long lead times before a specific cell population would be suitably matured for use, researchers are now pursuing alternative methods to promote maturation in cultured muscle cells.
Anisotropy
Skeletal muscle tissues are exquisitely aligned in vivo to facilitate their biological function [36]. Specifically, skeletal muscle fibers are oriented uniaxially between tendons to enable directional contraction and skeletal manipulation [37]. Both skeletal and cardiac muscle cultures maintained on nanoscale topographies show enhanced cellular alignment over cells on unpatterned plastic [38, 39]. Furthermore, cells aligned in this manner undergo maturation in terms of MyHC isoform expression, sarcomere development, and muscle regulatory factor expression profiles [38, 40]. Chemical surface patterning using hydrophilic and hydrophobic self-assembled monolayers has also been used to promote the uniaxial alignment of cultured myogenic cells, and such surfaces have been shown to promote myotube maturation, characterized by greater levels of sarcomeric structural development [41]. Similarly, microcontact printing has proven effective for patterning skeletal muscle culture surfaces that promote alignment and rates of fusion for differentiating myotubes [42].
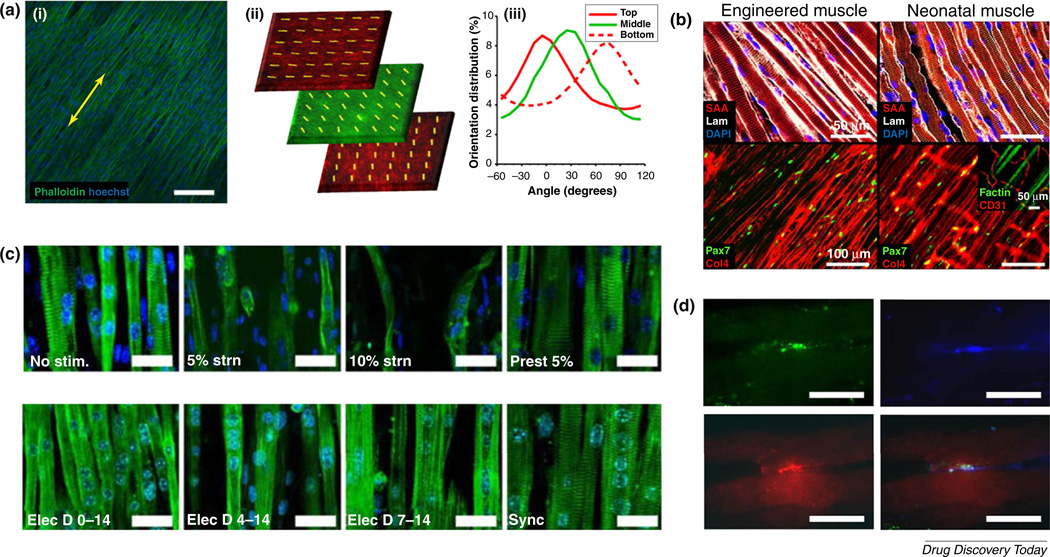
Experiments utilizing thermoresponsive nanofabricated substrates (TNFS) have enabled the production of aligned skeletal muscle cell sheets that can be detached from the culture dish by altering ambient temperatures [43] (Fig. 2a). Such patterned cell sheets retain their structural anisotropy once detached from their nanopatterned substrates. Moreover, stacking of multiple detached sheets at varying angles demonstrates that individual cell sheets maintain their internal alignment for at least 7 days, regardless of the orientation of neighboring sheets [43]. Such technology represents an exciting possibility for the generation of 3D, cell-dense tissue patches with controllable interlayer orientations. Application of this technology in conjunction with human skeletal muscle cell types represents a promising option for the development of more physiologically structured tissues with complex anatomical features. Such tissues could be of considerable utility for modeling diseases in which lateral connectivity between anisotropic muscle fibers and their surrounding matrix is impaired, as is the case in muscular dystrophy.
FIGURE 2.
Current skeletal muscle engineering strategies. (a) Thermoresponsive nanofabricated substrates enable the transfer of aligned muscle cells to new substrates with no loss of anisotropy (i). Yellow arrows indicate the direction of transferred cell sheet alignment. These sheets can be stacked to create 3D tissues with layer-specific orientations (ii). The presented image comprises confocal images taken from different layers of a z-stack, showing three membrane-stained nanopatterned cell sheets stacked together, each with a 45°-shift in orientation from the layer above. The top and bottom layers were stained with a red membrane dye, while the middle layer was stained with green membrane dye. Note the maintenance of orthogonal alignment after stacking (ii). Quantification of individual layer alignment was confirmed by actin fiber alignment analysis (iii). Such technology can be used to generate scaffold-free, cell-dense, oriented muscle tissues. (b) In vitro-engineered 3D skeletal muscles show high structural homology to native rodent tissues. (c) Electromechanical conditioning promotes myofiber hypertrophy and sarcomere development in cultured skeletal muscle fibers. Cells were stained for actinin (green) and nuclei (blue). (d) Co-culture of skeletal muscle fibers with motor neurons promotes the development of neuromuscular junction-like structures, thereby improving the biomimicry of the construct. Cultured cells were stained for synaptic vesicle protein 2 (green), β-III-tubulin (blue), and acetylcholine receptors (red) as markers of presynaptic terminals, axons, and postsynaptic terminals, respectively. Adapted from [43] (a), [21] (b), [29] (c), and [79] (d). Scale bars: 100 mm (a,b) and 20 mm in (c,d).
Substrate elasticity
Contractile cell types, such as skeletal muscle myotubes, are highly influenced by the mechanical properties of the surrounding tissue. When supplanted in vitro, the mechanical properties of the culture substrate likewise have a pivotal role in modulating the development of the seeded cells. A major issue for the long-term maintenance of skeletal muscle cultures is the rigidity of the culture substrate, given that even relatively immature muscle cells slowly increase both their force of spontaneous contractions and their passive tension, thereby pulling themselves off the surface. Studies have shown that substrate elasticities that mimic the native skeletal muscle milieu promote enhanced satellite cell proliferation and differentiation to myoblast precursors [26, 44]. Furthermore, modulation of substrate elasticity and biopolymer coatings has been found to exert significant effects on sarcomeric development in terminally differentiated myotubes [45–47]. Logically, a substrate stiffness that emulates that of the native tissue will produce optimal skeletal muscle maturation in terms of striated sarcomere development, thus emphasizing the importance of biomimicry.
Three-dimensionality
2D in vitro environments provide a fundamentally alien setting for cultured cells. The rigidity of the substrate, as mentioned above, negatively affects the development of cells, at least in part because the binding of cell surface receptors to substrate ligands only occurs on one side of the cell, leading to aberrant signaling from the cell surface to the nucleus. By contrast, 3D constructs enable binding of the cell to ECM proteins over the entire cell surface, creating a more biomimetic microenvironment for the seeded cell. Moreover, flexibility of the 3D matrix promotes cellular maturation [27], and application of uniaxial anchoring points can lead to the development of cellular anisotropy because cells reorganize along the lines of principal strain within the construct [22, 23] (Fig. 2b). Recent advances in bioprinting technology provide further options for the synthesis of complex 3D muscle tissues with a greater degree of reproducibility and uniformity for application in high-throughput drug screens and disease-modeling assays [48].
