Abstract
Therapeutic monoclonal antibodies are mainly produced in mammalian cells to date. However, unglycosylated antibody fragments can also be produced in the bacterium Escherichia coli which brings several advantages, like growth on cheap media and high productivity. One of the most popular E. coli strains for recombinant protein production is E. coli BL21(DE3) which is usually used in combination with the pET expression system. However, it is well known that induction by isopropyl β-d-1-thiogalactopyranoside (IPTG) stresses the cells and can lead to the formation of insoluble inclusion bodies. In this study, we revisited the pET expression system for the production of a novel antibody single-chain variable fragment (scFv) with the goal of maximizing the amount of soluble product. Thus, we (1) investigated whether lactose favors the recombinant production of soluble scFv compared to IPTG, (2) investigated whether the formation of soluble product can be influenced by the specific glucose uptake rate (qs,glu) during lactose induction, and (3) determined the mechanistic correlation between the specific lactose uptake rate (qs,lac) and qs,glu. We found that lactose induction gave a much greater amount of soluble scFv compared to IPTG, even when the growth rate was increased. Furthermore, we showed that the production of soluble protein could be tuned by varying qs,glu during lactose induction. Finally, we established a simple model describing the mechanistic correlation between qs,lac and qs,glu allowing tailored feeding and prevention of sugar accumulation. We believe that this mechanistic model might serve as platform knowledge for E. coli.
Electronic supplementary material
The online version of this article (doi:10.1007/s00253-016-7620-7) contains supplementary material, which is available to authorized users.
Keywords: Escherichia coli BL21(DE3), pET expression system, Lactose induction, Antibody fragment, Soluble protein, Mechanistic model
Introduction
Antibodies are used to treat a wide variety of human diseases. More than 35 monoclonal antibodies and antibody fragments have been commercialized, and around 240 therapeutic monoclonal antibodies and antibody fragments are in clinical trials (Lee and Jeong 2015). Since more than 1000 kg of these therapeutics are needed per year worldwide, there is an urge for cheap and fast production (Elvin et al. 2013; Lee and Jeong 2015; Liu 2014; Rodrigues et al. 2010; Walsh 2014). Due to the requirement of post-translational modifications, most therapeutic monoclonal antibodies and antibody fragments are produced in mammalian cells to date. However, there are many drawbacks such as glycan heterogeneity, low volumetric productivity, long cultivation times, expensive media, and the potential risk of virus contamination (Khan 2013; Lee and Jeong 2015). Thus, the prokaryotic organism Escherichia coli has been investigated as alternative host for the production of unglycosylated antibody fragments, mainly single-chain variable fragments (scFv), which are also suitable for antigen detection (Lee and Jeong 2015; Spadiut et al. 2014; Wals and Ovaa 2014). E. coli can be cultivated on inexpensive media to high cell densities and has a high growth rate; its genetics are very well characterized and an increasingly large number of cloning vectors and mutant host strains are available (e.g., Baeshen et al. 2015; Rosano and Ceccarelli 2014). The E. coli strain BL21(DE3) and its derivatives are by far the most used E. coli strains for recombinant protein production as they exhibit several biotechnological advantages compared to other E. coli strains, such as low acetate yield, high biomass yield, and reduced expression of proteases (Choi et al. 2006; Ferrer-Miralles et al. 2015; Rosano and Ceccarelli 2014). Usually, the well-known pET expression system is used in combination with E. coli BL21(DE3) (Studier and Moffatt 1986). The lac operon can be induced by allolactose and its molecular mimic isopropyl β-d-1-thiogalactopyranoside (IPTG) (Neubauer et al. 1992). IPTG is a very strong inducer that is not metabolized by E. coli, which is why one point addition is sufficient. Thus, IPTG is usually used in industrial production processes with E. coli BL21(DE3). However, IPTG is known to put a high metabolic burden on the cells resulting in the formation of inactive aggregates of the recombinant target protein, known as inclusion bodies (IBs). Thus, lactose has been studied as alternative inducer. Lactose was found to be as effective as IPTG, to increase cell fitness, to reduce IB formation, and to enhance the formation of soluble recombinant product (Bashir et al. 2015; Fruchtl et al. 2015; Gombert and Kilikian 1998; Neubauer et al. 1992; Pei et al. 2011; Zou et al. 2014). However, lactose is metabolized by E. coli making stable induction more complicated as it has to be continuously supplied (Striedner et al. 2003). In a previous study, it was nicely shown that lactose metabolism strongly depends on the available amount of glucose (Kremling et al. 2015). However, a potential mechanistic correlation between glucose and lactose uptake has not been investigated yet.
In this study, we used E. coli BL21(DE3) and the pET expression system for the production of a novel scFv (Stadlmann et al. 2015). We hypothesized that induction by lactose increases the amount of soluble product compared to IPTG. Thus, we (1) tested and compared IPTG and lactose as inducer, (2) investigated whether the formation of soluble product can be influenced by the specific uptake rate of glucose during induction with lactose, and (3) determined a mechanistic correlation between the specific uptake rates of lactose and glucose.
Materials and methods
Strain
E. coli BL21(DE3) (Life technologies, Carlsbad, CA, USA) and the pET28a(+) expression system were used for production of the recombinant scFv which describes an engineered IgY fragment against PT-gliadin useful for the treatment of celiac disease (Stadlmann et al. 2015).
Bioreactor cultivations
Media
A defined minimal medium according to DeLisa et al. (1999) supplemented with 0.02 g/L kanamycin and different amounts of glucose and lactose (Table 1) was used for all cultivations.
Table 1.
Sugar concentrations in different DeLisa media
| Component | Pre-culture | Batch | Feed glucose | Feed lactose |
|---|---|---|---|---|
| C6H12O6·H2O (g/L) | 8.8 | 22.0 | 275 | – |
| C12H22O11·H2O (g/L) | – | – | – | 210 |
Pre-culture
A 500-mL sterile DeLisa pre-culture medium were inoculated from frozen stocks (1.5 mL, −80 °C) and incubated in a 2500-mL High-Yield shake flask in an Infors HR Multitron shaker (Infors, Bottmingen, Switzerland) at 37 °C and 230 rpm for 20 h. Then a 4500-mL DeLisa-batch medium in the bioreactor was inoculated with 500 mL of pre-culture.
