Abstract
The ability to rationally target disease-causing mutations has been made possible with programmable nucleases with the clustered, regularly interspaced short palindromic repeats/Cas9 system representing a facile platform for individualized gene-based medicine. In this study we employed footprint-free reprogramming of fibroblasts from a patient with mutations to the Fanconi anemia I (FANCI) gene to generate induced pluripotent stem cells (iPSCs). This process was accomplished without gene complementation and the resultant iPSCs were able to be gene corrected in a robust manner using the Cas9 nickase. The self-renewing iPSCs that were maintained under feeder-free conditions were differentiated into cells with characteristics of definitive hematopoiesis. This defined and highly efficient procedure employed small molecule modulation of the hematopoietic differentiation pathway and a vascular induction technique to generate hematopoietic progenitors. In sum, our results demonstrate the ability to induce patient-derived FA cells to pluripotency for patient-specific therapeutic cell derivation.
Introduction
Genome engineering for precision sequence alteration holds tremendous potential for personalized autologous therapies. Such an approach employs programmable nucleases for DNA break induction that can be repaired by homology-directed repair (HDR). In this context, an exogenous DNA molecule can serve as the repair template allowing for seamless, site-specific, permanent sequence modification. Further, the in situ modification maintains gene expression under the control of the endogenous regulatory elements. These considerations are differential from gene therapy that uses full-length, functional cDNA copies of a gene and exogenous regulatory sequences that typically drive sustained, high-level gene expression in a manner not subject to cellular regulation. Similar to gene editing, gene therapy can result in permanent and stable expression when integrating vectors are employed. However, the integration profile of viral vectors shows a bias for areas of transcriptional activity [1] that can make insertional mutagenesis a significant risk [2,3]. This risk can be further magnified in disorders such as Fanconi anemia (FA) that is characterized by deficiencies in the DNA repair pathway that may promote a premalignant phenotype less tolerant to genomic perturbation.
FA is a complex disease caused by mutations to one of 15–16 FA pathway genes: FANCM, whose implication in FA is not definitive, and bona fide FA genes: FANCA, FANCB, FANCC, FANCD1/BRCA2, FANCD2, FANCE, FANCG, FANCI, FANCJ/BRIP1, FANCL, FANCN/PALB2, FANCO/RAD51C, FANCP/SLX4, and FANCQ/ERCC4/XPF [4–7]. Sequence mutations to these genes result in an inability to repair interstrand DNA cross-links causing developmental anomalies, increased rates of tumorigenesis, and bone marrow failure [8]. Treatment by hematopoietic cell transplant (HCT) can be curative for the lethal bone marrow manifestations of the disease; however, the therapeutic benefits do not extend to the physical abnormalities. Furthermore, allogeneic HCT in FA patients is associated with significant risks due to the pre and post-transplant treatment procedures [9] including the possibility for a magnified incidence of tumor occurrence [10]. Toward advancing autologous therapies, and with recognition of potential vector-associated risks, early stage (Phase I/II) gene therapy trials have been implemented. A main advantage of this is the mitigation of transplant-associated complications associated with HCT from an unrelated donor. However, the low numbers of hematopoietic progenitor cells available for modification remains a significant hurdle. One highly desirable solution to this would be to engineer autologous induced pluripotent stem cells (iPSCs) capable of forming engraftable hematopoietic progenitors. To date, however, the poor reprogramming efficiency of FA cells has restricted the realization of this potential [11]. Further, despite intense efforts, a true human iPSC-derived hematopoietic progenitor capable of in vivo engraftment has not yet been demonstrated. Therefore, we established a line of investigation utilizing cells from an FA patient (complementation group FANCI, chosen because of the central position of FANCI in the FA/BRCA pathway) to determine whether genome modification, improved reprogramming methodologies, and advances in cellular engineering could be synergized to address gaps in the fields of FA biology and transplant medicine.
Toward assessing the engineering capacity of FANCI cells, we employed the highly efficient nonintegrating Sendai virus reprogramming methodology [12] to generate iPSCs from primary fibroblasts. These iPSCs were highly permissive to gene editing using the clustered, regularly interspaced short palindromic repeats (CRISPR)/Cas9 platform. This two-component system is comprised of a short guide RNA (gRNA) transcript and the Cas9 protein [13]. Cas9 can function as a double-stranded DNA nuclease or a single-stranded DNA nickase. Of note, we have demonstrated previously that nicking preferentially promotes HDR in FANCC cells [14]. Here too we observed the ability of the Cas9 nickase to mediate FANCI gene correction in patient-derived iPSCs, while the parental fibroblast cells were recalcitrant to gene editing. Using gene-corrected iPSCs we assessed their hematopoietic differentiation capability by performing directed differentiation in vitro. By combining modulation of the fate determinants of primitive and definitive hematopoiesis with a supportive endothelial coculture system, we were able to generate a population of CD34+CD38− cells. This phenotype is consistent with cord blood-derived cells [15] capable of engrafting and collectively represents an advance in cellular engineering and translational application for FA therapy.
Materials and Methods
Research subject line generation and culture
Primary fibroblasts were derived from a 4 mm skin punch biopsy collected from a pediatric patient given the designation FA-28. The cell line was cultured under hypoxic conditions and maintained in complete Dulbecco's modified Eagle's medium with 20% fetal bovine serum (FBS), 0.1 mg/mL each of penicillin and streptomycin, 10 ng/mL each of epidermal growth factor and fibroblast growth factor, 100 U/mL nonessential amino acids, and 1× antioxidant supplement (Sigma-Aldrich, St. Louis, MO). This study was performed in accordance with the principles for research on human subjects set forth by the Declaration of Helsinki and was preceded by parental informed consent and University of Minnesota Institutional Review Board approval.
CRISPR/Cas9 reagent construction
The Cas9 nuclease and nickase plasmids were a gift from Dr George Church (Addgene plasmids #41815 and #41816 [16], Cambridge, MA) and the FANCI-specific gRNAs were synthesized using a 60mer oligonucleotides in the following manner: First, an “empty” SpCas9 gRNA plasmid was constructed by cloning the U6 promoter, SpCas9 sgRNA scaffold sequence, and an intervening EcoRI restriction site into the pCR4 TOPO vector (Invitrogen, Carlsbad, CA). DNA sequences of the 60mer oligos are listed below and contain the 20 base pairs (bp) upstream of EcoRI, the 19 bp FANCI target sequence, and the 20 bp downstream of EcoRI. An additional 20 bp of homology were included on either end by polymerase chain reaction (PCR) amplification of the template with universal primers Fwd (5′-TTTCTTGGCTTTATATATCTTGTGGAAAGGACGAAACACC-3′) and Rev (5′-ATTTTAACTTGCTATGCTGTTTCCAGCATAGCTCTAAAAC-3′). The 99 bp PCR products were then cloned into the linearized pCR4 U6 gRNA vector by Gibson Assembly [17].
Template oligonucleotide sequences (5′-3′)
s1: GTGGAAAGGACGAAACACCG actgtacaaaggctgctta GTTTTAGAGCTATGCTGGAA
as1: GTGGAAAGGACGAAACACCG cagcctttgtacagtctga GTTTTAGAGCTATGCTGGAA
as2: GTGGAAAGGACGAAACACCG aacaactttgaagaactaa GTTTTAGAGCTATGCTGGAA
1461 donor construction
The left and right donor arms were amplified from the human genome with primers Left Fwd (5′-TTTCTTCTCCCTGCAGCAAT-3′) and Left Rev (5′-TACTGATTCTATATATACTTTTTCTCCAAT-3′) generating a 1,126 bp fragment and Right Fwd (5′-TGCCAAAACTGAGGTTTAACTG-3′) and Right Rev (5′-AATAAAACTCATTCCCCTCAACAA-3′) generating a 1,052 bp fragment. Donor arms were assembled into pcDNA3.1/V5-His plasmid (Invitrogen) containing a floxed PGK-puromycin selection cassette by enzyme digest and T4 DNA ligation (New England Biolabs, Ipswich, MA). A gBlock gene fragment (Integrated DNA Technologies, Coralville, IA) with overlapping homology was added immediately upstream of the right arm by Gibson Assembly for introduction of the corrective base and four silent mutations, including a knockout HindIII site. To facilitate construction of the puromycin and puromycin-free AAV donors, a 196 bp Ultramer (Integrated DNA Technologies) containing left arm 5′ homology and right arm 3′ homology (95 bp each) separated by a unique restriction site (6 bp) was added to pAAV-mCherry-MCS by PCR amplification and T4 DNA ligation. The full left arm→puromycin→right arm sequence was excised as a single fragment from the pcDNA backbone by restriction digest and shuttled into the pAAV vector by way of Gibson Assembly. Similarly, the puromycin-free AAV donor was constructed by Gibson Assembly of PCR-amplified left and right arms and an intervening gBlock with overlapping homology for introduction of the corrective base and 30 silent mutations, including a knockout HindIII site.