Two additional advantages of 3D tissue modeling, apart from improved maturation, are the ease of integrating complex electro-mechanical conditioning protocols (discussed below), and the ability to measure contractile force output directly [49, 50]. The latter readout enables real-time analysis of functional development in these engineered tissues, theoretically facilitating high-throughput screening of muscle responses to drug treatment, or the comparison of functional characteristics between wild-type and dystrophic engineered muscles [51].
Studies have shown that doping of engineered 3D muscle matrices with factors capable of promoting muscle maturation is a viable option for further advancing cellular development in vitro. For example, investigations into cells grown on surfaces treated with Arg-Gly-Asp (RGD) peptides, which promote integrin binding to the substrate, have shown improved expression of differentiated skeletal muscle markers by up to 50% [52–54]. An indepth review of different scaffold matrix options for muscle engineering and a discussion of the various factors with potential to improve the biomimicry of the engineered tissue niche are beyond the scope of this review, but are available elsewhere [55].
Electromechanical conditioning
As is the case for exercise in vivo, physical stimulation regimens have the potential to promote the maturation of functional performance in engineered muscle tissues. The application of controlled electrical stimulation to induce active muscle contraction, and mechanical stimulation (stretch) to alter passive muscle loading, are used to mimic work, and have been conclusively shown to promote the maturation of muscle cells in vitro [56, 57].
Mechanical loading regimens have been found to cause significant alterations in skeletal muscle structure and function through the activation of numerous intracellular signaling mechanisms, including the mitogen-activated protein kinase/extracellular signal-related kinase (MAPK)/(ERK), reactive oxygen species (ROS), and Rho pathways [56, 58, 59]. Similarly, both uniaxial static and cyclic stretch are known to have beneficial effects on engineered skeletal muscle construct survival, alignment, hypertrophy, and differentiation. Specifically, Candiani et al. [60] demonstrated that application of a biomimetic stretch regimen (2-day steady ramp stretch to 3.3%, then cyclic 5-pulse, 0.5 Hz, 3.4% burst stretches) induced an eightfold increase in MyHC protein expression compared with static control constructs. Similarly, Moon et al. showed that application of 10% cyclic stretch to human myogenic precursors for 1–3 weeks promoted cellular alignment, differentiation, and contractile properties [61]. Mechanical loading has also been shown to elicit skeletal muscle fiber switching in cultured myotubes toward a slow/oxidative phenotype [62], demonstrating how careful control of the physical niche can reproduce environmental conditions that impact the native slow:fast twitch fiber ratio.
Analogous to mechanical stimuli, exposure of muscle cells to long-term electrical stimulation is another component of exercise-in-a-dish, and similarly encourages enhanced structural and functional development in the cultured cells. Electrical stimulation fosters myofiber maturation by altering mitochondrial density, MyHC expression, metabolic activity, and contractile force generation [22, 63]. Furthermore, modulation of stimulation regimens to mimic firing patterns in fast and slow motor neurons promotes fiber-type switching in cultured skeletal myotubes [64, 65]. As an alternative to electrical stimulation, recent work with optogenetics demonstrated successful expression of light-activated ion channels in cultured muscle cells, facilitating light activation of engineered muscle contraction [66]. Such technologies enable continuous long-term stimulation of cultured cells without the confounding build-up of toxic byproducts within the medium. Light activation of transfected muscle cells facilitates not only the selective activation of specific cells in culture for targeted stimulation studies, but also numerous possibilities based on the generation of light-activated bioactuators for use in pumps and robotics applications [67].
The obvious step of combining electrical stimulation patterns with mechanical stretch regimens, while simultaneously enabling real-time monitoring of contractile performance, has also been attempted. For example, Liao and colleagues studied cultured rodent myotubes subjected to 20-V, 1-Hz electrical stimuli combined with 5 and 10% cyclic stretch [29] (Fig. 2c). Measurement of myofiber striation, protein expression for muscle maturation markers, and contractile force demonstrated the positive effect that this combined stimulation package had on cultured cells above each stimulus alone and against unstimulated controls, thus demonstrating the additive benefit of coupling electromechanical maturation strategies.
To mimic a patient’s life with a debilitating neuromuscular disease, mechanical loading regimens also offer the potential to exacerbate in vitro phenotypes when investigating dystrophic cells. A recent study utilizing a microfluidic-assisted cyclic mechanical stimulator to monitor membrane permeability in an in vitro model of DMD reported that, while static wild-type and DMD cells exhibited little difference in membrane leakage, mechanically stretched cells showed significantly different phenotypes, with greater permeability recorded from DMD cells [68]. Such data highlight the importance of biomimetic physical cues, not only for promoting physiologically relevant tissue development, but also for helping to stratify disease phenotypes between healthy and dystrophic cells within in vitro environments.
Matrix doping/medium supplementation
The addition of growth factors and other media supplements to induce greater levels of cellular maturation is a common technique across all cell types and can be particularly effective for muscle cell cultures. For example, insulin-like growth factor 1 (IGF-1) significantly improves myoblast differentiation and subsequent myotube maturation in culture, producing larger, more structurally developed fibers with more nuclei per fiber [69, 70]. Addition of neural agrin to culture medium has also been shown to upregulate dystrophin expression, enhance acetylcholine cluster formation, and improve twitch force production in engineered skeletal muscle tissues [71]. Treatment of skeletal muscle cultures with testosterone increased myogenic commitment and promoted IGF-1 upregulation [72], while chronic exposure of myotubes to creatine led to significant improvements in the functional parameters of the cultured cells [73]. Similarly, transforming growth factor (TGF)-β treatment has been shown to improve peak twitch and tetanic contractile activity, as well as sarcomere development and organization, in scaffold-free engineered skeletal muscle tissues [74]. Studies have also investigated the addition of a variety of neuronal growth factors to muscle cultures, and demonstrated that such supplementation can significantly enhance myotube differentiation and sarcomeric development in cultured skeletal muscle cells [75].
Co-culture
Finally, integration of skeletal muscle cultures with appropriate supporting cell types promotes cellular maturation, while simultaneously improving the biomimicry of the culture platform through the inclusion of other cell types present in the native tissue. The inclusion of motor neurons is critical to engineered skeletal muscle development because complete maturation of the tissue in vivo is dependent on innervation by infiltrating neuronal growth cones. Numerous studies have demonstrated that co-culture of muscle with motor neurons upregulates MyHC expression and improves contractile function and calcium handling [76–78]. In addition, the engineering of functional neuromuscular junctions in vitro [31, 79, 80] (Fig. 2d) could facilitate the modeling of peripheral neuropathies, such as amyotrophic lateral sclerosis and Charcot-Marie-Tooth disease [81, 82]. These conditions are currently incurable, so high-fidelity models of their pathologies may prove beneficial in advancing therapeutic development.
Integration of endothelial cells with skeletal muscle constructs improves vascular development within the engineered tissue, and improves engraftment upon in vivo implantation [83, 84]. Co-culture of muscle constructs with tendon tissue does not significantly improve the functional development of the cultured myocytes, but does produce 3D tissue that more closely resembles native counterparts and results in myotendinous junctions able to withstand physiological strain rates [37]. While co-culture with certain subsets of muscle-adjacent tissues might not inherently improve the development and maturation of cultured skeletal muscle, there is little doubt that their resemblance to native tissue gives them greater utility for downstream applications, such as drug screening. For example, to adequately model dystrophic skeletal muscle, it would be beneficial to include the connections of the myofibers with the ECM, given that one of the functions of dystrophin is to anchor the internal actin cytoskeleton to the surrounding connective tissue. Compounds that can augment ECM connections (possibly by altering gene expression or by some other mechanism) and circumvent the need for dystrophin might be attractive drug candidates.
