Significance
Tudor domain-containing protein 3 (TDRD3), a multidomain scaffold protein functions as an epigenetic reader on nuclear chromatin and binds with fragile X mental retardation protein on mRNA. It forms a conserved complex with topoisomerase 3β (Top3β), a type IA topoisomerase, and participates in both transcription and translation. The mechanism of how TDRD3 acts as Top3β’s partner in regulating these cellular processes is unknown. Here, we demonstrated that TDRD3 is able to stimulate Top3β’s DNA and RNA topoisomerase catalytic activities, through binding and stabilizing single-stranded regions in DNA and RNA substrates. Because these regions are the preferred site for Top3β, TDRD3 therefore acts as a regulator to provide access of the enzyme to these nucleic acid substrates and to act upon them.
Keywords: Tudor domain-containing domain 3, DNA topoisomerases, RNA topoisomerases, RNA circles, RNA duplex
Abstract
Topoisomerase 3β (Top3β) can associate with the mediator protein Tudor domain-containing protein 3 (TDRD3) to participate in two gene expression processes of transcription and translation. Despite the apparent importance of TDRD3 in binding with Top3β and directing it to cellular compartments critical for gene expression, the biochemical mechanism of how TDRD3 can affect the functions of Top3β is not known. We report here sensitive biochemical assays for the activities of Top3β on DNA and RNA substrates in resolving topological entanglements and for the analysis of TDRD3 functions. TDRD3 stimulates the relaxation activity of Top3β on hypernegatively supercoiled DNA and changes the reaction from a distributive to a processive mode. Both supercoil retention assays and binding measurement by fluorescence anisotropy reveal a heretofore unknown preference for binding single-stranded nucleic acids over duplex. Whereas TDRD3 has a structure-specific binding preference, it does not discriminate between DNA and RNA. This unique property for binding with nucleic acids can have an important function in serving as a hub to form nucleoprotein complexes on DNA and RNA. To gain insight into the roles of Top3β on RNA metabolism, we designed an assay by annealing two single-stranded RNA circles with complementary sequences. Top3β is capable of converting two such single-stranded RNA circles into a double-stranded RNA circle, and this strand-annealing activity is enhanced by TDRD3. These results demonstrate that TDRD3 can enhance the biochemical activities of Top3β on both DNA and RNA substrates, in addition to its function of targeting Top3β to critical sites in subcellular compartments.
Higher-order structural complexities in nucleic acids can be brought about by the folding and intertwining through inter- and intramolecular base pairing. They are impediments to the transactions of genetic information, including replication, transcription, recombination, and translation, which involve the unwinding and rewinding of base-paired regions to access encoded information (1–3). DNA topoisomerases are nature’s tools to resolve the topological entanglements in nucleic acids. They are ubiquitous in nature, first discovered in bacteria in 1971 (4), and in mammalian cells in 1972 (5), and are characterized by a mechanism involving the formation of a covalent protein/DNA adduct to generate a transient and reversible break allowing for topological transformation (6). Based on whether the strand passage is through a protein-mediated single-stranded gate or a double-stranded gate, the enzymes are classified into type I or type II topoisomerases, respectively. Both types are further classified into A and B families based on their structural and mechanistic features (7). All topoisomerases are essential for the growth and development of an organism, suggesting that they have critical but distinct functions in all cells.
Ever since their discovery, there is a fascination for whether these critical DNA enzymes may play a role in the RNA world. Early experiments involving the knotting of a single-stranded circle in trefoil formation (8) and protein-linked RNA cleavage (9, 10) suggest the biochemical feasibility of this idea. The interest is further boosted by recent experiments demonstrating that human and Drosophila topoisomerase 3β (Top3β) is associated with mRNA in cytoplasm and has RNA topoisomerase activity (11, 12). The deficiency in this enzyme is attributed to one of the causative mutations for schizophrenia and cognitive disorder (11, 12) and results in abnormal synaptic structures in Drosophila (12).
How RNA or DNA topoisomerase activity can affect neurological functions remains to be elucidated. However, there are interesting insights to be gained by considering the interacting partners of Top3β. A mediator protein with multiple domains for interprotein interactions, TDRD3 is a key partner for Top3β in both the nucleus and cytoplasm (13). TDRD3 can recognize dimethylated arginine in proteins including inner core histones and RNA polymerase II, thus targeting its partner proteins to the transcriptionally active chromatin, especially at their 5′ end (14, 15). Because the knockdown or deletion of TDRD3 results in R-loop formation near the promoter regions, it is plausible that the absence of Top3β near the transcriptional start site can lead to a buildup of excessive negative supercoiling, thus stabilizing the R-loop structure (13). TDRD3 also associates with the partner protein fragile X mental retardation protein (FMRP) in the cytoplasm. Missing FMRP can cause fragile X mental retardation, a major cause of congenital mental disorder (16). FMRP is preferentially localized in the coding regions of mRNA and plays a role in inhibiting the translation of mRNA relevant to synaptic functions in neuronal cells (17). It is thus suggested that Top3β may also have a role in translation through its RNA topoisomerase activity in modulating the complex structures of folded mRNA (12). Whereas Top3β can have important functions in two processes of gene expression in nucleus and cytoplasm through its interactions with TDRD3, it is unclear whether TDRD3 has any additional role in regulating Top3β activity, beyond its targeting function of directing Top3β to critical sites in subcellular compartments.
Our results presented here show that TDRD3 can promote both DNA and RNA topoisomerase activities, likely through stabilizing the complex of the enzyme bound to the single-stranded regions of the substrate, which is also the site where type IA enzymes carry out strand passage reactions. The finding that TDRD3 has a structure-specific preference for single-stranded nucleic acids, but does not discriminate between DNA and RNA, adds an aspect to the scaffolding function of TDRD3 in the assembly of the nucleoprotein complex. A mediator protein that is able to interact with specific proteins and to target to the preferred structure of nucleic acids can have a versatile role serving as a hub for assembling stable complexes in chromatin or polysomes.
With an increasing interest in RNA topoisomerase activity, it is important to develop biochemical assays other than the trefoil formation of circular RNA. Using two single-stranded circular RNA substrates with complementary sequences, we have demonstrated that Top3β and other type IA enzymes can mediate the formation of double-stranded circular RNA through the intermolecular stranded passage activity of these enzymes. This circle-annealing assay poses a less stringent requirement for the sequence and structure of the substrate RNA compared with the knotting assay, and can thus broaden our capability to test the effect of an RNA sequence or structure on RNA topoisomerase activity. In addition, this versatile biochemical method to prepare an interesting class of molecules of circular duplex RNA has unique potential for investigating the functions of other RNA enzymes, including topoisomerases.
Results
Stimulation of Top3β Relaxation Activity by TDRD3.
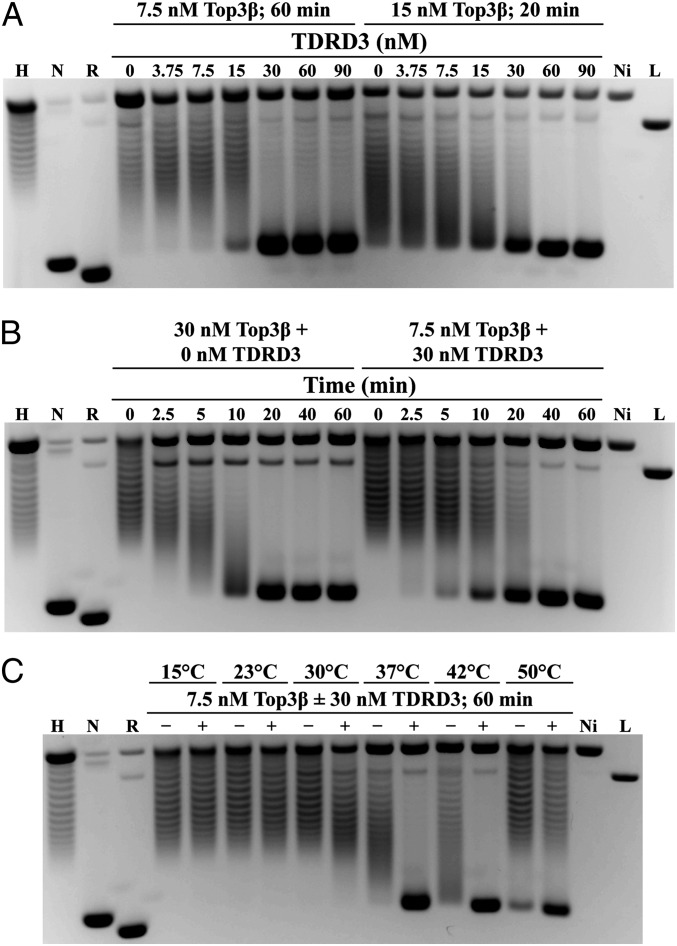
A sensitive method for detecting the effect of TDRD3 on Top3β’s activity is the relaxation of hypernegatively supercoiled DNA by Drosophila Top3β, where the DNA is relaxed to a supercoiling state that is slightly more negatively supercoiled than the plasmid DNA (18). In reactions with 7.5 nM Top3β, increasing the concentration of TDRD3 stimulated the conversion of hypernegatively supercoiled substrate to a final product with a mobility just behind the negatively supercoiled plasmid DNA (Fig. 1A). When the Top3β concentration was doubled, an approximately twofold higher concentration of TDRD3 was required to duplicate the same stimulation pattern, suggesting stoichiometry-dependent stimulatory effect of TDRD3. In both cases, reactions became saturated, but without the DNA reaching a fully relaxed state, when TDRD3 and Top3β reached a similar stoichiometric ratio. The data suggest a possible protein–protein interaction between Top3β and TDRD3.
Fig. 1.