iPSC generation
Uncorrected fibroblasts were reprogrammed into iPSCs as described [18,19], using Sendai virus for delivery of reprogramming factors [12]. As part of Quality Assurance/Quality Control evaluations, karyotype, gene expression, and immunofluorescence were performed as previously described [18–21]. iPSCs were cultured in feeder-free mTeSR1 (STEMCELL Technologies, Vancouver, BC) on Geltrex-coated (Thermo Fisher Scientific, Waltham, MA) tissue culture plates at 37°C under hypoxic conditions. Sendai virus clearance [22] was assessed on reverse transcribed RNA using the following forward (F) and reverse (R) primers: b-actinF: GGCACCCAGCACAATGAAGATCAA b-ActinR: AGGATGGCAAGGGACTTCCTGTAA, (Sendai virus genome: SeV) SeVF: GGATCACTAGGTGATATCGAGC, SeVR: ACCAGACAAGAGTTTAAGAGATATGTA, KlfF: TTCCTGCATGCCAGAGGAGCCC, KlfR: AATGTATCGAAGGTGTCTCAA. mycF: TAACTGACTAGCAGGCTTGTCG and mycR: TCCACATACAGTCCTGGATGATGATG, (Sendai Klf, Oct3/4, Sox2; KOS) KOSF: ATGCACCGCTACGACGTGAGCGC, KOSR: ACCTTGACAATCCTGATGTGG. PCR conditions were as follows: 95°C × 2 min and 35 cycles of 95°C × 1 min, 58°C × 40 s, and 68°C for 1 min using AccuPrime DNA polymerase (ThermoFisher Scientific, Waltham, MA).
Gene transfer
For 293T transfections, 500 ng each of Cas9 nuclease and gRNA were delivered with Lipofectamine 2000 (Invitrogen). For all other cell types 10 μg of the 1461 pcDNA puromycin plasmid donor was delivered with 1 μg each of Cas9 nickase and gRNA for Neon electroporation (1,500 V, 20 ms pulse width, and a single pulse, Invitrogen, Valencia, CA). Gene correction of iPSCs was achieved by Neon transfection using 1,100 V, 20 ms pulse width, and a single pulse. Cas9 nickase and gRNA were delivered at 500 ng each with either 5 μg of 1461 AAV puromycin plasmid donor or 5 μg of 1461 AAV puromycin-free plasmid donor. For 12–24 h before gene transfer, iPSCs were treated with 10 μM Rho-associated protein kinase (ROCK) inhibitor, Y-27632 (Selleck Chemicals, Houston, TX). All cells were incubated at 30° C for 48 h following gene transfer, excluding iPSCs that were consistently maintained at 37°C under hypoxic conditions.
Surveyor nuclease
293T genomic DNA was isolated 72 h following gRNA and Cas9 nuclease or nickase transfection and was PCR amplified with 1461 87F: GCAGTGTAATCGGTTTCTTCC-3′) and 1461 764R: AACTGGTTGCAAATGCCTCT-3′) or 3058F: TCCCACACCTTTCTGATAACC and 3058R: TTTCTACCTCCACCCCCAGT for on target assessment. Off-target sites were chosen using the MIT CRISPR Design Tool (http://crispr.mit.edu) and predicted off-target sites in genes were assessed in the 293 s from above with the following primers: UBE2 V1F: CGTTAGTGGGCCAGTAGCAT, and UBE2 V1R: ATTGCTGCGTCTTTCCACTT, SDCBP2F: GTCAGGGGACCAAGGATTC, SDCBP2R: TATCCAGGTTGCCTCCAGTC, APPBP2F: TCCAGGCTGGTCTGAAACTC, APPBP2R: TGCAAAATAGAATGACCACCA, IPPF: TTCCACATTAGTGCATGAGTGA, IPPR: GCAGGTCCCATTTCTTGTGT, ELMOD3F: TTCTTTTGGGTTTCCATTGC, ELMOD3R: GGCCCTTAGCTGATCCTTTC, ADCY7F: AGGTGGGATGGTGAAGGAAT, ADCY7R: AGGAACATCCCTGTCCTCCT, PSMD14F: CCACCAGCAGTGTGTGAGAG, PSMD14R: AACAGCCACACAGTCATTGC, ARID4AF: CATTTTATCATATCAGGACATCAGTC, ARID4AR: CGCCCAGTCACAAAATAAAC, GALNT8F: CGGGGCATAAAGACAAAGAA, GALNT8R: CTTCTCGTTGCCTGCACATA, CCDC88AF:GCTCTGACATGCCAAAACAA,CCDC88AR:TGAAGCCTTTTGTGGTAGGAA.
PCR conditions were as follows: 95°C × 3 min followed by 35 cycles of 95°C × 1 min, 58°C × 40 s, and 68°C × 1 min with AccuPrime DNA polymerase (ThermoFisher Scientific, Waltham, MA). PCR products were incubated with Surveyor nuclease (Integrated DNA Technologies) and resolved on a 10% TBE PAGE gel (Invitrogen) [14,23]. Rates of activity were quantified by densitometry using the ImageJ application [23,24]. All gel images used the same exposure times.
Selection
Cells were selected in bulk in 0.3 μg/mL puromycin (EMD Millipore, Billerica, MA) 4 and 6 days after gene transfer, respectively. Resistant fibroblasts were plated at low density for monoclonal expansion as previously described [14,20]. Bulk iPSCs receiving the 1461 AAV puromycin plasmid donor were selected 7 days after gene transfer in increasing amounts of puromycin escalated at each passage starting at 0.3 μg/mL. Bulk iPSCs transfected with the 1461 AAV puromycin-free plasmid donor were subjected to three progressive rounds of mitomycin C selection lasting 48 h each and beginning 16 days after gene transfer. The concentrations used were 10, 14, and 16 nM mitomycin C (Sigma-Aldrich).
Cell correction molecular screening
To screen for correction by puromycin donor, the donor-genome junction was amplified using internal primer F39 (5′-AGCCTCGACTGTGCCTTCTA-3′) and external primer R2339 (5′-TCACTGAGGCGCAATGATTA-3′) downstream of the right donor arm within the endogenous locus. For correction by puromycin-free donor, R2339 was used along with internal 7F (5′-GAGGCCTTCGACTACCTGAG-3′) to prime off the corrective base and 6 and of the 30 silent mutations. PCR products were TOPO cloned into the pCR4 vector for confirmation of HDR by Sanger sequencing. cDNA correction was detected by amplification with Exon 15 Fwd (5′-ACCTGCTTTCAAATATCGTCATGT-3′) and Exon 17 Rev (5′-CTGACTGACACTGAGAGACTGA-3′), followed by HindIII restriction digest. HindIII-resistant amplicons were cloned and sequenced as described above.
Differentiation of iPSCs to CD34
Human iPSC lines were maintained on Matrigel or Geltrex coated plasticware with feeder free, serum free culture conditions in mTeSR™1 media (STEMCELL Technologies, Vancouver, BC). iPSC and derivative cells were cultured under hypoxic conditions (2% O2). Unless otherwise noted, all cytokines were from R&D Systems (Minneapolis, MN). For embryoid body (EB) generation, hiPSCs were cultured at 80–90 % confluency, followed by EB formation as follows: undifferentiated hiPSCs were dissociated with Accutase (STEMCELL Technologies, Vancouver, BC) and aggregates were resuspended in APEL medium (STEMCELL Technologies, Vancouver, BC), supplemented with bone morphogenic protein-4 (BMP-4; 25 ng/mL) and basic fibroblast growth factor (bFGF; 5 ng/mL) and replated on non-tissue culture treated 10 cm2 dishes. At 42–48 hours, developing EBs were collected and resuspended in APEL-differentiation media containing BMP-4 and bFGF, at the same concentration as above, along with 3 μM CHIR99021 (Stemgent, Lexington, MA) and 6 μM SB431542 (Selleck Chemicals, Houston, TX). Twenty-four hours later the EBs were collected and resuspended in 40% APEL + 60% STEMSPAN II (STEMCELL Technologies, Vancouver, BC) media containing vascular endothelial growth factor (20 ng/mL), bFGF (5 ng/mL), IL-3 (20 ng/mL), Flt3L (20 ng/mL) and stem cell factor (SCF; 100 ng/mL) and cultured for a further 5–6 days. At day 8/9 EBs were harvested, washed once with PBS and dissociated using Accutase (Innovative Cell Technologies, San Diego, CA) and 0.25% Trypsin-ethylenediaminetetraacetic acid (EDTA; ThermoFisher Scientific, Waltham, MA) until no visible clumps remained. CD34+ cells were isolated using the EasySepTM CD34+ isolation kit (STEMCELL Technologies, Vancouver, BC).
Differentiation of CD34 to T cell progenitors
Dissociated CD34 cells from EBs were plated on OP-9 DL9 stromal cells that were at ∼70% confluency in alpha-MEM media supplemented with SCF (100 ng/mL), IL7 (5 ng/mL), and Flt3 (10 ng/mL) for 7 days, at which point the cells were passed to new OP9 without SCF. The cells were maintained until flow cytometric analysis.
