Abstract
Phototherapy with ultraviolet (UV) light is a standard treatment for psoriasis, yet the mechanisms underlying the therapeutic effects are not well understood. Studies in human and mouse keratinocytes and in the skin tissues from human patients and mice showed that UV treatment triggers ubiquitination and downregulation of the type I interferon (IFN) receptor chain IFNAR1, leading to suppression of IFN signaling and an ensuing decrease in the expression of inflammatory cytokines and chemokines. The severity of imiquimod-induced psoriasiform inflammation was greatly exacerbated in skin of mice deficient in IFNAR1 ubiquitination (Ifnar1SA). Furthermore, these mice did not benefit from UV phototherapy. Pharmacologic induction of IFNAR1 ubiquitination and degradation by an antiprotozoal agent halofuginone also relieved psoriasiform inflammation in wild type but not in Ifnar1SA mice. These data identify downregulation of IFNAR1 by UV as a major mechanism of the UV therapeutic effects against the psoriatic inflammation and provide a proof of principle for future development of agents capable of inducing IFNAR1 ubiquitination and downregulation for the treatment of psoriasis.
Keywords: psoriasis, interferon, IFNAR1, phototherapy, skin inflammation
Introduction
Treatment using skin exposure to UV light (phototherapy) is a common and often effective therapeutic approach for the management of psoriasis (Koo, 1999). While immunosuppression stemming from the death of inflammatory leukocytes and/or alterations in cytokine signaling is thought to be responsible for UV action, the specific mechanisms underlying the efficacy of phototherapy are only beginning to be understood (Tartar et al., 2014, Wong et al., 2013, Lowes et al., 2007). To some extent, the progress in this field is hindered by a limited number of immunocompetent animal models of psoriatic inflammation that are responsive to UV treatment. A mouse model, where a psoriasiform skin inflammation is induced by topical application of a cream containing imiquimod, a ligand for the Toll-like receptor-7, has been described (van der Fits et al., 2009). Treatment of these mice with narrow-band UVB was shown to alleviate the imiquimod-induced inflammation. Importantly, one of the pathways altered in this model was that of type I interferon (IFN) signaling, which was also suppressed in human psoriasis patients treated with UV (Racz et al., 2011).
IFN (including IFNα, IFNβ, etc) are well characterized anti-viral, anti-tumorigenic, and pro-inflammatory cytokines that interact with their specific receptor consisting of the IFNAR1 and IFNAR2 chains. Ligand binding activates Janus kinases and increases the transactivation of the signal transducers and activators of transcription (STAT1/2) to promote the expression of IFN-stimulated genes (ISG, reviewed in (Platanias, 2005, Piehler et al., 2012, Fuchs, 2013). Several lines of evidence suggest the importance of IFN in the pathogenesis of psoriasis. First, iatrogenic development or exacerbation of psoriasis associated with the use of pharmacologic IFN was reported in patients who received IFN to treat hepatitis C, multiple sclerosis, or malignant melanoma (Afshar et al., 2013, Tas and Atsu, 2015, Kolb-Maurer et al., 2015). Second, induction of the IFN-stimulated gene expression signature has been described in samples from psoriasis patients (Baechler et al., 2006, Kim and Krueger, 2015). Third, inhibition of the IFN pathway attenuates skin inflammation in a mouse xenograft model of psoriasis (Nestle et al., 2005) and Irf2 knockout mice that display high levels of IFN signaling develop psoriasis-resembling skin inflammation (Hida et al., 2000).
However, there is a controversy regarding the role of IFN in the mouse model of imiquimod-induced psoriasiform skin inflammation. While some reports suggest the role of IFN in this model (Ueyama et al., 2014, Grine et al., 2015), others showed that knockout of Ifnar1 does not preclude IMQ-induced inflammation (Wohn et al., 2013). Cell surface levels of the IFNAR1 receptor chain determine the magnitude and duration of all know effects of IFN on cells, including anti-tumorigenic and pro-inflammatory effects of IFN (reviewed in (Piehler et al., 2012, Coccia et al., 2006, Uze et al., 2007)).
In human and mouse cells, the levels of IFNAR1 are primarily regulated by phosphorylation-dependent IFNAR1 ubiquitination, endocytosis, and degradation (Kumar et al., 2007, Kumar et al., 2004, Kumar et al., 2003). Phosphorylation of IFNAR1 on specific serine residues (Ser535 in human IFNAR1 and Ser526 in mouse IFNAR1) is required for recruitment of the ubiquitination machinery; this phosphorylation can be induced either by IFN or by non-ligand stimuli (reviewed in (Fuchs, 2013)). The latter stimuli that trigger IFNAR1 phosphorylation include inflammatory cytokines (Huangfu et al., 2012, Bhattacharya et al., 2014) and amino acid starvation, which acts via activation of the general control nonderepressible-2 (GCN2) kinase (Bhattacharya et al., 2013). These stimuli initiate signaling pathways that converge on activation of the p38 stress-activated protein kinase, which mediates IFNAR1 phosphorylation and ensuing ubiquitination and degradation (Bhattacharya et al., 2010, Bhattacharya et al., 2011).
Intriguingly, the exposure of mammalian cells to UV was shown to activate GCN2 kinase and ensuing phosphorylation of the translation regulator eIF2α (Deng et al., 2002) and activation of p38 (Hazzalin et al., 1996, Price et al., 1996). Here we describe studies revealing that UV stimulates IFNAR1 phosphorylation, ubiquitination, and downregulation in a human keratinocyte cell line and in mouse primary keratinocytes. Studies in skin from human and mice also demonstrate that UV treatment downregulates IFNAR1 and suppresses IFN signaling. Importantly, mice harboring homozygous knocked-in Ifnar1S526A allele (thereafter termed “SA”), which were shown to be insensitive to phosphorylation-dependent downregulation of IFNAR1 (Bhattacharya et al., 2014), display a grossly exacerbated psoriasiform inflammation and are resistant to UV therapy. Finally, administration of the GCN2-inducing anti-protozoan drug halofuginone is sufficient to trigger IFNAR1 ubiquitination and degradation and to alleviate psoriatic inflammation in the skin of wild type but not Ifnar1SA mice. These data identify downregulation of IFNAR1 by UV as a major mechanism of UV therapeutic effects and suggest a novel psoriasis treatment strategy involving further development of agents capable of inducing IFNAR1 ubiquitination and downregulation.
Results
UV triggers downregulation of IFNAR1 and ensuing suppression of IFN signaling
The mechanisms of suppression of IFN-driven gene expression signature reported in the skin of psoriasis patients treated with UV (Racz et al., 2011) are yet to be delineated. Given that IFNAR1 levels are the major determinant that regulates the responsiveness of cells and tissues to IFN (Piehler et al., 2012, Fuchs, 2013), we analyzed these levels in human normal skin before and after UV treatment. This analysis revealed a UV-induced decrease in IFNAR1 protein levels in the epidermis and/or dermis of human skin (Figure 1a and Table S1). Treatment of normal mouse skin with UV also led to a transient decrease in IFNAR1 protein (Figure 1b). Importantly, levels of IFNAR1 mRNA was not affected by UV exposure neither in human (Figure 1c) nor in mouse samples (Figure 1d) suggesting that UV induces post-transcriptional downregulation of IFNAR1. Exposure to UV in mammalian cells was shown to activate GCN2 kinase and ensuing phosphorylation of the eIF2α translation regulator (Deng et al., 2002). Activation of GCN2 and p38 protein kinase in response to amino acid deficit can also stimulate the ligand-independent phosphorylation, ubiquitination, and downregulation of IFNAR1 (Bhattacharya et al., 2013, Bhattacharya et al., 2011, Bhattacharya et al., 2010). Thus, we sought to determine the effect of UV on post-translational modifications of IFNAR1.
