Abstract
We review half a century of research on Cucumis melo resistance to Aphis gossypii from molecular to field levels. The Vat gene is unique in conferring resistance to both A. gossypii and the viruses it transmits. This double phenotype is aphid clone-dependent and has been observed in 25 melon accessions, mostly from Asia. It is controlled by a cluster of genes including CC-NLR, which has been characterized in detail. Copy-number polymorphisms (for the whole gene and for a domain that stands out in the LLR region) and single-nucleotide polymorphisms have been identified in the Vat cluster. The role of these polymorphisms in plant/aphid interactions remains unclear. The Vat gene structure suggests a functioning with separate recognition and response phases. During the recognition phase, the VAT protein is thought to interact (likely indirectly) with an aphid effector introduced during cell puncture by the aphid. A few hours later, several miRNAs are upregulated in Vat plants. Peroxidase activity increases, and callose and lignin are deposited in the walls of the cells adjacent to the stylet path, disturbing aphid behavior. In aphids feeding on Vat plants, Piwi-interacting RNA-like sequences are abundant and the levels of other miRNAs are modified. At the plant level, resistance to aphids is quantitative (aphids escape the plant and display low rates of reproduction). Resistance to viruses is qualitative and local. Durability of NLR genes is highly variable. A. gossypii clones are adapted to Vat resistance, either by introducing a new effector that interferes with the deployment of plant defenses, or by adapting to the defenses it triggered. Viruses transmitted in a non-persistent manner cannot adapt to Vat resistance. At population level, Vat reduces aphid density and genetic diversity. The durability of Vat resistance to A. gossypii populations depends strongly on the agro-ecosystem, including, in particular, the presence of other cucurbit crops serving as alternative hosts for adapted clones in fall and winter. At the crop level, Vat resistance decreases the intensity of virus epidemics when A. gossypii is the main aphid vector in the crop environment.
Keywords: NLR resistance gene, durability, melon, Cucumis melo, Aphis gossypii, resistance deployment, resistance to insects, resistance to viruses
Introduction
Host-plant resistance is an effective, environmentally friendly means of controlling insect pests, including aphids. Here, we consider plant resistance to be a heritable trait, displaying genotype-dependent variability within a plant species. Resistance to aphids has been described in several crops (Dogimont et al., 2010; Smith and Chuang, 2014). This resistance is controlled by one or several genes, which may be recessive or dominant. Resistance deters aphids from the crop, and affects their biotic potential, including their growth, development, and reproduction. So resistance is generally detected through these central aphid life history traits, rather than by a visible plant phenotype. The melon Vat gene is unique among the known resistance genes in that it has a pleiotropic effect as it also confers resistance to the viruses transmitted by aphids.
Melon crops are primarily colonized by only one aphid species, the melon aphid Aphis gossypii, a cosmopolitan aphid species. This aphid causes stunting and severe leaf-curling, and heavy colonization can result in plant death. Aphids also excrete honeydew onto the leaves and fruits. This sticky sweet substance acts as an ideal growth medium for sooty mold, which greatly decreases fruit quality. Moreover, A. gossypii is an efficient vector for viruses, contributing to the spread of diseases.
Resistance to A. gossypii in melon was first observed in the mid-20th century (Ivanoff, 1944). In 1967, an American team of entomologists and plant geneticists began a systematic study of resistance to A. gossypii in melon. They focused on the Indian line PI 371795, later called PI 414723, which suffers only mild attacks in the field (Kishaba et al., 1971; Bohn et al., 1972). In controlled no-choice tests, few aphids survive on this line, and the fecundity of those that do is low (Kishaba et al., 1971). This resistance is a dominant trait in PI 414723, and is controlled by a major gene and several minor genes (Kishaba et al., 1976). A French team of virologists and plant geneticists studied the resistance of the Korean line PI 161375 to Cucumber mosaic virus (CMV) in detail. They discovered an original phenotype of this line: complete resistance to CMV when the aphid A. gossypii inoculated the plant with the virus. Moreover, A. gossypii aphids departed from PI 161375 plants. These two phenotypes cosegregated in PI 161375 and were controlled by a single dominant gene (Pitrat and Lecoq, 1980). Complete resistance to CMV was also observed in PI 414723 when CMV was introduced into the plant by the aphid A. gossypii (Pitrat and Lecoq, 1982). PI 414723 and PI 161375 thus have similar features: resistance to CMV when A. gossypii inoculates the plant with the virus cosegregating with resistance to A. gossypii controlled by a single dominant gene (Pitrat and Lecoq, 1982). In both lines, the resistance to viruses is expressed only if the aphid inoculating the plant with the virus is A. gossypii. PI 161375 and PI 414723 plants are susceptible to viruses when other aphid species, such as A. citricola, A. craccivora, A. fabae, and Myzus persicae, inoculate the plant with viruses, or if viruses are introduced mechanically (Lecoq et al., 1979, 1980; Romanow et al., 1986). The resistance to viruses when A. gossypii inoculated the plant is also fully effective against unrelated viruses (Lecoq et al., 1980). The gene controlling this double phenotype has been named Vat, for ‘virus aphid transmission’ (Pitrat and Lecoq, 1982).
Several hundreds of accessions were tested for their effect on the aphid traits (Pitrat et al., 1996; Fergany et al., 2011). These large screenings have suggested that about 5% of accessions display resistance to colonization by A. gossypii. Among them, only a small number have been tested for the double phenotype characteristic of Vat, resistance to virus and resistance to aphids. Up to now, the double phenotype has been identified in 25 melon lines (Pitrat and Lecoq, 1980; Soria et al., 2000; Thomas et al., 2012b; Boissot et al., 2016). These melon accessions or lines originate from Asia, Africa, America, and Europe.
Two independent breeding programs were conducted early on, to transfer resistance to A. gossypii into cultivars, with the transfer of resistance from PI 161375 into Charentais-type melons and resistance from PI 414723 into Western Shipper–type melons. Consistent with the cosegregation of resistance to melon aphid and resistance to viruses, which were introduced by melon aphids, the inbred lines obtained in both programs also displayed resistance to viruses when the melon aphid inoculated the plant (Kishaba et al., 1992; Boissot et al., 2016). Margot became the first melon cultivar declared resistant to the melon aphid A. gossypii to be listed in the French catalog in 1987. Since then, 110 Charentais-type cultivars declared resistant to this aphid have been released in France (GEVES data). Melons are cultivated in the South East (SE) and South West (SW) of France, and on two islands of the Lesser Antilles (LA). Given the commercial success of some of the resistant cultivars, about 80% of the melon crops cultivated in SE France since 2000 are thought to have carried this resistance (Boissot et al., 2016).
Since these seminal studies were conducted, the molecular structure of the Vat gene has been elucidated, its double phenotype has been investigated at the cellular level, and its effect on the behavior and life-history traits of the aphid has been studied. Its spectrum of activity against the clonal diversity of A. gossypii has been studied in the laboratory, and its efficacy and the durability of these effects have been studied in situ. All these points will be reviewed after a short presentation of the three protagonists: Cucumis melo, A. gossypii and the viruses transmitted by A. gossypii.
Cucumis melo, Aphis gossypii and the Viruses it Transmits to Melon
Cucumis melo Shares a Number of Features Specific to Cucurbits, but is Genetically Isolated in its Family
Cucumis melo is one of the principal species from the Cucurbitaceae family. Asia is its geographic region of origin and it belongs to the C. melo/C. callosus-C. trigonus complex, which diverged 3 million years ago (Mya) from an Australian sister species, C. picrocarpus (Sebastian et al., 2010). This clade diverged from the lineage leading to cucumber (C. sativus) about 10 Mya. A highly effective reproductive barrier now isolates C. melo from most of its relatives, with successful crosses reported only with C. hystrix (Chen and Adelberg, 2000). Based on data for polymorphism at simple sequence repeat (SSR) markers, C. melo split into two main genetic clusters (Table 1), the first containing four groups (A, B, C, D) and the second containing three groups (E, F, and G; Serres-Giardi and Dogimont, 2012). These data, together with findings for chloroplast polymorphisms (Tanaka et al., 2013), suggest that there were two or three domestication events, one in Asia, another in Africa or Western Asia, and a third in Africa (Pitrat, 2013).
Table 1.
List of melon lines exhibiting the double phenotype, resistance to aphids and resistance to viruses when the aphids inoculate the plant, and multilocus genotypes (MLGs) of Aphis gossypii clones revealing that phenotype.
| Characteristic of melon Accessions |
Characteristics of aphid clonesc |
|||||||
|---|---|---|---|---|---|---|---|---|
| Genetic groupsa | Botanical groupsb | Asia | Africa | America | Europe | I | II | III |
| (I) A | Inodorus | Anso 77 | CUCU3 | NM1 | ||||
| (I) A | Inodorus | Invernizo 8427 | NM1 | |||||
| (I) A | Reticulatus | PI 224770 | NM1 | |||||
| (I) B | Flexuosus | Fegouss 1 | NM1 | |||||
| (I) B | unknown | San Ildefonso | CUCU3 | NM1 | ||||
| (I) C | unknown | Durgapura Madhu | C9 | NM1 | ||||
| (I) D | Makuwa | Kanro Makuwa 1 | C9 | NM1 | ||||
| (I) D | Makuwa | Kanro Makuwa 2 | C9 | NM1 | ||||
| Unknown | Momordica | AM 51 | C9, CUC1, GWD | CUCU3 | NM1 | |||
| (II) E | Momordica | PI 414723 | C9 | NM1 | ||||
| (II) E | Wild | PI 482398 | C9, GWD | CUCU3 | NM1 | |||
| (II) E | Wild | HSD2455 | CUCU3 | |||||
| (II) F | Acidulus | PI 482420 | C9 | NM1 | ||||
| (II) F | Acidulus | 90625 | NM1 | |||||
| (II) F | Acidulus | PI 164723 | NM1 | |||||
| (II) F | Chito | Meloncillo | NM1 | |||||
| (II) G | Chinensis | Chenggam | NM1 | |||||
| (II) G | Chinensis | Miel Blanc | NM1 | |||||
| (II) G | Chinensis | PI 161375 | CUCU3 | NM1 | ||||
| (II) G | Chinensis | PI 255478 | C9 | NM1 | ||||
| (II) G | Chinensis | PI 266935 | NM1 | |||||
| (II) G | Conomon | Shiro Uri Okayama | NM1 | |||||
| (II) G | Makuwa | K 5442 | C9 | NM1 | ||||
| (II) G | Makuwa | Ginsen Makuwa | C9 | NM1 | ||||
| (II) G | Makuwa | Shirokawa Nashi Makuwa | NM1 | |||||
aAccording to Serres-Giardi and Dogimont (2012). bAccording to Pitrat et al. (2000). cAccording to Thomas et al. (2016).
Cucumis melo is now found throughout the world and, like many crops, cultivated melons display extensive phenotypic polymorphism, defining botanical groups, whereas wild melons display low levels of phenotypic polymorphism (Pitrat et al., 2000). The first evidence of C. melo domestication date to just after 3000 BC, in China and Egypt (Pitrat, 2003). Diversification after domestication is controlled mostly by recessive traits, such as sex expression, fruit shape, vein tracts, number of placentas, a gelatinous sheath around the seeds, and white flesh color, whereas disease resistance is mostly conferred by dominant genes (Pitrat, 2013). Melon is now an important fruit crop, with 16 commercial melon types identified by the Organization for Economic Co-operation and Development (OECD) guidelines on the basis of fruit characteristics (shape, skin color and surface characteristics, color of the flesh and dehiscence of the peduncle). Twenty-five to 30 million tons of melon have been produced annually over the last 10 years, with about half of this total in China (FAOSTAT database1). Melon has been subject to intense selection, and its isolation in the genus Cucumis has led to reclaim the broad diversity present in both cultivated and wild forms (Pitrat, 2013). Twenty to 30 new melon cultivars have been added to French catalogs annually since 2000 (GEVES data2).
Melon is a diploid species with a relatively small genome (450 Mb) that has recently been fully sequenced (Garcia-Mas et al., 2012). It has 12 chromosomes, and, like all cucurbits, its genome displays no evidence of recent duplication since the eudicot paleotriplication event. It has a small number of resistance genes, only 81 putative NLR genes were identified (Garcia-Mas et al., 2012), possibly reflecting an unusual adaptive strategy in cucurbits potentially involving specific mechanisms of disease resistance gene regulation or the characteristic vascular structure of these plants. Cucurbits have an unusual vascular structure, with two types of phloem: the fascicular phloem is located in the main vascular bundles, and the extra-fascicular phloem is peripheral to the fascicular phloem, dispersed throughout the cortical tissue of the stems and petioles (Zhang et al., 2010). The fascicular phloem is mostly involved in sugar transport, whereas the extra-fascicular phloem may be involved in signaling and the transport of other metabolites.
