Abstract
HNF4A mutations cause increased birth weight, transient neonatal hypoglycaemia and maturity onset diabetes of the young (MODY). The most frequently reported HNF4A mutation is p.R114W (previously p.R127W) but functional studies have shown inconsistent results, there is lack of co-segregation in some pedigrees and an unexpectedly high frequency in public variant databases. We confirm that p.R114W is a pathogenic mutation with an odds ratio of 30.4 (95% CI: 9.79 – 125, P=2x10-21) for diabetes in our MODY cohort compared to controls. p.R114W heterozygotes do not have the increased birth weight of patients with other HNF4A mutations (3476g vs. 4147g, P=0.0004) and fewer patients responded to sulfonylurea treatment (48% vs. 73%, P=0.038). p.R114W has reduced penetrance; only 54% of heterozygotes developed diabetes by age 30 compared to 71% for other HNF4A mutations. We re-define p.R114W as a pathogenic mutation causing a distinct clinical subtype of HNF4A MODY with reduced penetrance, reduced sensitivity to sulfonylurea treatment and no effect on birth weight. This has implications for diabetes treatment, management of pregnancy and predictive testing of at-risk relatives. The increasing availability of large-scale sequence data is likely to reveal similar examples of rare, low-penetrance MODY mutations.
Introduction
Heterozygous loss of function HNF4A mutations cause maturity onset diabetes of the young (MODY)[1]. HNF4A is a transcription factor important in the function of the pancreatic β-cell. Patients with HNF4A mutations present with a common phenotype of increased birth weight (median increase of 790g) and occasional neonatal hypoglycaemia (15%)[2]. HNF4A MODY patients are typically sulfonylurea sensitive and a genetic diagnosis is important because it determines the best treatment. Mutations in HNF4A account for 10% of genetically confirmed MODY cases[3]. The most commonly reported HNF4A mutation is p.R114W (NM_175914.4:c.340C>T p.Arg114Trp, rs137853336, previously described as p.R127W[4]). In our MODY cohort it accounts for 30/176 (17.1%) HNF4A cases, while in two Italian cohorts 5/6(83.3%) HNF4A cases were caused by p.R114W [5, 6].
Several pieces of evidence made us question whether p.R114W truly causes MODY. p.R114W is present in the ExAC database[7] in 7/32198 European (non-Finnish) samples when we would only expect 0.35 HNF4A MODY cases based on a population frequency of MODY of 108 per million and HNF4A mutations only accounting for 10% of MODY cases[3]. Another source of doubt over the pathogenicity of p.R114W comes from published families. The original p.R114W family[4] contains two phenocopies diagnosed at 11 and 36 years of age and a p.R114W heterozygote who was not diagnosed with diabetes until age 90. Shanker et. al[8] presented the case of a digenic pedigree of HNF1A p.G292fs and p.R114W; however p.R114W was inherited from a clinically unaffected mother (at age 46) who did not have a dominant family history of diabetes. There is also controversy over the functional effect of the mutation. Navas et. al[9] assessed the transcriptional transactivation of p.R114W to be the same as wild type, concluding that it was a polymorphism. Two later studies suggested a 30% and 50% reduction in activity compared to wild type [10, 11].
In this study we show that p.R114W is a pathogenic mutation causing MODY-like diabetes, but with reduced penetrance and a distinct clinical phenotype.
Methods
MODY cohort
Between 1996 and 2016, 2289 probands with a clinical suspicion of MODY were referred for genetic testing to the Molecular Genetics Laboratory at the Royal Devon and Exeter Hospital. Clinical information was provided on a standardised referral form by the clinician at the time of referral for genetic testing, this included a question on sulfonylurea sensitivity. The latest version of the referral form can be found at http://diabetesgenes.org/content/mody.
30 probands and 51 family members with p.R114W were identified by Sanger sequencing of HNF4A. One additional proband was heterozygous for p.R114W and a pathogenic HNF1A mutation, p.R159Q; they were excluded from analysis to avoid confounding results. 100 probands and 224 family members with other pathogenic HNF4A mutations were identified. Pedigrees demonstrating co-inheritance were defined as those where p.R114W was inherited from a mutation positive parent with diabetes.