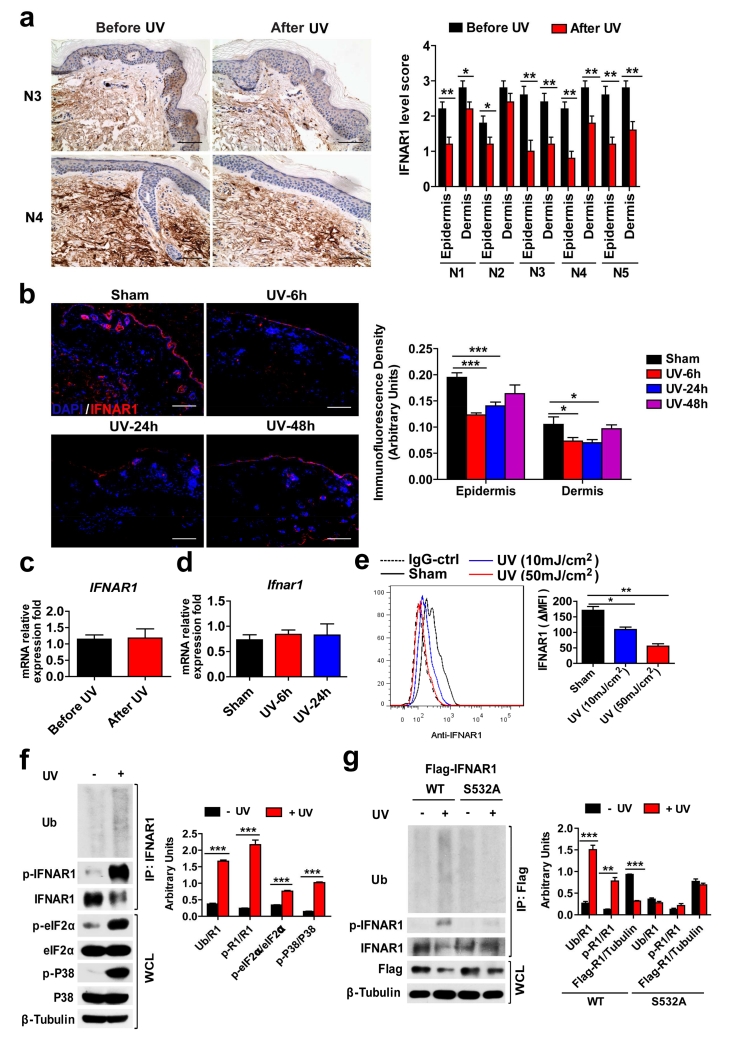
Figure 1. UV downregulates IFNAR1 in human and mouse keratinocytes and skin tissues.
(a) Representative IFNAR1 IHC staining (left) of human normal skin before and 24 h after UV treatment (#3 and 4) and quantification of these data from all samples (right). Scale bar 100 μm. Data are shown as the means ± SEM (n=5). *, P <0.05; **, P < 0.01.
(b) immunofluorescent analysis (left) and its quantification (right) of normal skin sections from non-irradiated (sham), and UV-irradiated WT mice 6, 24, and 48 h after treatment stained with DAPI (blue) and anti-IFNAR1 (red). Scale bar 100 μm. Data are shown as the means ± SEM (n=6).*, P <0.05; ***, P <0.001.
(c) The relative IFNAR1 mRNA level in human normal skin before and 24 h after UV treatment. Data are shown the means± SEM (n=5).
(d) The relative Ifnar1 mRNA level in the skin from non-irradiated (sham), and UV-irradiated WT mice 6 and 24 h after treatment. Data are shown as the means ± SEM (n=6).
(e) Representative FACS analysis of the cell surface IFNAR1 level (left) and the quantified MFI of IFNAR1 (normalize to IgG-Ctrl staining cells, right) in non-irradiated (sham) and UV-irradiated HaCaT cells 3 h after treatment. Data are shown the means± SEM (n=3). *, P <0.05; **, P < 0.01.
(f) Representative Immunoblotting of immunoprecipitated IFNAR1 and whole cell lysates (WCL) (left) and the quantification of the ratio of indicated band intensity (right) from non-irradiated and UV-irradiated HaCaT cells 1 h after treatment. Data are shown the means± SEM (n=3). ***, P <0.001.
(g) Representative Immunoblotting of immunoprecipitated IFNAR1 and whole cell lysates (WCL) (left) and the quantification of the ratio of indicated band intensity (right) from HaCaT cells transiently expressed Flag-IFNAR1 (wild type or S532A mutant) 1 h after UV treatment or without irradiation. Data are shown the means± SEM (n=3). **, P < 0.01; ***, P <0.001.
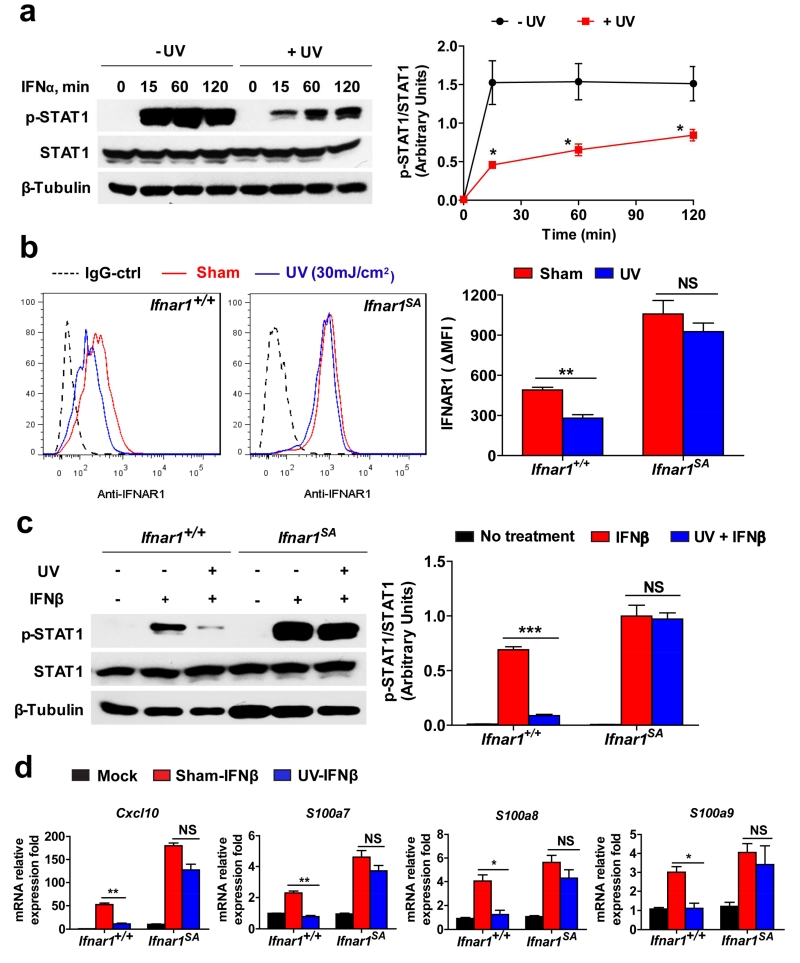
Cell surface levels of IFNAR1 on HaCaT human keratinocytes was noticeably decreased after UV treatment in vitro (Figure 1e). UV treatment of these cells also stimulated phosphorylation of eIF2α and activation of p38 kinase. Furthermore, exposure to UV resulted in an increased phosphorylation of IFNAR1 on Ser532 (known to be p38-dependent and critical for the subsequent ligand-independent phosphorylation of IFNAR1 on Ser535 (Bhattacharya et al., 2010, Bhattacharya et al., 2011) and increased IFNAR1 ubiquitination (Figure 1f). Compared to wild type FLAG-tagged recombinant IFNAR1, its S532A mutant expressed in HaCaT cells was less sensitive to UV-induced ubiquitination (Figure 1g) suggesting that UV treatment induces the phosphorylation-dependent ubiquitination and downregulation of IFNAR1. Consistent with the important role of IFNAR1 levels in the regulation of IFN responses, pre-treatment of HaCaT cells with UV attenuated phosphorylation of STAT1 in response to human IFNα (Figure 2a) suggesting that UV can suppress IFN signaling. To determine whether this mechanism can also function in primary cells, we used mouse keratinocytes from wild type (Ifnar1+/+) mice, or their littermates harboring the homozygous knocked-in Ifnar1SA that lacks the critical Ser residue (Ser526) and that are insensitive to phosphorylation-dependent ubiquitination and downregulation of IFNAR1 (Bhattacharya et al., 2014). Primary keratinocytes isolated from these mice were found to be responsive to IFN treatment as seen from analysis of changes in the expression of inflammatory genes known to be involved in the pathogenesis of psoriatic inflammation (including Cxcl10, S100A7, S100A8, and S100A9, Figure S1). Importantly, Ifnar1SA keratinocytes were insensitive to UV-induced downregulation of IFNAR1 (Figure 2b), UV-mediated suppression of STAT1 phosphorylation (Figure 2c), and expression of IFN-inducible genes (Figure 2d). Collectively, these results suggest that downregulation of IFNAR1, at least in part underlies the UV-induced suppression of IFN signaling.
