Abstract
Gene expression changes in the functional absence of a specific RecQ protein, and how that relates to disease outcomes including cancer predisposition and premature aging in RecQ helicase associated syndromes, are poorly understood. Here we describe detailed experimental strategy for identification of RECQ1-regulated transcriptome that led us to uncover a novel association of RECQ1 in regulation of cancer cell migration and invasion. We initiated a focused study to determine whether RECQ1, the most abundant RecQ protein in humans, alters gene expression and also investigated whether RECQ1 binds with G4 motifs predicted to form G-quadruplex structures in the target gene promoters. Rescue of mRNA expression of select RECQ1-downregulated genes harboring G4 motifs required wild-type RECQ1 helicase. However, some RECQ1-regulated genes are also regulated by BLM and WRN proteins regardless of the presence or absence of G4 motifs. The approach described here is applicable for systematic comparison of gene expression signatures of individual RecQ proteins in isogenic background, and to elucidate their participation in transcription regulation through G-quadruplex recognition and/or resolution. Such strategies might also reveal molecular pathways that drive the pathogenesis of cancer and other diseases in specific RecQ deficiency.
Keywords: Gene expression, RecQ, Helicase, G4, cell migration, cell invasion
1. Introduction
Compelling evidence from the growing body of work suggests that activities of the RecQ gene family may influence carcinogenesis [1–3]. In humans, the RecQ helicase family consists of five known members: RECQ1, BLM, WRN, RecQ4, and RecQ5β. Genetic defects in WRN, BLM and RecQ4 give rise to rare diseases Bloom Syndrome, Werner Syndrome and Rothmund-Thomson/ RAPADILINO/ Baller-Gerold Syndromes, respectively. These diseases are each distinguished by clinical features ranging from growth defects (Bloom Syndrome and Rothmund Thomson Syndrome) to premature aging (Werner Syndrome); however, increased genomic instability and predisposition to cancer is a hallmark of RecQ-related diseases [1–3]. Most recently, whole genome sequencing efforts have revealed that mutations in RECQ1 (also known as RECQL or RECQL1) gene predispose individuals to familial breast cancer [4,5]. Thus, RECQ1 is now classified as a breast cancer susceptibility gene. The vast majority of studies on RecQ helicases have focused on investigating their roles in cancer predisposition diseases through DNA repair. In contrast, whether or not these important DNA binding proteins have gene regulatory functions and how that relates to cancer predisposition and other diseases is largely unknown.
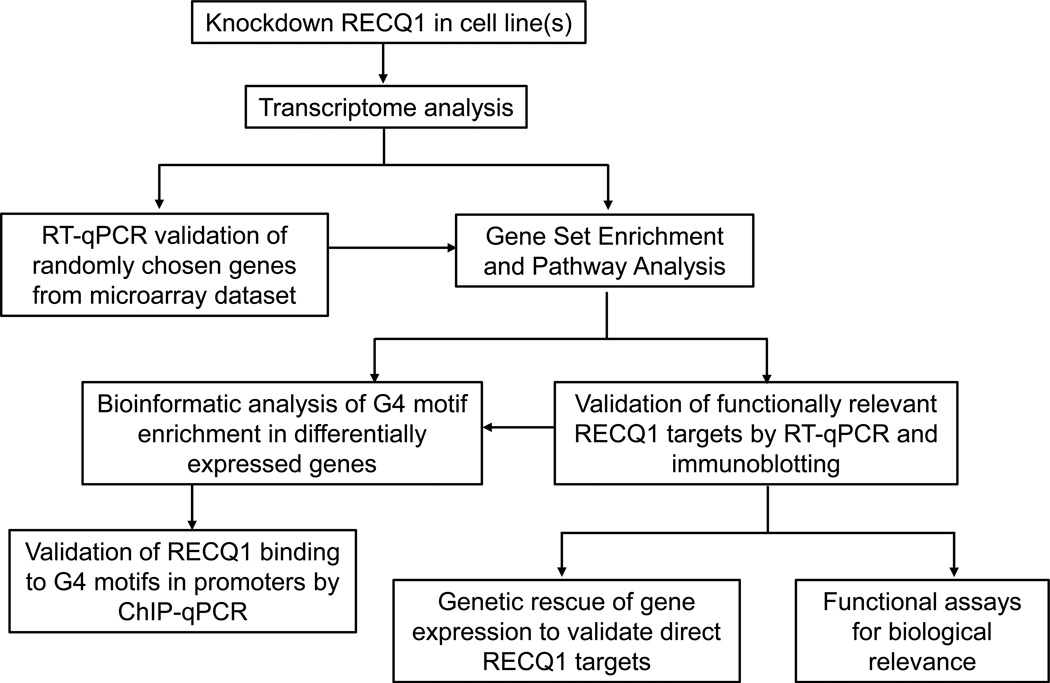
Our previous work demonstrated that the transient loss of RECQ1, the most abundant of the five human RecQ proteins, is sufficient to cause genomic instability and reduce proliferation of cancer cells [6–8]. Subsequent studies elucidated that RECQ1 helicase is a major player in maintaining replication fork progression under stress [9–12]. We have found that RECQ1 is enriched at specific genomic regions that pose significant challenge to replication and transcription, and are hotspots for instability and mutagenesis in cancer [12,13]. We reasoned that RECQ1 could be a multifunctional protein due to its high abundance and specific interactions with chromatin. For instance, a RECQ1 homolog in Neurospora mediates posttranscriptional gene silencing [14]. Rat RECQ1 was identified in a piRNA protein complex important for gene silencing [15]. A recent study implicated human RECQ1 in accurate transcription directed by the HomoID box element-containing promoters [16]. To investigate a potential role of human RECQ1 in gene regulation, we initiated a focused analysis of genome-wide changes in gene expression upon RECQ1 knockdown in HeLa (cervical adenocarcinoma) cells that are frequently used to investigate genome stability functions of RECQ1, and highly invasive MDA-MB-231 breast cancer cells which are widely used for molecular and functional analyses of cell migration, invasion and metastasis (Figure 1) [17].
Figure 1. Experimental approach to determine gene regulatory functions of RECQ1.
We utilized siRNA knockdown of RECQ1 and microarray analysis in our original study [17]. Functions of RECQ1 (or other RecQ proteins) can also be investigated following shRNA-mediated knockdown or CRISPR/Cas9- mediated genetic knockout in cell lines. This approach can also be adapted for transcriptome sequencing (RNA-Seq).