TDRD3 stimulates the relaxation activity of Top3β on hypernegatively supercoiled DNA substrates. Topological states of the plasmid DNA were: hypernegatively supercoiled (H), negatively supercoiled (N), relaxed circle (R), nicked circle (Ni), and linearized (L). (A) Titration assay of TDRD3 concentrations showed that relaxation activity of Top3β was stimulated in both a time- and concentration-dependent manner. (B) Time course assay showed that relaxation by Top3β alone occurs in a distributive manner, and the presence of TDRD3 in the reactions changes the relaxation mode to a processive one. (C) Top3β- and TDRD3-catalyzed relaxation activity was temperature dependent.
To gain insight into the mechanistic basis for stimulatory effect of TDRD3, we examined the kinetic behavior of Top3β relaxation with and without TDRD3 (Fig. 1B). With Top3β alone, intermediate products consisting of DNA molecules with different supercoiling densities were observed at early time points before the reaction was completed with the appearance of the final product. Thus, the enzyme showed a distributive mode of relaxation, wherein intermediates precede the appearance of the final product. In contrast, in reactions with both Top3β and TDRD3, the relaxation of Top3β changed to a processive mode without the accumulation of intermediates. It is plausible that TDRD3 can enhance the binding of Top3β to the DNA substrate and allow the reaction to proceed to completion before releasing its binding to DNA.
Because Top3β, like other type IA enzymes, prefers DNA substrates with underwound regions, the effect of TDRD3 in promoting the binding of Top3β to DNA may be mediated through this structure. We tested this hypothesis by examining whether its stimulatory effect is also favored under conditions promoting the generation of underwound regions in DNA. Indeed the stimulatory effect of TDRD3 is more apparent when the reactions were carried at 37 °C, 42 °C, and 50 °C, but not at lower temperatures (Fig. 1C), indicating that an increase in the reaction temperature promotes the stimulatory effect of TDRD3. Complete substrate relaxation was seen when incubation was performed at 37 °C and 42 °C, with an incomplete reaction at 50 °C possibly due to partial enzyme inactivation at this elevated temperature. Therefore, one possible mechanism for how TDRD3 regulates the activity of Top3β is through binding and stabilizing single-stranded regions in plasmid substrates, and thereby recruits the enzyme to these sites.
Retention of Negative Supercoiling by TDRD3 in Plasmid DNA.
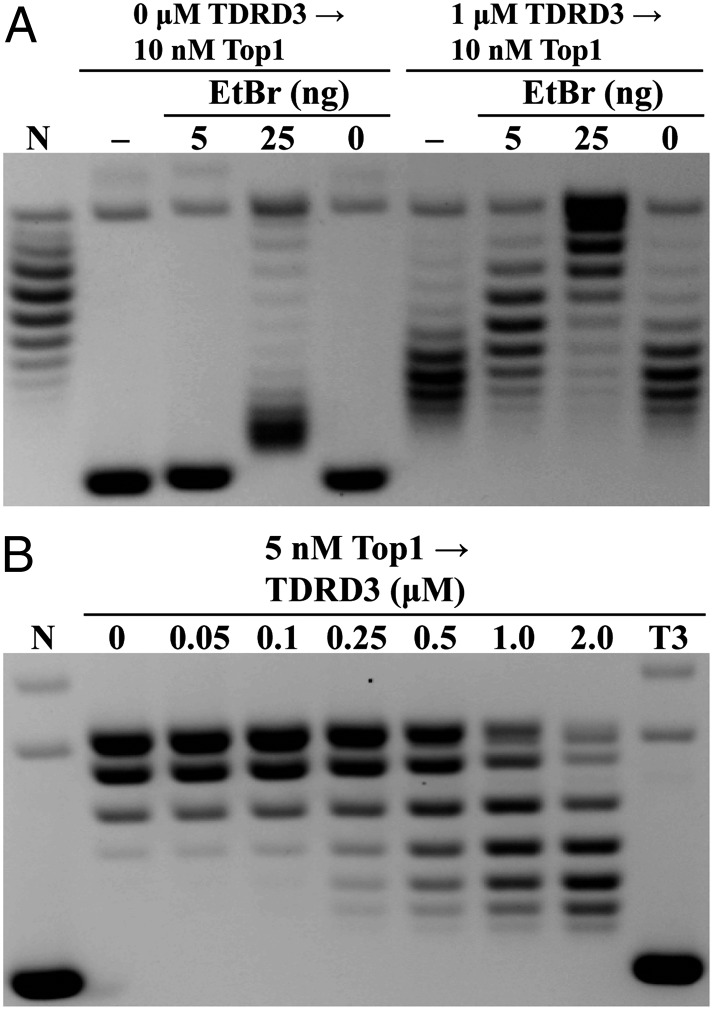
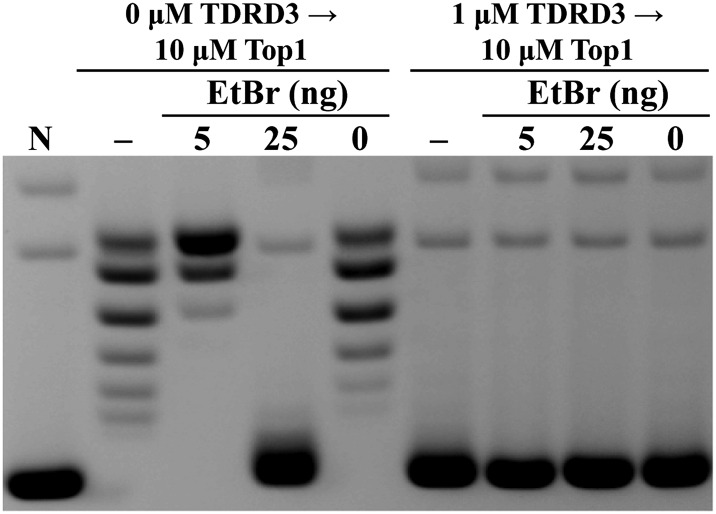
Given the proposed mechanism that TDRD3 stimulates Top3β’s activity through its binding to the single-stranded region in the plasmid DNA, one would expect it to retain negative supercoiling in the presence of a topoisomerase to remove unconstrained supercoiling. We examined whether TDRD3 could alter local DNA topology by using two approaches to assay the negative supercoiling retention (for schematic diagrams of the design of these experiments, see Fig. S1 A and B). In these experiments, we used Drosophila topoisomerase I (Top1), a type IB enzyme, which provides an efficient swivel to remove unconstrained supercoiling in the circular DNA substrates. We first used negatively supercoiled plasmid pUC19 DNA as a substrate to demonstrate the retention of plasmid negative supercoiling by TDRD3, as shown by the reduction in linking numbers of the final products, in contrast to control reactions with Top1 only (Fig. 2A). However, an alternative interpretation would be that the retention of negative supercoiling in the presence of TDRD3 was due to its inhibition of Top1 relaxation activity. To demonstrate that Top1 remained active and capable of removing supercoiling in reactions containing TDRD3, we added the DNA intercalator ethidium bromide (EtBr) to these reactions to introduce a dose-dependent unwinding of DNA. Additional negative supercoiling due to the presence of ethidium was observed in the reactions with TDRD3. These reaction products were analyzed by gel electrophoresis with chloroquine to help reveal the relative linking deficiencies among the samples, thus demonstrating the gradual reduction of linking numbers due to the presence of ethidium and the effect of the addition of TDRD3. Analysis of identical samples with gel electrophoresis without chloroquine supports the notion that TDRD3 retains DNA supercoiling and that the addition of ethidium further introduces more negative supercoiling to these DNAs (Fig. S2). These results suggest that Top1 can efficiently remove DNA supercoiling regardless of the presence of TDRD3.
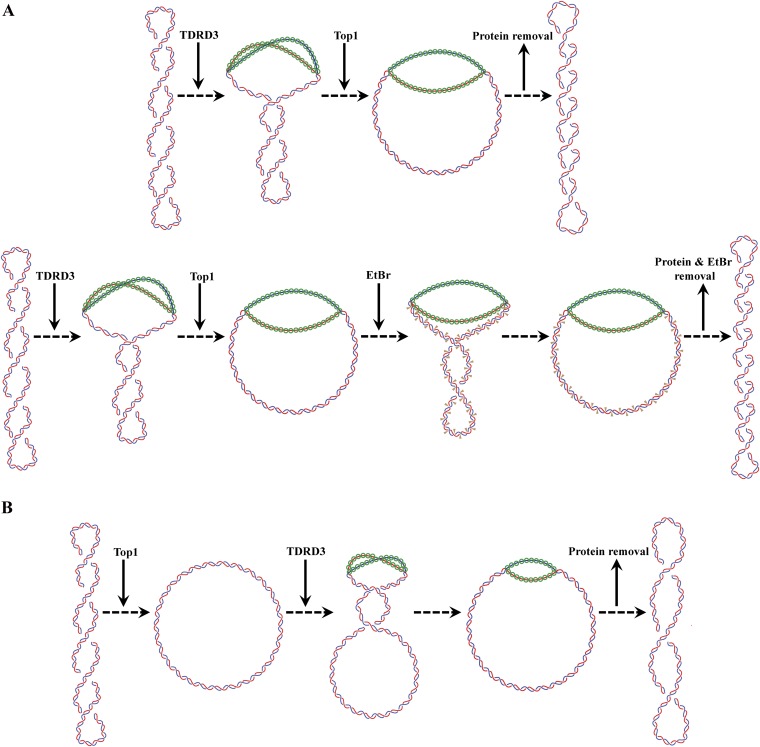
Fig. S1.
