Significance
We report autosomal recessive mutations in the enzyme glutamate pyruvate transaminase 2 (GPT2) in a neurological syndrome involving intellectual disability, reduced brain growth, and progressive motor symptoms. We show that the mutations inactivate the enzyme. GPT2 catalyzes the reversible addition of an amino group from glutamate to pyruvate, yielding alanine and α-ketoglutarate. The GPT2 gene demonstrates expression in brain postnatally, and the protein localizes to mitochondria. As in humans, Gpt2-null mice exhibit reduced brain growth. Furthermore, mutant mouse brains show abnormal metabolite levels, including in pathways involving amino acid metabolism, the TCA cycle, and neuroprotective mechanisms. Our study identifies GPT2 as an important mitochondrial enzyme in disease that has general relevance to developmental and potentially to neurodegenerative mechanisms.
Keywords: GPT2, intellectual and developmental disability, mitochondria, metabolomics, spastic paraplegia
Abstract
Mutations that cause neurological phenotypes are highly informative with regard to mechanisms governing human brain function and disease. We report autosomal recessive mutations in the enzyme glutamate pyruvate transaminase 2 (GPT2) in large kindreds initially ascertained for intellectual and developmental disability (IDD). GPT2 [also known as alanine transaminase 2 (ALT2)] is one of two related transaminases that catalyze the reversible addition of an amino group from glutamate to pyruvate, yielding alanine and α-ketoglutarate. In addition to IDD, all affected individuals show postnatal microcephaly and ∼80% of those followed over time show progressive motor symptoms, a spastic paraplegia. Homozygous nonsense p.Arg404* and missense p.Pro272Leu mutations are shown biochemically to be loss of function. The GPT2 gene demonstrates increasing expression in brain in the early postnatal period, and GPT2 protein localizes to mitochondria. Akin to the human phenotype, Gpt2-null mice exhibit reduced brain growth. Through metabolomics and direct isotope tracing experiments, we find a number of metabolic abnormalities associated with loss of Gpt2. These include defects in amino acid metabolism such as low alanine levels and elevated essential amino acids. Also, we find defects in anaplerosis, the metabolic process involved in replenishing TCA cycle intermediates. Finally, mutant brains demonstrate misregulated metabolites in pathways implicated in neuroprotective mechanisms previously associated with neurodegenerative disorders. Overall, our data reveal an important role for the GPT2 enzyme in mitochondrial metabolism with relevance to developmental as well as potentially to neurodegenerative mechanisms.
Intellectual and developmental disabilities (IDDs) occur in 2% of people worldwide and rank first in the United States as a cause of lifelong disability (1). IDD can be accompanied by various associated features, including motor disability. Hereditary spastic paraplegia (HSP) is a unique form of motor disability that is characterized by progressive spasticity and weakness of the lower extremities (2). In addition, postnatal microcephaly may occur with IDD and complicated forms of HSP, and likely reflects failures in processes driving postnatal brain growth such as neuronal arborization, synaptogenesis, and gliogenesis (3).
In this study, we present the discovery of loss-of-function mutations in the gene encoding the enzyme glutamate pyruvate transaminase 2 (GPT2) in autosomal recessive IDD with postnatal microcephaly and motor findings consistent with progressive spastic paraplegia. GPT2 [also known as alanine transaminase 2 (ALT2) or alanine aminotransferase 2 (ALAT2)] is one of two related enzymes that catalyze the reversible addition of an amino group from glutamate to pyruvate, yielding alanine and α-ketoglutarate. GPT2 and highly related GPT are among several transaminases that regulate critical metabolic processes, including amino acid metabolism and the tricarboxylic acid (TCA) cycle. Glutamate is also a substrate in the synthesis of glutathione, an important antioxidant in cells (4). We show that GPT2 is localized to mitochondria and is expressed at increasing levels in brain during early postnatal development corresponding to periods of circuit development.
Mitochondria play an essential role in neurons given the high energy demands of these cells, and mitochondria are particularly abundant at synapses, where they mediate synaptic growth, energetics, signaling, and protection (5). Mitochondrial dysfunction is involved in both human neurodevelopmental as well as neurodegenerative disease (6–8). The TCA cycle in mitochondria functions in both energy production as well as in biosynthetic processes. TCA cycle intermediates may be extracted for biosynthetic metabolism through a process termed cataplerosis, and TCA intermediates are replenished via anaplerosis (9).
In addition to identification of mutations in GPT2 in human neurological disease, we have developed a mouse model of Gpt2 deficiency. Akin to the human phenotype, we see reduced brain growth in this mouse model. Metabolomics studies in Gpt2-null mouse brains reveal reduced TCA metabolites consistent with a defect in anaplerosis. We also identify metabolomic signatures reflecting abnormalities in amino acid metabolism, as well as in neuroprotective mechanisms previously implicated in neurodegenerative disease. Overall, our data support an important role for GPT2 in mitochondria-based metabolism and neurological disease with both developmental and progressive features.
Results
Clinical Analyses of Pedigrees with Intellectual Disability and Postnatal Microcephaly.
As a contribution to understanding the genetic underpinnings of neurological disease, we have studied two large consanguineous kindreds with a neurodevelopmental condition. Affected members in families shown in Fig. 1A share a phenotype that includes intellectual disability with postnatal microcephaly and variable, progressive spasticity (see also ref. 10). In total, our studies included 14 affected individuals. All affected individuals were born after uneventful pregnancies without asphyxia. Head circumference was normal at birth for all seven individuals for whom data were available, but affected individuals subsequently developed microcephaly postnatally, ranging from 2.8 to 6.8 SDs below the mean (Table 1 and SI Appendix, Table S1). The majority (88%) of affected individuals were reported to be hypotonic during infancy. All 9 cases with data available (of 14 total cases) were able to walk by the age of 3 y. All had delayed speech and showed oral–motor dysfunction. Five (36%) had a history of seizures, with three (MC9426, MC9429, and MI1002) having been diagnosed with various types of epilepsy syndromes, including one with Lennox–Gastaut syndrome, and the other two individuals each reporting a single seizure episode. Full detailed clinical descriptions are provided in SI Appendix, Table S1.
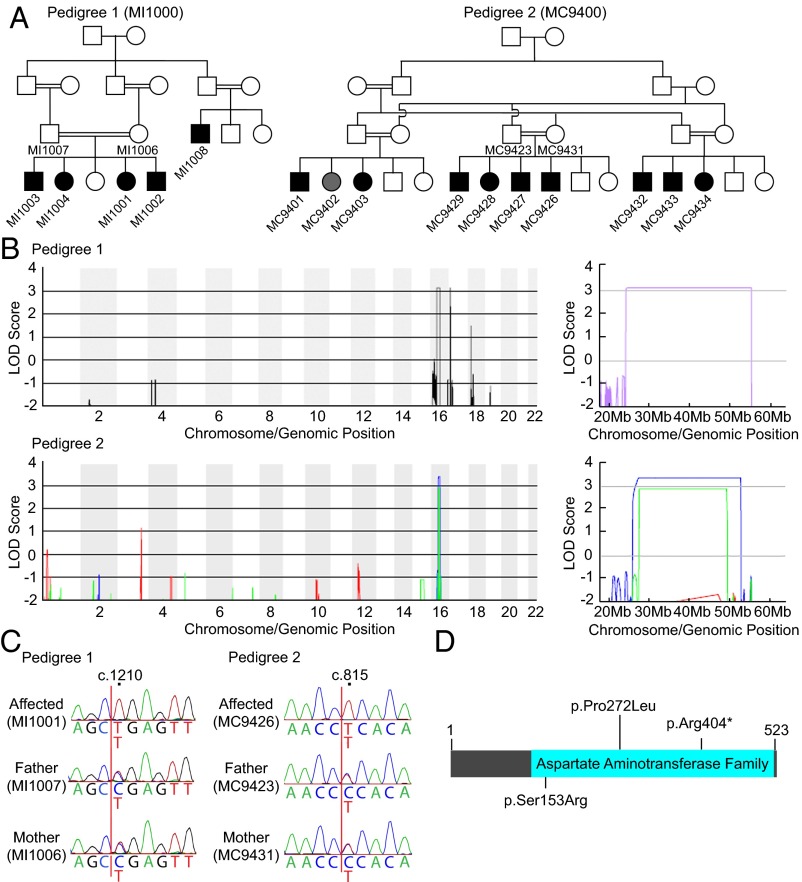
Fig. 1.
