Abstract
Importance
There are well-documented racial disparities in outcomes for African American patients with clear cell renal cell carcinoma (ccRCC). Despite a dramatic improvement in overall survival in white patients since the advent of targeted therapy, survival for African Americans with advanced ccRCC has not changed. There is little known about potential racial differences in tumor biology of ccRCC.
Objective
To determine if there are racial differences in the somatic mutation rate and gene expression of ccRCC tumors from white and African American patients.
Design, Setting, and Participants
Overall, 438 patients with ccRCC were identified through The Cancer Genome Atlas (TCGA) clear cell kidney (KIRC) dataset (419 white and 19 African American patients). The GSE25540 dataset containing 135 patients (125 white and 10 African American patients) was used for validation. Tumor samples were collected from numerous cancer centers and were examined for racial differences in somatic mutation rates and RNA expression. Racial differences in somatic mutation rates and RNA expression were examined.
Main outcomes and Measures
The comparison of somatic mutation rates and differences in RNA expression in white and African American patients with ccRCC.
Results
Overall, 419 ccRCC tumor data sets from non-Hispanic white patients and 19 from non-Hispanic African American patients were identified through the publically available TCGA KIRC data set, and a validation set of 125 white and 10 African American ccRCC patient tumors was identified from the publicly available GSE25540 data set. African American patients were significantly less likely than white patients to have VHL mutations (2 of 12 [17%] vs 175 of 351 [50%], respectively; P = 04) and were enriched in the ccB molecular subtype (79% in African American vs 45% in white patients; P = 005), a molecular subtype that carries a worse prognosis. It was found that RNA expression analysis revealed relative down-regulation of hypoxia-inducible factor (HIF) and vascular endothelial growth factor (VEGF)-associated pathways in African American patients compared with white patients.
Conclusions and Relevance
African American patients have less frequent VHL inactivation, are enriched in the ccB molecular subtype, and have decreased up-regulation of HIF-associated gene signatures than white patients. These genomic differences would predict decreased responsiveness to VEGF-targeted therapy and are a biologically plausible contributing factor to the worse survival of African American patients with ccRCC, even in the targeted therapy era.
Renal cell carcinoma (RCC) is the eighth most commonly diagnosed cancer in the United States. It accounts for nearly 64000 new cancer cases and over 13 000 deaths per year in the United States alone.1 Multiple studies from the pretargeted therapy era demonstrate that African American patients with RCC have inferior overall survival compared with white patients regardless of age, sex, stage, histologic subtype, or surgical treatment.2-4 Despite this documented survival disparity, there is no data examining genomic or transcriptomic differences of clear cell RCC (ccRCC) in African American patients vs white patients.
The last decade has produced a wealth of knowledge about the molecular drivers of RCC, including the comprehensive molecular characterization of ccRCC by The Cancer Genome Atlas (TCGA).5 A hallmark event in the development of ccRCC is loss of heterozygosity of the von Hippel-Lindau (VHL) tumor suppressor gene. VHL inactivation occurs at a rate of 52% to 82% in sporadic ccRCC5 and leads to stabilization of the α subunit of the hypoxia-inducible factor (HIF) family of transcription factors. Stabilization of HIFα subunits results in their heterodimerization with HIFβ subunits and a transcriptionally active complex whose target genes are intimately involved in tumor angiogenesis. The vascular endothelial growth factor (VEGF) pathway in particular is a key transcriptional HIF target and is targeted by a number of the currently US Food and Drug Administration-approved therapies. In addition, several other genes, such as PBRM1, SETD2, and BAP1, are also mutated in ccRCC patient tumors.6 Herein, we investigate potential genetic differences in ccRCC that are associated with race.
Methods
The Cancer Genome Atlas and Validation Data and Analysis
RNA sequencing, somatic mutation, and copy number data from the TCGA kidney clear cell (KIRC) data set were downloaded from the publically available TCGA data portal (https://tcga-data.nci.nih.gov/tcga/), and somatic mutation data was downloaded from an independent and publicly available data set from Peña-Llopis et al6 (GSE25540 data set).