Variants were classified as pathogenic if they were nonsense, frameshift or essential splice site mutations. Missense mutations had to be reported in the literature as pathogenic in three or more families.
Type 2 diabetes cases and control samples
The type 2 diabetes cases (N=9185) and controls (N=12890) were all of white European ancestry and came from three sources. Clinical characteristics are provided in Supplementary Table 1.
Of these, 6763 cases and 7073 controls were from the Genetics of Diabetes Audit and Research in Tayside Scotland (GoDARTS), described previously[12].
2100 were cases from the Diabetes UK Warren 2 repository[13] and 1519 controls were from the UK Blood Services Collection of Common Controls. The cases include probands from the Warren 2 sib-pair, trios and duos resources and additional cases diagnosed between 35 and 65 years of age, described previously[14]. The controls were part of the Wellcome Trust Case Control Consortium, described previously[15].
770 cases and 4849 controls came from the Exeter Ten Thousand study (EXTEND) (www.exeter10000.org), which recruited volunteers over the age of 18 years living within 25 miles of Exeter, UK, and the Exeter Family Study of Child Health (EFSOCH), described previously[16].
Genotyping
Genotyping p.R114W in the type 2 diabetes case control study samples was performed by LGC genomics (Middlesex, UK) using a KASP assay. We included 5 positive control samples to ensure adequate genotype clustering. The genotyping success rate was >95% in all cohorts and there were no discrepancies amongst 758 duplicate pairs.
Haplotype analysis
The haplotype context of p.R114W were assessed in a subset of 34 patients by genotyping SNPs flanking the mutation; based on methods described previously[17]. See Supplementary Table 2 for list of SNPs.
Computational analyses
Birth weights were corrected for gestational age using the 1990 British child growth reference data[18]. Statistical analysis was performed in Stata® (version 14); tests specified in text. T-tests assumed unequal variances. The Kaplan-Meier plot records the proportion of heterozygotes who are diabetic at each age where data is available, for those patients who did not develop diabetes age was censored at age of referral. The effect of p.R114W was assessed using in silico tools run using Annovar[19] and Alamut (Rouen, France).
