Introduction
Dysregulation of lipid metabolism is a hallmark of metabolic disease and cancer. While it is not yet clear how changes in lipid amounts and composition affect tumor cell growth, evidence indicates that these changes play a critical role in driving oncogenesis. [1,2] Given the strong epidemiological link between obesity and cancer, identification of the key molecular targets downstream of molecules that regulate lipid metabolism and link these disease states is needed to facilitate the development of new therapeutic strategies.
Phosphatase and tensin homologue (PTEN) is one of the most commonly mutated tumor suppressors in human cancer. PTEN functions as a lipid phosphatase; it contains a tensin-like domain and a catalytic domain that dephosphorylates phosphatidylinositol-3,4,5-trisphosphate (PtdIns [3,4,5]P3). Loss of PTEN results in the accumulation of this substrate at the cellular membrane, leading to recruitment of AKT and activation of numerous downstream signaling pathways that support cell growth and survival [3,4]. Extensive studies suggest that decreased PTEN activity also leads to metabolic reprogramming of cells to facilitate the synthesis of macromolecules needed to drive cell growth [5]. This is accomplished, in part, by the ability of PTEN to regulate both glucose and lipid metabolism. While the central role of PTEN in glucose metabolism has been well defined [6–9], less is known about how PTEN affects lipid metabolism [10,11]. Here we discuss emerging evidence supporting the idea that PTEN-mediated repression of genes that encode tRNAs and rRNAs is needed to suppress oncogenic transformation and maintain lipid and energy homeostasis. This involves Maf1, a transcription factor that functions downstream of PTEN to coordinately restrain RNA synthesis, lipid biosynthesis, and tumor growth.
PTEN signaling pathways affect tRNA and rRNA synthesis
While the primary function of the PTEN/PI3K/AKT signaling pathway is to control cellular metabolism [1,2], metabolic reprogramming through PTEN and its downstream targets appears to be a pre-requisite for rapid growth of cancer cells. Studies on the molecular functions of PTEN have primarily focused on the PTEN-controlled signaling pathways that alter the expression of protein-coding genes or non-coding (nc) RNAs, transcribed by RNA polymerase (pol) II [3,4]. However, the majority of cellular RNAs are synthesized by RNA pols I and III, whose major products are tRNAs and rRNAs. These RNAs, which constitute 85% of total cellular RNA, are essential for protein synthesis [12]. While the genes encoding these RNAs were long thought to serve constitutive rather than regulatory functions, many studies have now revealed that the increased expression of these transcripts is in fact a hallmark of transformed cells and human tumors [13–15]. Subsequent studies demonstrated that these RNA pol I- and III-mediated transcription processes are exquisitely regulated through various oncogenic and tumor suppressor proteins, including PTEN [15–20].
PTEN was first demonstrated to repress RNA pol III-dependent transcription through its lipid phosphatase activity and its ability to inhibit PI3K/AKT and downstream effectors, target of rapamycin complex 1 (TORC1) and S6 kinase (S6K) [20]. Analysis of the RNA pol III-specific transcription components revealed that PTEN expression leads to changes in the TBP (TATA-binding protein)-containing complex, TFIIIB. PTEN changes the phosphorylation state of this complex, disrupting the formation of the TFIIIB complex and its recruitment to promoters [20]. PTEN was similarly shown to repress RNA pol I-dependent transcription through its inhibition of the PI3K/TORC1/S6K pathway [21]. In this case, PTEN acts to dissociate TBP from the promoter, thereby disrupting the RNA pol I-specific TBP-containing SL-1 complex. In addition, inhibition of this pathway results in inactivation of the RNA pol I-specific factors, TIF-1A/RRN3 [22] and UBF [23], further contributing to transcription repression. Thus, multiple RNA pol I and III transcription components are subject to PTEN/PI3K/TORC1 modulation (Figure 1, Key figure). This highlights the multiple mechanisms that are used for restraining the synthesis of tRNAs and rRNAs in order to ultimately control cellular growth properties.
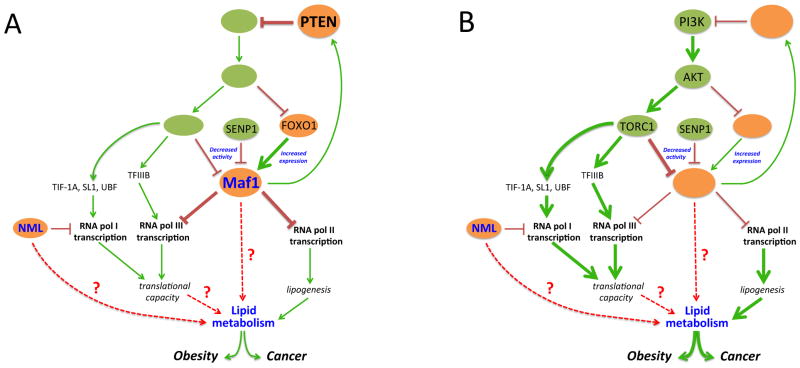
KEY FIGURE, Figure 1.
PTEN coordinately regulates RNA and lipid metabolism through Maf1
PTEN acts through its downstream signaling targets to repress transcription from all three nuclear RNA polymerases. PTEN controlled signaling events target Maf1, a global repressor of RNA pol III-dependent transcription and a selective repressor of RNA pol II-mediated transcription. TORC1 and SENP1 regulate PTMs on Maf1 to negatively regulate its activity while FOXO1 positively regulates Maf1 expression. Maf1-mediated changes in gene expression coordinate the repression of both lipid biogenesis and the translational capacity of cells. In mouse models, the loss of either NML or Maf1, negative regulators of RNA pol I- and pol III-dependent transcription, respectively, leads to transcription induction and dysregulation of lipid metabolism. Maf1 represents a novel molecular link between RNA and lipid metabolism that may explain the established association between obesity and cancer. However, more work is needed to understand how Maf1 and NML contribute to lipid homeostasis and whether Maf1-mediated repression of RNA pol III-mediated transcription contributes to lipid metabolism. (A) In cells with functional PTEN, PI3K/AKT/TORC1 signaling is inhibited while FOXO1 is activated. Inactivation of mTORC1 limits the expression of RNA pol I and III transcribed genes by reducing the activity of their transcription components. The inability of TORC1 to phosphorylate Maf1 leads to enhanced Maf1 activity while the activation of FOXO1 leads to enhanced Maf1 expression. Maf1 also induces the expression of PTEN, leading to further inactivation of AKT. The overall increase in cellular Maf1 activity coordinates repression of its RNA pol II and III target genes to limit translational capacity and lipogenesis. Maintaining repression of these processes may suppress the development of cancer and metabolic diseases such as obesity. (B) Inactivation or loss of PTEN results in constitutive activation of PI3K and AKT. The resultant activation of mTORC1 leads to induction of both RNA pol I- and III-dependent transcription through enhanced activity of transcription factors. The activation of TORC1 also causes a reduction in Maf1 activity while the inactivation of FOXO1 results in a decrease in Maf1 expression. This overall decrease in Maf1 leads to a derepression of Maf1 targeted RNA pol II and III transcribed genes leading to enhanced translational capacity and lipogenesis. Dysregulation of these processes and altered RNA and lipid metabolism may contribute to the development of obesity and cancer.
