Significance
The solute carrier (SLC) 4 transporters are membrane proteins that control bicarbonate transport in human red blood cells and regulate borate transport in plants and yeast. Previously, one member of the SLC4 family, human Band 3, had its crystal structure determined, which showed it in an outward-open state. We report here what is, to our knowledge, the second crystal structure of an SLC4 protein, the plant borate transporter Bor1. Critically, the structure is in an occluded state open to neither side of the membrane. Because it is in a new state, we are able to compare our model with other related structures and deduce structural transitions that provide alternating access to both sides of the membrane for Bor1 and related transporters.
Keywords: X-ray structure, SLC4 transporter, Bor1, Band 3, membrane protein
Abstract
Boron is essential for plant growth because of its incorporation into plant cell walls; however, in excess it is toxic to plants. Boron transport and homeostasis in plants is regulated in part by the borate efflux transporter Bor1, a member of the solute carrier (SLC) 4 transporter family with homology to the human bicarbonate transporter Band 3. Here, we present the 4.1-Å resolution crystal structure of Arabidopsis thaliana Bor1. The structure displays a dimeric architecture in which dimerization is mediated by centralized Gate domains. Comparisons with a structure of Band 3 in an outward-open state reveal that the Core domains of Bor1 have rotated inwards to achieve an occluded state. Further structural comparisons with UapA, a xanthine transporter from the nucleobase-ascorbate transporter family, show that the downward pivoting of the Core domains relative to the Gate domains may access an inward-open state. These results suggest that the SLC4, SLC26, and nucleobase-ascorbate transporter families all share an elevator transport mechanism in which alternating access is provided by Core domains that carry substrates across a membrane.
The defining feature of transporters is the ability to carry specific molecules across a membrane. The solute carrier (SLC) group comprises a diverse array of transporters grouped into at least 52 families based on function and sequence homology (1). The SLC4 family is termed the bicarbonate transporters and is subdivided into sodium-coupled cotransporters and anion exchanger subclasses. The SLC4 anion exchangers transport ions in an electroneutral manner, most commonly transporting bicarbonate in exchange for chloride. In addition to bicarbonate transporters, the SLC4 transporters include borate efflux transporters, originally discovered in plants (2, 3). Boron is an essential plant micronutrient that is taken up from the soil and participates in the formation of esters found in plant cell walls. Specifically, borate diesters cross-link a primary cell wall component, pectic polysaccharide rhamnogalacturonan II (RG-II) and, thus, contribute to plant cell wall stability (4, 5). In excess levels, however, boron is toxic to plants. The regulation of boron by transporters is therefore important for plant viability and has implications for worldwide agriculture. Indeed, there are ongoing efforts to engineer plants that are tolerant of either high or low boron levels in soil (6–8). The transport and regulation of boron levels is regulated partly by Bor1, a boron exporter that loads xylem, such that boron is transported from roots to shoots and leaves (3). The precise chemical nature boron takes during transport is not known, but is commonly assumed to be borate, an anionic form of boric acid. Bor1 is active in plants under limiting borate conditions, but is degraded under high concentrations of borate to avoid accumulation of toxic boron levels in plant shoots (9). Although the transporter function and regulation of Bor1 in response to excess borate have been defined, the mechanism by which Bor1 transports borate, and which ions it couples to transport, remains unclear.
The archetypal SLC4 anion exchanger is Band 3, also known as SLC4A1 or anion exchanger 1 (AE1). Band 3 is the most abundant membrane protein in human red blood cells (10), and reversibly exchanges bicarbonate and chloride ions in an electroneutral manner. In tissues, CO2 diffuses into red blood cells and is converted to bicarbonate, which is exported in exchange for chloride ions. In lungs, the partial pressure of CO2 is lower and the process is reversed, thus driving cellular respiration. Decades of biochemical characterization have provided a wealth of information about Band 3 topology and multimerization (11–15), as well as the identification of amino acid residues likely involved in substrate transport (16–23). However, our understanding of transport by SLC4 anion exchangers remains limited by a paucity of structural data, for the SLC4 family in general and Bor1 in particular. Recently, the first crystal structure of the transporter domain of an SLC4 protein, human Band 3, was reported in an outward-open state (24). To better understand the structural transitions that control substrate translocation by SLC4 transporters, we determined the structure of C-terminally truncated Arabidopsis thaliana Bor1 (residues 1–645) in a previously unobserved state. Like the Band 3 structure, Bor1 is a dimer, with each monomer comprised of two domains, the Core and the Gate, and dimerization mediated by the Gate domains. Unlike Band 3, however, we observe Bor1 in an occluded configuration, in which the Core domains have rotated inward toward the Gate domains. Our structure helps define the conformational landscape used by SLC4 transporters in the course of a transport cycle.
Results
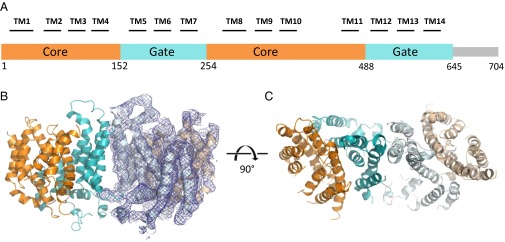
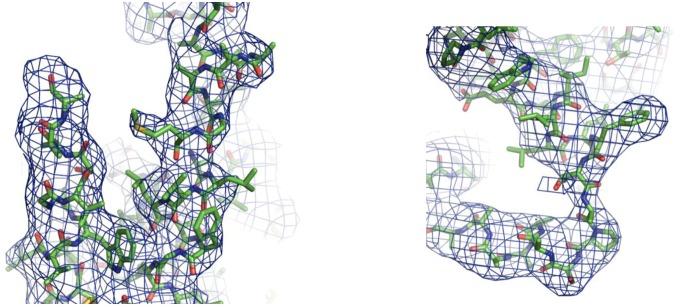
A. thaliana Bor1 (AtBor1) was overexpressed and purified from Saccharomyces cerevisiae. Initially, crystals of the full-length 704-residue AtBor1 could be grown but with diffraction limited to ∼7-Å resolution. One impediment to determining a structure of any macromolecular complex is the presence of natively unfolded regions. Secondary structure predictors suggested that the C-terminal region of AtBor1 may be unfolded, which led us to make a series of C-terminal truncations and test for their effect on crystal diffraction. Superior crystals were obtained with a construct, termed AtBor11–645, in which the last 59 residues were removed. AtBor11–645 crystallized in space group P42212 and permitted the collection of diffraction data to 4.1-Å resolution. The data are anisotropic and extend to an overall resolution of 4.1 × 4.1 × 5.4 Å (25). Subsequent molecular replacement searches using the Gate and the Core domains of human Band 3 produced a single molecular replacement solution with a monomer in the asymmetric unit, and this solution recapitulated the dimer around a crystallographic twofold axis, further validating the solution. After restrained refinement in Refmac5 (26), the model was refined to an Rwork/Rfree of 35.9/39.1% with good stereochemistry (Methods, Fig. 1, and Table 1). We were able to build 399 of 645 residues, or 62% of the primary sequence. The defined structure begins at residue 33 and ends at residue 586. It contains the 14 expected transmembrane helices (TMs), with several loops connecting them left out because they could not be reliably fit to electron density. The most significant difference in sequence between AtBor1 and Band 3 is that Bor1 has a nearly 100-residue insertion between TMs 10 and 11, which appears disordered in our structure. The loops we were able to fit to electron density were built as poly-alanine segments. To show our structure is as free from model bias as possible, we calculated a simulated annealing composite omit map, which shows continuous electron density for all 14 TM helices (Fig. 1B). The 2Fo–Fc electron density is observable for some of the bulkiest side chains (Fig. S1).