Results
p.R114W is enriched in MODY cohort compared to controls and patients with type 2 diabetes
In our MODY cohort we identified 30/2289 (1.3%) probands with p.R114W of which 26/1696 (1.5%) are white European. This compares to 4/12890 (0.03%) controls (OR 49.4, 95% CI: 17.1 - 194, P=3.7x10-37). p.R114W is present in 7/32198 (0.02%) European (non-Finnish) samples in ExAC[7], comparable to our controls (OR 1.43, 95% CI: 0.301- 5.62, P=0.52). To assess if p.R114W predisposes to type 2 diabetes rather than MODY we genotyped 9185 individuals with type 2 diabetes. There are 26/1696 p.R114W heterozygotes amongst the white Europeans in our MODY cohort compared to 9/9185 (0.1%) patients with type 2 diabetes which gives an odds ratio of 15.6 (95% CI: 7.08 - 38.0, P=2x10-21). The odds ratio of prevalence in the type 2 diabetes samples versus controls is 3.16 (95% CI: 0.88 - 14.0, P=0.05; Supplementary Table 3). Overall these results suggest that p.R114W predominantly predisposes to MODY-like rather than type 2 diabetes. Co-inheritance of p.R114W and diabetes is not consistent with high penetrance
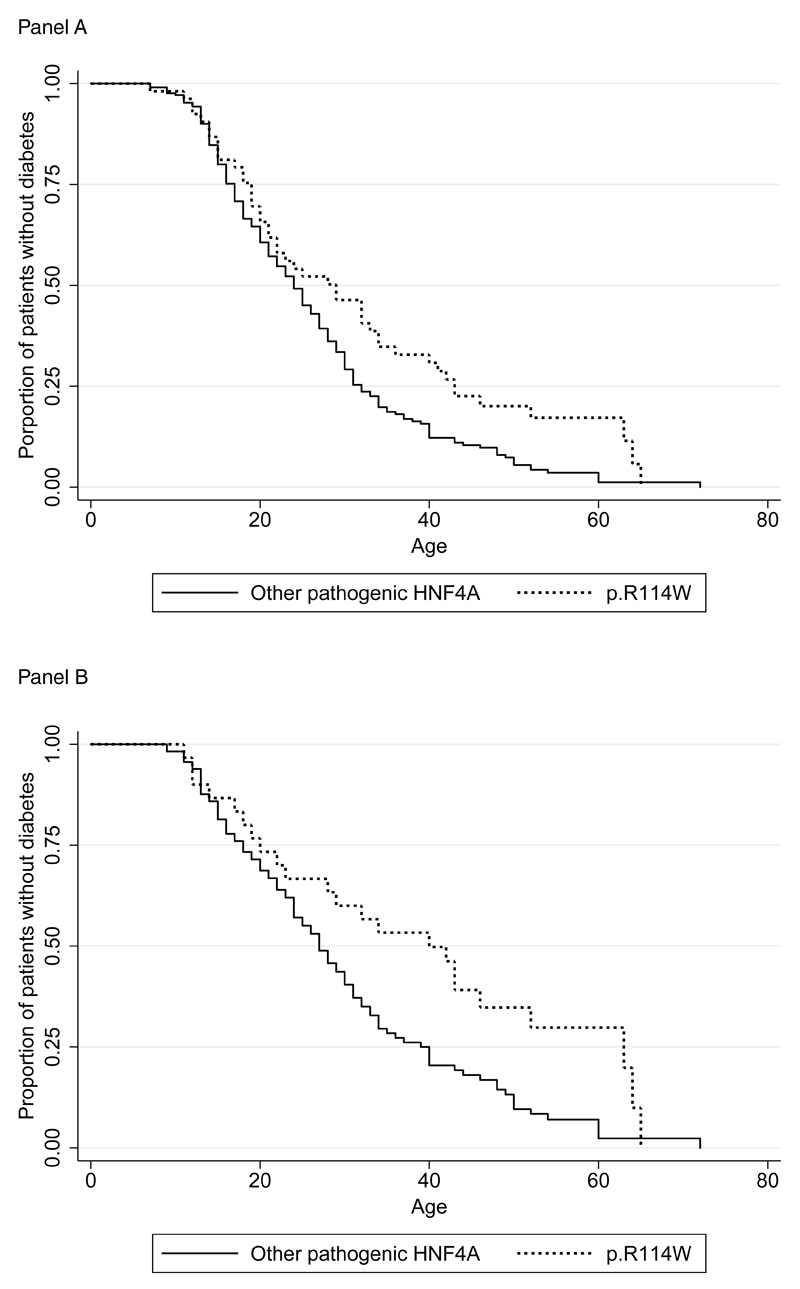
Co-inheritance studies were possible in 14/30 pedigrees, in which p.R114W showed co-inheritance with diabetes in 10/14 (Supplementary Figure 1). In the 4 families where p.R114W did not demonstrate co-inheritance with diabetes there are 4 non-diabetic p.R114W heterozygotes aged 36-53. 82% of individuals heterozygous for other HNF4A mutations are expected to be diabetic by age 36 (Figure 1A). In addition to the previously reported p.R114W/HNF1A digenic pedigree[8] we identified a proband with p.R114W and a pathogenic HNF1A mutation, p.R159Q. While the majority of families tested demonstrate co-inheritance consistent with a pathogenic mutation, the presence of multiple non-diabetic family members with p.R114W aged >30 years is not consistent with the high penetrance typical of HNF4A mutations. 46% of p.R114W heterozygotes were not diabetic at age 30 compared to 29% for other HNF4A mutations (Figure 1A). Patients with p.R114W are 34.2% less likely to develop diabetes compared to other HNF4A mutations across the age range in our cohort (stcox, P=0.013).
Figure 1.