A. gossypii Glover: A Biotype Specializing on Cucurbits
The A. gossypii group diverged from other aphids 12 to 25 Mya, during the radiation period of its host plants (Hyojoong et al., 2011). Within this group, species diversification may have been a rapid and recent process, as suggested by phylogenetic trees based on morphological characters (Kim et al., 2010) and the inability of differentiating between species on the basis of mitochondrial DNA COI/COII (Coeur d’acier et al., 2007). The mitochondrial Cytb and nuclear sodium channel para-type (SCP) genes can be used to distinguish the species A. gossypii Glover from related species native to North America, Europe, and Asia (Carletto et al., 2009a; Hyojoong et al., 2011; Lagos-Kutz et al., 2014). Hereafter, we will use the term A. gossypii to refer to A. gossypii Glover.
Aphis gossypii is a cosmopolitan species that is extremely polyphagous, colonizing hundreds of plant species (Ebert and Cartwright, 1997). In northern areas, at latitudes above 30°N, A. gossypii produces sexual morphs in the fall, which produce eggs that diapause on its primary hosts (Kring, 1959; Takada, 1988; Ferrari and Nicoli, 1994). These primary hosts differ between geographic areas, with Rose of Sharon (Hibiscus syriacus) frequently identified as a host plant in Asia, Europe, and America. In spring and summer, A. gossypii becomes a pest on crops, its secondary hosts, on which it reproduces clonally. In intertropical areas, A. gossypii reproduces clonally all year round. Thus, depending on the area of melon production, A. gossypii populations may consist of a mixture of strictly clonal lineages and lineages derived from sexual reproduction or of strictly clonal lineages only.
Aphis gossypii, currently named the cotton-melon aphid, is a pest for several crops, including melon, marrow, zucchini, potato, eggplant, cotton, ornamental hibiscus, and citrus fruit trees. Like all aphids, A. gossypii carries the bacterium Buchnera aphidicola as an obligate endosymbiont providing several essential nutrients (Douglas, 2003) and the phenotypic plasticity in host plant use by A. gossypii may be related to the size of the B. aphidicola population (Zhang et al., 2016). Many other facultative endosymbionts have been detected in aphid species and shown to play a role in species ecology (Oliver et al., 2010); however, facultative endosymbionts appear to be rare in A. gossypii (Carletto et al., 2008).
A small number of Cytb sequence polymorphisms differentiate three haplotypes of A. gossypii collected on crops and plants from the Cucurbitaceae, Malvaceae, Solanaceae, and Rosaceae in Africa, South America, Australia, and Europe (Carletto et al., 2009a). All individuals collected from cucurbits belong to the same haplotype. A small number of mitochondrial DNA sequence polymorphisms between the Cytb and 16S genes distinguish two biotypes of A. gossypii specializing on cotton and cucurbits in North China (Wang et al., 2016). At the end of the 1990s, a set of SSR markers (SSRs) was developed to assess A. gossypii diversity (Vanlerberghe-Masutti et al., 1999). Several hundred multilocus genotypes (MLGs), defined on the basis of allelic combinations at eight SSR markers, have since been described. The largest set of MLGs was identified in a study of spring migrants in France and the Lesser Antilles; they formed seven genetic clusters (Thomas et al., 2012a, 2016). All individuals collected from colonies on melon shared MLGs distributed between three clusters (later named in the manuscript I, II, and III). In data analyzed with the same set of reference clones (Brévault et al., 2008; Charaabi et al., 2008; Carletto et al., 2009b; Chen et al., 2013; Thomas et al., 2016), 75 MLGs were observed in colonies collected from cucurbits in Asia, Africa, Europe, Australia, and Caribbean islands, four of which — C4, C11, C9 and NM1 — were observed in at least two geographic areas suggesting that these clusters contain individuals specializing on cucurbits.
Biological studies have been conducted at the laboratory and field levels to assess the strength of the host specialization of A. gossypii biotypes. The results of these studies can be related with genetic knowledge. Many laboratory host-transfer experiments have been conducted with plants from the Cucurbitaceae (cucumber), Malvaceae, (cotton, okra, and hibiscus), Solanaceae (eggplant and sweet pepper), and Rutaceae (citrus plants and Chinese prickly ash) (Guldemond et al., 1994; Liu et al., 2008; Carletto et al., 2009b; Satar et al., 2013; Wu et al., 2013; Xu et al., 2014; Wang et al., 2016). Overall, the results obtained suggest that on one hand cucurbit biotypes poorly colonize plants from other plant families, if not at all, with the exception of H. syriacus and on the other hand, most, if not all, biotypes specialized on other crops poorly colonize cucurbits. Host switching in the field has been inferred from molecular markers. Studies conducted in different agricultural environments in Africa and China in which cucurbits are present together with cotton, and citrus or Solanaceous crops have confirmed that lineages specializing on cucurbits cannot easily switch to other crops (Brévault et al., 2008; Charaabi et al., 2008; Wang et al., 2016).
Taking into account genetic, ecological and lab experiment data, specializing on cucurbits of a part of A. gossypii species is fairly clear. All clones able to colonize cucurbits form an ecological group called the Cucurbit host-race (Carletto et al., 2009b), they are assigned to the genetic clusters I, II, and III (Thomas et al., 2016). The history of co-evolution between A. gossypii and cucurbits merits further investigation, particularly as the role of cucurbit’s distinctive phloem structure for A. gossypii specialization on these plants.
The Viruses Transmitted to Melon by A. gossypii Belong to Three Families
More than 70 virus species have been reported to attack cucurbits (Lecoq and Katis, 2014). Some cause severe epidemics in melon crops worldwide. Five of these species are transmitted by aphids, including the melon aphid. These viruses may or may not persist in the vector. Non-persistent viruses are acquired and transmitted to the plant during brief probes (lasting less than 1 min), do not require a latent period in the vector and are retained in the vector for only short periods of time (aphids remain viruliferous for only a few hours). Persistent viruses are acquired during phloem punctures for feeding (over periods of several hours or even days); they have a latent period and are retained for long periods in the vector (aphids often remain viruliferous for life). Further information about the transmission of plant viruses by insects is available from a recent review (Fereres and Raccah, 2015).
Non-persistent viruses include the CMV, belonging to genus Cucumovirus, family Bromoviridae. This virus has a worldwide distribution, and has been observed to infect more than 1200 species from more than 100 plant families (Jacquemond, 2012). This virus may have the widest host range of any known plant virus. It can be transmitted by more than 80 aphid species. CMV causes typical mosaic symptoms on melon leaves, plant stunting, mottle or mosaic on fruits, and yield losses.
Several potyviruses (family Potyviridae) attacking melon crops are also non-persistent (Lecoq and Desbiez, 2012). They are transmitted by 20 to 40 aphid species. Watermelon mosaic virus (WMV) is observed worldwide. It can infect more than 170 plant species. On melon leaves, it induces mosaic, vein banding and deformation, such as blisters, filiformis and size reduction. On fruits, it induces severe discoloration, with slight deformation in some cases. Zucchini yellow mosaic virus (ZYMV) is also distributed worldwide, but has a smaller host range than CMV or WMV (only 11 families). It induces vein clearing, yellowing, with blisters and enations on leaves and severe stunting. On fruits, ZYMV induces mosaic or necrotic cracks, marbling and hardening of the flesh. Moreover isolates belonging to the pathotype F induce wilting in melons carrying the Fn gene (Risser et al., 1981) instead of mosaic in melons carrying the Fn+ allele. The Fn gene (for Flaccida necrosis) is present in numerous melon accessions. Papaya ringspot virus (PRSV) mostly infects tropical and Mediterranean cucurbit crops. Its host range is restricted to cucurbits and a few other plant species, such as papaya. On melon, it causes severe mosaic, blistering, and malformations on leaves. Fruits may also display various degrees of discoloration and deformation.
Persistent viruses include the Cucurbit aphid-borne yellows virus (CABYV) a member of genus Polerovirus, family Luteoviridae (Lecoq and Desbiez, 2012). This virus infects many cucurbits, beet, lettuce and many weed species. It is transmitted by a small number of aphid species (M. persicae and Macrosiphum euphorbiae are additional vectors). It induces yellowing of the older leaves, but complete discoloration of the plant is observed with some melon cultivars. Its effect on yield is less marked than other viruses infecting melon, particularly as it has no effect on fruit quality, instead inducing flower abortions and reducing the number of fruits per plant.
Keep in Mind Some Features When Considering the Double Phenotype
The double-resistance phenotype elicited by A. gossypii has been identified in all seven genetic groups in C. melo (Table 1). It has been identified in wild accessions from Africa, PI 482398 and HSD2455, both of which have some cultivated characteristics, but it has not yet been identified in wild accessions from Asia. Conversely, most of the accessions and landraces displaying the double phenotype are native to Asia. The double phenotype has been observed in all botanical groups of this species in Asia. We use the term ‘Vat melon line’ here to refer to any accession or line displaying this double phenotype in a study.
The aphid clones used in bioassays characterizing the double phenotype are only rarely mentioned. Molecular markers for their identification are available (Vanlerberghe-Masutti et al., 1999) but are still only rarely used to characterize the A. gossypii clones used in bioassays. A DNA-reference clone set, at least from clones belonging to the three cucurbit clusters, should be established by the scientific community and made available. Aphids assigned to clusters I and III are the most frequently used (Table 1). No data have been published concerning the capacity of clones that are not able to colonize Cucurbits, to elicit resistance to viruses in Vat melon.
Resistance to viruses when the melon aphid inoculates the plant with virus has been documented principally for CMV. For example, all the accessions mentioned in Table 1 displayed the double phenotype when using this virus in the bioassays. In PI 161375 and PI 414723, the resistance to viruses is fully effective against other potyviruses, such as PRSV (formerly known as, WMV1), WMV (formerly known as, WMV2) (Lecoq et al., 1980), and ZYMV (formerly known as, MYSV) (Risser et al., 1981; Kishaba et al., 1992; Soria et al., 2000), when the melon aphid inoculates the plants. Like CMV, these viruses have a non-persistent mode of transmission. While not formally tested, it is likely that this large spectrum of resistance to viruses transmitted in a non-persistent manner is common to all accessions displaying resistance to CMV transmission from A. gossypii. For other virus species transmitted in a persistent manner, such as CABYV, no laboratory data have ever been published.
From Vat Gene to Vat Cluster
The Vat locus was mapped to C. melo’s linkage group V using segregating populations from a cross between the susceptible line Védrantais and the resistant accession PI 161375 (Pitrat, 1991; Baudracco-Arnas and Pitrat, 1996). It was localized to a subtelomeric position on a saturated map combining two recombinant inbred populations resulting from crosses between Védrantais and two resistant accessions PI 161375 and PI 414723 (Périn et al., 2002). In early 2000, a map-based strategy was used to isolate the Vat gene. This approach involved the use of 6000 plants from a back-cross population derived from Védrantais and PI 161375. Recombination events within the terminal region of linkage group V were screened and recombinant plants were phenotyped for resistance to aphids. A physical map encompassing the Vat gene was obtained by screening a melon bacterial artificial chromosome (BAC) library constructed from PI 161375, and the genomic sequence spanning the Vat region was annotated. A comparison of molecular data and phenotypic data for resistance to melon aphid and resistance to viruses when the melon aphid inoculated the viruses showed that the Vat gene was a single functional locus conferring both types of resistance (Pauquet et al., 2004). Nine of the 14 of the back-cross progeny displaying recombination in the genomic sequence spanning the Vat region are presented in Figure 1, with their phenotype. The Vat gene is 6-kb long, and consists of five exons and four introns. It encodes a predicted 1467-amino acid protein presumed to be located in the cytoplasm (Dogimont et al., 2014). This protein belongs to the coiled-coil (CC)-nucleotide binding site (NBS)-leucine-rich repeat (LRR) family (Figure 2). Only three other genes conferring resistance to aphids or other hemipterans, Mi-1 in tomato and Bph14 and Bph26 in rice are known to encode proteins from the NLR family (Rossi et al., 1998; Nombela et al., 2003; Casteel et al., 2006; Du et al., 2009; Tamura et al., 2014). For confirmation of the effect of the Vat resistance allele, an 11-kb DNA fragment harboring Vat’s coding region, promoter, and 3′-flanking region, was introduced into two susceptible C. melo lines by Agrobacterium-mediated transformation (Dogimont et al., 2014). Four lines derived from independent transformation events were obtained and all lines displayed high levels of resistance to NM1 melon aphids and complete resistance to viruses when the NM1 aphids inoculate the transgenic plants with CMV, WMV, and ZYMV.