Fig. 1.
Overview of Bor1 structure. (A) Schematic of AtBor1 construct. The Core domain is indicated in orange, and the Gate domain in teal. Approximate positions of TM helices are indicated by black lines and numbered. (B) Side view of Bor1 dimer, with cytoplasmic side on the bottom. One monomer is surrounded by a simulated annealing composite omit map, contoured at 1.0 σ. (C) Top-down view of Bor1 dimer, rotated 90° from B.
Table 1.
Data collection and refinement statistics
| Data collection and refinement | AtBor11–645 |
| Data collection | |
| Space group | P42212 |
| Cell dimensions | |
| a, b, c (Å) | 184.56, 184.56, 89.83 |
| α, β, γ (°) | 90, 90, 90 |
| Resolution (Å) | 20–4.1 |
| Rpim | 0.046 (0.555) |
| I/σ(I) | 19.3 (1.1) |
| CC1/2 | 0.998 (0.619) |
| Completeness (%) | 96.3 (87.6) |
| Redundancy | 12.0 (7.7) |
| Refinement | |
| Resolution (Å) | 20–4.1 |
| No. of reflections | 11,319 |
| Rwork/Rfree | 35.9/39.1 |
| No. of atoms | |
| Protein | 2,873 |
| B factors | |
| Protein | 257.9 |
| Rmsd | |
| Bond lengths (Å) | 0.011 |
| Bond angles (°) | 1.384 |
The reported data are merged from two isomorphous crystals. Values in parentheses are for the highest-resolution shell.
Fig. S1.
Representative electron density shows features for some side chains. Displayed in blue is a 2Fo–Fc map, contoured at 1.0 σ. Most residue side chains are not visible, although some of the most electron-dense residues, such as Trp, Phe, and Met residues displayed here, appear surrounded by density and aided with registry placement.
Each Bor1 monomer recapitulates a fold seen in Band 3 (24), and in the more distantly related nucleobase-ascorbate transporter (NAT) proteins UraA and UapA (27, 28). Additionally, the SLC26 family has been observed to display the same overall fold, as shown in a recent structure of SLC26Dg (29). As in those structures, Bor1 consists of two distinct domains, the Gate and the Core. The Gate comprises six TMs (5–7 and 12–14, residues 152–254 and 489–586), and provides the entire dimerization interface, which buries 731 Å2 of surface area per monomer. The Core comprises eight TMs (1–4 and 8–11, residues 33–151 and 291–486) and contains the putative substrate-binding site (Fig. 1).
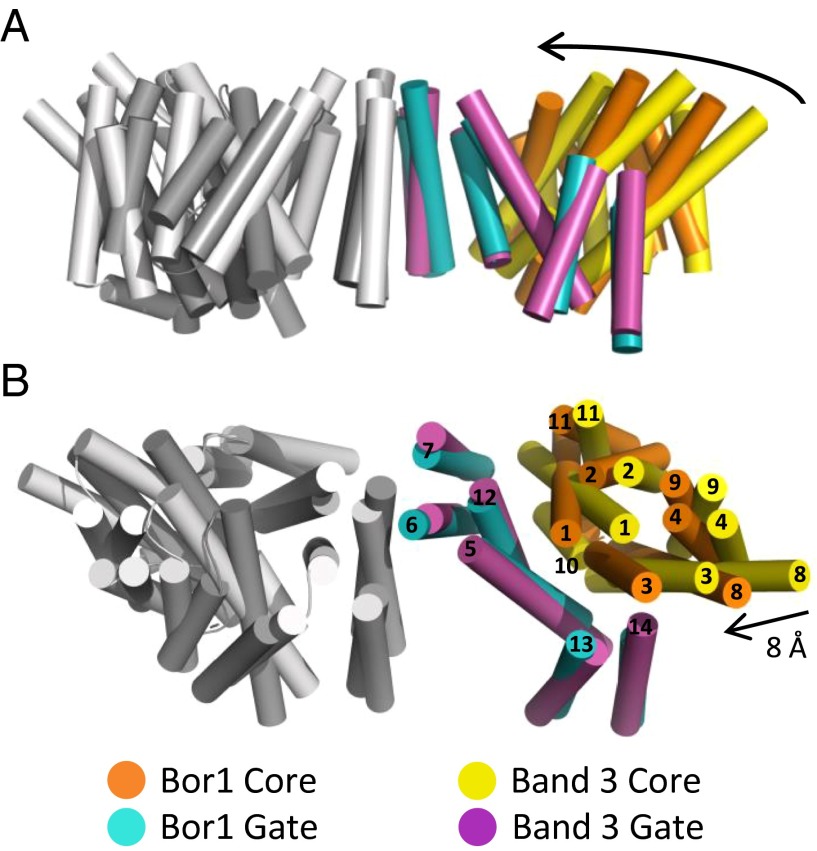
Comparisons of large-scale structural rearrangements of Bor1 domains are surprisingly informative about the quaternary motions that Bor1 is likely to undergo in the course of a transport cycle. A superposition of Bor1 and Band 3 reveals that the TMs of the Gate domains are in essentially the same position (Cα rmsd = 1.6 Å), whereas the Core domains of Bor1 are rotated inward toward the dimer symmetry axis (Fig. 2). In particular, the Core TM helices most proximal to the Gate domain—TMs 1, 3, and 8—are rotated inward toward the Gate domain and downward toward intracellular side. The extracellular-facing ends of those three helices each moved inward toward the Gate by ∼8 Å. Because of this observed rigid-body movement, Bor1 is not in an outward-open state as Band 3, but rather occupies an occluded state.