2. Description of method
2.1. Cell culture and transfection
HeLa and MDA-MB-231 were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM) containing 10% fetal bovine serum (FBS). Stable shRNA-mediated knockdown of RECQ1 in HeLa and MDA-MB-231 cells was achieved using a lentiviral system as described [9]. All cells were cultured in a humidified atmosphere containing 5% CO2 at 37°C and routinely checked for mycoplasma contamination (Sigma, catalog no. MP0035). On-Target plus SMARTpool small interfering RNAs (siRNAs) against RECQ1 (NM_032941), WRN (NM_000553), BLM (NM_000057), and non-targeting control (CTL) were purchased from Dharmacon (catalog nos. L-013597-00-0005, L-010378-00-0005, L-007287-00-0005, and D-001810-10-05, respectively). We have previously established the specificity of the siRNA pool [18]. All siRNA transfections were performed by reverse transfection at a final concentration of 20 nM using Lipofectamine RNAiMAX (Invitrogen, catalog no. 13-778-075) as instructed by the manufacturer. For the complementation assays, plasmid transfections for shRNA-resistant RECQ1 expression vectors [9] were performed using Lipofectamine 2000 (Invitrogen, catalog no. 11668019) as instructed by the manufacturer.
2.2. Validation of knockdown by RT-qPCR and immunoblotting
Forty-eight hours after siRNA transfections, total RNA was isolated and whole cell protein lysates were prepared. We consistently observed significant knockdown of RECQ1 mRNA (>80%) and protein (>75%), as measured by reverse transcription followed by quantitative real-time PCR (RT-qPCR) and immunoblotting. Total RNA from cultured cells was isolated by using Aurum Total RNA Mini Kit (Bio-Rad, catalog no. 7326820) as directed by the manufacturer. For RT-qPCR analysis, 1 µg of total RNA was reverse transcribed using the iScript RT kit (Bio-Rad, catalog no. 1708841), and qPCR was performed using iTaq Universal SYBR Green (Bio-Rad, catalog no. 1725121) as directed by manufacturer. To assess the efficiency of knockdown at the protein level, whole-cell lysates were prepared by using radioimmunoprecipitation assay (RIPA) buffer containing protease inhibitor cocktail (Roche, catalog no. 04693132001), and the protein concentration was measured using bicinchoninic acid (BCA) protein quantification kit (Thermo Scientific, catalog no. PI23227). Ten microgram of total protein per lane was loaded on a SDS-PAGE gel and RECQ1 was detected by immunoblotting using anti-RECQ1 antibody (Santa Cruz, catalog no. SC-25547) at 1:1000 dilution. Fifty microgram of total protein per lane was loaded to detect WRN and BLM using anti-WRN (Santa Cruz, catalog no. SC-5629) at 1:500 dilution and anti-BLM (Bethyl Lab, catalog no. A300–110A) antibodies at 1:1000 dilution, respectively. The loading control GAPDH was detected using 1:1000 dilution of anti-GAPDH antibody (Cell Signaling, catalog no. 2118S).
2.3. Transcriptome analysis by microarrays
Forty-eight hour after siRNA transfections in HeLa or MDA-MB-231 cells, total RNA was isolated from triplicate wells of a six well plate using RNeasy minikit (Qiagen, catalog no. 74104). For RNA quality assessment, the RNA integrity number (RIN) was determined for isolated total RNA after running the samples on a Bioanalyzer (Agilent 2100 bioanalyzer). The concentration of the RNA samples was determined using a Nanodrop spectrophotometer (Thermo Scientific). For microarrays, 250 ng total RNA was labeled using Ilumina TotalPrep RNA amplification kit (Applied Biosystems, catalog no. 4393543) as recommended by the manufacturer. Microarrays were performed using a HumanHT-12 v4 Expression Bead Chip kit (Illumina, catalog no. BD-103-0204) and analyzed with the R/Bioconductor packages (Lumi.limma). To identify genes differentially expressed upon knockdown of RECQ1 in MDA-MB-231 cells, we used cut-off of 1.42-fold (>30% down-regulation as compared to control siRNA transfected cells) and also analyzed the microarray data based on false discovery rate (adjusted p<0.05). Knockdown of RECQ1 in MDA-MB-231 cells resulted in many more down-regulated genes as compared to up-regulated genes suggesting that RECQ1 may primarily act as a positive regulator of gene expression in MDA-MB-231 cells. Notably, knockdown of RECQ1 did not significantly alter the mRNA levels of other members of the RECQ family including WRN, BLM, RecQ4 and RecQ5β further confirming that RECQ1 does not regulate the expression of these RecQ proteins and demonstrating the specificity of RECQ1 siRNAs. Using the same criteria that was used for analysis in MDA-MB-231 cells, the number of up-regulated and down-regulated genes was comparable in HeLa cells. We compared the list of differentially expressed genes to examine the overlap between RECQ1-regulated genes in these two cell lines and found that 91 genes were down-regulated and 36 were up-regulated in both MDA-MB-231 and HeLa cells [17]. Our results indicated that though the majority of gene expression changes resulting from RECQ1 knockdown could be cell-type specific, a subset of these changes are common between HeLa and MDA-MB-231 cells.
2.4. Validation of RECQ1-regulated genes by RT-qPCR
To validate the microarray results we randomly selected a subset of differentially regulated genes and measured their mRNA expression in CTL and RECQ1-knockdown cells by RT-qPCR [17]. Consistent with the microarray results, the levels of CD55 and FST mRNAs increased significantly whereas CDK6, CENPA, EBNA1BP2, PTK2, SSR1 and TGFBI mRNAs were significantly down-regulated upon knockdown of RECQ1 in both MDA-MB-231 and HeLa cells. Some RECQ1-activated genes such as CENPA, PTK2, ITGA2, ITGA3 and TGFBR2 were expressed at higher levels in MDA-MB-231 cells which also has higher RECQ1 protein levels than HeLa cells. In contrast, other RECQ1 targets such as EBNA1BP2 and CDK6 were not differentially expressed between HeLa and MDA-MB-231 cells suggesting that the regulation of RECQ1 targets genes in these two diverse cell lines may be mediated by complex mechanisms [17].