Figure 2. UV inhibits IFN signaling.
(a) Representative immunoblotting analysis of STAT1 phosphorylation (left) and the quantification (the ratio of band intensity of phospho-STAT1 to total STAT1, right) in non-irradiated and UV-irradiated HaCaT cells treated with IFNα (100 IU/mL) for the indicated time points. Data are shown the means± SEM (n=3). *, P <0.05.
(b) Representative FACS analysis of the cell surface IFNAR1 level (left) and the quantified MFI of IFNAR1 (normalize to IgG-Ctrl staining cells, right) in non-irradiated (sham) and UV-irradiated primary keratinocytes isolated from neonatal WT (Ifnar1+/+) and Ifnar1SA mice 3 h after treatment. Data are shown the means± SEM (n=3). **, P < 0.01.
(c) Representative immunoblotting analysis of STAT1 phosphorylation (left) and the quantification (the ratio of band intensity of phospho-STAT1 to total STAT1, right) in non-irradiated and UV-irradiated primary keratinocytes (Ifnar1+/+ and Ifnar1SA) treated with IFNβ (1000 IU/mL) for 15 min. Data are shown the means± SEM (n=3). ***, P < 0.001.
(d) Relative (to mock treated cells, average value 1.0) mRNA expression levels of indicated genes in non-irradiated (sham) and UV-irradiated primary keratinocytes (Ifnar1+/+ and Ifnar1SA) treated with IFNβ (1000 IU/mL) for 6 h. Data are shown as the means ± SEM (n=3). *, P <0.05; **, P < 0.01.
Downregulation of IFNAR1 attenuates the severity of psoriasiform inflammation and determines the efficacy of its treatment by phototherapy
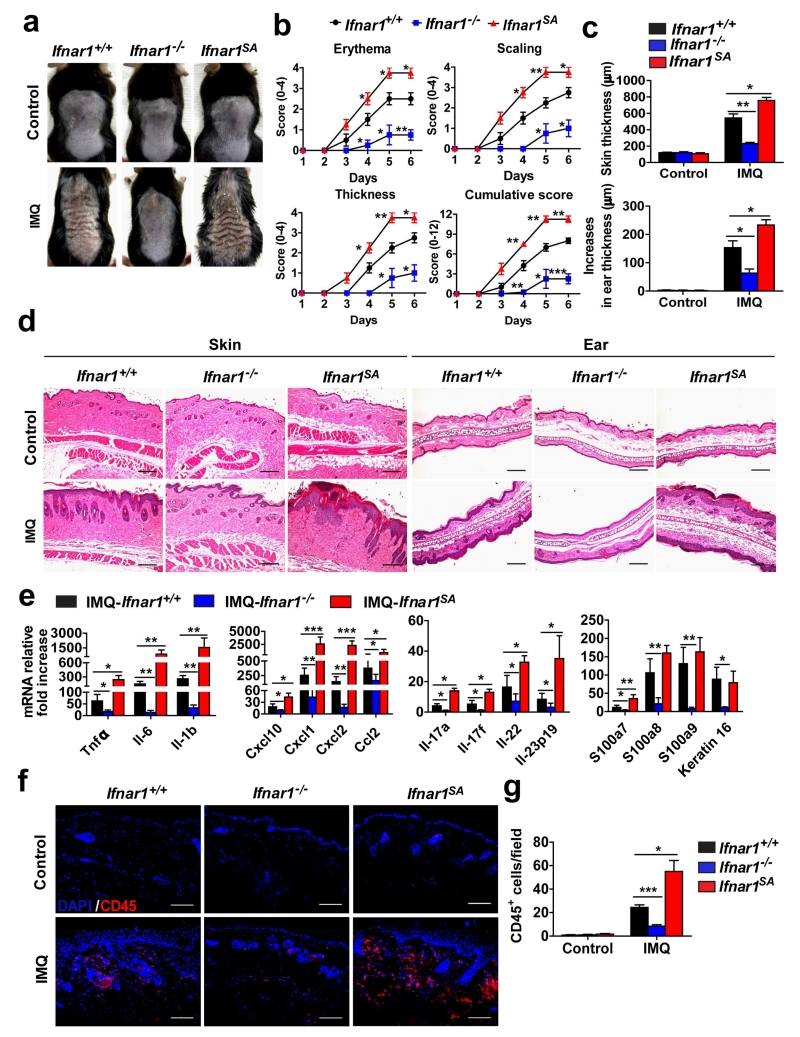
We next sought to determine whether IFNAR1 downregulation contributes to the therapeutic effects of UV using a mouse model of imiquimod-induced psoriasiform inflammation that was shown previously to be responsive to UV phototherapy (Racz et al., 2011). After the imiquimod exposure, wild type mice displayed erythema, scaling, and increase in the thickness of the dorsal skin and ears (Figure 3a-c), histological signs of inflammation, and increased expression of inflammatory cytokines (Figure 3d and e). Consistent with the report of Wohn et al (Wohn et al., 2013), mice lacking Ifnar1 were still capable of developing psoriasiform inflammation; however, our data clearly revealed that the severity of this inflammation was notably reduced upon Ifnar1 ablation (Figure 3). Similar results were reported by Grine and colleagues (Grine et al., 2015).
Figure 3. IFNAR1 levels determine the severity of psoriasiform skin inflammation.
Ifnar1+/+, Ifnar1−/−, and Ifnar1SA mice were treated with IMQ cream for 5 consecutive days on the shaved back skin and ear. Skin and ear tissue samples were harvested on day 6.
(a) Phenotypical presentation of WT (Ifnar1+/+), IFNAR1 deficient (Ifnar1−/−), and IFNAR1 SA (Ifnar1SA) mice on day 6 after IMQ treatment or no treatment (Control) of the shaved back skin.
(b) Time course of the scores in WT (Ifnar1+/+), IFNAR1 deficient (Ifnar1−/−), and IFNAR1 SA mice (Ifnar1SA). Data show the means ± SEM (n=4-5). *, P <0.05; **, P < 0.01. ***, P<0.001.
(c) The skin and ear thickness of WT (Ifnar1+/+), IFNAR1 deficient (Ifnar1−/−), and IFNAR1 SA mice (Ifnar1SA) after IMQ treatment or no treatment (Control). Data show the means ± SEM (n=4-5). *, P <0.05; **, P < 0.01.
(d) H&E staining of skin and ear tissue from WT (Ifnar1+/+), IFNAR1 deficient (Ifnar1−/−), and IFNAR1 SA mice (Ifnar1SA) after IMQ treatment or no treatment (Control). Individual experiments were conducted 3 times with similar results, with 1 representative shown for each group. Scale bars (100 μm).
(e) Relative fold increase (IMQ treated over untreated) of mRNA levels of indicated genes in the skin tissues of WT (Ifnar1+/+), IFNAR1 deficient (Ifnar1−/−), and IFNAR1 SA mice (Ifnar1SA) mice on day 6. Data are shown the means ± SEM (n=4-5). *, P <0.05; **, P < 0.01. ***, P<0.001.
(f) Immunofluorescent analysis of skin sections stained with DAPI (blue) and anti-CD45 (red) on day 6. Individual experiments were conducted 3 times with similar results, with 1 representative shown for each group. Scale bars (100 μm).
(g) Quantitative analysis of CD45-positive cells in each section shown in f. Data are shown the means ± SEM (n=5). *, P <0.05; ***, P<0.001.
Given that inflammatory cytokines themselves can induce p38 activity and downregulation of IFNAR1 (Bhattacharya et al., 2014), we hypothesized that inflammation-driven IFNAR1 ubiquitination and degradation may partially counteract the pro-inflammatory effects of IFN in wild type animals. Indeed, levels of IFNAR1 in the epidermis and dermis of imiquimod-treated Ifnar1+/+ mice were notably lower compared to those in the skin compartments of Ifnar1SA animals (Figure S2). Importantly, skin inflammation in the latter animals was greatly exacerbated as was evident from evaluation of clinical criteria (Figure 3a-c), histopathological alterations in dorsal skin and ear (Figure 3d), and additional analysis of leukocyte infiltration (Figure 3f-g). In addition, the fold increase (IMQ treated over non-treated) in the expression of genes encoding relevant inflammatory cytokines including TNFα, IL-6, IL-1β, IL-17, IL-22, and IL-23, and chemokine genes (e.g. Cxcl10, Cxcl1, etc) was greater in the skin of Ifnar1SA animals (Figure 3e). These results strongly suggest that IFN signaling plays an important role in the pathogenesis of imiquimod-induced psoriasiform inflammation and that downregulation of IFNAR1 attenuates the severity of this inflammation.