Batch and fed-batch cultivations
Batch and fed-batch cultivations were done in a stainless steel Satorius Biostat Cplus bioreactor (Satorius, Göttingen, Germany) with a working volume of 10 L. The bioreactor was aerated with a mixture of pressurized air and pure oxygen at 1.5 vvm and agitated constantly at 1000 rpm. Dissolved oxygen (dO2) was monitored with a fluorescence dissolved oxygen electrode Visiferm DO425 (Hamilton, Reno, NV, USA) and kept above 40 % throughout all cultivations by varying the ratio of pressurized air to pure oxygen. pH was monitored with an EasyFerm electrode (Hamilton, Reno, NV, USA) and maintained constant at pH 7.2 by addition of NH4OH (12.5 %). Base consumption was determined gravimetrically. CO2 and O2 concentrations in the off-gas were monitored by a DASGIP GA gas analyzer (Eppendorf, Hamburg, Germany). All process parameters were adjusted and logged by the process information management system Lucullus (Biospectra, Schlieren, Switzerland).
The batch phase was conducted at 35 °C and yielded a biomass concentration of 8–9 g dry cell weight (DCW) per liter. After depletion of glucose, visible by a drop in the CO2 off-gas signal, a fed-batch to generate a biomass was conducted. We fed at a constant specific glucose uptake rate (qs,glu) of 0.2 g/g/h. When the final DCW reached 25 g/L, the culture was induced by IPTG or lactose, respectively. During non-induced fed-batch and induction with IPTG, DCW in the bioreactor was estimated using a soft-sensor tool (Wechselberger et al. 2013). During induction with lactose, DCW was calculated assuming a constant biomass yield (YX/S = 0.37 g/g, own unpublished data). The feed rate was adjusted to maintain a constant qs,glu and was calculated according to Eq. 1.
| 1 |
- F
Feed rate (g/h)
- qs
Specific substrate uptake rate (g/g/h)
- X
DCW concentration (g/L)
- V
Reactor volume (L)
- W
Amount of substrate per feed (g/g)
Cultivation strategy
In this study, cultivations following a standard procedure comprising three phases (batch, non-induced fed-batch, induced fed-batch) as well as dynamic experiments (pulses and shifts) were carried out. Applying dynamic process conditions to accelerate strain characterization and bioprocess development is a common approach in our working group (Dietzsch et al. 2011a, 2011b; Jazini and Herwig 2011; Spadiut et al. 2013; Zalai et al. 2012). An overview of the different cultivations and their respective goals is shown in Supplementary Table S1. Induction was either performed by 0.5 mM IPTG or by lactose which was applied either as pulses or by continuous feeding. In these cultivations, the lactose concentration in the media was kept in excess between 5 and 15 g/L.
Sampling
Samples were taken at the beginning and the end of the batch and the non-induced fed-batch. During induction, sampling was performed at the beginning and the end of each shift/pulse and every hour during fed-batch cultivations. Specific product formation rates and final product yields are given for an induction phase of approximately 4 h for all cultivations. DCW was determined by centrifugation (4500g, 4 °C, 10 min) of 5 mL cultivation broth, washing the obtained cell pellet with a 0.1 % NaCl solution and subsequent drying at 105 °C for 48 h. Optical density at 600 nm (OD600) was measured in the photometer Genesys 20 (Thermo Scientific, Waltham, MA, USA). Samples were diluted with deionized water to stay within the linear range of the photometer (OD600 0.1–0.8). A linear correlation between OD600 and DCW was established to verify and, if necessary, correct the DCW estimation of the soft-sensor (Eq. 2).
| 2 |
- DCW
Biomass dry cell weight (g/L)
- OD600
Optical density at 600 nm
Substrate and metabolite quantification
Cell-free samples of the cultivation broth were analyzed for concentrations of substrates and metabolites by HPLC (Agilent Technologies, Santa Clara, CA, USA) with a Supelcogel C-610 H ion exchange column (Sigma-Aldrich, St. Louis, MO, USA) and a refractive index detector (Agilent Technologies, Santa Clara, CA, USA). The mobile phase was 0.1 % H3PO4 with a constant flow rate of 0.5 mL/min, and the system was run isocratically at 30 °C.
Product quantification
Cells were harvested (4500g, 4 °C, 10 min), resuspended, and diluted in Tris buffer (100 mM, 10 mM EDTA, pH 7.4) to a DCW concentration of 5 g/L and subsequently homogenized in an EmusiflexC3 Homgeniziser (Avestin, Ottowa, ON, USA) at 1500 bar for five passages. After centrifugation (14,000g, 4 °C, 10 min), soluble protein (SP) was recovered in the supernatant and IBs in the pellet.
Cell debris from 0.5 mg DCW was resuspended in 1× Laemmli buffer, and the supernatant was diluted with 2× Laemmli buffer before the samples were heated at 95 °C for 10 min. Ten microliters of each sample were loaded onto pre-cast SDS gels (8–16 %) (GE Healthcare, Little Chalfont, UK). Gels were run in an Amersham ECL Gel Box, a horizontal mini-gel system (GE Healthcare, Little Chalfont, UK) for 90 min at 140 V and stained with Coomassie Blue. On every gel, three BSA standards (0.5 μg, 1.5 μg, 3 μg per lane) were applied. The protein bands were evaluated densitometrically using the software Image Lab (Bio-Rad, Hercules, CA, USA). Calibrations always gave an R2 above 0.96 and allowed the quantification of SP and IBs. This method has been used in over 700 publications to date and is known to give precise data (e.g., Matusica et al. 2016; Wahlang et al. 2016; Xu et al. 2016).