Fig. 2.
Comparison with Band 3 shows Core domains rotate relative to Gate domains. Side view (A) and (B) top-down view of Bor1 and Band 3. One monomer each of Bor1 and Band 3 are in gray. The other monomer of Bor1 and Band 3 are each represented as cylinders and colored as indicated. For clarity, only the 14 TM helices are displayed. Black arrows indicate inward motion of Band 3 Core domain (yellow) relative to the Bor1 Core (orange).
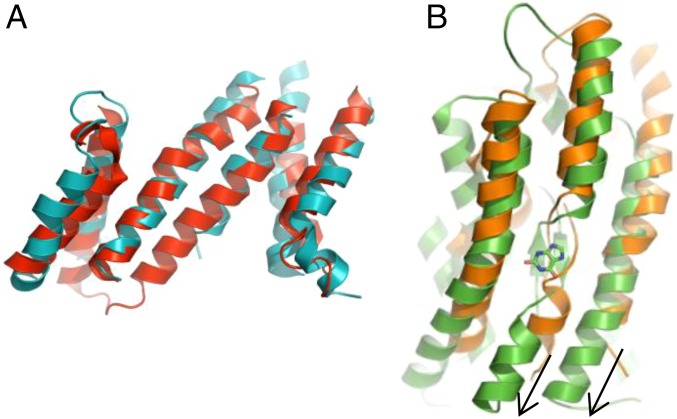
It has been previously observed that the distantly related NAT family shares a similar fold with SLC4 transporters despite sharing only ∼10% sequence identity. Whereas the Band 3 structure was determined in an outward-open state, NAT transporters UapA and UraA were determined in inward-open states with substrates bound (27, 28). Superposition of the TMs of the Gate domains of Bor1 and UapA shows a reasonably close alignment, with a Cα rmsd of 3.1 Å (Fig. 3A). The positioning of the Core domains with respect to the Gates, however, reveals further conformational differences with Bor1 (Fig. 3B). In Bor1, the Core is rotated “down” toward the cytoplasmic side relative to Band 3, whereas in UapA, the Core is further down toward the cytoplasmic side relative to Bor1. The putative substrate-binding site moves downward toward the intracellular side by ∼5 Å. Because of this vertical transition, the substrate-binding site of UapA is solvent-exposed and, thus, in an inward-open state. The superpositions of Bor1 with the outward-open Band 3 and the inward-open UapA thus appear sufficient to explain how the Core domains might move relative to the Gate domains to provide alternating access.
Fig. 3.
Comparison with open-inward UapA shows downward movement of Core domain. (A) Superposition of Bor1 (teal) and UapA (red) Gate domains, viewpoint from the Core. (B) Differences between Bor1 (orange) and UapA (green) Core domains, viewpoint from the Gate. The substrate bound to UapA, xanthine, is shown in green sticks. Black arrows show the downward movement of TM8 and TM10 of UapA relative to Bor1.
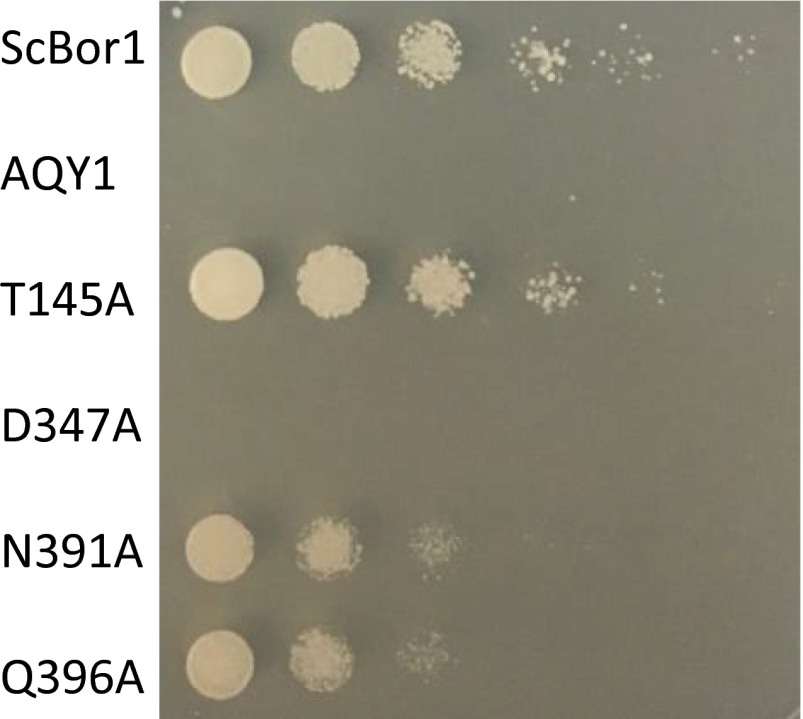
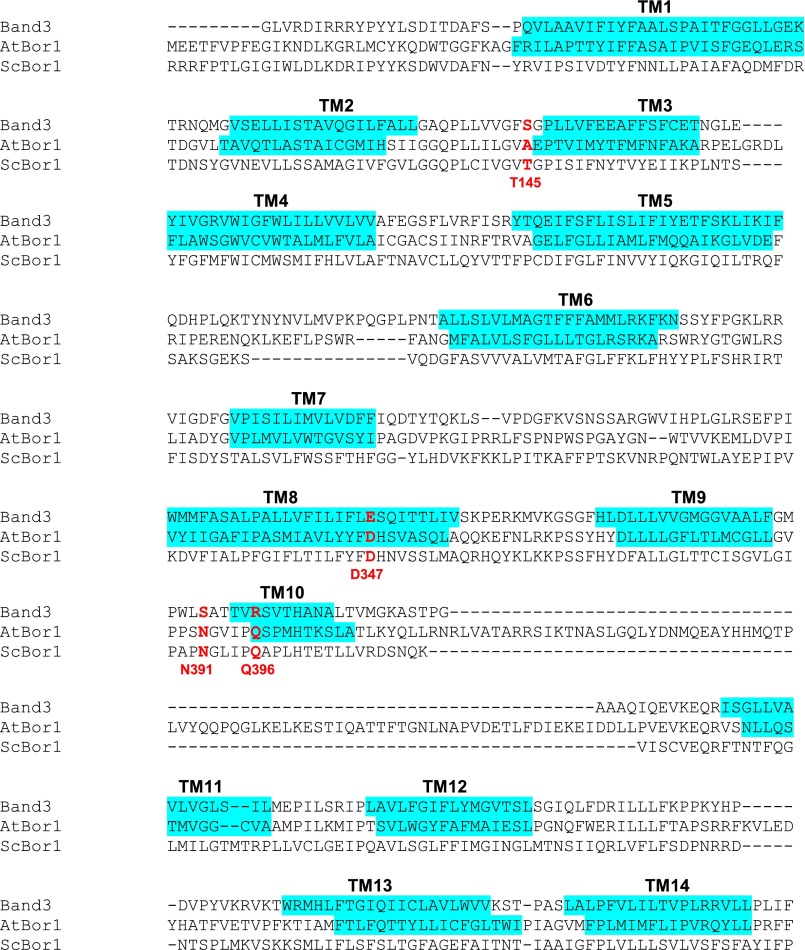
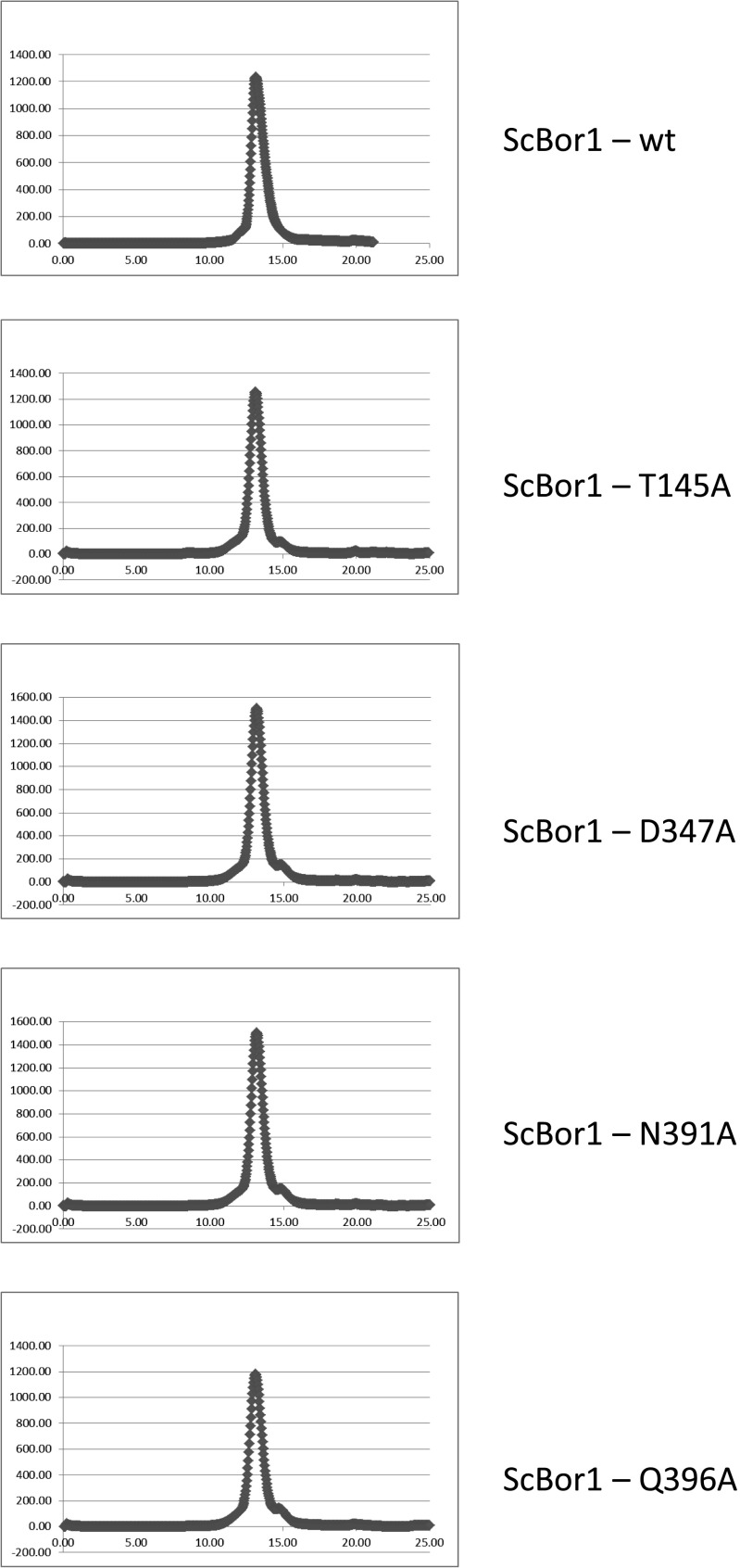