FIGURE 1.
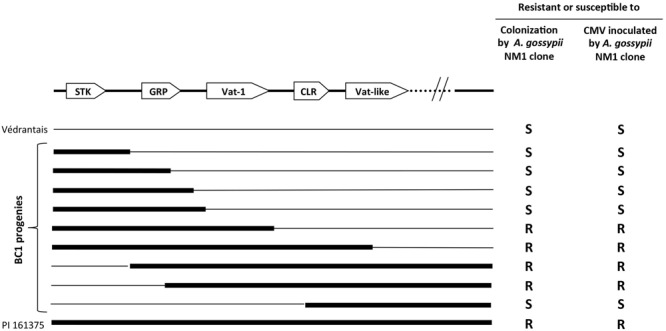
Genomic map spanning the Vat region in 9 Back-Cross 1 plants derived from a cross between PI 161375 and Védrantais displaying segregation for resistance to Aphis gossypii and resistance to viruses transmitted by A. gossypii. STK, GRP, and CLR indicate serine-threonine kinase, glycine-rich protein and copia-like retroelement proteins, respectively. Adapted from (Dogimont et al., 2014).
FIGURE 2.
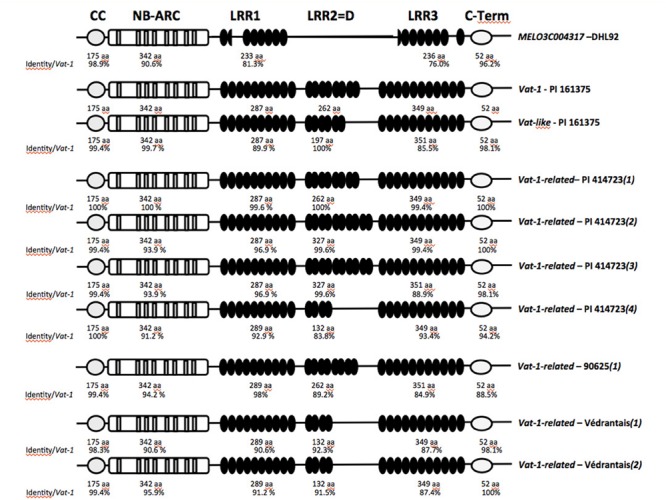
Schematic diagrams of predicted Vat protein domains encoded by the Vat-1 gene and polymorphisms detected in related sequences. Adapted from Dogimont et al. (2008a, 2014) and González et al. (2014).
All the bioassays conducted to identify Vat in the melon genome to date have used NM1. This clone has been used since the early studies by the French team and has provided the most clear-cut differentiation between susceptible and resistant accessions for both resistance to melon aphid and resistance to the viruses introduced into the plant by melon aphid (Boissot et al., 2016). In a study investigating whether the Vat allele of PI 161375 had a specific aphid clone effect or a much broader effect, one of the transgenic lines was tested with a set of A. gossypii clones from the cucurbit host-race. The bioassay used assessed the resistance to viruses transmitted by the aphid. In the transgenic line, the resistance to viruses introduced by an aphid was aphid clone-specific. Surprisingly, for some clones used for inoculation purposes, resistance to the virus was expressed in the native line, PI 161375 but not in the transgenic line (Table 2). These remarkable differences reveal that at least one other gene is involved in the resistance elicited by some A. gossypii clones in PI 161375 (Boissot et al., 2016). In accordance with general rules for the naming of genes, it has been suggested that the gene isolated from PI 161375 (Dogimont et al., 2014) should be renamed Vat-1, and the additional gene Vat-2. There may be allelic series for both these loci (Boissot et al., 2016).
Table 2.
Pattern of resistance to Cucumber Mosaic Virus (CMV) inoculated by six A. gossypii clones on PI 161375, from which Vat-1 was isolated, TR3, the transgenic line in which it was introduced and Margot a line in which aphid resistance from PI 161375 was introduced by classical breeding.
| C6 | GWD | CUC1 | C9 | GWD2 | NM1 | |
|---|---|---|---|---|---|---|
| PI 161375 | S | R | R | R | R | R |
| TR3 | S | S | I | R | R | R |
| Margot | S | R | R | R | R | R |
R, resistant; S, susceptible; I, intermediate.
It is not clear from the results presented above whether Vat-1 and Vat-2 form a cluster. This point has been investigated indirectly. PI 161375 is a Korean line harboring Vat-1 and Vat-2; it belongs to the Chinensis botanical group (Table 1). Vat-1 was introgressed from this line into a Charentais line (Cantalupensis group). The process of introgression consists in a first crossing between a Charentais line and PI 161375 and after, backcrossing the aphid-resistant progeny with the Charentais line, referred to as the recurrent line. The bioassays, to select plants resistant to aphids from each back-cross progeny, used the aphid clone NM1. Remarkably, the spectrum of resistance to viruses transmitted by aphids in PI 161375 was found to be conserved in the line Margot, which was obtained after seven back-crosses, (Table 2) and therefore Margot carries Vat-2 (Boissot et al., 2016). This suggests that Vat-1 and Vat-2 are probably very tightly linked. We will therefore use the name ‘Vat’ for the region containing Vat-1 and Vat-2.
Several genomics studies have focused on the region containing Vat. Genes conferring resistance to various pathogens are located in the vicinity of Vat: resistance to Podosphaera xanthii (Yuste-Lisbona et al., 2001; Perchepied et al., 2005a), Cucumber vein yellowing virus (Ibn Oaf, 2012), the Fn gene (Pitrat and Lecoq, 1982) triggering plant necrosis in response to some isolates of ZYMV (Risser et al., 1981), and the quantitative trait loci (QTL) FomV-2 conferring partial resistance to Fusarium oxysporum f. sp. melonis (Perchepied et al., 2005b). The density of resistance genes in melon is highest in the region containing Vat (Garcia-Mas et al., 2012); 28 genes of the NLR family have been identified in a 1-Mb region containing Vat (González et al., 2014). Characterization of four C. melo accessions displaying resistance to viruses when different A. gossypii clones inoculated the plants has identified Vat-1-related sequences (protein identity over 80%; Figures 1 and 2).
These sequences display polymorphisms within all parts of the gene (Figure 2). In the LRR part of Vat, two types of polymorphism are observed: single-nucleotide polymorphisms (SNP) and length polymorphisms. The length polymorphisms occur in a specific domain, domain D or LRR2 (Figure 2) (Dogimont et al., 2008a, 2014). This domain consists of near-perfect repeats of 65 amino acids. The Vat-1-related sequences contain two to five repeats. The repeats are 83.1–89.2% identical (Dogimont et al., 2008a). PI 161375 has a Vat-1-related sequence with three repeats known as Vat-like. Vat-like is located 17 kb from Vat-1 (Figure 1) and is not involved in the resistance elicited by the NM1 clone (Dogimont et al., 2008b). In PI 414723, a line exhibiting resistance to several clones, four Vat-1-related sequences have been identified. One of these sequences has only a few SNPs relative to Vat-1. Two sequences have five repeats in LRR2, and both are strong candidates for the control of resistance to P. xanthii (Dogimont et al., 2008b). Both these Vat-related sequences have few SNPs relative to the sequence of Vat-1. The fourth Vat-1-related sequence has two repeats in LRR2 and more SNPs relative to Vat-1 than the other Vat-1-related sequences. In 90625, a line exhibiting resistance restricted to only one clone, NM1 (Boissot et al., 2016), only one Vat-1-related sequence has been identified. This sequence contains four repeats in LRR2 and several SNPs. In Védrantais, a Charentais line resistant to viruses when only one aphid clone, C4, inoculate the plant (Boissot et al., 2016), two Vat-1-related sequences with several SNPs relative to the sequence of Vat-1 and two repeats in LRR2 have been identified. Finally, the cadre of Vat-like genes in each accession may not be complete, since no complete genome sequences are available for these accessions. Involvement of these Vat-1-related sequences in the aphid resistance has not been demonstrated. Actually, the melon reference genome was built from a line (DHL92) given as susceptible to A. gossypii (González et al., 2014), but accurate double-phenotypic data, with a set of characterized clones, are missing for that line. This line has a Vat-1 homolog, MELOC004317, shorter (1038 aa) than the reference one (1467 aa), in particular the LRR2 part is fully absent (Figure 2) (González et al., 2014).
The presence of large numbers of NLR genes in close proximity to Vat, including Vat-like genes, makes assembly difficult and even unsatisfactory when sequences are obtained for small fragments. Sequencing studies of a set of accessions are required with longer fragments for an accurate assembly of the area. Comprehensive cross-comparison between molecular and phenotypic data is therefore required to obtain a full understanding of the genetic control of resistance to A. gossypii aphids and the viruses they transmit. The use of transgenic lines, even if difficult to obtain (Chovelon et al., 2011), will clearly help us to decipher the role of each locus in this cluster. As soon as candidates are identified, transformation with these candidates could be used to validate their roles in the resistance spectrum. This approach could provide new opportunities for genomic selection for resistance in melon.
Molecular Responses in the Vat Melon/A. gossypii Aphid Interaction
The Vat-1 gene belongs to the NLR gene family. According to the general framework developed for this category of resistance genes, its functioning involves separate recognition and response phases (Figure 3A). In this case, the recognition phase involves perception of an aphid effector by the plant’s VAT protein, which may be direct or indirect. The vast majority of plant-pathogen effectors have been shown to interact indirectly with a NLR protein of their host. To date, modality of the interaction between aphid effectors and NLR proteins of their host is still unknown. This interaction should occur in the cytoplasm of cells (the predicted location of the VAT protein), and the effector molecule must therefore be delivered to the plant cell. Aphid mouthparts enclose two flexible stylets that the insect drives into the plant tissue to puncture the phloem, so that they can feed on plant sap. On their way to the phloem, the aphid stylets bend around the cells, and the salivary channel ejects a salivary gel that forms a sheath around the stylets. Along the way to their destination, the aphid stylets briefly puncture cells, into which the salivary channel ejects a watery saliva (Tjallingii, 2006). The effector is probably injected into the cells during these puncture events. No aphid effector capable of interacting with an NLR protein has yet been identified. A transcriptomic approach has been used to identify candidates involved in the virulence of A. gossypii on Vat melon (Dutartre-Fricaux et al., 2014). Genetically similar virulent and avirulent clones were used, and a head-reference transcriptome of more than 33000 contigs was generated by de novo assembly. This reference transcriptome has been used to search for candidate effector genes based on their differential expression and/or presenting sequence polymorphisms between virulent and avirulent aphid clones.
FIGURE 3.
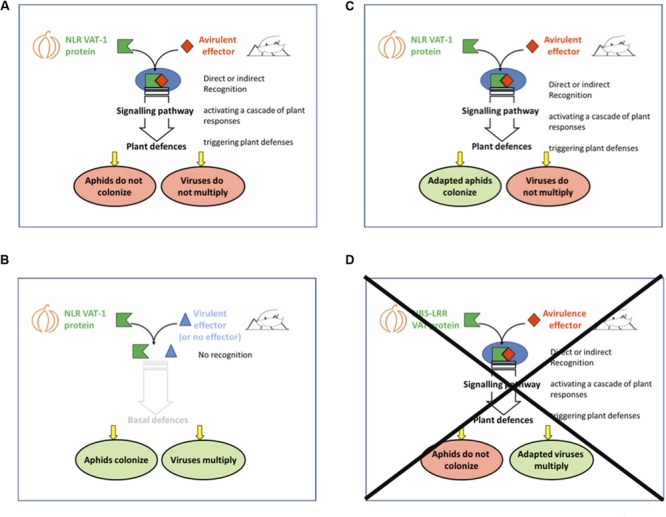
Models of A. gossypii/Vat-melon plant interaction based on interaction (direct or not) between the VAT protein and the avirulence effector (Boissot et al., 2016). The three cases observed were: (A) resistance to aphids and viruses, (B) susceptibility to aphids and viruses, (C) susceptibility to aphids and resistance to viruses. (D) A fourth outcome, resistance to aphids and susceptibility to viruses, was not observed.