To evaluate the efficacy of UV treatment, we first established the minimal response dose (Figure S3) and then treated mice with either imiquimod and sham irradiation, or imiquimod and UV irradiation. Although UV treatment did not affect the overall serum levels of IFNα (Figure S4a), this treatment notably suppressed the expression of IFN-inducible genes (including Ifitm1, Isg15, etc) in mouse skin (Figure S4b). Importantly, similar to data obtained in normal human and mouse skin (Figure 1a and b), UV treatment dramatically decreased the levels of IFNAR1 in imiquimod-treated inflamed skin from wild type but not in Ifnar1SA mice (Figure S4c). These data suggest that by comparing UV responsiveness in Ifnar1+/+ and Ifnar1SA animals (which exhibit a similar minimal response dose, Figure S3), it would be possible to determine whether downregulation of IFNAR1 is required for the therapeutic efficacy of UV treatment.
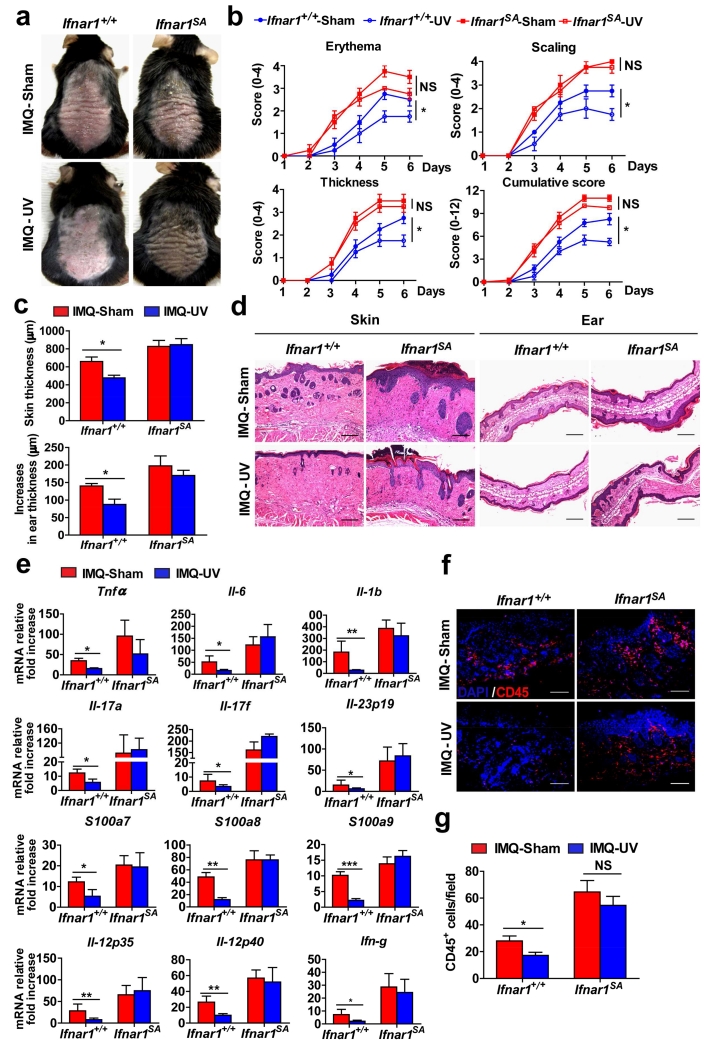
Consistent with data reported previously (Racz et al., 2011), UV treatment alleviated the course of psoriasiform inflammation in wild type animals as evident from clinical evaluation and scores (Figure 4a-c), histopathologic analysis (Figure 4d), expression of inflammatory cytokines (including IFNγ and IL-12 (Walters et al., 2003)) and markers of inflammation (Figure 4e), and leukocyte infiltration (Figure 4f and g). Remarkably, little (if any) anti-inflammatory effects were seen in imiquimod-treated Ifnar1SA mice (Figure 4). These data suggest that downregulation of IFNAR1 is required for a therapeutic efficacy of UV treatment against psoriatic inflammation.
Figure 4. IFNAR1 downregulation is required for efficient UV therapy.
Ifnar1+/+ and Ifnar1SA mice were treated with IMQ cream for 5 consecutive days on the shaved back skin and ear, and then irradiated or not (sham) with UV each day after IMQ treatment. Skin and ear tissue samples were harvested on day 6.
(a) Phenotypical presentation at day 6 of non-irradiated (sham) IMQ-treated, and UV-irradiated IMQ-treated WT (Ifnar1+/+) and IFNAR1 SA (Ifnar1SA) mice.
(b) Time course of the scores in non-irradiated (sham) IMQ-treated, and UV-irradiated IMQ-treated WT (Ifnar1+/+) and IFNAR1 SA mice (Ifnar1SA). Data show the means ± SEM (n=4-6). *, P <0.05.
(c) The skin and ear thickness of non-irradiated (sham) IMQ-treated, and UV-irradiated IMQ-treated WT (Ifnar1+/+) and IFNAR1 SA mice (Ifnar1SA). Data show the means ± SEM (n=4-6). *, P <0.05.
(d) H&E staining of skin and ear tissue from non-irradiated (sham) IMQ-treated, and UV-irradiated IMQ-treated WT (Ifnar1+/+) and IFNAR1 SA mice (Ifnar1SA). Individual experiments were conducted 3 times with similar results, with 1 representative shown for each group. Scale bars (100 μm).
(e) Relative mRNA fold increase (over non-IMQ treated mice) at day 6 of indicated genes in the skin tissues of non-irradiated (sham) IMQ-treated, and UV-irradiated IMQ-treated WT (Ifnar1+/+) and IFNAR1 SA mice (Ifnar1SA). Data show the means ± SEM (n=4-6). *, P <0.05; **, P < 0.01. ***, P<0.001.
(f) Immunofluorescent analysis of skin sections at day 6 stained with DAPI (blue) and anti-CD45 (red) from non-irradiated (sham) IMQ-treated, and UV-irradiated IMQ-treated WT (Ifnar1+/+) and IFNAR1 SA (Ifnar1SA) mice. Individual experiments were conducted 3 times with similar results, with 1 representative shown for each group. Scale bars (100 μm).
(g) Quantitative analysis of CD45-positive cells in each section shown in f. Data are shown the means ± SEM (n=5).*, P <0.05.