Given that dysregulation of RNA pol I- and III-dependent gene activity may be important drivers of cancer development (see Box 1), is targeting these transcription processes a viable strategy for cancer therapy? Genetically engineered mouse models of leukemia and lymphoma revealed strong sensitivity and selectivity in tumor cell death relative to normal cells with treatment of the RNA pol I inhibitor, CX-5461 [24]. Currently, clinical trials of this small molecule are underway in patients with lymphoma and leukemia [25]. Work on hematological malignancies, which are sensitive to inhibition of RNA pol I transcription, has so far revealed that aberrant myc expression and p53 status are key predictors of sensitivity to RNA pol I inhibition therapy [24,26]. However, further work will be needed to determine the feasibility of this approach in other tumor types and to learn how dysregulation of other proteins, such as PTEN, may affect efficacy. Together, these studies form a compelling argument that the aberrant activation of RNA pol I- and RNA pol III-specific factors by oncogenic signaling pathways and the inactivation of tumor suppressors drives cancer development and that targeting these processes may provide effective new therapeutic strategies to treat cancer.
BOX 1. Regulation of RNA pol III-mediated transcription, protein synthesis, and oncogenesis.
Although dysregulation of the RNA pol III-dependent transcription process is a hallmark of cancer cells [13–15], it is important to understand whether this actually contributes to cancer development. Evidence so far indicates that the observed increases in transcription contribute to oncogenic transformation [63,64]. One study revealed that c-myc-transformed cells required increases in RNA pol III dependent-transcription for anchorage-independent growth and tumor formation in mice [65]. So how might changes in this transcription process and its products drive oncogenesis? While the specific changes in RNA pol III transcription that lead to cellular transformation have not been clearly elucidated, emerging evidence supports the idea that alterations in tRNA gene expression and activity leads to changes in the cellular composition of functional tRNAs [66]. Analysis of tRNA pools showed that there are marked differences in types and amounts of tRNAs in different tissues [67,68]. In proliferating cells, the tRNAs that are induced are typically repressed in differentiating cells, and these tRNAs contain anticodons corresponding to a codon-usage signature that encompasses proliferation-related genes [69]. Furthermore, tRNAiMet overexpression in human cell lines can independently produce significant changes in tRNA expression profiles [70]. New evidence also indicates that the maturation of tRNA transcripts is a key regulatory step that can affect the population of functional tRNAs in fission yeast. When tRNA synthesis is induced, at least one tRNA modification enzyme, Trm1, becomes limiting [71]. This results in hypomodification of certain tRNAs and impact tRNA function in translation. Collectively, these studies support the idea that changes in RNA pol III-dependent transcription can lead to selective alterations in the abundance and activities of tRNAs. These changes in tRNAs, together with the mRNA codon-usage demand, could then result in selective changes in protein translation. In addition to tRNAs, alterations in other RNA pol III transcribed products that regulate protein processing (7SL RNA), mRNA splicing (U6 RNA) and transcription elongation (7SK RNA) could also conceivably lead to changes in the population of functional proteins. A large body of work has identified specific changes in translation initiation factors in cancer cells that alter mRNA translation [72,73]. Thus, there are multiple mechanisms by which protein synthesis may be dysregulated in cancer cells, including changes in the synthesis of RNA pol I and III products, to drive cancer development.
The transcription factor, Maf1, functions as a tumor suppressor
Maf1 is a central repressor of genes transcribed by RNA pol III. It functions to restrict the synthesis of these cellular RNA products in response to low nutrient availability and cellular stress [27,28]. Maf1 associates with RNA pol III both on and off the DNA and inhibits pre-initiation complex assembly by polymerase molecules that have dissociated from the DNA template [29,30]. The fact that Maf1 represses RNA pol III gene promoters is well established and conserved in all eukaryotes. Less well defined is Maf1’s ability to regulate transcription of RNA pol II transcribed genes [31–35]. Under certain growth conditions, deletion of Maf1 results in decreased transcription of gluconeogenetic genes in S. cerevisiae. However it is unclear whether Maf1 functions directly or indirectly on these genes [36]. Mammalian Maf1 occupies the promoters and inhibits transcription of select RNA pol II-dependent growth promoting genes that contain Elk-1 binding sites [31]. In this case, recruitment of Maf1 to these promoters was reciprocal with that of Elk-1. Studies in C. elegans and mammalian cells showed that Maf1 inhibits expression of the TBP gene, a central transcription initiation factor that is used by all nuclear RNA pols [31,33]. Maf1-mediated changes in TBP expression can indirectly affect genes that are sensitive to the concentration of TBP, including RNA pol I-dependent genes [31], although this is context-dependent [37]. Maf1 was also reported to induce transcription of the PTEN promoter [35]. In another example, the effect of Maf1 on specific RNA pol II genes involved changes in RNA pol III and Maf1 recruitment to a promoter-associated short interspersed element (SINE) [34]. However Maf1 does not contain a canonical DNA binding domain and there is no direct demonstration of Maf1 alone binding to DNA. Thus, the mechanism by which Maf1 is recruited to RNA pol II-dependent promoters is not known. Genome-wide detection of mammalian Maf1 in chromatin has been problematic and one study detected Maf1 at RNA pol III promoters but not RNA pol II promoters [38]. Collectively, current studies indicate that Maf1 is bound to both RNA pol II- and III-dependent promoters but more studies are needed to elucidate what contacts specify Maf1 recruitment to these promoters and whether common or distinct mechanisms are used.
