Abstract
Replicative helicases work closely with the replicative DNA polymerases to ensure that the genomic DNA is copied in a timely and error free manner. Both in prokaryotes and eukaryotes, the helicase and DNA polymerase enzymes are functionally and physically coupled at the leading strand replication fork and rely on each other for optimal DNA strand separation and synthesis activities. In this review, we describe pre-steady state kinetic methods to quantify the base pair unwinding-synthesis rate constant, a fundamental parameter to understand how the helicase and polymerase help each other during leading strand replication. We describe a robust method to measure the chemical step size of the helicase-polymerase complex that determines how the two motors are energetically coupled while tracking along the DNA. The 2-aminopurine fluorescence-based methods provide structural information on the leading strand helicase-polymerase complex, such as the distance between the two enzymes, their relative positions at the replication fork, and their roles in fork junction melting. The combined information garnered from these methods informs on the mutual dependencies between the helicase and DNA polymerase enzymes, their stepping mechanism, and their individual functions at the replication fork during leading strand replication.
Keywords: Replicative helicase, DNA polymerase, pre-steady state kinetics, base pair unwinding-synthesis rate constant, chemical step size, structural biology
1. Introduction
Helicases are motor proteins that use the energy from NTP hydrolysis for directional translocation on nucleic acids. This translocation activity of helicases drives numerous nucleic acid metabolic processes in the cell. For example, helicases that move unidirectionally along single-stranded (ss) DNA separate the strands of double-stranded (ds) DNA and aid in DNA replication and repair, whereas those tracking along dsDNA catalyze DNA recombination and genome packaging reactions [1–7]. Given their involvement and importance in practically all nucleic acid metabolic pathways, methods have been developed to understand the fundamental properties of helicases [8–12]. However, in almost all cases, DNA or RNA helicases work in conjunction with associated proteins that affects helicase activities [13, 14]; therefore, it is essential to study the biochemical properties of helicases not only in isolation, but also in the context of the partner proteins.
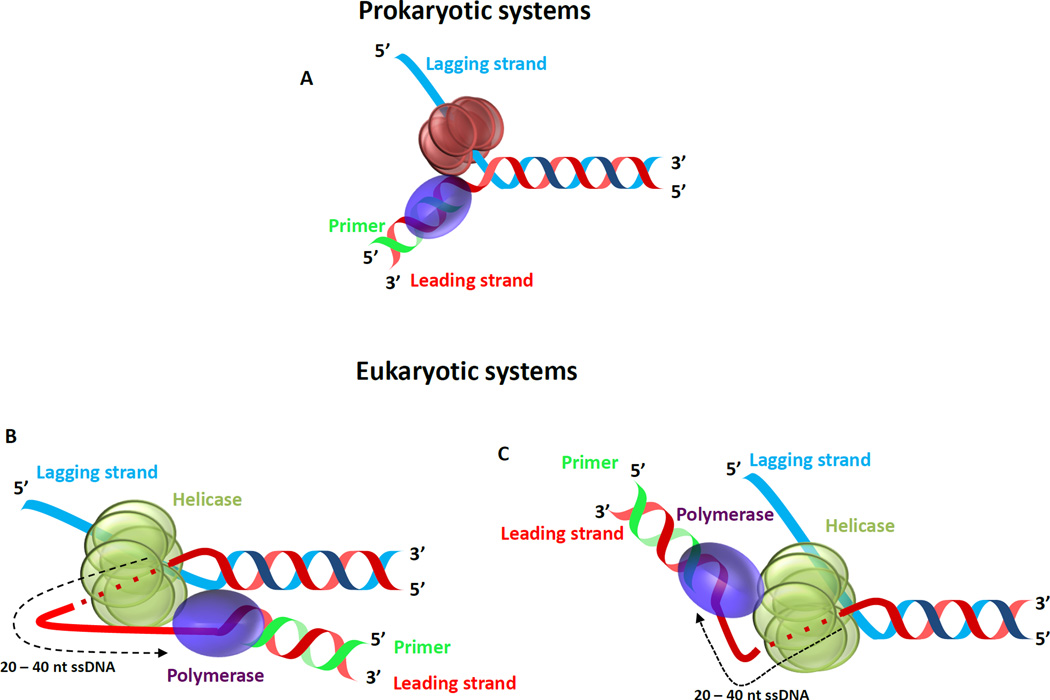
Replicative helicases are ring-shaped hexameric proteins that work with replicative DNA polymerases (DNAP) to catalyze leading-strand DNA synthesis (Figure 1). In prokaryotic systems such as T7 bacteriophage and E. coli, the replicative helicase translocates in the 5’ to 3’ direction and is bound to the lagging strand. Therefore, the helicase and leading strand DNAP in prokaryotes are bound to opposite strands of the DNA (Figure 1A). On the other hand, eukaryotic replicative helicases translocate in the 3’ -> 5’ direction and thus bind to the same strand as the leading strand DNAP (Figure 1B and C). Because the path of the DNA is not known, further studies are required to determine the relative positions of the helicase and DNAP in relation to the fork junction [15]. The helicase and DNAP interact with several accessory proteins, including the polymerase processivity factor, single strand DNA binding protein (SSB), and primase to ensure timely replication of both the strands of the genomic DNA [14, 16–19].
Figure 1. Helicase - DNA polymerase at the replication fork.
(A) Cartoon showing the hexameric ring shaped replicative helicase and the replicative DNA polymerase enzymes bound to a replication fork DNA that represents an intermediate structure during leading strand replication. (B & C) Two proposed models for the architecture of the eukaryotic replisome based on single particle EM studies [15]. Depending on the orientation of the helicase with respect to the fork junction (If the C-terminal domain of the helicase is facing the fork junction the leading strand DNAP is present ahead as in (B) or if the N-terminal domain of the helicase is facing the fork junction the leading strand DNAP is behind the helicase as in (C). Further studies that capture the DNA path are required to identify the correct orientation of the system.
The focus of this review is to describe pre-steady state kinetics and biophysical methods to quantify the coupled activities of the helicase and the leading strand DNAP complex that drives replication fork progression during leading strand DNA synthesis. We describe two pre-steady state kinetic assays to measure the rate constant of DNA base pair (bp) unwinding and synthesis, which is a basic kinetic parameter that when quantified as a function of DNA stability, DNA length, and dNTP concentrations provides a deeper mechanistic understanding of the functional coupling between the helicase and DNAP enzymes. We describe a new way to measure the chemical step-size of the helicase-DNAP complex, which is defined as the number of NTPs hydrolyzed by the helicase for every new base pair unwound and synthesized. The step-size is a fundamental parameter of a translocating motor protein, and in this case, provides information on the energetics and coordinated stepping mechanism of the helicase and DNAP motors at the replication fork. Finally, we focus on methods that provide structural information, such as the positions of the helicase and DNAP with respect to each other on the replication fork and with respect to the fork junction. The 2-aminopurine (2-AP) fluorescence-based methods described here are powerful for determining the relative roles of the helicase and DNAP in replication fork unwinding. Most of these methods were developed using the T7 bacteriophage replisome as a model system (a prokaryotic system), but the methods can be adapted and in principle applied to other prokaryotic and eukaryotic replication complexes.
1.1. Coupled activities of the helicase and DNAP at the replication fork
Replicative helicases and DNAPs on their own move with a fast rate on ssDNA templates, but slow down considerably when the template is dsDNA and needs to be unwound [14] (and references within). The unwinding rate constant of an isolated helicase decreases with increasing stability or GC-content in the dsDNA [20–23]. However, partnering the replicative helicase with the replicative DNAP not only increases the rate constant of DNA unwinding, but the unwinding activity becomes independent of the DNA stability and sequence. Thus, the DNAP helps the helicase by increasing the efficiency of DNA unwinding [24–28]. Reciprocally, the helicase stimulates the strand displacement synthesis activity of the DNAP [26, 27, 29]. The replicative DNAP has limited strand displacement DNA synthesis activity on dsDNA template [27, 29, 30]. The strand displacement activity of the DNAP is stimulated by SSB, but requires high dNTP concentrations to reach values observed with the partnering replicative helicase [29]. On the other hand, coupling DNAP with the replicative helicase enables fast strand displacement DNA synthesis at lower and physiologically appropriate dNTPs concentrations.
The isolated helicase and DNAP enzymes have limited processivity (nucleotide traveled per binding event), whereas the helicase-DNAP complex is highly processive. Part of the reason for the high processivity of the helicase-DNAP complex is physical interactions between the two enzymes. The interdependency between the activities of helicase and DNAP has been observed in many prokaryotic replisomes, including T7, T4, E. coli, human mitochondrial replication systems [24, 25, 28] and more recently in eukaryotic systems [31].
2. Methods to measure the rate constant of base pair melting and DNA synthesis by the helicase-DNAP complex
Given that both helicase and DNAP influence each other’s unwinding-synthesis rates, it is important to develop methods to measure the rate constant of base pair unwinding-synthesis by the helicase-DNAP complex. To measure the kinetics of single base pair unwinding and synthesis, one can monitor the extension of a DNA primer on a replication fork substrate with single nucleotide resolution using sequencing polyacrylamide gel analysis. The kinetics of processive nucleotide addition provides the kinetics of unwinding, because the DNAP depends on the helicase to unwind extended regions of dsDNA beyond ~5 bp [27, 28]. This assay (gel-based) has single base pair resolution, but it is not a high throughput assay; meaning, it is tedious; therefore, we have developed a real time fluorescence-based assay which is comparatively high throughput [29]. Similar assays have been used to measure the unwinding activity of helicases [32, 33]. The fluorescence-based assay does not monitor the primer extension reaction; instead, it measures the separation of the two DNA strands in real time. The analysis of the kinetics of DNA strand separation provides the average bp unwinding-synthesis rate constant of the helicase-DNAP complex.
The DNA unwinding-synthesis reactions by the helicase-DNAP are generally fast and occur in the millisecond time scale; therefore, rapid mixing methods are necessary. The stopped-flow instrument can be used when there is an absorbance or fluorescence change accompanying the reaction. Without such optical changes, one can use radiolabeled substrates and rapid mixing quench-flow instrument, wherein reactions are quenched after predefined time intervals and the labeled products are analyzed subsequently (gel-based assay).
Both the discontinuous gel-based analysis assay and the real time stopped-flow assay and their advantages and disadvantages are described in detail below. The protocols were developed using the T7 bacteriophage helicase and DNAP enzymes and can be modified for the specific enzyme(s) under study.