Although the development of complex multi-cell-type co-cultures improves function, maturation, and better recreates native tissues, it is noteworthy that, as the number of cell types increases in a given culture platform, maintaining optimal culture conditions for each cell type becomes more difficult. To date, no more than four human cell types have been successfully maintained in co-culture and skeletal muscle was not part of this cohort [85, 86]. Given the highly specialized nutritional requirements of many human cell types, elucidation of the optimal culture conditions at peak maturation would be difficult [87]. Furthermore, multiple cell types might in fact confound the interpretation of drug studies, because discerning exactly which cell type is responding to the treatment might be very difficult. Increased complexity arising from multi-cell-type co-cultures should only be considered if the specific interaction between the cell types is the focus of the inquiry (e.g. neuromuscular junction formation between motor neurons and myotubes for the study of peripheral neuropathies) or if the inclusion of supporting cell types is found to promote a maturation effect unachievable by alternative methods. Otherwise, the increased difficulty in developing optimal culture parameters will likely prove more of a hindrance than a help in the search to create more adult-like muscle cells and engineered tissues.
Induced pluripotent stem cells in skeletal muscle modeling
Patients with muscular dystrophy are clinically heterogeneous and, thus, lose the ability to walk over a wide age range [88]. This variation is caused by the fact that mutations in the dystrophin gene range from null alleles (severe forms) to partially functional alleles causing BMD [89] and contribute to substantial variability in patient responsiveness to a given therapy [90]. In addition, the effect of genetic modifiers, such as TGF-β-binding protein 4, can further increase variability in phenotype, even in patients carrying the same underlying genetic mutation [89, 91]. The range of symptomatic progression between patients with varied genetic backgrounds highlights how such differences can have a significant impact on overall pathology. There is currently no model of dystrophic skeletal muscle that is capable of accounting for such patient-to-patient variability before moving to clinical trials.
Given that over 1000 different mutations in the dystrophin gene have been discovered so far in patients with DMD alone, patient-specific dystrophic myotubes derived from human iPSCs would be clinically transformative. To that end, researchers have successfully created an iPSC-based disease-in-a-dish model of DMD cardiomyopathy starting from the urine of patients [92–94]. Two reasons motivated these efforts to model the cardiomyopathy before the skeletal muscle pathology. First, the cardiomyopathy is what ultimately shortens the lives of patients with DMD. Second, experimental protocols to differentiate stem cells to cardio-myocytes are far ahead of similar small-molecule approaches to generate skeletal muscle. However, a landmark study using the mdx mouse model of DMD reported that myotubes could be differentiated from embryonic stem cells (ESCs) and that they exhibit an abnormal branched phenotype [95]. This result corroborated an earlier study with DMD patient biopsy-derived primary myoblasts showing defective myogenic differentiation [96]. Recently, work on the mdx mouse model provided evidence that DMD pathogenesis can be identified at the satellite cell stage. Dumont et al. reported that dystrophin is expressed in activated satellite cells, where it has a role in regulating symmetric cell division. Thus, dystrophin deficiency leads to a reduction in the generation of myogenic progenitors and, therefore, impaired muscle regeneration [97]. These studies suggest that skeletal muscle stem and/or progenitor cells, as well as myotubes, derived from dystrophic human iPSC lines should exhibit disease-related phenotypes. However, exactly how this will manifest within in vitro models of skeletal muscle has yet to be investigated in detail.
The differentiation of myoblasts and more mature myotubes from human iPSCs would be of substantial benefit to disease modeling, toxicity testing, and drug discovery efforts, because it would constitute a nearly inexhaustible source of muscle cells from individual patients. Accomplishing this would fulfill three essential disease-modeling needs: (i) enable the transfer of patient-specific genetic defects to the in vitro model, including potential genetic modifiers that have confounded or escaped study in currently available approaches; (ii) allow the physiology and biochemistry of the disease phenotype in the muscle cells on the dish to be compared with the clinical symptoms of the patient who donated the starting sample; urine in the case above; and (iii) facilitate the identification of a clinically relevant disease-specific readout, amenable to high-throughput screening, that can be attenuated or even reversed by potential therapeutic compounds. As mentioned above, methods for producing high-purity iPSC-derived cardiac muscle cells are well established [98, 99], although robust techniques for generating human iPSC-derived myoblasts capable of differentiating into functional skeletal muscle myotubes are less comprehensively available. Most efforts directed at deriving myogenic cells from human iPSCs to date have been based on the overexpression of myogenic transcription factors, such as Paired Box 3 (PAX3), PAX7, or Myogenic Differentiation 1 (MYOD1), by viral gene delivery. For example, Abujarour et al. [100] overexpressed MYOD1 in human iPSCs from patients with DMD, and the resulting myogenic cells efficiently formed myotubes, a result that was contrary to previous studies with primary biopsied myoblasts from patients with DMD [101–103] (Fig. 3a). Although this approach does produce myogenic cells, the random integration of viral DNA can limit cell proliferation, potentially mask disease-related phenotypes, or worse result in disease-causing insertional mutagenesis events. In addition, such protocols typically rely on nonhuman-derived media supplements, and are time consuming, in some cases taking over 5 months to reach a suitable endpoint [104, 105]. Recently, defined protocols for directing human iPSCs down the myogenic lineage using a chemical compound (CHIR99031) were reported, and have proven effective at pushing rodent and human stem cells to adopt a myogenic program, emphasizing a critical role for WNT signaling activation during the early stages of somite induction [95, 106] (Fig. 3b). These defined myogenic protocols lead to the formation of robust myotubes in vitro [95, 107, 108]. However, the contractile functionality of these human iPSC-derived myotubes has yet to be demonstrated, and an in depth study of the maturation state of these cells has not yet been performed. In addition, it remains to be seen whether a suitable purification strategy with which to isolate expandable skeletal muscle stem and/or progenitor cells from disease-specific human iPSCs can be integrated with these techniques to produce highly pure myogenic populations for downstream applications, as is commonly implemented for iPSC-derived cardiomyocyte production. For large-scale chemical compound-screening studies, such methods will be vital to ensure the generation of reliable and reproducible muscle cell populations.
FIGURE 3.
Induced pluripotent stem cell models of muscular dystrophy. (a) Structural and morphological characterization of Duchenne muscular dystrophy (DMD)- and Becker muscular dystrophy-specific myotubes in response to treatment with insulin-like growth factor 1 (IGF-1) and Wnt7a (potential therapeutics for treatment of muscular dystrophy in humans). (b) Muscle fibers differentiated from human induced pluripotent stem cells (iPSCs) stained for titin (i, ii) and fast myosin heavy chain (MyHC; iii). Note the presence of multinuclear myotubes and defined sarcomeric structures. Adapted from [100] (a) and [95] (c). Scale bars: 200 mm (a,bi,iii) and 20 mm (bii).
In addition to obtaining patient iPSCs for modeling muscular dystrophies, gene-editing techniques might provide further potential to expand disease-modeling capacities using edited wild-type iPSCs. Recent work demonstrated the use of CRISPR/Cas9 gene editing to create dystrophin-null cardiac muscle cells with similar phenotypes to patient iPSCs [92, 93]. The establishment of more robust methods for producing skeletal muscle myoblasts from iPSCs could open similar strategies for producing dystrophin-null myotubes for in vitro analysis. Recent research showed that CRISPR/Cas9 editing in a mouse model of DMD can be used to partially correct the in vivo phenotype [109]. Such results provide evidence for the potential to edit myotube genotypes to correct or exacerbate the phenotype for in vitro studies.