Results
In this study, we revisited the pET expression system in E. coli BL21(DE3) for the production of a novel scFv. We wanted to (1) prove that lactose favors the recombinant production of soluble scFv compared to IPTG, (2) investigate if the formation of soluble product can be influenced by qs,glu during lactose induction, and (3) determine a mechanistic correlation between qs,lac and qs,glu.
Induction by IPTG vs. lactose
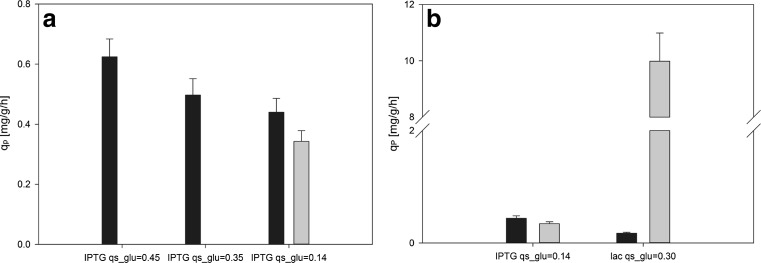
We tested and compared IPTG and lactose as inducers for the pET expression system. Slow uptake of glucose and thus a low specific growth rate (μ) was described to result in higher amount of SP (Shin et al. 1998). Thus, we wanted to investigate whether this strategy was also suitable for the expression of the novel scFv and conducted three fed-batch cultivations with different qs,glu during induction with 0.5 mM IPTG (Fig. 1a, Table 2).
Fig. 1.
a Specific product formation rate (q P) at different q s,glu during IPTG induction. Black bars indicate specific production rate of inclusion bodies (q P,IB), and gray bars specific production rate of soluble product (q P,SP). b Specific product formation rate (q P) at different q s,glu during IPTG or lactose induction. Black bars indicate specific production rate of inclusion bodies (q P,IB), and gray bars specific production rate of soluble product (q P,SP)
Table 2.
Strain physiological parameters of E. coli BL21(DE3) producing a scFv via a pET expression system during either IPTG or lactose induction
| Inducer | q s,glu (g/g/h) | μ (1/h) | q P,IB (mg/g/h) | Std. dev (mg/g/h) | q P,SP (mg/g/h) | Std. dev (mg/g/h) | q P,acetate (mg/g/h) | Std. dev (mg/g/h) | IB-titer (mg/g) | SP-titer (mg/g) |
|---|---|---|---|---|---|---|---|---|---|---|
| IPTG | 0.45 | 0.16 | 0.62 | 0.059 | 0.00 | 3.11 | 0.006 | 2.50 | 0.00 | |
| IPTG | 0.35 | 0.08 | 0.50 | 0.055 | 0.00 | 2.65 | 0.005 | 1.99 | 0.00 | |
| IPTG | 0.14 | 0.03 | 0.44 | 0.046 | 0.34 | 0.036 | 0.24 | 0.001 | 1.76 | 1.37 |
| Lactose | 0.30 | 0.10 | 0.17 | 0.021 | 9.99 | 0.899 | 0.00 | 0.69 | 39.55 |
At qs,glu of 0.45 g/g/h and 0.35 g/g/h only IBs were formed during IPTG induction. IB formation decreased when feeding at lower qs,glu. At qs,glu of 0.14 g/g/h, soluble scFv was produced. Apparently the amount of SP and IBs was strongly linked to qs,glu and thus μ. Furthermore, by decreasing the feeding rate from qs,glu 0.45 to 0.14 g/g/h, the acetate formation rate was reduced more than 10-fold from 3.11 to 0.24 mg/g/h. It is known that the formation of acetate has a negative impact on cell growth and recombinant protein production and should thus be kept at a minimum to ensure high product quality (De Mey et al. 2007). Our findings that a low μ leads to higher production of SP as well as lower acetate formation are in good agreement with literature (Hellmuth et al. 1994; Sanden et al. 2005; Shin et al. 1998). Nevertheless, more than half of the scFv was still found as insoluble and inactive IBs. A further decrease of qs,glu to potentially increase the amount of SP was not feasible as it would have led to slow growth and thus very long process times. Therefore, the alternative inducer lactose which is described to enhance cell fitness and to increase the amount of SP (Donovan et al. 1996) was tested (Fig. 1b, Table 2). Even though qs,glu was 0.30 g/g/h during lactose induction and thus more than 2-fold higher than in the only experiment with IPTG which gave SP (qs,glu = 0.14 g/g/h) around 30-fold more soluble scFv was produced. Furthermore, the acetate concentration was below the detection limit when lactose was used as inducer (Table 2).
Tunability of recombinant protein production by varying qs,glu during lactose induction
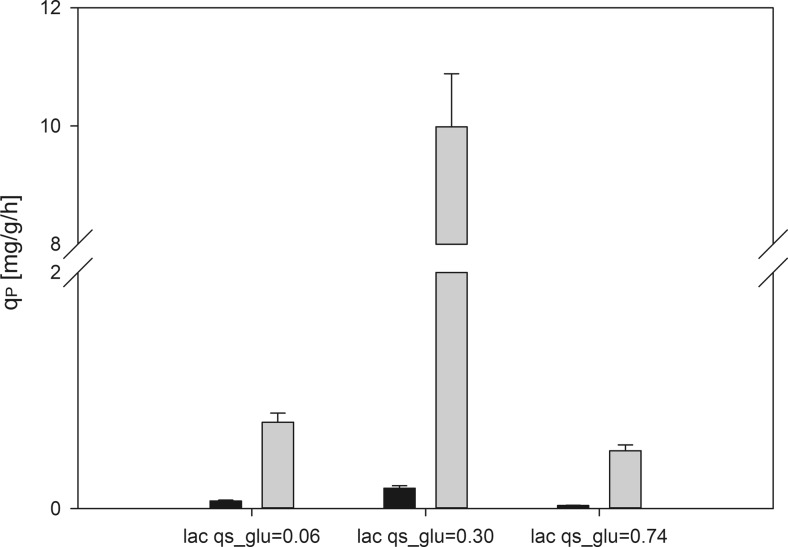
Furthermore, we investigated the potential tunability of recombinant protein production by testing three different qs,glu during induction with lactose, which was always present in excess (Fig. 2).