2.1 Design of the synthetic replication fork DNA substrate
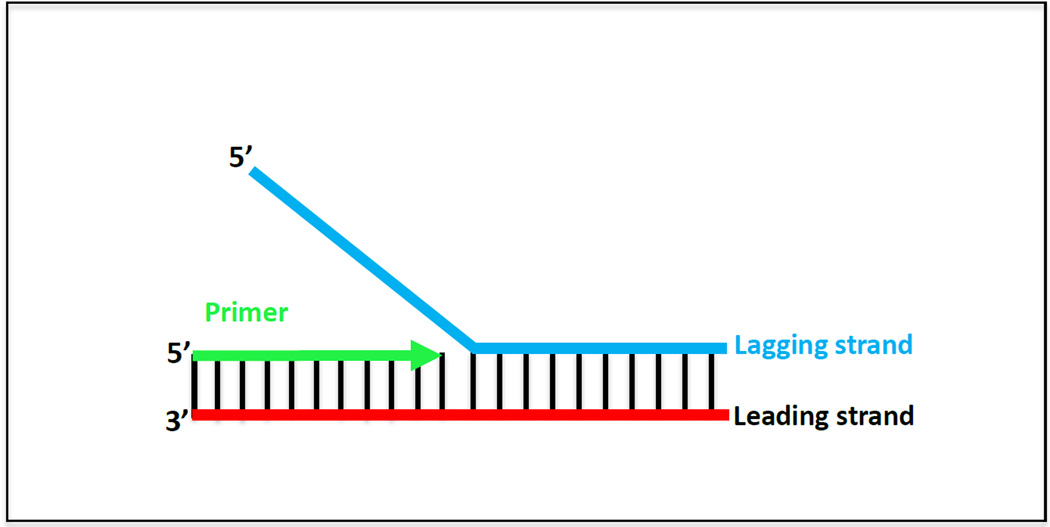
To accurately measure the unwinding-synthesis kinetics and to determine the base pairs unwound-synthesized per second by the helicase-DNAP, short preformed replication fork DNAs are used that mimic a unidirectional replication fork (Figure 2). The linear dsDNA region to be unwound-copied has to be sufficiently long (between 30 – 60 bp or longer) and stable to ensure that the two strands do not fall apart in the absence of enzymes. When designing the substrates, sequences that form secondary structures and sequences the helicase and DNAP may specifically interact with must be avoided. To study the effects of dsDNA stability on the activities of the helicase and DNAP, the sequence of the dsDNA region can be designed to contain varying percentages of GC bp (0% GC-content least stable and 100% GC most stable).
Figure 2. Replication Fork Substrate.
Cartoon of the replication fork DNA mimic made from chemically synthesized oligodeoxynucleotides. The lagging strand DNA is shown in blue, the leading strand DNA in red and the primer strand in green.
The ssDNA overhangs in the fork DNA enable loading of the helicase and DNAP. For prokaryotic systems (such as T4 and T7 helicases and E. coli DnaB), the 5’-ssDNA overhang serves as a platform for the assembly of the hexameric helicase. The length of the 5’ and 3’ overhangs should be experimentally optimized by comparing the speed and processivity of unwinding-synthesis using overhangs of different lengths for the specific helicase and DNAP under study. The primer to be extended by the DNAP is annealed to the 3’-ssDNA overhang (Figure 2). The T7 helicase requires a 35-nt 5’ overhang [34] and the T7 DNAP functions well with a 24-nt primer, although longer primers can be used. This works generally for most prokaryotic replisomes, but an estimate for the primer length can be obtained from footprinting experiments or from existing crystal structures of the DNAP and adjusted for optimal primer extension.
Similar synthetic fork substrates can be used to study eukaryotic helicases and DNAPs [35]. The eukaryotic replicative helicases translocate in the 3’ -> 5’ direction and hence bind to the same strand as the leading strand DNAP. Thus the DNA substrates are designed to contain ssDNA region between the primer template junction and fork junction to accommodate the helicase at the replication fork. While this design of DNA substrate has been used to study the functional coupling between the eukaryotic helicase and DNAP, it is currently unclear whether the helicase or the DNAP is the leading motor in eukaryotic systems [15].
2.2. Gel-based assay to measure the single base pair unwinding-synthesis rate constant of the helicase-DNAP
2.2.1 Fork DNA
The replication fork substrate is created by mixing purified deoxyoligonucleotide strands, corresponding to the lagging and leading strands, and the primer strand in stoichiometric amounts. Oligonucleotides are typically purified by denaturing PAGE, extracted from the gel by electro-elution, and ethanol-precipitated. The absorbance of the oligonucleotide is measured at 260 nm and the concentration is calculated using the extinction coefficients. For the gel-based assays, the 5’-end of the primer in the fork DNA is radiolabeled with [γ-32Pi] or the primer is synthesized with a fluorophore (commonly, fluorescein) covalently attached to the 5’ end (Figure 3A). To generate the replication fork substrate, the oligonucleotides are mixed together in Tris-EDTA (TE) buffer (pH 8.0) + 50 mM NaCl, heated to 95 °C for 10 minutes and slowly cooled to room temperature. The correct ratio of the strands is determined by titrating the leading and lagging strands and checking the products on a non-denaturing PAGE using the Stains-All dye (Sigma Aldrich) to visualize the DNA.
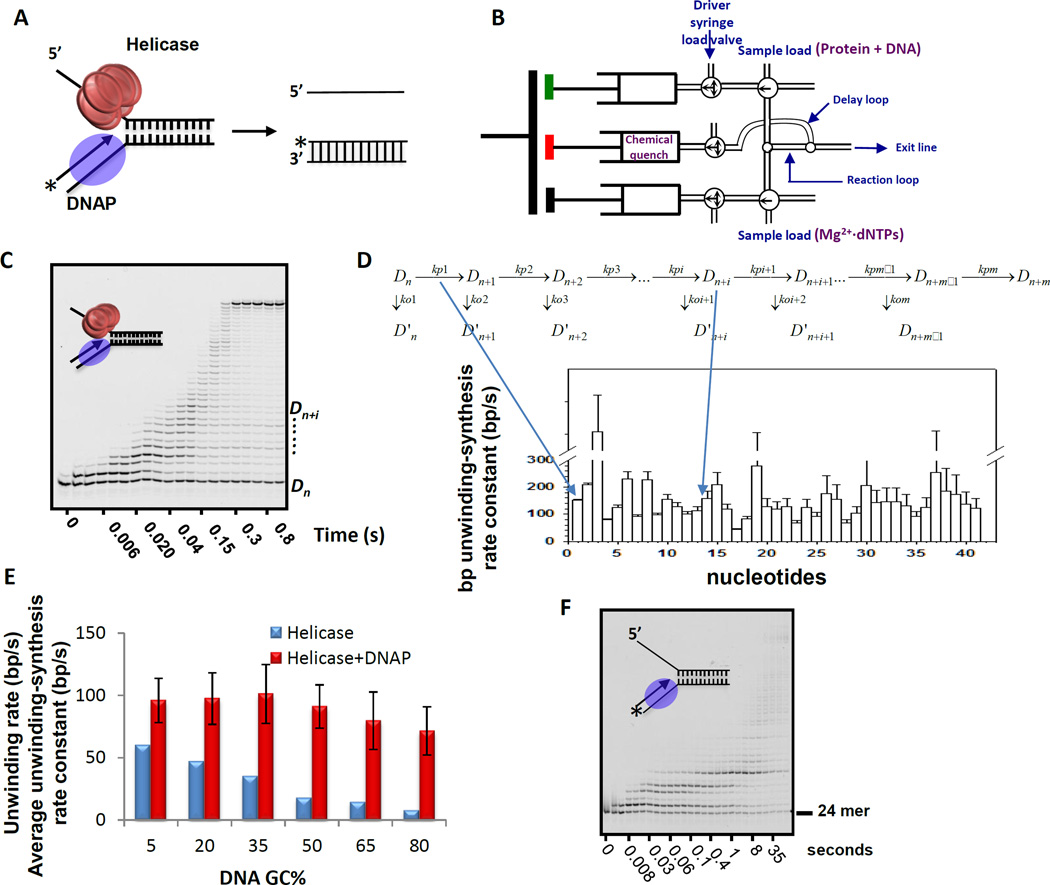
Figure 3. Gel-based assay to determine the base pair unwinding-synthesis rate constant of the helicase-DNAP complex.
(A) The helicase-DNAP complex is assembled on the replication fork DNA. The 5’-end of the 24-mer primer in the fork DNA is labeled with a fluorophore or γ[32Pi]. The unwinding-synthesis reaction initiated with dNTPs is monitored by following the extension of the labeled DNA primer. (B) Schematic of the rapid quench-flow instrument for rapid mixing and quenching of the reactions with millsecond time resolution. The duration of the reaction is changed by adjusting the volume of the delay line and the flow rate through the delay loop. (C) Image of polyacrylamide/urea sequencing gel showing extension of the 24-mer DNA primer by T7 helicase-DNAP (1 mM dNTPs) at 18 °C. The T7 helicase is the product of T7 gp4, which contains both helicase and primase activities[58]. T7 DNAP is a 1:1 complex of polymerase T7 gp5 and processivity factor E. coli thioredoxin[59]. (D) The polymerization model is used to globally fit the formation and decay of each primer extension product to obtain the individual base pair unwinding-synthesis rate constants. The kp1, kp2, etc. are the rate constants for the formation of Dn+1, Dn+2, etc. extension products, respectively. The ko1, ko2, etc. are the rate constants for the dissociation of the DNAP from the Dn+1, Dn+2, etc. complex intermediates. The computer fitting provides the rate constants for the individual nucleotide addition reactions (kp1, kp2, etc.) from which one can calculate the average single base pair unwinding synthesis rate constant. (E) The average unwinding-synthesis rate constant of the helicase-DNAP is compared to the helicase’s rate constant of unwinding on fork DNA substrates with 5–80% GC-content. (F) The sequencing gel image shows the strand-displacement DNA synthesis activity of the isolated T7 DNAP (1 mM dNTPs) at 18 °C.
2.2.2 Rapid quench-flow assay
The reactions are carried out in a temperature controlled rapid quench-flow instrument (Kintek Corp, Austin, TX) (Figure 3B). The quench-flow apparatus has three syringes of which two are drive syringes that mix the reaction components whereas the third syringe contains a chemical-quench to stop the reaction after defined time intervals. The reaction time is adjusted by changing the length of the delay loop. A circulating water bath is used to maintain the chamber at the desired reaction temperature.
The helicase and DNAP are preassembled on the fork DNA (with radiolabeled or fluorophore-labeled primer) typically in equimolar amounts (20 to 100 nM each of DNA and proteins) and loaded in one syringe of the instrument (Figure 3B). T7 helicase requires dTTP to form a ring around DNA and does not require Mg2+ for assembly; hence, it is preassembled on the fork DNA using dTTP without Mg2+ [36]. Leaving out Mg2+ and adding 2 mM EDTA to chelate contaminating divalent metal ions assures that helicase assembly occurs without dTTP hydrolysis. Most helicases use ATP and may require special conditions for assembly and loading on the DNA [37] and these requirements must be considered during the pre-incubation of the enzymes with the DNA. The lack of Mg2+ also prevents DNA synthesis by the DNAP in the assembly mixture.
The reactions are initiated by rapidly mixing equal volumes of the pre-incubated enzyme-DNA complex and a mixture of dNTPs and Mg2+, and after specified time intervals, the reactions are stopped using a chemical quench (such as EDTA). The quenched reactions are mixed with the sequencing gel-loading dye, and the products are resolved on a high-resolution polyacrylamide gel (24% acrylamide/7 M urea) with 1.5× TBE buffer (Figure 3C). Using a 1:6 ratio of sample to dye, boiling the sample, and heating the gel-running buffer on the top chamber of the sequencing gel apparatus helps in improving the resolution, especially for high GC-content DNA substrates.