At the molecular plant level, the response phase is thought to involve the activation of a signaling cascade, leading to the rapid accumulation of reactive oxygen species (ROS) and defense hormones. In a Vat melon line, the levels of miRNAs involved in the post-transcriptional regulation of gene expression change rapidly after puncture by A. gossypii (Sattar et al., 2012b). Within 12 h of the infestation by melon aphids, 23 families of miRNAs display modulations. Their potential targets suggest a physiological function in disease and stress responses (5), phytohormone perception and signaling (11), miRNA biogenesis (2), and plant growth and development (Sattar et al., 2016). Ethylene, jasmonic acid and auxin have been identified as potential defense hormones in Vat plants infested with A. gossypii (Anstead et al., 2010; Sattar et al., 2016). In two Vat melon lines, peroxidase activity was found to increase within 10 min of aphid puncture (Sarria Villada et al., 2009). Callose, a polysaccharide usually laid down at plasmodesmata, is deposited within 20 min of aphid infestation, and lignin, a macromolecule derived from phenyl propanoids essential for cell wall thickening, is deposited 4.5 h after aphid infestation. The plasma membrane is damaged and the cells collapse. Callose and lignin are deposited in the wall of cells adjacent to the stylet sheath. These reactions do not occur in non-Vat plants. These data clearly illustrate the massive transcriptional reprogramming induced by A. gossypii infestation in Vat-resistant melon plants, triggering a wide range of plant defense responses (Dogimont and Boissot, 2016; Sattar et al., 2016).
At the molecular aphid level, changes in gene expression were investigated in A. gossypii feeding on Vat and non-Vat plants. There is an unexpectedly high abundance of 27 nt-long sRNA sequences in aphids feeding on Vat plants (Sattar et al., 2012a). These sRNAs belong to the Piwi-interacting RNA (piRNA) family. This class of sRNAs is absent from plants. Their biogenesis in animals is still poorly understood. They have been shown to be involved in the silencing of transposable elements exclusively in animal gonads (Vodovar and Saleh, 2012), facilitating short-term adaptation. Their role in A. gossypii remains unknown, but may relate to the lifting of maternal effects. Such effects are observed, for example, in aphids collected from cotton, which have low rates of reproduction in the first generation after transfer onto eggplant, but higher rates in subsequent generations (Satar et al., 2013). Eighty-one conserved microRNAs (miRNAs), 12 aphid-specific miRNAs, and nine novel candidate miRNAs have also been identified (Sattar et al., 2012a). These candidate miRNAs have been shown to be differentially regulated between aphids feeding on Vat and non-Vat plants and may affect their reproductive rates as described below.
Effect of Vat Plant Responses on A. gossypii Behavior and Biology and the Virus they Transmit
Aphid feeding is disrupted on Vat plants. Electrical-Penetration graph (EPG), in which the pathway of aphid stylets from epidermis to phloem can be followed, have been conducted on several Vat melon lines, with different genetic backgrounds and on melon aphid clones originating from different geographic areas (Kennedy et al., 1978; Chen et al., 1997; Klingler et al., 1998; Boissot et al., 2000; Garzo et al., 2002). The journey of the stylets through the mesophyll to the phloem takes from 90 to 140 min in non-Vat plants (Kennedy et al., 1978; Chen et al., 1997; Klingler et al., 1998; Boissot et al., 2000; Garzo et al., 2002), but is disrupted in Vat plants. The observed cellular response seems to occur after the aphid stylets have punctured plant cells rather than during the intercellular penetration of plant tissues by the stylets (Sarria Villada et al., 2009), consistent with the hypothesis that recognition occurs after the delivery of the effector to the cell. Cytological studies have shown that there are more stylet sheaths in Vat plants than in non-Vat plants (Kennedy et al., 1978), suggesting that early mesophyll cell puncture by A. gossypii may be more frequent in Vat plants. The stylets take longer to reach the phloem in Vat plants than in non-Vat plants and are less likely to reach their final destination in Vat plants than in non-Vat plants. Prior exposure of Vat plants to A. gossypii feeding does not modify the expression of this resistance (Chen et al., 1997).
Findings on Vat melon suggest that the plant responses elicited by short cell punctures either hinder the passage of the stylets between cells due to the deposition of callose and/or lignin in the cell walls, or deter the aphid from progressing further into the tissues, through an oxidative burst detected by aphid after brief periods of ingestion following the release of saliva into the cell. Moreover, melon aphids reaching the phloem of Vat plants do not remain there to feed (less than 10 min, if at all; Kennedy et al., 1978; Chen et al., 1997; Klingler et al., 1998; Boissot et al., 2000; Garzo et al., 2002). This suggests that feeding may be difficult in these plants, possibly due to phloem clogging, although this plant reaction has not yet been described in Vat melon. Few quantitative differences in phloem sap have been identified between Vat and non-Vat plants that might explain the deterrence of A. gossypii (Chen et al., 1997). None of the studies, EPG or histological studies, investigating these aspects took into account the dual phloem structure of cucurbits and the nature of the phloem in which A. gossypii is able to establish feeding.
Aphids escape from Vat plants. In free-choice tests, winged aphids are less numerous on Vat plants 24 h after being offered a choice of plants (Kennedy and Kishaba, 1977), and wingless aphids are less numerous on Vat leaf disks from 30 min after being offered a choice of leaf disks (Garzo et al., 2002). Without choice, i.e., only one plant accession is available, wingless aphids walk away from the plant in the 2–3 days after their deposition. This behavior has been observed on several Vat melon lines, with aphid clones originating from different geographic areas that probably displayed marked genetic differences (Pitrat and Lecoq, 1982; Garzo et al., 2001; Boissot et al., 2010; Thomas et al., 2012b). Based on the timing of these events, we can conclude that early plant responses, occurring rapidly after cell puncture by the aphid, have an immediate effect on aphid behavior.
Aphids poorly reproduce on Vat plants. When wingless aphids are encaged on Vat plants, they display low rates of reproduction, mostly due to a longer pre-reproductive period and a smaller number of progeny (Klingler et al., 1998; Garzo et al., 2002; Boissot et al., 2010). This lower reproductive rate may be directly due to poor, disrupted feeding on the contents of phloem (Kennedy et al., 1978; Chen et al., 1997; Klingler et al., 1998; Boissot et al., 2000; Garzo et al., 2002). This hypothesis is supported indirectly by the observation that aphids produce far less honeydew when feeding on Vat plants than on non-Vat plants (Klingler et al., 1998). The resistance factor reducing the reproductive rate is not transmitted through grafting (Kennedy and Kishaba, 1977), consistent with the notion that feeding is somehow difficult, rather than with the phloem being toxic. The rate of aphid reproduction is quantitatively affected by Vat and QTLs (Kishaba et al., 1976; Boissot et al., 2010; Thomas et al., 2012b).
Vat plants display particularly high levels of resistance to viruses when aphids inoculated unrelated viruses transmitted in the non-persistent mode: 100% of the non-Vat plants displayed symptoms, whereas only 0 to a few per cent of Vat plants had symptoms, with this small number of plants displaying full symptoms (Lecoq et al., 1979; Kishaba et al., 1992; Boissot et al., 2016). When Vat resistance was first discovered, it was thought to block virus transmission, and Pitrat and Lecoq therefore named the gene responsible Vat, for ‘virus aphid transmission’ (Pitrat and Lecoq, 1982). Nevertheless, A. gossypii was subsequently shown to acquire the virus from Vat plants and to transmit it to non-Vat plants (Romanow et al., 1986), calling this hypothesis into question. According to the resistance to transmission hypothesis, a plant factor blocks the virus in the stylet (a molecule or a particular pH). The aphid must, therefore, first ingest material from the plant, before it egests saliva into the cells and delivers the viruses. All EPG studies have shown that, after puncturing cells, the aphid first salivates and then ingests the contents of the cell (Martin et al., 1997). Further studies have failed to demonstrate resistance to transmission due to retention of the virus in the stylets.
Vat resistance to viruses has to be considered in the general framework described for NLR resistance (Boualem et al., 2016). The VAT protein of non-Vat plants (i.e., carrying a ‘susceptible’ Vat allele) cannot recognize (directly or otherwise) the aphid effectors and viruses delivered to the cell by the aphid. This lack of recognition leads to systemic viral infection. In Vat plants, the resistant isoform of VAT recognizes an effector molecule from the aphid. This recognition induces resistance mechanisms limiting the replication and movement of the virus. The micro-oxidative burst triggered by aphid puncture in Vat plants (Sarria Villada et al., 2009) is thought to block the viruses in the inoculated cell or in neighboring cells. Callose deposit at plasmodesmata may help to contain virus particles in the inoculated or neighboring cells. The response is local: when a Vat plant is first inoculated with CMV by A. gossypii, CMV superinoculation with M. persicae on the same leaf leads to systemic infection (Mistral and Boissot, 2016). In the absence of the aphid effector, the recognition phase does not occur when the Vat plant is infected with viruses. In this case, the viruses replicate and move around the plant, establishing a systemic virus infection.
Virus aphid transmission (the initial name from which the Vat acronym is derived) does not provide an accurate picture of the action of the Vat gene product according to recent data. It would, therefore, be more appropriate to consider Vat to stand for ‘virus aphid triggered.’
In Vat plants, no resistance to viruses transmitted in the persistent mode has ever been reported. As CABYV is restricted to phloem cells, the aphid must reach the phloem cells and feed for long enough to acquire virus particles. Mechanical inoculation is not possible for this virus, suggesting that effective inoculation is dependent on the delivery of the virus directly into the phloem. As A. gossypii rarely reaches the phloem of Vat plants (Kennedy et al., 1978; Chen et al., 1997; Klingler et al., 1998; Boissot et al., 2000; Garzo et al., 2002), CABYV acquisition and inoculation should be disrupted in Vat plants, but this point needs to be investigated.
Ability of A. gossypii and the Viruses it Transmits to Adapt to Vat
The LRR domain of the Vat-1 gene is subject to diversifying selection (Dogimont et al., 2014). This selection responds to the diversifying selection acting on the avirulence gene, as frequently reported for other avirulence genes in plant pathogens (Rouxel and Balescent, 2010). This model describes the general framework for the molecular arms race between plants and pathogens. An aphid clone adapted to a given Vat allele would either not deliver an ‘avirulent’ effector to the plant (deletion) or would deliver a ‘virulent’ effector that is not recognized by the VAT protein. In both cases, the expected phenotype would be colonization of Vat plant by the clone, and susceptibility to viruses introduced into the plant by that aphid clone (Figure 3B).
This model has been challenged by testing transgenic lines containing the Vat-1 gene, for resistance to both A. gossypii and CMV introduced into the plant by six A. gossypii clones (C6, C9, CUC1, GWD, GWD2, and NM1) (Boissot et al., 2016). The phenotypes for five of the six clones were consistent with the general model (Figures 3A,B): the clones were either unable to fully colonize the Vat-1 transgenic line and triggered resistance to CMV (NM1, C9, Figure 3A), or they fully colonized the Vat-1 transgenic line and did not trigger resistance to CMV (C6, CUC1, GWD, Figure 3B). These five clones belonged to the three clusters corresponding to the cucurbit host-race (Table 1). The phenotypes observed with one clone (GWD2) were not consistent with the general model, with the clone triggering resistance to viruses but nevertheless being able to colonize the transgenic plants (Figure 3C).
Unlinking of resistance to viruses triggered by the aphid and resistance to aphids was confirmed by testing eleven Vat lines identified from the natural range of diversity in melon with nine clones, the six previously described and CUC6, CUC3, and C4 (Boissot et al., 2016). Only 52 of the 117 interactions characterized, considering results on transgenic lines and natural Vat lines, were consistent with the general model (Figures 3A,B). It has been hypothesized that the decoupling of the resistances to aphids and viruses (Figure 3C) results from aphid adaptation, making it possible for the aphid to colonize the plant even if plant defenses are elicited. The Vat phenotype proved to be a highly powerful tool for investigating a phenomenon never before studied for plant/pathogen interactions. This new model for adaptation to NLR resistance is revealed by the double phenotype, which can be used to follow the resistance process at two levels: recognition, and the efficacy of the plant defenses triggered.