The TCGA RNaseq data set was log2 transformed and median-centered. Two-class significance analysis of microarrays (using 2-fold change and false discovery rate = 0) was performed to generate race-specific gene lists.7 The significant genes and corresponding fold changes as determined by significance analysis of microarrays were analyzed by Ingenuity Pathway Analysis (Ingenuity Systems) for predicted pathway activation and/or inhibition. Gene Set Enrichment Analysis (Broad Institute) was performed comparing ccRCC tumors from African American patients vs white patients (ethnicity defined by the TCGA) against Molecular Signature Database c2.all.v4.0. The mutations in the 9 most commonly mutated genes—VHL, PBRM1, SETD2, KDM5C, PTEN, BAP1, MTOR, TP53, and PI3KCA7—were analyzed between African American patients and white patients. Patients were then classified into the previously described RNA subtypes of ccRCC—ccA and ccB—based on patterns of differential gene expression using prediction analysis of microarray.8,9
Statistical Analysis
Differences in somatic mutation rates by race were compared using χ2 or Fisher exact test where applicable. Fisher exact test was used to compare prevalence of the ccA and ccB molecular subtypes between races.
Results
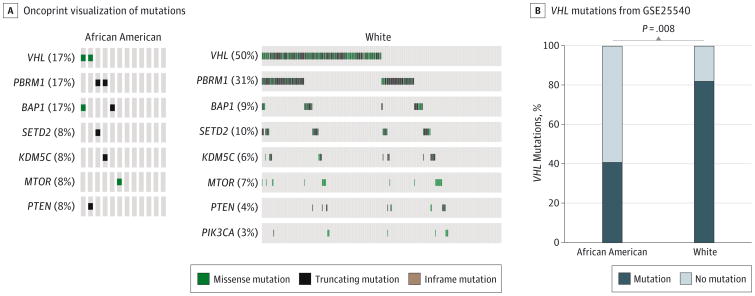
We first investigated whether there are racial differences in the mutational spectrum of ccRCC tumors comparing data from African American patients (n = 19) and white patients (n = 419) from the TCGA ccRCC KIRC data set. Overall, 175 of 351 (50%) white patients had VHL mutations, whereas 2 of 12 (17%) African American patients had VHL mutations (P = .04) (Figure 1A) (eTable 1 in the Supplement). In contrast, there were no racial differences in the mutational frequency of other TCGA KIRC–defined significantly mutated genes (Figure 1A) (eTable 1 in the Supplement). These results were validated for VHL, PBRM1 and BAP1 in an independent data set verifying the lower prevalence of VHL mutation in African American patients (Figure 1B) (eTable 1 in the Supplement)6
Figure 1. Gene Mutations in Clear Cell Renal Cell Carcinoma Tumors by Data Set Source and Race.
A, Mutations in significantly mutated genes according to the The Cancer Genome Atlas kidney clear cell data sets are shown by race, B, Frequency of VHL mutations in an independent data set6 are shown by race.
Given the lower frequency of VHL mutations in African American patients, we investigated the RNA expression of the VEGF ligands and VEGF receptors to evaluate for downstream effects of VHL loss. In the TCGA KIRC data set, 419 white patients and 19 African American patients were available for analysis. In the TCGA data set, African American patients had significantly lower expression of the VEGFA ligand (eFigure 1A in the Supplement), as well as the FLT-1 (VEGFR1) and KDR (VEGFR2) receptors (eFigure 1B in the Supplement). Furthermore, significance analysis of microarrays and subsequent ingenuity pathway analysis of upstream regulators predicted that numerous genes associated with HIF (EPAS1, HIF1A, ARNT, and CREB1) transcriptional regulation were up-regulated in white patients (eTable 2 in the Supplement). Finally, several VEGF and HIF signatures were negatively enriched in African American patients as determined by Gene Signature Enrichment Analysis (eFigure 3 and eTable 3 in the Supplement). In aggregate, these findings indicate distinct differences in the biology of ccRCC in African American patients compared with white patients and suggest that ccRCC in African American patients may be less dependent upon HIF and VEGF signaling.