Pharmacologic elimination of IFNAR1 as the means for treating the psoriatic inflammation
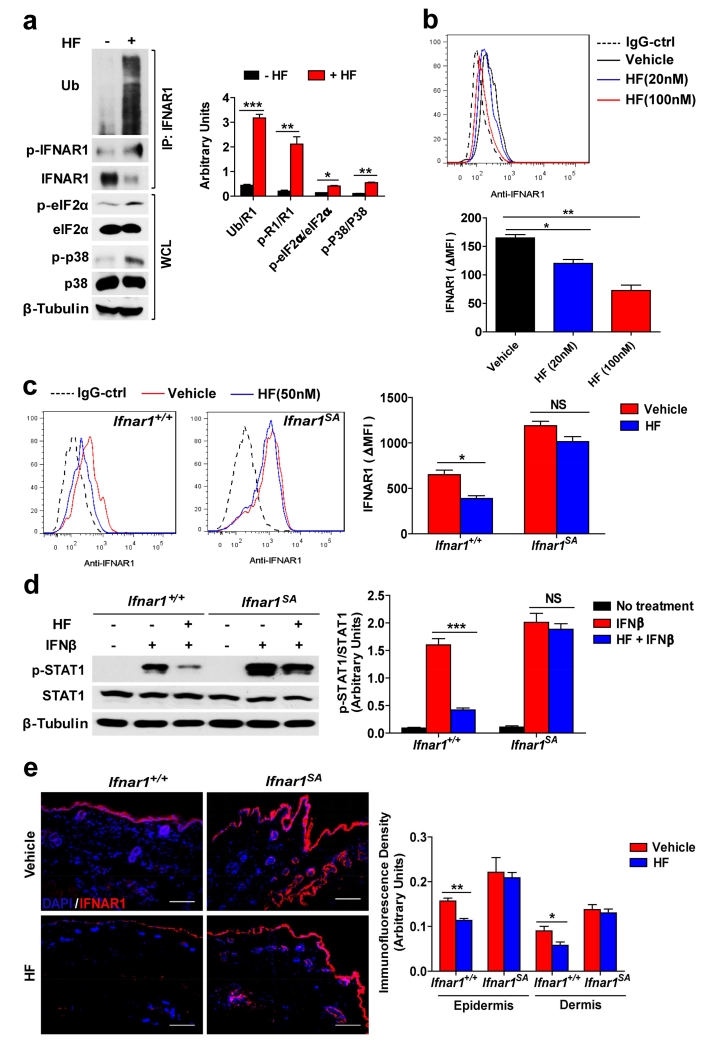
The effects of UV on skin are pleiotropic and affect numerous signaling, transcriptional, and cellular events (Chen et al., 2014) beyond its ability to downregulate IFNAR1. We sought to determine whether IFNAR1 downregulation triggered by another stimulus may be sufficient to elicit a therapeutic response against psoriatic inflammation. To this end, we used the anti-protozoan agent halofuginone, which was shown to induce Gcn2 activities (Peng et al., 2012). Treatment of human keratinocytes with halofuginone stimulated phosphorylation of eIF2α and activation of p38 kinase (Figure 5a), ubiquitination of IFNAR1 (Figure 5a), and downregulation of cell surface IFNAR1 level (Figure 5b). In murine primary keratinocytes, halofuginone-induced downregulation of cell surface IFNAR1 and suppression of IFN-stimulated STAT1 phosphorylation was seen in wild type cells but not in cells derived from Ifnar1SA mice (Figure 5c and d). In vivo administration of halofuginone led to downregulation of IFNAR1 protein (but not mRNA) in mouse splenocytes (Figure S5a and b). Finally, this treatment notably reduced IFNAR1 levels in normal skin of Ifnar1+/+ mice but not in Ifnar1SA animals (Figure 5e). Collectively, these results suggest that halofuginone is capable of stimulating IFNAR1 ubiquitination and eliciting the downregulation of IFNAR1 and suppression of IFN signaling.
Figure 5. The anti-protozoan drug halofuginone (HF) downregulates IFNAR1.
(a) Representative immunoblotting analysis of immunoprecipitated IFNAR1 and whole cell lysates (WCL) (left) and the quantification of the ratio of indicated band intensity (right) from vehicle- and HF-treated HaCaT cells 3 h after treatment. Data are shown the means± SEM (n=3). *, P <0.05; **, P < 0.01. ***, P<0.001.
(b) Representative FACS analysis of cell surface IFNAR1 level (above) and the quantified MFI of IFNAR1 (normalize to IgG-Ctrl staining cells, below) in vehicle- and HF-treated HaCaT cells 6 h after treatment. Data are shown the means± SEM (n=3). *, P <0.05; **, P < 0.01.
(c) Representative FACS analysis of cell surface IFNAR1 level (left) and the quantified MFI of IFNAR1 (normalize to IgG-Ctrl staining cells, right) in vehicle- and HF-treated primary keratinocytes isolated from neonatal WT (Ifnar1+/+) and Ifnar1SA mice 6 h after treatment. Data are shown the means± SEM (n=3). *, P <0.05.
(d) Representative immunoblotting analysis of STAT1 phosphorylation (left) and the quantification (the ratio of band intensity of phospho-STAT1 to total STAT1, right) in vehicle- and HF-treated primary keratinocytes (Ifnar1+/+ and Ifnar1SA) treated with IFNβ (1000 IU/mL) for 15 min. Data are shown the means± SEM (n=3). **, P < 0.01.
(e) Ifnar1+/+ and Ifnar1SA mice were injected intraperitoneally with 10% DMSO (vehicle) or 250 μg/kg of HF. Immunofluorescent analysis of skin sections stained with DAPI (blue) and anti-IFNAR1 (red) 12 h after HF injection (left) and its quantification (right). Scale bar 100 μm. Data are shown the means± SEM (n=4).*, P <0.05; **, P < 0.01.
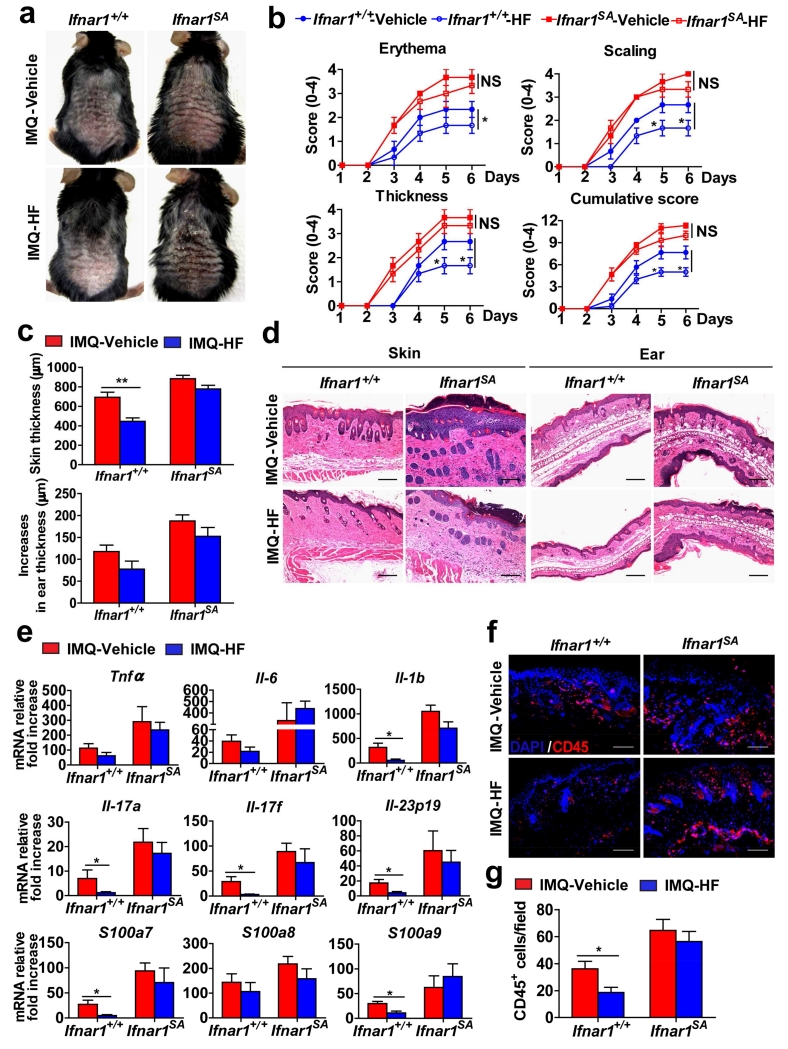
Remarkably, halofuginone treatment also decreased the levels of IFNAR1 in the skin of imiquimod-treated wild type mice (Figure S6). Examination of these data revealed a lesser thickening of epidermis in the skin of mice that received both imiquimod and halofuginone. Analysis of clinical and histolopathologic characteristics, as well as expression of inflammatory cytokines revealed that treatment with halofuginone leads to a notable alleviation of the imiquimod-induced psoriasiform inflammation (Figure 6). Importantly, Ifnar1SA animals that did not respond to halofuginone with a decrease in skin IFNAR1 levels (Figure S6) were also impervious to the ability of this drug to reduce skin inflammation (Figure 6). Together, these data suggest that pharmacologic elimination of IFNAR1 is sufficient for alleviating skin inflammation and provide a proof of principle for repurposing agents such as halofuginone (or developing novel agents) that stimulate IFNAR1 ubiquitination and degradation for the treatment of psoriasis.
Figure 6. Downregulation of IFNAR1 by HF attenuates the severity of psoriatic inflammation.
Ifnar1+/+ and Ifnar1SA mice were treated with IMQ cream for 5 consecutive days on the shaved back skin and ear, and injected intraperitoneally with 10% DMSO (vehicle) or 250 μg/kg of HF every other day, starting on the first day after IMQ treatment. Skin and ear tissue samples were harvested on day 6.