Identification of GPT2 mutations in large kindreds with recent shared ancestry affected by IDD with postnatal microcephaly. (A) Pedigree 1 (MI1000) is from Pakistan. Pedigree 2 (MC9400) is from the Sultanate of Oman. Filled symbols indicate affected individuals. Shaded symbol (MC9402) indicates a similarly affected individual, who was found not to have a homozygous GPT2 mutation. We have described the clinical presentation of pedigree 2 (MC9400) in a prior publication (10). (B) Genome-wide LOD scores for pedigree 1 show a region of linkage on chromosome 16q11.2, with a maximum multipoint LOD score of 3.13. Linkage analysis of pedigree 2 was performed for each of the three branches. Branch 1 (red) did not show a region of statistically significant linkage given heterozygous individual MC9402 with phenocopy, whereas both branch 2 (blue) and branch 3 (green) indicate linkage to chromosome 16q11.2 with LOD scores of 3.38 and 2.90, respectively. Magnified LOD score graphs for chromosome 16q are shown on the Right. (C) Sanger sequencing of GPT2 confirmed the identified mutations in pedigree 1 (c.1210C>T, p.Arg404*) and pedigree 2 (c.815C>T, p.Pro272Leu). Positions of mutations are according to NM_133443.3 (cDNA) and NP_597700.1 (protein). (D) Schematic diagram of conserved domains in GPT2. The positions of the mutations with a previously reported mutation (p.Ser153Arg) are shown (14).
Table 1.
Phenotypic features of 14 affected individuals with GPT2 mutations
| Phenotype | No. of positive cases | Frequency, % |
| Microcephaly | 14/14 | 100 |
| Postnatal microcephaly | 7/7 | 100* |
| Intellectual disability | 14/14 | 100 |
| Hypotonia during infancy | 7/8 | 88* |
| Oral–motor dysfunction | 12/12 | 100* |
| Hyperreflexia | 11/13 | 85* |
| Hypertonia | 12/13 | 92* |
| Joint contractures | 3/14 | 21 |
| Ability to walk by age 3 | 9/9 | 100* |
| Subsequent deterioration in walking ability | 10/12 | 83* |
| Spastic diplegia/paraplegia | 10/13 | 77* |
| Seizures | 5/14 | 36 |
Based on cases with data available.
The majority (83%) of the affected individuals showed subsequent difficulty in walking. Over time, motor examination was remarkable for hypertonia and hyperreflexia. In most cases (77%), lower extremities were more severely affected, thus presenting as spastic diplegia or spastic paraplegia. In some individuals, hand movements were clumsy, tremulous, and poorly coordinated. Results from brain imaging studies [computed tomography or magnetic resonance imaging (MRI)] were obtained for 10 individuals and showed no obvious structural malformation other than microcephaly, except for 1 individual who had reduced white matter volume and a thin corpus callosum (SI Appendix, Fig. S1).
Identifying GPT2 Mutations by Linkage Mapping and Whole-Exome Sequencing.
Pedigrees 1 and 2 showed strong linkage to a locus on chromosome 16q (Fig. 1B). In pedigree 1, genome-wide linkage analysis of the larger branch (affected individuals: MI1001, MI1002, MI1003, and MI1004) revealed a region of linkage on chromosome 16q with a maximum logarithm of odds (LOD) score of 3.13 (Fig. 1B). The region was delineated by SNP markers rs198188 to rs12917822 (chr16: 24,136,201–55,165,407). In pedigree 2, LOD scores were calculated for each of the three branches of the family separately due to complexity of the pedigree. Branches 2 and 3 showed evidence of linkage to the same region of chromosome 16q. The regions were delineated by SNP markers rs205162 to rs6498968 for branch 2 (chr16: 25,596,637–52,653,785) and rs17720179 to rs1567522 for branch 3 (chr16: 27,188,853–49,302,826), with maximum LOD scores of 3.38 and 2.91, respectively (Fig. 1B). In branch 1, two of the affected individuals (MC9401 and MC9403) showed linkage to this region.
Whole-exome sequencing (WES) of all affected individuals in pedigree 1 revealed a single shared homozygous, nonsense variant (c.1210C>T, p.Arg404*) in the GPT2 gene (RefSeq NM_133443.3) in the linkage interval. There were no other rare, loss-of-function mutations shared by all affected members of the pedigree in the interval or elsewhere in the genome. The homozygous GPT2 gene variant was confirmed by Sanger sequencing and segregated with the disease in pedigree 1 (Fig. 1C). All members unaffected for microcephaly in the pedigree were heterozygous. The candidate variant was not present in the National Heart, Lung, and Blood Institute (NHLBI) Exome Variant Server or in dbSNP. In pedigree 2, using WES of three affected individuals (MC9401, MC9428, and MC9432), we identified a homozygous missense variant in the GPT2 gene (c.815C>T, p.Pro272Leu) as the only shared candidate variant in the linkage interval (Fig. 1C). All affected individuals were homozygous, and no unaffected individuals were found to be homozygous. Individual MC9402, who showed a microcephalic phenotype, was heterozygous, and thereby likely represents a phenocopy. This p.Pro272Leu variant was not found in the NHLBI Exome Variant Server or in dbSNP, and was predicted to be damaging by PolyPhen-2 (11), SIFT (12), and Provean (13), suggesting its pathogenicity (SI Appendix, Table S2). Since our discovery of the mutations above, a single homozygous missense change in GPT2 (c.459C>G, p.Ser153Arg) was reported in two siblings with a similar phenotype, including microcephaly and developmental delay (14) (Fig. 1D).
Human Mutations in GPT2 Lead to Reduced Protein Levels and Enzyme Activity.
We studied the protein stability and enzymatic activity of the identified human mutations, and our results are consistent with a loss-of-function mechanism. We expressed each of the three mutated transcripts in HeLa cells and studied protein levels using Western blotting with an antibody raised to the full-length GPT2 (Fig. 2A). We detected a control, full-length protein at the predicted size of V5His-tagged GPT2 (62.8 kDa, arrow). For the truncating mutation p.Arg404*, we detected a scant quantity of the predicted truncated protein (44.4 kDa, arrowhead), consistent with nonsense-mediated decay of the mRNA and/or an unstable protein. Less than 4% of control protein was detected (quantified in Fig. 2B). The missense mutations p.Ser153Arg and p.Pro272Leu were expressed at 37.6 ± 6.3% and 10 ± 2.2% of control levels, respectively, reflecting protein instability (Fig. 2B). Structural predictions for the missense changes also suggest deleterious effects (SI Appendix, Fig. S2).
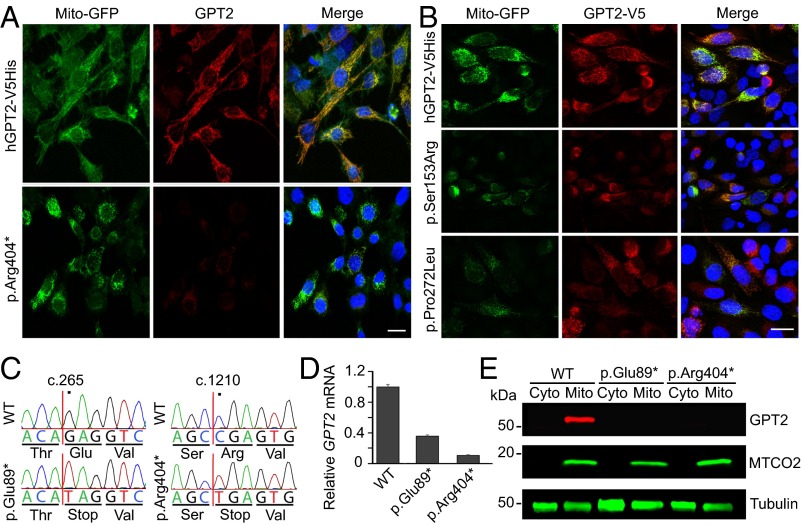
Fig. 2.