2.2.3 Analysis and data fitting to determine the rate constant of bp unwinding-synthesis
The DNA primer extension products are resolved on a high-resolution denaturing polyacrylamide gel and visualized using a Typhoon phosphorimager (GE Healthcare). Each band at each time point is quantified using the ImageQuant software (GE healthcare). To obtain the single bp unwinding-synthesis rate constant, the formation and decay of each primer extension product at every time point is globally fit to the polymerization model (Figure 3D) using gfit [38] in MATLAB, as described in detail previously [39] or using the Kintek explorer software (KinTek Corp).
This analysis provides the single bp unwinding-synthesis rate constant (kp1, kp2, etc. in Figure 3D) from which one can calculate the average rate constant of unwinding-synthesis over the given length of dsDNA. T7 helicase-DNAP catalyzes unwinding-synthesis reaction with an average rate constant of ~100 nt/s over 40 bp dsDNA, and this is constant and nearly independent of the GC-content from 5% to 50% in the dsDNA (Figure 3E - red bars). The rate constant slightly decreases when the GC-content in the dsDNA increases to 65% and 80%. We compare the average unwinding-synthesis rate constants of the helicase-DNAP to the bp unwinding rate constants of the isolated helicase (Figure 3E – blue bars). The isolated helicase’s rate constant is highly sensitive to the GC-content, whereas the helicase-DNAP’s is mildly sensitive to the GC-content.
2.2.4 Unwinding-synthesis rate constant – A quantitative measure of mutual dependency of helicase and DNAP
Without T7 helicase, the T7 DNAP stalls after adding about 5 nucleotides to the DNA primer (Figure 3F). This indicates that T7 DNAP requires the helicase to copy long stretches of dsDNA, and thus the DNAP depends on the helicase for processive strand displacement DNA synthesis. Similarly, the isolated T7 helicase can unwind the fork DNA, but the unwinding kinetics are slow (Figure 3F – blue bars), and moreover the rate constant of unwinding decreases with increasing GC-content in the dsDNA When T7 helicase is coupled with the T7 DNAP, the two are able to unwind-copy longer stretches of dsDNA and at a faster rate constant resembling the observed leading strand synthesis rate constants ([40]. The enhancement from helicase-DNAP coupling on the 5% GC-rich dsDNA is about 2-fold, but the enhancement on 65% and 80% GC-rich DNA is up to 8-fold, showing that the coupling between helicase and DNAP is more critical when the DNA sequence is GC-rich. These set of experiments demonstrate that accurate measurement of the bp unwinding/synthesis rate constants provides quantitative evidence for the mutual dependency between the helicase and DNAP enzymes. Further insights into the coupling mechanism is obtained when the bp unwinding-synthesis kinetics are measured as a function of dNTPs concentration (Section 2.3.4).
2.3 Real-time fluorescence-based assay to measure the kinetics of unwinding-synthesis by helicase-DNAP
2.3.1 Fork DNA
The leading and lagging strands of the replication fork substrate are chemically attached to a fluorophore-quencher pair (such as fluorescein and black hole quencher-1 (BHQ-1)). The fluorescein is attached at the 3’ end of the lagging strand and BHQ-1 at the 5’ end of the leading strand (Figure 4A). When the fork DNA is duplexed, the probes at the blunt end of the fork DNA are in close proximity and the fluorescence is quenched. When the fork DNA is unwound and the leading strand is fully copied, the fluorophore strand is separated from the quencher strand and the fluorescence increases. The fluorophore-labeled oligonucleotides can be obtained commercially from Biosearch Technologies (Novato, CA) or IDT (Coralville, IA). During the preparation of the fork DNA, the quencher-labeled strand is added in slight excess to ensure that the fluorescence of the fluorophore labeled strand is completely quenched. The correct annealing ratio is determined by titrating the two DNA strands and checking the annealed product by non-denaturing PAGE and staining the DNA with the Stains-All (Sigma Aldrich) dye.
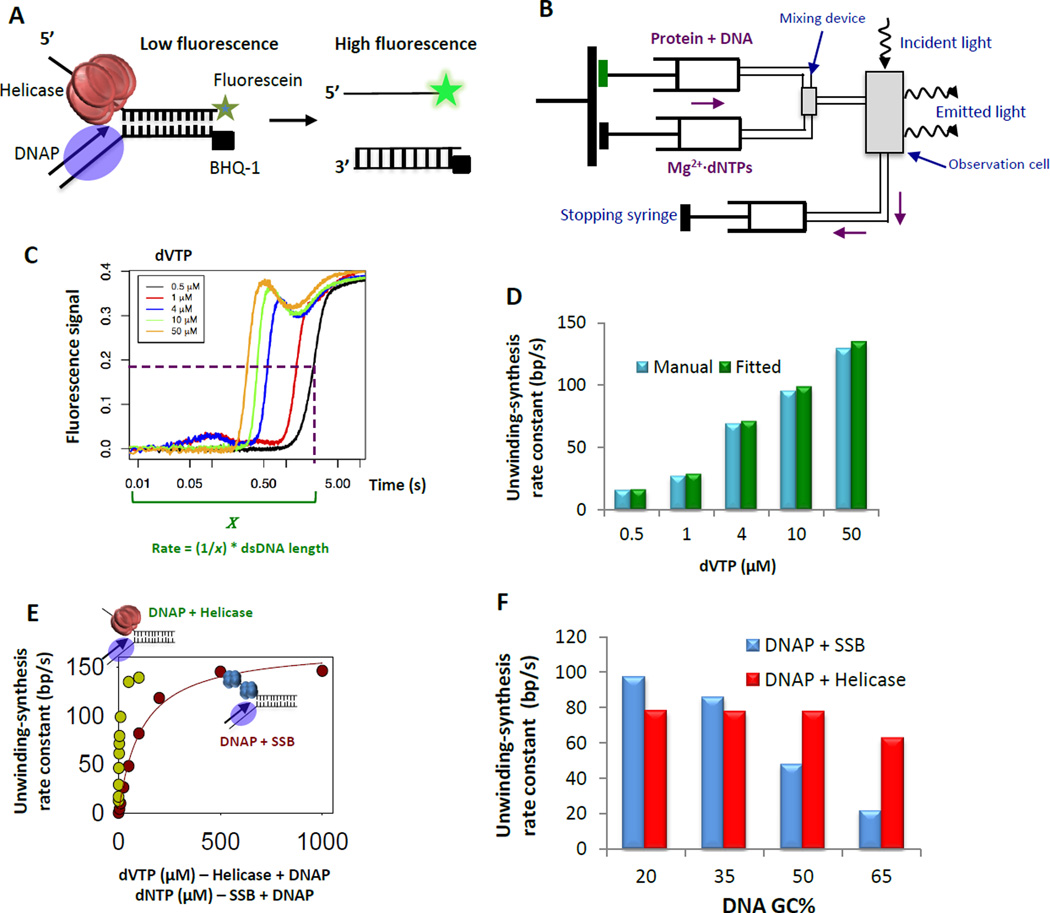
Figure 4. Stopped-flow fluorescence assay to determine the base pair unwinding synthesis kinetics.
(A) The replication fork DNA is labeled with the fluorescein fluorophore at the 3’ end of the lagging strand and BHQ-1 quencher at the 5’ end of the leading strand. The fluorescence intensity is quenched when the replication fork DNA is duplexed and increases when the strands are separated by the unwinding-synthesis activity of the helicase-DNAP (B) Schematic of the stopped-flow instrument (KinTek Instrument) that allows rapid mixing of small volumes of reactants and real-time fluorescence intensity measurements with close to 1 ms dead time under temperature controlled conditions. (C) Kinetic traces of 50% GC fork DNA strand separation reaction at 18 °C by the unwinding-synthesis activities of the helicase-DNAP at increasing concentrations of dVTP (dATP + dCTP + dGTP) and 500 µM dTTP (for helicase assembly). The lag represents the time taken to unwind-copy the dsDNA region of the fork DNA, and this lag time decreases as the dNTP concentration (DNAP substrate) increases. The dip in fluorescence signal following the initial increase is potentially due to interaction of the fluorophore with the proteins as they reach the end of the DNA strand [23]. The ‘x’ is time to reach half-maximal fluorescence increase. (D) The unwinding-synthesis rate constants can be estimated by fitting the kinetic data in panel C to the stepping model (green bars) or by multiplying the inverse of the lag time (x) and the dsDNA length (blue bars). (E) Unwinding rate constants of T7 DNAP with SSB (red circles) or with helicase (green circles) with increasing concentrations of dNTPs. (E) The unwinding rate constants of T7 DNAP with SSB (blue bars) or with helicase (red bars) as a function of increasing GC content (5–65%) in the dsDNA region.
2.3.2 Stopped-flow assay
The real-time unwinding-synthesis assays are carried out in a stopped-flow instrument (Kintek Corp, Austin, TX) (Figure 4B). The stopped-flow apparatus has two motor controlled syringes that hold the reaction components, a rapid mixing device that rapidly mixes small volumes of the reactants, and an observation cell where the measurements are made. The circulating water bath is set up at a constant reaction temperature of choice as changes in temperature affect both the enzyme activity and the fluorescence signal. The excitation wavelength and emission filter to be used depends on the properties of the fluorophore attached to the DNA substrate. An excitation wavelength of 480 nm and a 515 nm long pass cut-off filter were used for fluorescein-BHQ-1 fluorophore-quencher pair.
As described in Section 2.2.2, the enzymes are pre-assembled on the DNA substrate prior to reaction initiation. Typically, equimolar amounts of enzymes and DNA substrate (in the range of 10 to 100 nM) are good starting concentrations. Excess enzymes can bind non-specifically to the fluorophore or the quencher and affect the fluorescence signal in unexpected ways. Hence, the enzyme concentrations and the incubation time for assembly should be optimized experimentally. The unwinding-synthesis reaction is initiated by rapidly mixing the pre-assembled enzyme fork DNA mixture from one syringe and a mixture of dNTPs, Mg2+ and excess of ssDNA (dT90) trap from the second syringe, and the fluorescence is measured in real time. The dT90 ssDNA binds to the dissociated helicase molecules preventing them from rebinding the fork or the unwound strands. The resulting fluorescence versus time kinetic traces show a lag followed by an exponential increase in fluorescence (Figure 4C). In this case, the enzymes arriving at the end of the fork DNA quenches the fluorescein fluorescence; hence a dip in fluorescence is observed before the fluorescence plateaus.
2.3.3. Fitting the real-time unwinding-synthesis kinetics
The fluorescence based unwinding-synthesis assay is an all-or-none assay, meaning that the fluorescence increase is observed only after the entire length of the dsDNA is unwoundcopied. Therefore, the fluorescence kinetic trace shows an initial lag that represents the time to unwind-copy the dsDNA region before the strands become fully separated. This type of lag as opposed to a lag from a slow initiation step can be verified by measuring the kinetics with fork DNAs of increasing dsDNA lengths [20]. If the lag time truly represents the time taken to unwind the dsDNA, the lag time should increase with increasing dsDNA length in the fork DNA. If the unwinding-synthesis reaction occurs through synchronized separation of the lagging strand and copying of the leading strand, then the fluorescence intensity should increase with a steep slope. A shallow slope indicates enzyme populations unwinding-synthesizing with different kinetics and this could be due to many reasons, including multiple populations of the initial complex or stalling during unwinding-synthesis [38, 39].