Supercoiling retention assay with negatively supercoiled (A) or relaxed (B) DNA substrates. (A) Drosophila TDRD3 was added first to the reaction for DNA binding before the addition of Drosophila Top1 to relax topologically unconstrained supercoiling. Final reaction products after protein removal have a deficiency in linking number compared with the negatively supercoiled substrate (Upper row). Addition of ethidium bromide to the reaction generates products with more linking deficiency (Lower row). (B) Top1 was added first to relax the negatively supercoiled substrate, followed by the addition of TDRD3 to bind to single-stranded regions to produce slightly positively supercoiled intermediates and further Top1 relaxation to yield the final product. Following protein removal, the final reaction product will have a slight linking deficiency compared with the fully relaxed DNA.
Fig. 2.
TDRD3 demonstrates retention of negative supercoiling in plasmid substrates. (A) Addition of TDRD3 generates final products with greater linking number deficiency than control reactions without TDRD3. The extent of negative supercoiling retention was manifested by using different concentrations of EtBr. Final reaction products were analyzed in the presence of chloroquine to distinguish differences in negative supercoiling densities (−, reaction was stopped immediately after Top1 incubation). (B) TDRD3 generates final products with linking number deficiency from Top1-relaxed pUC19 DNA, compared with control reactions with Top1 only. The extent of supercoiling retention was dependent on TDRD3 concentrations. (N, negatively supercoiled substrates; T3, 2 µM TDRD3 without Top1.)
Fig. S2.
Retention of negative supercoiling by TDRD3. Experiments were carried out under similar experimental conditions as described in Fig. 2A, except the final reaction products were analyzed with a native gel. Under these conditions, DNA products from reactions with TDRD3 all migrated with mobility comparable to the native plasmid DNA (N), consistent with the notion that they were negatively supercoiled.
To further demonstrate the potential supercoiling retention ability of TDRD3, we first used Top1 to relax the DNA, and then compared the supercoiling differences after incubating with or without TDRD3. The idea is that because there was no supercoiling in DNA before the addition of TDRD3, any change of supercoiling must be due to the effect of TDRD3 (Fig. S1B). This experiment can minimize the possible inhibitory effect of TDRD3 on Top1 relaxation activity. However, because the starting DNA substrate is relaxed and contains very few underwound regions, one would expect that the supercoiling retention by TDRD3 does not reach the same extent as was observed with negatively supercoiled DNA (Fig. 2A). This is born out in the experiments showing the dose-dependent retention of negative supercoiling (Fig. 2B). The DNA unwinding is at much lower levels compared with results using plasmid DNA as a starting substrate. These results demonstrate that, whereas TDRD3 cannot actively unwind DNA, it can bind with higher affinity at the underwound region in the plasmid DNA. Through its known interaction with Top3β, TDRD3 can thereby target Top3β to its preferred DNA structure and carry out strand passage reaction.
Preference of TDRD3 in Binding Single-Stranded Nucleic Acids.
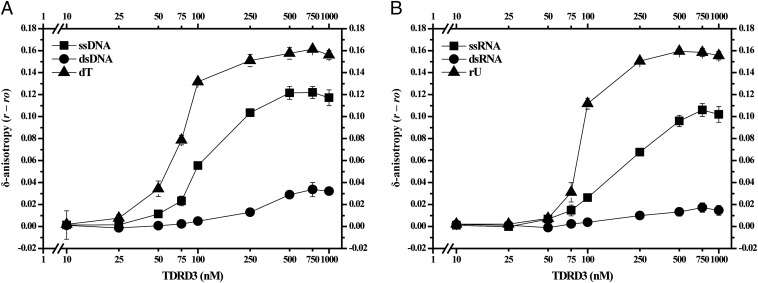
The retention of negative supercoiling assayed here by the topological shift method supports the idea that TDRD3 can preferentially associate with a single-stranded region of DNA, it does not rule out the possibility of other topological changes such as left-handed DNA wrapping associated with protein binding. We therefore directly monitored the binding affinities of TDRD3 with single- vs. double-stranded nucleic acids using fluorescence anisotropy to detect the binding to DNA and RNA oligomers tagged with a fluorophore. The temperature of the reactions was reduced from 37 °C to 25 °C to minimize fraying of double-stranded oligonucleotides. We observed anisotropy changes that were associated with the binding of TDRD3 to distinct structures of fluorescein-labeled DNA and RNA oligomers (Fig. 3 A and B). A single-stranded 73-nucleotide DNA with mixed sequence showed both a larger δ-anisotropy and a lower K1/2 (the protein concentration giving half-maximal binding) than did the same sequence in duplex form, indicating a higher affinity of TDRD3 for the single-stranded form (Fig. 3A). By the same criteria, the homopolymeric sequence (dT)73, showed higher affinity with TDRD3 than did the single-stranded DNA with a mixed sequence. When tested with an RNA oligonucleotide of similar sequence and length, we observed the same order of preference for TDRD3 binding with (rU)73 having the highest affinity and duplex RNA having the lowest (Fig. 3B). Fluorescence anisotropy experiments therefore indicated a preference for TDRD3 binding to nucleic acids with the least secondary structure, whether they are DNA or RNA. These results demonstrating that TDRD3 appears to bind RNA as well as it does to DNA, together with recent work that showed human Top3β possesses RNA topoisomerase activity (12), led us to investigate in more detail the effect of TDRD3 on the RNA topoisomerase activity of Top3β.
Fig. 3.
TDRD3 shows preference to bind single-stranded DNA and RNA oligomers. (A) TDRD3 binds to 73-bp duplex DNA, 73-nucleotide ssDNA, and (dT)73 oligomers with apparent increasing affinity; half-maximal binding (K1/2) under these conditions was seen with ∼300 nM, 110 nM, and 70 nM TDRD3 for duplex DNA, ssDNA, and the dT oligomers, respectively. (B) The binding of TDRD3 to RNA oligomers showed a pattern similar as the DNA substrates, with higher affinity for (rU)73 oligomers compared with ssRNA oligomers. Very little binding to duplex RNA was detected. Half-maximal binding was seen with ∼190 nM for ssRNA, and 90 nM TDRD3 for the rU oligomer. The average and the SDs of δ-anisotropy values for each TDRD3 concentration were calculated from triplicate measurements (n = 3).
Synthesis of Single-Stranded RNA Circles Through Self-Splicing of a Group I Intron Sequence and Its Use as a Substrate for RNA Topoisomerase.
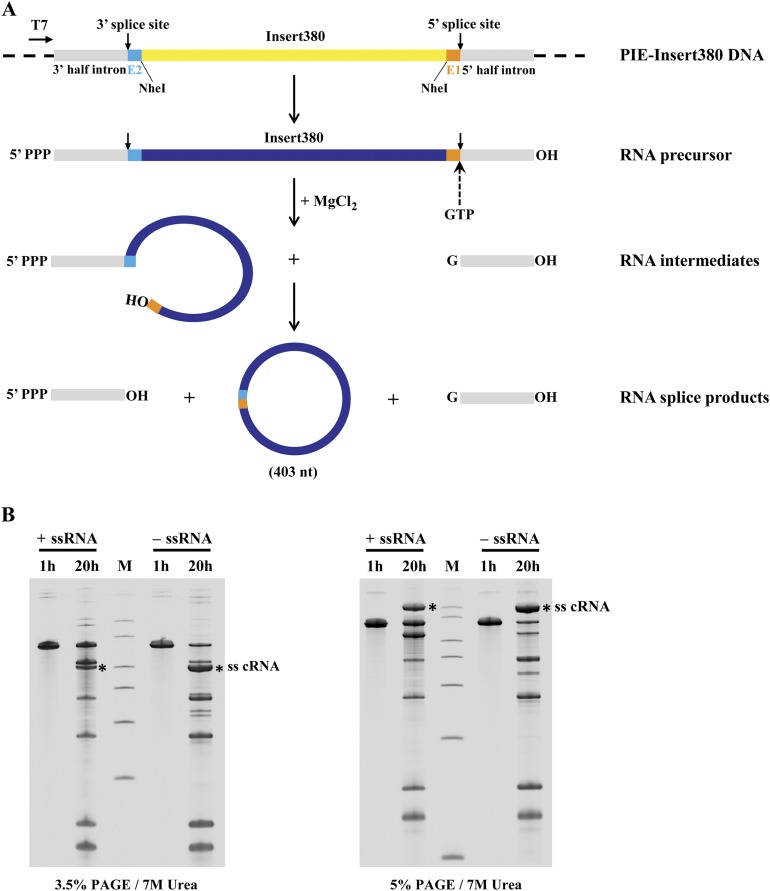
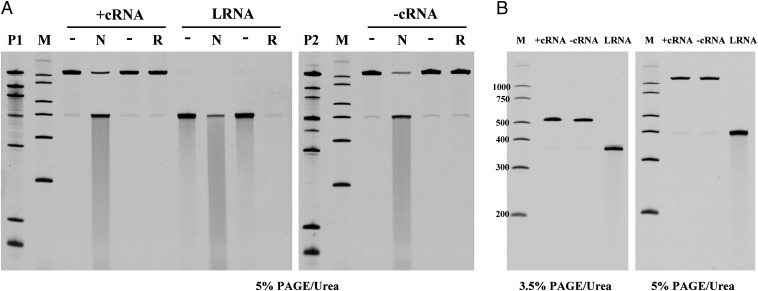
High binding affinity to single-stranded RNAs by TDRD3 suggests that it may also play a role in Top3β-mediated activity on the RNA substrate. Two groups have shown Escherichia coli Top3 and human Top3β have RNA topoisomerase activity capable of interconverting single-stranded RNA circles to knots (8, 12), but this activity depends on a substrate sequence enabling a folded secondary structure. To gain insight into the RNA topoisomerase activity catalyzed by Top3β, we prepared complementary single-stranded RNA circles that could be annealed to form a substrate with a linking number of zero. Topoisomerase activity on this substrate would be predicted to generate linked RNA circles. The single-stranded circles were generated with a reengineered self-splicing group I intron (19, 20) containing the sequence of a permuted intron–exon (PIE) version of the Anabaena pre-tRNA group I intron (21). Two constructs, PIE–insert380+ and PIE–insert380−, contain a 380-bp fragment in opposite orientation flanked by the autocatalytic splice sites (Fig. S3A). The in vitro transcripts from these two constructs can generate two single-stranded RNA circles with complementary sequences. A 676-nt-long precursor RNA was observed after a 1 h transcription and various spliced products were detected after a 20-h incubation (Fig. S3B). Candidate bands from each construct were isolated from 3.5% denaturing polyacrylamide gel (Fig. S3B) and further characterized by mild alkaline degradation, RNase R treatment, and gel mobility with respect to markers. Alkaline treatment of gel-purified circular RNA candidates (+cRNA and −cRNA) produced fragments with a discrete shift in mobility (Fig. 4A), in contrast to a smear of fragments produced from the linear form (LRNA). The nicked product of each circular RNA migrates close to the position of the corresponding LRNA marker. We further used a 3′-to-5′ exoribonuclease, RNase R, specific for the degradation of linear RNAs, to confirm its circularity. Both +cRNA and −cRNA are resistant to RNase R, whereas LRNA is completely degraded (Fig. 4A). Lastly, the electrophoretic mobility of cRNAs is expected to be more sensitive to the gel concentration than LRNA. With electrophoresis on 3.5% and 5% denaturing gels, cRNA has a much slower migration on a higher percentage (5%) gel, but the control LRNA shows mobility similar to the 400-nt marker of RNA ladder on both gel conditions (Fig. 4B). These in-vitro–generated single-stranded RNA circles validated by three independent methods are now useful as a substrate for monitoring RNA topoisomerase activity.