Fig. 2.
Specific product formation rate (q P) at different q s,glu during lactose induction. Black bars indicate specific production rate of inclusion bodies (q P,IB), and gray bars specific production rate of soluble product (q P,SP)
Figure 2 shows that both the formation of SP and IBs strongly depended on qs,glu during lactose induction. There was a clear optimum at qs,glu = 0.30 g/g/h. At qs,gluc of 0.06 g/g/h 14-fold less SP was produced and also IB formation was reduced 3-fold. We speculate that at this low qs,glu, the cells basically only had enough energy for maintenance metabolism but not for recombinant protein production. However, we were rather surprised to see reduced production of both SP and IB also at the higher qs,glu of 0.74 g/g/h. Thus, we analyzed qs,lac at the respective qs,glu. As shown in Table 3, qs,lac was strongly dependent on qs,glu. At qs,glu of 0.74 g/g/h, only 0.02 g/g/h lactose were metabolized. We speculate that this amount of inducer was too low to guarantee full induction, thus resulting in reduced productivity.
Table 3.
Strain physiological parameters of E. coli BL21(DE3) producing a scFv via a pET expression system during lactose induction at different q s,glu
| Inducer | q s,glu (g/g/h) | q s,lac (g/g/h) | q P,IB (mg/g/h) | Std. dev (mg/g/h) | q P,SP (mg/g/h) | Std. dev. (mg/g/h) | q P,acetate (mg/g/h) | IB-titer (mg/g) | SP-titer (mg/g) |
|---|---|---|---|---|---|---|---|---|---|
| Lactose | 0.06 | 0.09 | 0.06 | 0.007 | 0.73 | 0.077 | 0.00 | 0.26 | 2.92 |
| 0.30 | 0.08 | 0.17 | 0.021 | 9.99 | 0.899 | 0.00 | 0.69 | 39.55 | |
| 0.74 | 0.02 | 0.03 | 0.003 | 0.49 | 0.049 | 0.00 | 0.12 | 1.96 |
Mechanistic correlation between qs,lac and qs,glu
Motivated by our observation of an apparent mechanistic correlation between qs,lac and qs,glu (Table 3), we performed several cultivations to shed more light on this physiological correlation. First, we performed a batch cultivation with excess of both glucose and lactose. In this experiment, we observed the well-known phenomenon of carbon catabolite repression (Stulke and Hillen 1999) meaning that as long as glucose was present in excess, no lactose was taken up (Supplementary Fig. S1). Only when glucose was depleted, lactose was metabolized at a very slow rate of 0.05 g/g/h, which is in agreement to literature (Kremling et al. 2015).
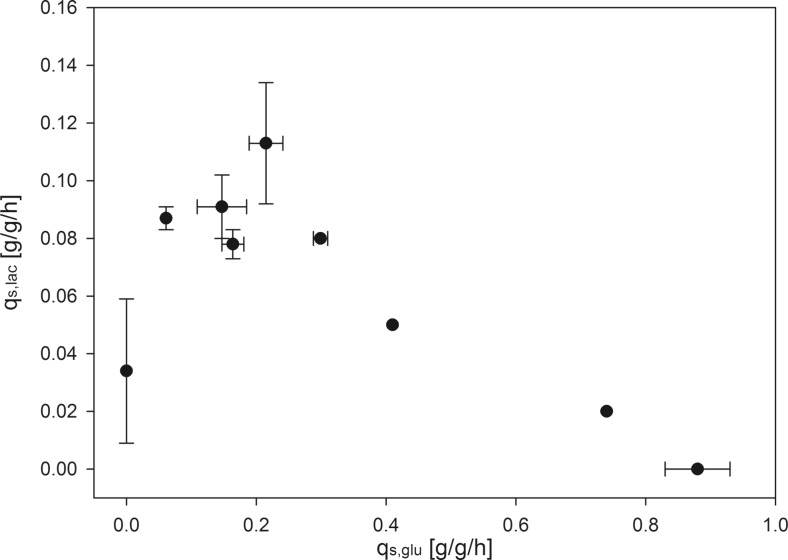
On the contrary, at limiting amounts of glucose but an excess of lactose, we observed a much higher qs,lac (Table 3). Several subsequent cultivations revealed that qs,lac was in fact a function of qs,glu (Fig. 3, Table 4).
Fig. 3.
q s,lac as a function of q s,glu for the recombinant E. coli strain producing scFv with the pET expression system
Table 4.
Experimentally evaluated q s,lac at respective q s,glu for the recombinant E. coli strain producing scFv with the pET expression system
| q s,glu (g/g/h) | Std. dev (g/g/h) | q s,lac (g/g/h) | Std. dev (g/g/h) |
|---|---|---|---|
| 0.00 | – | 0.03 | 0.025 |
| 0.06 | – | 0.09 | 0.004 |
| 0.15 | 0.038 | 0.09 | 0.011 |
| 0.16 | 0.017 | 0.08 | 0.005 |
| 0.22 | 0.026 | 0.11 | 0.021 |
| 0.30 | 0.011 | 0.08 | – |
| 0.41 | – | 0.05 | – |
| 0.74 | – | 0.02 | – |
| 0.88 | 0.050 | 0.00 | – |
As shown in Fig. 3, qs,lac increased with increasing qs,glu having a maximum at qs,glu of 0.2–0.25 g/g/h, before it decreased again. We used these findings to generate a simple mechanistic model describing qs,lac as a function of qs,glu. Such a mechanistic model would greatly facilitate bioprocess development as only a few experiments would be required to determine the correlation between qs,lac and qs,glu. Thus, lactose accumulation and resulting osmotic stress can be reduced (Shiloach and Fass 2005) and tailored feeding and induction are possible. We hypothesized that the correlation between qs,lac and qs,glu was described by two phenomena, namely (1) qs,lac depended Monod-like on qs,glu until a certain maximum was reached, before (2) qs,lac decreased at high qs,glu which was treated similarly to the phenomenon of substrate inhibition (Sivakumar et al. 1994). To describe this correlation, we adapted the model proposed by Han and Levenspiel (1988) (Eq. 3).