The unwinding-synthesis kinetics as measured by the fluorescence change is fit to the stepping model to obtain the average single bp unwinding-synthesis rate constant (Figure 4C)[38]. The same stepping model was used to determine the base pair unwinding rate constant of the isolated helicase, and its theory and protocol are reviewed in detail [38, 39]. A quick estimate of the unwinding-synthesis rate constant can also be obtained from the time corresponding to the mid-point of the increasing portion of the fluorescence trace (Figure 4C – time x). The rate constant in this case is obtained by multiplying the inverse of the time (1/x) and the length of the dsDNA region.
Where x is the time corresponding to the mid-point of the increasing portion of the fluorescence trace. Other software such as GraFit [41] can also be used to fit the lag and subsequent increase in fluorescence. The manual and fitted rate constants are very close in values (Figure 4D).
2.3.4 Roles of the helicase and DNAP in leading strand DNA synthesis
The real-time unwinding-synthesis stopped flow assay provides a relatively easy way to explore dNTP (substrates of DNAP) concentration effects, offering deeper insights into the mechanism of helicase-DNAP coupling. As shown by the gel-assay, the isolated T7 DNAP cannot fully unwind-copy the 40 bp fork DNA (Figure 3F), but this reaction is efficient in the presence of T7 helicase or E. coli SSB [27]. The unwinding-synthesis rate constants of both the helicase-DNAP and SSB-DNAP increase in a hyperbolic manner with increasing dNTP concentrations, and fitting the data to the Michaelis Menton kinetics provides the maximum unwinding-synthesis rate constant (kcat) and the apparent Kd of dNTPs (Figure 4D). The fittings show that in the presence of SSB, T7 DNAP can unwind-copy the dsDNA with a fast maximum rate constant of 166 bp/s just like in the presence of helicase, but with an apparent dNTPs Kd of 124 µM, which is 24 times higher than with the helicase (~4 µM) (Figure 4E). As detailed below, such insights cannot be obtained from kinetic measurements at a single dNTP concentration. Similarly, the kinetics of fork DNAs of different GC-content show that the SSB-supported strand displacement DNA synthesis is sensitive to the GC-content of the DNA, whereas the helicase-supported activity is GC-independent (Figure 4F). Additionally, the real time assay can be used to explore how dTTP concentrations (substrate of the helicase) affect the kinetics of unwinding by the helicase in the presence of SSB or DNAP [29].
The dNTPs and GC-content dependences of the unwinding-synthesis kinetics are critical in understanding the role of the helicase in leading strand DNA synthesis. The SSB can support strand-displacement synthesis by the DNAP, but achieving fast rates of DNA synthesis requires high dNTP concentrations. On the other hand, fast DNA synthesis are achieved in the presence of the helicase at low dNTP concentrations. This is because the helicase is able to efficiently trap the unwound junction bases by close association with the DNAP [29].
2.4. Advantages and Disadvantages of real time fluorescence-based and the discontinuous gel-based assay for measuring the unwinding-synthesis kinetics
The fluorescence and gel-based assays are complementary. The fluorescence-based method has the advantage of being real-time and high-throughput. The ability to rapidly measure DNA strand separation kinetics in real time makes it highly suitable for measuring the base pairs unwound-synthesized per second and obtaining information such as the effect of substrate concentrations on the observed rates, which is more tedious to measure using the gel-based assays. However, the fluorescence assay is an all-or-none assay that does not directly measure DNA synthesis, but uses DNA unwinding as a read-out to measure DNA synthesis The gel-based assay is discontinuous, but monitors DNA synthesis directly and unwinding indirectly. The biggest advantage of the gel-based assay is that it measures intermediate DNA synthesis products and thus can be applied to address questions such as replisome stalling and lesion bypass [42]. The gel-based assay also determines the fraction of primer extended by the helicase-DNAP and informs on the amount of correctly assembled complexes on the fork DNA.
A prerequisite for all pre-steady state assays is that the enzymes must be pre-assembled on the DNA substrate without catalysis prior to initiation of the reaction. While most DNAPs can be assembled without incorporation by leaving out dNTPs in the reaction mixture, many helicases require special conditions for assembly such as additional loaders and may not always be amenable to assembly without hydrolysis.
3. Method to measure the chemical step-size (bp translocated/NTP hydrolyzed) of the helicase-DNAP
The number of base pairs unwound-synthesized by the helicase-DNAP complex for every NTP/dNTP hydrolyzed by the helicase is defined as the chemical step-size which provides the energetics of the stepping reaction. Helicase translocation on the DNA is powered by NTP/dNTP hydrolysis to NDP/dNDP and Pi and the DNAP translocation is powered by the incorporation of dNMP into the primer accompanied by the release of PPi. The DNAP translocates in steps of one nucleotide, meaning it extends the primer one nucleotide at a time. However, the step-size of the helicase, meaning the number of base pairs unwound for every cycle of NTP/dNTP hydrolysis needs to be determined. Knowing how many base pairs are unwound-synthesized at each cycle of NTP hydrolysis enables us to understand the coordinated stepping mechanism of the two motors on the DNA during leading strand synthesis. Accurate determination of the chemical step size also addresses the question of whether the chemical step-size changes when the dsDNA is AT-rich versus GC-rich [27].
The chemical step-size of an isolated helicase (bp unwound/NTP hydrolyzed) is determined by taking a ratio of the base pair unwinding rate constant and the NTP hydrolysis rate constant [20]. This method has an inherent problem in that it is difficult to measure the unwinding and NTP hydrolysis kinetics under the same reaction conditions. This is because DNA unwinding is measured at low fork DNA concentration (~1–5 nM) to prevent the separated DNA strands from reannealing. However, at such low DNA concentrations, the NTP hydrolysis activity is not reliably measured, because the amount of NDP+Pi released is below the detection limit. Hence, the NTP hydrolysis kinetics is independently measured at high DNA and helicase concentrations (~1–2 µM) that are not amenable for measuring unwinding rates. The different experimental conditions used for measuring the NTP hydrolysis and DNA unwinding rates could lead to inaccurate estimates of the chemical step-size potentially due to differences in active enzyme concentrations in the two reactions. The following ‘one-pot’ assay overcomes this problem by measuring the DNA unwinding-synthesis and NTP hydrolysis kinetics in a single reaction mixture.
3.1 One-pot assay to measure the bp/NTP chemical step-size of the helicase-DNAP
To measure the kinetics of DNA unwinding-synthesis and NTP hydrolysis in the same reaction mixture, the kinetics of DNA synthesis is used to report on the kinetics of DNA unwinding. The DNA synthesis kinetics can be measured accurately and since T7 DNAP has limited strand-displacement synthesis activity in the absence of the helicase, the observed rate constants of DNA synthesis correspond to the DNA unwinding rate constants. In this assay, the leading strand DNA is unwound and copied into dsDNA, which means that the lagging strand cannot reanneal with the leading strand. Hence, the bp/NTP measurements can be performed at high DNA and enzyme concentrations (2 µM each), which allows for accurate measurement of the NTP hydrolysis kinetics.
T7 helicase prefers dTTP and dATP as the fuel for unwinding; hence, radiolabeled dNTPs are sufficient to measure both the DNA synthesis and dNTP hydrolysis activities. If the helicase uses ATP, the experiments are carried out in the presence of a mixture of radiolabeled ATP and radiolabeled dNTPs. The helicase hydrolyzes [α32P]dNTP (or NTP) into [α32P]dNDP (or NDP) and Pi and the DNAP incorporates [α32P]dNMP in the newly synthesized DNA (Figure 5A). From the amount of dNDPs produced and dNMPs incorporated into the growing primer over the reaction time, both NTP hydrolysis and DNA synthesis kinetics are simultaneously measured to calculate the chemical step-size of the helicase-DNAP. As mentioned above, measuring the NTP hydrolysis and DNA synthesis kinetics in a single reaction overcomes the inaccuracies of the traditional method.
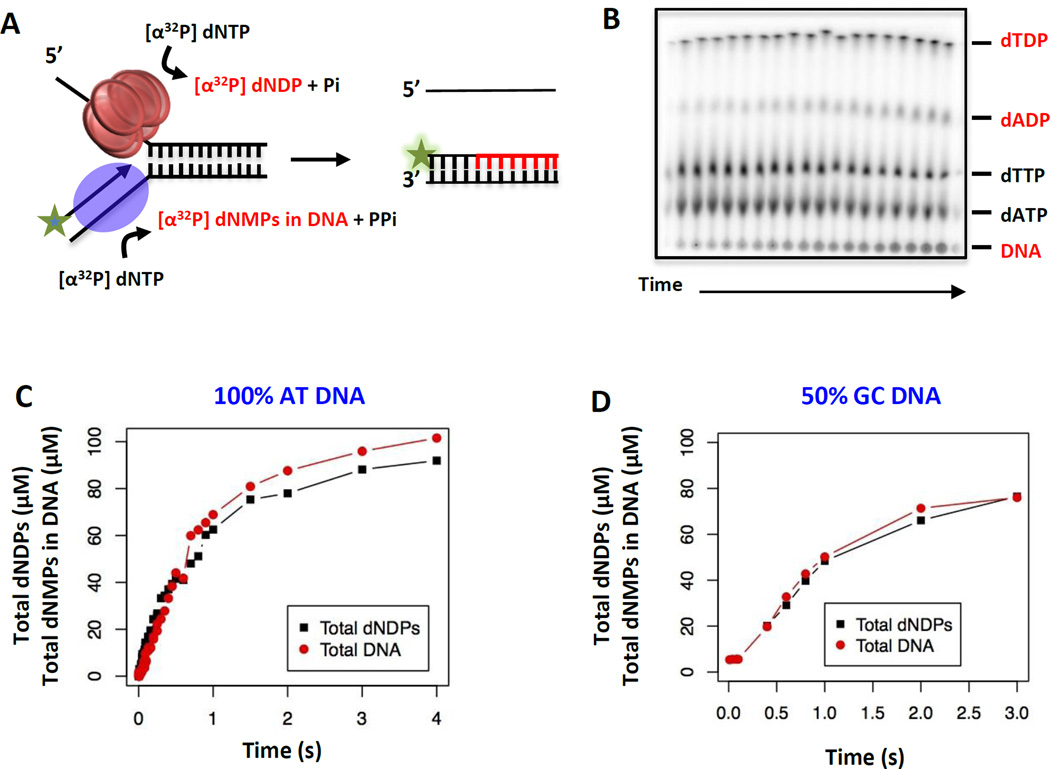
Figure 5. One-pot assay to measure the chemical step size (bp/NTP hydrolyzed) of the helicase-DNAP catalyzing leading strand DNA synthesis.
(A) One-pot assay to measure the base pairs unwound-copied by the T7 helicase-DNAP and dNTP hydrolyzed by the helicase using [α32P]dNTPs, which quantify the kinetics of dNTP hydrolysis to dNDP and the incorporation of dNMP into the DNA. (B) Thin-layer chromatography (PEI-cellulose) shows the separation of [α32P]dNTPs from the corresponding [α32P]dNDPs and DNA (labeled with [α32P]dNMPs). (C) Kinetics of total dNDPs production and total dNMPs incorporation into the DNA by the helicase-DNAP on the 100% AT-rich fork DNA substrate. (D) Kinetics of total dNDPs production and total dNMPs incorporated on the 50% GC-rich DNA substrate.