Fig. S3.
Generation of single-stranded RNA circles from group I permuted intron–exon (PIE) construct. (A) Self-splicing pathway based on group I PIE sequence to produce a single-stranded RNA circle. PIE–insert380 plasmid consists of two conserved splice sites, partial exons (E1 and E2), and split introns from the Anabaena group I PIE sequence, as well as a 380-bp insert at NheI cloning sites under the control of T7 promoter. PIE–insert380 generated transcript with in vitro transcription is capable of self-splicing to produce circular RNA. The 3′-OH of GTP attached to 5′-phosphate of the 5′-half intron with the release of the circular exon having a 3′, 5′-phosphodiester linkage at the 3′-splice site. Various products could form, including a 403-nt single-stranded RNA circle. (B) Time course of the production of self-spliced, single-stranded RNA circles. Full lengths of plus and minus RNA transcripts were produced 1 h after in vitro transcription, and RNA splicing products including single-stranded RNA circles (ss cRNA*) were detected at 20 h posttranscription. The mobility difference of circular RNA is exaggerated as the percentage of acrylamide increases, confirming its nonlinear structures. All RNA samples were fractionated by TBE-urea gel electrophoresis.
Fig. 4.
Splicing of PIE–insert380 and identification of single-stranded RNA circles. (A) Single-stranded circular (+cRNA or −cRNA) and LRNA were isolated from spliced products (P1 and P2). Gel-purified circular RNA candidates were treated in nicking conditions (N) or RNase R digestion (R) to validate the circular structure. With partial alkali hydrolysis, cRNA produced predominantly LRNA, compared with the smear pattern produced from LRNA. The resistance to RNase R digestion confirmed the nonlinear structure of +cRNA and −cRNA. Samples without treatments were shown as controls (−). (B) cRNA showed a larger mobility difference relative to markers (M) between 3.5% (Left) and 5% (Right) denaturing polyacrylamide gels, verifying the predicted circular structure. Control LRNA showed similar mobility on both gels.
Effect of TDRD3 on RNA Strand-Annealing Activity by Top3β.
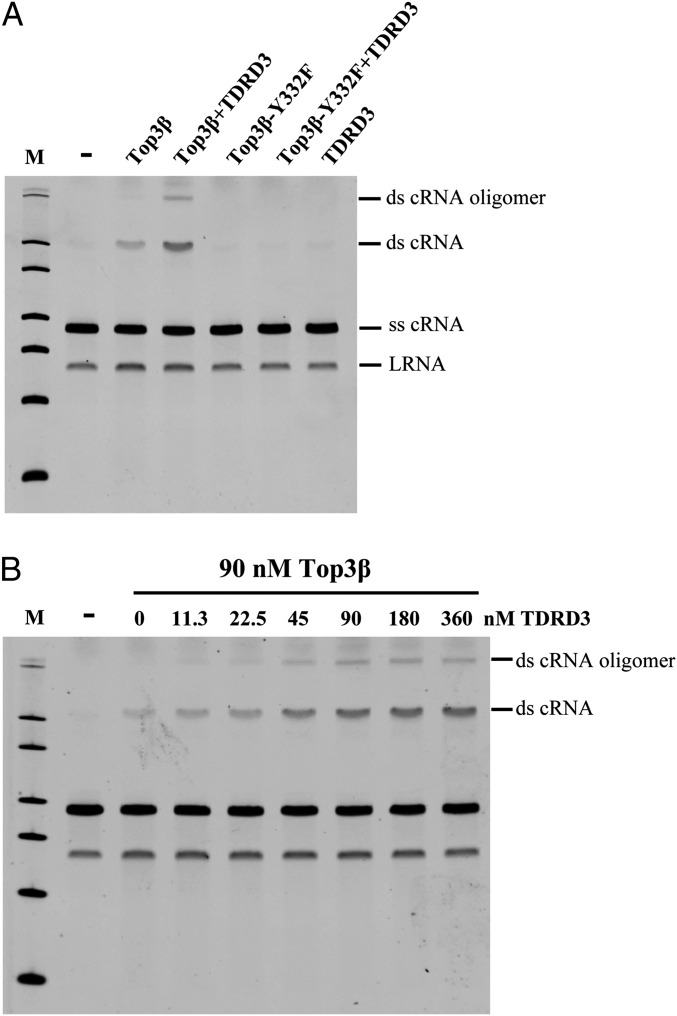
To date, the only method to assay the catalytic activity of RNA topoisomerase is to detect the interconversion of an open circle and trefoil knot (8). In this study, we designed another assay by monitoring strand annealing between two complementary single-stranded RNA circles (ss cRNA). By analyzing the reaction products from incubating RNA circles with Top3β and/or TDRD3 in a denaturing gel, we showed that Top3β is capable of generating a product band with slower migration compared with the substrates (Fig. 5A). The structure of this product was later identified as a double-stranded RNA circle (ds cRNA) by electron microscopy (Fig. 6). Moreover, the presence of TDRD3 can enhance Top3β’s activity, resulting in an increase in the formation of a double-stranded RNA circle. The active site mutant Top3β-Y332F failed to generate any products, indicating that annealing of single-stranded RNA circles depends on the strand passage activity of Top3β. The stimulation of RNA topoisomerase activity of Top3β by TDRD3 is dose dependent, with the stoichiometry reaching a plateau near 2:1, similar to its effect on DNA topoisomerase activity (Fig. 5B).
Fig. 5.
TDRD3 stimulates RNA strand-annealing activity catalyzed by Top3β. (A) Single-stranded +cRNA and −cRNA were incubated with 0 (−) or 90 nM Top3β. Top3β catalyzes the conversion of single-stranded cRNA into double-stranded cRNA, and the presence of 90 nM TDRD3 enhanced Top3β’s RNA topoisomerase activity. The active site mutant (Top3β-Y332F) showed no strand annealing activity. LRNA is a breakdown product of single-stranded RNA circles. (B) Titration of TDRD3 concentrations showed a dose-dependent stimulation in RNA strand-annealing activity catalyzed by Top3β.
Fig. 6.
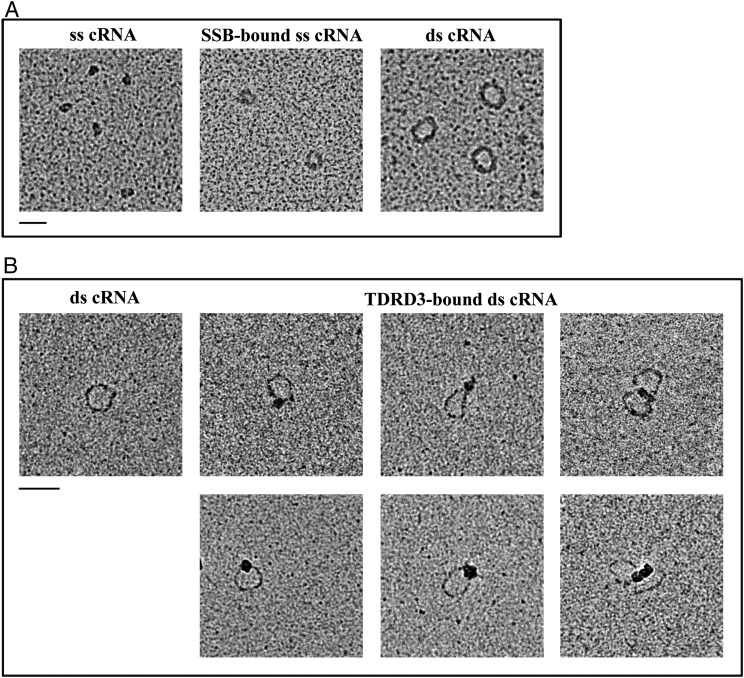
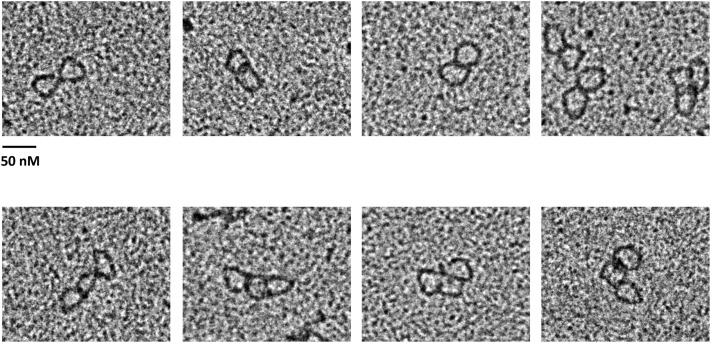
Visualization of RNA circles by electron microscopy. (A) EM images of single-stranded RNA circles (collapsed structures), partially opened single-stranded RNA circles bound by SSB, and double-stranded RNA circles generated by Top3β, shown Left, Middle, and Right, respectively. The molar ratio of ss cRNA to SSB is 1:65. (B) Double-stranded RNA circles bound by TDRD3. The molar ratio of ds cRNA to TDRD3 is 1:10. The Left image shows ds cRNA without protein. (Scale bars, 50 nm.)