Kaplan-Meier plot comparing the proportion of p.R114W heterozygotes that are diabetic to patients with other pathogenic HNF4A mutations (at each age where data is available). Panel A includes all probands and family members – the hazard ratio for diabetes is 0.659 (P=0.013) comparing p.R114W heterozygotes to patients with other HNF4A mutations. Panel B includes only family members to remove the effect of ascertainment bias in the probands – the hazard ratio for diabetes is 0.553 (P=0.013) comparing p.R114W heterozygotes to patients with other HNF4A mutations.
Multiple independent origins of the p.R114W mutation
Identifying a variant on multiple haplotype backgrounds provides evidence for pathogenicity by excluding the presence of a separate, unobserved pathogenic mutation on the same haplotype. Analysis of flanking SNPs suggests that p.R114W arose on at least two haplotype backgrounds (maximum shared haplotype <368kb; Supplementary Table 2).
p.R114W heterozygotes have a distinct phenotype compared to patients with other HNF4A mutations
Raised birth weight is a clinically important feature of HNF4A MODY as it impacts on pregnancy management[2]. p.R114W heterozygotes show no increase in birth weight compared to the general population (mean SDS -0.032, 95% CI:-0.67 - 0.61) and a significant decrease compared to patients with other HNF4A mutations (t-test, P=0.0003) (Table 1). The age of diagnosis of diabetes is later, at a median of 34 years of age compared to 24 years for other HNF4A mutations (t-test, P=0.018), which is consistent with p.R114W having a lower penetrance. Clinician reported sensitivity to sulfonylureas is also lower than for other HNF4A mutations (47.6% compared to 72.9%, t-test, P=0.038). An analysis of just the white European subset gave similar results (Supplementary Table 4).
Table 1.
Characteristics of carriers of p.R114W compared to carriers of other pathogenic HNF4A mutations.
| p.R114W mutation | Other pathogenic HNF4A mutations | P | |
|---|---|---|---|
| Probands | 30 | 100 | N/A |
| Heterozygous family members | 27 | 134 | N/A |
| Birth weight corrected to 40 weeks gestation (g) (Mean (95% CI)) | 3476 (3160,3792) (n=23) | 4147 (3985,4309) (n=84) | 0.0004 * |
| Standard deviation scores of birth weight (Mean (95% CI)) | -0.032 (-0.67,0.61) (n=23) | 1.36 (1.03,1.68) (n=84) | 0.0003 * |
| Birth weight corrected to 40 weeks gestation >4000g | 5 (n=23) | 45 (n=84) | 0.009 † |
| Age of diagnosis of diabetes in probands (years) (Median (Q1,Q3)) | 21 (15,32) (n=30) | 21 (16,28) (n= 96) | 0.63 * |
| Age of diagnosis of diabetes in family members (years) (Median (Q1,Q3)) | 34 (20,50) (n=21) | 24 (16,34) (n=95) | 0.018 * |
| Duration of diabetes (years) (Median (Q1,Q3)) | 5 (0,14.5) (n=51) | 7 (1,19) (n=191) | 0.025 * |
| Initial treatment of diabetic patients (Diet/OHA/Ins) | 10/19/16 (n=45) | 47/50/59 (n=156) | N/A |
| Diabetic patients treated with sulfonylureas at some time during diabetes | 21 | 70 | N/A |
| Patients reported as being sulfonylurea sensitive | 10 (47.6%) | 51 (72.9%) | 0.038 † |
| BMI of diabetic patients (Median (Q1,Q3)) | 22.65 (22.975,29.2) (n=40) | 23.9 (21.945,27) (n=128) | 0.97 * |
| Median HbA1c of diabetic patients (IFCC & DCCT) | 65.0mmol/mol 8.1% (n=31) | 57.4mmol/mol 7.4% (n=151) | 0.97 * |
t-test
fisher’s exact test
Discussion
p.R114W is a pathogenic mutation causing MODY-like diabetes, but with reduced penetrance
p.R114W is enriched in our MODY cohort and demonstrates partial co-inheritance with diabetes. However individuals with the mutation do not always present with the disease – the mutation therefore has reduced penetrance. It is possible that secondary genetic or environmental factors may be necessary to cause disease. p.R114W affects a conserved base according to its GERP score, is a conserved amino acid (down to Fruit fly) and is predicted to be deleterious by in silico tools including SIFT and Polyphen (Supplementary Table 5). This supports the case that it is a pathogenic variant.