3.1.1 DNA substrate design and experimental protocol for determining chemical step size
The fork DNA substrates used in this assay are similar to the synthetic replication fork mimics (section 2.1). The experiments are carried out in a rapid quench-flow instrument (KinTek Corporation) at a set temperature (Figure 3B). The helicase and DNAP are assembled on the fork DNA as described in section 2.2.2 and the reactions are initiated with Mg2++dNTPs spiked with [α32P]dNTPs and quenched with formic acid (4M) after specific time intervals (ms to s). Use of formic acid as the quench provides optimal migration of the reaction products on the Thin Layer Chromatography (TLC) plate.
The radiolabeled DNA product, dNTPs and dNDPs are resolved from each other on PEI-cellulose TLC plate (EMD Millipore) (Figure 5B). It is necessary to experimentally determine the optimal buffer conditions for separating the different dNTPs and the hydrolysis products. In our experience, two dNTPs can be optimally resolved in a single TLC plate. Hence, depending on the composition of the dsDNA region to be copied, the reaction should be split into two parts and spiked with different combinations of [α32P]dNTPs. For example, if the dsDNA region of the replication fork substrate is 100% AT, only dATP and dTTP are required for DNA synthesis. Because the T7 helicase preferentially uses dTTP for assembly and unwinding, it is sufficient to spike the reaction with [α32P]dATP and [α32P]dTTP and the products can be resolved in a single TLC. However, if the dsDNA region to be copied contains all four dNTPs, the reaction mixture should be split into two parts; one part spiked with [α32P]dATP and [α32P]dTTP and the other part is spiked with [α32P]dGTP and [α32P]dCTP and analyzed on separate TLC plates developed with 0.5–0.6 M potassium phosphate buffer (pH 3.4). Similarly, if the helicase under study uses ATP as its preferred substrate, the spiked NTPs and TLC conditions have to be appropriately modified.
The highly charged DNA molecules stay close to the origin of spotting on the TLC whereas the dNDPs and unhydrolyzed dNTPs migrate faster (Figure 5B). The amount of dNDPs produced over the reaction time period provides the dNTP hydrolysis kinetics. The amount of labeled dNMPs incorporated into DNA provides the DNA synthesis kinetics and indirectly the unwinding kinetics. It is important to carry out a control experiment without Mg2+ to correct for background hydrolysis of dNTPs. It is also important to have a pre-quenched sample to correct for impurities in the [α32P] dNTPs. To check for processive dNMP incorporation, the DNA synthesis reactions are analyzed on a high-resolution polyacrylamide gel. The TLC and gel are exposed to a phosphorimager screen and scanned using a Typhoon phosphorimager (GE Healthcare). The dNDP, dNTP and DNA intensities on the TLC and gel are quantified using ImageQuant (GE Healthcare).
3.1.2. Data Analysis to determine the chemical step size of helicase-DNAP
The total µM amount of dNMPs incorporated by the DNAP is described by the following equation.
Where, ‘background corrected counts in the DNA spot’ is obtained from DNA counts, ‘total counts’ is the sum of all the background corrected counts in the TLC lane and the ‘total input dNTP concentration’ is the sum of independent concentrations of the spiked dNTPs in the reaction. For example, if the reaction is spiked with [α32P]dATP and [α32P]dTTP and it contains 200 µM each of unlabeled dNTPs, the input dNTP concentration would be 400 µM. It is important to correct for impurities in the labeled dNTPs and background hydrolysis using background counts obtained from reactions carried out in the absence of Mg2+.
The total µM amount of dNDPs produced is given by the equation below.
When two TLCs are run for the same experiment, the total dNDPs and total dNMPs in DNA is the sum of the two TLCs.
The total µM amount of dNDPs and the total µM dNMPs incorporated into the DNA are plotted as a function of reaction time (Figure 5C and 5D). Based on the overlap between the dNDP and dNMP plots, one can identify the chemical step-size of the helicase-DNAP. If the helicase hydrolyzes more than one or less than one dNTP for every dNMP incorporated by the DNAP, the plots of the dNDPs and dNMPs as a function of time will not overlap. However, if the helicase hydrolyzes one dNTP for every dNMP incorporated by the DNAP, the plots will overlap (Figure 5C and 5D).
3.2 Energetics of single base pair unwinding-synthesis during leading strand synthesis
The one-to-one correlation between the amount of dNDPs produced and dNMPs incorporated by the T7 helicase-DNAP complex on the 100% AT-rich dsDNA (Figure 5C) indicates that the chemical step-size is one bp/NTP. The same one bp/NTP chemical step-size is observed with the 50% GC-rich fork DNA (Figure 5D). One bp/NTP has also been observed in many non-hexameric helicases [43–46] and thus appears to be a general feature of helicases. It is interesting that the hydrolysis of one NTP, which results in 10–12 kcal/mol free energy, is coupled to unwind only one base pair which is stabilized by ~1.5–2.0 kcal/mol. These results indicate that the unidirectional translocation of the helicase-DNAP is an energetically costly process. The chemical step-size of one bp/NTP for the helicase-DNAP complex provides insights into the physical step-size of the helicase. The DNAP translocates one nucleotide at a time; therefore, the one-to-one coupling between DNA synthesis and NTP hydrolysis implies that the helicase translocates on the DNA one nucleotide at a time or the physical step-size of the helicase is one nucleotide. The coupled helicase and DNAP motors are moving forward in steps of one nucleotide during leading-strand replication. The chemical step-size of one bp/NTP does not change with DNA stability, which indicates that one bp/NTP is a fundamental property of the T7 helicase-DNAP.
4. Methods to determine the relative positions of helicase and DNAP at the replication fork
To date there are no high-resolution structures of the helicase-DNAP complex on the replication fork DNA. Low-resolution electronic microscopy (EM) and Small Angle X-ray Scattering (SAXS) cannot provide the precise positions of the helicase and DNAP with respect to the replication fork junction at a single base pair resolution. The biochemical methods described below show several ways to map the precise positions of the two enzymes at the replication fork.
4.1. Exonuclease footprinting to map the position of helicase at the replication fork
Exonuclease digestion is widely used to footprint proteins bound to specific regions on nucleic acids [47]. The replication fork substrate has both dsDNA and ssDNA regions (Figure 2); hence, one type of exonuclease enzyme is insufficient to ensure complete digestion of the DNA. Exonuclease III digests one strand of the dsDNA molecule from the 3’ end whereas Exonuclease T digests ssDNA in the same direction. Thus, by using a combination of the two enzymes, the footprint of proteins on both ss and ds DNA regions of the fork DNA substrate is determined in one reaction. In this case, if the 5’ end of the lagging strand in the replication fork substrate is labeled with [γ32Pi] or a fluorophore, then the length of the DNA fragment after treatment with a mixture of Exonuclease III and Exonuclease T provides the position of the helicase in relation to the 5’ end of the lagging strand (Figure 6A).
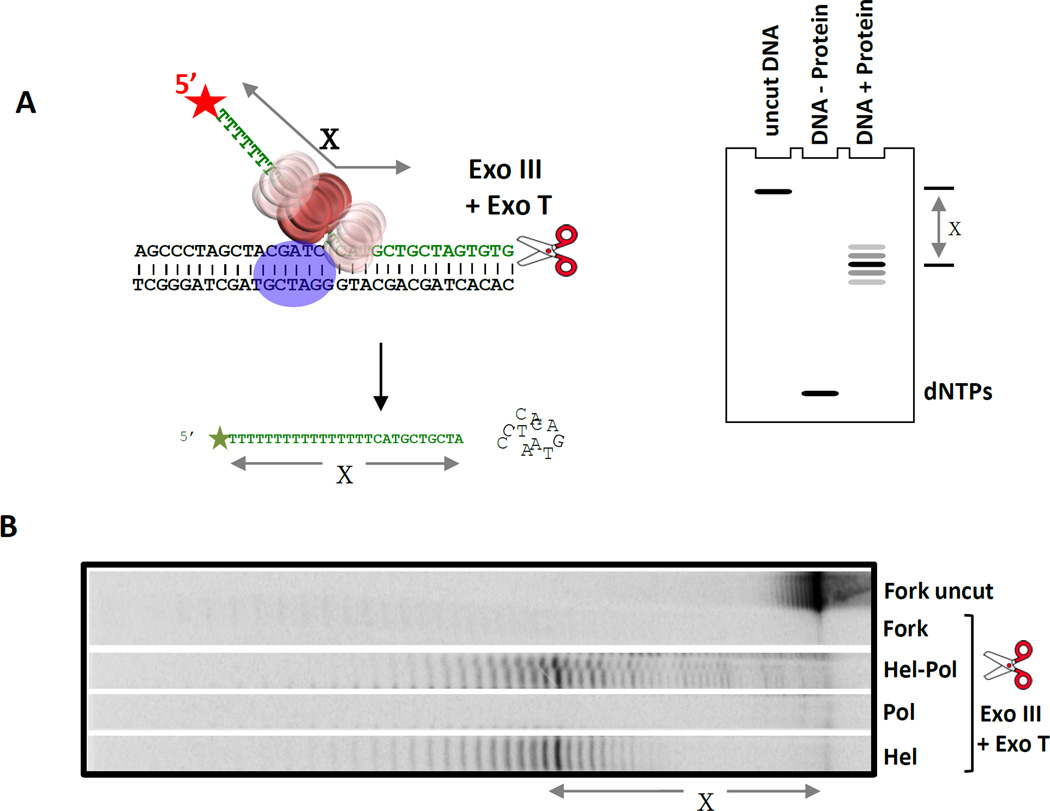
Figure 6. Exonuclease footprinting to determine the position of T7 helicase on the replication fork substrate in complex with T7 DNAP.
(A) The 5’ end of the lagging DNA strand in the fork DNA (with a 36-nt ssDNA tail) is labeled to visualize the protected DNA fragments after digestion with a combination of Exo III and Exo T enzymes. The simulated gel cartoon shows the expected length of the protected DNA fragments when the helicase is bound at the fork junction. The length of the protected DNA fragments (‘X’) is determined by analysis on a high resolution sequencing gel. If X is less than 36-nt, then the helicase is positioned away from the fork junction, if X is 36-nt then the downstream boundary of the helicase is at the fork junction, and if X is greater than 36-nt then the helicase is positioned downstream of the fork junction. (B) Sequencing gel shows the products of ExoIII + ExoT digestion (X = 38-nt) in the presence and absence of proteins (T7 helicase and T7 helicase-DNAP). The length of the protected DNA fragments with the helicase and helicase-DNAP are similar indicating that the position of the helicase on the fork DNA does not change in the presence of the DNAP. The prominent 38-nt central band indicates that the helicase is within 2-nt downstream of the fork junction [21].
4.1.1. Experimental protocol for exonuclease footprinting
The helicase and helicase-DNAP are stalled on the replication fork substrate prior to exonuclease digestion. An important criterion for exonuclease footprinting is that the half-life of the enzyme-DNA complex should be longer than the time required for the exonuclease reaction. The conditions required for assembling the helicase without translocation may vary between enzymes and need to be determined for the specific helicase of interest. The helicase can be assembled on the DNA using non-hydrolyzable NTP analog, e.g., T7 helicase is assembled on the DNA without translocation using dTMP-PCP (a non-hydrolyzable dTTP analog). The helicase-DNA complex with this analog has nM Kd and a life-time of several hours [48]. The DNAP can be stalled on the DNA by leaving out dNTPs or by using ddNTP terminated primer that cannot be extended by the DNAP.