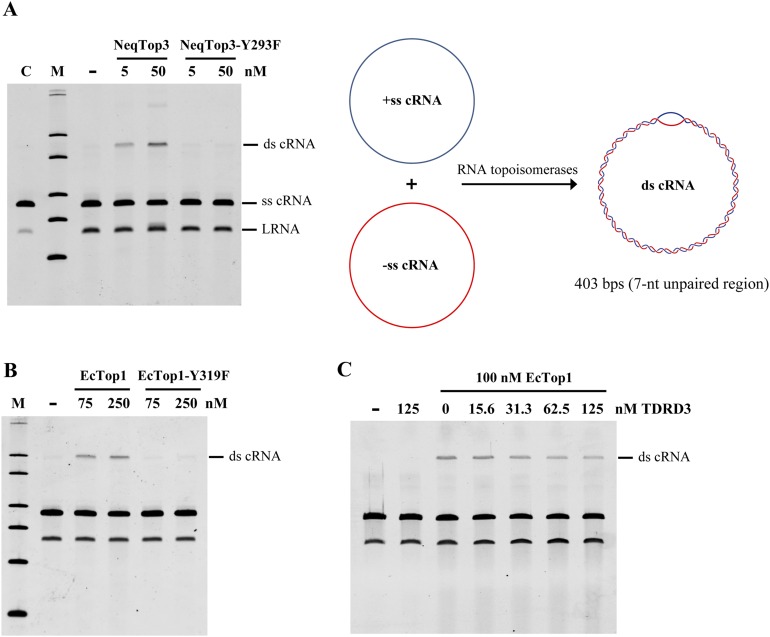
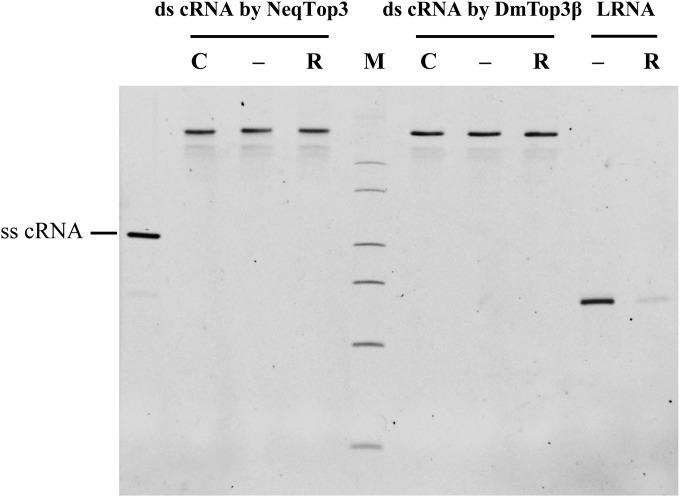
To test if this RNA annealing activity can be used to examine the RNA topoisomerase activity in other type IA enzymes, we tested with the prototype type IA topoisomerase, E. coli Top1 (EcTop1) and the newly identified hyperthermophile Nanoarchaeum equitans Top3 (NeqTop3). Both NeqTop3 and EcTop1 have ability to generate double-stranded RNA circles (Fig. S4 A and B), and again the active site mutants (EcTop1-Y319F and NeqTop3-Y293F) showed no activity. TDRD3 has no stimulatory effect on EcTop1’s activity, indicating that the specificity of interaction between TDRD3 and Top3β may be critical for its stimulation (Fig. S4C). RNA products generated by Top3β and NeqTop3 were resistant to RNase R digestion under conditions in which linear RNA was completely degraded (Fig. S5), confirming that the RNA strands remained intact and circular after treatment.
Fig. S4.
RNA strand-annealing activity of type IA topoisomerases. NeqTop3 (A) and EcTop1 (B) have similar RNA topoisomerase activity, as assayed by the conversion of single-stranded to double-stranded RNA circles (schematic diagram in A). The tyrosine mutants (NeqTop3-Y293F and EcTop1-Y319F) showed no strand-annealing activity as controls. (C) A slight inhibition of EcTop1’s activity was observed with an increasing amount of TDRD3. RNA substrates without incubation (lane C), and reaction products without adding the enzyme (−) are shown as controls.
Fig. S5.
RNase R treatment validates the circular structure of double-stranded RNA circles. Gel-purified ds cRNA generated by NeqTop3 or DmTop3β were treated with (R) or without (−) RNase R at 37 °C for 2 h to confirm the circularity of their structure. LRNA was included as a positive control to show its sensitivity to RNase R digestion, along with the RNA samples without incubation (C). All RNA samples were analyzed on a 3.5% TBE-urea PAGE without heat denaturation.
Direct Visualization of Double-Stranded RNA Circles by Electron Microscopy.
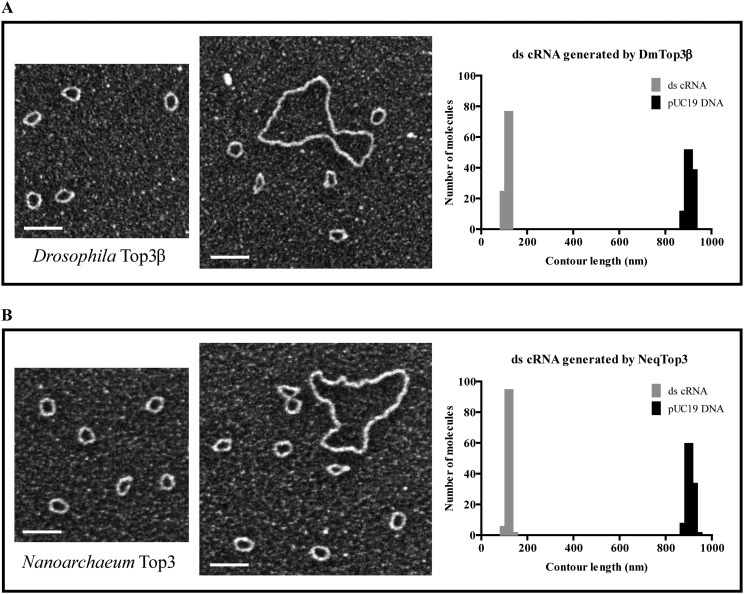
The structures of the circular RNA substrate and topoisomerase-mediated annealing products were further characterized by EM (Fig. 6A). The 403-nt-long single-stranded RNA circles appeared as discrete collapsed blobs, presumably reflecting the formation of secondary structures in RNA. In the presence of single-stranded nucleic acid binding protein (SSB), the single-stranded RNA-containing nucleoprotein complex appeared as small circles. The presumptive double-stranded circular RNA product generated by Top3β was purified from denaturing gels and visualized by cytochrome c spreading EM. Larger circles were detected with a contour length expected for double-stranded RNA (Fig. 6A and Fig. S6A). These data are consistent with the conclusion that Top3β is able to convert single-stranded RNA circles into a double-stranded form. We also examined the RNA products generated by NeqTop3, and observed that RNA circles generated by NeqTop3 are similar to those from Top3β (Fig. S6B). We measured the contour length of RNA samples, and that from pUC19 DNA as the length marker for comparison, with the circumference distribution shown in Fig. S6 A and B. The measured contour length of pUC19 with 2,686 bp is 917 nm, and those for dsRNA circles generated by Top3β and NeqTop3 are 124 nm and 128 nm, respectively, comparable to the estimated length of 113 nm based on a molecule with 403 bp and the known helical rise of A-RNA being 0.28 nm (22, 23). In addition to double-stranded RNA circles, Top3β was able to generate a higher molecular weight RNA product upon TDRD3 stimulation (Fig. 5). The structure of this product determined by EM appeared as oligomers of double-stranded RNA circles (Fig. S7), indicating that Top3β has the ability to conjoin two or more double-stranded RNA circles to form interlinked multirings. Our results showed that TDRD3 can efficiently bind to single-stranded DNA and RNA, and may be recruited to the junction of single-stranded and double-stranded regions. For the double-stranded RNA circles produced here, there is an expected 7-nt single-stranded bubble because the same splice site junction sequence is present in both senses of single-stranded circular RNA (Fig. S3). We therefore incubated double-stranded RNA circles with TDRD3, and visualized the RNA–protein complex by direct-mounting EM. We found that TDRD3 binds to a single site in an RNA circle, and also sometimes between two of them (Fig. 6B). The direct examination by EM further demonstrated that Top3β possesses an RNA topoisomerase activity to anneal two circular molecules, and this activity is promoted by TDRD3 via its association with single-stranded regions in the substrate, thus providing access to Top3β.
Fig. S6.
Dark-field EM images and size distribution of double-stranded RNA circles generated by Drosophila Top3β and N. equitans Top3. Gel-purified ds cRNA generated from Top3β (A) and NeqTop3 (B) were visualized by dark-field transmission EM. Small circles represent double-stranded RNA with big circles of pUC19 plasmid DNA as a length marker. Histograms of the size distribution of ds cRNA generated from Top3β and NeqTop3 were made from a total of 100 molecules from each sample and pUC19 DNA marker. The average contour lengths of ds cRNA from Top3β and NeqTop3 are 124 nm and 128 nm, respectively, along with the lengths of pUC19 being 916 nm and 917 nm in both samples. (Scale bars, 100 nm.)