There is some evidence that p.R114W is a hypomorphic mutation with transactivation activity reduced but not absent in vitro[10, 11]. p.R114W is in the DNA binding domain of HNF4A. Chandra et al. demonstrated that it had reduced DNA binding affinity compared to wild type but with a smaller effect than that observed for other HNF4A mutations[20]. The previously reported HNF4A p.R76W is also in the DNA binding domain and causes renal features in addition to the classical HNF4A symptoms [24]. This suggests that the difference in DNA binding affinity may be the cause of the atypical sets of phenotypes presented by these two mutations, by dictating which target genes the HNF4A transcription factor is able to interact with.
Other hypomorphic rare MODY gene variants include the HNF1A p.G319S missense/splicing variant in the Oji Cree[21, 22] and the hypomorphic p.E508K variant in Latinos [23]. There is also some evidence that p.T130I is a low-frequency variant predisposing to type 2 diabetes in Europeans[24]. These variants are associated with an increased risk of type 2 diabetes whereas p.R114W appears to predominantly cause MODY-like diabetes in both Caucasian and Japanese pedigrees.
p.R114W has no effect on birth weight
p.R114W heterozygotes have a distinct clinical presentation of HNF4A MODY-like diabetes with normal birth weight. The p.R76W HNF4A mutation causes a unique phenotype of renal Fanconi syndrome in addition to the common HNF4A diabetic phenotype[25], but still causes increased birth weight. p.R114W appears to be having an effect in the adult but not the fetus. One speculative mechanism for this would be a lower threshold of HNF4A activity required in the fetus for healthy function.
Reclassifying the clinical features of p.R114W will impact patient treatment and family management
p.R114W has a distinct profile of clinical features. This means that specific recommendations for patient management are required. In contrast to offspring who inherit other HNF4A mutations, fetal inheritance of p.R114W will not confer a high risk of macrosomia. Whilst sulfonylurea treatment is successful for some patients, overall response is lower compared to patients with other HNF4A mutations. The reduced penetrance of this mutation is important for genetic counselling of at-risk relatives undergoing predictive tests since the chance of presenting with diabetes is no longer based simply on the odds of inheriting the mutation; not all heterozygotes will develop diabetes and those that do are likely to have a later onset.
Ascertainment bias affects estimates of penetrance
This study is based on the largest cohort of families with p.R114W included in a single study, giving us the clearest view of the characteristics of the mutation. However, there is a risk of ascertainment bias due to the referral criteria for MODY genetic testing. Probands were selected based on diabetic symptoms, age of diagnosis and, importantly, a family history of diabetes. This means we are at risk of overestimating the penetrance of p.R114W. However this will be the case for all MODY genes, making comparisons of penetrance fair. Ascertainment bias is one of the reasons it is hard to establish accurate estimates of penetrance for rare diseases. Publically available sequencing data (such as ExAC[7] and the 1000 genomes project[26]) can be used as controls to help generate unbiased estimates of penetrance. However in the case of MODY, using ExAC as a control has the limitation that it contains some individuals with type 2 diabetes, a subset of these could have misdiagnosed MODY. Removing these would only strengthen our conclusions.
Identifying p.R114W on two distinct haplotypes demonstrates that while it is possible for there to be a founder effect in Europeans, it cannot explain all cases of p.R114W. The fact that the majority of p.R114W patients are European could also be attributed to ascertainment bias as the majority of our cohort were recruited in the UK.
The pathogenicity of MODY mutations found in ExAC should be questioned
The estimated prevalence of MODY in the United Kingdom is 108 per million population[3]. The most frequently reported individual MODY mutation accounts for 15% of MODY cases[27]. Based on this frequency we would expect to see the commonest individual MODY mutation in ExAC European (non-Finnish) at a frequency of only 0.52, thus any MODY mutation seen multiple times in ExAC is unexpectedly common and should have its pathogenicity and penetrance re-evaluated.