This ongoing work suggests that iPSC-derived skeletal muscle cells will soon be more widely available, and the capacity to produce purified myotubes reliably from dystrophic sources will likely follow soon after. As such, the development of engineered patient-specific human muscular dystrophy models with which to evaluate novel treatment modalities ahead of animal studies represents a tangible goal for the near future.
Current in vitro models of muscular dystrophy
Given the lack of robust methods for generating human iPSC-derived skeletal muscle myoblasts, relatively few models of human skeletal muscle diseases currently exist. Primary fibroblasts from patients with DMD or BDM have been differentiated successfully into fusion-competent myoblasts and these cells were shown to be capable of further differentiation into MyHC-positive myotubes [100]. Although these cells were used to test muscular dystrophy therapies currently in preclinical and clinical stages of development [IGF-1 and wingless-type MMTV integration site family, member 7A (Wnt7a); Fig. 3a], little evaluation of in vitro phenotype stratification between wild-type and diseased cells was performed. Therefore, it is not yet clear whether such cells offer a viable source of dystrophic cells for downstream in vitro phenotypic screening assays.
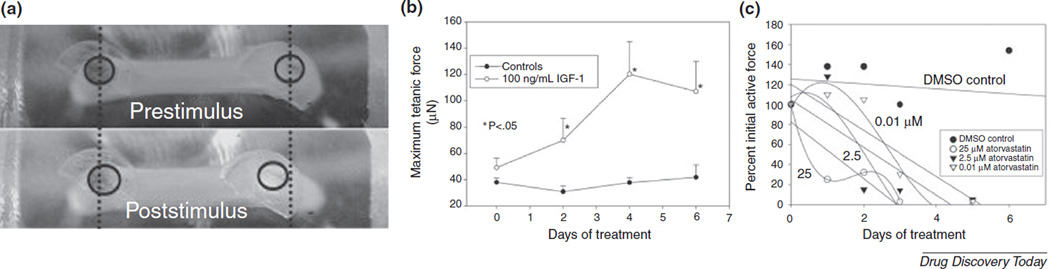
A 3D tissue-engineered model of DMD has also been produced using primary cells derived from the mdx mouse [51]. This study generated a high-throughput (96-well) 3D contractility assay using microengineered artificial muscles and used it to screen potential therapeutic drug candidates for their capacity to improve dystrophic muscle contractile function. Eleven of the tested compounds significantly improved tetanic contractile function in these constructs compared with placebo-treated controls, demonstrating the potential utility of this disease model for therapeutic screening. However, no significant difference in contractile properties was observed between wild-type and dystrophic constructs. Given that fetal MyHC expression patterns were observed in these cells, it is possible the constructs had not matured to a point where phenotypic differences were present, highlighting the need to use suitable maturation strategies for effective development in late-onset disease models. Microengineered muscle has been used more generally as a screening platform for evaluating the physiological effect of drugs on muscle performance (Fig. 4). Such data highlight the potential widespread utility of such technology, beyond specific studies of dystrophic muscle, for evaluating muscle performance in response to different experimental conditions.
FIGURE 4.
Measurement of engineered muscle contraction as a screening system for evaluating drug effects in vitro. (a) Example of post displacement in response to a maximum tetanic electrical stimulus. Prestimulus and 1-s poststimulus are shown to demonstrate how movement of posts in response to contraction can be used to evaluate force. The circles in the photographs mark the post top images. (b) Quantification of tetanic force contraction over a 6-day insulin-like growth factor 1 (IGF-1) treatment period using the model described in (a). (c) Measurement of the effects of atorvastatin on muscle contractile function. Atorvastatin was shown to decrease engineered muscle active force generation in a time–concentration-dependent manner. Adapted from [50].
Despite their limitations, these studies highlight the potential for engineered muscle tissues to uncover important mechanisms behind the initiation and progression of dystrophic pathology. The advantages of high throughput, increased reproducibility, reduced cost, and the defined nature of engineered muscle platforms over animal models provide sufficient incentive to utilize and optimize these technologies for disease modeling and drug discovery applications (Fig. 5). As engineered muscle maturation methods improve, and iPSC-derived myoblasts become more readily available, the importance of engineered skeletal muscle tissues for preclinical screening and basic science studies will likely expand rapidly.
FIGURE 5.
Schematic illustration highlighting how development of microphysiological skeletal muscle models can help drive improvements in the care of patients with dystrophy. Abbreviation: iPSC, induced pluripotent stem cells.
Current limitations and future perspectives
Perhaps the most obvious limitation of current methods to generate mature skeletal muscle is that no single strategy has yet been able to promote complete maturation of engineered constructs to a state indistinguishable from native adult human muscle tissue. It seems likely that combinatorial methods, integrating multiple exogenous stimuli, will be necessary to achieve these ends. Studies have shown that combining electrical stimulation with mechanical strain improves skeletal muscle maturation over and above what is achievable with either stimulus alone [29, 110]. Similarly, studies into the sequential addition of multiple growth factors describe the increased benefit of supplementation to cultures of skeletal muscle myotubes [75]. Likewise, the addition of neuronal factors has been shown to promote the spontaneous contractile activity of cultured human myotubes, and a lack of contraction is observed when these factors are omitted [111]. Using current technology, there is no substitute for combinatorial maturation methods to generate fully adult tissues. However, the integration of multiple signals increases the overall cost, feasibility, and reproducibility of the culture model, potentially eliminating the widespread adoption of such systems. Integration of the fewest maturation signals that still achieve efficient and consistent differentiation as well as maximal maturation will ensure widespread adoption of such systems and, therefore, preclinical and eventual clinical utility. Which combined stimuli are necessary and sufficient to produce muscular tissues with adequate maturity for disease modeling and predictive drug screening applications have yet to be established, but are important as these platforms move toward commercial realization.
Another factor limiting the effective implementation of human engineered muscle models is an understanding of what a mature in vitro phenotype will entail. Most researchers agree that functional outputs for skeletal muscle, in terms of contractile force, conduction velocities, and action potential waveforms, mature over time, and adult profiles are needed to model tissue function effectively [21, 33]. However, if such functionality can be achieved using human cell sources that still exhibit immature gene expression profiles, is this sufficient for effective modeling purposes? In other words, if maturity remains incomplete in engineered muscle tissue, which factors are most important to push toward adult-like values, and which can be forfeited?
A third consideration associated with the use of iPSCs in muscle engineering is the inherent variability between patient-derived lines. Human stem cells have long been known to exhibit significant interline variability in terms of doubling time, transcriptional profile, germ layer predisposition, and propensity to form teratomas [112]. Part of this variability can be attributed to different culture techniques between labs. However, the biggest contributor is the enormous genetic variation inherent in the population that is completely independent of the disease under study. Despite the understanding that this variation exists, little has been done to characterize the extent of this variability, or to develop the means to screen potential cell sources in an efficient, cost-effective, and high-throughput manner. Variability between lines is a significant roadblock to application of patient-based iPSC disease modeling because it might prevent effective comparisons between cell types, limiting the general conclusions that can be drawn for a given condition. This is why most laboratories have embraced the use of CRISPR/Cas9 technology to genetically engineer the desired mutation in an iPSC line from a normal donor [92, 113–116]. This approach creates a matched set of cell lines, with the only difference being the CRISPR-introduced mutation. One disadvantage of this approach is that one or a few ‘representative’ mutations (usually null alleles) causing a disease are chosen, with the assumption that a therapeutic intervention for that mutation can also be applied to all patients regardless of their mutations. It remains to be seen whether this will be true. However, if consistent in vitro phenotypes resulting from engineered mutations can be achieved, then the use of these models for informing decisions on the efficacy and/or toxicity of a drug is attainable in the near future. The genetic engineering approach, coupled with the development of comprehensive screening metrics with which to evaluate different patient-derived iPSC lines, will work in concert to ensure consistency and comparability across studies utilizing different cell sources.