Mutations in GPT2 lead to reduced protein levels and enzyme activity. (A) Western blot results of mutated GPT2 protein. Lysates of HeLa cells transfected with human control GPT2 (hGPT2-V5His), p.Arg404*, p.Ser153Arg, p.Pro272Leu, or vector-only constructs were probed with anti-GPT2 antibody. All of the three GPT2 mutants show reduced protein levels compared with control GPT2 (hGPT2-V5His). Arrow indicates the expected GPT2 protein band; arrowhead indicates the truncated GPT2 protein band. (B) Quantification of Western blots. (Left) Overexpressed GPT2 bands were normalized to tubulin. (Right) Overexpressed GPT2 level in each lane was normalized to the GPT2 control (hGPT2-V5His). n = 6 replicates. Data are presented as means ± SEMs. ***P < 0.001. (C) GPT enzyme functional assay indicated that mutations in GPT2 lead to loss of the enzyme activity. HeLa cells were transfected with human control GPT2 (hGPT2-V5His), p.Arg404*, p.Ser153Arg, p.Pro272Leu, or vector-only constructs. Lysates were used to analyze the GPT activity. (Left) GPT activity was measured by pyruvate production after either a 20- or 40-min incubation with substrate at 37 °C. All three GPT2 mutants had similar pyruvate production as the vector control. (Right) GPT activity was measured in cell lysate after incubating with substrate at 37 °C for 40 min and is represented as enzyme activity (in milliunits) per milligram of tested sample. Only the overexpressed GPT2 control protein showed significant high enzyme activity. n = 4 replicates. Data are presented as means ± SEMs. ***P < 0.001.
To substantiate further the interpretation that these mutations lead to loss of enzyme function, we tested the enzymatic activity of mutant proteins directly. We expressed constructs for full-length GPT2 protein, as well as for each of the three mutant alleles, in HeLa cells. We subsequently made protein extract from cells and tested GPT2 enzyme activity. In the cell lines expressing each of the mutant alleles, including the missense mutations, enzyme activity relative to control was substantially diminished (to background levels) (Fig. 2C). Thus, the mutations in GPT2 are associated with reductions or loss of protein as well as loss of enzyme activity. Even in the case of the missense mutations, wherein there was a small level of intact protein, the enzyme activity of protein with these missense mutations was not detectable above background levels.
GPT2 Expression Increases in Postnatal Developing Brain.
To evaluate the role of GPT2 in developing brain, we first measured protein and enzyme activity in developing mouse brains. In normal mice, we found highest levels of Gpt2 protein in the early postnatal period using Western blotting (SI Appendix, Fig. S3A). Total Gpt enzyme activity was detectable at postnatal day 1 (P1), yet enzyme activity was found to increase nearly 10-fold by P18 (SI Appendix, Fig. S3B), correlating with increases in Gpt2 protein levels. In published RNA-seq data from cell lineages purified from mouse brain, Gpt2 mRNA expression was threefold higher than that for the related enzyme Gpt in neurons [8.9 vs. 3.0 fragments per kilobase of exons per million fragments mapped (FPKM), respectively] (SI Appendix, Fig. S3C) (15). Gpt2 also has a high level of expression (far exceeding Gpt expression) in oligodendrocyte precursor cells (137.7 vs. 4.8 FPKM, respectively) and newly formed oligodendrocytes (48.7 vs. 2.6 FPKM, respectively) (SI Appendix, Fig. S3D) (15).
In humans, through analyses of the Allen Institute’s Developmental Transcriptome Dataset, we observed that GPT2 mRNA is broadly expressed across all parts of the human brain throughout development and into adulthood, with highest levels in the early postnatal years (SI Appendix, Fig. S4A). Interestingly, although mRNA levels for GPT2 and for GPT are correlated, GPT2 mRNA levels are generally >20-fold higher than GPT levels in human brain (SI Appendix, Fig. S4B). Based on these data, peak levels of GPT2 expression correlate with major periods of synaptogenesis and myelination in both mouse and human developing postnatal brain (16–19).
GPT2 Protein Is Localized to Mitochondria.
GPT2 harbors a strong mitochondrial-localization sequence, which is not present in GPT (SI Appendix, Fig. S5A). None of the missense mutations appeared to interfere with the predicted mitochondrial localization using PSORT II predictions (20) (SI Appendix, Fig. S5A). Exogenously expressed GPT2 wild-type (WT) protein demonstrated a very high degree of costaining with mitochondrial markers, such as Mito-GFP (Fig. 3A, Top). By comparison, expression of the p.Arg404* mutant protein produced very little protein (Fig. 3A, Bottom); however, the small amount of protein that was discernible (by increasing the imaging parameters to maximal sensitivity) localized to mitochondria (SI Appendix, Fig. S5B). The exogenously expressed missense proteins, either the p.Ser153Arg or p.Pro272Leu, both demonstrated higher levels of protein than the truncating mutation but reduced levels of staining relative to the control (Fig. 3B). This reduced amount of staining did colocalize with the mitochondrial stains, yet we expect these enzymes to be inactive, as shown previously (Fig. 2C).
Fig. 3.
GPT2 localizes to mitochondria. (A) HeLa cells coexpressing Mito-GFP (a mitochondrial marker, green) and either normal, human GPT2 (hGPT2-V5His) or human GPT2 with the p.Arg404* mutation were immunostained with anti-GPT2 antibody (red). Under the same imaging parameters, p.Arg404* showed a decrease in protein expression level. Immunostaining for GPT2 showed high colocalization with mitochondrial markers for control protein. (B) HeLa cells coexpressing Mito-GFP and C-terminal V5-tagged human GPT2 control (hGPT2-V5His) or mutations p.Ser153Arg or p.Pro272Leu were stained with anti-V5 antibody (red). Under the same imaging parameters as control, the images of cells expressing mutant GPT2 demonstrate reduced protein levels yet maintain mitochondrial localization in the residual signal. (Scale bar, 10 μm.) (C–E) GPT2-mutant HEK293FT cells were generated by CRISPR-Cas9 genome editing. (C) Sanger sequencing traces of WT HEK293FT cells and the GPT2-mutant HEK293FT cell lines are shown. The mutant cell lines harbor homozygous nonsense mutations in the GPT2 gene (c.265G>T, p.Glu89*; c.1210C>T, p.Arg404*). Positions of the variants are according to NM_133443.3 (cDNA) and NP_597700.1 (protein). (D) Graphed are the results of qRT-PCR analysis of WT HEK293FT cells and the two lines of GPT2-mutant cells (p.Glu89*, p.Arg404*). For all samples, GPT2 mRNA was normalized to 18S rRNA. (E) Immunoblot analysis of WT HEK293FT cells and the GPT2 mutant cells (p.Glu89*, p.Arg404*). In WT cells, GPT2 is absent from the cytosolic fraction (Cyto) and highly enriched in the mitochondrial fraction (Mito). In cells expressing either of the two mutants, GPT2 is absent from both fractions. Tubulin and MTCO2 (cytochrome c oxidase subunit II) are shown as a loading control marker and a mitochondrial marker, respectively.
Next, we established two GPT2-mutant HEK293FT cell lines using CRISPR-Cas9 genome editing (Fig. 3C and SI Appendix, Table S3). We introduced homozygous nonsense mutations (c.265G>T, p.Glu89*; and c.1210C>T, p.Arg404*), the latter of which is the same variant as we identified in pedigree 1. We confirmed by quantitative reverse transcription–PCR (qRT-PCR) that cells containing either of these two GPT2 mutations showed significant decreases in GPT2 mRNA compared with WT cells (Fig. 3D). Western blot analysis of fractionated lysates from WT and mutant HEK293FT cells revealed that endogenous GPT2 localized to the mitochondrial fraction in WT cells and was absent from the cytoplasmic fraction, whereas GPT2 was undetectable in either fraction in mutant cells (Fig. 3E).
Gpt2-Null Mice Demonstrate Reduced Postnatal Brain Growth.