(a) Phenotypical presentation at day 6 of IMQ- and vehicle-treated, and IMQ- and HF-treated WT (Ifnar1+/+) and IFNAR1 SA (Ifnar1SA) mice.
(b) Time course of the scores in IMQ- and vehicle-treated, and IMQ- and HF-treated WT (Ifnar1+/+) and IFNAR1 SA mice (Ifnar1SA). Data show the means ± SEM (n=4-5). *, P <0.05.
(c) The skin and ear thickness of IMQ- and vehicle-treated, and IMQ- and HF-treated WT (Ifnar1+/+) and IFNAR1 SA mice (Ifnar1SA). Data show the means ± SEM (n=4-5). **, P <0.01.
(d) H&E staining of skin and ear tissue from IMQ- and vehicle-treated, and IMQ- and HF-treated WT (Ifnar1+/+) and IFNAR1 SA mice (Ifnar1SA). Individual experiments were conducted 3 times with similar results, with 1 representative shown for each group. Scale bars (100 μm).
(e) Relative mRNA fold increase (over non-IMQ treated mice) at day 6 of indicated genes in the skin tissues of IMQ- and vehicle-treated, and IMQ- and HF-treated WT (Ifnar1+/+) and IFNAR1 SA mice (Ifnar1SA). Data show the means ± SEM (n=4-5). *, P <0.05.
(f) Immunofluorescent analysis at day 6 of skin sections stained with DAPI (blue) and anti-CD45 (red) from IMQ- and vehicle-treated, and IMQ- and HF-treated WT (Ifnar1+/+) and IFNAR1 SA (Ifnar1SA) mice. Individual experiments were conducted 3 times with similar results, with 1 representative shown for each group. Scale bars (100 μm).
(g) Quantitative analysis of CD45-positive cells in each section shown in f. Data are shown the means± SEM (n=5). *, P <0.05.
Discussion
Biochemical, pharmacologic and genetic studies described here reveal an important role of IFN in the pathogenesis of psoriasis. Although correlative evidence from human studies (Afshar et al., 2013, Tas and Atsu, 2015, Kolb-Maurer et al., 2015, Baechler et al., 2006) and results obtained in human psoriatic skin grafted onto immunocompromised mice (Nestle et al., 2005) suggested the role of IFN in psoriatic inflammation, the mechanistic importance of IFN signaling remained controversial based on variable strength of attenuation of the imiquimod-induced inflammation in Ifnar1-null mice (Grine et al., 2015, Wohn et al., 2013). The latter phenotype might have been underestimated because it was compared with wild type mice where wild type IFNAR1 can undergo accelerated inflammation-induced ubiquitination and downregulation. A similar scenario of masking the importance of IFNAR1-driven signaling due to inflammatory eliminative signaling and rapid proteolytic turnover of IFNAR1 has been reported in experiments that utilized the experimental models of pancreatitis, hepatitis, and septic shock (Bhattacharya et al., 2014). We propose that inflammation-induced IFNAR1 downregulation may partially counteract the contribution of IFN to the inflammation itself. Our current data demonstrate a dramatic exacerbation of clinical score, histopathological inflammatory alterations, and expression of inflammatory cytokines in mice deficient in IFNAR1 ubiquitination and downregulation (Figure 3). These results strongly suggest an important role of IFN in psoriasiform inflammation and indicate that agents designed to stimulate IFNAR1 elimination may elicit a therapeutic effect against psoriasis.
Phototherapy has been a standard of care for psoriasis treatment for decades (Koo, 1999). Prompted by a commonality in activation of the GCN2-p38 pathway by UV and by amino acid starvation (shown previously to induce the ubiquitination and degradation of IFNAR1 (Bhattacharya et al., 2013, Bhattacharya et al., 2011), we investigated the effects of UV on IFNAR1 levels and signaling. Given that UV treatment downregulates the expression of inflammatory cytokines in mouse skin (Figure 4e) it is unlikely that these cytokines mediate the UV-induced downregulation of IFNAR1 in the context of phototherapy of psoriatic inflammation. These results together with experiments demonstrating a rapid ubiquitination and downregulation of IFNAR1 and suppression of downstream activation of STAT1 in human and mouse keratinocytes exposed to UV in vitro (Figure 1-2) suggest that UV directly activates the signaling pathways that trigger elimination of IFNAR1. Whereas our studies were initially prompted by UV-GCN2 connection, our results neither prove the role of GCN2 in UV-induced IFNAR1 downregulation nor exclude the participation of other pathways in this downregulation.
In addition, unabated IFNAR1 levels and signaling in cells and tissues harboring the ubiquitination-deficient Ifnar1SA suggest that ubiquitination and downregulation of IFNAR1 represent a critical mechanism that mediates the UV-induced suppression of the IFN pathway. Furthermore, psoriatic inflammation in the skin of Ifnar1SA animals was refractory to phototherapy (Figure 4). These results identify downregulation of IFNAR1 by UV as a major mechanism of UV therapeutic effects against psoriasis.
Characterization of the IFN pathway as a contributing element in the pathogenesis of psoriasis argues for new approaches of targeting IFN signaling to achieve therapeutic effects. Whereas pharmacologic neutralization of IFNα using an antibody-based agent (Sifalimumab) neither suppressed IFN gene expression signature nor exhibited clinical activity against the established psoriasis plaques (Bissonnette et al., 2010), it is important to note that this antibody was designed neither to affect activity of either other types of IFN (e.g. IFNβ) nor to interfere with the IFN receptor. It is plausible that IFNβ and/or other types of Type I IFN may also need to be neutralized for efficient therapeutic efforts. Another approach aiming to topically inhibit Janus kinases in the skin is emerging as promising (Kwatra et al., 2012), despite potential intrinsic limitations due to the long-term risks of inhibiting signaling locally elicited by numerous cytokines (including immunosuppressive ones such as IL-10) the effects of which require Janus kinase activity.
The results presented here provide a proof of principle for pharmacological mimicking or enhancement of the effects of UV by stimulating IFNAR1 downregulation to alleviate psoriatic inflammation. Our data show that the anti-protozoan agent halofuginone, capable of inducing the GCN2-p38 pathway, stimulates a robust downregulation of IFNAR1 in vitro and in vivo (Figure 5). Administration of halofuginone elicited a notable suppression of skin inflammation and improvement of clinical score in the imiquimod model of psoriasiform inflammation in wild type mice. Importantly, these effects were not observed in Ifnar1SA animals (Figure 6), consistent with an essential role of downregulation of IFNAR1 for the therapeutic effects of halofuginone. These results provide a foundation for further development and/or repurposing of agents capable of inducing IFNAR1 ubiquitination and downregulation for psoriasis treatment. While halofuginone did not exhibit major toxicities in animal studies or human volunteers and limited number of patients with chronic graft-versus-host disease or scleroderma (Pines and Spector, 2015), its potential for psoriasis therapy needs to be rigorously investigated in the future.
Materials and Methods
Human skin samples
All experiments using human samples were approved by the Institutional Review Board of the VAMC of Philadelphia. The human skin biopsy samples were collected from normal skin of five psoriasis patients before and 24 h after ultraviolet irradiation. Written and informed consent was obtained from all patients in this study. The characteristics of their skin are described in Table S1. A Berger solar simulator with a 150 watt lamp (Solar Light Company, Inc., Glenside, PA; model number 16S-150/300 using a 150 Watt lamp) was used as the source of ultraviolet radiation. This solar simulator emits UVA1, UVA2, and UVB light in a ratio similar to that of sunlight (0.0036% UVC, 0.016% UVB, 96.63% UVA (11.28% UVA2 and 88.72% UVA1). The current was calibrated before each use. Before each use the machine’s surface power density in mW/cm2 was measured at the simulator’s aperture, and this was used to calculate the seconds necessary to deliver the target dosages.
Mice
All animal procedures were approved by the Institution Animal Care and Use Committee (IACUC, protocols # 803995 and 804688) of the University of Pennsylvania. Eight to ten weeks old male mice including wild type (Ifnar1+/+), Ifnar1−/− mice (Roth-Cross et al., 2008) (a generous gift of S. Weiss) and Ifnar1SA (Ifnar1tm1.1Syfu) mice (Zheng et al., 2011) were used in this study. All mice were on 100% C57Bl/6 background (Ifnar1SA were generated in C57Bl/6 embryonic stem cells (Liu et al., 2009) and maintained in this background; Ifnar1−/− mice were back-crossed into C57Bl/6 background for more than 20 generations) and were homozygous for the indicated Ifnar1 alleles.