2.5. Gene Ontology and Network Analysis
Gene ontology (GO) analysis identifies biological processes that are significantly overrepresented in a gene set. We first selected genes that were differentially expressed at least 1.5-fold (p<0.05) in our microarrays and overrepresented biological processes in the upregulated or downregulated gene set were identified using Ingenuity pathways (http://www.ingenuity.com) [17]. Cellular proliferation was the most significantly over-represented process in the RECQ1-down-regulated gene set which was also consistent with the previously reported reduced proliferation upon knockdown of RECQ1 in various cell lines [6,19]. Although a role of RECQ1 in regulation of cell migration and invasion was not known, we found that the top 10 over-represented processes in both HeLa and MDA-MB-231 cells also included cellular movement and cell morphology. This data suggested that RECQ1 may play a role in regulating cell migration and invasion by maintaining the expression of multiple genes involved in these biological processes, and this effect is not cell-type specific. Among the RECQ1-activated genes are CDK6, CENPA, DICER1, GAS6, SMAD3, IRS1, MPR2, TGFBR2, YES1 (cell cycle progression and proliferation) and ITGA2, ITGA3, ITGB4, RHOB, EZR, VIM, PTK2 (cell adhesion, migration and morphology) [17]. Direct gene network analysis (ingenuity pathways) indicated that many of these above-mentioned genes form a highly connected gene network through protein-protein or protein-DNA interactions [17]. These analyses suggested that RECQ1 may maintain the functionality of a network of genes and multiple pathways to promote proliferation and cellular movement. These results led us to perform functional assays and demonstrate that silencing RECQ1 significantly reduced cell migration (~60%) and invasion (~65%) of MDA-MB-231 cells [17].
2.6. Bioinformatic analysis of G4 motif abundance
RecQ proteins are structure specific helicases that unwind a range of substrates including secondary DNA structures [2,20]. Among the DNA substrates that have been shown to be bound by RECQ1, are the G-quadruplex structures which are formed in vivo by the sequence motif G≥3NxG≥3NxG≥3NxG≥3 (G4 motifs) and function in essential cellular processes including DNA replication and transcription [21–23]. Since G4 motifs are enriched in the promoter regions of many genes, we searched for the G4 motif in the promoters (−1kb to +300bp from transcription start site) of 502 genes down-regulated upon knockdown of RECQ1 in MDA-MB-231 cells [17]. Among the 19,395 promoters of all the protein-coding genes in the human genome, the promoters of 9682 genes (49.9%) had a G4 motif whereas 53.5% of the RECQ1-down-regulated genes had a G4 motif in their promoter.
2.7. Chromatin Immunoprecipitation (ChIP) to determine RECQ1 binding to G4 containing sequences in vivo
We next tested whether RECQ1 binds with the G4 motifs in the promoters of its target genes by ChIP-qPCR of select genes that were down-regulated upon RECQ1 knockdown (CDK6, EBNA1BP2, EZR, ITGA2, ITGA3 and RRAS) [17]. Cells were cultured overnight at a density of 1 × 107 per 15 cm diameter dish. Chromatin and proteins were cross-linked by incubating cells in 1% formaldehyde for 15 min at room temperature and the reaction was stopped by 10 min incubation with 125 mM glycine. Cells were collected and washed sequentially with solution I (10 mM HEPES [pH 7.5], 10 mM EDTA, 0.5 mM EGTA, 0.75% Triton X-100) and solution II (10 mM HEPES [pH 7.5], 200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA). The cell pellets were resuspended in 2 ml lysis buffer (25 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.1% SDS, 1% Triton X-100, 0.5% deoxycholate freshly supplemented with 1× protease inhibitor cocktail (Roche)) and sonicated on ice by 10 s pulses at 30% of maximal power on a Misonix 2000 sonicator (Misonix). This sonication method consistently yielded chromatin fragments corresponding to an average DNA length of 400–1000 bp as assessed on a 1.5% agarose gel. After centrifugation at 20,000× g for 15 min to remove any debris, the supernatant was pre-cleared with 5 µl protein-G-sepharose/salmon sperm DNA beads (Millipore, catalog no. 16–201) at 4°C for 1 h. For each immunoprecipitation, 600 µl (equivalent of 3 × 106 cells) of the pre-cleared chromatin was incubated overnight at 4°C with 3 µg of RECQ1 antibody (Bethyl Lab, catalog no. A300–450A) which we had previously confirmed for immunoprecipitation specificity by immunoblotting. Reaction containing an equivalent amount of rabbit IgG (Vector Lab, catalog no. I-1000) was included as the background control. 10% of the pre-cleared chromatin was set aside as input control. Antibody-chromatin complexes were pulled down by adding 50 µl of protein-G-sepharose/salmon sperm DNA beads and incubated for 2 h at 4°C. The beads were washed for 10 min each with the following solutions: lysis buffer (as mentioned above), high-salt wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl [pH 8.1], 500 mM NaCl), LiCl wash buffer (250 mM LiCl, 1% NP-40, 1% deoxycholate, 1 mM EDTA, 10 mM Tris-HCl [pH 8.0]), and TE buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA). Finally, DNA was eluted with elution buffer (1% SDS, 100 mM NaHCO3). Eluates were incubated at 65°C for overnight with the addition of 5 M NaCl to a final concentration of 200 mM to reverse the formaldehyde cross-linking and digested at 55°C for 3 h with proteinase K at a final concentration of 50 µg per ml. Following phenol/chloroform extraction and ethanol precipitation, sheared DNA fragments served as template in qPCR analysis. qPCR were performed using Taq Universal SYBR Green Supermix (Bio-Rad, catalog no. 1725121) with technical triplicates, and threshold cycle numbers (Ct) were determined with a CFX96 real time PCR detection system (Bio-Rad). Fold enrichment of the targeted genomic sequences were calculated over IgG as: fold enrichment = 2−(CtIP − CtIgG), where CtIP and CtIgG are mean threshold cycles of PCR done in triplicates on DNA samples immunoprecipitated with specific antibody and control IgG, respectively. We used a genomic locus from human chromosome 3 and 17 as negative controls and that from c-myc served as a positive control for G4 motif. The sequences of the qPCR primers are listed in Table 1. All qPCR reactions were also checked by melt curve analyses and agarose gel electrophoresis to confirm the presence of a single specific product. We observed significant enrichment of RECQ1 to the G4 motifs in the promoters of CDK6, EBNA1BP2, EZR, ITGA2, ITGA3 and RRAS genes [17].
Table 1.