We generated mice with a germline disruption of the Gpt2 gene (SI Appendix, Fig. S6) and observed decreases in postnatal brain growth akin to the human phenotype. Homozygous mutant mice were born in the expected Mendelian ratio (SI Appendix, Table S4). Western blotting of brain lysates demonstrated that Gpt2 protein was abolished in the mutant mice (SI Appendix, Fig. S6C). Also of note, postnatal brains of homozygous mutant mice at P18 showed significantly reduced total brain Gpt enzyme activity (SI Appendix, Fig. S6D). Homozygous mutant brain size was generally indistinguishable at birth, and, behaviorally, mutants were similar to control littermates for the first 2 wk postnatally. However, at approximately P18, homozygous mutant mice began to demonstrate reduced motor activity. Subsequent to the emergence of reduced motor behavior, homozygous mutant mice grew sickly and died generally between P18 and P26 (SI Appendix, Table S4). Given the expression of the β-galactosidase gene from the endogenous Gpt2 promoter in the targeted allele, we were able to study the distribution of Gpt2 expression in the heterozygous mouse brain. We found that, in P78 heterozygous mice, Gpt2 expression was widely distributed throughout the brain (SI Appendix, Fig. S7). Staining was particularly apparent in the prefrontal cortex, striatum, hippocampus, and granular cell layer of the cerebellum.
We compared brain area between littermate homozygous mutant (n = 12) and WT animals (n = 7) at P18–P22 (Fig. 4). Gpt2-null brains showed a small but consistent and significant reduction in cortical area relative to control (P < 0.03) (Fig. 4B). Furthermore, analysis of areas and lengths of different brain regions showed a consistent and significant decrease in brain size in Gpt2-null animals compared with control animals (Fig. 4 C and D). For example, comparison of combined areas of cortex (CX), midbrain (MB), and cerebellum (CB) demonstrated a 13% decrease in area (P < 0.0015). We further observed that the number of SV2-positive puncta representing synapses were significantly reduced (30% less than WT, P = 0.005) at 10–14 d in vitro (Fig. 4 E and F). Based on these findings, we conclude that GPT2 has an important role in brain growth during postnatal development and potentially in synapse development.
Fig. 4.
Gpt2 deficiency causes a reduction in brain size in mouse. (A) Panels depict two representative brain pairs from littermate animals for each genotype at P21 (Top) and P22 (Bottom). Left panels show WT perfused brains. Right panels show Gpt2-null brains (MUT). Measurements were taken of the cortex (CX), the midbrain (MB), the cerebellum (CB), the left hemisphere (LH), and the right hemisphere (RH). Ruler demarcations reflect 1-mm scale. (B) Cortical area presented as a percentage of total brain area is shown for WT animals and for Gpt2-null animals (MUT). **P < 0.03. (C) Graphed is a comparison of area in square centimeters in four different brain regions: LH (**P < 0.0025), RH (**P < 0.0015), CX+MB+CB (*P < 0.0015), and MB+CB (n.s., P < 0.1). (D) Graphed is a comparison of length measurements between WT and Gpt2-null (MUT) animals. Measurements are depicted in the brain image on the Right. We found significant differences in width across two hemispheres (L1, green line) (*P < 0.015), in the midline anterior–posterior distance (L2, red line) (**P < 0.0015), in the diagonal length of the left hemisphere (L3, blue line) (***P < 0.0002), and in the right hemisphere (L4, yellow line) (****P < 0.000001). n = 7 WT and 12 MUT animals for all brain size measurements. Data are presented as means ± SEMs. Statistical analysis was conducted using Student’s t test. (E) Gpt2-null hippocampal neurons show reduced synapses compared with neurons from WT littermates at 10 DIV, as visualized by SV2 staining (green). Neurons were immunostained with antibodies against SV2 and MAP2 (red). (Scale bar, 10 μm.) (F) The graph depicts quantification of SV2 densities per 135-μm2 region of field of view in WT and MUT neurons. Data are presented as means ± SEMs. n = 4 WT animals and 3 MUT animals across 3 litters. ***P = 0.005.
Amino Acid Metabolism Is Defective in Gpt2-Null Mouse Brains.
To identify metabolic defects in neural tissue in vivo, we investigated Gpt2-null mouse brains for derangements in metabolite levels. We analyzed 276 metabolites from P18 acute brain preparations from Gpt2-null and WT mice using targeted mass spectrometry (MS)-based metabolomics and metabolite set enrichment analysis (MSEA) (Fig. 5A). The top three pathways identified were as follows: protein biosynthesis [P value = 0.0000000644, false discovery rate (FDR) = 0.00000515] (largely reflecting changes in amino acid metabolism); citric acid cycle (P value = 0.00000733, FDR = 0.000293); and glutathione metabolism (P value = 0.0000142, FDR = 0.000379) (Fig. 5A and SI Appendix, Table S5).
Fig. 5.
Metabolomics profile of postnatal Gpt2-null mouse brain. (A) Overrepresentation analysis of metabolomics in brains from Gpt2-null (MUT) compared with WT Gpt2 mice at P18. The reference library contained 88 metabolite sets for normal metabolic pathways. Metabolites showing significant increases or decreases (compared with control on Student’s t test P value less than 0.05) were flagged. The hypergeometric test was used to determine whether flagged metabolites from specific pathways were significantly overrepresented. Red, FDR < 0.01; green, 0.01 < FDR < 0.03; blue, 0.03 < FDR < 0.05. (B–E) Box plots of metabolites, with whiskers representing minimum and maximum values. The peak intensity areas were normalized by a pooled reference sample from the WT Gpt2 group (probabilistic quotient normalization), and auto (unit) scaling was performed whereby the data were mean-centered and divided by the SD of each group. Unpaired Student’s t test was performed assuming equal group variance. The box plots in each of the panels show the following: (B) metabolites from the primary GPT enzyme reaction, (C) amino acids, (D) TCA cycle intermediates, and (E) metabolites involved in neuroprotective mechanisms. n = 6 WT animals and 6 MUT animals. * = 0.01 < raw P < 0.05, ** = 0.001 < P < 0.01, *** = P < 0.001. Red stars denote a corresponding FDR below 0.05. Also refer to SI Appendix, Table S5, for MSEA data; SI Appendix, Table S6, for full set of metabolite Student’s t tests; and SI Appendix, Table S7, for raw data. †Arginine is a conditionally essential amino acid.
At the level of primary metabolites in the GPT reversible reaction in Gpt2-null brains, alanine showed a strong decrease (fold change = 0.885, FDR = 0.008) (Fig. 5B and SI Appendix, Table S6). We corroborated these brain findings of alanine depletion by direct measure of alanine secretion and by isotope tracing experiments in Gpt2-null murine embryonic fibroblasts (MEFs) (Fig. 6). Gpt2-null MEFs exhibited near-complete loss of alanine secretion into the media (Fig. 6A). Gpt2 deletion also substantially reduced transfer of 15N from [α-15N]glutamine to alanine (Fig. 6B), as would be expected if Gpt2 rather than Gpt1 were the major isoform exchanging amino groups between glutamate and α-ketoglutarate. When cultured in medium supplemented with uniformly labeled [U-13C]glucose, the Gpt2-null MEFs also had reduced 13C labeling in alanine, whereas lactate labeling was conserved (Fig. 6C, two left panels). Therefore, taken together, metabolomic and direct isotope tracing measures indicate that Gpt2 is the major Gpt isoform responsible for alanine production and secretion.
Fig. 6.