Evidence supports that Maf1 is an important downstream effector of PTEN’s tumor suppressor function. Maf1 overexpression suppresses the anchorage-independent growth of PTEN-deficient human U87 glioblastoma cells with changes in cytoskeletal organization [31]. In addition, Maf1 overexpression in HT-29 colon carcinoma cells results in a decrease in both anchorage-independent growth and tumorigenicity in athymic mice [32]. Diminished nuclear Maf1 protein expression was also observed in several PTEN-negative human liver and prostate tumors compared to normal tissue [32]. Further analyses showed that Maf1 expression is frequently down-regulated in different subtypes of primary human liver cancers [35]. However, mice that are completely deficient for Maf1 were not prone to developing cancers [37]. Maf1 represses AKT signaling by increasing PTEN expression and this contributes to its ability to inhibit liver cancer cell proliferation [35]. Thus, Maf1 both represses the expression of genes that promote an oncogenic state, and it enhances PTEN expression. Together, these results demonstrate that Maf1 functions as a tumor suppressor.
Maf1 regulation
Several studies have revealed that mammalian Maf1 activity is regulated through post-translational modifications (PMTs). Phosphorylation of mammalian Maf1 TORC1 renders it less repressive of transcription [39–41]. Small ubiquitin-related modifier (SUMO) modification of Maf1 results in repressed transcription and cellular growth. Sentrin-specific protease 1 (SENP1) is the deSUMOylase that regulates this SUMO modification [42]. As SENP1 is overexpressed in a variety of human cancers [43], Maf1 could be an important SENP1 target that contributes to its oncogenic properties. In addition to PMTs, Maf1 expression is also subject to regulation by PTEN. Compared with PTEN-expressing cells, PTEN-deficient mouse tissues and cancer cell lines displayed decreased Maf1 protein expression. PTEN regulation of Maf1 was shown to involve the PI3K/AKT and forkhead box protein O1 (FOXO1) signaling pathways, through which changes in Maf1 expression led to reciprocal changes in RNA pol III-dependent transcription [33]. Importantly, this regulatory pathway is conserved in evolution. Loss of expression of the PTEN ortholog abnormal dauer formation (DAF)-18 in C. elegans or the FOXO ortholog DAF-16 also resulted in alterations in Maf1 expression, with converse changes in RNA pol III transcription [33]. These studies elaborate a new role for FOXO1 in repressing RNA pol III-dependent transcription and uncover another mechanism by which PTEN inhibits this transcription process through its ability to positively control Maf1 expression. Thus, FOXO1 and mTORC1 are important downstream targets of PTEN that play opposing roles in regulating the cellular activity of Maf1 (Figure 1).
Roles of PTEN targets, mTORC1 and FOXO1, in lipid biosynthesis
As a major negative regulator of PI3K/AKT signaling, PTEN plays a fundamental role in both lipid and glucose metabolism [1,2]. The increased de novo lipogenesis, prevalent in tumor cells, has been less studied. Increased cancer cell proliferation requires rapid biosynthesis of lipids to generate new membranes and serve other structural roles. Lipids themselves also serve as signaling molecules and can have a direct effect on cancer cell growth. Furthermore, lipids serve as substrates for PMTs. Collectively, lipids play a prominent role in promoting tumorigenesis [44]. Lipid biosynthesis or lipogenesis occurs by inducing the expression of the lipogenic enzymes. These include fatty acid synthase (Fasn), a rate-limiting enzyme for lipid biosynthesis, and acetyl-coA carboxylase (Acc). Emerging studies demonstrate that inhibition of these enzymes limits the growth of cancer cells and reduces tumorigenesis in various models [45,46]. As a result, small molecule inhibitors of Fasn are currently under development as anticancer therapeutic agents [47–49].
Two key downstream targets of AKT, mTORC1 and FOXO1, play critical and opposing roles in lipogenesis by controlling the expression of stearyl receptor element binding protein (SREBP), a key transcription factor that promotes lipogenic enzyme gene induction. While FOXO1 represses SREBP expression [50], its role in lipid metabolism in mouse models is still ambiguous. AKT phosphorylates FOXO1 at three sites, preventing its nuclear localization and target gene regulation. However, expression of different mutant FOXO1 proteins, which results in nuclear accumulation of FOXO1, displayed opposing phenotypes. Expression of FOXO1 mutated at a single AKT phosphorylation site, S253, led to an increase in intracellular lipid accumulation and fatty liver disease. However, possible changes in lipogenesis were not evaluated [51]. In contrast, mutation of all three AKT phosphorylation sites led to suppression of lipogenic gene expression [52]. The role of FOXO1-mediated regulation of Fasn also differs depending on whether AKT signaling is induced [53]. Overall, the current view is that the transcriptional repression function of FOXO1 is needed to suppress lipogenesis. When FOXO1 is phosphorylated, together with either inactivation or deletion of AKT, lipids can accumulate. In contrast, activation of mTORC1 promotes lipogenesis. This is mediated by the ability of TORC1 to stimulate SREBP expression, processing, and its nuclear localization [54]. However, deletion of the negative regulator of mTORC1, tuberous sclerosis proteins 1 and 2 (TSC1/2), and the resultant hyperactivation of mTORC1, are not sufficient to trigger SREBP activation when AKT is not induced [55]. Together, these results indicate that there are unidentified signals that dictate the ability of FOXO1 and mTORC1 to regulate lipogenesis and that more studies are needed to elucidate their roles in lipid metabolism.