IMQ-induced psoriasiform inflammation
Aldara cream (CLIPPER HUMAN PHARMS) containing 5% imiquimod was applied to the shaved dorsal skin and to the ears daily for 5 days. Control mice also were shaved but otherwise were left untreated. The severity of inflammation of the back skin was scored as described previously (van der Fits et al., 2009). The ear thickness and skin thickness was measured using a micrometer. Mice were euthanized on day 6. Mouse dorsal skin and ear tissue were fixed in 4% PFA. Tissue sections were stained with H&E.
UV treatment
For mice irradiation, mice were treated daily with imiquimod on the shaved back skin for 5 days. Starting on the first day of the experiment, mice were irradiated with a FS20 T12/UVB lamp (National Biological Corporation, spectrum centered at roughly 311 nm, equipped with a filter to exclude UVC) daily one hour after imiquimod treatment. The starting applied UV dose was 70% minimal response doses (around 400mJ/cm2) on the first day and it was increased each time by 5%. Mice were sacrificed on day 6. The samples were taken from the back skin. For cell irradiation, the culture medium was removed, and the cells were washed with PBS for one time. Each 10 cm cell plate contained 500 μl PBS covered with a filter and were irradiated with the UVB lamp at 30-50mJ/cm2. After UV exposure, the PBS was removed and replaced with culture medium, and the cells were incubated until harvesting.
Plasmids, antibodies and other reagents
HaCaT cells (ATCC) were cultured in DMEM contained with 10% FBS. Human Flag-IFNAR1 expression plasmids (wild type and S532A mutant) were described previously (Bhattacharya et al., 2010). Recombinant human IFNα2b was from Roche. Recombinant murine IFNβ was from PBL. Halofuginone was from Toronto Research Chemicals Inc.
Antibodies and immuno-techniques
Commercially available antibodies against pSTAT1, STAT1, p-eIF2α, p-p38, p38 (Cell Signaling Technology), eIF2α (Biosources), p38 (Santa Cruz Biotechnology), β-tubulin (Cell Signaling Technology), and ubiquitin (P4D1, Santa Cruz Biotechnology) were purchased. Cell lysis, immunoprecipitation (IP), and immunoblotting procedures were described previously (Liu et al., 2009). Antibodies against total human IFNAR1 (Goldman et al., 1999) and phosphorylated IFNAR1 (Bhattacharya et al., 2010, Bhattacharya et al., 2011, Kumar et al., 2004) were described previously. The immunoblotting band intensity was measured by Image J.
Immunohistochemistry Staining
Paraffin-embedded skin sections were immunostained with anti-human IFNAR1 (Sigma-Aldrich) according to the instructions of Dako’s EnVision Flex Kits (High pH antigen retrieval solution). Five sections were analyzed per subject. The IFNAR1 levels in the skin sections were scored as described in Ref (Messina et al., 2008). Briefly, we calculated both the percentage of stained cells and the intensity of staining as follows: 0=no stained cells; 1=1% to 25% stained cells; 2=26%-50% stained cells; 3=more than 50% cells, and the intensity 0=negative staining; 1=weak staining; 2=moderate staining; 3=strong staining. The sum of these was the composite score to quantify the IFNAR1 level: 0 (0), 1 (1-2), 2 (3-4), 3 (5-6).
Flow cytometry
Cell surface levels of IFNAR1 in human and mouse cells were determined by flow cytometry following staining cells with anti-hIFNAR1 (AA3, 1:1000 (Goldman et al., 1999)), subsequently goat anti mouse IgG conjugated with Alexa Fluor 488 (1:1000, Life Technologies) or anti-mIFNAR1 conjugated with PE (Biolegend, 1:500). The data were analyzed with FlowJo Software.
Immunofluorescence
The frozen skin sections were fixed with acetone, blocked with 5% goat serum with 1% BSA-PBS, stained with biotin-anti mouse CD45 (1:100, Biolegend), anti-mouse IFNAR1 (1:100, Sino Biological Inc), and subsequently streptavidin conjugated Alexa Fluor 594 (1:500, Biolegend) or goat anti-rabbit conjugated Alexa Fluor 594 (1:500, Life Technologies). The slides were mounted with DAPI (Life Technologies) and visualized with a fluorescence microscope (Olympus BX51). Quantification of CD45 positive cells was determined by counting five different fields per section, and the IFNAR1-immunofluorescence density was analyzed by Image J.
Mouse primary keratinocytes culture
Isolation of keratinocytes from neonatal mice and their culture conditions were as described previously (Lichti et al., 2008). Briefly, the newborn mice within 3 days of birth were sacrificed. The tail and legs were snipped off, and the body was decontaminated by dipping several times in sterile H2O, 70% ethanol, sterile PBS in sequence. The whole skin was removed from the body and incubated in 0.25% trypsin-EDTA at 4°C overnight. The next day, we separated the epidermis and dermis, tease the epidermal sheet into small pieces, and dissociated keratinocytes by mechanical shearing of rotating. The keratinocytes were cultured at 34°C with 8% CO2 in K-SFM (Life Technologies) plain medium supplemented with Chelated FBS (4%), mEGF (10 ng/mL), BPE (50 μg/mL), calcium (45 μM), and antibiotics.
Quantitative Real-time PCR
Human skin total RNA was extracted from paraffin embedded skin sections by using PureLink™ FFPE RNA Isolation Kit (Invitrogen). Mouse skin total RNA was extracted from whole biopsies from the back skin by using Trizol (Invitrogen) and the RNeasy kit (Qiagen) following sacrifice. Using 1 μg of total RNA template, cDNA was prepared using SuperScript II reverse transcriptase (Invitrogen) and oligo (dT) primers. Tnfa, Il6, ll1b, Cxcl10, Cxcl1, Cxcl2, Ccl2, Il17a, Il17f, Il22, Il23, S100a7, S100a8, S100a9, keratin16, Il-12p35, Il-12p40, Ifng, Ifitm1, Stat1, Isg15, Ifnar1, and Gapdh mRNA levels were measured using real-time quantitative PCR analysis with the ABI PRISM 7700 sequence detection system (Applied Biosystems). Sequences for the PCR primers are shown in Table S2.
Statistical analyses
All data are presented as means ± SEM of at least three independent experiments or from a representative experiment of three independent experiments. Statistical analyses were carried out using GraphPad Prism software. A two-tailed unpaired t test was used to evaluate significance; P values <0.05 were considered statistically significant.
Supplementary Material
Acknowledgements
This work was supported by the NIH/NCI RO1 CA092900 (to S.Y.F. and H.R.), PO1 CA165997 grant (to S.Y.F.), and CA016583 grant (to J.T.S.). Additional support from T32 CA009140 (to K.V.K.) is also appreciated. We thank Hee Joo Kim for help with experiments; Susan Weiss for reagents, and the members of Fuchs, Diehl, and Koumenis labs (at the University of Pennsylvania) for critical suggestions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions
S.Y.F., J.G., J.T.S., V.P.W., and H.R. designed the research; J.G., M.G., X.Y., K.V.K., C.M.M., M.S., and H.R. performed the experiments and interpreted the data; S.Y.F., J.G., J.T.S. and D.P.B. wrote the manuscript with the help of all authors.
Conflict of interest
All authors declare no competing financial interests.