There is no ligand bound in our structure, but as in the case with Band 3 (24), UraA (27), UapA (28), and SLC26Dg (29), the substrate-binding site likely resides where the ends of the shortened TM helices TM3 and TM10 pass each other. Although our model was improved by keeping most side chains present, the resolution precludes commenting specifically on their conformations and contacts. To determine experimentally which residues might be involved in borate transport, we used a genetic complementation assay challenging the growth of S. cerevisiae on plates supplemented with boric acid. Yeast are a boron-tolerant organism, and the expression of S. cerevisiae Bor1 has been shown to enable yeast growth when challenged with boric acid (30). BOR1 was deleted in S. cerevisiae and shown to be complemented by transforming with the wild-type BOR1 (Fig. 4). Consistent with previous studies (7), transforming with AtBor1 fails to complement and rescue growth; this is suspected to be because AtBor1 exports borate at lower concentrations (9), which are too low to cause toxicity to S. cerevisiae. Analysis of the Band 3 structure, in conjunction with a multiple sequence alignment of human Band 3, ScBor1, and AtBor1 enabled identification of residues in Bor1 possibly involved in substrate binding (Fig. S2). Because we could not complement with AtBor1, we mutated the homologous residues in ScBor1: T145, D347, N391, and Q396. Upon mutating these residues to alanine, complementation assays show that the D347A mutant completely abolishes growth (Fig. 4). D347 is homologous to E681 in human Band 3, a residue critical for substrate transport (17, 18, 20). Additionally, the N391A and Q396A mutants reduce growth relative to BOR1. A T145A mutant, however, does not impact growth relative to BOR1. Importantly, wild-type ScBor1 and all mutants could be expressed and purified under identical conditions, and showed no abnormal migration behavior by size-exclusion chromatography (Fig. S3), indicating that the loss of complementation displayed by the mutants is not due to defects in expression, folding, aggregation, or sorting to the plasma membrane. Thus, the data suggest that the ScBor1 Core domain residues D347, N391, and Q396 are important for substrate transport. These results do not preclude the possibility that other residues in either the Gate or Core domains could be involved in substrate binding and transport; rather, the complementation data represent experimental identification of residues in a borate transporter that may be important for transport activity.