Maf1 regulates lipid metabolism
The idea that FOXO1 and mTORC1 signaling molecules control Maf1 expression and activity, respectively, suggested that Maf1 might regulate the levels of intracellular lipids. When Maf1 is knocked-down in mammalian cell lines, an increase in both the expression of the lipogenesis enzymes Fasn and Acc and intracellular lipid accumulation is observed [32]. Consistent with these results, mice fed a high carbohydrate diet to induce insulin signaling and lipogenesis displayed a significant decrease in Maf1 expression in the liver. Overexpression of Maf1 in the livers of these mice via adenoviral delivery blocked increases in Fasn, Acc and triglycerides. The ability of Maf1 to control intracellular lipid accumulation is conserved. In yeast, Maf1 affects both glucose and lipid homeostasis [56]. Manipulation of Maf1 expression in C. elegans resulted in altered lipid storage in the intestine and hypodermis [33]. This is at least partially a result of altered expression of Fasn and Acc, which occurred concomitantly with the expression of RNA pol III transcripts. As lipid homeostasis is controlled through additional mechanisms that involve degradation and transport, further analysis will be needed to determine whether Maf1 also regulates other aspects of lipid metabolism. Thus, Maf1 is an important downstream target of TORC1 and FOXO1 that contributes to their ability to regulate intracellular lipids. FOXO1 represses lipogenesis by positively regulating Maf1 expression [32,33] and by negatively controlling SREBP expression [50]. In contrast, mTORC1 induces lipogenesis by phosphorylating Maf1 to repress its activity [39–41] and by inducing SREBP expression [54]. Together, these studies identify a new mechanism by which FOXO1 and mTORC1 coordinately control lipogenesis through Maf1 and define a new evolutionarily conserved role for Maf1 as an important downstream effector of these signaling proteins in the regulation of intracellular lipids (Figure 1).
On a molecular level, how does Maf1 negatively regulate lipogenesis? Examination of genes known to promote lipid biogenesis revealed that Maf1 repressed the expression of the lipogenic enzymes Fasn and Acc [32,33]. In mammalian cells, Maf1-mediated repression was shown to involve its recruitment to the Fasn gene promoter without affecting the expression or occupancy of SREBP [32]. Thus, even under conditions in which SREBP expression is induced, Maf1 is able to restrain the activation of the lipogenic genes. While this function of Maf1 is, at least in part, responsible for its ability to regulate intracellular lipid accumulation, the contributions of other Maf1 targeted genes remain to be determined. It will be particularly important to examine whether specific Maf1-mediated changes in RNA pol III-dependent transcription are involved in regulating intracellular lipids.
Lessons from mouse models
Recent studies reported that a whole body knockout of Maf1 in mice resulted in a lean phenotype [37]. The Maf1-deficient mice were found to be resistant to high fat diet-induced obesity and fatty liver disease. Energy expenditure is increased via several mechanisms. Maf1-deficient mice exhibited increased synthesis of precursor tRNAs in a variety of tissues without significant changes in the total amounts of mature tRNAs. Although increases in lipogenesis were observed in the livers of these mice, there was also an increase in lipid consumption through enhanced autophagy, resulting in futile cycling of hepatic lipids [37]. The study suggests that given the high-energy demand for the increased synthesis of these macromolecules, Maf1-deficient cells do not accumulate lipids to the extent seen in wild-type cells. Thus, metabolic efficiency is compromised in the mice lacking Maf1. Interestingly, increased Maf1 expression in C. elegans and decreased intracellular lipid accumulation resulted in reduced fecundity and reproductive success, whereas the same was observed in mice when Maf1 was inactivated [33,37]. These results identify another biological function for Maf1 in reproduction that may be a consequence of its ability to control lipid homeostasis. Mouse models in which Maf1 is inactivated in a tissue-specific manner are needed to identify potential non-cell-autonomous effects of Maf1. Notably, in Drosophia, down-regulation of Maf1 resulted in accelerated development and increased body size. This was due to a non cell-autonomous effect of Maf1 inhibition in the fat body that led to enhanced systemic insulin signaling [57].
Studies have identified energy-dependent nucleolar silencing complex (eNoSC) as an important negative regulator of RNA pol I-transcribed genes that senses cellular energy status to control rRNA synthesis [58]. Upon nutrient deprivation, the energy-dependent change in the NAD+/NADH ratio regulates eNoSC to epigenetically repress rRNA synthesis by establishing heterochromatin [58]. In doing so, eNoSC protects cells from energy deprivation-dependent apoptosis. The nucleomethylin (NML) component of this complex is required for silencing of rDNA genes. Similar to the phenotype observed with the whole body knockout of Maf1, liver-specific knockout of NML led to protection from diet-induced fatty liver and obesity in mice [59]. Given that the rate-limiting step for ribosome biosynthesis is rRNA transcription, which demands major energy expenditure, repression of rRNA synthesis is critical in order to maintain metabolic efficiency. Together, studies from the Maf1- and NML-deficient mouse models support the idea that both proteins coordinately repress genes that are major consumers of nucleotides, which are energetically costly to synthesize. By repressing the synthesis of these RNAs, this then allows for the accumulation of lipids and energy storage. Loss of these proteins, and the resulting high transcription rate of these genes, prevents appropriate control of intracellular energy consumption.
The discovery that Maf1-deficient mice cannot accumulate intracellular lipids under a high fat diet to the same extent as wild-type mice appears incongruent with the results demonstrating that Maf1 inhibits intracellular lipid accumulation in mice fed a high carbohydrate diet. However, there are several potential explanations for these seemingly opposing results. A complete whole body deletion of Maf1 may have different effects compared with a tissue-specific deletion or knockdown of Maf1. The lack of Maf1 during embryonic development could also affect the differentiation of cells that store lipids and result in altered lipid metabolism. Furthermore, long-term versus transient Maf1 deficiency and the different high carbohydrate/high fat diets that might contribute to these phenotypic differences need to be further examined.
Concluding Remarks
Recent studies have defined a new role for Maf1, a key downstream target of PTEN and an established global repressor of RNA pol III-transcribed genes, in lipid metabolism. Maf1 is regulated by PTEN and its downstream targets mTORC1 and FOXO1, and represses genes that dictate translational capacity and the synthesis of lipids de novo. Given that both of these processes are highly dysregulated in cancer, Maf1 is likely a critical component that coordinately represses these gene expression pathways to suppress tumorigenesis (Figure 1). Current studies indicate that both Maf1 and nicotinamide mononucleotide adenylyltransferase (NMNAT1), a positive effector of eNoSC activity, may function as tumor suppressors. Maf1 expression is diminished in human liver cancers [32, 35] while the NMNAT1 locus frequently undergoes heterozygous deletion in human cancers [60]. The ability of Maf1 to control intracellular lipids further suggests that it may also be dysregulated in metabolic diseases, although studies are necessary to assess this possibility. Given the well-established link between obesity and cancer [61,62], and the role of Maf1 in lipid metabolism and oncogenesis, Maf1 likely serves as a molecular link between these diseases.