Primers used for ChIP-qPCR detection of RECQ1 binding to G4 motifs
| Gene name | Primers for ChIP-qPCR | |
|---|---|---|
| CDK6 | Forward | AAGGAGTCTCGGTTGGAAACGG |
| Reverse | GTGGCGAACACCCCTTTCTCCCAC | |
| EBNA1BP2 | Forward | GAGGGTACAAAGGAAAAGGCAG |
| Reverse | CCAAACAGTTCTGCTGAAGAC | |
| EZR | Forward | GAACACTCAGTGGGGAGGATC |
| Reverse | CTTTCCCCGCCGGGTATCCCTG | |
| ITGA2 | Forward | CTGTACGGGAGCCAAGGTGCGGT |
| Reverse | GTTTCTGGGCAGCTCCTGCAG | |
| ITGA3 | Forward | CAGATGCGCCAATGCCAGGTC |
| Reverse | CAAAGGCTTAGATTTCCCCT | |
| RRAS | Forward | TGCTCATGTCGCCACCGCTGCTG |
| Reverse | CTCCTCCCTTTCTTTGGCCCGTC | |
| Negative CTL 1 | Forward | TAGGCTGGAGGTCGTGGTTA |
| Reverse | CGGCGCTTTCGGATTAACT | |
| Negative CTL 2 | Forward | CTGATGAGCCATGAGGGCGATG |
| Reverse | ATTGGTCCTGTGGTTGGAAACAG | |
| Positive CTL (c-MYC) | Forward | CTACGGAGGAGCAGCAGAGAAAG |
| Reverse | GTGGGGAGGGTGGGGAAGGT | |
2.8. Rescue of mRNA expression of select RECQ1-downregulated genes by RECQ1 helicase
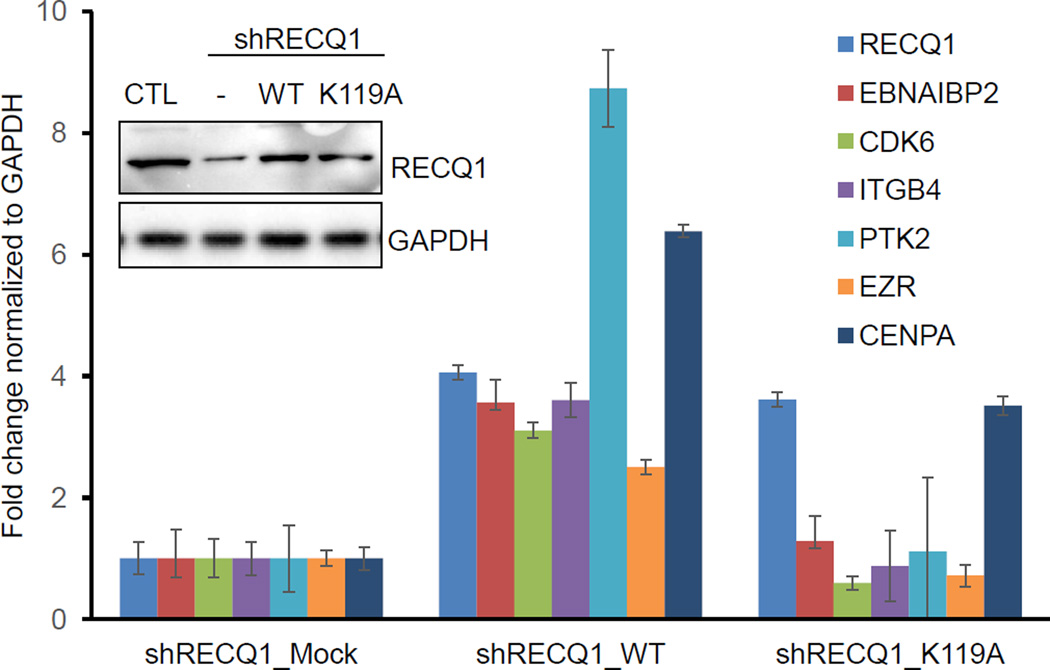
Using MDA-MB-231 cells stably transduced with a CTL (luciferase) or RECQ1 shRNA and mock-transfected with a shRNA resistant RECQ1 expression plasmid, we were able to rescue the expression of select RECQ1-activated genes [17]. Here, we asked whether this rescue of genes containing G4 motifs in their promoter regions required RECQ1 helicase activity. To test this, RECQ1-shRNA transduced MDA-MB-231 cells were mock-transfected or transfected with a RECQ1-shRNA resistant plasmid expressing either the wild-type (WT) or a helicase-dead (K119A) RECQ1 protein [24]. Expression of RECQ1 protein was confirmed by immunoblotting (Figure 2). We performed RT-qPCR to determine the expression of 5 RECQ1-activated genes containing G4 motifs in their promoter regions (CDK6, EBNA1BP2, EZR, ITGB4, and PTK2). The sequences of the qPCR primers are listed in Table 2. We found that reintroduction of WT-RECQ1 upregulated expression of all 5 genes that were downregulated in RECQ1-shRNA knockdown cells whereas the helicase-dead K119A mutant RECQ1 failed to restore the mRNAs levels of these genes (Figure 2). In contrast, mRNA expression of CENPA that lacks G4 motif in its promoter was upregulated by both WT-RECQ1 and K119A mutant RECQ1 (Figure 2). In light of the reports that the recombinant RECQ1 protein did not resolve oligonucleotide based model G-quadruplex substrates [25–27], this is an unexpected result. However, G-quadruplex structures are diverse and their conformations depend on a variety of factors including the number and positions of G in the DNA sequence [21]. Given that RECQ1 is enriched at G4 motifs in the promoter of these genes, our results may indicate potential for RECQ1 unwinding of polymorphic G-quadruplex structures in the genome, the requirement of RECQ1 helicase activity to unwind the intermediates that are generated during structural transition of a G4 motif containing duplex DNA to a quadruplex or during the process of transcription in intact cells. Alternatively, yet unknown accessory factor(s) may enable RECQ1 helicase unwind G-quadruplex structures in vivo. These possibilities remain to be explored and are not mutually exclusive.
Figure 2. Rescue of mRNA expression of select RECQ1-regulated genes harboring G4 motifs in their promoter by wild-type RECQ1.