REFERENCES
- Afshar M, Martinez AD, Gallo RL, Hata TR. J Eur Acad Dermatol Venereol. 2013;27:771–8. doi: 10.1111/j.1468-3083.2012.04582.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baechler EC, Batliwalla FM, Reed AM, Peterson EJ, Gaffney PM, Moser KL, Gregersen PK, Behrens TW. Immunol Rev. 2006;210:120–37. doi: 10.1111/j.0105-2896.2006.00367.x. [DOI] [PubMed] [Google Scholar]
- Bhattacharya S, HuangFu WC, Dong G, Qian J, Baker DP, Karar J, Koumenis C, Diehl JA, Fuchs SY. Oncogene. 2013;32:4214–21. doi: 10.1038/onc.2012.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya S, HuangFu WC, Liu J, Veeranki S, Baker DP, Koumenis C, Diehl JA, Fuchs SY. J Biol Chem. 2010;285:2318–25. doi: 10.1074/jbc.M109.071498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya S, Katlinski KV, Reichert M, Takano S, Brice A, Zhao B, Yu Q, Zheng H, Carbone CJ, Katlinskaya YV, Leu NA, McCorkell KA, Srinivasan S, Girondo M, Rui H, May MJ, Avadhani NG, Rustgi AK, Fuchs SY. EMBO Mol Med. 2014;6:384–97. doi: 10.1002/emmm.201303236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya S, Qian J, Tzimas C, Baker DP, Koumenis C, Diehl JA, Fuchs SY. J Biol Chem. 2011;286:22069–76. doi: 10.1074/jbc.M111.238766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissonnette R, Papp K, Maari C, Yao Y, Robbie G, White WI, Le C, White B. J Am Acad Dermatol. 2010;62:427–36. doi: 10.1016/j.jaad.2009.05.042. [DOI] [PubMed] [Google Scholar]
- Chen H, Weng QY, Fisher DE. J Invest Dermatol. 2014;134:2080–5. doi: 10.1038/jid.2014.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coccia EM, Uze G, Pellegrini S. Cell Mol Biol (Noisy-le-grand) 2006;52:77–87. [PubMed] [Google Scholar]
- Deng J, Harding HP, Raught B, Gingras AC, Berlanga JJ, Scheuner D, Kaufman RJ, Ron D, Sonenberg N. Curr Biol. 2002;12:1279–86. doi: 10.1016/s0960-9822(02)01037-0. [DOI] [PubMed] [Google Scholar]
- Fuchs SY. J Interferon Cytokine Res. 2013;33:211–25. doi: 10.1089/jir.2012.0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman LA, Zafari M, Cutrone EC, Dang A, Brickelmeier M, Runkel L, Benjamin CD, Ling LE, Langer JA. J Interferon Cytokine Res. 1999;19:15–26. doi: 10.1089/107999099314379. [DOI] [PubMed] [Google Scholar]
- Grine L, Dejager L, Libert C, Vandenbroucke RE. J Immunol. 2015;194:5094–102. doi: 10.4049/jimmunol.1403015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazzalin CA, Cano E, Cuenda A, Barratt MJ, Cohen P, Mahadevan LC. Curr Biol. 1996;6:1028–31. doi: 10.1016/s0960-9822(02)00649-8. [DOI] [PubMed] [Google Scholar]
- Hida S, Ogasawara K, Sato K, Abe M, Takayanagi H, Yokochi T, Sato T, Hirose S, Shirai T, Taki S, Taniguchi T. Immunity. 2000;13:643–55. doi: 10.1016/s1074-7613(00)00064-9. [DOI] [PubMed] [Google Scholar]
- Huangfu WC, Qian J, Liu C, Liu J, Lokshin AE, Baker DP, Rui H, Fuchs SY. Oncogene. 2012;31:161–72. doi: 10.1038/onc.2011.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Krueger JG. Dermatol Clin. 2015;33:13–23. doi: 10.1016/j.det.2014.09.002. [DOI] [PubMed] [Google Scholar]
- Kolb-Maurer A, Goebeler M, Maurer M. Int J Mol Sci. 2015;16:14951–60. doi: 10.3390/ijms160714951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo JY. J Dermatol. 1999;26:723–33. doi: 10.1111/j.1346-8138.1999.tb02083.x. [DOI] [PubMed] [Google Scholar]
- Kumar KG, Barriere H, Carbone CJ, Liu J, Swaminathan G, Xu P, Li Y, Baker DP, Peng J, Lukacs GL, Fuchs SY. J Cell Biol. 2007;179:935–50. doi: 10.1083/jcb.200706034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar KG, Krolewski JJ, Fuchs SY. J Biol Chem. 2004;279:46614–20. doi: 10.1074/jbc.M407082200. [DOI] [PubMed] [Google Scholar]
- Kumar KG, Tang W, Ravindranath AK, Clark WA, Croze E, Fuchs SY. EMBO J. 2003;22:5480–90. doi: 10.1093/emboj/cdg524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwatra SG, Dabade TS, Gustafson CJ, Feldman SR. J Drugs Dermatol. 2012;11:913–8. [PubMed] [Google Scholar]
- Lichti U, Anders J, Yuspa SH. Nat Protoc. 2008;3:799–810. doi: 10.1038/nprot.2008.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, HuangFu WC, Kumar KG, Qian J, Casey JP, Hamanaka RB, Grigoriadou C, Aldabe R, Diehl JA, Fuchs SY. Cell Host Microbe. 2009;5:72–83. doi: 10.1016/j.chom.2008.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowes MA, Bowcock AM, Krueger JG. Nature. 2007;445:866–73. doi: 10.1038/nature05663. [DOI] [PubMed] [Google Scholar]
- Messina JL, Yu H, Riker AI, Munster PN, Jove RL, Daud AI. Cancer Control. 2008;15:196–201. doi: 10.1177/107327480801500302. [DOI] [PubMed] [Google Scholar]
- Nestle FO, Conrad C, Tun-Kyi A, Homey B, Gombert M, Boyman O, Burg G, Liu YJ, Gilliet M. J Exp Med. 2005;202:135–43. doi: 10.1084/jem.20050500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng W, Robertson L, Gallinetti J, Mejia P, Vose S, Charlip A, Chu T, Mitchell JR. Sci Transl Med. 2012;4:118ra11. doi: 10.1126/scitranslmed.3002629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piehler J, Thomas C, Garcia KC, Schreiber G. Immunol Rev. 2012;250:317–34. doi: 10.1111/imr.12001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pines M, Spector I. Molecules. 2015;20:573–94. doi: 10.3390/molecules20010573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platanias LC. Nat Rev Immunol. 2005;5:375–86. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- Price MA, Cruzalegui FH, Treisman R. EMBO J. 1996;15:6552–63. [PMC free article] [PubMed] [Google Scholar]
- Racz E, Prens EP, Kurek D, Kant M, de Ridder D, Mourits S, Baerveldt EM, Ozgur Z, van IWF, Laman JD, Staal FJ, van der Fits L. J Invest Dermatol. 2011;131:1547–58. doi: 10.1038/jid.2011.53. [DOI] [PubMed] [Google Scholar]
- Roth-Cross JK, Bender SJ, Weiss SR. J Virol. 2008;82:9829–38. doi: 10.1128/JVI.01199-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tartar D, Bhutani T, Huynh M, Berger T, Koo J. J Drugs Dermatol. 2014;13:564–8. [PubMed] [Google Scholar]
- Tas F, Atsu N. Cutan Ocul Toxicol. 2015:1–2. doi: 10.3109/15569527.2015.1020545. [DOI] [PubMed] [Google Scholar]
- Ueyama A, Yamamoto M, Tsujii K, Furue Y, Imura C, Shichijo M, Yasui K. J Dermatol. 2014;41:135–43. doi: 10.1111/1346-8138.12367. [DOI] [PubMed] [Google Scholar]
- Uze G, Schreiber G, Piehler J, Pellegrini S. Curr Top Microbiol Immunol. 2007;316:71–95. doi: 10.1007/978-3-540-71329-6_5. [DOI] [PubMed] [Google Scholar]
- van der Fits L, Mourits S, Voerman JS, Kant M, Boon L, Laman JD, Cornelissen F, Mus AM, Florencia E, Prens EP, Lubberts E. J Immunol. 2009;182:5836–45. doi: 10.4049/jimmunol.0802999. [DOI] [PubMed] [Google Scholar]
- Walters IB, Ozawa M, Cardinale I, Gilleaudeau P, Trepicchio WL, Bliss J, Krueger JG. Arch Dermatol. 2003;139:155–61. doi: 10.1001/archderm.139.2.155. [DOI] [PubMed] [Google Scholar]
- Wohn C, Ober-Blobaum JL, Haak S, Pantelyushin S, Cheong C, Zahner SP, Onderwater S, Kant M, Weighardt H, Holzmann B, Reizis B, Becher B, Prens EP, Clausen BE. Proc Natl Acad Sci U S A. 2013;110:10723–8. doi: 10.1073/pnas.1307569110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong T, Hsu L, Liao W. J Cutan Med Surg. 2013;17:6–12. doi: 10.2310/7750.2012.11124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Qian J, Carbone CJ, Leu NA, Baker DP, Fuchs SY. Blood. 2011;118:4003–6. doi: 10.1182/blood-2011-06-359745. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.