While Maf1 and components of eNoSC may play central roles in suppressing an oncogenic state, Maf1- or NML-deficiency prevents diet-induced obesity in mice. Thus, it is not yet clear whether strategies to enhance or inhibit Maf1 or NML function will be useful for treating metabolic disease or cancer. Clearly more studies are needed to understand the complex roles of these proteins and the contexts in which they function to regulate lipid metabolism and oncogenesis. Nevertheless, compelling evidence supports the idea that the ability of rRNA and tRNA synthesis to control biosynthetic capacity, once thought to be limited to protein synthesis, now encompasses a critical new function in lipid metabolism. Future studies are needed to determine how changes in the synthesis of these RNAs, through perturbations of the PTEN/PI3K signaling pathway, can influence intracellular lipid accumulation and metabolic disease.
TRENDS BOX.
While epidemiological links exists between obesity and cancer, the basis for this is unclear. Maf1 represents a novel link between lipid metabolism and oncogenesis, providing a molecular explanation for the strong association between these two diseases.
PTEN and downstream targets FOXO1 and mTORC1 play critical roles in lipid biogenesis. Maf1, a global repressor of tRNA and 5S rRNA transcription, is a target of these signaling events and key player in their ability to control intracellular lipid accumulation.
While tRNAs and rRNAs were thought to possess only constitutive functions for protein translation, new studies suggest that their tight regulation is a critical determinant for cells to be able to store lipids. Restraining their synthesis is important for maintaining metabolic efficiency and lipid homeostasis.
Acknowledgments
We would like to thank R. Maxson and members of the laboratory of D. Johnson for helpful discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Menendez JA, Lupu R. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat Rev Cancer. 2007;7:763–777. doi: 10.1038/nrc2222. [DOI] [PubMed] [Google Scholar]
- 2.Baenke F, Peck B, Miess H, Schulze A. Hooked on fat: the role of lipid synthesis in cancer metabolism and tumour development. Dis Model Mech. 2013;6:1353–1363. doi: 10.1242/dmm.011338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Song MS, Salmena L, Pandolfi PP. The functions and regulation of the PTEN tumour suppressor. Nat Rev Mol Cell Biol. 2012;13:283–296. doi: 10.1038/nrm3330. [DOI] [PubMed] [Google Scholar]
- 4.Worby CA, Dixon JE. PTEN. Annu Rev Biochem. 2014;83:641–669. doi: 10.1146/annurev-biochem-082411-113907. [DOI] [PubMed] [Google Scholar]
- 5.Ortega-Molina A, Serrano M. PTEN in cancer, metabolism, and aging. Trends Endocrinol Met. 2013;24:184–189. doi: 10.1016/j.tem.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gottlob K, Majewski N, Kennedy S, Kandel E, Robey RB, Hay N. Inhibition of early apoptotic events by Akt/PKB is dependent on the first committed step of glycolysis and mitochondrial hexokinase. Genes Dev. 2001;15:1406–1418. doi: 10.1101/gad.889901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pastorino JG, Hoek JB, Shulga N. Activation of glycogen synthase kinase 3beta disrupts the binding of hexokinase II to mitochondria by phosphorylating voltage-dependent anion channel and potentiates chemotherapy-induced cytotoxicity. Cancer Res. 2005;65:10545–10554. doi: 10.1158/0008-5472.CAN-05-1925. [DOI] [PubMed] [Google Scholar]
- 8.Sun Q, Chen X, Ma J, Peng H, Wang F, Zha X, Wang Y, Jing Y, Yang H, Chen R, Chang L, Zhang Y, Goto J, Onda H, Chen T, Wang MR, Lu Y, You H, Kwiatkowski D, Zhang H. Mammalian target of rapamycin up-regulation of pyruvate kinase isoenzyme type M2 is critical for aerobic glycolysis and tumor growth. Proc Natl Acad Sci USA. 2011;108:4129–4134. doi: 10.1073/pnas.1014769108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cordero-Espinoza L, Hagen T. Increased concentrations of fructose 2,6-bisphosphate contribute to the Warburg effect in phosphatase and tensin homolog (PTEN)-deficient cells. J Biol Chem. 2013;288:36020–36028. doi: 10.1074/jbc.M113.510289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stiles B, Wang Y, Stahl A, Bassilian S, Lee WP, Kim YJ, Sherwin R, Devaskar S, Lesche R, Magnuson MA, Wu H. Liver-specific deletion of negative regulator Pten results in fatty liver and insulin hypersensitivity. Proc Natl Acad Sci USA. 2004;101:2082–2087. doi: 10.1073/pnas.0308617100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Z, Li J, Bi P, Lu Y, Burcham G, Elzey BD, Ratliff T, Konieczny SF, Ahmad N, Kuang S, Liu X. Plk1 phosphorylation of PTEN causes a tumor-promoting metabolic state. Mol Cell Biol. 2014;34:3642–3661. doi: 10.1128/MCB.00814-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paule MR, White RJ. Survey and summary: transcription by RNA polymerases I and III. Nucleic Acids Res. 2000;28:1283–1298. doi: 10.1093/nar/28.6.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ruggero D, Pandolfi PP. Does the ribosome translate cancer? Nat Rev Cancer. 2003;3:179–192. doi: 10.1038/nrc1015. [DOI] [PubMed] [Google Scholar]
- 14.White RJ. RNA polymerases I and III growth control cancer. Nat Rev Mol Cell Biol. 2005;6:69–78. doi: 10.1038/nrm1551. [DOI] [PubMed] [Google Scholar]