We speculate that individuals of some clones adapted to Vat defenses they trigger (e.g., GWD2). If these aphids infest a Vat plant successfully, they must accept the Vat plant and reproduce at a high rate on it. EPG has revealed that Vat affects the exploratory behavior of the aphid on the plant, but this effect is quantitative, with some aphids reaching the phloem of Vat plants. Individuals of a clone adapted to Vat defenses probably reach the phloem more often than those of a non-adapted clone. A ‘classical avenue’ of research will involve comparison of the transcriptomes of adapted and non-adapted clones puncturing Vat plants, to track the aphid genes involved in this adaptation. We propose to explore a new avenue of research: does the dual phloem structure of cucurbits, and of melon in particular, play a role in this adaptation?
It is possible a priori that viruses transmitted in the non-persistent mode can overcome Vat-mediated resistance. In this scenario, viral variants may multiply and leave the punctured cells before the defense mechanisms are fully effective, resulting in the development of systemic infections. The expected phenotype would be ‘susceptibility to viruses introduced by an aphid clone that is incapable of colonizing Vat plants’ (Figure 3D). To date, this double phenotype has never been observed in a transgenic line carrying the Vat-1 gene, suggesting that viral adaptation may not occur (Boissot et al., 2016). Consistent with these findings, experimental evolution experiments with viruses on Vat-plants have been unsuccessful. Sequential virus transmissions from infected Vat melon plants to healthy Vat melon plants were established with two aphid clones and three viruses, CMV, ZYMV, and WMV. None of these viruses evolved in response to the resistance triggered by these two clones, even when four sequential virus transmissions could be done (Boissot et al., 2016). These results strongly suggest that viruses transmitted in the non-persistent mode do not readily adapt to the Vat resistance triggered by A. gossypii.
Effect of Vat on A. gossypii at the Population Level and its Durability
Aphid density is lower on Vat plants than on non-Vat plants (Thomas et al., 2016). With the aim of quantifying the effect of Vat at crop level, we compiled bibliographic data for a density index for Vat and non-Vat plants grown in field experiments conducted at the same location (Schoeny et al., 2014; Thomas et al., 2016). The aphid density index was 44% lower on Vat plants (Figure 4). This index is related to aphid density per m2 over the entire cropping period, by an exponential relationship (y = 1463e0.1088x with r2 = 0.72, n = 304). Therefore, for non-Vat plant indexes of 90, 50, and 30, aphid density is reduced by factors of 50, 11, and 4, respectively, on Vat plants.
FIGURE 4.
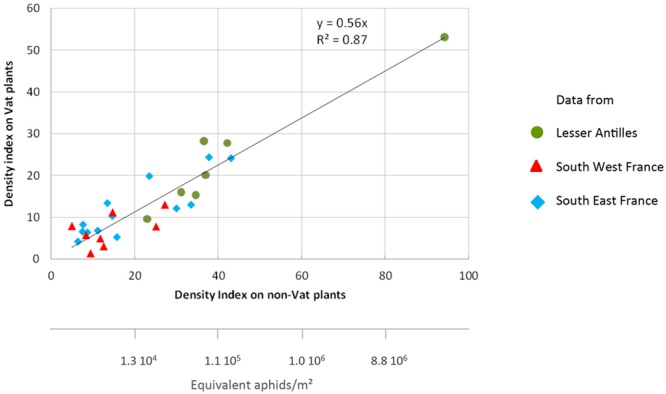
Aphid densities on Vat and non-Vat plants grown in 28 fields in three melon production areas from 2008 to 2014. The data shown are from (Schoeny et al., 2014; Thomas et al., 2016).
There are four key phases in the dynamics of crop infestation by aphids: visiting by winged aphids, infestation with the wingless nymphs they generate, development into colonies, and production of winged individuals for dispersal. Melon crops are visited by spring migrant aphids of numerous species. The proportion of A. gossypii among the visiting aphids and the genetic composition of the A. gossypii spring migrant population depend on geographic area (Thomas et al., 2016). Only some of the A. gossypii spring migrants generate progeny (Figure 5), mostly specializing on cucurbits (i.e., belonging to the Cucurbits I, II, and III genetic clusters). This selection leads to a significant decrease in clonal richness between the spring migrant and wingless populations on melon plants (Thomas et al., 2012a), and this decrease continues during subsequent steps, reflecting differences in fitness or competition between clones on melons (Thomas et al., 2016) (Figure 5).
FIGURE 5.
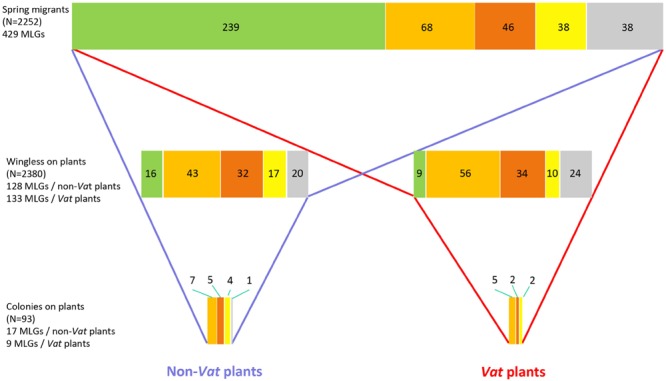
Reduction of the clonal richness of A. gossypii populations during the infestation of melon crops in France. Clonal richness at each step of the infestation is represented by the length of the stripe. Colors within the stripe represent different genetic clusters of A. gossypii populations, with the size of each rectangle proportional to the number (indicated within) of MLGs assigned to the genetic cluster. The data are available from the Dryad Digital Repository: http://dx.doi.org/10.5061/dryad.gf54q
 Cucurbit I,
Cucurbit I,  Cucurbit II,
Cucurbit II,  Cucurbit III,
Cucurbit III,  other clusters,
other clusters,  Not assigned at 75%.
Not assigned at 75%.
The effect of Vat on the first step of infestation has never been reported at field level. However, Vat plants were found to be less attractive for winged aphids than non-Vat plants in greenhouse experiments based on artificial infestation (Kennedy and Kishaba, 1977). The effect of Vat on subsequent phases of infestation has been characterized in open-field melon crops under natural infestation conditions, but only in French melon production areas and in the Lesser Antilles (Thomas et al., 2016). The wingless populations on Vat plants have a genetic composition different from that of the populations on non-Vat plants, as clonal richness and clonal diversity decreased during infestation. Aphids from group III of the cucurbit host-race are eliminated in favor of aphids from group I. In French production areas, the third step, colony development, is erratic on melon crops, and, in the presence of Vat, this step is very rarely observed, generally in only a few aphid genotypes from group I or II. The fourth phase, the production of winged individuals for dispersal, is density dependent (Dixon, 1985), and therefore very rare on Vat plants. The populations of A. gossypii occurring in French melon crops contain a large proportion of aphids from genetic group III, and the decrease in density and diversity on Vat plants probably reflects selection rather than competition. Consistent with this hypothesis, a laboratory study showed that Vat-mediated resistance affected the population growth of 90% of group III clones, but only 40% of those from group I or II (Lombaert et al., 2009). Only group I aphids are present in the Lesser Antilles, where the third and fourth steps are regularly reached, even on Vat plants. The effect of Vat on aphid clonal richness is illustrated in Figure 5. Only four clones, CUC1, CUC6, GWD, and C6, have regularly been identified in colonies on Vat plants in field experiments (Thomas et al., 2016) and studied in laboratory experiments (Boissot et al., 2016). C6 does not trigger resistance to CMV and probably has a virulent effector not recognized by Vat, whereas CUC1, CUC6, and GWD trigger resistance to CMV and are adapted to plant response.
An ecological genetics analyze of melon-aphid dynamics has been applied in three different agricultural systems over the last decade, to predict the durability of Vat resistance to A. gossypii (Thomas et al., 2016). It appears that A. gossypii is evolving at a regional level in response to the deployment of Vat melon crops. For example, two different bottlenecks affect the dynamics of adapted clones in melon-producing areas, due to (i) the low levels of dispersal morph production on Vat melon and (ii) the winter extinction of clones. The low levels of dispersal morph production result from the containment of populations at levels of crowding below that required to induce the production of winged morphs. Winter extinction occurs due to the absence of other cucurbit crops to serve as hosts between two melon crop cycles, limiting the maintenance of Vat-adapted clones. In melon-producing areas without bottlenecks (such as the Lesser Antilles), resistance is predicted to be not durable. In areas in which both types of bottlenecks occur (such as South-West France), resistance is predicted to be durable. In South-East France, only one of the two bottlenecks occurs, and cucurbits are cultivated almost year-round. Moreover, in South-East France Vat melons have been cultivated at a large scale since 2000, and Vat resistance is now jeopardized by the emergence of adapted clones. These findings suggest that, for a cosmopolitan pest, such as A. gossypii, decisions concerning resistance deployment should take into account the genetic structure of the pest population at regional scale, the availability of winter host plants for adapted biotypes between crop cycles (Thomas et al., 2016) and the allele composition of the Vat cluster.
The manipulation of agricultural systems to increase the durability of Vat resistance through winter extinction does not appear to be feasible. However, it may be possible to increase durability by preventing the production of dispersal morphs from adapted clones. Different ways of achieving this aim have been investigated. The use of strips of flowering plants sown close to Vat melon crops to attract natural enemies has been investigated (Schoeny et al., 2014), but the efficacy of this approach will need to be confirmed over several years. Alternatively, QTLs decreasing the production of dispersal morphs on Vat plants could be sought. The identification of such QTLs is probably feasible in melon accessions displaying resistance to aphids sensu stricto (no elicitation of resistance to virus by A. gossypii). This phenotype, like classical phenotypes of resistance to aphids described in other crops, has already been observed in the natural range of melon diversity (Boissot et al., 2016). QTLs controlling this phenotype could be combined with Vat resistance in a melon breeding program.
Effect of Vat on Viruses at the Population Level and Durability of Resistance to Viruses
The effect of Vat on virus epidemics is poorly documented. Field experiments were conducted in France in the late 1970s to compare the development of CMV in Vat accession PI 161375 and a non-Vat melon cultivar (Lecoq and Pitrat, 1983). CMV progression curves had the same general “S” shape, with a steep slope, but disease onset was always earlier in the susceptible plots, with symptoms observed 12–24 days later in Vat plots than in non-Vat plots. This evaluation continued into the 1980s, with the resistant cultivar Virgos (Lecoq and Pitrat, 1989). In accordance with previous results, resistance delayed the CMV epidemic development and greatly decreased the rate of disease increase. It should be borne in mind that PI 161375 and Virgos carry composite resistance to CMV: Vat and the oligogenic and recessive resistance to ‘common’ CMV strains (Guiu-Aragones et al., 2014). It is, therefore, difficult to determine the actual contribution of Vat to the control of CMV epidemics. These experiments were more informative for WMV, because ‘resistance to common CMV strains’ is not effective against WMV. In Vat melon plots, WMV epidemics were delayed slightly (by about 5 days), with no significant reduction of the rate of disease increase (Lecoq and Pitrat, 1989).
Recent studies in South-East France compared virus epidemic development in melon lines differing only by the presence/absence of Vat (Schoeny et al., 2014; Boissot et al., 2015). In most field trials, Vat had a significant effect on CMV epidemics, mostly by reducing the rate of disease increase. It had no effect on WMV epidemics, probably because A. gossypii is not the principal aphid vector of this virus.
The partial effect of Vat on CMV epidemics is consistent with Vat resistance being elicited by only a proportion of the viruliferous aphids visiting melon crops. Indeed, more than 80 aphid species are able to transmit CMV, therefore viruliferous aphids belonging to these species trigger epidemics when they visit Vat and non-Vat melon crops. Moreover within A. gossypii, it remains unclear whether A. gossypii not belonging to the Cucurbit host-race can elicit resistance to CMV resistance. Nevertheless, the partial effect of Vat on CMV epidemics remains significant, probably because A. gossypii is one of the most efficient vectors of CMV based on laboratory experiments.
Finally, the use of Vat to control the spread of non-persistent viruses in melon crops is dependent on the importance of viruliferous A. gossypii relative to other vector species in the spread of the virus in the crop. The effect of Vat is not sufficient for the full control of virus epidemics in crops, but the broad spectrum of this effect and the inability of viruses to adapt to it (Boissot et al., 2016) have made this type of resistance much of a ‘holy grail’ for plant breeders. In the postgenomic era, it may be possible to edit this resistance gene to make it possible for any aphid species to trigger resistance or for resistance to occur without the need for aphid triggering.