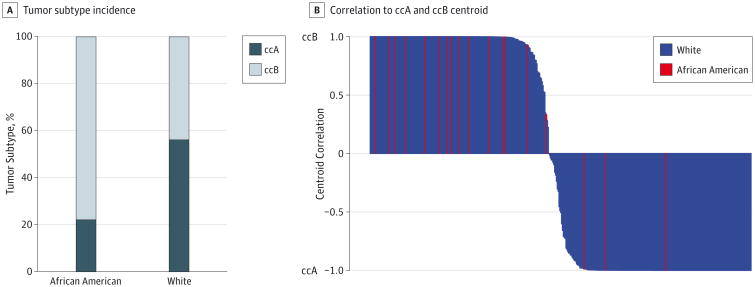
Clear cell renal cell carcinoma is a heterogenous disease that can be classified into 2 distinct molecular subtypes, ccA and ccB,8 with the ccB subtype associated with a worse survival.8,9 Classification of the TCGA KIRC tumors into ccA and ccB subtypes showed that African Americans had significant enrichment for the ccB subtype (15 of 19 [79%] African American patients have the ccB subtype compared with 190 of 419 [45%] white patients; P = 005) (Figure 2A) (eFigure 3 in the Supplement) and in general had high correlation to the ccB centroid (Figure 2B). This predominance of the ccB subtype in African American patients and its association with worse survival suggests that the intrinsic biology of ccRCC in African American patients may contribute to their well-documented worse survival outcomes.
Figure 2. Incidence and Correlation to Centroid of Clear Cell Renal Cell Carcinoma Tumor Subtype.
A, Visual representation of the incidence of ccA and ccB RNA subtypes of clear cell renal cell carcinoma tumor subtype by race, B, Distance and correlation to centroid of tumor subtype (ccA and ccB) by race.
Discussion
This is the first report to our knowledge to examine genomic differences between ccRCC in African American patients vs white patients. Our results indicate that ccRCC tumors from African American patients exhibit a lower rate of VHL mutation and a correspondingly lower level of HIF and VEGF pathway activation. We therefore postulate that a significantly larger proportion of tumors from African American patients may have a HIF-independent and VEGF-independent propensity for aggressiveness, resulting in resistance to the commonly used VEGF-targeted therapies. While our results point toward a biologic rationale for the lack of improvement in survival of African American patients since the advent of targeted therapy,10 we recognize that there are also a host of other potential factors (eg, access to health care, time to diagnosis, and appropriate treatment) that likely contribute to the racial disparities seen in advanced ccRCC. Nonetheless, the results presented herein provide insight into the potential role that genomic variation plays in the disparity observed between races.
Finally, why African American patients with ccRCC have less frequent VHL mutations is unclear. One possibility is that the development of ccRCC in African Americans may have a different etiology. For example, patients with end-stage renal disease have a higher incidence of RCC than the general population,11 and ccRCC tumors developing in patients with end-stage renal disease have lower rates of 3p loss or VHL mutation.12,13 Given higher rates of end-stage renal disease in African Americans (likely a consequence of a higher incidence and earlier onset of hypertension),14 it is plausible that the lower rate of VHL mutation in ccRCC tumors from African Americans may be the result of increased rates of end-stage renal disease. Comprehensive population-based studies will be needed to define correlations between race and etiologic heterogeneity.
Conclusions
Despite an impressive improvement in overall survival in white patients since the advent of VEGF pathway–targeted therapy, the survival for African American patients with advanced ccRCC has not changed. We present evidence of distinct differences in tumor biology between African American patients and white patients that would predict for less responsiveness to VEGF-targeted therapy. While this is a plausible biologic explanation for the lack of improved survival in African American patients, it is likely that disparities in health care delivery contribute to survival differences as well.
Supplementary Material
Key Points.
Question
Are there genomic differences between clear cell renal cell carcinomas (ccRCC) that arise in African American patients and white patients that may contribute to the disparity in survival outcomes observed between races?
Findings
Using data from the The Cancer Genome Atlas clear cell kidney project, we found that African American patients have a significantly lower rate of mutations of the von Hippel-Lindau tumor suppressor gene, as well as decreased hypoxia-inducible factor activation. In addition, African American patients are enriched for a previously defined RNA subtype (ccB) that is associated with worse survival outcomes.
Meaning
There are distinct racial differences in the biology of ccRCC tumors that may partially explain the well-documented racial disparity in survival outcomes of African American patients with advanced ccRCC.