Defective alanine production and glutamine-dependent anaplerosis in Gpt2-null cells. Immortalized MEF cell lines were prepared from WT and Gpt2-null mice (MUT) and investigated in culture. (A) Total alanine abundance in the medium was measured by HPLC. Alanine secretion in the MUT cells was scarcely detectable. (B) Graphed are fractional enrichments of extracellular alanine during culture with [α-15N]glutamine. Levels of transfer of radioisotope from glutamine to alanine are substantially reduced in the Gpt2-null cells. (Schematic Right) 15N (green) is transferred from [α-15N]glutamine, via [α-15N]glutamate, to [15N]alanine. This pathway requires activity of a glutamate pyruvate transaminase, such as Gpt2. (C) Shown are mass isotopologue distributions for alanine, lactate, malate, and citrate after culture with [U-13C]glucose and unlabeled glutamine. Altered label distributions, including reduced alanine m+3, and enhanced malate m+3 and citrate m+5 fractions, are consistent with suppressed glutamine-dependent anaplerosis (22). (Schematic Right) [U-13C]Glucose gives rise to [U-13C]pyruvate (pyruvate m+3) via the glycolytic pathway. This pyruvate can be converted to lactate m+3 or alanine m+3. Conversion of pyruvate to alanine involves the Gpt1 (cytosol)- or Gpt2 (mitochondrion)-dependent transfer of an amino group to pyruvate from glutamate. A large reduction of labeling of alanine from glucose in Gpt2-deficient cells implies that Gpt2 is required for alanine synthesis. (D) Shown are mass isotopologue distributions for malate and citrate after culture with unlabeled glucose and [U-13C]glutamine. Reduced direct transfer of radioisotope from glutamine to malate and citrate demonstrates suppressed glutamine-dependent anaplerosis in Gpt2-null cells. (Schematic Right) [U-13C]Glutamine generates fully labeled α-ketoglutarate via glutamate. The resulting α-ketoglutarate can proceed around the oxidative TCA cycle as shown, giving rise to malate m+4 and citrate m+4. In addition, α-ketoglutarate may become reductively carboxylated through the addition of one unlabeled carbon, resulting in citrate m+5 (49). Loss of Gpt2, which catalyzes the conversion of glutamate to α-ketoglutarate, is predicted to reduce labeling of metabolites downstream of α-ketoglutarate during culture with [U-13C]glutamine. It would also increase the contribution of glucose carbon to TCA cycle metabolites, as described in ref. 22. Data are presented as the means ± SDs for three parallel cultures, with results shown for one representative experiment. *P < 0.05, **P < 0.01, ***P < 0.001 by unpaired, two-sample Student’s t test.
Although the alanine pool was depleted, several other amino acid pools were substantially elevated in P18 brain, particularly essential amino acids, including phenylalanine (fold change = 1.641, FDR = 0.024) (Fig. 5C and SI Appendix, Table S6). Notably urea was substantially decreased (fold change = 0.246, FDR = 0.017), which suggests a decrease in production of amine groups, consistent with dysregulation of total cellular aminotransferase activity and/or nitrogen disposal perhaps due to decreases in alanine secretion.
Additional Abnormal Metabolomics Signatures in Gpt2-Null Mouse Brains: Anaplerosis and Neuroprotective Mechanisms.
In addition to defects in amino acid metabolism, P18 Gpt2-null mouse brains exhibit abnormal metabolomics signatures involving TCA cycle and neuroprotective mechanisms (Fig. 5A and SI Appendix, Table S5). With regard to the TCA cycle, five of eight TCA cycle intermediates were substantially decreased (Fig. 5D and SI Appendix, Table S6), including the following: citrate (fold change = 0.655, P = 0.015), isocitrate (fold change = 0.868, P = 0.0064), succinate (fold change = 0.759, P = 0.009), fumarate (fold change = 0.881, P = 0.002), and malate (fold change = 0.735, P = 0.001). Surprisingly, ATP and GTP levels were somewhat elevated, as was acetyl-CoA.
Depletion of TCA cycle intermediates in brain is consistent with a defect in anaplerosis, the metabolic process involved in replenishing TCA cycle intermediates used during cellular anabolism and biosynthesis. Because Gpt enzyme activity converts glutamate to α-ketoglutarate, this function has been proposed to promote glutamine-dependent anaplerosis (21). Overall, labeling of TCA cycle intermediates from [U-13C]glucose was enhanced by Gpt2 deletion (Fig. 6C). Altered label distributions included enhanced malate m+3 and citrate m+5 fractions, which is consistent with suppressed glutamine-dependent anaplerosis and increased carboxylation of pyruvate (22). These altered labeling patterns are expected if reduced entry of glutamine-derived carbon into the TCA cycle via α-ketoglutarate is offset by an increased contribution of 4-carbon intermediates from glucose. Culture in [U-13C]glutamine confirmed a reduced contribution of glutamine-dependent anaplerosis to TCA cycle intermediates in Gpt2-deficient cells (Fig. 6D).
Finally, we found in P18 Gpt2-null brains that several metabolites related to neuroprotective mechanisms are misregulated (Fig. 5E): glutathione (fold change = 0.715, FDR = 0.038), folate (fold change = 0.383, FDR = 0.024), and cysteine (fold change = 0.609, FDR = 0.083) levels were reduced; whereas cystathionine levels were substantially elevated (fold change = 2.409, FDR = 0.00036). Elevations of cystathionine were also found in newborn P0 mouse brain (fold change = 1.701, FDR = 0.029) (SI Appendix, Table S8). We conducted metabolomics analysis on newborn (P0) brain from mutant animals compared with combined WT and heterozygous littermates. Although Gpt2 protein expression undergoes substantial up-regulation in the postnatal period (SI Appendix, Fig. S3A), we reasoned that analysis at this early time point could pinpoint the most proximal metabolic derangements resulting from loss of Gpt2. At P0, only four metabolites demonstrated misregulated levels adhering to a conservative FDR correction. Among these were two metabolites observed also at the P18 time point: alanine (fold change = 1.573, FDR = 0.000165), a primary metabolite in the Gpt2 reaction, and also interestingly, cystathionine (fold change = 1.701, FDR = 0.029). Of note, alanine was up-regulated at P0, whereas by contrast it was significantly down-regulated at P18. The other two misregulated metabolites that withstood FDR correction were hydroxyproline (fold change = 1.767, FDR = 0.000335) and sarcosine (fold change = 1.337, FDR = 0.0153). Both of these metabolites are involved in amino acid metabolism. Overall, our data support a role for Gpt2 in mitochondrial function and various metabolic pathways, including amino acid metabolism, anaplerotic processes involved in TCA cycle intermediates, and pathways implicated in neuroprotective mechanisms (e.g., pathways involving the metabolites cysteine, cystathionine, glutathione, and folate).
Discussion
In this study, we report rare, homozygous mutations in the gene encoding the enzyme GPT2 in large consanguineous pedigrees segregating with a neurological phenotype involving IDD, postnatal microcephaly, and spastic paraplegia with progressive features. We present one extended kindred with a LOD score greater than 3.1 (pedigree 1, MI1000) segregating a homozygous nonsense GPT2 mutation (p.Arg404*). Another large extended kindred of three branches (pedigree 2, MC9400) segregates a deleterious, homozygous missense GPT2 mutation (p.Pro272Leu) with LOD scores greater than 3.3 and 2.9 in branches 2 and 3, respectively. An individual of pedigree 2 (MC9402, branch 1), who similarly presented with microcephaly and IDD, was not homozygous for the GPT2 variant. Because this family is highly consanguineous, it is likely that this individual has an additional genetic mutation. During the course of our work, a third homozygous, deleterious missense GPT2 mutation (p.Ser153Arg) was reported in a large pedigree with recent shared ancestry and three affected individuals presenting with a similar phenotype (14).
In biochemical studies, we demonstrate that the human mutations are loss of function. For the p.Arg404* mutant protein, we demonstrate that the truncated GPT2 protein is not made or is unstable in expression studies in cell lines. We also show here that the p.Pro272Leu and p.Ser153Arg proteins are unstable and, through structural predictions, likely have deleterious effects on protein folding (SI Appendix, Fig. S2). All mutant GPT2 proteins, p.Arg404*, p.Pro272Leu, and p.Ser153Arg, are enzymatically inactive, consistent with loss-of-function, autosomal recessive mutations.
The clinical phenotype of the affected individuals associated with GPT2 mutations presented herein is generally uniform. Postnatal microcephaly and IDD were noted in all affected individuals, most had oral motor difficulties, and several had seizures. These clinical findings are highly consistent with those of the individuals reported in ref. 14. There is no clear difference in severity between individuals with truncating mutations and missense mutations, consistent with our biochemical data that all mutations lead to a severe or complete loss of GPT2 enzyme function.