Concluding remarks
Advances in tissue-engineering strategies have led to significant improvements in the biomimicry of engineered skeletal muscle in recent years. Combined with emerging iPSC-related technologies for differentiating human skeletal muscle cells, the potential for generating predictive congenital disease models of human muscle tissues is in its infancy but rapidly expanding. However, to recapitulate late-onset disease phenotypes and adult muscle physiologies accurately, further work is required to drive the development and maturation of cultured constructs. As our ability to achieve more adult-like physiologies improves, the capacity for tissue-engineered skeletal muscle to augment current animal models and drive forward our understanding of dystrophic disease mechanisms will likewise expand.
Acknowledgments
This work was supported by Muscular Dystrophy Association (MDA) research grants to D-H.K. (Award ID 255907) and to G.L. (Award ID 381465), as well as a National Institutes of Health R21 Grant (R21AR064395) (to D-H.K.), a New York Stem Cell Foundation award (to G.L.), and an International Collaborative R&D Program Grant with KIAT funded by the MOTIE (N0000894) awarded to D-H.K. D-H.K also thanks the Department of Bioengineering at the University of Washington for the new faculty startup fund.
Biography

Deok-Ho Kim is currently a Professor in the Department of Bioengineering at the University of Washington. He received his PhD in Biomedical Engineering from The Johns Hopkins University. His current research interests cover multiscale biomimetic materials/devices, functional tissue engineering, microscale stem/tumor cell niche engineering, and cell mechanobiology. He has published over 130 peer-reviewed journal articles and referenced conference proceedings, book chapters and patent applications, and given over 100 national and international invited lectures.
References
- 1.McGreevy JW, et al. Animal models of Duchenne muscular dystrophy: from basic mechanisms to gene therapy. Dis. Model Mech. 2015;8:195–213. doi: 10.1242/dmm.018424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van der Worp HB, et al. Can animal models of disease reliably inform human studies? PLoS Med. 2010;7:e1000245. doi: 10.1371/journal.pmed.1000245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Greek R, Menache A. Systematic reviews of animal models: methodology versus epistemology. Int. J. Med. Sci. 2013;10:206–221. doi: 10.7150/ijms.5529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Partridge TA. The mdx mouse model as a surrogate for Duchenne muscular dystrophy. FEBS J. 2013;280:4177–4186. doi: 10.1111/febs.12267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.MacIntosh BR, et al. Skeletal Muscle: Form and Function. Human Kinetics; 2006. [Google Scholar]
- 6.Alberts B, et al. Molecular Biology of the Cell. Garland Science; 2007. [Google Scholar]
- 7.Sciote JJ, Morris TJ. Skeletal muscle function and fibre types: the relationship between occlusal function and the phenotype of jaw-closing muscles in human. J. Orthod. 2000;27:15–30. doi: 10.1093/ortho/27.1.15. [DOI] [PubMed] [Google Scholar]
- 8.Gomes AV, et al. The role of troponins in muscle contraction. IUBMB Life. 2002;54:323–333. doi: 10.1080/15216540216037. [DOI] [PubMed] [Google Scholar]
- 9.Ervasti JM. Costameres: the Achilles’ heel of Herculean muscle. J. Biol. Chem. 2003;278:13591–13594. doi: 10.1074/jbc.R200021200. [DOI] [PubMed] [Google Scholar]
- 10.Nigro V, Piluso G. Spectrum of muscular dystrophies associated with sarcolemmal-protein genetic defects. Biochim. Biophys. Acta. 2015;1852:585–593. doi: 10.1016/j.bbadis.2014.07.023. [DOI] [PubMed] [Google Scholar]
- 11.Millay DP, et al. Calcium influx is sufficient to induce muscular dystrophy through a TRPC-dependent mechanism. Proc. Natl. Acad. Sci. U.S.A. 2009;106:19023–19028. doi: 10.1073/pnas.0906591106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Millay DP, et al. Genetic and pharmacologic inhibition of mitochondrial-dependent necrosis attenuates muscular dystrophy. Nat. Med. 2008;14:442–447. doi: 10.1038/nm1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goonasekera SA, et al. Enhanced Ca(2)(+) influx from STIM1-Orai1 induces muscle pathology in mouse models of muscular dystrophy. Hum. Mol. Genet. 2014;23:3706–3715. doi: 10.1093/hmg/ddu079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Piccolo F, et al. Intracellular accumulation and reduced sarcolemmal expression of dysferlin in limb-girdle muscular dystrophies. Ann. Neurol. 2000;48:902–912. [PubMed] [Google Scholar]
- 15.Wallace GQ, McNally EM. Mechanisms of muscle degeneration, regeneration, and repair in the muscular dystrophies. Annu. Rev. Physiol. 2009;71:37–57. doi: 10.1146/annurev.physiol.010908.163216. [DOI] [PubMed] [Google Scholar]
- 16.Kharraz Y, et al. Understanding the process of fibrosis in Duchenne muscular dystrophy. Biomed. Res. Int. 2014;2014:965631. doi: 10.1155/2014/965631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ogura Y, et al. Therapeutic potential of matrix metalloproteinases in Duchenne muscular dystrophy. Front. Cell Dev. Biol. 2014;2:11. doi: 10.3389/fcell.2014.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li W, et al. Progression and variation of fatty infiltration of the thigh muscles in Duchenne muscular dystrophy, a muscle magnetic resonance imaging study. Neuromuscul. Disord. 2015;25:375–380. doi: 10.1016/j.nmd.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 19.Uhl EW, Warner NJ. Mouse models as predictors of human responses: evolutionary medicine. Curr. Pathobiol. Rep. 2015;3:219–223. doi: 10.1007/s40139-015-0086-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nam K-H, et al. Biomimetic 3D tissue models for advanced high-throughput drug screening. J. Lab. Automat. 2015;20:201–215. doi: 10.1177/2211068214557813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Juhas M, et al. Biomimetic engineered muscle with capacity for vascular integration and functional maturation in vivo . Proc. Natl. Acad. Sci. U. S. A. 2014;111:5508–5513. doi: 10.1073/pnas.1402723111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van der Schaft DW, et al. Engineering skeletal muscle tissues from murine myoblast progenitor cells and application of electrical stimulation. J. Vis. Exp. 2013:e4267. doi: 10.3791/4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith AS, et al. Characterization and optimization of a simple, repeatable system for the long term in vitro culture of aligned myotubes in 3D. J. Cell. Biochem. 2012;113:1044–1053. doi: 10.1002/jcb.23437. [DOI] [PubMed] [Google Scholar]