RECQ1 protein levels were measured by immunoblotting from MDA-MB-231 cells transduced with a CTL or RECQ1 shRNA and mock transfected (−) or transfected with a shRNA resistant plasmid expressing wild-type RECQ1 (WT) or helicase-dead (K119A) mutant RECQ1. GAPDH was used as loading control. Expression of RECQ1 mRNA in shRNA knockdown cells mock transfected (shRECQ1_mock) or transfected with WT (shRECQ1_WT) or mutant (shRECQ1_K119A) was also measured by RT-qPCR. The effect of RECQ1 rescue on the mRNA expression of six RECQ1-regulated genes was measured by RT-qPCR. CDK6, EBNA1BP2, EZR, ITGB4, and PTK2 contain G4 motif in their promoter regions, whereas a G4 motif is absent in CENPA. Error bars represent standard deviation from experiment done in triplicates.
Table 2.
Primers used for gene expression analysis by RT-qPCR
| Gene | Primers for qPCR | |
|---|---|---|
| RECQ1 | Forward | CAATGGCTGGAAAGGAGGTA |
| Reverse | CAGAGTTAAAAGCAGCCCTGGT | |
| WRN | Forward | GGATCAGCACAGTCAGAAAATGTTCT |
| Reverse | GGATAGATTCAGTTTCCTAAGTTCACC | |
| BLM | Forward | GGATCCTGGTTCCGTCCGC |
| Reverse | CCTCAGTCAAATCTATTTGCTCG | |
| EBNA1BP2 | Forward | AGAAAGGCTCAAAGTGGAACA |
| Reverse | CATGACACCAACAGAAGGGAT | |
| CDK6 | Forward | CGTGGTCAGGTTGTTTGATG |
| Reverse | CGGGCTCTGGAACTTTATCC | |
| ITGB4 | Forward | CCACCGAGCCCTTCCT |
| Reverse | GCCGCCTGCCTCCA | |
| PTK2 | Forward | TATATGAGTCCAGAGAATCCAG |
| Reverse | GCTTCACAATATGAGGATGGT | |
| EZR | Forward | GAACAGACCTTTGGCTTGGA |
| Reverse | CACTCCAAGGAAAGCCAATC | |
| CENPA | Forward | TCCGAAAGCTTCAGAAGAGC |
| Reverse | AGGCGTCCTCAAAGAGATGA | |
| ITGA2 | Forward | GCCAATAATCCAAGAGTTGTGTT |
| Reverse | TATTTTCTTGCATATTGAATTGCTTC | |
| ITGA3 | Forward | GCCCACCTGGTGTGACTTCT |
| Reverse | CGTGGTACTTGGGCATGATCT | |
| TGFBR2 | Forward | ATGACGAACTGCAGCATCAC |
| Reverse | GGGTCATGGCAAACTGTCTC | |
| RRAS | Forward | CAAACTTACCAGGGACCAGTG |
| Reverse | TTTCCAGGTCCTGAGCAATC | |
| SMAD3 | Forward | CACCAGGATGAACCACAGCT |
| Reverse | GGCTCGGCAGTAGGTAACTG | |
| GAPDH | Forward | CCCTTCATTGACCTCAACTA |
| Reverse | CCAAAGTTGTCATGGATGAC | |
| B-actin | Forward | ACCAACTGGGACGAT ATGGAGAAGA |
| Reverse | TACGACCAGAGGCATACAGGGACAA | |
2.9. Evaluating the expression levels of select RECQ1 targets in MDA-MB-231 cells following knockdown of WRN and BLM
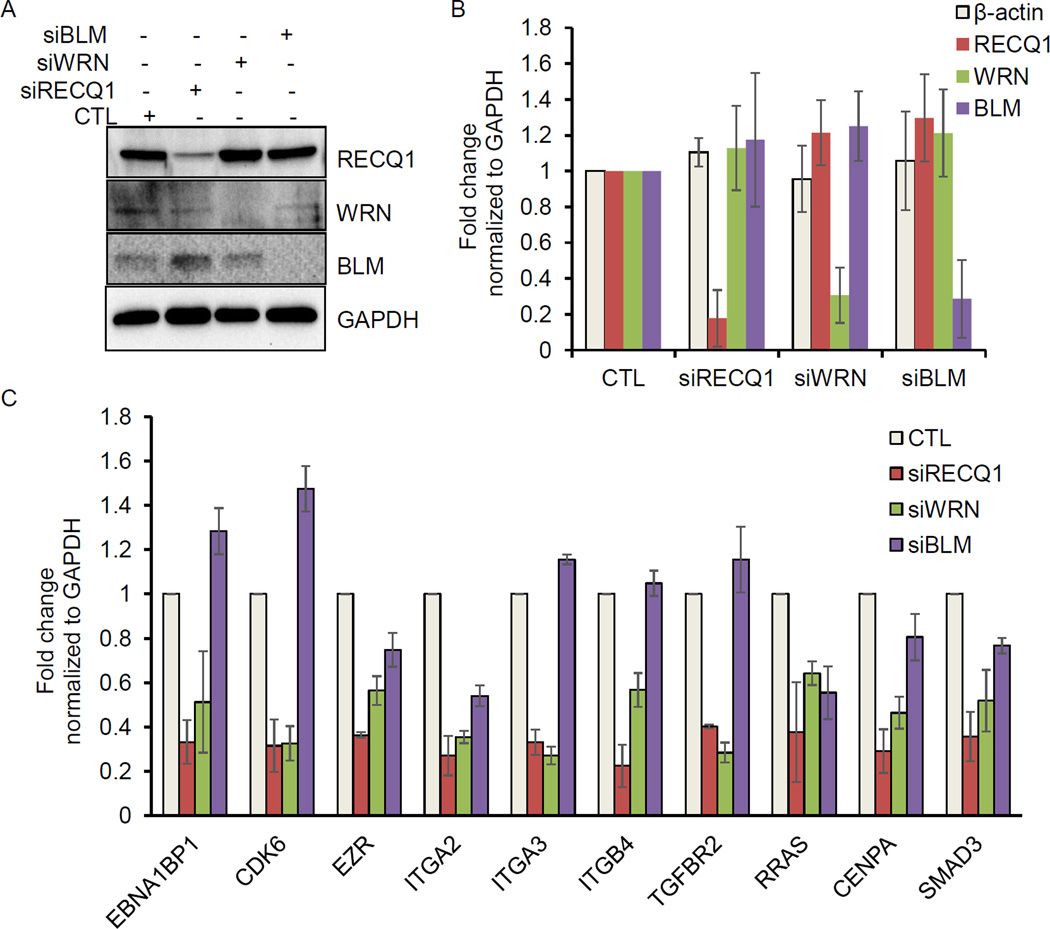
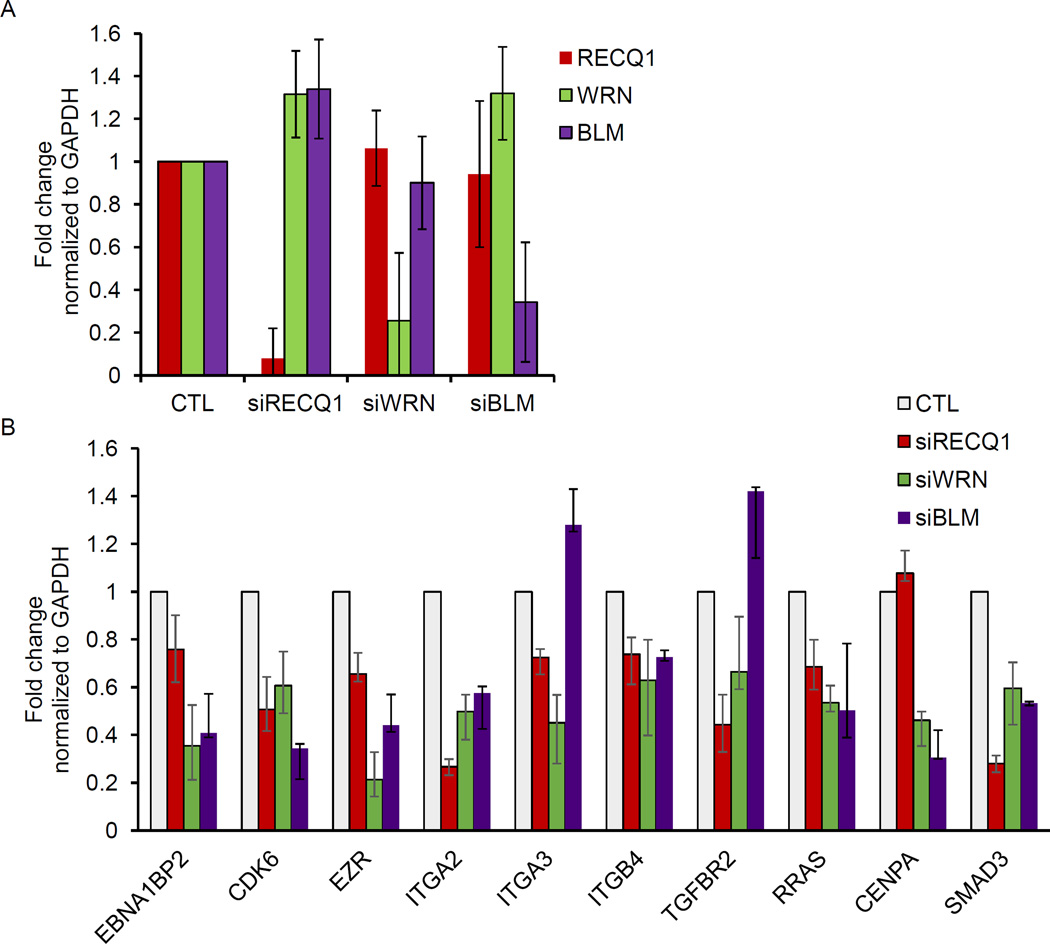