Fig. S7.
Identification of double-stranded RNA products with a slower migration by EM. Under the stimulation of TDRD3, Top3β-generated ds cRNA with a higher molecular weight was purified from the denaturing PAGE and observed by EM using a drop-spreading method. The contour length of each single circle is around 120 nm, indicating that multiple ds cRNAs are interlocked to form dimer or oligomer structures. (Scale bar, 50 nm.)
Discussion
TDRD3 is a mediator protein known to participate in multiple nucleoprotein complexes through its interactions with key cellular proteins. It can function in two cellular compartments, the nucleus and the cytoplasm, regulating gene expression via transcription and translation (14). TDRD3 can bind to the dimethylated arginine present in core histones and RNA polymerase II, allowing it to target to the actively transcribed chromatin, especially near the promoter region (14, 15). TDRD3 can also interact with FMRP and target to mRNA (24, 25). Top3β is one of the TDRD3-interacting proteins (11–13). However, its effect on the biochemical activities of topoisomerase has not been thoroughly investigated. We show here that TDRD3 can promote both the DNA and the RNA topoisomerase activities of Top3β.
We used relaxation of hypernegatively supercoiled DNA as a sensitive and facile assay for Top3β. Earlier work demonstrated that Top3β has modest relaxation activity toward plasmid DNA (26), while maintaining robust activity for the hypernegatively supercoiled DNA (18), presumably because of the presence of underwound single-stranded regions that serve as the preferred binding site for Top3β. With this assay, we showed that TDRD3 can enhance Top3β relaxation activity and shift the reaction from a distributive to processive mode, suggesting that it can stabilize the enzyme-bound DNA complex until the excessive supercoiling is removed. This effect is likely caused by the preference of TDRD3 to bind single-stranded nucleic acids, also the known binding site for a type IA topoisomerase like Top3β. The N-terminal domain of TDRD3 harbors an intact oligonucleotide/oligosaccharide binding (OB) fold (24, 25). This motif has been proposed to bind single-stranded DNA/RNA or function as a protein-interacting interface (27, 28). Interestingly, this region had been shown to interact with Top3β (11–13); therefore, it is plausible that the OB fold of TDRD3 is responsible for stimulating the enzyme’s activities. Whereas TDRD3 is able to bind many key proteins through its OB, ubiquitin-associated (UBA), and Tudor domains (12, 13, 24, 25), the structure specificity in nucleic acids binding demonstrated here enables TDRD3 to serve more effectively as a scaffold for assembling nucleoprotein complexes. TDRD3 can associate with the promoter region of the actively transcribed genes and target Top3β to resolve R-loop structures associated with the movement of a transcriptional fork (13). Because the R loop can expose a single-stranded DNA region, it is a structure that may facilitate the recruitment of the TDRD3/Top3β complex. Depletion of TDRD3 in Tdrd3−/− primary mouse embryonic fibroblast cells can result in DNA strand breaks and the formation of γ-H2AX foci (13). Because single-stranded DNA regions are known to be vulnerable to DNA damage and strand breaks, the recruitment of TDRD3 to this region may help assemble a repair machinery for maintaining genome stability.
More than half of the cellular TDRD3 is localized in the cytoplasm (24), where it can associate with FMRP, and is present in the stress granules that are the storage compartment for mRNA during cellular stress (24, 25). TDRD3 can also associate with Top3β in cytoplasm, and the ternary complex of Top3β/TDRD3/FMRP can be localized to stress granules and mRNA in polysomes (11, 12). Because circularized messenger ribonucleoproteins (mRNPs) were found in stress-induced cellular compartments (29), Top3β and TDRD3 might be involved in compacting this complex through topological alterations of mRNA. Nevertheless, the biochemical function of Top3β/TDRD3/FMRP ternary complex remains a question of intense interest. Top3β deficiencies have been linked to schizophrenia and cognitive impairment (11) and to abnormal synaptic structures in Drosophila (12). Because Top3β has been shown to possess RNA topoisomerase activity, this ternary complex may regulate the translation of long mRNA in neuronal cells through modulating the structural complexity of RNA. Interestingly, our data here also showed that TDRD3 can function beyond being a mediator for binding to partner proteins. TDRD3 is a nucleic acid binding protein that does not discriminate between DNA and RNA and has a strong preference for single-stranded structure. This unique binding preference may enable TDRD3 to play critical roles in controlling the structure of chromatin and mRNA through its interaction with Top3β. Its single-stranded specificity can allow TDRD3 to target to the cellular compartments that require its presence: the promoter region of actively transcribed chromatin and long mRNA being translated by the passage of ribosomes. In addition, Top3β as a type IA enzyme also prefers to carry out its strand passage action at the single-stranded region, and in doing so, its activity can be enhanced through interaction with TDRD3.
There has been continuing interest in addressing the question of whether a DNA topoisomerase can also function as an RNA enzyme, and if so, how these two activities are regulated. RNA topoisomerase activity has been demonstrated in E. coli Top3 and human Top3β through the knotting of an RNA circle (8, 12) and in vaccinia Top1, human Top1, and E. coli Top3 with cleavage assays (9, 10). Because the trefoil formation assay requires the specific structure of two pairs of intramolecular base-paired regions in the RNA circle, it will be important to develop an alternative approach with a less stringent sequence requirement. We demonstrated here the application of the group I intron self-splicing sequence to generate an RNA circle and the intermolecular annealing of two circles with complementary sequences to form double-stranded circles. Because the rewinding of two circular molecules depends absolutely on the presence of an efficient swivel with strand passage activity, it provides a robust and sensitive assay for RNA topoisomerase. With this assay, we detected RNA enzyme activity in DNA topoisomerases isolated from diverse sources, including E. coli Top1, N. equitans Top3, and Drosophila Top3β. Interestingly, with Drosophila Top3β, we could also observe stimulation of RNA circle annealing activity by TDRD3. The stimulation of DNA and RNA topoisomerase activities by TDRD3 is specific to Top3β, but not other type IA enzymes such as N. equitans Top3 and E. coli Top1. A possible mechanism proposed here is, following the binding of TDRD3 to Top3β and its nucleic acids substrates via the conserved OB-fold domain, TDRD3 might be able to stabilize Top3β-bound nucleic acid complex that subsequently stimulates the enzyme’s activity. Alternatively, TDRD3 might be competing with other type IA enzymes to bind to single-stranded nucleic acids, leading to the inhibition of the enzymes’ activity. The prototype type IA topoisomerase, E. coli Top1 demonstrates this phenomenon, particularly in reactions with relatively high TDRD3 concentrations. Our results support the notion that TDRD3 and Top3β can function on chromatin in nuclei and on mRNA in cytoplasm. TDRD3 may promote these two activities by targeting the single-stranded region in nucleic acid substrates, which is the preferred site for Top3β to carry out strand passage.
With the unique RNA topoisomerase reaction described here, we have the possibility of generating double-stranded circular RNA molecules incorporating any desired sequence in these molecules. The presence of double-stranded RNA circles has not been detected in nature. However, recent results indicate that single-stranded RNA circles can be made from sequences of multiple genomic loci (30–32). Whereas the exact molecular function of the circular RNAs remains to be elucidated, there is a growing awareness of the ubiquitous nature of the presence of circular RNA and its functional importance. There is a preponderance of circular RNA in neuronal cells, and furthermore, some of these RNAs have specific intracellular location (33–35). The proposed molecular functions of circular RNAs include sponging microRNA and interacting with mRNA for the regulation of gene expression (36–39). The association and dissociation of these RNAs with a circular counterpart can be facilitated by an RNA topoisomerase, especially if the partner RNA is long and has an extensively folded structure. TDRD3-stimulated, strand-annealing activity of Top3β might be involved in this function, although the mechanism on how the proteins will be recruited and how the binding to complex RNA structures occur remain unknown. Facile methods to generate circular single-stranded and double-stranded RNA may provide avenues to investigate the molecular functions of these regulatory molecules.
Materials and Methods
Plasmid Relaxation Assays of Drosophila Top3β and TDRD3.
Hypernegatively supercoiled pUC19 DNA (300 ng) were incubated with the indicated amount of Top3β and TDRD3 in 40 mM Hepes-KOH pH 7.5, 1 mM MgCl2, and 50 μg/mL BSA at 37 °C (18) for 60 min. Reactions were stopped by adding to final concentrations of 15 mM EDTA, 0.3% SDS, and 1.0 mg/mL proteinase K, heated at 45 °C for 30 min. Electrophoresis with 1.0% Tris/borate/EDTA (TBE) agarose gel was carried out in the presence of 0.75 μg/mL EtBr for 18 h at 30 V.
Negative Supercoiling Retention Assays of Drosophila TDRD3.
Two approaches were used here with either supercoiled DNA (i) or relaxed DNA (ii) as the binding substrate. Ionic buffer conditions were based on previous studies (18, 40, 41), with modifications. (i) A total of 300 ng negatively supercoiled pUC19 DNA was incubated with 1 µM TDRD3 in 40 mM Hepes-KOH pH 7.5, 5 mM MgCl2, and 50 μg/mL BSA at 45 °C for 5 min, followed by incubation at 37 °C for 10 min after adding 10 nM Top1. Reactions were either stopped or allowed to proceed by adding either 5 or 25 ng EtBr. Following a 10-min incubation at 37 °C, reactions were stopped and phenol was extracted to separate EtBr and protein from DNA. (ii) A total of 5 nM Top1 with 300 ng negatively supercoiled pUC19 DNA was incubated in the presence of 10 mM Tris⋅HCl pH 7.9, 50 mM KCl, 10 mM MgCl2, 0.1 mM EDTA, and 50 µg/mL BSA for 5 min at 37 °C, followed by reactions at 45 °C for 10 min with TDRD3 at the indicated concentrations up to 2 µM. Reactions were stopped as mentioned above and electrophoresed on 1.0% TBE agarose gel with or without 30 µM chloroquine at 30 V for 18 h.