Acknowledgments
Funding/Support: This study was supported by the American Association for Cancer Research Kure It Program (W.Y.K.) and the Department of Defense (grant No. CA120297) (B.K.).
Role of the Funder/Sponsor: The funders/sponsors had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Additional Contributions: We acknowledge the members of the Kim Laboratory for useful discussions.
Footnotes
Supplemental content at jamaoncology.com
Author Contributions: Dr Krishnan and Kardos had full access to all of the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Study concept and design: Krishnan, Rose, Kardos, Kim.
Acquisition, analysis, or interpretation of data: Krishnan, Rose, Kardos, Milowsky, Kim.
Drafting of the manuscript: Krishnan, Rose, Kardos, Kim.
Critical revision of the manuscript for important intellectual content: Krishnan, Rose, Kardos, Milowsky, Kim.
Statistical analysis: Krishnan, Kardos.
Administrative, technical, or material support: Rose. Study supervision: Rose, Milowsky, Kim
Conflict of Interest Disclosures: None reported
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65(1):5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Chow WH, Shuch B, Linehan WM, Devesa SS. Racial disparity in renal cell carcinoma patient survival according to demographic and clinical characteristics. Cancer. 2013;119(2):388–394. doi: 10.1002/cncr.27690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berndt SI, Carter HB, Schoenberg MP, Newschaffer CJ. Disparities in treatment and outcome for renal cell cancer among older black and white patients. J Clin Oncol. 2007;25(24):3589–3595. doi: 10.1200/JCO.2006.10.0156. [DOI] [PubMed] [Google Scholar]
- 4.Tripathi RT, Heilbrun LK, Jain V, Vaishampayan UN. Racial disparity in outcomes of a clinical trial population with metastatic renal cell carcinoma. Urology. 2006;68(2):296–301. doi: 10.1016/j.urology.2006.02.036. [DOI] [PubMed] [Google Scholar]
- 5.Cancer Genome Atlas Research Network. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499(7456):43–49. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peña-Llopis S, Vega-Rubín-de-Celis S, Liao A, et al. BAP1 loss defines a new class of renal cell carcinoma. Nat Genet. 2012;44(7):751–759. doi: 10.1038/ng.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li J, Tibshirani R. Finding consistent patterns: a nonparametric approach for identifying differential expression in RNA-Seq data. Stat Methods Med Res. 2013;22(5):519–536. doi: 10.1177/0962280211428386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brannon AR, Reddy A, Seiler M, et al. Molecular stratification of clear cell renal cell carcinoma by consensus clustering reveals distinct subtypes and survival patterns. Genes Cancer. 2010;1(2):152–163. doi: 10.1177/1947601909359929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brooks SA, Brannon AR, Parker JS, et al. ClearCode34: A prognostic risk predictor for localized clear cell renal cell carcinoma. Eur Urol. 2014;66(1):77–84. doi: 10.1016/j.eururo.2014.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vaishampayan U, Vankayala H, Vigneau FD, et al. The effect of targeted therapy on overall survival in advanced renal cancer: a study of the national surveillance epidemiology and end results registry database. Clin Genitourin Cancer. 2014;12(2):124–129. doi: 10.1016/j.clgc.2013.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Breda A, Lucarelli G, Rodriguez-Faba O, et al. Clinical and pathological outcomes of renal cell carcinoma (RCC) in native kidneys of patients with end-stage renal disease: a long-term comparative retrospective study with RCC diagnosed in the general population. World J Urol. 2015;33(1):1–7. doi: 10.1007/s00345-014-1248-y. [DOI] [PubMed] [Google Scholar]
- 12.Hughson MD, Schmidt L, Zbar B, et al. Renal cell carcinoma of end-stage renal disease: a histopathologic and molecular genetic study. J Am Soc Nephrol. 1996;7(11):2461–2468. doi: 10.1681/ASN.V7112461. [DOI] [PubMed] [Google Scholar]
- 13.Chow WH, Dong LM, Devesa SS. Epidemiology and risk factors for kidney cancer. Nat Rev Urol. 2010;7(5):245–257. doi: 10.1038/nrurol.2010.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lackland DT. Racial differences in hypertension: implications for high blood pressure management. Am J Med Sci. 2014;348(2):135–138. doi: 10.1097/MAJ.0000000000000308. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.