Unlike RECQ1, WRN and BLM helicases have demonstrated G4 resolving activities in vitro [2,26]. Thus, we asked whether these two RecQ proteins impact the expression of select RECQ1-target genes. To test this in an isogenic background, we transfected MDA-MB-231 cells with control siRNAs and siRNAs against RECQ1, WRN or BLM. Immunoblotting from whole-cell lysates prepared 48 hour post-transfection verified specific and efficient knockdown of each of these proteins (Figure 3A). Knockdown of RECQ1, WRN, and BLM mRNA was also confirmed by RT-qPCR (Figure 3B). Next, we took a candidate gene based approach and performed RT-qPCR to determine changes in mRNA expression of 10 genes that we had identified and validated to be downregulated in RECQ1 knockdown cells [17]. Of these 10 candidate genes, 8 genes contain predicted G4 motifs in their promoter regions (EBNA1BP2, CDK6, EZR, ITGA2, ITGA3, ITGB4, TGFBR2 and RRAS) whereas the promoter of 2 genes (CENPA and SMAD3) lack G4 motif. The sequences of the qPCR primers are listed in Table 2. Our results revalidated RECQ1-regulated genes and revealed that WRN or BLM knockdown in MDA-MB-231 also modulated mRNA expression of these genes regardless of the presence or absence of G4 motifs (Figure 3C). Remarkably, CDK6 was downregulated in RECQ1 and WRN knockdown cells whereas it was modestly upregulated in BLM knockdown MDA-MB-231 cells (Figure 3C). In this subset of candidate RECQ1-regulated genes, we noted diverse effects of WRN and BLM knockdown. Expression of 5 out of 10 genes (EBNA1BP2, CDK6, ITGA3, ITGB4, and TGFBR2) was significantly downregulated upon WRN knockdown but remained unaltered or modestly upregulated upon BLM knockdown (Figure 3C). Next, we validated these results in another breast cancer cell line, MCF7 (Figure 4). As compared to control siRNA transfected MCF7 cells, RECQ1 knockdown in MCF7 cells downregulated expression of 9 out of 10 genes (EBNA1BP2, CDK6, EZR, ITGA2, ITGA3, ITGB4, TGFBR2, RRAS and SMAD3) whereas the expression of CENPA did not change. Expression of 8 out of these 10 candidate genes was downregulated upon WRN or BLM knockdown in MCF7 cells, whereas expression of ITGA3 and TGFBR2 was modestly upregulated upon BLM knockdown (Figure 4B). Noteworthy, genes identified as differentially expressed in fibroblasts of Werner and Bloom Syndrome patients are largely non-overlapping [28]. Altogether, this points to both shared and unique targets of RECQ1, WRN and BLM indicating potentially diverse mechanism of gene regulation through these three helicases.
Figure 3. Depletion of WRN or BLM in MDA-MB-231 cells alters mRNA expression of select RECQ1-regulated genes.