Fig. 4.
Complementation assay identifies Core residues involved in borate transport. S. cerevisiae BOR1 serves as the positive control, and the aquaporin AQY1 is the negative control. Among BOR1 mutants tested, D347A completely eliminates growth, T145A grows essentially as effective as wild-type BOR1, and N391A and Q396A are reduced relative to WT BOR1 (all yeast numbering).
Fig. S2.
Sequence alignment of HsBand3, AtBor1, and ScBor1. TM helices are highlighted in cyan. Marked in red are four residues identified from the Band 3 structure as possible substrate-interacting side chains, subsequently mutated to alanine in ScBor1 (ScBor1 numbering) for complementation assays.
Fig. S3.
Size-exclusion chromatography for ScBor1 and mutants tested for complementation. All mutants express and migrate similarly to the wild-type protein.
Discussion
The alternating access mechanism for transport was proposed by Jardetzky 50 y ago (31). The underlying idea is that a substrate binds a cavity from one side of the membrane, triggers a conformational change, and then exits from a cavity facing the other side of the membrane. This basic idea has stood up remarkably well over time. Transporters can be grouped into three basic mechanisms: rocker switch, rocking bundle, and elevator (32). The rocker switch mechanism operates much as Jardetzky described, with two sides of a transporter moving around an immobile substrate-binding site. In the rocking bundle model, the substrate-binding site remains immobile while one domain of the protein moves around a less labile domain. The difference between an elevator transporter mechanism and a rocking-bundle transport model can be subtle. Three hallmark signs of a bona fide elevator mechanism are the following: (i) a relatively rigid, immobile scaffolding domain; (ii) a mobile carrier domain that contains all or nearly all substrate binding; and (iii) a vertical displacement of the substrate-binding site (32). The NAT transporter UapA has been proposed to function as an elevator transporter (28). Bor1, and the SLC4 family in general, also appear to meet these requirements. Both Bor1 and Band 3 structures were determined in the absence of substrate. However, UraA and UapA each were determined with the presence of their substrates, uracil and xanthine, respectively. In both cases, contacts with ligand are mediated by residues in the Core domain. Although the Band 3 structure did not have substrate bound, its likely substrate-coordinating residues are suspected through structural comparisons with UraA and UapA and mutagenesis studies (17, 18, 20, 24). They, along with the Bor1 residues identified through genetic assays we describe here, also belong solely to the Core domain. The data collectively suggest that substrates in SLC4, SLC26, and NAT transporters are bound by the Core domain and not the Gate domain.
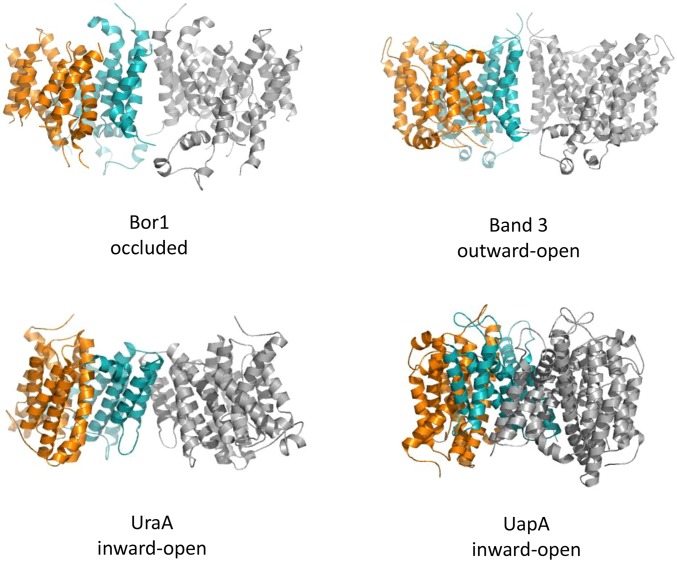
The identity of the Gate as a mostly rigid scaffolding domain also appears to fulfill the description of an elevator mechanism. The NAT transporter UraA was the first protein of this fold to have its structure determined. It also served as the origin of the naming of the Gate and Core domains, which have since been adopted in Band 3, UapA, and now Bor1. The UraA structure was reported as a monomer, which is present in its asymmetric unit. However, crystal symmetry of that structure shows that UraA, too, dimerizes through its Gate domain (Fig. S4), a feature that is better appreciated now with the subsequent addition of other structures to the literature.
Fig. S4.
Dimeric assemblies for Bor1, Band 3, UraA, and UapA. In each case, one monomer is in gray, whereas the other is displayed with its Gate domain in teal and its Core domain in orange.
The SLC26 family also shares a similar fold to UraA, UapA, Bor1, and Band 3. Unlike the other structures, the fumarate transporter SLC26Dg crystallized as a monomer (29). However, prior biochemical studies of other members of the SLC26 family suggest the family is ordinarily comprised of dimers (33, 34). Dimerization by the Gate domain thus appears to be conserved among each of the SLC4, SLC26, and NAT transporter families. It is unclear whether each of the two Cores of Bor1 may move around the Gates independently of one another, or whether there is cooperativity between the two. However, transport studies of Band 3 show that one monomer may transport while the other is blocked by an inhibitor, suggesting that Band 3 monomers operate independently (35, 36). Additionally, in the case of the trimeric amino acid transporter and elevator transporter archetype Gltph, individual subunits sample states independently of each other (37–39).