The assembled helicase or helicase-DNAP fork complex is treated with the exonuclease mix (exo III + exo T). The optimal concentration of the exonuclease enzymes is determined by carrying out exonuclease digestion of free DNA substrate at varying concentrations of the enzyme mix. The concentration of enzymes at which complete digestion of the DNA substrate is observed is used in the reaction. The reactions are quenched after defined time intervals, resolved on a denaturing polyacrylamide sequencing gel, and the gel is scanned using a phosphorimager. If the protein-DNA interactions are not strong, exonucleases can displace the proteins and hence obscure the interpretation of the results. It is necessary to carry out a time course to check for such events.
4.1.2. Position of the helicase with respect to the fork junction in the leading strand complex
If the downstream edge of the helicase is at the replication fork junction, the length of the protected DNA fragment is expected to be the length of the 5’-end ssDNA overhang (Figure 6A). If the helicase has interactions with the downstream dsDNA region, the protected DNA fragment will be longer than the length of the 5’-end ssDNA overhang. If the helicase is bound at a position upstream of the replication fork junction, the protected DNA fragment will be smaller than the length of the 5’-end ssDNA overhang of the fork. A comparison of the digestion profiles of the fork DNA bound to helicase and helicase-DNAP informs on any changes in the position of the helicase in the presence of the DNAP. The data in Figure 6B shows that the downstream edge of T7 helicase is at the replication fork junction, and this position of the helicase at the fork junction does not change in the presence of T7 DNAP.
One can map the downstream edge of the DNAP on the replication fork by applying the same method using a 5’-3’ exonuclease such as T7 exonuclease. Below are outlined other methods to map the position of the DNAP with respect to the replication fork junction (Section 4.2).
4.2. Mapping the position of the DNAP with respect to the replication fork using interstrand cross-linked replication fork DNA or stalled helicase
To determine the position of the active site of the DNAP with respect to the fork junction in an actively synthesizing helicase-DNAP complex, one can walk the helicase-DNAP complex to a specific position on the fork DNA by preventing further translocation (e.g. by using an interstrand DNA cross-link) (Figure 7A). Alternatively, the helicase can be stalled at the fork junction some distance downstream of the primer-end using a non-hydrolyzable dTTP (NTP) analog (dTMP-PCP) (Figure 7B). The length of the primer extension products provides the position of the DNAP active site with respect to the fork junction. In both methods (interstrand cross-link or stalled helicase), it is important to carry out the experiment in the time period of the primer-extension reaction using rapid kinetic methods and not perform an end-point assay. In the end-point assay, which is typically carried out manually and takes many seconds or minutes, one cannot ensure that the helicase remains bound to the fork DNA in the mapping period.
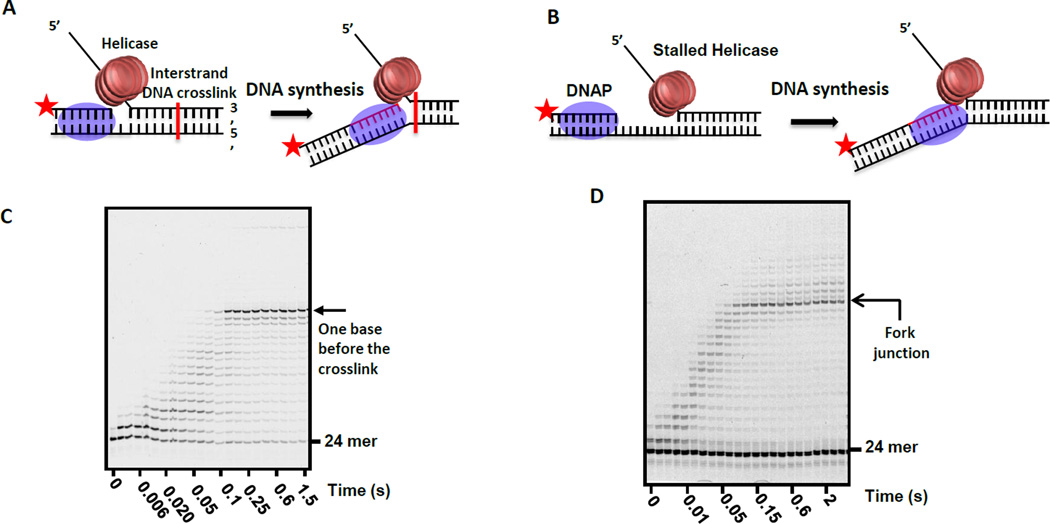
Figure 7. Mapping the position of the DNAP active site on the replication fork with the helicase.
(A) DNA substrate with an interstrand transplatin DNA crosslink positioned approximately in the middle of the dsDNA region of the fork substrate. Extension of the labeled primer by the helicase-DNAP complex is carried out in the presence of all dNTPs to map the position of the DNAP active site with respect to the transplatin crosslink. (B) DNA substrate with 20-nt ssDNA gap between the primer end and fork junction. T7 helicase is stalled at the fork junction using dTMP-PCP, a non-hydrolysable dTTP analog, and the extension of the primer by the DNAP to the point of the stalled helicase is followed using a labeled primer. (C) High resolution sequencing gel shows the time course of primer extension. T7 DNAP is able to reach all the way to the crosslink point. (D) High resolution sequencing gel shows the time course of the extension of the primer up to the fork junction without stalling at any upstream positions.
4.2.1. Substrate design to map the active site of the DNAP at the replication fork
For mapping the position of the DNAP using the interstrand DNA cross-link, a single transplatin crosslink is introduced at a specific position approximately in the middle of the dsDNA region of the replication fork substrate (Figure 7A). The primer is radiolabeled or fluorescently labeled at the 5’ end to visualize DNA extension with single nucleotide resolution. The transplatin cross-linked dsDNA fork substrate is prepared as described in [27]. The oligonucleotide sequence of the lagging strand in the transplatin cross-linked substrate should be pyrimidine rich with a single stretch of GAG in the entire sequence. It is necessary to have two G bases in the sequence, because formation of the interstrand cross-link is a two-step reaction of which the first step involves formation of an intrastrand cross-link between the two G bases. It is also important to include a nucleotide between the two G bases in the sequence because transplatin, due to steric effects, cannot form an intrastrand cross-link between adjacent G bases in a sequence [49].
4.2.2. Experimental protocol for mapping the position of DNAP at the replication fork
The primer extension reaction is carried out using a rapid quench-flow instrument (Figure 3B). The primer extension reaction acts as the read out for reporting the position of the DNAP active site. For the experiments with the transplatin cross-linked fork DNA, the helicase and DNAP are preassembled on the fork DNA in the presence of dTTP but without Mg2+, and the primer extension reactions are initiated by rapidly mixing with dNTPs and Mg2+. For the experiments with the stalled helicase, the helicase and DNAP are preassembled on the DNA substrate in the presence of Mg-dTMP-PCP and Mg2+ and the DNA synthesis reaction is initiated with dNTPs and quenched after preset time intervals (ms to s) with a chemical quench (such as EDTA). The primer extension products are resolved on a high resolution sequencing gel and quantitated using the ImageQuant software.
4.2.3. Position of the DNAP active site of the in the leading strand complex
If the active site of the DNAP is located many bases behind the helicase in the leading strand complex, then primer extension will stop that many bases before the interstrand cross-link. In this case, the primer was extended all the way to the interstrand DNA cross-link within millisecond time scale, which indicates that T7 DNAP travels on the fork DNA with its active site close to the fork junction (Figure 7C). Similarly, when the T7 helicase is stalled at the fork junction using dTMP-PCP, the arriving T7 DNAP extends the primer all the way to the junction base pair within milliseconds (Figure 7D). The DNAP stalls at the fork junction for several seconds, and at longer times, it continues past the fork junction to catalyze strand-displacement DNA synthesis with a low efficiency. These experiments indicate that the active site of the DNAP in the leading strand complex is very close to the replication fork junction where the helicase is also bound. While this may be a general feature of prokaryotic systems [26], the situation in the eukaryotic systems must be different, because the helicase and DNAP are present on the same strand; thus, both enzymes cannot be juxtaposed at the fork junction (Figure 1B,C).
5. 2-AP Fluorescence method to determine the precise positions of the helicase and DNAP at the fork junction and their roles in unwinding the fork junction
The above mapping studies indicate that the active sites of the helicase and DNAP bound to the opposite strands of the replication fork are in close proximity to the fork junction. This suggests that both helicase and DNAP are in a position to influence the unwinding of the junction base pair. The textbook model of DNA replication posits that the role of the helicase is to unwind the dsDNA and the role of the DNAP is to copy the unwound strands. Recent single molecule and ensemble studies provide a consensus model where the DNAP actively contributes to the unwinding process during DNA replication [26, 27, 29]. Fluorescent base analogs such as 2-aminopurine (2-AP), pyrolo-dC and 6-MI can monitor base pair unwinding and unstacking and have been used to study various nucleic acid enzymes [50, 51].
The 2-AP is a fluorescent base analog of adenine, which forms a normal base pair with thymine. However, the fluorescence intensity of 2-AP is quenched when it is base paired and stacked with the adjacent bases. The fluorescence intensity of 2-AP increases when the base pair is unwound and the stacking interactions are lost. By systematically incorporating a single 2-AP in the replication fork at defined positions, one can measure not only the relative contribution of the helicase and DNAP to junction base pair unwinding, but also map the positions of their active sites with respect to the fork junction, as outlined below.
5.1. Substrate design and preparation for 2-AP fluorescence assays
A single 2-AP is incorporated into the lagging strand sequence of the fork DNA, such that 2-AP is part of the first (junction 2-AP) or the second base pair (internal 2-AP) of the fork junction (Figure 8A). To determine the positions of the active sites of the helicase and DNAP with respect to the fork junction, the junction 2-AP is moved one to five nucleotides downstream of the primer-end by introducing ssDNA gap between the primer-end and fork junction ranging from zero to four nucleotides (Figure 8A). To minimize differential effects of neighboring bases on the observed fluorescence intensity of 2-AP, the oligonucleotides of the fork DNA are designed in such a way that the bases adjacent to the 2-AP are the same irrespective of the position of the 2-AP in the replication fork substrate. By manipulating the sequence of the complementary strand, the position of the 2-AP in the dsDNA region is modified [29, 52]. Additionally, by using primers of different lengths, the position of the primer-template junction, and hence the DNAP active site, in relation to the fork junction can be modified. The 2-AP modified and unmodified oligonucleotides can be purchased from IDT and gel-purified. The replication fork substrates with 2-AP at different positions are made by annealing the appropriate combination of oligonucleotides.