Differentiation of CD34 to hematopoietic progenitors
Single cell CD34 cells at a density of 1 × 106 were plated on VeraVec™ cells (Angiocrine Bioscience, New York, NY) that were at 80% confluency in StemSpan II media supplemented with: IGF-1 (25 ng/mL), BMP-4 (10 ng/mL), SCF (200 ng/mL), Flt3 L (10 ng/mL), IL-6 (10 ng/mL), thrombopoietin (30 ng/mL), IL-11 (5 ng/mL), bFGF (5 ng/mL), VEGF (5 ng/mL), IL-3 (30 ng/mL), and erythropoietin (2 IU/mL). Cocultures were maintained for 9 days, at which point they were analyzed by flow cytometry.
Colony forming unit assay
Cells were placed in MethoCult according to the manufacturer's instructions (STEMCELL Technologies). Colonies were enumerated by an experienced analyst using light microscopy at low magnification (4 × objective).
Flow cytometry
All antibodies were acquired from eBiosciences (San Diego, CA), and FACS was performed on a FACSAria instrument (Beckton-Dickinson, Franklin Lakes, NJ).
Cellular isolation
Antigen-specific cell isolation was performed using the EasySep magnetic separation system (STEMCELL Technologies).
Results
FANCI gene, CRISPR/Cas9 targeting, and experimental approach
The FANCI gene is located at 15q26.1 (Fig. 1A) that encompasses ∼75 kb of genomic sequence and encodes a 150 kd protein that associates with FANCD2 to form a complex that localizes to sites of DNA damage [7,25]. Two FANCI compound heterozygous mutations were present in cells acquired from a male patient: the c.1461 T > A mutation in exon 15 causing a premature stop codon and the c.3058 + 4A>G intron mutation that, similar to other FA genes [26], likely causes a splicing abnormality that prevents functional protein production.
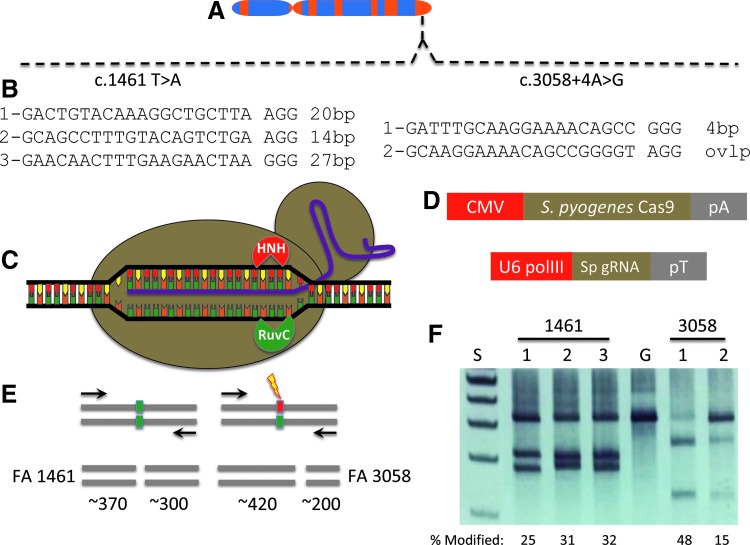
FIG. 1.
FANCI gene and CRISPR/Cas9 targeting. (A) FANCI locus. The FANCI gene at 15q26.1 with the compound heterozygous c.1461 A>T and c.3058 + 4A>G gene mutations shown. (B) CRISPR/Cas9 gene targeting. Guide RNA candidates for the 1461 mutation are shown at left and the 3058 targets are shown at right. A space demarcates the target sequence from the PAM and the distance from the guide RNA to the specific mutation is indicated in base pairs (bp). (C) Streptococcus pyogenes (Sp) CRISPR/Cas9 architecture. A guide RNA transcript is shown in purple that contacts the target sequence in the context of an -NGG PAM. The Cas9 nuclease (tan circles) unwinds the DNA and the HNH and RuvC nuclease domains each cleave one of the DNA strands (D) CRISPR/Cas9 gene delivery platforms. The human codon optimized Cas9 nuclease was expressed from a plasmid DNA under the control of the cytomegalovirus (CMV) promoter and a bovine growth hormone polyadenylation signal (pA). The guide RNA (Sp gRNA) was expressed in trans from the U6 polymerase III promoter and a poly-thymidine termination signal (pT). (E) Surveyor nuclease. CRISPR/Cas9 FANCI gene candidates were introduced into 293T cells and the target locus was amplified (primers are shown as arrows). Amplicon length for the 1461 product was 678 bp and for the 3,058 site it was 619 bp. Unmodified (ie, non-nuclease targeted) alleles are indicated with a green box. Nuclease modification (lightning bolt) result in nonhomologous endjoining repair with small sequence alterations (red box). Denaturation and renaturation results in heteroduplex formation between modified and unmodified sequences that are cleaved into predictable sized fragments by the Surveyor enzyme. (F) FANCI candidate reagent activity. A representative polyacrylamide gel of three experiments is shown for the three 1461 and two 3058 reagents with cleavage products observed. At left are the molecular weight standards (S) and the lane labeled “G” is the GFP-treated control with an absence of DNA fragmentation. Below the gel are the cleavage rates from a representative experiment calculated from a constant exposure utilized across experiments. CRISPR, clustered, regularly interspaced short palindromic repeats; PAM, protospacer adjacent motif.
The gene sequence proximal to each mutation was analyzed for permissive target sites for the CRISPR/Cas9 system derived from Streptococcus pyogenes [27]. S. pyogenes Cas9 recognizes sequences in the context of a three bp–NGG sequence termed a protospacer adjacent motif (PAM) [13] and targeting sites proximal to the respective mutations were identified (Fig. 1B). Five gRNA candidates were constructed for testing with the Cas9 nuclease that possesses helicase activity and has two functional domains (HNH and RuvC), each responsible for cleaving one strand of DNA (Fig. 1C). The Cas9 plasmid and a second plasmid encoding the specific gRNA (Fig. 1D) were introduced into 293T cells and analyzed for functionality using the Surveyor nuclease method [23]. This assay detects target-specific activity by virtue of Cas9 nuclease-mediated insertions and deletions resulting in amplicon cleavage by the Surveyor enzyme (Fig. 1E) [23] with all five candidates showing robust FANCI gene targeting activity (Fig. 1F). Based on these data, we implemented an experimental procedure summarized in Fig. 2 to correct the FANCI gene mutation in primary cells. A primary fibroblast cell line was derived from a skin punch biopsy (Fig. 2A) and this population of cells served as an experimental template for CRISPR/Cas9 gene correction (Fig. 2B) using a plasmid-based donor (Fig. 2C).
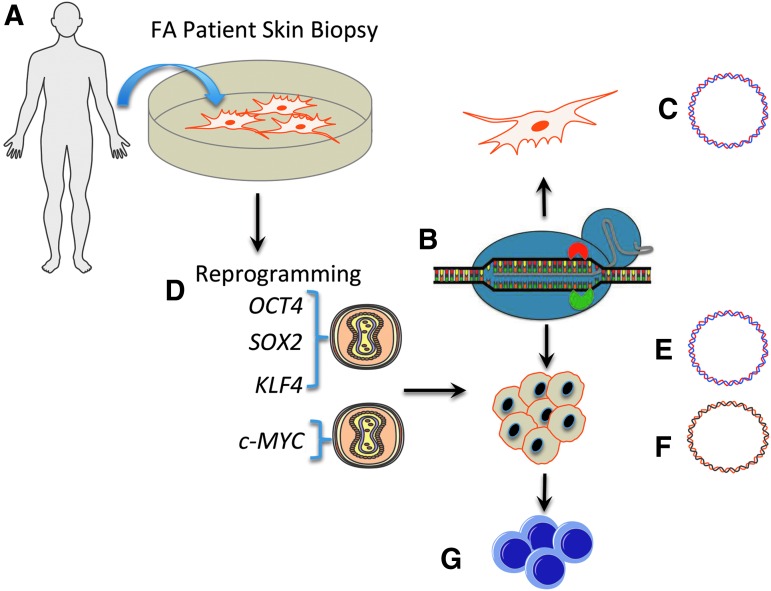
FIG. 2.
Experimental schema. (A) Cell line derivation. Fibroblasts from a patient with a compound heterozygous FANCI gene mutation (c.1461 A>T, c.3058 + 4A>G) were obtained via a minimally invasive skin punch biopsy. (B) CRISPR/Cas9 gene targeting. Fibroblasts were employed for genome modification by introduction of a Cas9 nickase plasmid (inactivated HNH domain is shown by red half circle and the active RuvC domain is shown in green). Co-delivered were a gRNA plasmid and a homologous recombination repair template containing the corrected 1461 base and a puromycin selection cassette borne on a plasmid (C). (D) Cellular reprogramming. Primary FANCI fibroblasts were transduced with Sendai viral vectors containing OCT4, SOX2, and KLF4 in combination and c-myc alone. (E, F) iPSC gene correction. A CRISPR/Cas9 nickase, guide RNA and homologous recombination donor plasmid with (E) or without (F) an exogenous puromycin gene were introduced into iPSC. Selection was facilitated by addition of (E) puromycin or (F) mitomycin C. (G) Hematopoietic lineage derivation. Gene corrected cells were employed for in vitro assessment of hematopoietic progenitor development. iPSC, induced pluripotent stem cell.