| 3 |
- qs,lac
Specific lactose uptake rate (g/g/h)
- qs,lac,max
Maximum specific lactose uptake rate (g/g/h)
- qs,glu
Specific glucose uptake rate (g/g/h)
- qs,glu,crit
Critical specific glucose uptake rate up to which lactose is consumed (g/g/h)
- qs,lac,noglu
Specific lactose uptake rate at qs,glu = 0 (g/g/h)
- KA
Affinity constant for the specific lactose uptake rate (g/g/h)
- m, n
Type of inhibition (noncompetitive, uncompetitive, competitive)
The unknown parameters in this model are the maximum specific lactose uptake rate (qs,lac,max), the critical specific glucose uptake rate up to which lactose is consumed (qs,glu,crit), the affinity constant (KA), the two constants n and m indicating the type of inhibition (noncompetitive, uncompetitive, competitive; (Han and Levenspiel 1988)) and the specific lactose uptake rate at zero glucose uptake (qs,lac,noglu). For the parameter identification, the Nelder-Mead simplex method in MATLAB (Lagarias et al. 1998) was used to minimize the objective function (Eq. 4) which describes the distance between the experimental data and the predicted values by the model. For points, where no standard deviation was available, the mean of all standard deviations was taken.
| 4 |
- S
objective function
- qs,lac,meas,i
ith measurement of qs,lac
- qs,lac,model,i
predicted qs,lac at timepoint of ith measurement
- σi
standard deviation of the ith data point
Based on our observations, we defined that qs,lac is greater than zero when no glucose is consumed. Furthermore, parameter values must have a mechanistic meaning, which is why we assumed qs,lac, KA, and n to be positive and even constrained them further (e.g., qs,lac < qs,glu,crit, KA < 1, n > 0, etc.). To analyze the model and to evaluate the impact of the model parameters, we performed a sensitivity analysis, where we increased or decreased the parameters by 20 % (an example is shown in Supplementary Fig. S2).
The sensitivity analysis revealed the parameter m to have almost no impact on the curve. Furthermore, the parameter estimation revealed that this parameter is almost zero (6.2 × 10−9). This is in accordance with our assumption of a noncompetitive inhibition which is described by m = 0 (Han and Levenspiel 1988). Thus, we set m to zero and simplified the model (Eq. 5).
| 5 |
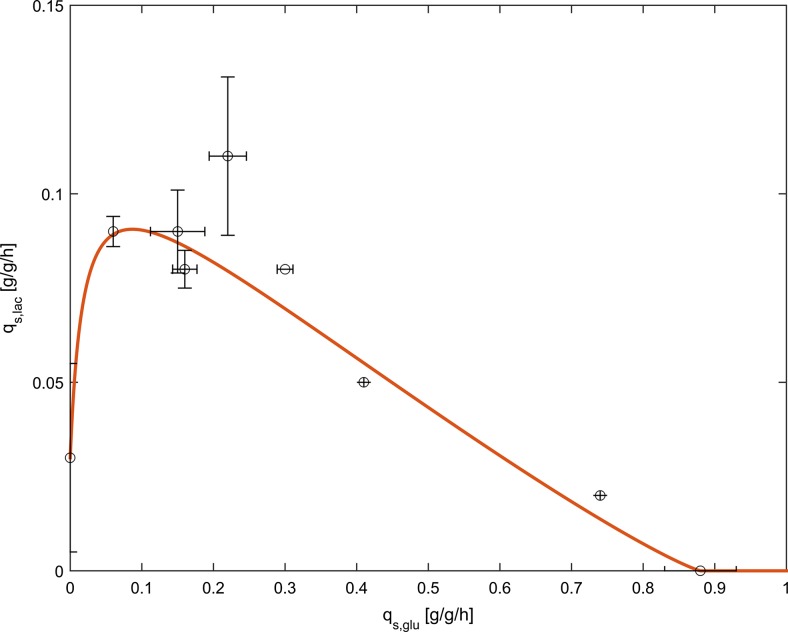
Additionally, the sensitivity analysis showed that n, qs,lac,max, KA, qs,lac,noglu, and qs,glu,crit had a significant impact on the curve. However, the impact of n was rather small and even a potential error of 20 % would lead to only small deviations of the curve (Supplementary Fig. S2). We used our experimental data for the recombinant E. coli strain producing the novel scFv and fitted the mechanistic model to the data using Eq. 5. The curve with the estimated parameters fitted the data with a normalized root mean square error (NRMSE) of 10.3 % and a coefficient of variation (CV) of 18.6 %. Furthermore, all parameters showed physiologically reasonable values. The resulting curve and the corresponding parameter values are shown in Fig. 4 and Table 5, respectively.
Fig. 4.
Optimal fit with parameters q s,lac,max = 0.088 g/g/h, q s,glu,crit = 0.88 g/g/h, q s,lac,noglu = 0.034 g/g/h, K A = 0.019 g/g/h, n = 1.16
Table 5.
Parameters of optimal fit for the mechanistic correlation between qs,lac and q s,glu for the recombinant E. coli strain producing scFv with the pET expression system
| q s,lac,max (g/g/h) | q s,glu,crit (g/g/h) | q s,lac,noglu (g/g/h) | K A (g/g/h) | n (−) |
|---|---|---|---|---|
| 0.088 | 0.88 | 0.034 | 0.019 | 1.16 |
Discussion
In this study, we used a pET expression system and an E. coli BL21(DE3) strain for the production of a novel antibody single-chain variable fragment. Since we wanted to maximize the amount of soluble product and reduce the formation of IBs, we (1) analyzed whether lactose favors the recombinant production of soluble scFv compared to IPTG, (2) investigated whether the formation of soluble product can be influenced by qs,glu during lactose induction, and (3) determined the mechanistic correlation between qs,lac and qs,glu.