MDA-MB-231 cells were transfected with CTL, WRN, BLM or RECQ1 siRNAs for 48 hour and the extent of knockdown was determined by (A) measuring their protein levels by immunoblotting. GAPDH was used as loading control; and (B) measuring their mRNA levels by RT-qPCR normalized to GAPDH. The housekeeping gene β-actin was used as negative control. (C) MDA-MB-231 cells were transfected with CTL or specific RecQ siRNAs (WRN, BLM or RECQ1) for 48 hour and changes in mRNA expression of a subset of RECQ1-regulated genes was validated by RT-qPCR normalized to GAPDH. EBNA1BP2, CDK6, EZR, ITGA2, ITGA3, ITGB4, TGFBR2, and RRAS contain G4 motif in their promoter regions, whereas G4 motif is absent in CENPA and SMAD3. Error bars in B and C represent standard deviation from at least 3 experiments.
Figure 4. Validation of select RECQ1-regulated gene expression changes in MCF-7 cells.
MCF-7 cells were transfected with CTL, WRN, BLM or RECQ1 siRNAs for 48 hour. (A) Knockdown efficiency was determined by measuring their mRNA levels by RT-qPCR normalized to GAPDH. (B) Changes in mRNA expression of a subset of RECQ1-regulated genes were validated by RT-qPCR normalized to GAPDH. Error bars represent standard deviation from 3 experiments.
3. Discussion
Functional defects in RecQ helicases predispose to cancer development. The tumor suppressor roles of RecQ proteins are mediated through their genome caretaker roles [29,30] but the evidence that RecQ proteins directly regulate tumorigenesis is scarce. Our results provide novel evidence for the involvement of RECQ1 in regulation of gene expression in addition to its role in DNA damage repair. Our findings indicate that RECQ1 contributes to tumor development and progression, in part, by regulating the expression of key genes that promote cancer cell migration, invasion and metastasis. The promoters of genes downregulated upon RECQ1 silencing were significantly enriched for G4 motifs which are associated with potential to form G-quadruplex structures. Binding of endogenous RECQ1 to the G4 motifs in the promoters of select downregulated genes suggests that RECQ1 may modulate gene expression by regulating the in vivo stability of G-quadruplex structures. Elegant studies from the Maizels, Harris, and Monnat labs have very recently identified gene regulatory functions of XPB, XPD, BLM, and WRN helicase [31–33]. Regulation of gene expression by these helicases significantly correlated with the presence of G4 motif. This is consistent with an earlier study from the Johnson lab that reported altered gene expression in the Werner and Bloom Syndrome associated with the G4 forming sequences [34]. Similar to XPB, but unlike BLM or WRN, RECQ1 binds but does not unwind G-quadruplex. Inability of a helicase-dead RECQ1 to rescue the expression of select genes containing G4 motifs in their promoter regions provides first, although preliminary, indication that RECQ1 regulation of cancer-associated genes may involve its helicase activity. Collectively these studies suggest that binding to G4 motifs and modulating their stability may be a common mechanism of transcriptional regulation by these distinct DNA helicases. We propose that mechanistically diverse interaction of distinct RecQ helicases with specialized DNA structures regulates specific genes and molecular pathways.
Understanding how the loss of a specific RecQ protein may promote genomic instability and cancer susceptibility is essential to uncover how individual RecQ homologs work in a given cell and in the context of human health. We report here the beginning of a systematic effort to identify unique and common genes and pathways regulated by RECQ1, WRN, and BLM by profiling gene expression changes upon their individual depletion with specific siRNAs in isogenic cellular background. The advantage of transient knockdown with siRNAs is enrichment of direct target genes of these helicases instead of secondary genetic changes that may appear over time in cells with chronic loss of RECQ1, WRN, or BLM. Combining gene expression changes upon individual depletion of each of these RecQ proteins with the identification of their genome-wide binding sites should help identify direct targets of these helicases. Gene expression profiling after transient loss of WRN or BLM may identify unique molecular signatures and pathways that drive the pathogenesis of cancer and other diseases in Werner and Bloom Syndrome. Comparison of RECQ1-regulated transcriptome with that of WRN or BLM may reveal potentially novel relevance of RECQ1 in development and disease.
RECQ1 is overexpressed and amplified in many clinical cancer samples [12]. Recent analysis of publicly available gene expression data also associated RECQ1 expression with tumor progression and prognosis in hematological malignancies [35]. In silico, RECQ1 mRNA expression is associated with cancer cell migration, invasion and metastasis across a panel of human tumor cell lines (NCI-60) derived from nine different tissues of origin [36]. In tissue culture, RECQ1-depleted MDA-MB-231 cells have reduced invasion and migration [17]. Global gene expression changes upon RECQ1 knockdown in MDA-MB-231 cells suggested that RECQ1 enhances the expression of multiple genes that play key roles in cell migration, invasion, and metastasis. Moreover, the expression of a subset of RECQ1-activated genes identified in our study positively correlated with RECQ1 expression in breast cancer patients in TCGA dataset [17]. It has been shown that silencing RECQ1 in normal cells does not significantly affect proliferation but RECQ1 knockdown in multiple cancer cell lines results in reduced proliferation [19]. Comparing and contrasting gene expression changes upon RECQ1 knockdown in a panel of non-tumorigenic versus tumorigenic cell lines may reveal RECQ1-regulated gene networks essential for normal cellular function and cancer progression. Delineating what aspects of RECQ1 catalytic functions contribute to the observed cellular phenotypes, and how this is regulated is critical to establish its biological functions. Overall, such experiments should provide novel insight into the broader roles of RecQ proteins in human health and their specific functions in gene regulation.
Highlights.
Experimental approach for identifying RECQ1-regulated transcriptome is described.
A strategy to find novel cellular functions of RECQ1 through its target genes.
Transcription regulation by RecQ helicases involves G-quadruplex structures.
Systematic comparison of RecQ-regulated gene expression signatures is proposed.