Concerning persistent viruses, Vat steadily and significantly decreases CABYV epidemics, mostly by delaying them (Schoeny et al., 2014; Boissot et al., 2015). A. gossypii is the principal vector of CABYV so, even though this aspect has not yet been investigated in the laboratory, it appears likely that Vat affects CABYV transmission by decreasing both acquisition and inoculation rates. The effect of Vat on CABYV population genetic diversity has not yet been documented.
Vat resistance has only a partial effect on virus epidemics in melon and is not used in that aim by growers. As a matter of fact resistance to viruses sensu stricto needs to be integrated in cultivars to control virus epidemics in crops. This type of resistance to viruses has been identified in C. melo species, but has generally been little used in plant breeding programs (Pitrat, 2016). If deployed at a large scale in melon crops, such resistance would exert a selection pressure on viruses, placing the durability of the resistance at risk. The utility of combining Vat with resistance to viruses sensu stricto has been investigated for CMV and CABYV (Boissot et al., 2015). The epidemic data obtained for Vat and non-Vat melon crops in South-East France have been integrated into a mathematical model of the evolutionary and epidemiological processes shaping the dynamics of a virus population in a landscape composed of a seasonal cultivated compartment and a reservoir compartment containing virus throughout the year (Fabre et al., 2015). Various agro-ecological systems were considered, mimicking the situation of melon crops in South-East France. The deployment of resistance to viruses sensu stricto combined with Vat would probably be beneficial for CABYV control, but the potential benefit remains uncertain (although certainly not negative) for the long-term control of CMV. Another modeling study has suggested that the maintenance of low-population aphid populations could prevent the emergence of highly virulent CMV+N-satRNA isolates (Betancourt et al., 2016).
Conclusion
Finally, since the description of the ‘aphid side’ of the pleiotropic phenotype of Vat in the late 1960s, each decade has contributed to improvements in our knowledge and use of this amazing gene. The ‘virus side’ of the pleiotropic phenotype was elucidated in the late 1970s, with the breeding and deployment of the first Vat cultivars in the 1980s, and mapping in the 1990s. The assignment of this gene to the NLR gene family in the first decade of the 21st century provided clues to its mode of action, which is now at least partially understood. A succession of new technologies over this period provided new insight into the pleiotropic phenotype of Vat. Our knowledge of the genetic diversity of A. gossypii has also been refined over time. A. gossypii genetic diversity presents a major challenge to Vat resistance in the field, but also provides us with opportunities to extend our knowledge of the mechanisms underlying Vat resistance.
The A. gossypii/melon interaction can be investigated within the broader A. gossypii/cucurbit interaction, for at least two points. First, the double phenotype conferred by Vat makes it possible to investigate this interaction over the subgroup of A. gossypii constituting the Cucurbit host-race. How diverse are the A. gossypii strains able to elicit resistance to viruses? The VAT protein probably interacts with ligands present in the A. gossypii species or in the A. gossypii group. Once the avirulence factor interacting with the VAT protein has been identified, it will be possible to perform genetic diversity studies on that factor. Second, the particular structure of the phloem in cucurbits may play a key role in the specialization of A. gossypii, an insect feeding on plant sap, on cucurbits and in adaptation to Vat resistance, which decreases the access of A. gossypii to the phloem.
The double phenotype can also be used as a tool for ‘reading’ the recognition phase independently of the response phase, whether this response is considered in terms of the response of the plant, or that of the aphid. Promising preliminary results have been obtained with this approach, and the double phenotype could be more extensively used for such studies. The observation that some A. gossypii clones trigger resistance responses in Vat plants and are adapted to this response provides new insight into the capacity of pests and pathogens to adapt to NLR-mediated resistance in plants. The general framework for resistance mediated by such genes is that, within a pest/pathogen species, a clone/isolate is considered to have adapted if it does not trigger NLR-mediated resistance (i.e., there is no recognition phase). The models proposed for Vat/A. gossypii interaction suggest that aphid clones are adapted to Vat plants either because their avirulence factors do not trigger resistance or because they can colonize the plants even if plant defenses are triggered. Does this second mechanism exist in other NLR plant resistance/pest or pathogen interactions? If so, it would complicate the identification of avirulence factors, because adapted and non-adapted pests/pathogens could have identical avirulence effectors interacting (directly or indirectly) with the protein encoded by the resistance gene. It would also call into question the validity of durability modeling approaches based exclusively on gene-for-gene interaction.
The second way in which A. gossypii clones can adapt, such that the A. gossypii clones can colonize Vat plants whilst eliciting resistance to viruses, appears to be the most common mechanism in A. gossypii populations developing colonies on Vat crops, raising questions about the evolutionary advantages of such a mode of adaptation. This type of adaptation increases the chances of A. gossypii being able to colonize a melon crop free of viruses (because the viruses transmitted by this aphid are blocked in Vat plants), probably leading to the production of a larger number of progeny. Further studies are required to assess the advantages of these two types of adaptation.
Many different studies of Vat resistance have been carried out, revealing the considerable utility of this gene for addressing research questions and the limitations of the use of resistance genes in agriculture. One key question remains: is this gene really unique among the genes conferring resistance to aphids? No other aphid resistance gene has been reported to confer resistance to viruses transmitted in the non-persistent mode. However, it is not clear how many of the many known aphid resistance genes have been tested for effects on viruses.
Author Contributions
NB conceived the review and built the figures and tables. AS contributed to the introduction’, the presentation of viruses transmitted by A. gossypii to melon crops, to molecular features to the effect of Vat at the whole-plant level, Vat effect on viruses populations and conclusion. She revised the entire manuscript. FV-M contributed to the presentation of A. gossypii and the part on Vat effect on A. gossypii populations. She revised the entire manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer IK and handling Editor declared their shared affiliation, and the handling Editor states that the process nevertheless met the standards of a fair and objective review.
Acknowledgments
We thank LW for inviting us to contribute to this special Frontier issue. We would also like to thank Michel Pitrat, Hervé Lecoq, and Catherine Dogimont for the exciting discussions we had on Vat. Michel Pitrat and Véronique Chovelon also reviewed part of this manuscript, and we would like to thank them for that. We also wish to thank Gérard Febvay from the French ‘Biologie adaptative des pucerons et organismes associés’ (BAPOA) network for his weekly bibliographic review, which is so useful, making it easier to keep up to date with progress in this field at all times.
Footnotes
References
- Anstead J., Samuel P., Song N., Wu C., Thompson G. A., Goggin F. (2010). Activation of ethylene-related genes in response to aphid feeding on resistant and susceptible melon and tomato plants. Entomol. Exp. Appl. 134 170–181. 10.1111/j.1570-7458.2009.00945.x [DOI] [Google Scholar]
- Baudracco-Arnas S., Pitrat M. (1996). A genetic map of melon (Cucumis melo L) with RFLP, RAPD, isozyme, disease resistance and morphological markers. Theor. Appl. Genet. 93 57–64. 10.1007/BF00225727 [DOI] [PubMed] [Google Scholar]
- Betancourt M., Fraile A., Milgroom M. G., García-Arenal F. (2016). Aphid vector population density determines the emergence of necrogenic satellite RNAs in populations of Cucumber mosaic virus. J. Gen. Virol. 97 1453–1457. 10.1099/jgv.0.000435 [DOI] [PubMed] [Google Scholar]
- Bohn G. W., Kishaba A. N., Toba H. H. (1972). Mechanisms of resistance to melon aphid in a muskmelon line. HortScience 7 281–282. [Google Scholar]
- Boissot N., Pavis C., Guillaume R., Lafortune D., Sauvion N. (2000). “Insect resistance in Cucumis melo accession 90625,” in Proceedings of the 7th EUCARPIA Meeting on Cucurbit Genetics and Breeding eds Katzir N., Paris H. (Leuven: International Society for Horticultural Science; ) 297–304. [Google Scholar]
- Boissot N., Schoeny A., Millot P., Wipf-Scheibel C., Nozeran K., Lecoq H., et al. (2015). Gérer les épidémies virales en combinant résistances aux virus et résistances aux pucerons : quel intérêt ? Innov. Agron. 46 127–135. [Google Scholar]
- Boissot N., Thomas S., Chovelon V., Lecoq H. (2016). NBS-LRR-mediated resistance triggered by aphids: viruses do not adapt; aphids adapt via different mechanisms. BMC Plant Biol. 16:25 10.1186/s12870-016-0708-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boissot N., Thomas S., Sauvion N., Marchal C., Pavis C., Dogimont C. (2010). Mapping and validation of QTLs for resistance to aphids and whiteflies in melon. Theor. Appl. Genet. 121 117–125. 10.1007/s00122-010-1287-8 [DOI] [PubMed] [Google Scholar]
- Boualem A., Dogimont C., Bendahmane A. (2016). The battle for survival between viruses and their host plants. Curr. Opin. Virol. 17 32–38. 10.1016/j.coviro.2015.12.001 [DOI] [PubMed] [Google Scholar]
- Brévault T., Carletto J., Linderme D., Vanlerberghe-Masutti F. (2008). Genetic diversity of the cotton aphid Aphis gossypii in the unstable environment of a cotton growing area. Agric. For. Entomol. 10 1–9. [Google Scholar]
- Carletto J., Blin A., Vanlerberghe-Masutti F. (2009a). DNA-Based discrimination between the sibling species Aphis gossypii Glover and Aphis frangulae Kaltenbach. System. Entomol. 34 307–314. 10.1111/j.1365-3113.2008.00458.x [DOI] [Google Scholar]
- Carletto J., Gueguen G., Fleury F., Vanlerberghe-Masutti F. (2008). Screening the bacterial endosymbiotic community of sap-feeding insects by terminal-restriction fragment length polymorphism analysis. Entomol. Exp. Appl. 129 228–234. 10.1111/j.1570-7458.2008.00760.x [DOI] [Google Scholar]
- Carletto J., Lombaert E., Chavigny P., Brevault T., Lapchin L., Vanlerberghe-Masutti F. (2009b). Ecological specialization of the aphid Aphis gossypii Glover on cultivated host plants. Mol. Ecol. 18 2198–2212. 10.1111/j.1365-294X.2009.04190.x [DOI] [PubMed] [Google Scholar]
- Casteel C. L., Walling L. L., Paine D. P. (2006). Behavior and biology of the tomato psyllid, Bactericera cockerelli, in response to the Mi-1.2 gene. Entomol. Exp. Appl. 121 67–72. 10.1111/j.1570-8703.2006.00458.x [DOI] [Google Scholar]