An important aspect of our work is that we have had the opportunity to follow patients longitudinally. Thereby, we are able to extend the neurological phenotype, which appears to include progressive features in addition to developmental disease. With regard to motor findings, we find that hypotonia during infancy gives way to hypertonia more notably in the lower extremities (spastic diplegia or paraplegia). Among the 14 affected individuals studied herein, ∼80% showed slowly progressive spastic diplegia or paraplegia with hyperreflexia and hypertonia as one of the major features. Although rapid regression is not noted, motor difficulties seem to be slowly progressive in many of the cases, consistent with our metabolomics studies that suggest abnormalities in neuroprotective mechanisms. Defects in such mechanisms have been previously implicated in hereditary spastic diplegia (23).
The acquisition of neurological features during early postnatal life indicates the brain’s requirement for GPT2 during this critical period of brain development. Postnatal brain development is characterized by the formation, refinement, and maintenance of circuitry involving neuronal arborization, synaptogenesis, oligodendrocyte expansion, and myelination (3). We find GPT2 expression to increase in brain during the early postnatal period and to be expressed in both neurons and glia. Prior studies have indicated strong neuronal expression and weaker or absent glial expression in adult human brain (24); however, data presented here support a role for GPT2 in both neurons and glia, at least during early postnatal development (SI Appendix, Fig. S3). Our finding that GPT2 expression reaches highest levels in the early postnatal period both in human and in mouse brain, concurrent with peaks in synaptogenesis (17, 18, 25), suggests a role in this process. We also observe decreases in brain growth postnatally both in human and in mouse. Our initial findings support defects in synapse formation (Fig. 4 E and F), again consistent with failures in synaptogenesis perhaps arising from mitochondrial dysfunction. Importantly, we demonstrate here that GPT2 is localized to mitochondria. GPT2 has a strong mitochondrial localization signal, whereas GPT does not. The two isoforms of GPTs, GPT and GPT2, show different spatial expression patterns. GPT2 is highly expressed in the CNS. There is also a possibility that other defects in postnatal brain development are involved, including abnormal oligodendrocyte development. In humans, myelination increases rapidly between 6 and 24 mo postnatally and may increase more gradually into adolescence (16, 26). A minority of patients have shown a reduced white matter volume on brain MRI [participant MI1003; SI Appendix, Table S1; and also an affected individual in the study by Celis et al. (14)]. Interestingly, in mouse, we observe strong Gpt2 expression in oligodendrocyte precursor cells that undergo rapid expansion in the early postnatal period (19, 27–29).
It is appealing to speculate that disease progression in the postnatal period may be prevented by intervention, particularly in the setting of abnormalities of amino acid metabolism. GPTs serve an important role in a variety of metabolic functions, including amino acid metabolism, the urea cycle, and the TCA cycle, in addition to roles in neurotransmitter (both glutamate and GABA) synthesis. Glutamate, in turn, is also a substrate for the synthesis of glutathione, a key element in the cellular machinery to protect against oxidative damage and cell death (30, 31). Depletion of alanine levels is the strongest signal observed of the primary metabolites in the brain. Similarly, in patient cerebrospinal fluid, decreases in alanine were seen (14). Interestingly, we do not see strong changes in glutamate. Elevations of glutamate at the synapse would be predicted to be particularly excitotoxic (32); however, we do not see strong changes in total brain glutamate in newborn mice or at P18. Future studies may examine specific cell types or tissue compartments (i.e., the synapse) more precisely for alterations in metabolites or damage. Given the lack of spatial resolution, our current experiments make it difficult to interpret a role for GPT2 in the glutamate–glutamine cycle at the synapse. Indeed, the GPT enzyme has been used experimentally and in therapeutic models to reduce excitotoxic glutamate levels through a proposed scavenging approach (33).
In the postnatal brain at P18, our experiments seem to indicate that Gpt2 serves to provide a source of alanine in the brain. Also, given the very low urea levels, Gpt2 activity may play an important role in nitrogen metabolism perhaps through generation of alanine. At P18, by contrast to decreases in alanine, Gpt2-null brains demonstrate elevations in several essential amino acids, including elevations in phenylalanine, which have been implicated in other encephalopathies, notably phenylketonuria, and may be modifiable by diet. We also observed elevations in glycine, which is also associated with progressive encephalopathy, intellectual disability, and seizures (34).
We also observe metabolic defects indicating abnormalities in glutamine-dependent anaplerosis of TCA cycle intermediates. GPT2 facilitates glutamine’s ability to serve as an anaplerotic precursor by enabling the conversion of glutamate to α-ketoglutarate in the mitochondria. Our findings are similar to a previous study in which GPT activity was observed to be required for glutamine to stimulate both respiration and growth of KRAS-transformed cancer cells in culture (35). The importance of the role of GPT2 and anaplerosis in neurons and glia warrants further investigation.
Importantly, a metabolic signature previously associated with neurodegenerative disease was observed in Gpt2-null brain. Cystathionine elevation is among the strongest signals in the metabolomics dataset with statistically significant elevations at both P0 and P18. Recently, several studies have implicated the accumulation of this product, as well as decreases in cysteine, which are also observed in Gpt2-null brains, in neurodegenerative disease. More specifically, through deficiency in cystathionine γ-lyase and mechanisms involving both redox homeostasis and sulfhydration, this metabolic pathway has been pinpointed in both Huntington’s disease (36, 37) and Parkinson’s disease (38), as well as other neurological diseases with mitochondrial defects. Other statistically significant perturbations in P18 brain supporting abnormalities in neuroprotective mechanisms include reductions in glutathione and folate. In summary, we have identified mutations in the mitochondrial enzyme GPT2 in a human neurological disorder, wherein mutations lead to metabolic dysfunctions that have general relevance to developmental as well as potentially to neurodegenerative mechanisms and disease.
Subjects and Methods
Human Studies.
Human subject research was conducted according to protocols approved by the institutional review boards of Boston Children’s Hospital, Beth Israel Deaconess Medical Center, and Butler Hospital, and equivalent committees of the other participating institutions. Written informed consent was obtained from all participants or their legal guardians.
Genome-Wide SNP Genotyping, Linkage Analysis, and WES.
DNA was genotyped using either Affymetrix Genome_Wide Human SNP Array 6.0 (MI1000) or Illumina HumanOmniExpress BeadChip array (MC9400). Affymetrix genotype calls were made using Birdsuite 1.5.5 (39) and Illumina calls by GenomeStudios version 2011.1 (Illumina). Genome-wide LOD scores were calculated using high-quality SNPs. PLINK, version 1.07 (40), was used to reduce linkage disequilibrium between markers. MERLIN, version 1.1.2 (41), was used to remove genotyping errors and to calculate LOD scores, assuming an autosomal recessive mode of inheritance with 100% penetrance and a disease allele prevalence of 0.0001. WES was performed using the Agilent Sure-Select Human All Exon or All Exon, version 5, capture array. Samples were sequenced on an Illumina HiSeq 2500. Sequence data were analyzed using GATK and variants annotated using ANNOVAR (42).
DNA Constructs and Antibodies.
Human GPT2 gene (GenBank: BC062555.1) was cloned into pcDNA3.1/V5-His-TOPO mammalian expression vector to generate C-terminal V5His-tagged phGPT2-V5His. GPT2 mutants p.Arg404*, p.Ser153Arg, and p.Pro272Leu were constructed using QuikChange site-directed mutagenesis (Agilent Technologies). The pTag GFP2-Mito vector was from Evrogen. Antibodies used were as follows: rabbit anti–C-terminal GPT2 antibody (SAB140991; Sigma-Aldrich), rabbit anti–full-length GPT2 antibody (16757-1-AP; Proteintech), mouse anti-V5 antibody (R960-25; Life Technologies), mouse anti-SV2 antibody (SV2-c; Developmental Studies Hybridoma Bank), chicken anti-MAP2 antibody (AB-15452; Millipore), mouse anti-Tubulin antibody (ab56676; Abcam), and rabbit anti-MTCO2 antibody (ab109739; Abcam).
GPT Enzyme Activity Assay.
GPT activity was analyzed using the alanine aminotransferase (ALT) Assay Kit (Abcam). Mouse brains or cells were homogenized in ice-cold ALT Assay buffer and centrifuged. Protein concentrations of supernatants were determined by bicinchoninic acid assay. GPT activity was detected using 30 μg of brain extracts or 3.5 μg of cell extracts.