Acknowledgments
This work was funded by the NIGMS/NIH grant 5SC1GM093999-06 to Sudha Sharma and the Intramural Research Program of the National Institutes of Health, National Cancer Institute, and Center for Cancer Research to Ashish Lal. We also acknowledge infrastructure support from the NIMHD/NIH under award number G12MD007597.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest: None
Financial Disclosures: None
References
- 1.Croteau DL, Popuri V, Opresko PL, Bohr VA. Annu. Rev. Biochem. 2014;83:519–552. doi: 10.1146/annurev-biochem-060713-035428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brosh RM., Jr Nat. Rev. Cancer. 2013;13:542–558. doi: 10.1038/nrc3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Monnat RJ., Jr Semin. Cancer Biol. 2010;20:329–339. doi: 10.1016/j.semcancer.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cybulski C, Carrot-Zhang J, Kluzniak W, Rivera B, Kashyap A, Wokolorczyk D, et al. Nat. Genet. 2015;47:643–646. doi: 10.1038/ng.3284. [DOI] [PubMed] [Google Scholar]
- 5.Sun J, Wang Y, Xia Y, Xu Y, Ouyang T, Li J, et al. PLoS Genet. 2015;11:e1005228. doi: 10.1371/journal.pgen.1005228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sharma S, Brosh RM., Jr PLoS One. 2007;2:e1297. doi: 10.1371/journal.pone.0001297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sami F, Sharma S. Comput. Struct. Biotechnol. J. 2013;6:e201303014. doi: 10.5936/csbj.201303014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sharma S, Brosh RM., Jr Cell. Cycle. 2008;7:989–1000. doi: 10.4161/cc.7.8.5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berti M, Ray Chaudhuri A, Thangavel S, Gomathinayagam S, Kenig S, Vujanovic M, et al. Nat. Struct. Mol. Biol. 2013;20:347–354. doi: 10.1038/nsmb.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Popuri V, Croteau DL, Brosh RM, Jr, Bohr VA. Cell. Cycle. 2012;11:4252–4265. doi: 10.4161/cc.22581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berti M, Vindigni A. Nat. Struct. Mol. Biol. 2016;23:103–109. doi: 10.1038/nsmb.3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu X, Parvathaneni S, Hara T, Lal A, Sharma S. Mol. Cancer. 2013;12 doi: 10.1186/1476-4598-12-29. 29-4598-12-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sami F, Lu X, Parvathaneni S, Roy R, Gary RK, Sharma S. Biochem. J. 2015;468:227–244. doi: 10.1042/BJ20141021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cogoni C, Macino G. Science. 1999;286:2342–2344. doi: 10.1126/science.286.5448.2342. [DOI] [PubMed] [Google Scholar]
- 15.Lau NC, Seto AG, Kim J, Kuramochi-Miyagawa S, Nakano T, Bartel DP, et al. Science. 2006;313:363–367. doi: 10.1126/science.1130164. [DOI] [PubMed] [Google Scholar]
- 16.Contreras-Levicoy J, Moreira-Ramos S, Rojas DA, Urbina F, Maldonado E. Gene. 2012;505:318–323. doi: 10.1016/j.gene.2012.05.059. [DOI] [PubMed] [Google Scholar]
- 17.Li XL, Lu X, Parvathaneni S, Bilke S, Zhang H, Thangavel S, et al. Cell. Cycle. 2014;13:2431–2445. doi: 10.4161/cc.29419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sharma S, Phatak P, Stortchevoi A, Jasin M, Larocque JR. DNA Repair (Amst) 2012;11:537–549. doi: 10.1016/j.dnarep.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Futami K, Kumagai E, Makino H, Goto H, Takagi M, Shimamoto A, et al. Cancer. Sci. 2008;99:71–80. doi: 10.1111/j.1349-7006.2007.00647.x. [DOI] [PubMed] [Google Scholar]
- 20.Sharma S. J. Nucleic Acids. 2011;2011:724215. doi: 10.4061/2011/724215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maizels N, Gray LT. PLoS Genet. 2013;9:e1003468. doi: 10.1371/journal.pgen.1003468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huppert JL, Balasubramanian S. Nucleic Acids Res. 2007;35:406–413. doi: 10.1093/nar/gkl1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rhodes D, Lipps HJ. Nucleic Acids Research. 2015;43:8627–8637. doi: 10.1093/nar/gkv862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sharma S, Sommers JA, Choudhary S, Faulkner JK, Cui S, Andreoli L, et al. J. Biol. Chem. 2005;280:28072–28084. doi: 10.1074/jbc.M500264200. [DOI] [PubMed] [Google Scholar]
- 25.Popuri V, Bachrati CZ, Muzzolini L, Mosedale G, Costantini S, Giacomini E, et al. J. Biol. Chem. 2008;283:17766–17776. doi: 10.1074/jbc.M709749200. [DOI] [PubMed] [Google Scholar]
- 26.Mendoza O, Bourdoncle A, Boule JB, Brosh RM, Jr, Mergny JL. Nucleic Acids Res. 2016:1989–2006. doi: 10.1093/nar/gkw079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu Y, Shin-ya K, Brosh RM., Jr Mol. Cell. Biol. 2008;28:4116–4128. doi: 10.1128/MCB.02210-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smestad JA, Maher LJ., 3rd BMC Med. Genet. 2015;16 doi: 10.1186/s12881-015-0236-4. 91-015-0236-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakayama H. Oncogene. 2002;21:9008–9021. doi: 10.1038/sj.onc.1205959. [DOI] [PubMed] [Google Scholar]
- 30.Hickson ID. Nat. Rev. Cancer. 2003;3:169–178. doi: 10.1038/nrc1012. [DOI] [PubMed] [Google Scholar]
- 31.Nguyen GH, Tang W, Robles AI, Beyer RP, Gray LT, Welsh JA, et al. Proc. Natl. Acad. Sci. U.S.A. 2014;111:9905–9910. doi: 10.1073/pnas.1404807111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gray LT, Vallur AC, Eddy J, Maizels N. Nat. Chem. Biol. 2014;10:313–318. doi: 10.1038/nchembio.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tang W, Robles AI, Beyer RP, Gray LT, Nguyen GH, Oshima J, et al. Hum. Mol. Genet. 2016 doi: 10.1093/hmg/ddw079. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson JE, Cao K, Ryvkin P, Wang LS, Johnson FB. Nucleic Acids Res. 2010;38:1114–1122. doi: 10.1093/nar/gkp1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Viziteu E, Kassambara A, Pasero P, Klein B, Moreaux J. Biomark Res. 2016;4 doi: 10.1186/s40364-016-0057-4. 3-016-0057-4. eCollection 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sharma S. Front. Genet. 2014;5:426. doi: 10.3389/fgene.2014.00426. [DOI] [PMC free article] [PubMed] [Google Scholar]