In fact, we showed that lactose allowed a much higher production of soluble product compared to IPTG even when μ was increased. This outcome is in good agreement not only with several recent studies that also showed the benefits of using lactose as inducer compared to IPTG for other recombinant products (Bashir et al. 2015; Fruchtl et al. 2015; Zou et al. 2014) but also for antibody fragments (Donovan et al. 2000; Sanchez-Arreola et al. 2013). However, data on potential tunability of recombinant protein production by changing qs,glu and thus μ during lactose induction are scarce to date.
Thus, we investigated a potential tunability of recombinant protein production and found that the formation of soluble product could be tuned by varying qs,glu during lactose induction. Furthermore, we observed a mechanistic correlation between qs,glu and qs,lac in these experiments which motivated us to analyze this phenomenon in more detail.
In fact, we determined a mechanistic correlation between qs,lac and qs,glu and established a simple model. The shape of this curve can be explained by different phenomena. At high qs,glu, no lactose is taken up which can be explained by the phenomenon called carbon catabolite repression. When qs,glu decreases, cAMP gets formed inside the cell. cAMP binds to the catabolite activator protein (CAP) which undergoes a change in conformation and binds to the promoter region of the lac operon. Consequently, transcription of the lac operon’s genes involving lactose permease and β-galactosidase is initiated and lactose can be taken up. However, in the absence of glucose, there is hardly any lactose taken up. The transport of lactose into the cell is ATP related (Voet et al. 2010). Without glucose metabolism, basically no ATP is generated, which is why this transport can barely happen (Kremling et al. 2015; Luo et al. 2014; Mayer et al. 2014; Siegal 2015).
The outcomes of this study can be used for strain characterization and fast bioprocess development. The values for the parameters qs,lac,noglu and qs,glu,crit, which have a high impact on the curve, can be easily determined by simple batch cultivations. To determine the other parameters (qs,lac,max, KA, and n), we recommend performing two to three experiments where lactose is provided in excess and qs,glu is adjusted to values below qs,glu,crit. In the optimal case, one of these qs,glu values corresponds to the maximum of qs,lac. However, if this is not the case, qs,lac,max is simply underestimated, leaving no risk for lactose accumulation. By performing these few experiments, enough data can be gathered to establish the mechanistic model. The mechanistic model can then be used to interpolate unknown qs,lac values to qs,glu points and thus allows fed-batch fermentations at different qs,glu preventing unwanted lactose accumulation. Furthermore, qs,lac can be adjusted within the feasible range allowing to tune the production of soluble protein, as we showed for the novel scFv (Table 3). By this approach, the production of soluble and active protein can be significantly increased.
Summarizing this work emphasizes the applicability of lactose, a cheap, nontoxic waste product, as inducer for the production of soluble recombinant proteins in E. coli. In future studies, we will investigate if the mechanistic model established in this study describes platform knowledge applicable to different E. coli strains. We suggest that by performing two-batch experiments and two to three fed-batches at different qs,glu and concomitant lactose excess, enough data are available to fit the model and obtain the mechanistic correlation for basically any E. coli strain. Furthermore, we will test if induction by lactose triggers expression on a cellular level or if E. coli subpopulations are generated.
Electronic supplementary material
(PDF 7255 kb)
Acknowledgments
Open access funding provided by TU Wien (Vienna | Austria). The authors acknowledge Sciotec Diagnostic Technologies for providing the used strain.
Author contributions
DJW, LV, CH, and OS designed the study. DJW, LV, and BE conducted the experiments and analyzed the data. DJW and SU developed the model. DJW and OS wrote the paper.
Compliance with ethical standards
Ethical approval
The authors declare that they have no conflict of interest. This article does not contain any studies with human participants or animals performed by any of the authors.
References
- Baeshen MN, Al-Hejin AM, Bora RS, Ahmed MM, Ramadan HA, Saini KS, Baeshen NA, Redwan EM. Production of biopharmaceuticals in E. coli: current scenario and future perspectives. J Microbiol Biotechnol. 2015;25:953–962. doi: 10.4014/jmb.1412.12079. [DOI] [PubMed] [Google Scholar]
- Bashir H, Ahmed N, Khan MA, Zafar AU, Tahir S, Khan MI, Khan F, Husnain T. Simple procedure applying lactose induction and one-step purification for high-yield production of rhCIFN. Biotechnol Appl Biochem. 2015 doi: 10.1002/bab.1426. [DOI] [PubMed] [Google Scholar]
- Choi JH, Keum KC, Lee SY. Production of recombinant proteins by high cell density culture of Escherichia coli. Chem Eng Sci. 2006;61:876–885. doi: 10.1016/j.ces.2005.03.031. [DOI] [Google Scholar]
- De Mey M, De Maeseneire S, Soetaert W, Vandamme E. Minimizing acetate formation in E. coli fermentations. J Ind Microbiol Biotechnol. 2007;34:689–700. doi: 10.1007/s10295-007-0244-2. [DOI] [PubMed] [Google Scholar]