- 15.White RJ. RNA polymerases I and III, non-coding RNAs and cancer. Trends Genet. 2008;24:622–629. doi: 10.1016/j.tig.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 16.Dumay-Odelot H, Durrieu-Gaillard S, Da Silva D, Roeder RG, Teichmann M. Cell growth- and differentiation-dependent regulation of RNA polymerase III transcription. Cell Cycle. 2010;9:3687–3699. doi: 10.4161/cc.9.18.13203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rana T, Misra S, Mittal MK, Farrow AL, Wilson KT, Linton MF, Fazio S, Willis IM, Chaudhuri G. Mechanism of down-regulation of RNA polymerase III-transcribed non-coding RNA genes in macrophages by Leishmania. J Biol Chem. 2011;286:6614–6626. doi: 10.1074/jbc.M110.181735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fairley JA, Mitchell LE, Berg T, Kenneth NS, von Schubert C, Silljé HH, Medema RH, Nigg EA, White RJ. Direct regulation of tRNA and 5S rRNA gene transcription by Polo-like kinase 1. Mol Cell. 2012;45:541–552. doi: 10.1016/j.molcel.2011.11.030. [DOI] [PubMed] [Google Scholar]
- 19.Gjidoda A, Henry RW. RNA polymerase III repression by the retinoblastoma tumor suppressor protein. Biochim Biophys Acta. 2013;1829:385–392. doi: 10.1016/j.bbagrm.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Woiwode A, Johnson SA, Zhong S, Zhang C, Roeder RG, Teichmann M, Johnson DL. PTEN represses RNA polymerase III-dependent transcription by targeting the TFIIIB complex. Mol Cell Biol. 2008;28:4204–4214. doi: 10.1128/MCB.01912-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang C, Comai L, Johnson DL. PTEN represses RNA polymerase I transcription by disrupting the SL1 complex. Mol Cell Biol. 2005;25:6899–6911. doi: 10.1128/MCB.25.16.6899-6911.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mayer C, Zhao J, Yuan X, Grummt I. mTOR-dependent activation of the transcription factor TIF-IA links rRNA synthesis to nutrient availability. Genes Dev. 2004;18:423–434. doi: 10.1101/gad.285504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hannan KM, Brandenburger Y, Jenkins A, Sharkey K, Cavanaugh A, Rothblum L, Moss T, Poortinga G, McArthur GA, Pearson RB, Hannan RD. mTOR-dependent regulation of ribosomal gene transcription requires S6K1 and is mediated by phosphorylation of the carboxy-terminal activation domain of the nucleolar transcription factor UBF. Mol Cell Biol. 2004;23:8862–8877. doi: 10.1128/MCB.23.23.8862-8877.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bywater MJ, Poortinga G, Sanij E, Hein N, Peck A, Cullinane C, Wall M, Cluse L, Drygin D, Anderes K, Huser N, Proffitt C, Bliesath J, Haddach M, Schwaebe MK, Ryckman DM, Rice WG, Schmitt C, Lowe SW, Johnstone RW, Pearson RB, McArthur GA, Hannan RD. Targeting RNA polymerase I with an oral small molecule CX-5461 inhibits ribosomal RNA synthesis and solid tumor growth. Cancer Cell. 2011;22:51–65. doi: 10.1158/0008-5472.CAN-10-1728. [DOI] [PubMed] [Google Scholar]
- 25.Quin JE, Devlin JR, Cameron D, Hannan KM, Pearson RB, Hannan RD. Targeting the nucleolus for cancer intervention. Biochim Biophys Acta. 2014;1842:802–816. doi: 10.1016/j.bbadis.2013.12.009. [DOI] [PubMed] [Google Scholar]
- 26.Poortinga G, Quinn LM, Hannan RD. Targeting RNA polymerase I to treat MYC-driven cancer. Oncogene. 2015;34:403–412. doi: 10.1038/onc.2014.13. [DOI] [PubMed] [Google Scholar]
- 27.Upadhya R, Lee J, Willis IM. Maf1 is an essential mediator of diverse signals that repress RNA polymerase III transcription. Mol Cell. 2002;10:1489–1494. doi: 10.1016/s1097-2765(02)00787-6. [DOI] [PubMed] [Google Scholar]
- 28.Boguta M. Maf1, a general negative regulator of RNA polymerase III in yeast. Biochim Biophys Acta. 2013;1829:376–384. doi: 10.1016/j.bbagrm.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 29.Cabart P, Willis IM. Facilitated recycling protects human RNA polymerase III from repression by Maf1 in vitro. J Biol Chem. 2008;283:36108–36117. doi: 10.1074/jbc.M807538200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vannini A, Ringel R, Kusser AG, Berninghausen O, Kassavetis GA, Cramer P. Molecular basis of RNA polymerase III transcription repression by Maf1. Cell. 2010;143:59–70. doi: 10.1016/j.cell.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 31.Johnson SS, Zhang C, Fromm J, Willis IM, Johnson DL. Mammalian Maf1 is a negative regulator of transcription by all three nuclear RNA polymerases. Mol Cell. 2007;26:367–379. doi: 10.1016/j.molcel.2007.03.021. [DOI] [PubMed] [Google Scholar]
- 32.Palian BM, Rohira AD, Johnson SAS, He L, Zheng N, Dubeau L, Stiles B, Johnson DL. Maf1 is a novel target of PTEN and PI3K signaling that negatively regulates oncogenesis and lipid metabolism. PLoS Genet. 2014;11:e1004789. doi: 10.1371/journal.pgen.1004789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khanna A, Johnson DL, Curran SP. Novel roles for Maf1 in the maintenance of lipid homeostasis. Cell Rep. 2014;9:2180–2191. doi: 10.1016/j.celrep.2014.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee YL, Li YC, Su CH, Chiao CH, Lin IH, Hsu MT. MAF1 represses CDKN1A through a Pol III-dependent mechanism. Elife. 2015;4:e06283. doi: 10.7554/eLife.06283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Y, Kwan TC, Wang S, Li X, Yang Y, Fu L, Huang W, Li M, Wang HY, Zheng SXF. MAF1 Suppresses AKT-mTOR Signaling and Liver Cancer through Activation of PTEN Transcription. Hepatology. 2016 doi: 10.1002/hep.28507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morawiec E, Wichtowska D, Graczyk D, Conesa C, Lefebvre O, Boguta M. Maf1, repressor of tRNA transcription, is involved in the control of gluconeogenetic genes in Saccharomyces cerevisiae. Gene. 2013;526:16–22. doi: 10.1016/j.gene.2013.04.055. [DOI] [PubMed] [Google Scholar]