Fluorescence Anisotropy Assays of Drosophila TDRD3 Binding to Nucleic Acids.
The 5′-carboxyfluorescein (FAM)-labeled 73 single-stranded DNA and similar RNA oligomers (ssDNA and ssRNA), the respective unlabeled complementary strands, dT and rA oligomers were purchased from Integrated DNA Technologies. The sequence of FAM-labeled 73 ssDNA was 5′-GGACTCTGCCTCAAGACGGTAGTCAACGTGACCAGCACTGACCCATTAGGGACCGTCCACCCGACGCCTCCTG-3′. dsDNA and dsRNA oligomers were generated by annealing respective complementary ssDNA or ssRNA strands. Anisotropy measurements were performed using spectrofluorometer Fluorolog-3 (Horiba Jovin Yvon) at 25 °C for 5 min, with 50 nM substrates, titrated against various concentrations of TDRD3 up to 1 μM, in 10 mM Tris⋅HCl pH 7.9, 50 mM KCl, 10 mM MgCl2, and 0.1 mM EDTA. The average and SD of δ-anisotropy values (r − ro) from triplicate measurements for each TDRD3 concentration were plotted to show the binding pattern of each nucleic acids substrates to the protein. TDRD3 concentrations that gave half-maximal binding (K1/2) were estimated both graphically and by fitting to the Hill equation for cooperative binding. Similar values were obtained from both methods. The values in the legend to Fig. 3 are obtained from curve fitting.
RNA Strand-Annealing Assays.
The assay contained 15 ng of single-stranded +cRNA and –cRNA, 40 mM Hepes-KOH (pH 7.5), 5 mM MgCl2, 50 μg/mL BSA, 7.5 mM DTT, and Top3β or TDRD3 with indicated concentrations. Reactions were performed at 37 °C for 1 h and stopped with 10 mM EDTA, 0.2% SDS, and 1 mg/mL proteinase K at 45 °C for 30 min. For EcTop1 and EcTop1-Y319F mutant, reactions were performed in 10 mM Tris⋅HCl (pH 8.0), 50 mM NaCl, 100 μg/mL gelatin, and 6 mM MgCl2 at 37 °C for 1 h. Reactions for NeqTop3 and NeqTop3-Y293F mutant were incubated at 56 °C for 0.5 h in 10 mM Tris⋅HCl (pH 8.0), 50 mM KCl, 10 mM MgCl2, 0.1 mM EDTA, and 50 μg/mL gelatin. All samples were denatured with formamide in loading buffer (95% formamide, 18 mM EDTA, 0.025% SDS, and xylene cyanol) at 90 °C for 4 min and run on 3% TBE-urea polyacrylamide gels at 150 V for 1 h followed by SYBR Gold staining (Invitrogen).
Electron Microscopy.
Gel-purified double-stranded RNA circles were examined by the cytochrome c drop-spreading method (42) and processed for tungsten shadowing. For visualization of TDRD3 and ds cRNA, purified ds cRNA were incubated with TDRD3 at a molar ratio of 1:10 in 40 mM Hepes-KOH (pH 7.5) and 1 mM MgCl2 at 37 °C for 20 min, fixed by 0.4% glutaraldehyde at room temperature for 5 min, followed by adding a final concentration of 100 mM Tris⋅HCl (pH 7.5) to terminate the fixation. The RNA–protein complex was incubated with spermidine buffer (2 mM spermidine HCl, 10 mM Tris⋅HCl (pH 7.5), 50 mM NaCl, 75 mM KCl, 2 mM MgCl2, and 0.5 mM CaCl2), mounted directly on glow-discharged carbon-coated 400-mesh copper grids (EM Sciences), then washed with a water/ethanol series, followed by rotary shadowcast with tungsten (43). Grids were imaged with a FEI Tecnai G2 F20 TEM instrument at 120 kV and a Gatan US1000 digital camera. To visualize ss cRNA, RNA was incubated with extreme thermostable SSB (New England Biolabs) at 50 °C for 20 min followed by fixation, gel filtration, and direct mounting.
SI Materials and Methods
Enzymes.
Wild-type and catalytic-dead mutant (Y332F) Drosophila Top3β proteins were expressed and purified as described previously (18, 40) except that single-stranded DNA-agarose resin was replaced with heparin–sepharose. Recombinant Drosophila Top1 was purified following a published procedure (44). N. equitans Top3 (NeqTop3) and its mutant (NeqTop3-Y293F) were purified with a published method (45). E. coli Top1 (EcTop1) and the mutant (EcTop1-Y319F) were provided by Yuk-Ching Tse-Dinh (Florida International University). All restriction endonucleases were purchased from New England Biolabs. RNase R was from Epicentre. MEGAscript T7 kit and TURBO DNase were from Ambion.
Expression and Purification of Drosophila TDRD3.
Drosophila TDRD3 protein-encoding region was cloned in pET23a vector (Novagen) and transformed into BL21(DE3)pLysS cells. Cells were grown at 37 °C in LB media with protein expression induced with 1 mM isopropyl β-d-thiogalactopyranoside and further grown at 18 °C for 16 h before harvesting. All purification buffers contained 1 mM phenylmethylsulfonyl fluoride, and 2.5 mM 2-mercaptoethanol. Cell pellets were resuspended in 50 mM Tris⋅HCl pH 7.9, 10 mM imidazole, 500 mM NaCl, 10% glycerol, 0.1 mg/mL lysozyme, and protease inhibitor mixture (Roche), and lysate was sonicated and centrifuged. Soluble fraction was loaded onto a 5-mL cobalt HiTrap TALON column for AKTA fast protein liquid chromatography (FPLC) (GE Healthcare). Protein was gradient eluted over 15 column volumes (CVs) from 20 to 500 mM imidazole in 50 mM Tris⋅HCl pH 7.9, 500 mM NaCl, and 10% glycerol. Peak fractions with TDRD3 eluted at 50 mM imidazole were pooled, diluted fivefold, and applied to a 5-mL sulfopropyl cation exchanger HiTrap SP column. Gradient elution over 6 CVs from 100 to 1,000 mM NaCl was performed with a buffer of 50 mM Tris⋅HCl pH 7.9, 100 mM NaCl, and 10% glycerol. TDRD3 peak fractions that eluted at 350 mM NaCl were pooled and fractionated with a 120-mL Superdex 200 10/300 GL column with a buffer of 50 mM Tris⋅HCl pH 7.9, 500 mM NaCl, and 5% glycerol. Peak fractions containing purified TDRD3 were pooled and stored in small aliquots at −80 °C.
Plasmid DNA Substrates.
Negatively supercoiled pUC19 DNA was purified from DH5α bacteria, with twice cesium chloride-ethidium bromide (CsCl-EtBr) equilibrium density ultracentrifugation, butanol extraction, and extensive dialysis in 10 mM Tris⋅HCl/0.1 mM EDTA (TE) buffer for storage. Hypernegatively supercoiled pUC19 DNA substrates (18) were prepared by incubating plasmid DNA with Drosophila Top1 in the presence of ethidium bromide (DNA base pair to ethidium bromide ratio of 2:1). The enzyme was then inactivated and the ethidium bromide was removed by organic extraction, and the DNA recovered by ethanol precipitation.
Permuted Intron–Exon Sequence (PIE) Constructs.
The plasmid PIE–insert380(+) containing the Anabaena pre-tRNA group I intron was modified from pRNP–RNA (21, 46). PIE–insert380(−) is similar to PIE–insert380(+) except with the complementary sequence as insert.
Preparation of Single-Stranded RNA Circles.
Single-stranded RNA precursor was transcribed using a MEGAscript T7 kit from a PIE–insert380 DNA template linearized with BamHI. The overnight transcripts were treated with two units of TURBO DNase at 37 °C for 2 h to remove DNA template. Because the buffer for in vitro transcription contains GTP and MgCl2, the self-splicing reaction occurred spontaneously after RNA precursor formation. RNA splicing products were analyzed by electrophoresis on a 3.5% TBE polyacrylamide gel containing 7 M urea. Gel slice containing single-stranded RNA circle was soaked in a buffer containing 500 mM NH4Ac (pH 8.0), 0.1% SDS, 1 mM EDTA, and 10 mM Mg(Ac)2 at 4 °C overnight. RNA was further purified by ethanol precipitation and resuspended in water. Plus and minus single-stranded RNA circles (+cRNA and −cRNA) were produced from the DNA templates of PIE–insert380(+) and PIE–insert380(−), respectively.
Nicking Assay.
A total of 75 ng of gel-purified single-stranded +cRNA and −cRNA was treated in the alkali nicking buffer (75 mM NaHCO3, pH 9, and 1 mM EDTA) at 90 °C for 3 min. After treatment, RNA samples were denatured in formamide loading buffer and analyzed by electrophoresis in 5% TBE-urea polyacrylamide gels.
RNase R Resistance Assay.
A total of 50 ng of gel-purified single-stranded +cRNA and −cRNA were treated with or without 4 units of RNase R at 37 °C for 1.5 h. The reactions were terminated by adding stop solution at 45 °C for 30 min and analyzed by 5% TBE-urea polyacrylamide gels. A total of 3 ng of gel-purified double-stranded RNA circles were digested with 10 units of RNase R at 37 °C for 2 h followed by electrophoresis in a 3.5% TBE-urea polyacrylamide.
Acknowledgments
We thank Dr. Yuk-Ching Tse-Dinh (Florida International University) for wild-type and mutant (Y319F) EcTop1 proteins and Hung-Hsun Shuai for technical assistance. This work was supported by Ministry of Science and Technology Taiwan Grant MOST-104-2321-B-001-012 and Academia Sinica intramural funding AS-105-TP-B04 (to T.-s.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1605517113/-/DCSupplemental.