- DeLisa MP, Li JC, Rao G, Weigand WA, Bentley WE. Monitoring GFP-operon fusion protein expression during high cell density cultivation of Escherichia coli using an on-line optical sensor. Biotechnol Bioeng. 1999;65:54–64. doi: 10.1002/(SICI)1097-0290(19991005)65:1<54::AID-BIT7>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Dietzsch C, Spadiut O, Herwig C. A dynamic method based on the specific substrate uptake rate to set up a feeding strategy for Pichia pastoris. Microb Cell Factories. 2011;10:14. doi: 10.1186/1475-2859-10-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzsch C, Spadiut O, Herwig C. A fast approach to determine a fed batch feeding profile for recombinant Pichia pastoris strains. Microb Cell Factories. 2011;10:85. doi: 10.1186/1475-2859-10-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan RS, Robinson C, Glick B. Review: optimizing inducer and culture conditions for expression of foreign proteins under the control of the lac promoter. J Ind Microbiol. 1996;16:145–154. doi: 10.1007/BF01569997. [DOI] [PubMed] [Google Scholar]
- Donovan RS, Robinson CW, Glick BR. Optimizing the expression of a monoclonal antibody fragment under the transcriptional control of the Escherichia coli lac promoter. Can J Microbiol. 2000;46:532–541. doi: 10.1139/cjm-46-6-532. [DOI] [PubMed] [Google Scholar]
- Elvin JG, Couston RG, van der Walle CF. Therapeutic antibodies: market considerations, disease targets and bioprocessing. Int J Pharm. 2013;440:83–98. doi: 10.1016/j.ijpharm.2011.12.039. [DOI] [PubMed] [Google Scholar]
- Ferrer-Miralles N, Saccardo P, Corchero JL, Xu Z, Garcia-Fruitos E. General introduction: recombinant protein production and purification of insoluble proteins. Methods Mol Biol. 2015;1258:1–24. doi: 10.1007/978-1-4939-2205-5_1. [DOI] [PubMed] [Google Scholar]
- Fruchtl M, Sakon J, Beitle R. Expression of a collagen-binding domain fusion protein: effect of amino acid supplementation, inducer type, and culture conditions. Biotechnol Prog. 2015;31:503–509. doi: 10.1002/btpr.2048. [DOI] [PubMed] [Google Scholar]
- Gombert AK, Kilikian BV. Recombinant gene expression in Escherichia coli cultivation using lactose as inducer. J Biotechnol. 1998;60:47–54. doi: 10.1016/S0168-1656(97)00185-5. [DOI] [PubMed] [Google Scholar]
- Han K, Levenspiel O. Extended monod kinetics for substrate, product, and cell inhibition. Biotechnol Bioeng. 1988;32:430–447. doi: 10.1002/bit.260320404. [DOI] [PubMed] [Google Scholar]
- Hellmuth K, Korz DJ, Sanders EA, Deckwer WD. Effect of growth rate on stability and gene expression of recombinant plasmids during continuous and high cell density cultivation of Escherichia coli TG1. J Biotechnol. 1994;32:289–298. doi: 10.1016/0168-1656(94)90215-1. [DOI] [PubMed] [Google Scholar]
- Jazini M, Herwig C. Effect of post-induction substrate oscillation on recombinant alkaline phosphatase production expressed in Escherichia coli. J Biosci Bioeng. 2011;112:606–610. doi: 10.1016/j.jbiosc.2011.08.013. [DOI] [PubMed] [Google Scholar]
- Khan KH. Gene expression in mammalian cells and its applications. Adv Pharm Bull. 2013;3:257–263. doi: 10.5681/apb.2013.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremling A, Geiselmann J, Ropers D, de Jong H. Understanding carbon catabolite repression in Escherichia coli using quantitative models. Trends Microbiol. 2015;23:99–109. doi: 10.1016/j.tim.2014.11.002. [DOI] [PubMed] [Google Scholar]
- Lagarias JC, Reeds JA, Wright MH, Wright PE. Convergence properties of the Nelder-Mead simplex method in low dimensions SIAM. J Opt. 1998;9:112–147. [Google Scholar]
- Lee YJ, Jeong KJ. Challenges to production of antibodies in bacteria and yeast. J Biosci Bioeng. 2015;120:483–490. doi: 10.1016/j.jbiosc.2015.03.009. [DOI] [PubMed] [Google Scholar]
- Liu JK. The history of monoclonal antibody development—progress, remaining challenges and future innovations. Ann Med Surg (Lond) 2014;3:113–116. doi: 10.1016/j.amsu.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, Zhang T, Wu H. The transport and mediation mechanisms of the common sugars in Escherichia coli. Biotechnol Adv. 2014;32:905–919. doi: 10.1016/j.biotechadv.2014.04.009. [DOI] [PubMed] [Google Scholar]
- Matusica D, Alfonsi F, Turner BJ, Butler TJ, Shepheard SR, Rogers ML, Skeldal S, Underwood CK, Mangelsdorf M, Coulson EJ. Inhibition of motor neuron death in vitro and in vivo by a p75 neurotrophin receptor intracellular domain fragment. J Cell Sci. 2016;129:517–530. doi: 10.1242/jcs.173864. [DOI] [PubMed] [Google Scholar]
- Mayer S, Junne S, Ukkonen K, Glazyrina J, Glauche F, Neubauer P, Vasala A. Lactose autoinduction with enzymatic glucose release: characterization of the cultivation system in bioreactor. Protein Expr Purif. 2014;94:67–72. doi: 10.1016/j.pep.2013.10.024. [DOI] [PubMed] [Google Scholar]
- Neubauer P, Hofmann K, Holst O, Mattiasson B, Kruschke P. Maximizing the expression of a recombinant gene in Escherichia coli by manipulation of induction time using lactose as inducer. Appl Microbiol Biotechnol. 1992;36:739–744. doi: 10.1007/BF00172185. [DOI] [PubMed] [Google Scholar]
- Pei XL, Wang QY, Li CL, Qiu XF, Xie KL, Huang LF, Wang AM, Zeng ZW, Xie T. Efficient production of a thermophilic 2-deoxyribose-5-phosphate aldolase in glucose-limited fed-batch cultivations of Escherichia coli by continuous lactose induction strategy. Appl Biochem Biotechnol. 2011;165:416–425. doi: 10.1007/s12010-011-9261-8. [DOI] [PubMed] [Google Scholar]