The unmanipulated primary fibroblasts were also reprogrammed to pluripotency using Sendai virus gene transfer of a four factor cocktail of OCT4, SOX2, KLF4, and c-MYC (OSKM) that were delivered in trans (Fig. 2D). Plasmid donor templates allowing for puromycin selection (Fig. 2E) or an exogenous marker sequence free template reliant on phenotypic correction-mediated resistance to mitomycin C (MMC; Fig. 2F) were assessed for their ability to modify cells. FANCI genotypically corrected were then utilized for the design and optimization of hematopoietic cell phenotype engineering (Fig. 2G).
Cellular reprogramming and FANCI gene correction
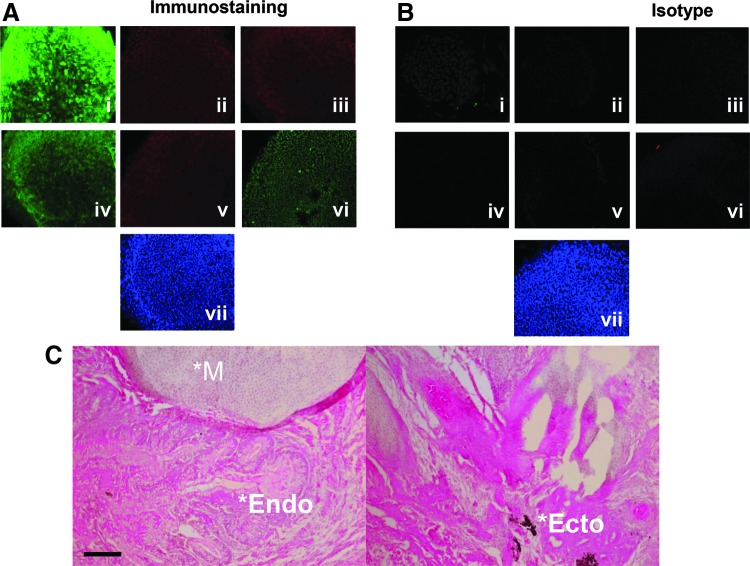
Previous studies have shown that FA cells are highly refractory to reprogramming to pluripotency using retroviral vectors expressing the OSKM factors [11,28]. One study observed better efficiency using lentiviral vectors and maximal rates when the FA phenotype was corrected [11]. In subsequent studies, Liu et al. successfully reprogrammed FANCA fibroblasts that were not complemented using nonintegrating reprogramming five factor delivery and p53 inhibition [29]. In this study we employed the Sendai virus platform to determine whether reprogramming FANCI fibroblasts to iPSCs could be accomplished. We observed expression of the hallmark pluripotency proteins: TRA-1-60, NANOG, SSEA-3, TRA-1-81, SSEA-4, and Oct3/4 by immunofluorescence microscopy (Fig. 3A and B). Implantation of iPSCs into immune-deficient animals resulted in teratoma development with tissues derived from all three germ layers (Fig. 3C). Importantly, both pre- and postreprogramming cell populations maintained normal karyotypes, and derivative iPSCs showed hallmark iPSC gene and epigenetic profiles and loss of Sendai virus genome particles (Supplementary Figs. S1–S3; Supplementary Data are available online at www.liebertpub.com/scd). These data show the pluripotent potential of these iPSCs that were successfully reprogrammed without integration of genotoxic viral vectors.
FIG. 3.
iPSC characterization. (A) FANCI iPSC immunofluorescence. iPSC were stained with antibodies to (i) TRA-1-60 (ii) Nanog (iii) SSEA-3, (iv) TRA-1-81 (v) SSEA-4, (vi) Oct3/4, and (vii) DAPI nuclear stain. (B) Isotype control for the same conditions as in (A). (C) In vivo teratoma assay. iPSC were injected into the flank of immune-deficient mice (n = 5) and the tumor mass was excised and analyzed by histopathology. A representative image showing mesoderm (*M), endoderm (*Endo), and ecotoderm (*Ecto) is shown. Images are with a 10× objective and black scale bar at lower left of (C) is 50 μM. At least three clones were derived and analyzed.
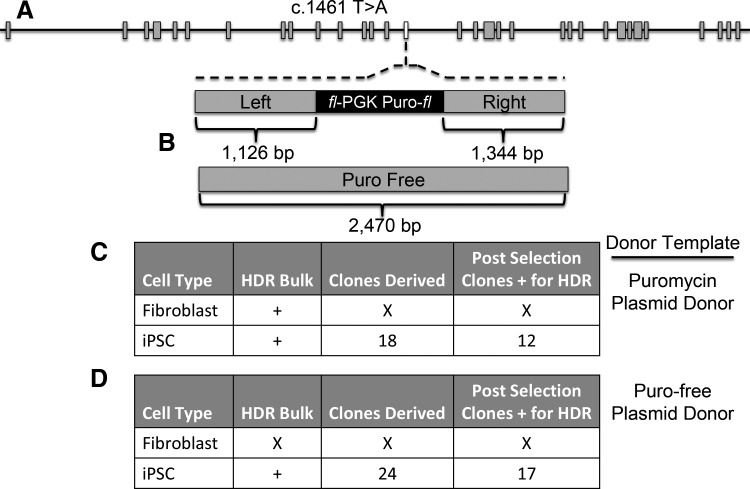
We next compared the ability of primary fibroblasts and iPSC-derived cells to undergo CRISPR/Cas9 gene editing. For this we focused on the c.1461 T > A mutation using gRNA candidate 2 (Figs. 1B and 4A) as the c.3058 + 4A>G locus showed a window of short interspersed nuclear elements that can complicate gene targeting and as heterozygous gene correction is sufficient for FA phenotype rescue. For HDR-based correction we constructed two donors: one with donor arms proximal to the mutation site flanking a floxed PGK-puromycin cassette and a second, puromycin marker-free template (Fig. 4B). As an important design consideration to prevent retargeting of the modified sequence, and to provide signature sequences for unambiguous detection, the donor templates each contained polymorphic substitutions (Supplementary Fig. S4). The repair templates, Cas9 nickase, and gRNA were delivered as plasmid DNA molecules by electroporation-based gene transfer into fibroblasts and iPSCs. HDR in a polyclonal population of bulk puromycin-selected fibroblasts was observed (Supplementary Fig. S5); however clonal cellular expansion was not possible due to senescence of the isolates (Fig. 4C). In contradistinction, the iPSCs treated similarly showed that 66% of the postselection analyzed clones had undergone HDR (Fig. 4C).
FIG. 4.
FANCI cell treatment and screening. (A) FANCI gene architecture. The FANCI gene exons are shown with larger gray boxes and the 1461 mutation occurs in exon 15 (white box). (B) Donor templates. Top: the floxed puromycin-based selection cassette (fl-PGK-puro-fl) contains the murine phosphoglycerate kinase 1 promoter and a bovine growth hormone polyadenylation sequence that is flanked by arms of homology of ∼1.1 kb (left) and ∼1.3 kb (right) in length. Bottom: a homologous donor with a length of ∼2.4 kb and no exogenous selection marker was also generated. (C, D) Homologous recombination frequencies in fibroblasts or iPSC utilizing differential donor templates. (C) Plasmid donor with puromycin selection gene. (D) Puromycin-free plasmid donor. In (C and D) the donor plasmids were electroporated into iPSC or primary fibroblasts along with the Cas9 nickase and FANCI 1461 gRNA and selection was performed by (C) puromycin or (D) mitomycin C. Tabular format for (C, D) indicate the cell type, whether evidence of homologous recombination was documented in the polyclonal bulk, postselection cellular population, the number of homogenous clones that could be derived, and the number of clones that showed conclusive evidence for HDR. “X” lettering indicates an absence of demonstrable HDR or an inability of cellular clones to be propagated due to senescence or toxicity. “+” indicates HDR documentation by PCR. HDR, homology-directed repair; PCR, polymerase chain reaction.
Because the FA phenotype is characterized by sensitivity to alkylating agents such as MMC, we also constructed a donor plasmid suitable for gene correction and subsequent selection by MMC. Primary fibroblasts showed a high degree of senescence when MMC was applied (unpublished observations), and we were unable to demonstrate HDR in this treatment group (Fig. 4D). In contrast, MMC addition to treated iPSCs showed selective outgrowth of cells, and 70% of the post-selection clones showed HDR (Fig. 4D). Taken together these results show the ability of the CRISPR/Cas9 system to mediate HDR in clonogenic iPSCs using plasmid-based donors that allow selective amplification of corrected cells (Fig. 4 and Supplementary Figs. S6 and S7).
Off-target analysis
An important consideration for the employment of CRISPR/Cas9 reagents for genome modification is the possibility for off-target activity at other loci that contain sequence homology to the intended target site. We analyzed the FANCI reagent using predictive in silico software (MIT CRISPR Design Tool) [27] and identified intragenic off-target candidates (Table 1) that were assessed by the Surveyor method. We observed no off-target effects at any of the predicted sites, suggesting a highly specific reagent at the resolution of the Surveyor assay (Table 1 and Supplementary Fig. S8).