- 37.Bonhoure N, Byrnes A, Moir RD, Hodroj W, Preitner F, Praz V, Marcelin G, Chua SC, Martinez-Lopez N, Singh R, Moullan N, Auwerx J, Willemin G, Shah H, Hartil K, Vaitheesvaran B, Kurland I, Hernandez N, Willis IM. Loss of the RNA polymerase III repressor MAF1 confers obesity resistance. Genes Dev. 2015;29:934–947. doi: 10.1101/gad.258350.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Orioli A, Praz V, Lhote P, Hernandez N. Human MAF1 targets and represses active RNA polymerase III genes by preventing recruitment rather than inducing long-term transcriptional arrest. Genome Res. 2016 doi: 10.1101/gr.201400.115. gr.201400.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shor B, Wu J, Shakey Q, Toral-Barza L, Shi C, Follettie M, Yu K. Requirement of the mTOR kinase for the regulation of Maf1 phosphorylation and control of RNA polymerase III-dependent transcription in cancer cells. J Biol Chem. 2010;285:15380–15392. doi: 10.1074/jbc.M109.071639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kantidakis T, Ramsbottom BA, Birch JL, Dowding SN, White RJ. mTOR associates with TFIIIC, is found at tRNA and 5S rRNA genes, and targets their repressor Maf1. Proc Natl Acad Sci USA. 2010;107:11823–11828. doi: 10.1073/pnas.1005188107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Michels AA, Robitaille AM, Buczynski-Ruchonnet D, Hodroj W, Reina JH, Hall MN, Hernandez N. mTORC1 directly phosphorylates and regulates human MAF1. Mol Cell Biol. 2010;30:3749–3757. doi: 10.1128/MCB.00319-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rohira AD, Chen CY, Allen J, Johnson DL. Covalent SUMO modification of Maf1 controls RNA polymerase III-dependent transcription repression. J Biol Chem. 2013;288:19288–19295. doi: 10.1074/jbc.M113.473744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Q, Xia N, Li T, Xu Y, Zou Y, Zuo Y, Fan Q, Bawa-Khalfe T, Yeh ET, Cheng J. SUMO-specific protease 1 promotes prostate cancer progression and metastasis. Oncogene. 2013;32:2493–2498. doi: 10.1038/onc.2012.250. [DOI] [PubMed] [Google Scholar]
- 44.Beloribi-Djefaflia S, Vasseur S, Guillaumond F. Lipid metabolic reprogramming in cancer cells. Oncogenesis. 2016;5:e189. doi: 10.1038/oncsis.2015.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Flavin R, Peluso S, Nguyen PL, Loda M. Fatty acid synthase as a potential therapeutic target in cancer. Future Oncol. 2010;6:551–562. doi: 10.2217/fon.10.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jones SF, Infante JR. Molecular Pathways: Fatty Acid Synthase. Clin Cancer Res. 2015;21:5434–5438. doi: 10.1158/1078-0432.CCR-15-0126. [DOI] [PubMed] [Google Scholar]
- 47.Chajes V, Cambot M, Moreau K, Lenoir GM, Joulin V. Acetyl-CoA carboxylase alpha is essential to breast cancer cell survival. Cancer Res. 2006;66:5287–5294. doi: 10.1158/0008-5472.CAN-05-1489. [DOI] [PubMed] [Google Scholar]
- 48.Orita H, Coulter J, Tully E, Kuhajda FP, Gabrielson E. Inhibiting fatty acid synthase for chemoprevention of chemically induced lung tumors. Clin Cancer Res. 2008;14:2458–2464. doi: 10.1158/1078-0432.CCR-07-4177. [DOI] [PubMed] [Google Scholar]
- 49.Oliveras G, Blancafort A, Urruticoechea A, Campuzano O, Gómez-Cabello D, Brugada R, López-Rodríguez ML, Colomer R, Puig T. Novel anti-fatty acid synthase compounds with anti-cancer activity in HER2+ breast cancer. Ann NY Acad Sci. 2010;1210:86–92. doi: 10.1111/j.1749-6632.2010.05777.x. [DOI] [PubMed] [Google Scholar]
- 50.Deng X, Zhang W, O-Sullivan I, Williams JB, Dong Q, Park EA, Raghow R, Unterman TG, Elam MB. FOXO1 inhibits sterol regulatory element-binding protein-1c (SREBP-1c) gene expression via transcription factors Sp1 and SREBP-1c. J Biol Chem. 2012;287:20132–20143. doi: 10.1074/jbc.M112.347211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Qu S, Altomonte J, Perdomo G, He J, Fan Y, Kamagate A, Meseck M, Dong HH. Aberrant Forkhead box O1 function is associated with impaired hepatic metabolism. Endocrinology. 2006;147:5641–5652. doi: 10.1210/en.2006-0541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang W, Patil S, Chauhan B, Guo S, Powell DR, Le J, Klotsas A, Matika R, Xiao X, Franks R, Heidenreich KA, Sajan MP, Farese RV, Stolz DB, Tso P, Koo SH, Montminy M, Unterman TG. FoxO1 regulates multiple metabolic pathways in the liver: effects on gluconeogenic, glycolytic, and lipogenic gene expression. J Biol Chem. 2006;281:10105–10117. doi: 10.1074/jbc.M600272200. [DOI] [PubMed] [Google Scholar]