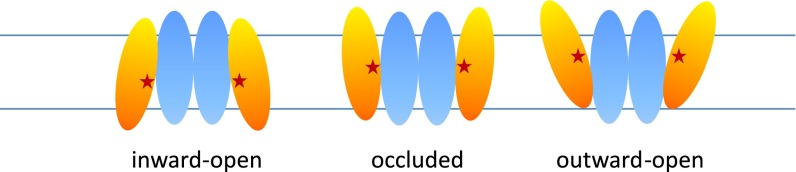
The combination of Core domain structural rearrangements and substrate binding residues together suggest that the SLC4, SLC26, and NAT families all use a conserved elevator transport mechanism (Fig. S5). In this scheme, the Core domains can move such that they are open to either the extracellular or intracellular sides, whereas the Gate domains remain relatively static. The vertical displacement of the substrate-binding site in the Bor1 occluded state to either the open-outward or open-inward states is approximately 5 Å each, or 10 Å total vertical displacement between the inward- and outward-facing states. This change is not as large as the 18 Å observed in the trimeric elevator transporter Gltph (40), or the 15 Å in the model of transport by VcINDY (41). Rather, a 10-Å change compares more with the 10-Å vertical displacement observed in the sodium/proton dimeric exchanger NapA, which is also proposed to function as an elevator transporter (42). Thus, the available evidence suggests that the SLC4, SLC26, and NAT family transporters all share a conserved elevator transport mechanism.
Fig. S5.
Elevator transport model for SLC4 anion exchangers. Three main states have been identified for SLC4 transporters. In the inward-open state, substrates (red stars) have access to the binding site in the Core domain (yellow). The Core domains vertically pivot through an occluded state to an outward-open state, at which point the substrate may exit the cell. The Gate domains (blue) remain rigid.
Methods
Protein Expression and Purification.
A 2-μm plasmid S. cerevisiae expression construct based on p423 GAL1 contained nucleotides coding for the A. thaliana borate transporter Bor1 (UniProt ID: Q8VYR7) with a C-terminal deca-histidine tag preceded by a thrombin cleavage site. Transformed S. cerevisiae (strain DSY-5) were grown at 30 °C in CSM-His to OD600 of ∼10. Protein expression was induced by the addition of 8% (wt/vol) galactose dissolved in 4× yeast extract-peptone media, to a final galactose concentration of 2%. Cells were harvested after 16 h shaking at 30 °C by spinning at 3,500 × g for 15 min. Yeast pellets were resuspended in a buffer containing 50 mM Tris pH 7.0, 1 mM EDTA, and 1 mM phenylmethylsulfonyl fluoride for protease inhibition. Cells were lysed by bead beating with 0.5-mm glass beads for six 1-min pulses separated by 2-min rest periods. The glass beads were filtered from the homogenate and washed with a 2× buffer for a final lysis buffer of 50 mM Tris pH 7.0, 700 mM NaCl, 10% glycerol, 1 mM EDTA, and 1 mM PMSF. The homogenate was centrifuged for 25 min at 18,000 × g, followed by sedimentation of membranes by ultracentrifugation at 185,000 × g for 150 min. Membranes were resuspended in 50 mM Tris pH 7.0, 500 mM NaCl, and 10% glycerol and frozen at −80 °C.
Membrane pellets were solubilized by the addition of 225 mg of n-dodecyl-β-d-maltoside (DDM) per gram of membrane. Critically, membranes were always resuspended in a volume of 15 mL of buffer per gram of membrane, such that 22 5mg of DDM per gram of membrane was a concentration of 1.5% DDM by wt/vol. Membranes were solubilized with a stir bar for 60 min at 4 °C. Unsolubilized material was removed by ultracentrifugation for 142,000 × g for 20 min. Imidazole pH 8.0 was added to 20 mM, and the sample was loaded onto a preequilibrated 5-mL Ni-NTA column by using a peristaltic pump. After loading, the column was washed with 50 mL containing 20 mM imidazole, and with another 50 mL containing 80 mM imidazole. Protein was eluted in a buffer containing 20 mM Tris pH 7.0, 200 mM Na2SO4, 10% glycerol, 250 mM imidazole, and 0.01% lauryl maltose neopentyl glycol (LMNG). The sample was concentrated by using 100-kDa cutoff Amicon concentrators and buffer exchanged to remove the imidazole. One-hundred units of bovine thrombin were added to remove the deca-His tag overnight at 4 °C. The following day, the sample was loaded onto a 5-mL Ni-NTA column and the flow-through containing cleaved protein was collected, concentrated, and loaded onto an S200 gel filtration column equilibrated in 20 mM Mes pH 6.5, 100 mM Na2SO4, and 0.01% LMNG. Peak fractions were collected and concentrated. A typical yield was approximately 1.5 mg per 1 L of starting media.
Crystallization and Structure Determination.
Crystals were grown at 20 °C by vapor diffusion by mixing 250 nL of 3–4 mg/mL protein with 100 nL of reservoir containing 9–11% (wt/vol) polyethylene glycol 3500, 200–350 mM Li2SO4 and 100 mM sodium citrate pH 5.6–6.0. Bipyramidal crystals with a final size of approximately 200 × 200 × 200 µm were obtained after 2–3 d of crystal growth. The three steps that most improved X-ray diffraction resolution were removing the last 59 C-terminal residues to make the 1–645 construct, switching detergent from DDM to LMNG, and dehydrating crystals. To dehydrate crystals, first the crystals were cryoprotected in a mother liquor solution supplemented with 25–30% glycerol. After looping the crystal, dehydration was achieved by holding the loop exposed to air for 10 s before flash-freezing in liquid nitrogen. Each of the three unit cell dimensions decreased by approximately 5%. Data were collected at the Advanced Light Source beamline 8.3.1. Datasets were processed by using HKL2000 in space group P42212. Molecular replacement was performed with PHASER and obtained a single solution when using two search components comprised of the Gate and Core domains of human Band 3 (PDB ID code: 4YZF) (24), which possesses 26% sequence identity and 56% sequence similarity to AtBor11–645. Iterative model building in Coot (43) and refinement in Refmac5 (26) gradually improved the model as judged by map quality and R factors. Refmac5 was run with jelly-body refinement (σ = 0.03), and with secondary structural restraints turned on. Because our data are low resolution, three modeling strategies were attempted: the human Band 3 starting solution, a poly-alanine model, and a model comprised of the A. thaliana Bor1 sequence based on the solution of Band 3 and modeled by Robetta (44). Judging by map quality and R factors, the Robetta model was the best fit. A multiple sequence alignment of Bor1 and Band 3 with secondary structure elements mapped onto them additionally guided the building and residue assignment (Fig. S2). The Fo–Fc difference density and composite omit maps permitted the building of some, but not all, of the missing loops, which were modeled as poly-alanine. The final model yielded a crystallographic R factor of 35.9% and a free R factor of 39.1%. MolProbity evaluation of the Ramachandran plot gave 88.0% in favored regions and 1.8% outliers. The overall MolProbity score of 2.33 is in the 99th percentile among proteins in comparable resolution (45). All structural figures were prepared by using PyMOL (46). For comparisons of Bor1 with either Band 3 or UapA, the Gate domains were superposed (i.e., aligned based on structure, and in a sequence-independent manner) by using Pymol. The composite structures were then aligned with the Gate domains to compare Core domain movements relative to the Gate domains.