Table 1.
Off-Target Sequence Analysis
| Target | Gene | Function | Surveyor (nuc/nick) |
|---|---|---|---|
| GACTGTACAAAGGCTGCTTA AGG | FANCI | DNA repair | +/− |
| GACAATAAAAAGGCTGCTTA TGG | UBE2 V1 | Ubiquitin conjugation | −/− |
| GGCTGTTTAAACGCTGCTTA AAG | SDCBP2 | Syndecan binding protein | −/− |
| AACTGAACAGAGGCTGCTTC AAG | APPBP2 | Amyloid binding protein | −/− |
| GACTGTCCCAAGGCTGCTGA TAG | IPP | Actin binding protein | −/− |
| TTCTGTCCAAAGGCTGCTCA CAG | ELMOD3 | GTPase activation | −/− |
| CACTGGACAGAGGCTGCTCA GGG | ADCY7 | Adenylyl cyclase | −/− |
| GACTGAACAAAGGCTGAATA CAG | PSMD14 | 26S proteasome subunit | −/− |
| GACTGTGAAAAGGCTGGTGA AAG | ARID4A | Nuclear protein/pRB binding | −/− |
| GACTTTACAAAACCTGTTTA CGG | GALNT8 | N-acetyl galactosaminyltransferase | −/− |
| GACTGTTAAAAGTCTCCTTA AAG | CCDC88A | Actin binding protein | −/− |
Intragenic off-target sites were identified by the CRISPR Design Tool that are shown at left with mismatches between the FANCI site in bold and underlined and genes and their functions are shown. At right are the results of the Surveyor analysis in Cas9 nuclease (“nuc”) or Cas9 nickase (“nick”) indicated by a +/− sign in regards to whether locus modification (ie, cleavage products generated by the Surveyor enzyme) was observed or not.
iPSC-directed differentiation
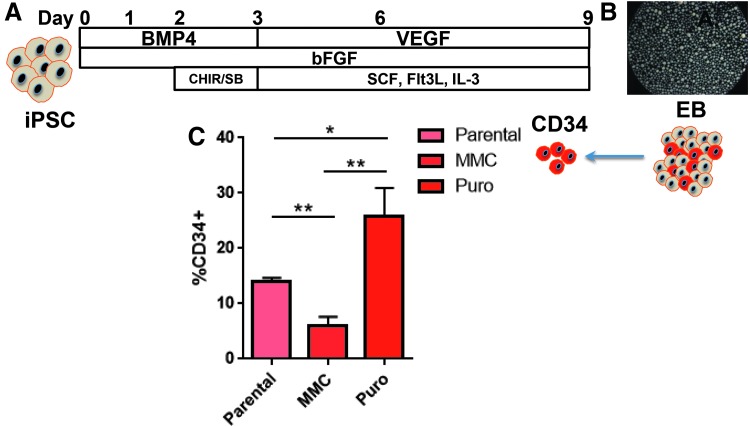
Hematopoiesis occurs in two waves: primitive and definitive, with the latter giving rise to long-term repopulating hematopoietic stem/progenitor cells (HSPCs) [30]. Efforts to date employing embryonic stem cells or iPSCs for HSPC generation have resulted in cells resembling the primitive hematopoietic fate [31]. More recent studies have shown the importance of the Wnt-β catenin pathway in driving mesodermal commitment to the definitive wave, while primitive hematopoiesis is reliant upon activin-nodal signaling [32]. As such, we hypothesized that EB formation under previously described conditions with mesodermal induction driven by addition of BMP-4 and bFGF could be augmented with the activin-nodal inhibitor SB431542 and the GS3Kβ inhibitor CHIR99021 [32–35] to drive the cells toward a definitive phenotype (Fig. 5A).
FIG. 5.
EB CD34 cell generation from gene corrected FANCI iPSC. (A) Culture conditions. iPSC under defined conditions that included basic fibroblast growth factor (bFGF), bone morphogenic protein-4 (BMP-4), vascular endothelial growth factor (VEGF) along with hematopoietic cytokines Flt-3 ligand, stem cell factor (SCF), and interleukin-3 (IL-3) and SB431542 and CHIR99021 were formed into (B) EB that adopted a round, uniform shape. (C) CD34 from FANCI EB. The CD34 cells were purified from EBs from uncorrected parental FA, MMC, or puromycin selected, gene corrected cells. Percentage of CD34 from total EB cellular fraction is shown. Data are representative from at least three experiments and *p ≤ 0.05 and **p ≤ 0.01. EB, embryoid body.
Hematopoietic commitment and progenitor expansion within embryoid bodies (EBs) was subsequently mediated by exposure to VEGF, SCF, Flt-3 ligand, and IL-3 (Fig. 5A). EB formation (Fig. 5B) under these conditions resulted in CD34 cell production with a noted (∼2-fold) increase when gene-corrected cells that had been selected with puromycin were employed (Fig. 5C). These data suggested that MMC may negatively impact EB and/or CD34 formation, and to test this we treated iPSCs derived from a patient with an intact FA DNA repair pathway with MMC for 48 h before EB formation. We observed a marked decrease in EB formation in the MMC group with diminished cellularity as determined by trypan blue exclusion (mean MMC group = 1.2 × 106 ± 3 × 105 and mean untreated group = 5 × 106 ± 1 × 106). As a result, fewer CD34 cells, representing <10% of the EB cellular population, were able to be isolated (data not shown).
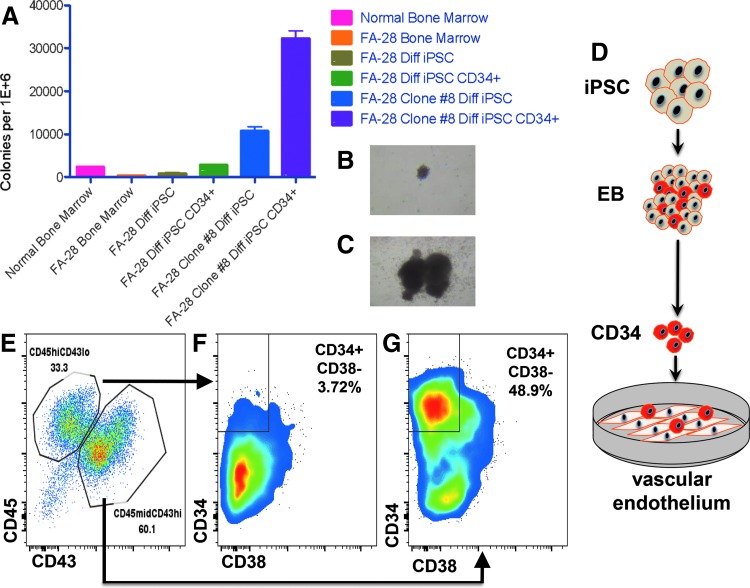
To test the colony-forming unit (CFU) potential of gene-corrected cells, mixed and pure populations of cells were placed in methylcellulose for CFU assays (Fig. 6A). Bone marrow aspirates containing populations of progenitors and stroma showed higher levels of CFU in normal than in FA patients (Fig. 6A). Similarly, uncorrected FA cells as EBs containing CD34 positive and negative cells, and pure populations of CD34+ EB-derived cells, showed diminished CFU potential compared to a representative clone of corrected cells obtained using a puromycin-based HDR donor (Fig. 6A). In addition, the CFU morphology of uncorrected parental cells (Fig. 6B) was consistently smaller than corrected cells (Fig. 6C).
FIG. 6.
Hematopoietic potential of gene corrected FA iPSC. (A) Methylcellulose colony forming unit assay. Total bone marrow (eg, mixed cellular population) from normal or a FANCI patient are shown at left. The remaining columns are iPSC cells. FA28 Diff iPSC and Diff iPSC CD34 are populations of cells that underwent CD34 EB formation and were placed in methycellulose in bulk (Diff iPSC) or were CD34 purified (Diff iPSC CD34+). Likewise, a corrected clone (number 8) was treated identically. Data are represented as number of colonies per 1 million plate cells. All data from the figure are from at least three independent experiments. (B and C) Hematopoietic colonies. Photomicrograph of BFU-E from uncorrected FA (B) or gene corrected (C) cells is shown from colonies in (A). (D) CD34 cell generation and culture on VeraVec™ vascular endothelium. Heterogeneous EBs generated over the course of 8 days in prohematopoietic cytokines (listed in Fig. 5) contain CD34+ cells (shown in red) that were purified to homogeneity and added to VeraVec E4ORF1 gene transformed vascular endothelium for 7 days. (E–G) FACS analysis of CD34+ cells. (E) CD45/CD43 analysis showing a CD45highCD43low and CD43highCD45midrange expression level cell populations. Each distinct population was fractionated for CD34 CD38 expression. The CD43 low population correlated with a CD34+CD38+ phenotype (F) and the CD43 high population showed expression of hematopoietic progenitor markers CD34+ CD38− (G). Image is representative of five independent experiments using puromycin selected, gene-corrected iPSC colonies.