- Charaabi K., Carletto J., Chavigny P., Marrakchi M., Makni M., Vanlerberghe-Masutti F. (2008). Genotypic diversity of the cotton-melon aphid Aphis gossypii (Glover) in Tunisia is structured by host plants. Bull. Entomol. Res. 98 333–341. 10.1017/S0007485307005585 [DOI] [PubMed] [Google Scholar]
- Chen J., Adelberg J. W. (2000). Interspecific hybridization in Cucumis - Progress, problems, and perspectives. HortScience 35 11–15. [Google Scholar]
- Chen J. Q., Rahbé Y., Delobel B., Sauvion N., Guillaud J., Febvay G. (1997). Melon resistance to the aphid Aphis gossypii: behavioural analysis and chemical correlations with nitrogenous compounds. Entomol. Exp. Appl. 85 33–44. 10.1046/j.1570-7458.1997.00232.x [DOI] [Google Scholar]
- Chen Y., Vanlerberghe-Masutti F., Wilson L. J., Barchia I., Mcloon M. O., Smith T., et al. (2013). Evidence of superclones in Australian cotton aphid Aphis gossypii Glover (Aphididae:Hemiptera). Pest. Manag. Sci. 69 938–948. 10.1002/ps.3455 [DOI] [PubMed] [Google Scholar]
- Chovelon V., Restier V., Giovinazzo N., Dogimont C., Aarrouf J. (2011). Histological study of organogenesis in Cucumis melo L. after genetic transformation: why is it difficult to obtain transgenic plants? Plant Cell Rep. 30 2001–2011. 10.1007/s00299-011-1108-9 [DOI] [PubMed] [Google Scholar]
- Coeur d’acier A., Jousselin E., Martin J. F., Rasplus J. Y. (2007). Phylogeny of the Genus Aphis Linnaeus, 1758 (Homoptera: Aphididae) inferred from mitochondrial DNA sequences. Mol. Phylogenet. Evol. 42 598–611. 10.1016/j.ympev.2006.10.006 [DOI] [PubMed] [Google Scholar]
- Dixon A. F. G. (1985). Aphid Ecology. London: Chapman, Hall. [Google Scholar]
- Dogimont C., Bendahmane A., Chovelon V., Boissot N. (2010). Aphid resistance in cultivated crops: genetic and molecular bases and interaction with aphid populations. CR Biol. 333 566–576. 10.1016/j.crvi.2010.04.003 [DOI] [PubMed] [Google Scholar]
- Dogimont C., Boissot N. (2016). “Insect resistance in melon and its modification by molecular breeding,” in Functional Genomics and Biotechnology in Solanaceae and Cucurbitaceae Crops eds Ezura H., Arizumi T., Garcia-Mas J., Rose J. (Berlin: Springer; ) 199–219. [Google Scholar]
- Dogimont C., Boissot N., Chovelon V., Tual S., Grand X., Rittener V., et al. (2008a). “Aphid and virus resistance triggered by the CC-NBS-LRR Vat melon gene,” in Proceedings of the International Conference Genetic control of Plant Pathogenic Viruses and Their Vectors: Towards New Resistance Strategies Cadiz. [Google Scholar]
- Dogimont C., Chovelon V., Pauquet J., Boualem A., Bendahmane A. (2014). The Vat locus encodes for a CC-NBS-LRR protein that confers resistance to Aphis gossypii infestation and A. gossypii-mediated virus resistance. Plant J. 80 993–1004. 10.1111/tpj.12690 [DOI] [PubMed] [Google Scholar]
- Dogimont C., Chovelon V., Tual S., Boissot N., Rittener V., Giovinazzo N., et al. (2008b). “Molecular diversity at the Vat/Pm-W resistance locus in melon,” in Proceedings of the IXth EUCARPIA meeting on genetics and breeding of Cucurbitaceae: Cucurbitaceae 2008 ed. Pitrat M. (Avignon: INRA; ) 219–227. [Google Scholar]
- Douglas A. E. (2003). “Buchnera bacteria and other symbionts of aphids,” in Insect Symbiosis eds Bourtzis K., Miller T. A. (Boca Raton, FL: CRC Press; ) 23–38. [Google Scholar]
- Du B., Zhang W. L., Liu B. F., Hu J., Wei Z., Shi Z. Y., et al. (2009). Identification and characterization of Bph14, a gene conferring resistance to brown planthopper in rice. Proc. Natl. Acad. Sci. U.S.A. 106 22163–22168. 10.1073/pnas.0912139106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutartre-Fricaux L., Bouchet J.-P., Beucher C., Boissot N., Dogimont C. (2014). “Identification by a RNASeq approach of virulence gene candidates in the non-model species Aphis gossypii,” in Proceedings of the 2nd international Hemipteran-Plant interactions symposium Riverside. [Google Scholar]
- Ebert T. A., Cartwright B. (1997). Biology and ecology of Aphis gossypii Glover (Homoptera: Aphididae). Southwest. Entomol. 22 116–153. [Google Scholar]
- Fabre F., Rousseau E., Mailleret L., Moury B. (2015). Epidemiological and evolutionary management of plant resistance: optimizing the deployment of cultivar mixtures in time and space in agricultural landscapes. Evol. Appl. 8 919–932. 10.1111/eva.12304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fereres A., Raccah B. (2015). “Plant virus transmission by insects,” in eLS (Chichester: John Wiley & Sons; ) 1–12. 10.1002/9780470015902.a0000760.pub3 [DOI] [Google Scholar]
- Fergany M., Kaur B., Monforte A. J., Pitrat M., Rys C., Lecoq H., et al. (2011). Variation in melon (Cucumis melo) landraces adapted to the humid tropics of southern India. Genet. Resour. Crop Evol. 58 225–243. 10.1007/s10722-010-9564-6 [DOI] [Google Scholar]
- Ferrari R., Nicoli G. (1994). Life cycle and natural enemies of Aphis gossypii Glover: first observations. Inf. Fitopatol. 44 59–62. [Google Scholar]
- Garcia-Mas J., Benjak A., Sanseverino W., Bourgeois M., Mir G., Gonzalez V. M., et al. (2012). The genome of melon (Cucumis melo L.). Proc. Natl. Acad. Sci. U.S.A. 109 11872–11877. 10.1073/pnas.1205415109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzo E., Palacios I., Fereres A. (2001). “Characterization of melon germplasm resistant to Aphis gossypii Glover,” in Aphids in a New Millenium eds Hull M.é, Simon J.-C., Dedryver C.-A., Rispe C. (France: INRA; ), 441–447. [Google Scholar]
- Garzo E., Soria C., Gomez-Guillamon M. L., Fereres A. (2002). Feeding behavior of Aphis gossypii on resistant accessions of different melon genotypes (Cucumis melo). Phytoparasitica 30 129–140. 10.1007/BF02979695 [DOI] [Google Scholar]
- González V. M., Aventín N., Centeno E., Puigdomènech P. (2014). Interspecific and intraspecific gene variability in a 1-Mb region containing the highest density of NBS-LRR genes found in the melon genome. BMC Genomics 15:1131 10.1186/1471-2164-15-1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiu-Aragones C., Monforte A. J., Saladie M., Correa R. X., Jordi Garcia-Mas J., Martın-Hernandez A. M. (2014). The complex resistance to Cucumber mosaic cucumovirus (CMV) in the melon accession PI161375 is governed by one gene and at least two quantitative trait loci. Mol. Breed. 34 351–362. 10.1007/s11032-014-0038-y [DOI] [Google Scholar]
- Guldemond J. A., Tigges W. T., Devrijer P. W. F. (1994). Host Races of Aphis gossypii (Homoptera: Aphididae) on cucumber and chrysanthemum. Environ. Entomol. 23 1235–1240. 10.1093/ee/23.5.1235 [DOI] [Google Scholar]
- Hyojoong K., Seunghwan L., Yikweon J. (2011). Macroevolutionary patterns in the Aphidini aphids (Hemiptera: Aphididae): Diversification, host association, and biogeographic Origins. PLoS ONE 6:e24749 10.1371/journal.pone.0024749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibn Oaf H. S. A. (2012). Construction of Melon Linkage Map and Tagging of Cucumber Vein Yellowing Virus (CVYV) and Cucurbit Aphid Borne Yellows Virus (CABYV) Resistance Genes Using Simple Sequence Repeats (SSRs) Markers. Ph.D. thesis, University of Gezira; Wad Madani. [Google Scholar]
- Ivanoff S. S. (1944). Resistance of cantaloupes to downy mildew and melon aphid. J. Heredity 35 34–39. [Google Scholar]
- Jacquemond M. (2012). Cucumber mosaic virus. Adv. Virus Res. 84 439–504. 10.1016/B978-0-12-394314-9.00013-0 [DOI] [PubMed] [Google Scholar]
- Kennedy G. G., Kishaba A. N. (1977). Response of alate melon aphids to resistant and susceptible muskmelon lines. J. Econ. Entomol. 70 407–410. 10.1093/jee/70.4.407 [DOI] [Google Scholar]
- Kennedy G. G., Mc Lean D. L., Kinsley M. G. (1978). Probing behavior of Aphis gossypii on resistant and susceptible muskmelon. J. Econ. Entomol. 71 13–16. 10.1093/jee/71.1.13 [DOI] [Google Scholar]
- Kim H., Lee W., Lee S. (2010). Morphometric relationship, phylogenetic correlation and character evolution in the species-rich genus Aphis (Hemiptera: Aphididae). PLoS ONE 5:e11608 10.1371/journal.pone.0011608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishaba A. N., Bohn G. W., Toba H. H. (1971). Resistance to Aphis gossypii in muskmelon. J. Econ. Entomol. 64 935–937. 10.1093/jee/64.4.935 [DOI] [Google Scholar]
- Kishaba A. N., Bohn G. W., Toba H. H. (1976). Genetic aspects of antibiosis to Aphis gossypii in Cucumis melo from India. J. Am. Soc. Hortic. Sci. 101 557–561. [Google Scholar]
- Kishaba A. N., Castle S. J., Coudriet L., Mccreight J. D., Bohn G. W. (1992). Virus transmission by Aphis gossypii Glover to aphid-resistant and susceptible muskmelons. J. Am. Soc. Hortic. Sci. 117 248–254. [Google Scholar]
- Klingler J., Powell G., Thompson G. A., Isaacs R. (1998). Phloem specific aphid resistance in Cucumis melo line AR5: effects on feeding behaviour and performance of Aphis gossypii. Entomol. Exp. Appl. 86 79–88. 10.1046/j.1570-7458.1998.00267.x [DOI] [Google Scholar]
- Kring J. B. (1959). The life cycle of the melon aphid, Aphis gossypii glover, an example of facultative migration. Ann. Entomol. Soc. Am. 52 284–286. 10.1093/aesa/52.3.284 [DOI] [Google Scholar]
- Lagos-Kutz D., Favret C., Giordano R., Voegtlin D. J. (2014). Molecular and morphological differentiation between Aphis gossypii Glover (Hemiptera, Aphididae) and related species, with particular reference to the North American Midwest. Zookeys 459 49–72. 10.3897/zookeys.459.7850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecoq H., Cohen S., Pitrat M., Labonne G. (1979). Resistance to cucumber mosaic virus transmission by aphids in Cucumis melo. Phytopathology 69 1223–1225. 10.1094/Phyto-69-1223 [DOI] [Google Scholar]
- Lecoq H., Desbiez C. (2012). “Viruses of cucurbit crops in the mediterranean region: an ever-changing picture,” in Viruses and Virus Diseases of Vegetables in the Mediterranean Basin eds Lebenstein G., Lecoq H. (Charleston, SC: US Vegetable Laboratory; ) 68–114. [DOI] [PubMed] [Google Scholar]
- Lecoq H., Katis N. (2014). Control of cucurbit viruses. Adv. Virus Res. 90 255–296. 10.1016/B978-0-12-801246-8.00005-6 [DOI] [PubMed] [Google Scholar]
- Lecoq H., Labonne G., Pitrat M. (1980). Specificity of resistance to virus transmission by aphids in Cucumis melo. Ann. Phytopathol. 12 139–144. [Google Scholar]
- Lecoq H., Pitrat M. (1983). “Field experiments on the integrated control of aphid-borne viruses in muskmelon,” in Plant Virus Epidemiology eds Plumb R. T., Thresh J. M. (Oxford: Blackwell Scientific Publications; ) 169–176. [Google Scholar]
- Lecoq H., Pitrat M. (1989). “Effects of resistance on the epidemiology of virus diseases of cucurbits,” in Proceedings of the Cucurbitaceae 89: Evaluation and Enhancement of Cucurbit Germplasm Charleston ed. Thomas C. E. (Charleston, SC: USDA; ) 40–48. [Google Scholar]
- Liu X. D., Zhai B. P., Zhang X. X. (2008). Specialized host-plant performance of the cotton aphid is altered by experience. Ecol. Res. 23 919–925. 10.1007/s11284-007-0458-9 [DOI] [Google Scholar]
- Lombaert E., Carletto J., Piotte C., Fauvergue X., Lecoq H., Vanlerberghe-Masutti F., et al. (2009). Response of the melon aphid, Aphis gossypii, to host-plant resistance: evidence for high adaptative potential despite low genetic variability. Entomol. Exp. Appl. 133 46–56. 10.1111/j.1570-7458.2009.00904.x [DOI] [Google Scholar]
- Martin B., Collar J.-L., Tjallingii W. F., Fereres A. (1997). Intracellular ingestion and salivation by aphids may cause the acquisition and inoculation of non-persistently transmitted plant viruses. J. Gen. Virol. 78 2701–2705. 10.1099/0022-1317-78-10-2701 [DOI] [PubMed] [Google Scholar]