Genome Editing of GPT2 in HEK293FT Cells.
Genomic alterations in HEK293FT cells at the GPT2 locus were made using the CRISPR-Cas9 system (43). Twenty nucleotide guide RNA (gRNA) sequences were designed to target double-stranded breaks at respective GPT2 target loci. For each transfection, a 180-nt single-stranded oligonucleotide was cotransfected with gRNA and the CRISPR-Cas9 construct [pSpCas9(BB)-2A-GFP; Addgene] to create single base substitutions via nonhomologous recombination. At 48 h posttransfection, single cells were sorted with GFP signal by FACS. The targeted genomic region was sequenced (see SI Appendix, Table S3, for sequences used). GPT2 mRNA level was quantified by qRT-PCR using primers (Hs0037287_m1; Life Technologies).
Generation of Gpt2-Null Mice.
Gpt2 cryopreserved mutant embryos were established through resources at the Knockout Mouse Project at University of California, Davis (Project ID CSD24977). The background of the embryos is C57BL/6N, and these mice were fully backcrossed into a C57BL/6J line. The targeting construct involved a gene trap (splice acceptor) and LacZ-Neo cassette (SI Appendix, Fig. S6A). Heterozygous males and heterozygous females were mated to produce offspring of both genders and all genotypes. Animals were genotyped by PCR with forward primer CSD-gene-F (5′-CTA ACT TGT CCT GCA TGG TGT CAG C-3′) and reverse primer CSD-gene-ttR (5′-CTA CAA ACA CTG GAC CCA AAC GTC C-3′) for a WT band of 504 bp. Forward primer CSD-loxF (5′-GAG ATG GCG CAA CGC AAT TAA TG-3′) and reverse primer CSD-gene-R (5′-GGC TTT CTA CCA GGA GGA ACA GAG G-3′) yields a 283-bp mutant band (SI Appendix, Fig. S6B, Left). WT and mutant mRNA were detected using RT-PCR, with the forward primer Unitrap-F (5′-CTG AGG TAA TCC GAG CCA AC-3′) and reverse primer Unitrap-wtR (5′-CGC TCG TTT CTT AGC GTC TT-3′) detecting a WT band (114 bp) and the Unitrap-F primer and Unitrap-laczR primer (5′-TGG CGA AAG GGG GAT GTG-3′) detecting a 300-bp mutant band (SI Appendix, Fig. S6B, Right). LacZ staining was performed as described (44). All experiments involving mice were carried out in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals (45) under the protocols approved by the Brown University Institutional Animal Care and Use Committee.
Brain Size Measurement.
Matched littermate pairs collected between P18 and P22 were transcardially perfused with 4% (wt/vol) paraformaldehyde and imaged using a Leica MZ16F. ImageJ analysis software was used to measure the area of the cortical hemispheres, midbrain, and cerebellum. Length measurements were done using four defined starting points.
Hippocampal Cultures.
Hippocampal neurons were cultured as described (46). At 10–14 d in vitro (DIV), cells were immunostained with anti-SV2 and anti-MAP2 antibodies. MAP2-positive neurons were scored for SV2 puncta using ImageJ Particle Analysis.
Metabolomics.
Craniotomies were performed on P0 and P18 litters from Gpt2 heterozygous parents. Whole brains were used for metabolomics preparations performed as per Yuan et al. (47). Methanol extracts were dried and run by targeted tandem liquid chromatography (LC)-MS/MS. Briefly, samples were injected via hydrophilic interaction LC at high pH using HPLC coupled to a 5500 QTRAP mass spectrometer (AB/SCIEX). Metabolites were monitored using SRM mode for 300 transitions with positive/negative polarity switching. Peak areas for each detected metabolite were integrated using MultiQuant software (AB/SCIEX). The results were analyzed using MetaboAnalyst 3.0 (48).
Stable Isotope Tracing in MEFs.
MEFs were maintained in DMEM with 10% (vol/vol) FCS and immortalized by SV40 T antigen. Experiments were performed between 5 and 15 passages. Stable isotope tracing experiments were performed as described previously (22). Briefly, cultures were overlaid with DMEM containing either 10 mM [U-13C]glucose and 4 mM unlabeled glutamine, or 10 mM unlabeled glucose and 4 mM [U-13C]glutamine. After culturing for 6–8 h, the cells were rinsed in ice-cold normal saline and lysed with three freeze–thaw cycles in cold 50% (vol/vol) methanol. Lysates were centrifuged, and supernatants were evaporated and derivatized by tert-butyldimethylsilyl-methyltrifluoroacetamide (Sigma-Aldrich). One microliter of derivatized material was injected onto an Agilent 7890 gas chromatograph networked to an Agilent 5975 mass selective detector. For analysis of 15N labeling in extracellular alanine, MEFs were incubated with medium containing 10 mM unlabeled glucose and 4 mM [α-15N]glutamine for 8 h. An aliquot of medium was passed over an AG50 column (Bio-Rad), and then amino acids were eluted with 4 mL of 4 N ammonium hydroxide. These samples were evaporated and processed as above. Total alanine abundance in medium was measured by HPLC (Hitachi L8900).
Supplementary Material
Acknowledgments
We thank the families who participated in this study. E.M.M. and G.H.M. kindly acknowledge Christopher A. Walsh for generous mentorship and support. The study was supported by the following: Burroughs Wellcome Fund Career Award for Medical Scientists 1006815.01 (to E.M.M.); Manton Center for Orphan Disease Research, National Institute of Neurological Disorders and Stroke Grant R01NS035129, National Institutes of Health/Fogarty International Center Grant R21TW008223, Dubai Harvard Foundation for Medical Research, F. Hoffmann-La Roche Ltd., Qatar National Research Fund, and Boston Children’s Hospital Faculty Career Development Award (to G.H.M.); National Cancer Institute Grant R01CA157996 (to R.J.D.); the Suna and Inan Kirac Foundation (O.B.); and the Japan Foundation for Pediatric Research and the Japan Society for the Promotion of Science (T.N.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1609221113/-/DCSupplemental.