- 24.Kshitiz, et al. A nanotopography approach for studying the structure– function relationships of cells and tissues. Cell Adh. Migr. 2015;9:300–307. doi: 10.4161/cam.29359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bol M, et al. On the anisotropy of skeletal muscle tissue under compression. Acta Biomater. 2014;10:3225–3234. doi: 10.1016/j.actbio.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 26.Gilbert PM, et al. Substrate elasticity regulates skeletal muscle stem cell self-renewal in culture. Science. 2010;329:1078–1081. doi: 10.1126/science.1191035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hinds S, et al. The role of extracellular matrix composition in structure and function of bioengineered skeletal muscle. Biomaterials. 2011;32:3575–3583. doi: 10.1016/j.biomaterials.2011.01.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holmberg J, Durbeej M. Laminin-211 in skeletal muscle function. Cell Adh. Migr. 2013;7:111–121. doi: 10.4161/cam.22618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liao IC, et al. Effect of electromechanical stimulation on the maturation of myotubes on aligned electrospun fibers. Cell. Mol. Bioeng. 2008;1:133–145. doi: 10.1007/s12195-008-0021-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kostrominova TY, et al. Ultrastructure of myotendinous junctions in tendon-skeletal muscle constructs engineered in vitro . Histol. Histopathol. 2009;24:541–550. doi: 10.14670/hh-24.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morimoto Y, et al. Three-dimensional neuron-muscle constructs with neuromuscular junctions. Biomaterials. 2013;34:9413–9419. doi: 10.1016/j.biomaterials.2013.08.062. [DOI] [PubMed] [Google Scholar]
- 32.Belanger AY, McComas AJ. Contractile properties of human skeletal muscle in childhood and adolescence. Eur. J. Appl. Physiol. Occup. Physiol. 1989;58:563–567. doi: 10.1007/BF00418500. [DOI] [PubMed] [Google Scholar]
- 33.Smith AS, et al. A multiplexed chip-based assay system for investigating the functional development of human skeletal myotubes in vitro . J. Biotechnol. 2014;185:15–18. doi: 10.1016/j.jbiotec.2014.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Serena E, et al. Soft substrates drive optimal differentiation of human healthy and dystrophic myotubes. Integr. Biol. 2010;2:193–201. doi: 10.1039/b921401a. [DOI] [PubMed] [Google Scholar]
- 35.Martin NRW, et al. Factors affecting the structure and maturation of human tissue engineered skeletal muscle. Biomaterials. 2013;34:5759–5765. doi: 10.1016/j.biomaterials.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 36.Eisenberg BR, et al. Stereological analysis of mammalian skeletal muscleISoleus muscle of the adult guinea pig. J. Cell Biol. 1974;60:732–754. doi: 10.1083/jcb.60.3.732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Larkin LM, et al. Structure and functional evaluation of tendon-skeletal muscle constructs engineered in vitro . Tissue Eng. 2006;12:3149–3158. doi: 10.1089/ten.2006.12.3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang HS, et al. Nanopatterned muscle cell patches for enhanced myogenesis and dystrophin expression in a mouse model of muscular dystrophy. Biomaterials. 2014;35:1478–1486. doi: 10.1016/j.biomaterials.2013.10.067. [DOI] [PubMed] [Google Scholar]
- 39.Carson D, et al. Functionalized nanopatterned substrates for enhanced maturation in cultured human stem cell-derived cardiomyocytes. ACS Appl. Mater. Interface. 2016 http://dx.doi.org/10.1021/acsami.5b11671 (in press)
- 40.Yang HS, et al. Electroconductive nanopatterned substrates for enhanced myogenic differentiation and maturation. Adv. Healthcare Mater. 2015;5:137–145. doi: 10.1002/adhm.201500003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Molnar P, et al. Photolithographic patterning of C2C12 myotubes using vitronectin as growth substrate in serum-free medium. Biotechnol. Prog. 2007;23:265–268. doi: 10.1021/bp060302q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bajaj P, et al. Patterning the differentiation of C2C12 skeletal myoblasts. Integr. Biol. 2011;3:897–909. doi: 10.1039/c1ib00058f. [DOI] [PubMed] [Google Scholar]
- 43.Jiao A, et al. Thermoresponsive nanofabricated substratum for the engineering of three-dimensional tissues with layer-by-layer architectural control. ACS Nano. 2014;8:4430–4439. doi: 10.1021/nn4063962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lapin MR, et al. Substrate elasticity affects bovine satellite cell activation kinetics in vitro . J. Anim. Sci. 2013;91:2083–2090. doi: 10.2527/jas.2012-5732. [DOI] [PubMed] [Google Scholar]
- 45.Gribova V, et al. Effect of RGD functionalization and stiffness modulation of polyelectrolyte multilayer films on muscle cell differentiation. Acta Biomater. 2013;9:6468–6480. doi: 10.1016/j.actbio.2012.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Engler AJ, et al. Myotubes differentiate optimally on substrates with tissue-like stiffness: pathological implications for soft or stiff microenvironments. J. Cell Biol. 2004;166:877–887. doi: 10.1083/jcb.200405004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boonen KJ, et al. Essential environmental cues from the satellite cell niche: optimizing proliferation and differentiation. Am. J. Physiol. Cell Physiol. 2009;296:C1338–C1345. doi: 10.1152/ajpcell.00015.2009. [DOI] [PubMed] [Google Scholar]
- 48.Mandrycky C, et al. 3D bioprinting for engineering complex tissues. Biotechnol. Adv. 2015 doi: 10.1016/j.biotechadv.2015.12.011. http://dx.doi.org/10.1016/j.biotechadv.2015.12.011. [DOI] [PMC free article] [PubMed]
- 49.Vandenburgh H. High-content drug screening with engineered musculoskeletal tissues. Tissue Eng. Part B Rev. 2010;16:55–64. doi: 10.1089/ten.teb.2009.0445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vandenburgh H, et al. Drug-screening platform based on the contractility of tissue-engineered muscle. Muscle Nerve. 2008;37:438–447. doi: 10.1002/mus.20931. [DOI] [PubMed] [Google Scholar]
- 51.Vandenburgh H, et al. Automated drug screening with contractile muscle tissue engineered from dystrophic myoblasts. FASEB J. 2009;23:3325–3334. doi: 10.1096/fj.09-134411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Salimath AS, Garcia AJ. Biofunctional hydrogels for skeletal muscle constructs. J. Tissue Eng. Regen. Med. 2014 doi: 10.1002/term.1881. http://dx.doi.org/10.1002/term.1881. [DOI] [PubMed]
- 53.Wang P-Y, et al. Grooved PLGA films incorporated with RGD/YIGSR peptides for potential application on skeletal muscle tissue engineering. Colloids Surf. B: Biointerfaces. 2013;110:88–95. doi: 10.1016/j.colsurfb.2013.04.016. [DOI] [PubMed] [Google Scholar]
- 54.Wang PY, et al. The roles of RGD and grooved topography in the adhesion, morphology, and differentiation of C2C12 skeletal myoblasts. Biotechnol. Bioeng. 2012;109:2104–2115. doi: 10.1002/bit.24452. [DOI] [PubMed] [Google Scholar]
- 55.Cezar CA, Mooney DJ. Biomaterial-based delivery for skeletal muscle repair. Adv. Drug Deliv. Rev. 2015;84:188–197. doi: 10.1016/j.addr.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Niu A, et al. Src mediates the mechanical activation of myogenesis by activating TNFalpha-converting enzyme. J. Cell Sci. 2013;126:4349–4357. doi: 10.1242/jcs.125328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Player DJ, et al. Acute mechanical overload increases IGF-I and MMP-9 mRNA in 3D tissue-engineered skeletal muscle. Biotechnol. Lett. 2014;36:1113–1124. doi: 10.1007/s10529-014-1464-y. [DOI] [PubMed] [Google Scholar]