- Mistral P., Boissot N. (2016). “The resistance to virus triggered by aphid inoculation in Vat melon is not systemic,” in Proceedings of the 13th International Plant Virus Epidemiology Symposium Avignon. [Google Scholar]
- Nombela G., Williamson M., Muniz M. (2003). The root-knot nematode resistance gene Mi-1.2 of tomato is responsible for resistance against whitefly Bemisia tabaci. Mol. Plant Microbe Interact. 16 645–649. 10.1094/MPMI.2003.16.7.645 [DOI] [PubMed] [Google Scholar]
- Oliver K. M., Degnan P. H., Burke G. R., Moran N. A. (2010). Facultative symbionts in aphids and the horizontal transfer of ecologically important traits. Annu. Rev. Entomol. 55 247–266. 10.1146/annurev-ento-112408-085305 [DOI] [PubMed] [Google Scholar]
- Pauquet J., Burget E., Hagen L., Chovelon V., Menn A., Valot N., et al. (2004). “Map-based cloning of the Vat gene from melon conferring resistance to both aphid colonization and aphid transmission of several viruses,” in Proceedings of the Cucurbitaceae 2004 the 8th EUCARPIA meeting on Cucurbit genetics and breeding eds Lebeda A., Paris H. (Olomouc: Palaky University Olomouc; ) 325–329. [Google Scholar]
- Perchepied L., Bardin M., Dogimont C., Pitrat M. (2005a). Relationship between loci conferring downy mildew and powdery mildew resistance in melon assessed by quantitative trait loci mapping. Phytopathology 95 556–565. 10.1094/PHYTO-95-0556 [DOI] [PubMed] [Google Scholar]
- Perchepied L., Dogimont C., Pitrat M. (2005b). Strain-specific and recessive QTLs involved in the control of partial resistance to Fusarium oxysporum f. sp melonis race 1.2 in a recombinant inbred line population of melon. Theor. Appl. Genet. 111 65–74. 10.1007/s00122-005-1991-y [DOI] [PubMed] [Google Scholar]
- Périn C., Hagen L., De Conto V., Katzir N., Danin-Poleg Y., Portnoy V., et al. (2002). A reference map of Cucumis melo based on two recombinant inbred line populations. Theor. Appl. Genet. 104 1017–1034. 10.1007/s00122-002-0864-x [DOI] [PubMed] [Google Scholar]
- Pitrat M. (1991). Linkage groups in Cucumis melo L. J. Heredity 82 406–411. [Google Scholar]
- Pitrat M. (2003). “Melons, concombres et pastèques,” in Histoire de Légumes: des Origines à l’orée du XXIe siècle eds Pitrat M., Foury C. (Paris: INRA; ) 291–309. [Google Scholar]
- Pitrat M. (2013). Phenotypic diversity in wild and cultivated melons (Cucumis melo). Plant Biotechnol. 30 273–278. 10.5511/plantbiotechnology.13.0813a [DOI] [Google Scholar]
- Pitrat M. (2016). “Disease resistance in melon and its modification by molecular breeding techniques,” in Functional Genomics and Biotechnology in Solanaceae and Cucurbitaceae Crops eds Ezura H., Arizumi T., Garcia-Mas J., Rose J. (Berlin: Springer; ) 175–197. [Google Scholar]
- Pitrat M., Hanelt P., Hammer K. (2000). “Some comments on infraspecific classification of cultivars of melon,” in Proceedings of the 7th EUCARPIA Meeting on Cucurbit Genetics and Breeding eds Katzir N., Paris H. (Belgium: International Society for Horticultural Science; ) 29–36. [Google Scholar]
- Pitrat M., Lecoq H. (1980). Inheritance of resistance to cucumber mosaic virus transmission by Aphis gossypii in Cucumis melo. Phytopathology 70 958–961. 10.1094/Phyto-70-958 [DOI] [Google Scholar]
- Pitrat M., Lecoq H. (1982). Genetic relations between non-acceptance and antibiosis resistance to Aphis gossypii ine melon: search for linkage with other genes. Agronomie 2 503–508. 10.1051/agro:19820601 [DOI] [Google Scholar]
- Pitrat M., Risser G., Blancard D., Lecoq H. (1996). “Evaluation of a melon collection for disease resistances,” in Proceedings of the Cucurbits Towards 2000 VIth Eucarpia Meeting on Cucurbit Genetics and Breeding Maìlaga ed. Eucarpia N. (Algarrobo: Estacioìn Experimental “La Mayora”; ). [Google Scholar]
- Risser G., Pitrat M., Lecoq H., Rode J. C. (1981). Varietal susceptibility of melon to muskmelon yellow stunt virus (MYSV) and to its transmission by Aphis gossypii. Inheritance of the wilting reaction. Agronomie 1 835–838. 10.1051/agro:19811002 [DOI] [Google Scholar]
- Romanow L. R., Moyer J. W., Kennedy G. G. (1986). Alteration of efficiencies of acquisition and inoculation of watermelon mosaic virus 2 by plant resistance to the virus and to an aphid vector. Phytopathology 76 1276–1281. 10.1094/Phyto-76-1276 [DOI] [Google Scholar]
- Rossi M., Goggin F. L., Milligan S. B., Kaloshian I., Ullman D. E., Williamson V. M. (1998). The nematode resistance gene Mi of tomato confers resistance against the potato aphid. Proc. Natl. Acad. Sci. U.S.A. 95 9750–9754. 10.1073/pnas.95.17.9750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouxel T., Balescent M.-H. (2010). “Avirulence genes,” in eLS (Chichester: John Wiley & Sons; ). 10.1002/9780470015902.a0021267 [DOI] [Google Scholar]
- Sarria Villada E., Gonzales E. G., Lopez-Sese A. I., Fereres Castiel A., Gomez-Guillamon M. L. (2009). Hypersensitive response to Aphis gossypii Glover in melon genotypes carrying the Vat gene. J. Exp. Bot. 60 3269–3277. 10.1093/jxb/erp163 [DOI] [PubMed] [Google Scholar]
- Satar S., Kersting U., Yokomi R. (2013). Presence of two host races of Aphis gossypii Glover (Hemiptera: Aphididae) collected in Turkey. Ann. Appl. Biol. 162 41–49. 10.1111/j.1744-7348.2012.00578.x [DOI] [Google Scholar]
- Sattar S., Addo-Quaye C., Song Y., Anstead J. A., Sunkar R., Thompson G. A. (2012a). Expression of small RNA in Aphis gossypii and its potential role in the resistance interaction with melon. PLoS ONE 7:e48579 10.1371/journal.pone.0048579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattar S., Addo-Quaye C., Thompson G. A. (2016). miRNA-mediated auxin signalling repression during Vat-mediated aphid resistance in Cucumis melo. Plant Cell Environ. 39 1216–1227. 10.1111/pce.12645 [DOI] [PubMed] [Google Scholar]
- Sattar S., Song Y., Anstead J. A., Sunkar R., Thompson G. A. (2012b). Cucumis melo MicroRNA expression profile during aphid herbivory in a resistant and susceptible interaction. Mol. Plant Microbe Interact. 25 839–848. 10.1094/MPMI-09-11-0252 [DOI] [PubMed] [Google Scholar]
- Schoeny A., Boissot N., Lambion J., Wipf-Scheibel C., Mistral P., Gognalons P., et al. (2014). Conception d’associations en maraîchage de plein champ : exemple de production de melons associés à des bandes fleuries pour lutter contre les pucerons et les virus. Innov. Agron. 40 113–114. [Google Scholar]
- Sebastian P., Schaefer H., Telford I. R. H., Renner S. S. (2010). Cucumber (Cucumis sativus) and melon (C. melo) have numerous wild relatives in Asia and Australia, and the sister species of melon is from Australia. Proc. Natl. Acad. Sci. U.S.A. 107 14269–14273. 10.1073/pnas.1005338107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serres-Giardi L., Dogimont C. (2012). “How microsatellite diversity helps to understand the domestication history of melon,” in Proceedings of the Xth EUCARPIA Meeting on Genetics and Breeding of Cucurbitaceae: Cucurbitaceae 2012 eds Sari N., Solmaz I., Aras V. (Adana: Çukurova University; ) 254–263. [Google Scholar]
- Smith C. M., Chuang W.-P. (2014). Plant resistance to aphid feeding: behavioral, physiological, genetic and molecular cues regulate aphid host selection and feeding. Pest. Manag. Sci. 70 528–540. 10.1002/ps.3689 [DOI] [PubMed] [Google Scholar]
- Soria C., Diaz J. A., Moriones E., Gomez-Guillamon M. L. (2000). “Resistance to Aphis gossypii and to virus transmission by this aphid in melon,” in Proceedings of the 7th EUCARPIA Meeting on Cucurbit Genetics and Breeding eds Katzir N., Paris H. (Leuven: International Society for Horticultural Science; ) 305–312. [Google Scholar]
- Takada H. (1988). Interclonal variation in the photoperiodic response for sexual morph production of Japanese Aphis gossypii Glover (Hom, Aphididae). J. Appl. Entomol. 106 188–197. 10.1111/j.1439-0418.1988.tb00582.x [DOI] [Google Scholar]
- Tamura Y., Hattori M., Yoshioka H., Yoshioka M., Takahashi A., Wu J., et al. (2014). Map-based cloning and characterization of a brown planthopper resistance gene BPH26 from Oryza sativa L. ssp. indica cultivar ADR52. Sci. Rep. 4:5872 10.1038/srep05872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K., Akashi Y., Fukunaga K., Yamamoto T., Aierken Y., Nishida H., et al. (2013). Diversification and genetic differentiation of cultivated melon inferred from sequence polymorphism in the chloroplast genome. Breed. Sci. 63 183–196. 10.1270/jsbbs.63.183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas S., Boissot N., Vanlerberghe-Masutti F. (2012a). What do spring migrants reveal about sex and host selection in the melon aphid? BMC Evol. Biol. 12:47 10.1186/1471-2148-12-47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas S., Dogimont C., Boissot N. (2012b). Association between Aphis gossypii genotype and phenotype on melon accessions. Arthropod Plant Interact. 6 93–101. 10.1007/s11829-011-9155-2 [DOI] [Google Scholar]
- Thomas S., Vanlerberghe-Masutti F., Mistral P., Loiseau A., Boissot N. (2016). Insight into the durability of aphid resistance from the demo-genetic study of Aphis gossypii populations in melon crops. Evol. Appl. 9 756–768. 10.1111/eva.12382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjallingii W. F. (2006). Salivary secretions by aphids interacting with proteins of phloem wound responses. J. Exp. Bot. 57 739–745. 10.1093/jxb/erj088 [DOI] [PubMed] [Google Scholar]
- Vanlerberghe-Masutti F., Chavigny P., Fuller S. J. (1999). Characterization of microsatellite loci in the aphid species Aphis gossypii Glover. Mol. Ecol. 8 685–702. 10.1046/j.1365-294x.1999.00876.x [DOI] [PubMed] [Google Scholar]
- Vodovar N., Saleh M.-C. (2012). “Of insects and viruses: the role of small RNAs in insect defense,” in Advanced in Insect Physiology ed. Jockush E. L. (Cambridge, MA: Academic press; ) 1–36. [Google Scholar]
- Wang L., Zhang S., Luo J. Y., Wang C. Y., Lv L. M., Zhu X. Z., et al. (2016). Identification of Aphis gossypii Glover (Hemiptera: Aphididae) biotypes from different host plants in North China. PLoS ONE 11:e0146345 10.1371/journal.pone.0146345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W., Liang X. L., Zhao H. Y., Xu T. T., Liu X. D. (2013). Special plant species determines diet breadth of phytophagous insects: a study on host plant expansion of the host-specialized Aphis gossypii Glover. PLoS ONE 8:e60832 10.1371/journal.pone.0060832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T.-T., Ma T.-T., Liu X.-D. (2014). How does the host-specialized aphid deal with food deficiency? Insect Sci. 21 334–341. 10.1111/1744-7917.12114 [DOI] [PubMed] [Google Scholar]
- Yuste-Lisbona F. J., Capel C., Sarria E., Torreblanca R., Gómez-Guillamón M. L., Capel J., et al. (2001). Genetic linkage map of melon (Cucumis melo L.) and localization of a major QTL for powdery mildew resistance. Mol. Breed. 27 181–192. 10.1007/s11032-010-9421-5 [DOI] [Google Scholar]
- Zhang B. C., Tolstikov V., Turnbull C., Hicks L. M., Fiehn O. (2010). Divergent metabolome and proteome suggest functional independence of dual phloem transport systems in cucurbits. Proc. Natl. Acad. Sci. U.S.A. 107 13532–13537. 10.1073/pnas.0910558107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. C., Cao W. J., Zhong L. R., Godfray H. C., Liu X. D. (2016). Host plant determines the population size of an obligate symbiont (Buchnera aphidicola) in aphids. Appl. Environ. Microbiol. 82 2336–2346. 10.1128/AEM.04131-15 [DOI] [PMC free article] [PubMed] [Google Scholar]