- Rodrigues ME, Costa AR, Henriques M, Azeredo J, Oliveira R. Technological progresses in monoclonal antibody production systems. Biotechnol Prog. 2010;26:332–351. doi: 10.1002/btpr.348. [DOI] [PubMed] [Google Scholar]
- Rosano GL, Ceccarelli EA. Recombinant protein expression in Escherichia coli: advances and challenges. Front Microbiol. 2014;5:172. doi: 10.3389/fmicb.2014.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Arreola PB, Lopez-Uriarte S, Marichal-Gallardo PA, Gonzalez-Vazquez JC, Perez-Chavarria R, Soto-Vazquez P, Lopez-Pacheco F, Ramirez-Medrano A, Rocha-Pizana MR, Alvarez MM. A baseline process for the production, recovery, and purification of bacterial influenza vaccine candidates. Biotechnol Prog. 2013;29:896–908. doi: 10.1002/btpr.1749. [DOI] [PubMed] [Google Scholar]
- Sanden AM, Bostrom M, Markland K, Larsson G. Solubility and proteolysis of the Zb-MalE and Zb-MalE31 proteins during overproduction in Escherichia coli. Biotechnol Bioeng. 2005;90:239–247. doi: 10.1002/bit.20433. [DOI] [PubMed] [Google Scholar]
- Shiloach J, Fass R. Growing E. coli to high cell density—a historical perspective on method development. Biotechnol Adv. 2005;23:345–357. doi: 10.1016/j.biotechadv.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Shin CS, Hong MS, Kim DY, Shin HC, Lee J. Growth-associated synthesis of recombinant human glucagon and human growth hormone in high-cell-density cultures of Escherichia coli. Appl Microbiol Biotechnol. 1998;49:364–370. doi: 10.1007/s002530051183. [DOI] [PubMed] [Google Scholar]
- Siegal ML. Shifting sugars and shifting paradigms. PLoS Biol. 2015;13:e1002068. doi: 10.1371/journal.pbio.1002068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivakumar A, Srinivasaraghavan T, Swaminathan T, Baradarajan A. Extended monod kinetics for substrate inhibited systems. Bioprocess Eng. 1994;11:185–188. doi: 10.1007/BF00369628. [DOI] [Google Scholar]
- Spadiut O, Rittmann S, Dietzsch C, Herwig C. Dynamic process conditions in bioprocess development. Eng Life Sci. 2013;13:88–101. doi: 10.1002/elsc.201200026. [DOI] [Google Scholar]
- Spadiut O, Capone S, Krainer F, Glieder A, Herwig C. Microbials for the production of monoclonal antibodies and antibody fragments. Trends Biotechnol. 2014;32:54–60. doi: 10.1016/j.tibtech.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadlmann V, Harant H, Korschineck I, Hermann M, Forster F, Missbichler A. Novel avian single-chain fragment variable (scFv) targets dietary gluten and related natural grain prolamins, toxic entities of celiac disease. BMC Biotechnol. 2015;15:109. doi: 10.1186/s12896-015-0223-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Striedner G, Cserjan-Puschmann M, Potschacher F, Bayer K. Tuning the transcription rate of recombinant protein in strong Escherichia coli expression systems through repressor titration. Biotechnol Prog. 2003;19:1427–1432. doi: 10.1021/bp034050u. [DOI] [PubMed] [Google Scholar]
- Studier FW, Moffatt BA. Use of bacteriophage-T7 Rna-polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- Stulke J, Hillen W. Carbon catabolite repression in bacteria. Curr Opin Microbiol. 1999;2:195–201. doi: 10.1016/S1369-5274(99)80034-4. [DOI] [PubMed] [Google Scholar]
- Voet D, Voet JG, Pratt CW, Beck-Sickinger A, Hahn U. Lehrbuch der Biochemie. Weinheim: Wiley-VCH; 2010. [Google Scholar]
- Wahlang B, Prough RA, Falkner KC, Hardesty JE, Song M, Clair HB, Clark BJ, States JC, Arteel GE, Cave MC. Polychlorinated biphenyl-xenobiotic nuclear receptor interactions regulate energy metabolism, behavior, and inflammation in non-alcoholic-steatohepatitis. Toxicol Sci. 2016;149:396–410. doi: 10.1093/toxsci/kfv250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wals K, Ovaa H. Unnatural amino acid incorporation in E. coli: current and future applications in the design of therapeutic proteins. Front Chem. 2014;2:15. doi: 10.3389/fchem.2014.00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh G. Biopharmaceutical benchmarks 2014. Nat Biotechnol. 2014;32:992–1000. doi: 10.1038/nbt.3040. [DOI] [PubMed] [Google Scholar]
- Wechselberger P, Sagmeister P, Herwig C. Real-time estimation of biomass and specific growth rate in physiologically variable recombinant fed-batch processes. Bioprocess Biosyst Eng. 2013;36:1205–1218. doi: 10.1007/s00449-012-0848-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, Yang H, Sun M, Chen H, Jiang L, Zheng X, Ding G, Liu Y, Sheng Y, Cui D, Duan Y. 2,3′,4,4′,5-Pentachlorobiphenyl induces inflammatory responses in the thyroid through JNK and aryl hydrocarbon receptor-mediated pathway. Toxicol Sci. 2016;149:300–311. doi: 10.1093/toxsci/kfv235. [DOI] [PubMed] [Google Scholar]
- Zalai D, Dietzsch C, Herwig C, Spadiut O. A dynamic fed batch strategy for a Pichia pastoris mixed feed system to increase process understanding. Biotechnol Prog. 2012;28:878–886. doi: 10.1002/btpr.1551. [DOI] [PubMed] [Google Scholar]
- Zou C, Duan X, Wu J. Enhanced extracellular production of recombinant Bacillus deramificans pullulanase in Escherichia coli through induction mode optimization and a glycine feeding strategy. Bioresour Technol. 2014;172:174–179. doi: 10.1016/j.biortech.2014.09.035. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 7255 kb)