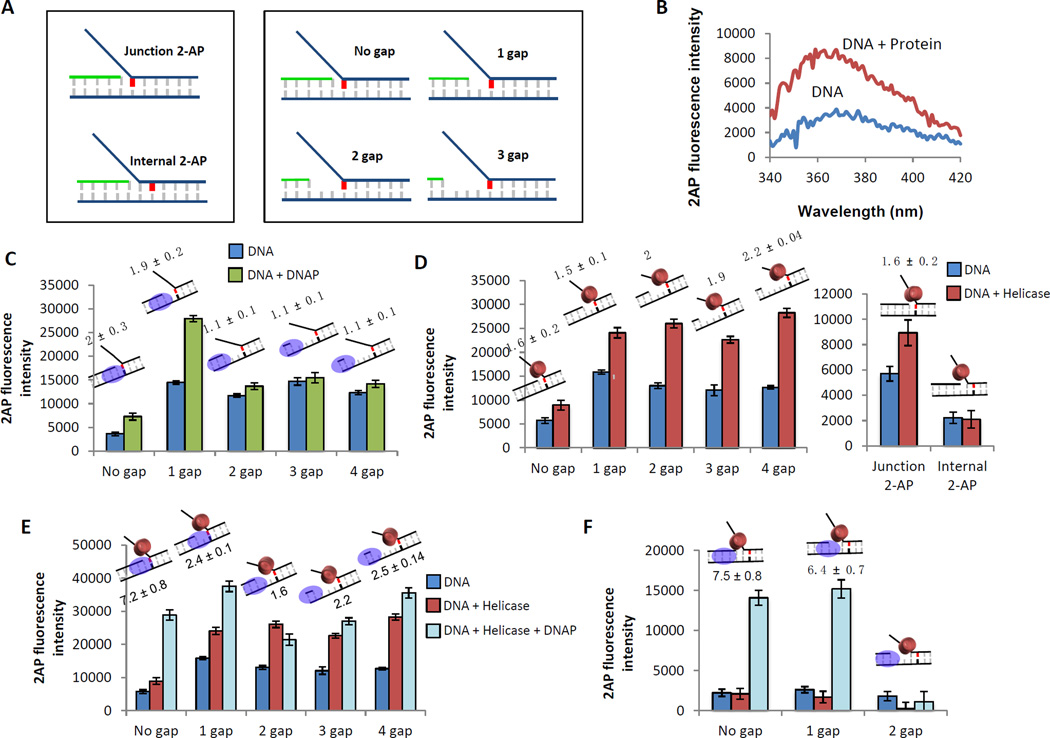
Figure 8. 2-AP fluorescence assay to determine the relative contribution of the helicase and DNAP to dsDNA unwinding.
(A) Schematics of the 2-AP labeled fork DNA substrates. A single 2-AP probe (indicated in red) is placed at the fork junction or one base pair downstream of the fork junction. The primer end is adjusted to generate fork DNA substrates with no gap, 1 gap, 2 gap, or 3 nt ssDNA gap between the primer end and fork junction. (B) Change in 2-AP fluorescence intensity on protein binding is monitored over a range of wavelengths. The change in 2-AP intensity on protein binding reflects on the effect of the protein on the 2-AP base pair. (C) Fluorescence intensities of 2-AP modified replication fork DNA with (green bars) and without (blue bars) T7 DNAP. Cartoons of the DNA substrate used in the experiment and the fold change of fluorescence intensity on protein binding are shown above each bar. (D) Fluorescence intensities of 2-AP modified replication fork DNA with (red bars) and without (blue bars) T7 helicase. (E) Fluorescence intensities of 2-AP modified replication fork DNA with (light blue bars) and without (blue bars) T7 helicase-DNAP. The red bars show intensity changes with T7 helicase alone. (F) Fluorescence intensities of 2-AP modified replication fork DNA with (light blue bars) and without (blue bars) T7 helicase-DNAP. The red bars are the effect of just T7 helicase binding in the absence of T7 DNAP.
5.2. Experimental protocol to measure 2-AP fluorescence for fork DNA melting studies
The steady-state fluorescence intensity of 2-AP upon helicase and DNAP binding is measured using a spectrofluorometer. To form stable protein-DNA complexes with the T7 helicase, non-hydrolysable dTTP analog (dTMP-PCP) is used, and dNTPs are left out to prevent DNA synthesis in the reaction mix. The 2-AP modified replication fork DNA is mixed with the individual enzymes or the helicase-DNAP complex and incubated for a predetermined time to reach equilibrium before measuring the fluorescence intensity. Fluorescence intensity measurements are taken for the buffer, buffer + 2-AP modified DNA and buffer + 2-AP DNA + protein(s). The change in fluorescence intensity on protein binding reflects on its influence on the 2-AP base (Figure 8B). The fork DNA is titrated with increasing concentrations of protein to determine the maximal fluorescence change and to choose the optimal enzyme concentration.
When using 2-AP as the probe, the sample is excited at 315 nm and the emission is monitored at 370 nm. Prior to selecting the emission wavelength, the emission spectra (340 – 420 nm) of the DNA and protein-DNA samples should be monitored (Figure 8B). Monitoring the spectra allows to observe any shifts in emission maxima if present. The samples should be maintained at a preset temperature during the course of the measurement. A control experiment with the unmodified replication fork DNA is required to correct for protein contribution to the fluorescence signal, because of the spectral overlap between the fluorescence of proteins and 2-AP fluorescence. Spectroscopy grade reagents and freshly collected double-distilled water help to minimize fluorescence noise from contaminants. The proteins and any nucleotides that are present in the reaction may contribute to the inner filter effect (Section 5.3) and this must be checked for and corrected when necessary.
5.3. Data Analysis and interpretation of the 2-AP fluorescence changes
If the helicase or DNAP binding to the fork DNA causes the 2-AP base to unstack, an increase in 2-AP fluorescence intensity is observed. Multiple measurements for the same sample are averaged to estimate instrument fluctuations. The 2-AP fluorescence intensity is corrected for (i) background fluorescence from the buffer, (ii) protein contributions and (iii) volume and inner filter effects, as shown below.
Where (2AP Intensity)obs is the observed fluorescence intensity, Vf is the total volume, Vo is the original volume, Absex is the absorbance of the sample at the excitation wavelength, and Absem is the absorbance of the sample at the emission wavelength.
The fold-change is then calculated as the ratio of the corrected 2-AP fluorescence with and without protein. The fold-change acts as a handle for comparing the differential effects of the helicase, DNAP, and helicase-DNAP on the 2-AP base. By designing DNA substrates with the 2-AP base at different positions, the base pair melting footprint of the independent and combined enzymes can be mapped [29].
There are a few things to keep in mind when interpreting the change in fluorescence intensity for DNA melting studies: (1) An increase in fluorescence intensity on protein binding does not necessarily indicate that the specific 2-AP base is unstacked. The increase in intensity could be due to unstacking of the adjacent base. (2) A change in the 2-AP fluorescence intensity can also be due to direct interactions of the protein residues with the 2-AP base, such as proximity of a tryptophan in the protein to the 2-AP base could lead to fluorescent quenching. Control experiments to detect such artifacts must be designed [29].
5.4. Contribution of T7 helicase and DNAP to junction base pair melting and their optimal positions at the replication fork
The 2-AP fluorescence based experiments show that T7 DNAP upon binding to the replication fork destabilizes two base pairs of the fork junction directly downstream from the primer-end (Figure 8C). The melting of the junction base pair by the T7 DNAP is observed when there is no gap or a one nucleotide gap between the primer-end and the fork junction, but not when the gap is two nucleotides or longer (Figure 8C). T7 DNAP binds to the primer-end; hence, the distance between the primer-end and fork junction is important in determining the influence of T7 DNAP on fork junction base pair unwinding. These experiments indicate that T7 DNAP is capable of unwinding the fork junction in the absence of DNA synthesis and can unwind up to the second base pair from the primer-end, creating two ssDNA template nucleotides. Consistent with this observation, the crystal structure [53](PDB ID: 2AJQ) shows that this length (2 nt) of unwound template DNA is required by the T7 DNAP to optimally interact with the primer-template junction.
Unlike T7 DNAP, the T7 helicase is not influenced by the distance between the primer-end and fork junction, instead it follows the fork junction and unwinds the first base pair at the fork junction irrespective of the distance between the fork junction and primer end (Figure 8D). Moreover, T7 helicase does not unwind the second base pair from the fork junction (internal 2-AP). This indicates that T7 helicase binds at the fork junction and destabilizes one base pair of the fork DNA.
When the helicase and DNAP are combined, there is enhanced unwinding of the fork junction base pairs as compared to the effect observed from isolated enzymes (Figure 8E and F). Enhanced junction unwinding is observed when there is no gap and a one nucleotide gap between the primer-end and fork junction, but not when the gap is two nucleotides or greater. This indicates that cooperative unwinding by the two enzymes is observed only when the DNAP active site is positioned no more than one nucleotide away from the fork junction.
Another replication fork construct that works well is where the 2-AP is introduced in the second base pair from the fork junction, and the fork junction contains a G:C base pair (Figure 8A-Internal 2-AP, 8F). In this case, when the G:C base pair at the fork junction is unwound, the fluorescence of 2-AP in the 2nd base pair increases from loss of stacking interactions with the first base pair. The advantage of this construct is the low background from thermal melting and contributions from the isolated helicase and DNAP. This construct also shows that the fluorescence increase by the combined enzymes is much greater compared to the increase observed with the isolated enzymes. Moreover, the fluorescence increase is observed when there is no gap or one nucleotide gap between the primer-end and the fork junction, but not when there are two nucleotides between the primer-end and fork junction (Figure 8F).
These experiments indicate that both the helicase and DNAP contribute to junction base pair unwinding. Importantly, the cooperativity in DNA unwinding by the combined helicase and DNAP enzymes is observed only when the helicase is no more than a single nucleotide ahead of the polymerase.
6. Concluding remarks
The methods described here provide both functional and structural insights into an important enzyme complex of the DNA replication machinery – the leading strand replication complex composed of the replicative helicase and leading strand DNAP. We describe methods to determine how the replicative helicase and DNAP are positioned at the fork junction and the distance between the two enzymes during normal replication. We describe methods to determine the energy consumption of the replicative helicase as it unwinds the DNA in the presence of the leading strand DNAP. We describe methods to determine the functional coupling between the two enzymes and their specific roles in DNA unwinding.
All the methods described in this review were developed using the T7 bacteriophage enzymes, a prokaryotic system, as a model system. The eukaryotic and prokaryotic replicative helicases have opposite polarity of DNA unwinding, but the methods in this review can be applied to the eukaryotic system provided the replisome enzymes can be preassembled on the DNA substrate to allow synchronous initiation. Presently, the complexity of the eukaryotic replication machinery poses substantial obstacles to obtaining a deeper mechanistic understanding of the replisome enzymes whereas complex mechanistic questions can be addressed with the prokaryotic systems. However, as more information becomes available about the individual proteins, the methods described here can be used to obtain foundational information about DNA replication in the eukaryotic system.
In addition to the ensemble methods described here to study helicase-DNAP coupling, recent years have seen the advent of several single molecule methods that track the activities of single helicase-DNAP complexes thereby overcoming an inherent limitation of ensemble methods that measure the average behavior of multiple species. Single molecule measurements are particularly useful in studying processes that occur in a subpopulation of molecules [11, 54]. Hence, these assays inform on alternate pathways and mechanisms. Single molecule methods have proven useful to study several aspects of DNA replication such as assembly of the replisome proteins, replication dynamics, mechanisms of individual replisome enzymes and coordination of leading and lagging strand replication, and all these have been reviewed in detail [55–57].