- 58.Ward CW, et al. Mechanical stretch-induced activation of ROS/RNS signaling in striated muscle. Antioxid. Redox Signal. 2014;20:929–936. doi: 10.1089/ars.2013.5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Matthews BD, et al. Cellular adaptation to mechanical stress: role of integrins, Rho, cytoskeletal tension and mechanosensitive ion channels. J. Cell Sci. 2006;119:508–518. doi: 10.1242/jcs.02760. [DOI] [PubMed] [Google Scholar]
- 60.Candiani G, et al. Cyclic mechanical stimulation favors myosin heavy chain accumulation in engineered skeletal muscle constructs. J. Appl. Biomater. Biomech. 2010;8:68–75. [PubMed] [Google Scholar]
- 61.Moon du G, et al. Cyclic mechanical preconditioning improves engineered muscle contraction. Tissue Eng. A. 2008;14:473–482. doi: 10.1089/tea.2007.0104. [DOI] [PubMed] [Google Scholar]
- 62.Parsons SA, et al. Genetic loss of calcineurin blocks mechanical overload-induced skeletal muscle fiber type switching but not hypertrophy. J. Biol. Chem. 2004;279:26192–26200. doi: 10.1074/jbc.M313800200. [DOI] [PubMed] [Google Scholar]
- 63.Nikolic N, et al. Electrical pulse stimulation of cultured human skeletal muscle cells as an in vitro model of exercise. PLoS ONE. 2012;7:e33203. doi: 10.1371/journal.pone.0033203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Khodabukus A, Baar K. Streptomycin decreases the functional shift to a slow phenotype induced by electrical stimulation in engineered muscle. Tissue Eng. A. 2014;21:1003–1012. doi: 10.1089/ten.TEA.2014.0462. [DOI] [PubMed] [Google Scholar]
- 65.Wehrle U, et al. Effects of chronic electrical stimulation on myosin heavy chain expression in satellite cell cultures derived from rat muscles of different fiber-type composition. Differentiation. 1994;58:37–46. doi: 10.1046/j.1432-0436.1994.5810037.x. [DOI] [PubMed] [Google Scholar]
- 66.Sakar MS, et al. Formation and optogenetic control of engineered 3D skeletal muscle bioactuators. Lab Chip. 2012;12:4976–4985. doi: 10.1039/c2lc40338b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Raman R, et al. Optogenetic skeletal muscle-powered adaptive biological machines. Proc. Natl. Acad. Sci. U. S. A. 2016;113:3497–3502. doi: 10.1073/pnas.1516139113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Michielin F, et al. Microfluidic-assisted cyclic mechanical stimulation affects cellular membrane integrity in a human muscular dystrophy in vitro model. RSC Adv. 2015;5:98429–98439. [Google Scholar]
- 69.Rommel C, et al. Mediation of IGF-1-induced skeletal myotube hypertrophy by PI(3)K/Akt/mTOR and PI(3)K/Akt/GSK3 pathways. Nat. Cell Biol. 2001;3:1009–1013. doi: 10.1038/ncb1101-1009. [DOI] [PubMed] [Google Scholar]
- 70.Shansky J, et al. Paracrine release of insulin-like growth factor 1 from a bioengineered tissue stimulates skeletal muscle growth in vitro . Tissue Eng. 2006;12:1833–1841. doi: 10.1089/ten.2006.12.1833. [DOI] [PubMed] [Google Scholar]
- 71.Bian W, Bursac N. Soluble miniagrin enhances contractile function of engineered skeletal muscle. FASEB J. 2012;26:955–965. doi: 10.1096/fj.11-187575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sculthorpe N, et al. Androgens affect myogenesis in vitro and increase local IGF-1 expression. Med. Sci. Sports Exerc. 2012;44:610–615. doi: 10.1249/MSS.0b013e318237c5c0. [DOI] [PubMed] [Google Scholar]
- 73.McAleer CW, et al. Mechanistic investigation of adult myotube response to exercise and drug treatment in vitro using a multiplexed functional assay system. J. Appl. Physiol. 2014;117:1398–1405. doi: 10.1152/japplphysiol.00612.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Weist MR, et al. TGF-beta1 enhances contractility in engineered skeletal muscle. J. Tissue Eng. Regen. Med. 2013;7:562–571. doi: 10.1002/term.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Das M, et al. Developing a novel serum-free cell culture model of skeletal muscle differentiation by systematically studying the role of different growth factors in myotube formation. In Vitro Cell. Dev. Biol. Anim. 2009;45:378–387. doi: 10.1007/s11626-009-9192-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Larkin LM, et al. Functional evaluation of nerve-skeletal muscle constructs engineered in vitro . In Vitro Cell. Dev. Biol. Anim. 2006;42:75–82. doi: 10.1290/0509064.1. [DOI] [PubMed] [Google Scholar]
- 77.Bach AD, et al. Expression of Trisk 51, agrin and nicotinic-acetycholine receptor epsilon-subunit during muscle development in a novel three-dimensional muscle-neuronal co-culture system. Cell Tissue Res. 2003;314:263–274. doi: 10.1007/s00441-003-0757-6. [DOI] [PubMed] [Google Scholar]
- 78.Dhawan V, et al. Neurotization improves contractile forces of tissue-engineered skeletal muscle. Tissue Eng. 2007;13:2813–2821. doi: 10.1089/ten.2007.0003. [DOI] [PubMed] [Google Scholar]
- 79.Smith AS, et al. A functional system for high-content screening of neuromuscular junctions. Technology. 2013;1:37–48. doi: 10.1142/S2339547813500015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Martin NR, et al. Neuromuscular junction formation in tissue-engineered skeletal muscle augments contractile function and improves cytoskeletal organization. Tissue Eng. A. 2015;21:2595–2604. doi: 10.1089/ten.tea.2015.0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kaus A, Sareen D. ALS patient stem cells for unveiling disease signatures of motoneuron susceptibility: perspectives on the deadly mitochondria, ER stress and calcium triad. Front. Cell Neurosci. 2015;9:448. doi: 10.3389/fncel.2015.00448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Saporta MA, et al. Axonal Charcot-Marie-Tooth disease patient-derived motor neurons demonstrate disease-specific phenotypes including abnormal electrophysiological properties. Exp. Neurol. 2015;263:190–199. doi: 10.1016/j.expneurol.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Criswell TL, et al. The role of endothelial cells in myofiber differentiation and the vascularization and innervation of bioengineered muscle tissue in vivo . Biomaterials. 2013;34:140–149. doi: 10.1016/j.biomaterials.2012.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Levenberg S, et al. Engineering vascularized skeletal muscle tissue. Nat. Biotechnol. 2005;23:879–884. doi: 10.1038/nbt1109. [DOI] [PubMed] [Google Scholar]
- 85.Mahler GJ, et al. Characterization of a gastrointestinal tract microscale cell culture analog used to predict drug toxicity. Biotechnol. Bioeng. 2009;104:193–205. doi: 10.1002/bit.22366. [DOI] [PubMed] [Google Scholar]
- 86.Zhang C, et al. Towards a human-on-chip: culturing multiple cell types on a chip with compartmentalized microenvironments. Lab Chip. 2009;9:3185–3192. doi: 10.1039/b915147h. [DOI] [PubMed] [Google Scholar]