To demonstrate that FA iPSCs under these conditions can form definitive cells of the hematopoietic compartment, we utilized a stringent procedure with parental cells to assess whether T progenitor cell formation was possible utilizing a coculture method on OP9 DL4 stromal cells (Supplementary Fig. S9). Toward optimizing conditions further, we extended our studies to include a vascular induction step that has been shown to promote multilineage progenitor production [36]. To accomplish this we generated CD34+ cells from EBs as above and placed them on a vascular endothelial cell feeder layer (Fig. 6D). This engineered cell line (termed VeraVec) was generated by adenoviral E4ORF1 transduction and provides supportive cues to developing cells [36]. Seven-day coculture of CD34+ cells and VeraVec cells resulted in two cell populations: CD45high/CD43low and CD45mid/CD43high (Fig. 6E). These cells were fractionated based on these parameters for analysis of CD34 and CD38 expression and we observed an emergent population of CD34+CD38− cells (Fig. 6F and G). This finding is highly significant as this cell subpopulation is prevalent in the definitive hematopoietic cells of the bone marrow [15] and these results demonstrate an efficient and robust methodology for iPSC-derived multilineage HSPCs.
Discussion
We present the following key findings: the ability to reprogram FANCI fibroblasts to pluripotency using Sendai viral, nonintegrating gene delivery; CRISPR/Cas9 mediated in situ gene correction; and the generation of a population of cells with characteristics of HSPCs using a novel differentiation methodology.
The reprogramming efficiency of FA cells is a significant hurdle in the field and was first reported by Raya et al. [28] and followed by Muller et al. [11]. Each study highlighted the requirement of an intact FA DNA repair pathway to accomplish iPSC generation and, of note, the reprogramming factors were delivered on integrating retro or lentiviral constructs. The highest reprogramming frequencies were observed using lentiviral transgenesis [11]. Following these studies Liu et al. demonstrated, using nonintegrating plasmid-based five factor reprograming factor delivery, that they could reprogram noncomplemented FANCA cells that also required p53 inhibition [29]. The reprogramming frequency was 10-fold higher by Liu et al. than other reports and the reason for this is unclear. One hypothesis is that viral vector gene expression in a stable fashion is incumbent on integration, a process that employs cellular DNA repair using endogenous factors. These factors can be differential with regard to the individual platforms [37,38] potentially indicating that in the DNA damage repair defective FA phenotype integrating vectors are suboptimal. Our results show that the nonintegrating Sendai virus was able to reprogram FANCI cells without complementation (Figs. 2 and 3) and while further experimentation directly assessing the platforms and complementation group deficiencies in a head-to-head manner are required, the generation of iPSCs proved crucial to effective gene repair in our study.
Programmable nucleases exist in multiple formats: zinc-finger nucleases (ZFNs), transcription activator-like effector nucleases, meganucleases, and the CRISPR/Cas9 system. Toward addressing the most commonly occurring FA gene mutation in the FANCA gene, Rio et al. utilized a ZFN pair to insert the FANCA cDNA by HDR into the AAVS1 safe harbor locus [6]. Therapeutic gene expression was mediated by the PGK promoter, resulting in sustained transgene expression representing a platform that would be applicable to any patient with FANCA gene mutations [6]. Previously, the present group has utilized the CRISPR/Cas9 D10A nickase derived from S. pyogenes to correct a prevalent FANCC gene mutation [14]. However, common to each study was the requirement of hTERT gene expression to facilitate gene repair in patient fibroblasts [6,14]. Gene-corrected FANCA fibroblasts were amenable to iPSC reprogramming by the Rio group while the FANCC cells were unable to be reprogrammed [6,14]. These studies differed in a key area related to the temporal aspects of gene correction. As stated, the Rio study generated cells with constitutive FANCA expression while we, both here and previously, targeted the disease-causing mutation in situ so that the endogenous command and control systems are retained (Figs. 1 and 4) [14]. Given that external DNA-damaging stimuli can result in FANCI gene upregulation [39], we consider this a critical and clinically relevant consideration. A potential downside to an individualized mutation targeting approach is that the highly heterogeneous FA mutation profile may make portions of this or other FA genes inaccessible to precision, site-specific nuclease design requirements. For the S. pyogenes Cas9, this design requirement is a target sequence containing an–NGG PAM, where “N” is any nucleotide [13]. Analysis of the compound heterozygote gene mutations present in this FANCI patient identified multiple candidates for our experimentation (Fig. 1). All of the candidates showed activity when delivered as plasmid DNA expression cassettes (Fig. 1F) and demonstrated the potent activity we have consistently observed with the S. pyogenes Cas9 [14,40]. Moreover, it shows that despite the putative recognition restrictions of Cas9, we were able to target sequence proximal to the mutation, an important consideration as HDR efficiency decreases as the distance from break site to target modification locus increases [41]. Relevant to this, the targeting capacity of the CRISPR/Cas9 system has been expanded with the discovery of alternate PAM variants through direct engineering or bacterial strain discovery [42–44], making larger regions of the genome accessible for genome editing.
We attempted to correct fibroblasts and iPSCs using CRISPR/Cas9 D10A plasmids with an inactive HNH domain (Figs. 1C and 2B) capable of nicking a single strand of DNA [45]. Further, we tested two donor platforms: one based on artificial (ie, puromycin) drug resistance and selection and a second MMC-based approach requiring correction of the cellular phenotype (Fig. 4B). The puromycin plasmid template showed demonstrable HDR in bulk populations of fibroblasts and iPSCs (Fig. 4C, D and Supplementary Figs. S5–S7). Clonal derivation of corrected fibroblasts was not possible; however, puromycin selection allowed recovery of 12 corrected iPSC clones (Fig. 4C). In non-puromycin donor treated fibroblasts we noted an inability to select modified cells using MMC as it triggered senescence (unpublished observations and Fig. 4D). In iPSCs we isolated 17 MMC-resistant clones that contained donor sequence modification (Fig. 4D and Supplementary Fig. S6). In total these results demonstrate the difficulty in mediating gene editing-based repair in primary, non-hTERT transformed FA fibroblasts; this is consistent with our previous observations and those of Rio et al. [6,14]. Conversely, iPSC gene-corrected clones were readily obtained when selective pressure was applied (Fig. 4). This consideration is important, as in the absence of selection, HDR rates in bulk-treated cells were sufficiently low to evade detection by our PCR strategy (data not shown). Thus, selection can streamline the obtainment of gene-corrected cells without extended screening of individual colonies. Further, given that cell lines in which the variables of gene transfer and poor ability to be clonally derived are removed, HDR rates can be <10% [14,46], making selection strategies important in the context of a chosen line of investigation.
Bone marrow failure is a life-threatening manifestation of FA, making gene modification of hematopoietic cells highly desirable. Direct HSPC gene correction has been demonstrated [47,48]; however, the depleted numbers of HSPCs in FA patients make them a difficult clinical and research tool to acquire. Further, the clinical status of individual patients will be highly variable, making standardized genome engineering in this context increasingly difficult. As such, the ability to engineer the genome of cells with direct or acquired therapeutic properties holds great promise for FA. Toward realizing this promise Raya et al. showed that FA iPSCs could be differentiated into CD34+CD45+ cells with a peak expression of ∼7% and ∼1% respectively, with an ability to form CFUs in vitro [28]. Likewise, the study by Rio et al. showed enhanced CFU potential from gene-corrected FA iPSCs and the following hematopoietic population frequencies: 12% CD34+CD43+, ∼7% CD34+CD45+, and ∼12% CD34−CD45+, respectively [6]. A study in the same year by Liu et al. demonstrated ∼9% CD34+CD43low, a marker for multipotent progenitors [49] in gene-corrected FANCA iPSCs [29]. In these studies Raya and Liu employed OP9 cells, a mouse stromal cell with the capacity to support hematopoiesis [50], to drive EB-derived CD34s to a hematopoietic phenotype [28,29]. More recently, Rio et al. used supplemental recombinant Wnt-3a and Wnt-11 protein in their differentiation schema [6]. Accumulating evidence has shown that the governance of fate determination between primitive and definitive hematopoiesis is modulated by activin-nodal and Wnt-β catenin signaling during the mesodermal phase of development [32]. As such, the inhibition of activin-nodal signaling and/or augmentation of the Wnt pathway results in enhanced definitive hematopoietic progenitor development from iPSCs [32]. Once derivative cells are specified for definitive lineage commitment, their subsequent phenotype can be directed by coculturing with cells providing niche developmental cues. For instance, T lymphoid generation is possible due to the instructive signals provided by the OP9 stromal cell layer expressing the Notch ligand Delta-like (OP9-DL4) [51,52], and we show acquisition of the CD5 and CD7 pan-T cell markers using a T cell biased differentiation protocol (Supplementary Fig. S9). To extend these foundational findings, we applied frontline differentiation procedures using, for the first time, tandem activin-nodal and Wnt signaling modulation with SB431542 and CHIR99021 (Figs. 5 and 6). The resultant CD34 cells were then cocultured on a supportive matrix of VeraVec E4ORF1 transgenic vascular endothelial cells [36]. This population of cells aids in recapitulating the instructive vascular niche and has allowed the acquisition of engraftment potential of human vascular endothelial cells [36]. After 1 week of coculture two populations of cells were observed: CD45highCD43low and CD45midCD43high (Fig. 6E). These findings are in agreement with previous data by Kennedy et al. using OP9 coculture methodologies that showed definitive CD43+CD45+ cells [52]. We further characterized the CD45midCD43high population and observed that they possessed properties consistent with definitive multipotent progenitors with engraftment properties as evidenced by a CD34+CD38− phenotype (Fig. 6G) [53]. As such, these data show that the acquisition of multilineage hematopoietic progenitors is possible using an in vitro vascular induction methodology. Our data are highly relevant and complement other studies utilizing VeraVec coculture to promote in vitro conditions for the specification of alternate cell types to transplantable and engraftable HSPCs and are additive to the cellular engineering field [36,54].