References
- 1.Centers for Disease Control and Prevention (CDC) State-specific rates of mental retardation—United States, 1993. MMWR Morb Mortal Wkly Rep. 1996;45(3):61–65. [PubMed] [Google Scholar]
- 2.Fink JK. Hereditary spastic paraplegia: Clinical principles and genetic advances. Semin Neurol. 2014;34(3):293–305. doi: 10.1055/s-0034-1386767. [DOI] [PubMed] [Google Scholar]
- 3.Gilmore EC, Walsh CA. Genetic causes of microcephaly and lessons for neuronal development. Wiley Interdiscip Rev Dev Biol. 2013;2(4):461–478. doi: 10.1002/wdev.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kelly A, Stanley CA. Disorders of glutamate metabolism. Ment Retard Dev Disabil Res Rev. 2001;7(4):287–295. doi: 10.1002/mrdd.1040. [DOI] [PubMed] [Google Scholar]
- 5.Sheng ZH. Mitochondrial trafficking and anchoring in neurons: New insight and implications. J Cell Biol. 2014;204(7):1087–1098. doi: 10.1083/jcb.201312123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burté F, Carelli V, Chinnery PF, Yu-Wai-Man P. Disturbed mitochondrial dynamics and neurodegenerative disorders. Nat Rev Neurol. 2015;11(1):11–24. doi: 10.1038/nrneurol.2014.228. [DOI] [PubMed] [Google Scholar]
- 7.Kang HC, Lee YM, Kim HD. Mitochondrial disease and epilepsy. Brain Dev. 2013;35(8):757–761. doi: 10.1016/j.braindev.2013.01.006. [DOI] [PubMed] [Google Scholar]
- 8.Sheng ZH, Cai Q. Mitochondrial transport in neurons: Impact on synaptic homeostasis and neurodegeneration. Nat Rev Neurosci. 2012;13(2):77–93. doi: 10.1038/nrn3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Owen OE, Kalhan SC, Hanson RW. The key role of anaplerosis and cataplerosis for citric acid cycle function. J Biol Chem. 2002;277(34):30409–30412. doi: 10.1074/jbc.R200006200. [DOI] [PubMed] [Google Scholar]
- 10.Rajab A, et al. An autosomal recessive form of spastic cerebral palsy (CP) with microcephaly and mental retardation. Am J Med Genet A. 2006;140(14):1504–1510. doi: 10.1002/ajmg.a.31288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Adzhubei IA, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4(7):1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 13.Choi Y, Sims GE, Murphy S, Miller JR, Chan AP. Predicting the functional effect of amino acid substitutions and indels. PLoS One. 2012;7(10):e46688. doi: 10.1371/journal.pone.0046688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Celis K, et al. Loss of function mutation in glutamic pyruvate transaminase 2 (GPT2) causes developmental encephalopathy. J Inherit Metab Dis. 2015;38(5):941–948. doi: 10.1007/s10545-015-9824-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Y, et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci. 2014;34(36):11929–11947. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brody BA, Kinney HC, Kloman AS, Gilles FH. Sequence of central nervous system myelination in human infancy. I. An autopsy study of myelination. J Neuropathol Exp Neurol. 1987;46(3):283–301. doi: 10.1097/00005072-198705000-00005. [DOI] [PubMed] [Google Scholar]
- 17.Huttenlocher PR, Dabholkar AS. Regional differences in synaptogenesis in human cerebral cortex. J Comp Neurol. 1997;387(2):167–178. doi: 10.1002/(sici)1096-9861(19971020)387:2<167::aid-cne1>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 18.Li M, et al. Synaptogenesis in the developing mouse visual cortex. Brain Res Bull. 2010;81(1):107–113. doi: 10.1016/j.brainresbull.2009.08.028. [DOI] [PubMed] [Google Scholar]
- 19.Mitew S, et al. Mechanisms regulating the development of oligodendrocytes and central nervous system myelin. Neuroscience. 2014;276:29–47. doi: 10.1016/j.neuroscience.2013.11.029. [DOI] [PubMed] [Google Scholar]
- 20.Horton P, Nakai K. Better prediction of protein cellular localization sites with the k nearest neighbors classifier. Proc Int Conf Intell Syst Mol Biol. 1997;5:147–152. [PubMed] [Google Scholar]
- 21.DeBerardinis RJ, et al. Beyond aerobic glycolysis: Transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci USA. 2007;104(49):19345–19350. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheng T, et al. Pyruvate carboxylase is required for glutamine-independent growth of tumor cells. Proc Natl Acad Sci USA. 2011;108(21):8674–8679. doi: 10.1073/pnas.1016627108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lo Giudice T, Lombardi F, Santorelli FM, Kawarai T, Orlacchio A. Hereditary spastic paraplegia: Clinical-genetic characteristics and evolving molecular mechanisms. Exp Neurol. 2014;261:518–539. doi: 10.1016/j.expneurol.2014.06.011. [DOI] [PubMed] [Google Scholar]
- 24.Lindblom P, et al. Isoforms of alanine aminotransferases in human tissues and serum—differential tissue expression using novel antibodies. Arch Biochem Biophys. 2007;466(1):66–77. doi: 10.1016/j.abb.2007.07.023. [DOI] [PubMed] [Google Scholar]
- 25.Glantz LA, Gilmore JH, Hamer RM, Lieberman JA, Jarskog LF. Synaptophysin and postsynaptic density protein 95 in the human prefrontal cortex from mid-gestation into early adulthood. Neuroscience. 2007;149(3):582–591. doi: 10.1016/j.neuroscience.2007.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barnea-Goraly N, et al. White matter development during childhood and adolescence: A cross-sectional diffusion tensor imaging study. Cereb Cortex. 2005;15(12):1848–1854. doi: 10.1093/cercor/bhi062. [DOI] [PubMed] [Google Scholar]
- 27.Kessaris N, et al. Competing waves of oligodendrocytes in the forebrain and postnatal elimination of an embryonic lineage. Nat Neurosci. 2006;9(2):173–179. doi: 10.1038/nn1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rowitch DH, Kriegstein AR. Developmental genetics of vertebrate glial-cell specification. Nature. 2010;468(7321):214–222. doi: 10.1038/nature09611. [DOI] [PubMed] [Google Scholar]
- 29.Takebayashi H, Ikenaka K. Oligodendrocyte generation during mouse development. Glia. 2015;63(8):1350–1356. doi: 10.1002/glia.22863. [DOI] [PubMed] [Google Scholar]
- 30.Franco R, Cidlowski JA. Apoptosis and glutathione: Beyond an antioxidant. Cell Death Differ. 2009;16(10):1303–1314. doi: 10.1038/cdd.2009.107. [DOI] [PubMed] [Google Scholar]
- 31.Snoke JE, Yanari S, Bloch K. Synthesis of glutathione from gamma-glutamylcysteine. J Biol Chem. 1953;201(2):573–586. [PubMed] [Google Scholar]
- 32.Lau A, Tymianski M. Glutamate receptors, neurotoxicity and neurodegeneration. Pflugers Arch. 2010;460(2):525–542. doi: 10.1007/s00424-010-0809-1. [DOI] [PubMed] [Google Scholar]
- 33.Boyko M, Gruenbaum SE, Gruenbaum BF, Shapira Y, Zlotnik A. Brain to blood glutamate scavenging as a novel therapeutic modality: A review. J Neural Transm (Vienna) 2014;121(8):971–979. doi: 10.1007/s00702-014-1181-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van Hove J, Coughlin CII, Scharer G. 2013 Glycine encephalopathy. GeneReviews, eds Pagon RA, et al. (University of Washington, Seattle, WA). Available at www.ncbi.nlm.nih.gov/books/NBK1357/. Accessed January 14, 2016.
- 35.Weinberg F, et al. Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity. Proc Natl Acad Sci USA. 2010;107(19):8788–8793. doi: 10.1073/pnas.1003428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paul BD, et al. Cystathionine γ-lyase deficiency mediates neurodegeneration in Huntington’s disease. Nature. 2014;509(7498):96–100. doi: 10.1038/nature13136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paul BD, Snyder SH. Neurodegeneration in Huntington’s disease involves loss of cystathionine γ-lyase. Cell Cycle. 2014;13(16):2491–2493. doi: 10.4161/15384101.2014.950538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vandiver MS, et al. Sulfhydration mediates neuroprotective actions of parkin. Nat Commun. 2013;4:1626. doi: 10.1038/ncomms2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Korn JM, et al. Integrated genotype calling and association analysis of SNPs, common copy number polymorphisms and rare CNVs. Nat Genet. 2008;40(10):1253–1260. doi: 10.1038/ng.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Purcell S, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin—rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30(1):97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 42.Wang K, Li M, Hakonarson H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ran FA, et al. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8(11):2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cepko C, Ryder E, Fekete DM, Bruhn S. 2006 Detection of β-Galactosidase and Alkaline Phosphatase Activities in Tissue (Department of Genetics, Harvard Medical School, Cambridge, MA). Available at genepath.med.harvard.edu/∼cepko/protocol/xgalplap-stain.htm. Accessed February 22, 2015.
- 45.National Research Council . Guide for the Care and Use of Laboratory Animals. 8th Ed National Academies Press; Washington, DC: 2011. [Google Scholar]
- 46.Ouyang Q, et al. Christianson syndrome protein NHE6 modulates TrkB endosomal signaling required for neuronal circuit development. Neuron. 2013;80(1):97–112. doi: 10.1016/j.neuron.2013.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yuan M, Breitkopf SB, Yang X, Asara JM. A positive/negative ion-switching, targeted mass spectrometry-based metabolomics platform for bodily fluids, cells, and fresh and fixed tissue. Nat Protoc. 2012;7(5):872–881. doi: 10.1038/nprot.2012.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xia J, Sinelnikov IV, Han B, Wishart DS. MetaboAnalyst 3.0—making metabolomics more meaningful. Nucleic Acids Res. 2015;43(W1):W251–W257. doi: 10.1093/nar/gkv380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mullen AR, et al. Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature. 2011;481(7381):385–388. doi: 10.1038/nature10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.