Complementation Assay.
BOR1 was deleted through one-step integration of knockout cassettes (47), resulting in a strain with the following genotype: MATalpha leu2 trp1-1 ura3-52 his3::GAL1-GAL4 pep4 prb1-1122 bor1 Δ::Kanmx. Cells were transformed with the same plasmid used for overexpression of protein, bearing HIS3 for selection and under inducible expression by the Gal1 promoter. The only difference in plasmids used in the experiment was whether it contained the negative control of aquaporin AQY1 or a mutation in BOR1 as indicated in Fig. 4. A single colony was picked and grown overnight in CSM-His media containing 2% raffinose. Ten microliters of cells were plated on CSM-His plates supplemented with 2% raffinose, 0.1% galactose, and 20 mM boric acid. Samples started at OD 0.5 and decreased by fivefold serial dilutions. Results were recorded after 5 d at 30 °C.
Acknowledgments
We thank J. Holton and G. Meigs for assistance with synchrotron data collection at the Advanced Light Source; and Janet Finer-Moore, Alex Kintzer, Jonny Leano, Yi-Liang Liu, Pawel Dominik, and James Fraser for critical reading of the manuscript. This work was supported by University of California Office of the President; Multicampus Research Programs and Initiatives Grant MR‐15-338599; the Program for Breakthrough Biomedical Research, which is partially funded by the Sandler Foundation, for support of beamline 8.3.1; and National Institutes of Health Grant R37 GM024485. B.H.T-S. was supported by an Alumni Fellow-sponsored Life Sciences Research Foundation fellowship.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 5L25).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1612603113/-/DCSupplemental.
References
- 1.Hediger MA, Clémençon B, Burrier RE, Bruford EA. The ABCs of membrane transporters in health and disease (SLC series): Introduction. Mol Aspects Med. 2013;34(2-3):95–107. doi: 10.1016/j.mam.2012.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Noguchi K, et al. bor1-1, an Arabidopsis thaliana mutant that requires a high level of boron. Plant Physiol. 1997;115(3):901–906. doi: 10.1104/pp.115.3.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takano J, et al. Arabidopsis boron transporter for xylem loading. Nature. 2002;420(6913):337–340. doi: 10.1038/nature01139. [DOI] [PubMed] [Google Scholar]
- 4.Kobayashi M, Matoh T, Azuma J. Two chains of Rhamnogalacturonan II are cross-linked by borate-diol ester bonds in higher plant cell walls. Plant Physiol. 1996;110(3):1017–1020. doi: 10.1104/pp.110.3.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O’Neill MA, Eberhard S, Albersheim P, Darvill AG. Requirement of borate cross-linking of cell wall rhamnogalacturonan II for Arabidopsis growth. Science. 2001;294(5543):846–849. doi: 10.1126/science.1062319. [DOI] [PubMed] [Google Scholar]
- 6.Miwa K, Takano J, Fujiwara T. Improvement of seed yields under boron-limiting conditions through overexpression of BOR1, a boron transporter for xylem loading, in Arabidopsis thaliana. Plant J. 2006;46(6):1084–1091. doi: 10.1111/j.1365-313X.2006.02763.x. [DOI] [PubMed] [Google Scholar]
- 7.Miwa K, et al. Plants tolerant of high boron levels. Science. 2007;318(5855):1417. doi: 10.1126/science.1146634. [DOI] [PubMed] [Google Scholar]
- 8.Mosa KA, et al. Enhanced boron tolerance in plants mediated by bidirectional transport through plasma membrane intrinsic proteins. Sci Rep. 2016;6(February):21640. doi: 10.1038/srep21640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takano J, Miwa K, Yuan L, von Wirén N, Fujiwara T. Endocytosis and degradation of BOR1, a boron transporter of Arabidopsis thaliana, regulated by boron availability. Proc Natl Acad Sci USA. 2005;102(34):12276–12281. doi: 10.1073/pnas.0502060102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fairbanks G, Steck TL, Wallach DF. Electrophoretic analysis of the major polypeptides of the human erythrocyte membrane. Biochemistry. 1971;10(13):2606–2617. doi: 10.1021/bi00789a030. [DOI] [PubMed] [Google Scholar]
- 11.Steck TL. Cross-linking the major proteins of the isolated erythrocyte membrane. J Mol Biol. 1972;66(2):295–305. doi: 10.1016/0022-2836(72)90481-0. [DOI] [PubMed] [Google Scholar]
- 12.Steck TL, Ramos B, Strapazon E. Proteolytic dissection of band 3, the predominant transmembrane polypeptide of the human erythrocyte membrane. Biochemistry. 1976;15(5):1153–1161. doi: 10.1021/bi00650a030. [DOI] [PubMed] [Google Scholar]
- 13.Reithmeier RA. Fragmentation of the band 3 polypeptide from human erythrocyte membranes. Size and detergent binding of the membrane-associated domain. J Biol Chem. 1979;254(8):3054–3060. [PubMed] [Google Scholar]
- 14.Lux SE, John KM, Kopito RR, Lodish HF. Cloning and characterization of band 3, the human erythrocyte anion-exchange protein (AE1) Proc Natl Acad Sci USA. 1989;86(23):9089–9093. doi: 10.1073/pnas.86.23.9089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Casey JR, Reithmeier RAF. Analysis of the oligomeric state of Band 3, the anion transport protein of the human erythrocyte membrane, by size exclusion high performance liquid chromatography. Oligomeric stability and origin of heterogeneity. J Biol Chem. 1991;266(24):15726–15737. [PubMed] [Google Scholar]
- 16.Jennings ML, Anderson MP. Chemical modification and labeling of glutamate residues at the stilbenedisulfonate site of human red blood cell band 3 protein. J Biol Chem. 1987;262(4):1691–1697. [PubMed] [Google Scholar]
- 17.Jennings ML, Smith JS. Anion-proton cotransport through the human red blood cell band 3 protein. Role of glutamate 681. J Biol Chem. 1992;267(20):13964–13971. [PubMed] [Google Scholar]
- 18.Tang XB, Fujinaga J, Kopito R, Casey JR. Topology of the region surrounding Glu681 of human AE1 protein, the erythrocyte anion exchanger. J Biol Chem. 1998;273(35):22545–22553. doi: 10.1074/jbc.273.35.22545. [DOI] [PubMed] [Google Scholar]