An additional finding we observed that is of importance to the FA field is the decreased efficiency of EB formation with an associated decrease in CD34 generation and poor ability to form CFUs when cells were selected with MMC (Fig. 5C and 6A). By using MMC pretreatment in iPSCs with an intact FA DNA repair pathway, we also observed diminished cellularity of the EBs (data referenced above) suggesting that MMC may impact the differentiation capabilities of iPSCs. Further experimentation to define the mechanism and related temporal effects of MMC on iPSCs is required and will further aid in selection strategies that allow isolation of gene-corrected cells with a high degree of plasticity able to serve as a foundation for directed therapeutic differentiation.
Finally, a crucial consideration, particularly in the genomic instability phenotype of FA, is the specificity of the candidate gene-editing reagent. Promiscuous cutting at off-target sites resulting in multiple DNA breaks can result in translocations and/or disruption of unintended sequences [40,55,56]. To mitigate the potential of off-target effects, we employed the Cas9 D10A nickase that has been shown to promote higher rates of HDR with minimal mutagenic nonhomologous endjoining (NHEJ) [14,46]. Indeed, treatment of cells with a Cas9 nuclease showed higher rates of NHEJ than did nickase-treated cells for the FANCI gene target (Table 1 and Supplementary Fig. S8). By assigning putative off-target sites using a predictive algorithm, we analyzed 10 off-target candidates contained within genes and did not observe appreciable modification at any of the sites (Table 1 and Supplementary Fig. S8). These findings suggest a specific reagent whose activity profile is restricted to the FANCI target. This fact, coupled with the use of a nicking version of Cas9, represents a maximally safe approach for FANCI genome editing.
In summary, these data highlight the robust nature in which iPSCs derived from a FANCI individual can be modified at the genome level to serve as an engineering template for therapeutic cell development. This stable and renewable population of cells holds tremendous potential for the entire spectrum of FA disease and its associated pathological manifestations as part of an autologous regenerative medicine approach.
Supplementary Material
Acknowledgments
We are grateful to Nancy Griggs Morgan and Kayla Cook for expert assistance in article preparation and editing. We are also thankful for the generosity of the Kidz1stFund, the Fanconi Anemia Research Fund, the Children's Cancer Research Fund, the Lindahl Family and the Corrigan Family. MJO is supported by 8UL1TR000114-02. JT is supported in part by R01 AR063070 and P01 CA065493. BRW is supported by NIH T32- HL007062. Research reported in this publication was supported by the National Center for Advancing Translational Sciences of the National Institutes of Health Award Number UL1TR000114 (MJO). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. DP is a major contributor along with BRW and MJO to conceiving, designing, and establishing the protocol to generate CD34+ hematopoietic cells. The authors are grateful to Chong Jai Kim, MD, PhD and the present work was partly supported by research funds from the National Research Foundation of Korea (NRF-2015K1A4A3046807).
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Cattoglio C, Facchini G, Sartori D, Antonelli A, Miccio A, Cassani B, Schmidt M, von Kalle C, Howe S, et al. (2007). Hot spots of retroviral integration in human CD34+ hematopoietic cells. Blood 110:1770–1778 [DOI] [PubMed] [Google Scholar]
- 2.Hacein-Bey-Abina S, Garrigue A, Wang GP, Soulier J, Lim A, Morillon E, Clappier E, Caccavelli L, Delabesse E, et al. (2008). Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J Clin Invest 118:3132–3142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hacein-Bey-Abina S, Von Kalle C, Schmidt M, McCormack MP, Wulffraat N, Leboulch P, Lim A, Osborne CS, Pawliuk R, et al. (2003). LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science 302:415–419 [DOI] [PubMed] [Google Scholar]
- 4.Osorio A, Bogliolo M, Fernandez V, Barroso A, de la Hoya M, Caldes T, Lasa A, Ramon y Cajal T, Santamarina M, et al. (2013). Evaluation of rare variants in the new fanconi anemia gene ERCC4 (FANCQ) as familial breast/ovarian cancer susceptibility alleles. Hum Mutat 34:1615–1618 [DOI] [PubMed] [Google Scholar]
- 5.Bogliolo M, Schuster B, Stoepker C, Derkunt B, Su Y, Raams A, Trujillo JP, Minguillon J, Ramirez MJ, et al. (2013). Mutations in ERCC4, encoding the DNA-repair endonuclease XPF, cause Fanconi anemia. Am J Hum Genet 92:800–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rio P, Banos R, Lombardo A, Quintana-Bustamante O, Alvarez L, Garate Z, Genovese P, Almarza E, Valeri A, et al. (2014). Targeted gene therapy and cell reprogramming in Fanconi anemia. EMBO Mol Med 6:835–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ceccaldi R, Sarangi P. and D'Andrea AD. (2016). The Fanconi anaemia pathway: new players and new functions. Nat Rev Mol Cell Biol 17:337–349 [DOI] [PubMed] [Google Scholar]
- 8.Bagby GC., Jr. (2003). Genetic basis of Fanconi anemia. Curr Opin Hematol 10:68–76 [DOI] [PubMed] [Google Scholar]
- 9.MacMillan ML, DeFor TE, Young JA, Dusenbery KE, Blazar BR, Slungaard A, Zierhut H, Weisdorf DJ. and Wagner JE. (2015). Alternative donor hematopoietic cell transplantation for Fanconi anemia. Blood 125:3798–3804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rosenberg PS, Greene MH. and Alter BP. (2003). Cancer incidence in persons with Fanconi anemia. Blood 101:822–826 [DOI] [PubMed] [Google Scholar]
- 11.Muller LU, Milsom MD, Harris CE, Vyas R, Brumme KM, Parmar K, Moreau LA, Schambach A, Park IH, et al. (2012). Overcoming reprogramming resistance of Fanconi anemia cells. Blood 119:5449–5457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujie Y, Fusaki N, Katayama T, Hamasaki M, Soejima Y, Soga M, Ban H, Hasegawa M, Yamashita S, et al. (2014). New type of Sendai virus vector provides transgene-free iPS cells derived from chimpanzee blood. PLoS One 9:e113052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA. and Zhang F. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339:819–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Osborn MJ, Gabriel R, Webber BR, DeFeo AP, McElroy AN, Jarjour J, Starker CG, Wagner JE, Joung JK, et al. (2015). Fanconi anemia gene editing by the CRISPR/Cas9 system. Hum Gene Ther 26:114–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ishikawa F, Livingston AG, Minamiguchi H, Wingard JR. and Ogawa M. (2003). Human cord blood long-term engrafting cells are CD34+ CD38. Leukemia 17:960–964 [DOI] [PubMed] [Google Scholar]
- 16.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE. and Church GM. (2013). RNA-guided human genome engineering via Cas9. Science 339:823–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA., 3rd and Smith HO. (2009). Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6:343–345 [DOI] [PubMed] [Google Scholar]
- 18.Tolar J, Xia L, Lees CJ, Riddle M, McElroy A, Keene DR, Lund TC, Osborn MJ, Marinkovich MP, Blazar BR. and Wagner JE. (2013). Keratinocytes from induced pluripotent stem cells in junctional epidermolysis bullosa. J Invest Dermatol 133:562–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tolar J, Xia L, Riddle MJ, Lees CJ, Eide CR, McElmurry RT, Titeux M, Osborn MJ, Lund TC, et al. (2011). Induced pluripotent stem cells from individuals with recessive dystrophic epidermolysis bullosa. J Invest Dermatol 131:848–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Osborn MJ, Starker CG, McElroy AN, Webber BR, Riddle MJ, Xia L, DeFeo AP, Gabriel R, Schmidt M, et al. (2013). TALEN-based gene correction for epidermolysis bullosa. Mol Ther 21:1151–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tolar J, Park IH, Xia L, Lees CJ, Peacock B, Webber B, McElmurry RT, Eide CR, Orchard PJ, et al. (2011). Hematopoietic differentiation of induced pluripotent stem cells from patients with mucopolysaccharidosis type I (Hurler syndrome). Blood 117:839–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lopez-Onieva L, Montes R, Lamolda M, Romero T, Ayllon V, Lozano ML, Vicente V, Rivera J, Ramos-Mejia V. and Real PJ. (2016). Generation of induced pluripotent stem cells (iPSCs) from a Bernard-Soulier syndrome patient carrying a W71R mutation in the GPIX gene. Stem Cell Res 16:692–695 [DOI] [PubMed] [Google Scholar]