- 87.Esch MB, et al. How multi-organ microdevices can help foster drug development. Adv. Drug Deliv. Rev. 2014;69–70:158–169. doi: 10.1016/j.addr.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Anderson MS, Kunkel LM. The molecular and biochemical basis of Duchenne muscular dystrophy. Trends Biochem. Sci. 1992;17:289–292. doi: 10.1016/0968-0004(92)90437-e. [DOI] [PubMed] [Google Scholar]
- 89.Vo AH, McNally EM. Modifier genes and their effect on Duchenne muscular dystrophy. Curr. Opin. Neurol. 2015;28:528–534. doi: 10.1097/WCO.0000000000000240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bello L, et al. Importance of SPP1 genotype as a covariate in clinical trials in Duchenne muscular dystrophy. Neurology. 2012;79:159–162. doi: 10.1212/WNL.0b013e31825f04ea. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Heydemann A, et al. Latent TGF-beta-binding protein 4 modifies muscular dystrophy in mice. J. Clin. Invest. 2009;119:3703–3712. doi: 10.1172/JCI39845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Guan X, et al. Dystrophin-deficient cardiomyocytes derived from human urine: new biologic reagents for drug discovery. Stem Cell Res. 2014;12:467–480. doi: 10.1016/j.scr.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Macadangdang J, et al. Nanopatterned human iPSC-based model of a dystrophin-null cardiomyopathic phenotype. Cell. Mol. Bioeng. 2015;8:320–332. doi: 10.1007/s12195-015-0413-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mack DL, et al. Disease-in-a-dish: the contribution of patient-specific induced pluripotent stem cell technology to regenerative rehabilitation. Am. J. Phys. Med. Rehabil. 2014;93(Suppl. 3):S155–S168. doi: 10.1097/PHM.0000000000000141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chal J, et al. Differentiation of pluripotent stem cells to muscle fiber to model Duchenne muscular dystrophy. Nat. Biotechnol. 2015;33:962–969. doi: 10.1038/nbt.3297. [DOI] [PubMed] [Google Scholar]
- 96.Sterrenburg E, et al. Gene expression profiling highlights defective myogenesis in DMD patients and a possible role for bone morphogenetic protein 4. Neurobiol. Dis. 2006;23:228–236. doi: 10.1016/j.nbd.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 97.Dumont NA, et al. Dystrophin expression in muscle stem cells regulates their polarity and asymmetric division. Nat. Med. 2015;21:1455–1463. doi: 10.1038/nm.3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lundy SD, et al. Structural and functional maturation of cardiomyocytes derived from human pluripotent stem cells. Stem Cells Dev. 2013;22:1991–2002. doi: 10.1089/scd.2012.0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lian X, et al. Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/beta-catenin signaling under fully defined conditions. Nat. Protoc. 2013;8:162–175. doi: 10.1038/nprot.2012.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Abujarour R, et al. Myogenic differentiation of muscular dystrophy-specific induced pluripotent stem cells for use in drug discovery. Stem Cells Transl. Med. 2014;3:149–160. doi: 10.5966/sctm.2013-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Delaporte C, et al. Comparison between the growth pattern of cell cultures from normal and Duchenne dystrophy muscle. J. Neurol. Sci. 1984;64:149–160. doi: 10.1016/0022-510x(84)90033-9. [DOI] [PubMed] [Google Scholar]
- 102.Hoffman EP, et al. Characterization of dystrophin in muscle-biopsy specimens from patients with Duchenne’s or Becker’s muscular dystrophy. New EnglJMed. 1988;318:1363–1368. doi: 10.1056/NEJM198805263182104. [DOI] [PubMed] [Google Scholar]
- 103.Jasmin G, et al. Impaired muscle differentiation in explant cultures of Duchenne muscular dystrophy. Lab. Invest. 1984;50:197–207. [PubMed] [Google Scholar]
- 104.Barberi T, et al. Derivation of engraftable skeletal myoblasts from human embryonic stem cells. Nat. Med. 2007;13:642–648. doi: 10.1038/nm1533. [DOI] [PubMed] [Google Scholar]
- 105.Borchin B, et al. Derivation and FACS-mediated purification of PAX3+/PAX7+ skeletal muscle precursors from human pluripotent stem cells. Stem Cell Rep. 2013;1:620–631. doi: 10.1016/j.stemcr.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mayeuf-Louchart A, et al. Notch regulation of myogenic versus endothelial fates of cells that migrate from the somite to the limb. Proc. Natl. Acad. Sci. U. S. A. 2014;111:8844–8849. doi: 10.1073/pnas.1407606111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Shelton M, et al. Robust generation and expansion of skeletal muscle progenitors and myocytes from human pluripotent stem cells. Methods. 2015 doi: 10.1016/j.ymeth.2015.09.019. http://dx.doi.org/10.1016/j.ymeth.2015.09.019. [DOI] [PubMed]
- 108.Shelton M, et al. Derivation and expansion of PAX7-positive muscle progenitors from human and mouse embryonic stem cells. Stem Cell Rep. 2014;3:516–529. doi: 10.1016/j.stemcr.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Long C, et al. Prevention of muscular dystrophy in mice by CRISPR/Cas9- mediated editing of germline DNA. Science. 2014;345:1184–1188. doi: 10.1126/science.1254445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Morgan KY, Black LD., 3rd Mimicking isovolumic contraction with combined electromechanical stimulation improves the development of engineered cardiac constructs. Tissue Eng. A. 2014;20:1654–1667. doi: 10.1089/ten.tea.2013.0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Guo X, et al. In vitro differentiation of functional human skeletal myotubes in a defined system. Biomater. Sci. 2014;2:131–138. doi: 10.1039/C3BM60166H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Osafune K, et al. Marked differences in differentiation propensity among human embryonic stem cell lines. Nat. Biotechnol. 2008;26:313–315. doi: 10.1038/nbt1383. [DOI] [PubMed] [Google Scholar]
- 113.Liu J, et al. CRISPR/Cas9 facilitates investigation of neural circuit disease using human iPSCs: mechanism of epilepsy caused by an SCN1A loss-of-function mutation. Transl. Psychiatry. 2016;6:e703. doi: 10.1038/tp.2015.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Oceguera-Yanez F, et al. Engineering the AAVS1 locus for consistent and scalable transgene expression in human iPSCs and their differentiated derivatives. Methods. 2015 doi: 10.1016/j.ymeth.2015.12.012. http://dx.doi.org/10.1016/j.ymeth.2015.12.012. [DOI] [PubMed]
- 115.Kang H, et al. CCR5 disruption in induced pluripotent stem cells using CRISPR/Cas9 provides selective resistance of immune cells to CCR5-tropic HIV-1 virus. Mol. Ther. Nucleic Acids. 2015;4:e268. doi: 10.1038/mtna.2015.42. [DOI] [PubMed] [Google Scholar]
- 116.Chen Y, et al. Engineering human stem cell lines with inducible gene knockout using CRISPR/Cas9. Cell Stem Cell. 2015;17:233–244. doi: 10.1016/j.stem.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Gillies AR, Lieber RL. Structure and Function of the Skeletal Muscle Extracellular Matrix. Muscle Nerve. 2011;44:318–331. doi: 10.1002/mus.22094. [DOI] [PMC free article] [PubMed] [Google Scholar]