- 53.He L, Hou X, Kanel G, Zeng N, Galicia V, Wang Y, Yang J, Wu H, Birnbaum MJ, Stiles BL. The critical role of AKT2 in hepatic steatosis induced by PTEN loss. Am J Pathol. 2010;176:2302–2308. doi: 10.2353/ajpath.2010.090931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Porstmann T, Santos CR, Griffiths B, Cully M, Wu M, Leevers S, Griffiths JR, Chung YL, Schulze A. SREBP activity is regulated by mTORC1 and contributes to Akt-dependent cell growth. Cell Metab. 2008;8:224–236. doi: 10.1016/j.cmet.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yecies JL, Zhang HH, Menon S, Liu S, Yecies D, Lipovsky AI, Gorgun C, Kwiatkowski DJ, Hotamisligil GS, Lee CH, Manning BD. Akt stimulates hepatic SREBP1c and lipogenesis through parallel mTORC1-dependent and independent pathways. Cell Metab. 2011;14:21–32. doi: 10.1016/j.cmet.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mierzejewska J, Chreptowicz K. Lack of Maf1 enhances pyruvate kinase activity and fermentative metabolism while influencing lipid homeostasis in Saccharomyces cerevisiae. FEBS Lett. 2016;590:93–100. doi: 10.1002/1873-3468.12033. [DOI] [PubMed] [Google Scholar]
- 57.Rideout EJ, Marshall L, Grewal SS. Drosophila RNA polymerase III repressor Maf1 controls body size and developmental timing by modulating tRNAiMet synthesis and systemic insulin signaling. Proc Natl Acad Sci U S A. 2012;109:1139–1144. doi: 10.1073/pnas.1113311109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Murayama A, Ohmori K, Fujimura A, Minami H, Yasuzawa-Tanaka K, Kuroda T, Oie S, Daitoku H, Okuwaki M, Nagata K, Fukamizu A, Kimura K, Shimizu T, Yanagisawa J. Epigenetic control of rDNA loci in response to intracellular energy status. Cell. 2008;133:627–639. doi: 10.1016/j.cell.2008.03.030. [DOI] [PubMed] [Google Scholar]
- 59.Oie S, Matsuzakim K, Yokoyamam W, Tokunagam S, Wakum T, Hanm SI, Iwasakim N, Mikogai A, Yasuzawa-Tanaka K, Kishimoto H, Hiyoshi H, Nakajima Y, Araki T, Kimura K, Yanagisawa J, Murayama A. Hepatic rRNA transcription regulates high-fat-diet-induced obesity. Cell Rep. 2014;7:807–820. doi: 10.1016/j.celrep.2014.03.038. [DOI] [PubMed] [Google Scholar]
- 60.Song T, Yang L, Kabra N, Chen L, Koomen J, Haura EB, Chen J. The NAD+ synthesis enzyme nicotinamide mononucleotide adenylyltransferase (NMNAT1) regulates ribosomal RNA transcription. J Biol Chem. 2013;288:20908–20917. doi: 10.1074/jbc.M113.470302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Calle EE, Rodriguez C, Walker-Thurmond K, Thun MJ. Overweight, obesity, and mortality from cancer in a prospectively studied cohort of US adults. N Engl J Med. 2003;348:1625–1638. doi: 10.1056/NEJMoa021423. [DOI] [PubMed] [Google Scholar]
- 62.Eheman C, Henley SJ, Ballard-Barbash R, Jacobs EJ, Schymura MJ, Noone A-M, Pan L, Anderson RN, Fulton JE, Kohler BA, Jemal A, Ward E, Plescia M, Ries LAG, Edwards BK. Annual Report to the Nation on the status of cancer, 1975–2008, featuring cancers associated with excess weight and lack of sufficient physical activity. Cancer. 2012;118:2338–2366. doi: 10.1002/cncr.27514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.White RJ. RNA polymerases I and III, non-coding RNAs and cancer. Trends Genet. 2008;24:622–629. doi: 10.1016/j.tig.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 64.Johnson DL, Johnson SA. Cell biology. RNA metabolism and oncogenesis. Science. 2008;320:461–462. doi: 10.1126/science.1158680. [DOI] [PubMed] [Google Scholar]
- 65.Johnson SA, Dubeau L, Johnson DL. Enhanced RNA polymerase III-dependent transcription is required for oncogenic transformation. J Biol Chem. 2008;283:19184–19191. doi: 10.1074/jbc.M802872200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Grewal SS. Why should cancer biologists care about tRNAs? tRNA synthesis, mRNA translation and the control of growth. Biochim Biophys Acta. 2015;1849:898–907. doi: 10.1016/j.bbagrm.2014.12.005. [DOI] [PubMed] [Google Scholar]
- 67.Dittmar KA, Goodenbour JM, Pan T. Tissue-Specific Differences in Human Transfer RNA Expression. PLoS Genet. 2006;2:e221. doi: 10.1371/journal.pgen.0020221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pavon-Eternod M, Gomes S, Geslain R, Dai Q, Rosner MR, Pan T. tRNA over-expression in breast cancer and functional consequences. Nucleic Acids Res. 2009;37:7268–80. doi: 10.1093/nar/gkp787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gingold H, Tehler D, Christoffersen NR, Nielsen MM, Asmar F, Kooistra SM, Christophersen NS, Christensen LL, Borre M, Sørensen KD, Andersen LD, Andersen CL, Hulleman E, Wurdinger T, Ralfkiær E, Helin K, Grønbæk K, Orntoft T, Waszak SM, Dahan O, Pedersen JS, Lund AH, Pilpel Y. A dual program for translation regulation in cellular proliferation and differentiation. Cell. 2014;158:1281–1292. doi: 10.1016/j.cell.2014.08.011. [DOI] [PubMed] [Google Scholar]
- 70.Pavon-Eternod M, Gomes S, Rosner MR, Pan T. Overexpression of initiator methionine tRNA leads to global reprogramming of tRNA expression and increased proliferation in human epithelial cells. RNA. 2013;19:461–466. doi: 10.1261/rna.037507.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Arimbasseri AG, Blewett NH, Iben JR, Lamichhane TN, Cherkasova V, Hafner M, Maraia RJ. RNA Polymerase III Output Is Functionally Linked to tRNA Dimethyl-G26 Modification. PLoS Genet. 2015;11:e1005671. doi: 10.1371/journal.pgen.1005671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Silvera D, Formenti SC, Schneider RJ. Translational control in cancer. Nat Rev Cancer. 2010;10:254–266. doi: 10.1038/nrc2824. [DOI] [PubMed] [Google Scholar]
- 73.Gao B, Roux PP. Translational control by oncogenic signaling pathways. Biochim Biophys Acta. 2015;1849:753–765. doi: 10.1016/j.bbagrm.2014.11.006. [DOI] [PubMed] [Google Scholar]