Conclusion
The most frequently reported HNF4A mutation, p.R114W, causes a distinct clinical phenotype of monogenic diabetes with reduced penetrance, no increase in birth weight and a lower likelihood of response to sulfonylureas. The increasing availability of large-scale sequence data is likely to reveal similar examples of rare, low-penetrance MODY mutations.
Supplementary Material
Acknowledgements
This work was supported by a Wellcome Trust Senior Investigator award to A.T.H. and S.E. A.T.H. is also supported by an NIHR Senior Investigator award. Additional support came from the University of Exeter and the NIHR Exeter Clinical Research Facility. M.N.W. is supported by MRC grant (MR/M005070/1). TWL is funded by the Wellcome Trust and the Royal Society (grant no. 105636/Z/14/Z). The genotyping was supported by the project for conceptual development of research organization 00064203/6001 (Ministry of Health, Czech Republic).This study makes use of data generated by the WTCCC. A complete list of the investigators who contributed to the generation of the data is available from www.wtccc.org.uk.
The views expressed are those of the authors and do not necessarily reflect those of the Wellcome Trust, the NHS, the National Institute for Health Research, or the Department of Health.
M.N.W is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
All authors declare that they have no conflict of interest.
Author Contributions
TWL, MNW, KC and KP carried out analysis and statistics. PD and SP carried out SNP genotyping. MS, KC, ADM, CNP and MIM collected patient data. KC, JALH and SE were responsible for the clinical diagnostic testing. ATH, SE, MNW and KC designed the study. TWL, KC, SE, ATH and MNW wrote the manuscript. All authors were involved in discussion, refinement of the manuscript and approved the final manuscript.
References
- 1.Yamagata K, et al. Mutations in the hepatocyte nuclear factor-4[alpha] gene in maturity-onset diabetes of the young (MODY1) Nature. 1996;384(6608):458–460. doi: 10.1038/384458a0. [DOI] [PubMed] [Google Scholar]
- 2.Pearson ER, et al. Macrosomia and hyperinsulinaemic hypoglycaemia in patients with heterozygous mutations in the HNF4A gene. PLoS Med. 2007;4(4):e118. doi: 10.1371/journal.pmed.0040118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shields BM, et al. Maturity-onset diabetes of the young (MODY): how many cases are we missing? Diabetologia. 2010;53(12):2504–2508. doi: 10.1007/s00125-010-1799-4. [DOI] [PubMed] [Google Scholar]
- 4.Furuta H, et al. Organization and partial sequence of the hepatocyte nuclear factor-4 alpha/MODY1 gene and identification of a missense mutation, R127W, in a Japanese family with MODY. Diabetes. 1997;46(10):1652–1657. doi: 10.2337/diacare.46.10.1652. [DOI] [PubMed] [Google Scholar]
- 5.Delvecchio M, et al. Low prevalence of HNF1A mutations after molecular screening of multiple MODY genes in 58 Italian families recruited in the pediatric or adult diabetes clinic from a single Italian hospital. Diabetes Care. 2014;37(12):e258–260. doi: 10.2337/dc14-1788. [DOI] [PubMed] [Google Scholar]
- 6.Ludovico O, et al. Identification and Clinical Characterization of Adult Patients with Multigenerational Diabetes Mellitus. PLoS One. 2015;10(8):e0135855. doi: 10.1371/journal.pone.0135855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lek M, et al. Analysis of protein-coding genetic variation in 60,706 humans. bioRxiv. 2015 doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shankar RK, et al. Digenic heterozygous HNF1A and HNF4A mutations in two siblings with childhood-onset diabetes. Pediatr Diabetes. 2013;14(7):535–538. doi: 10.1111/pedi.12018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Navas M, et al. Functional characterization of the MODY1 gene mutations HNF4(R127W), HNF4(V255M), and HNF4(E276Q) Diabetes. 1999;48(7):1459–1465. doi: 10.2337/diabetes.48.7.1459. [DOI] [PubMed] [Google Scholar]
- 10.Yang Q, et al. R127W-HNF-4alpha is a loss of function mutation but not a rare polymorphism and causes Type II diabetes in a Japanese family with MODY1. Diabetologia. 43:520–524. doi: 10.1007/s001250051338. [DOI] [PubMed] [Google Scholar]