- 23.Guschin DY, Waite AJ, Katibah GE, Miller JC, Holmes MC. and Rebar EJ. (2010). A rapid and general assay for monitoring endogenous gene modification. Methods Mol Biol 649:247–256 [DOI] [PubMed] [Google Scholar]
- 24.Hartig SM. (2013). Basic image analysis and manipulation in ImageJ. Curr Protoc Mol Biol Chapter 14:Unit14 15. [DOI] [PubMed] [Google Scholar]
- 25.Smogorzewska A, Matsuoka S, Vinciguerra P, McDonald ER, 3rd, Hurov KE, Luo J, Ballif BA, Gygi SP, Hofmann K, D'Andrea AD. and Elledge SJ. (2007). Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell 129:289–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Whitney MA, Saito H, Jakobs PM, Gibson RA, Moses RE. and Grompe M. (1993). A common mutation in the FACC gene causes Fanconi anaemia in Ashkenazi Jews. Nat Genet 4:202–205 [DOI] [PubMed] [Google Scholar]
- 27.Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, et al. (2013). DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol 31:827–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raya A, Rodriguez-Piza I, Guenechea G, Vassena R, Navarro S, Barrero MJ, Consiglio A, Castella M, Rio P, et al. (2009). Disease-corrected haematopoietic progenitors from Fanconi anaemia induced pluripotent stem cells. Nature 460:53–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu GH, Suzuki K, Li M, Qu J, Montserrat N, Tarantino C, Gu Y, Yi F, Xu X, et al. (2014). Modelling Fanconi anemia pathogenesis and therapeutics using integration-free patient-derived iPSCs. Nat Commun 5:4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Medvinsky A, Rybtsov S. and Taoudi S. (2011). Embryonic origin of the adult hematopoietic system: advances and questions. Development 138:1017–1031 [DOI] [PubMed] [Google Scholar]
- 31.Keller G, Kennedy M, Papayannopoulou T. and Wiles MV. (1993). Hematopoietic commitment during embryonic stem cell differentiation in culture. Mol Cell Biol 13:473–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sturgeon CM, Ditadi A, Awong G, Kennedy M. and Keller G. (2014). Wnt signaling controls the specification of definitive and primitive hematopoiesis from human pluripotent stem cells. Nat Biotechnol 32:554–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Madhu V, Dighe AS, Cui Q. and Deal DN. (2016). Dual inhibition of Activin/Nodal/TGF-beta and BMP signaling pathways by SB431542 and dorsomorphin induces neuronal differentiation of human adipose derived stem cells. Stem Cells Int 2016:1035374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Du J, Wu Y, Ai Z, Shi X, Chen L. and Guo Z. (2014). Mechanism of SB431542 in inhibiting mouse embryonic stem cell differentiation. Cell Signal 26:2107–2116 [DOI] [PubMed] [Google Scholar]
- 35.Wu Y, Ai Z, Yao K, Cao L, Du J, Shi X, Guo Z. and Zhang Y. (2013). CHIR99021 promotes self-renewal of mouse embryonic stem cells by modulation of protein-encoding gene and long intergenic non-coding RNA expression. Exp Cell Res 319:2684–2699 [DOI] [PubMed] [Google Scholar]
- 36.Sandler VM, Lis R, Liu Y, Kedem A, James D, Elemento O, Butler JM, Scandura JM. and Rafii S. (2014). Reprogramming human endothelial cells to haematopoietic cells requires vascular induction. Nature 511:312–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sakurai Y, Komatsu K, Agematsu K. and Matsuoka M. (2009). DNA double strand break repair enzymes function at multiple steps in retroviral infection. Retrovirology 6:114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baekelandt V, Claeys A, Cherepanov P, De Clercq E, De Strooper B, Nuttin B. and Debyser Z. (2000). DNA-Dependent protein kinase is not required for efficient lentivirus integration. J Virol 74:11278–11285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Walter D, Lier A, Geiselhart A, Thalheimer FB, Huntscha S, Sobotta MC, Moehrle B, Brocks D, Bayindir I, et al. (2015). Exit from dormancy provokes DNA-damage-induced attrition in haematopoietic stem cells. Nature 520:549–552 [DOI] [PubMed] [Google Scholar]
- 40.Osborn MJ, Webber BR, Knipping F, Lonetree CL, Tennis N, DeFeo AP, McElroy AN, Starker CG, Lee C, et al. (2016). Evaluation of TCR gene editing achieved by TALENs, CRISPR/Cas9 and megaTAL nucleases. Mol Ther 24:570–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kan Y, Ruis B, Lin S. and Hendrickson EA. (2014). The mechanism of gene targeting in human somatic cells. PLoS Genet 10:e1004251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kleinstiver BP, Prew MS, Tsai SQ, Nguyen NT, Topkar VV, Zheng Z. and Joung JK. (2015). Broadening the targeting range of Staphylococcus aureus CRISPR-Cas9 by modifying PAM recognition. Nat Biotechnol 33:1293–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kleinstiver BP, Prew MS, Tsai SQ, Topkar VV, Nguyen NT, Zheng Z, Gonzales AP, Li Z, Peterson RT, et al. (2015). Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature 523:481–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hou Z, Zhang Y, Propson NE, Howden SE, Chu LF, Sontheimer EJ. and Thomson JA. (2013). Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis. Proc Natl Acad Sci U S A 110:15644–15649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shen B, Zhang W, Zhang J, Zhou J, Wang J, Chen L, Wang L, Hodgkins A, Iyer V, Huang X. and Skarnes WC. (2014). Efficient genome modification by CRISPR-Cas9 nickase with minimal off-target effects. Nat Methods 11:399–402 [DOI] [PubMed] [Google Scholar]
- 46.Certo MT, Ryu BY, Annis JE, Garibov M, Jarjour J, Rawlings DJ. and Scharenberg AM. (2011). Tracking genome engineering outcome at individual DNA breakpoints. Nat Methods 8:671–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Genovese P, Schiroli G, Escobar G, Di Tomaso T, Firrito C, Calabria A, Moi D, Mazzieri R, Bonini C, et al. (2014). Targeted genome editing in human repopulating haematopoietic stem cells. Nature 510:235–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang J, Exline CM, DeClercq JJ, Llewellyn GN, Hayward SB, Li PW, Shivak DA, Surosky RT, Gregory PD, Holmes MC. and Cannon PM. (2015). Homology-driven genome editing in hematopoietic stem and progenitor cells using ZFN mRNA and AAV6 donors. Nat Biotechnol 33:1256–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vodyanik MA, Thomson JA. and Slukvin II. (2006). Leukosialin (CD43) defines hematopoietic progenitors in human embryonic stem cell differentiation cultures. Blood 108:2095–2105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gao J, Yan XL, Li R, Liu Y, He W, Sun S, Zhang Y, Liu B, Xiong J. and Mao N. (2010). Characterization of OP9 as authentic mesenchymal stem cell line. J Genet Genomics 37:475–482 [DOI] [PubMed] [Google Scholar]
- 51.Mohtashami M, Shah DK, Kianizad K, Awong G. and Zuniga-Pflucker JC. (2013). Induction of T-cell development by Delta-like 4-expressing fibroblasts. Int Immunol 25:601–611 [DOI] [PubMed] [Google Scholar]
- 52.Kennedy M, Awong G, Sturgeon CM, Ditadi A, LaMotte-Mohs R, Zúñiga-Pflücker JC. and Keller G. (2012). T lymphocyte potential marks the emergence of definitive hematopoietic progenitors in human pluripotent stem cell differentiation cultures. Cell Rep 2:1722–1735 [DOI] [PubMed] [Google Scholar]
- 53.Gilmore GL, DePasquale DK, Lister J. and Shadduck RK. (2000). Ex vivo expansion of human umbilical cord blood and peripheral blood CD34(+) hematopoietic stem cells. Exp Hematol 28:1297–1305 [DOI] [PubMed] [Google Scholar]
- 54.Gori JL, Butler JM, Chan YY, Chandrasekaran D, Poulos MG, Ginsberg M, Nolan DJ, Elemento O, Wood BL, et al. (2015). Vascular niche promotes hematopoietic multipotent progenitor formation from pluripotent stem cells. J Clin Invest 125:1243–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Frock RL, Hu J, Meyers RM, Ho YJ, Kii E. and Alt FW. (2015). Genome-wide detection of DNA double-stranded breaks induced by engineered nucleases. Nat Biotechnol 33:179–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gabriel R, Lombardo A, Arens A, Miller JC, Genovese P, Kaeppel C, Nowrouzi A, Bartholomae CC, Wang J, et al. (2011). An unbiased genome-wide analysis of zinc-finger nuclease specificity. Nat Biotechnol 29:816–823 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.