- 19.Müller-Berger S, et al. Roles of histidine 752 and glutamate 699 in the pH dependence of mouse band 3 protein-mediated anion transport. Biochemistry. 1995;34(29):9325–9332. doi: 10.1021/bi00029a007. [DOI] [PubMed] [Google Scholar]
- 20.Chernova MN, et al. Electrogenic sulfate/chloride exchange in Xenopus oocytes mediated by murine AE1 E699Q. J Gen Physiol. 1997;109(3):345–360. doi: 10.1085/jgp.109.3.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karbach D, Staub M, Wood PG, Passow H. Effect of site-directed mutagenesis of the arginine residues 509 and 748 on mouse band 3 protein-mediated anion transport. Biochim Biophys Acta - Biomembr. 1998;1371(1):114–122. doi: 10.1016/s0005-2736(98)00010-8. [DOI] [PubMed] [Google Scholar]
- 22.Jin XR, Abe Y, Li CY, Hamasaki N. Histidine-834 of human erythrocyte band 3 has an essential role in the conformational changes that occur during the band 3-mediated anion exchange. Biochemistry. 2003;42(44):12927–12932. doi: 10.1021/bi0350809. [DOI] [PubMed] [Google Scholar]
- 23.Barneaud-Rocca D, Etchebest C, Guizouarn H. Structural model of the anion exchanger 1 (SLC4A1) and identification of transmembrane segments forming the transport site. J Biol Chem. 2013;288(37):26372–26384. doi: 10.1074/jbc.M113.465989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arakawa T, et al. Crystal structure of the anion exchanger domain of human erythrocyte band 3. Science. 2015;350(6261):680–684. doi: 10.1126/science.aaa4335. [DOI] [PubMed] [Google Scholar]
- 25.Strong M, et al. Toward the structural genomics of complexes: Crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2006;103(21):8060–8065. doi: 10.1073/pnas.0602606103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murshudov GN, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu F, et al. Structure and mechanism of the uracil transporter UraA. Nature. 2011;472(7342):243–246. doi: 10.1038/nature09885. [DOI] [PubMed] [Google Scholar]
- 28.Alguel Y, et al. Structure of eukaryotic purine/H(+) symporter UapA suggests a role for homodimerization in transport activity. Nat Commun. 2016;7:11336. doi: 10.1038/ncomms11336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Geertsma ER, et al. Structure of a prokaryotic fumarate transporter reveals the architecture of the SLC26 family. Nat Struct Mol Biol. 2015;22(10):803–808. doi: 10.1038/nsmb.3091. [DOI] [PubMed] [Google Scholar]
- 30.Nozawa A, Takano J, Kobayashi M, von Wirén N, Fujiwara T. Roles of BOR1, DUR3, and FPS1 in boron transport and tolerance in Saccharomyces cerevisiae. FEMS Microbiol Lett. 2006;262(2):216–222. doi: 10.1111/j.1574-6968.2006.00395.x. [DOI] [PubMed] [Google Scholar]
- 31.Jardetzky O. Simple allosteric model for membrane pumps. Nature. 1966;211(5052):969–970. doi: 10.1038/211969a0. [DOI] [PubMed] [Google Scholar]
- 32.Drew D, Boudker O. Shared molecular mechanisms of membrane transporters. Annu Rev Biochem. 2016;85(1):060815–014520. doi: 10.1146/annurev-biochem-060815-014520. [DOI] [PubMed] [Google Scholar]
- 33.Compton EL, et al. Conserved structure and domain organization among bacterial Slc26 transporters. Biochem J. 2014;463(2):297–307. doi: 10.1042/BJ20130619. [DOI] [PubMed] [Google Scholar]
- 34.Detro-Dassen S, et al. Conserved dimeric subunit stoichiometry of SLC26 multifunctional anion exchangers. J Biol Chem. 2008;283(7):4177–4188. doi: 10.1074/jbc.M704924200. [DOI] [PubMed] [Google Scholar]
- 35.Macara IG, Cantley LC. Interactions between transport inhibitors at the anion binding sites of the band 3 dimer. Biochemistry. 1981;20(18):5095–5105. doi: 10.1021/bi00521a001. [DOI] [PubMed] [Google Scholar]
- 36.Macara IG, Cantley LC. Mechanism of anion exchange across the red cell membrane by band 3: Interactions between stilbenedisulfonate and NAP-taurine binding sites. Biochemistry. 1981;20(20):5695–5701. doi: 10.1021/bi00523a009. [DOI] [PubMed] [Google Scholar]
- 37.Georgieva ER, Borbat PP, Ginter C, Freed JH, Boudker O. Conformational ensemble of the sodium-coupled aspartate transporter. Nat Struct Mol Biol. 2013;20(2):215–221. doi: 10.1038/nsmb.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Verdon G, Boudker O. Crystal structure of an asymmetric trimer of a bacterial glutamate transporter homolog. Nat Struct Mol Biol. 2012;19(3):355–357. doi: 10.1038/nsmb.2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grewer C, et al. Individual subunits of the glutamate transporter EAAC1 homotrimer function independently of each other. Biochemistry. 2005;44(35):11913–11923. doi: 10.1021/bi050987n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reyes N, Ginter C, Boudker O. Transport mechanism of a bacterial homologue of glutamate transporters. Nature. 2009;462(7275):880–885. doi: 10.1038/nature08616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mulligan C, et al. The bacterial dicarboxylate transporter VcINDY uses a two-domain elevator-type mechanism. Nat Struct Mol Biol. 2016;23(3):256–263. doi: 10.1038/nsmb.3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Coincon M, et al. Crystal structures reveal the molecular basis of ion translocation in sodium/proton antiporters. Nat Struct Mol Biol. 2016;23(3):248–255. doi: 10.1038/nsmb.3164. [DOI] [PubMed] [Google Scholar]
- 43.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim DE, Chivian D, Baker D. Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res. 2004;32(Web Server issue):526–531. doi: 10.1093/nar/gkh468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 1):12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. The PyMOL Molecular Graphics System (Schrödinger, LLC), Version 1.8.
- 47.Longtine MS, et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14(10):953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]