- 11.Lausen J, et al. Naturally occurring mutations in the human HNF4α gene impair the function of the transcription factor to a varying degree. Nucleic Acids Research. 2000;28(2):430–437. doi: 10.1093/nar/28.2.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yaghootkar H, et al. Association Analysis of 29,956 Individuals Confirms That a Low-Frequency Variant at CCND2 Halves the Risk of Type 2 Diabetes by Enhancing Insulin Secretion. Diabetes. 2015;64(6):2279–2285. doi: 10.2337/db14-1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wiltshire S, et al. A Genomewide Scan for Loci Predisposing to Type 2 Diabetes in a U.K. Population (The Diabetes UK Warren 2 Repository): Analysis of 573 Pedigrees Provides Independent Replication of a Susceptibility Locus on Chromosome 1q. American Journal of Human Genetics. 2001;69(3):553–569. doi: 10.1086/323249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frayling TM, et al. Parent-offspring trios: a resource to facilitate the identification of type 2 diabetes genes. Diabetes. 1999;48(12):2475–2479. doi: 10.2337/diabetes.48.12.2475. [DOI] [PubMed] [Google Scholar]
- 15.Consortium, T.W.T.C.C. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447(7145):661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Knight B, Shields BM, Hattersley AT. The Exeter Family Study of Childhood Health (EFSOCH): study protocol and methodology. Paediatric and Perinatal Epidemiology. 2006;20(2):172–179. doi: 10.1111/j.1365-3016.2006.00701.x. [DOI] [PubMed] [Google Scholar]
- 17.Dusatkova P, et al. Ancestral mutations may cause a significant proportion of GCK-MODY. Pediatric Diabetes. 2012;13(6):489–498. doi: 10.1111/j.1399-5448.2011.00845.x. [DOI] [PubMed] [Google Scholar]
- 18.Cole TJ, Freeman JV, Preece MA. British 1990 growth reference centiles for weight, height, body mass index and head circumference fitted by maximum penalized likelihood. Statistics in Medicine. 1998;17(4):407–429. [PubMed] [Google Scholar]
- 19.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Research. 2010;38(16):e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chandra V, et al. Multidomain integration in the structure of the HNF-4alpha nuclear receptor complex. Nature. 2013;495(7441):394–398. doi: 10.1038/nature11966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hegele RA, et al. The Hepatic Nuclear Factor-1α G319S Variant Is Associated with Early-Onset Type 2 Diabetes in Canadian Oji-Cree. The Journal of Clinical Endocrinology & Metabolism. 1999;84(3):1077–1082. doi: 10.1210/jcem.84.3.5528. [DOI] [PubMed] [Google Scholar]
- 22.Harries LW, et al. Diabetes Susceptibility in the Canadian Oji-Cree Population Is Moderated by Abnormal mRNA Processing of HNF1A G319S Transcripts. Diabetes. 2008;57(7):1978–1982. doi: 10.2337/db07-1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Estrada K, et al. Association of a Low-Frequency Variant in HNF1A With Type 2 Diabetes in a Latino Population. JAMA. 2014;311(22):2305–2314. doi: 10.1001/jama.2014.6511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jafar-Mohammadi B, et al. A role for coding functional variants in HNF4A in Type 2 Diabetes susceptibility. Diabetologia. 2011;54(1):111–119. doi: 10.1007/s00125-010-1916-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hamilton AJ, et al. The HNF4A R76W mutation causes atypical dominant Fanconi syndrome in addition to a beta cell phenotype. J Med Genet. 2014;51(3):165–9. doi: 10.1136/jmedgenet-2013-102066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.The Genomes Project, C. A global reference for human genetic variation. Nature. 2015;526(7571):68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Colclough K, et al. Mutations in the genes encoding the transcription factors hepatocyte nuclear factor 1 alpha and 4 alpha in maturity-onset diabetes of the young and hyperinsulinemic hypoglycemia. Human mutation. 2013;34(5):669–685. doi: 10.1002/humu.22279. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.