Highlights.
Methods to investigate the functional coupling between the replicative helicase and DNA polymerase at the leading strand replication fork
Presteady state kinetic methods to measure the single base pair unwinding-synthesis rate constants of leading strand DNA synthesis
A single pot assay to determine the chemical step size or energy consumption of the helicase-DNA polymerase complex
Exonuclease footprinting, interstrand DNA crosslink, and 2-aminopurine fluorescence for positional analysis of helicase and DNA polymerase at the replication fork
Contributions of the helicase and DNA polymerase to fork junction base pair unwinding during leading strand DNA synthesis
Acknowledgments
We thank former and current Patel Lab members for developing and testing the methods described in this review. This work was supported by National Institute of Health (NIH) grant RO1 GM55310.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lohman TM, Tomko EJ, Wu CG. Nat Rev Mol Cell Biol. 2008;9:391–401. doi: 10.1038/nrm2394. [DOI] [PubMed] [Google Scholar]
- 2.Matson SW, Bean DW, George JW. Bioessays. 1994;16:13–22. doi: 10.1002/bies.950160103. [DOI] [PubMed] [Google Scholar]
- 3.Patel SS, Donmez I. J Biol Chem. 2006;281:18265–18268. doi: 10.1074/jbc.R600008200. [DOI] [PubMed] [Google Scholar]
- 4.Patel SS, Picha KM. Annu Rev Biochem. 2000;69:651–697. doi: 10.1146/annurev.biochem.69.1.651. [DOI] [PubMed] [Google Scholar]
- 5.Pyle AM. Annu Rev Biophys. 2008;37:317–336. doi: 10.1146/annurev.biophys.37.032807.125908. [DOI] [PubMed] [Google Scholar]
- 6.Trakselis MA. F1000Res. 2016;5 doi: 10.12688/f1000research.7509.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Briggs K, Fischer CJ. Biomol Concepts. 2014;5:383–395. doi: 10.1515/bmc-2014-0017. [DOI] [PubMed] [Google Scholar]
- 8.Dou SX, Xi XG. Methods. 2010;51:295–302. doi: 10.1016/j.ymeth.2010.04.017. [DOI] [PubMed] [Google Scholar]
- 9.Fischer CJ, Tomko EJ, Wu CG, Lohman TM. Methods Mol Biol. 2012;875:85–104. doi: 10.1007/978-1-61779-806-1_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nandakumar D, Patel S. In: Molecular Biophysics for the Life Sciences. Allewell N, Narhi LO, Rayment I, editors. New York: Springer; 2013. pp. 291–312. [Google Scholar]
- 11.Yodh JG, Schlierf M, Ha T. Q Rev Biophys. 2010;43:185–217. doi: 10.1017/S0033583510000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khan I, Sommers JA, Brosh RM., Jr DNA Repair (Amst) 2015;33:43–59. doi: 10.1016/j.dnarep.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garcia-Garcia C, Frieda KL, Feoktistova K, Fraser CS, Block SM. Science. 2015;348:1486–1488. doi: 10.1126/science.aaa5089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patel SS, Pandey M, Nandakumar D. Curr Opin Chem Biol. 2011;15:595–605. doi: 10.1016/j.cbpa.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun J, Shi Y, Georgescu RE, Yuan Z, Chait BT, Li H, O'Donnell ME. Nat Struct Mol Biol. 2015;22:976–982. doi: 10.1038/nsmb.3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Benkovic SJ, Valentine AM, Salinas F. Annu Rev Biochem. 2001;70:181–208. doi: 10.1146/annurev.biochem.70.1.181. [DOI] [PubMed] [Google Scholar]
- 17.Hamdan SM, Richardson CC. Annu Rev Biochem. 2009;78:205–243. doi: 10.1146/annurev.biochem.78.072407.103248. [DOI] [PubMed] [Google Scholar]
- 18.Johansson E, Dixon N. Cold Spring Harb Perspect Biol. 2013;5 doi: 10.1101/cshperspect.a012799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weller SK, Coen DM. Cold Spring Harb Perspect Biol. 2012;4:a013011. doi: 10.1101/cshperspect.a013011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Donmez I, Patel SS. EMBO J. 2008;27:1718–1726. doi: 10.1038/emboj.2008.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Galletto R, Jezewska MJ, Bujalowski W. J Mol Biol. 2004;343:101–114. doi: 10.1016/j.jmb.2004.07.056. [DOI] [PubMed] [Google Scholar]
- 22.Johnson DS, Bai L, Smith BY, Patel SS, Wang MD. Cell. 2007;129:1299–1309. doi: 10.1016/j.cell.2007.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lionnet T, Spiering MM, Benkovic SJ, Bensimon D, Croquette V. Proc Natl Acad Sci U S A. 2007;104:19790–19795. doi: 10.1073/pnas.0709793104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Delagoutte E, von Hippel PH. Biochemistry. 2001;40:4459–4477. doi: 10.1021/bi001306l. [DOI] [PubMed] [Google Scholar]
- 25.Kim S, Dallmann HG, McHenry CS, Marians KJ. Cell. 1996;84:643–650. doi: 10.1016/s0092-8674(00)81039-9. [DOI] [PubMed] [Google Scholar]
- 26.Manosas M, Spiering MM, Ding F, Croquette V, Benkovic SJ. Nucleic Acids Res. 2012;40:6187–6198. doi: 10.1093/nar/gks254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pandey M, Patel SS. Cell Rep. 2014;6:1129–1138. doi: 10.1016/j.celrep.2014.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stano NM, Jeong YJ, Donmez I, Tummalapalli P, Levin MK, Patel SS. Nature. 2005;435:370–373. doi: 10.1038/nature03615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nandakumar D, Pandey M, Patel SS. Elife. 2015;4 doi: 10.7554/eLife.06562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yuan Q, McHenry CS. J Biol Chem. 2009;284:31672–31679. doi: 10.1074/jbc.M109.050740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Georgescu RE, Langston L, Yao NY, Yurieva O, Zhang D, Finkelstein J, Agarwal T, O'Donnell ME. Nat Struct Mol Biol. 2014;21:664–670. doi: 10.1038/nsmb.2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ozes AR, Feoktistova K, Avanzino BC, Baldwin EP, Fraser CS. Nat Protoc. 2014;9:1645–1661. doi: 10.1038/nprot.2014.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tani H, Fujita O, Furuta A, Matsuda Y, Miyata R, Akimitsu N, Tanaka J, Tsuneda S, Sekiguchi Y, Noda N. Biochem Biophys Res Commun. 2010;393:131–136. doi: 10.1016/j.bbrc.2010.01.100. [DOI] [PubMed] [Google Scholar]
- 34.Ahnert P, Patel SS. J Biol Chem. 1997;272:32267–32273. doi: 10.1074/jbc.272.51.32267. [DOI] [PubMed] [Google Scholar]
- 35.Langston LD, Zhang D, Yurieva O, Georgescu RE, Finkelstein J, Yao NY, Indiani C, O'Donnell ME. Proc Natl Acad Sci U S A. 2014;111:15390–15395. doi: 10.1073/pnas.1418334111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Picha KM, Patel SS. J Biol Chem. 1998;273:27315–27319. doi: 10.1074/jbc.273.42.27315. [DOI] [PubMed] [Google Scholar]
- 37.Davey MJ, O'Donnell M. Curr Biol. 2003;13:R594–596. doi: 10.1016/s0960-9822(03)00523-2. [DOI] [PubMed] [Google Scholar]
- 38.Levin MK, Hingorani MM, Holmes RM, Patel SS, Carson JH. Methods Mol Biol. 2009;500:335–359. doi: 10.1007/978-1-59745-525-1_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pandey M, Levin MK, Patel SS. Methods Mol Biol. 2010;587:57–83. doi: 10.1007/978-1-60327-355-8_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pandey M, Syed S, Donmez I, Patel G, Ha T, Patel SS. Nature. 2009;462:940–943. doi: 10.1038/nature08611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leatherbarrow RJ. Horley, UK: Erithacus Software Limited; 2009. [Google Scholar]
- 42.Sun B, Pandey M, Inman JT, Yang Y, Kashlev M, Patel SS, Wang MD. Nat Commun. 2015;6:10260. doi: 10.1038/ncomms10260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Slatter AF, Thomas CD, Webb MR. Biochemistry. 2009;48:6326–6334. doi: 10.1021/bi900101h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ramanagoudr-Bhojappa R, Chib S, Byrd AK, Aarattuthodiyil S, Pandey M, Patel SS, Raney KD. J Biol Chem. 2013;288:16185–16195. doi: 10.1074/jbc.M113.470013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tomko EJ, Fischer CJ, Lohman TM. J Mol Biol. 2012;418:32–46. doi: 10.1016/j.jmb.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu J, Cheng W, Bustamante C, Oster G. J Mol Biol. 2010;404:439–455. doi: 10.1016/j.jmb.2010.09.047. [DOI] [PubMed] [Google Scholar]
- 47.Metzger W, Heumann H. In: DNA-Protein Interactions: Principles and Protocols. Moss T, editor. Totowa, NJ: Humana Press; 2001. pp. 39–47. [Google Scholar]
- 48.Hingorani MM, Patel SS. Biochemistry. 1993;32:12478–12487. doi: 10.1021/bi00097a028. [DOI] [PubMed] [Google Scholar]
- 49.Paquet F, Boudvillain M, Lancelot G, Leng M. Nucleic Acids Res. 1999;27:4261–4268. doi: 10.1093/nar/27.21.4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jose D, Weitzel SE, von Hippel PH. Proc Natl Acad Sci U S A. 2012;109:14428–14433. doi: 10.1073/pnas.1212929109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reha-Krantz LJ. Methods Mol Biol. 2009;521:381–396. doi: 10.1007/978-1-60327-815-7_21. [DOI] [PubMed] [Google Scholar]
- 52.Jose D, Datta K, Johnson NP, von Hippel PH. Proc Natl Acad Sci U S A. 2009;106:4231–4236. doi: 10.1073/pnas.0900803106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Doublie S, Tabor S, Long AM, Richardson CC, Ellenberger T. Nature. 1998;391:251–258. doi: 10.1038/34593. [DOI] [PubMed] [Google Scholar]
- 54.Sun B, Wang MD. Crit Rev Biochem Mol Biol. 2016;51:15–25. doi: 10.3109/10409238.2015.1102195. [DOI] [PubMed] [Google Scholar]
- 55.Perumal SK, Yue H, Hu Z, Spiering MM, Benkovic SJ. Biochim Biophys Acta. 2010;1804:1094–1112. doi: 10.1016/j.bbapap.2009.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stratmann SA, van Oijen AM. Chem Soc Rev. 2014;43:1201–1220. doi: 10.1039/c3cs60391a. [DOI] [PubMed] [Google Scholar]
- 57.van Oijen AM, Loparo JJ. Annu Rev Biophys. 2010;39:429–448. doi: 10.1146/annurev.biophys.093008.131327. [DOI] [PubMed] [Google Scholar]
- 58.Bernstein JA, Richardson CC. J Biol Chem. 1989;264:13066–13073. [PubMed] [Google Scholar]
- 59.Tabor S, Huber HE, Richardson CC. J Biol Chem. 1987;262:16212–16223. [PubMed] [Google Scholar]