References
- 1.Wang JC. Untangling the Double Helix. DNA Entanglement and the Action of the DNA Topoisomerases. Cold Spring Harbor Lab Press; Cold Spring Harbor, NY: 2009. [Google Scholar]
- 2.Wang JC. Cellular roles of DNA topoisomerases: A molecular perspective. Nat Rev Mol Cell Biol. 2002;3(6):430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- 3.Chen SH, Chan NL, Hsieh TS. New mechanistic and functional insights into DNA topoisomerases. Annu Rev Biochem. 2013;82:139–170. doi: 10.1146/annurev-biochem-061809-100002. [DOI] [PubMed] [Google Scholar]
- 4.Wang JC. Interaction between DNA and an Escherichia coli protein omega. J Mol Biol. 1971;55(3):523–533. doi: 10.1016/0022-2836(71)90334-2. [DOI] [PubMed] [Google Scholar]
- 5.Champoux JJ, Dulbecco R. An activity from mammalian cells that untwists superhelical DNA: A possible swivel for DNA replication (polyoma-ethidium bromide-mouse-embryo cells-dye binding assay) Proc Natl Acad Sci USA. 1972;69(1):143–146. doi: 10.1073/pnas.69.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tse YC, Kirkegaard K, Wang JC. Covalent bonds between protein and DNA. Formation of phosphotyrosine linkage between certain DNA topoisomerases and DNA. J Biol Chem. 1980;255(12):5560–5565. [PubMed] [Google Scholar]
- 7.Schoeffler AJ, Berger JM. DNA topoisomerases: Harnessing and constraining energy to govern chromosome topology. Q Rev Biophys. 2008;41(1):41–101. doi: 10.1017/S003358350800468X. [DOI] [PubMed] [Google Scholar]
- 8.Wang H, Di Gate RJ, Seeman NC. An RNA topoisomerase. Proc Natl Acad Sci USA. 1996;93(18):9477–9482. doi: 10.1073/pnas.93.18.9477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sekiguchi J, Shuman S. Site-specific ribonuclease activity of eukaryotic DNA topoisomerase I. Mol Cell. 1997;1(1):89–97. doi: 10.1016/s1097-2765(00)80010-6. [DOI] [PubMed] [Google Scholar]
- 10.DiGate RJ, Marians KJ. Escherichia coli topoisomerase III-catalyzed cleavage of RNA. J Biol Chem. 1992;267(29):20532–20535. [PubMed] [Google Scholar]
- 11.Stoll G, et al. Deletion of TOP3β, a component of FMRP-containing mRNPs, contributes to neurodevelopmental disorders. Nat Neurosci. 2013;16(9):1228–1237. doi: 10.1038/nn.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu D, et al. Top3β is an RNA topoisomerase that works with fragile X syndrome protein to promote synapse formation. Nat Neurosci. 2013;16(9):1238–1247. doi: 10.1038/nn.3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang Y, et al. Arginine methylation facilitates the recruitment of TOP3B to chromatin to prevent R loop accumulation. Mol Cell. 2014;53(3):484–497. doi: 10.1016/j.molcel.2014.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang Y, et al. TDRD3 is an effector molecule for arginine-methylated histone marks. Mol Cell. 2010;40(6):1016–1023. doi: 10.1016/j.molcel.2010.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sims RJ, 3rd, et al. The C-terminal domain of RNA polymerase II is modified by site-specific methylation. Science. 2011;332(6025):99–103. doi: 10.1126/science.1202663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bhakar AL, Dölen G, Bear MF. The pathophysiology of fragile X (and what it teaches us about synapses) Annu Rev Neurosci. 2012;35:417–443. doi: 10.1146/annurev-neuro-060909-153138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Darnell JC, et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146(2):247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilson TM, Chen AD, Hsieh T. Cloning and characterization of Drosophila topoisomerase IIIbeta. Relaxation of hypernegatively supercoiled DNA. J Biol Chem. 2000;275(3):1533–1540. doi: 10.1074/jbc.275.3.1533. [DOI] [PubMed] [Google Scholar]
- 19.Cech TR, Bass BL. Biological catalysis by RNA. Annu Rev Biochem. 1986;55:599–629. doi: 10.1146/annurev.bi.55.070186.003123. [DOI] [PubMed] [Google Scholar]
- 20.Cech TR. Self-splicing of group I introns. Annu Rev Biochem. 1990;59:543–568. doi: 10.1146/annurev.bi.59.070190.002551. [DOI] [PubMed] [Google Scholar]
- 21.Puttaraju M, Been MD. Group I permuted intron-exon (PIE) sequences self-splice to produce circular exons. Nucleic Acids Res. 1992;20(20):5357–5364. doi: 10.1093/nar/20.20.5357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arnott S, Hukins DW, Dover SD. Optimised parameters for RNA double-helices. Biochem Biophys Res Commun. 1972;48(6):1392–1399. doi: 10.1016/0006-291x(72)90867-4. [DOI] [PubMed] [Google Scholar]
- 23.Gast FU, Hagerman PJ. Electrophoretic and hydrodynamic properties of duplex ribonucleic acid molecules transcribed in vitro: Evidence that A-tracts do not generate curvature in RNA. Biochemistry. 1991;30(17):4268–4277. doi: 10.1021/bi00231a024. [DOI] [PubMed] [Google Scholar]
- 24.Goulet I, Boisvenue S, Mokas S, Mazroui R, Côté J. TDRD3, a novel Tudor domain-containing protein, localizes to cytoplasmic stress granules. Hum Mol Genet. 2008;17(19):3055–3074. doi: 10.1093/hmg/ddn203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Linder B, et al. Tdrd3 is a novel stress granule-associated protein interacting with the Fragile-X syndrome protein FMRP. Hum Mol Genet. 2008;17(20):3236–3246. doi: 10.1093/hmg/ddn219. [DOI] [PubMed] [Google Scholar]
- 26.Seki T, Seki M, Onodera R, Katada T, Enomoto T. Cloning of cDNA encoding a novel mouse DNA topoisomerase III (Topo IIIbeta) possessing negatively supercoiled DNA relaxing activity, whose message is highly expressed in the testis. J Biol Chem. 1998;273(44):28553–28556. doi: 10.1074/jbc.273.44.28553. [DOI] [PubMed] [Google Scholar]
- 27.Theobald DL, Mitton-Fry RM, Wuttke DS. Nucleic acid recognition by OB-fold proteins. Annu Rev Biophys Biomol Struct. 2003;32:115–133. doi: 10.1146/annurev.biophys.32.110601.142506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Flynn RL, Zou L. Oligonucleotide/oligosaccharide-binding fold proteins: A growing family of genome guardians. Crit Rev Biochem Mol Biol. 2010;45(4):266–275. doi: 10.3109/10409238.2010.488216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anderson P, Kedersha N. RNA granules: Post-transcriptional and epigenetic modulators of gene expression. Nat Rev Mol Cell Biol. 2009;10(6):430–436. doi: 10.1038/nrm2694. [DOI] [PubMed] [Google Scholar]
- 30.Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7(2):e30733. doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jeck WR, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19(2):141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9(9):e1003777. doi: 10.1371/journal.pgen.1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rybak-Wolf A, et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell. 2015;58(5):870–885. doi: 10.1016/j.molcel.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 34.Westholm JO, et al. Genome-wide analysis of drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Reports. 2014;9(5):1966–1980. doi: 10.1016/j.celrep.2014.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.You X, et al. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat Neurosci. 2015;18(4):603–610. doi: 10.1038/nn.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ashwal-Fluss R, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56(1):55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 37.Hansen TB, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 38.Memczak S, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 39.Zhang Y, et al. Circular intronic long noncoding RNAs. Mol Cell. 2013;51(6):792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 40.Wilson-Sali T, Hsieh TS. Generation of double-stranded breaks in hypernegatively supercoiled DNA by Drosophila topoisomerase IIIbeta, a type IA enzyme. J Biol Chem. 2002;277(30):26865–26871. doi: 10.1074/jbc.M204641200. [DOI] [PubMed] [Google Scholar]
- 41.Wilson-Sali T, Hsieh TS. Preferential cleavage of plasmid-based R-loops and D-loops by Drosophila topoisomerase IIIbeta. Proc Natl Acad Sci USA. 2002;99(12):7974–7979. doi: 10.1073/pnas.122007999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thresher R, Griffith J. Electron microscopic visualization of DNA and DNA-protein complexes as adjunct to biochemical studies. Methods Enzymol. 1992;211:481–490. doi: 10.1016/0076-6879(92)11026-f. [DOI] [PubMed] [Google Scholar]
- 43.Griffith JD, Christiansen G. Electron microscope visualization of chromatin and other DNA-protein complexes. Annu Rev Biophys Bioeng. 1978;7:19–35. doi: 10.1146/annurev.bb.07.060178.000315. [DOI] [PubMed] [Google Scholar]
- 44.Shaiu WL, Hsieh TS. Targeting to transcriptionally active loci by the hydrophilic N-terminal domain of Drosophila DNA topoisomerase I. Mol Cell Biol. 1998;18(7):4358–4367. doi: 10.1128/mcb.18.7.4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee SH, Siaw GE, Willcox S, Griffith JD, Hsieh TS. Synthesis and dissolution of hemicatenanes by type IA DNA topoisomerases. Proc Natl Acad Sci USA. 2013;110(38):E3587–E3594. doi: 10.1073/pnas.1304103110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Puttaraju M, Beebe JA, Niranjanakumari S, Been MD, Fierke CA. Generation and characterization of circular Bacillus subtilis RNase P RNA; activation by RNase P protein. Nucleic Acids Symp Ser. 1995;(33):92–94. [PubMed] [Google Scholar]