Significance
The transcriptional repressor protein Capicua (Cic) is a conserved regulator of organ growth and tissue patterning, and mutations in the CIC gene in humans result in the brain cancer oligodendroglioma. Cic activity is controlled by the receptor tyrosine kinase (RTK) signaling pathway. Here, we identify the kinase Minibrain (Mnb) and its adaptor Wings apart (Wap) as Cic regulators. Mnb and Wap bind to and phosphorylate the Cic protein, and inhibit the ability of Cic to repress gene expression. Mnb-dependent down-regulation of Cic is necessary for the proper growth of multiple organs and correct the patterning of tissues. Our results uncover a previously unknown mechanism of Cic regulation that acts in parallel to other growth-controlling pathways.
Keywords: minibrain, capicua, organ growth, DYRK1A, tissue patterning
Abstract
The transcriptional repressor Capicua (Cic) controls tissue patterning and restricts organ growth, and has been recently implicated in several cancers. Cic has emerged as a primary sensor of signaling downstream of the receptor tyrosine kinase (RTK)/extracellular signal-regulated kinase (ERK) pathway, but how Cic activity is regulated in different cellular contexts remains poorly understood. We found that the kinase Minibrain (Mnb, ortholog of mammalian DYRK1A), acting through the adaptor protein Wings apart (Wap), physically interacts with and phosphorylates the Cic protein. Mnb and Wap inhibit Cic function by limiting its transcriptional repressor activity. Down-regulation of Cic by Mnb/Wap is necessary for promoting the growth of multiple organs, including the wings, eyes, and the brain, and for proper tissue patterning in the wing. We have thus uncovered a previously unknown mechanism of down-regulation of Cic activity by Mnb and Wap, which operates independently from the ERK-mediated control of Cic. Therefore, Cic functions as an integrator of upstream signals that are essential for tissue patterning and organ growth. Finally, because DYRK1A and CIC exhibit, respectively, prooncogenic vs. tumor suppressor activities in human oligodendroglioma, our results raise the possibility that DYRK1A may also down-regulate CIC in human cells.
The high mobility group-box transcriptional repressor protein Capicua (Cic) has been identified as a key regulator of tissue patterning and organ growth in multiple developmental contexts (1, 2). In Drosophila, Cic controls anteroposterior and dorsoventral embryonic polarity, the subdivision of the lateral ectoderm, and pattern formation in several tissues (1, 3–6). In addition, Cic negatively regulates the growth of imaginal discs and the midgut (7, 8). In humans, a single Cic ortholog (CIC) has been implicated in the neurodegenerative disease spinocerebellar ataxia 1 (SCA1) (9), and recently mutations in CIC have been found in the majority of oligodendroglioma cases, suggesting that CIC is a tumor suppressor (10–12).
In both Drosophila and mammals, Cic functions as a primary sensor of signaling downstream of the receptor tyrosine kinase (RTK)/extracellular signal-regulated kinase (ERK) pathway (2, 5–8, 13–16). According to the current model, activation of RTK signaling results in the accumulation of doubly phosphorylated activated ERK, which directly binds to and phosphorylates Cic (5). ERK-mediated Cic phosphorylation leads to a rapid relief of repression of Cic target genes, followed by a slower export from the nucleus and eventual cytoplasmic degradation (13, 17). The molecular details of these processes are unknown, although apparently each of them contributes to the overall down-regulation of Cic activity. Cic is also involved in a mutual regulatory relationship with the Hippo pathway, although regulation of Cic in this context appears to take place at the RNA level (18).
Here, we present the identification of the kinase Minibrain (Mnb) (19, 20) and an adaptor protein, Wings Apart (Wap) (20, 21), as Cic regulators that cooperate to phosphorylate Cic and restrict its repressor activity. We show that Mnb/Wap and ERK target different regions of the Cic protein for phosphorylation, and that inhibition of Cic activity by Mnb and Wap is required for the growth of several organs and for correct patterning of the wing. Our data suggest that Mnb/Wap-dependent down-regulation of Cic occurs in parallel to the RTK/ERK and Hippo signaling pathways. We propose that Cic functions as an integrator of upstream developmental signals, which together tightly control its activity. This mechanism is necessary for the proper execution of tissue patterning and regulation of organ growth.
Results
Wap and Mnb Interact with Cic.
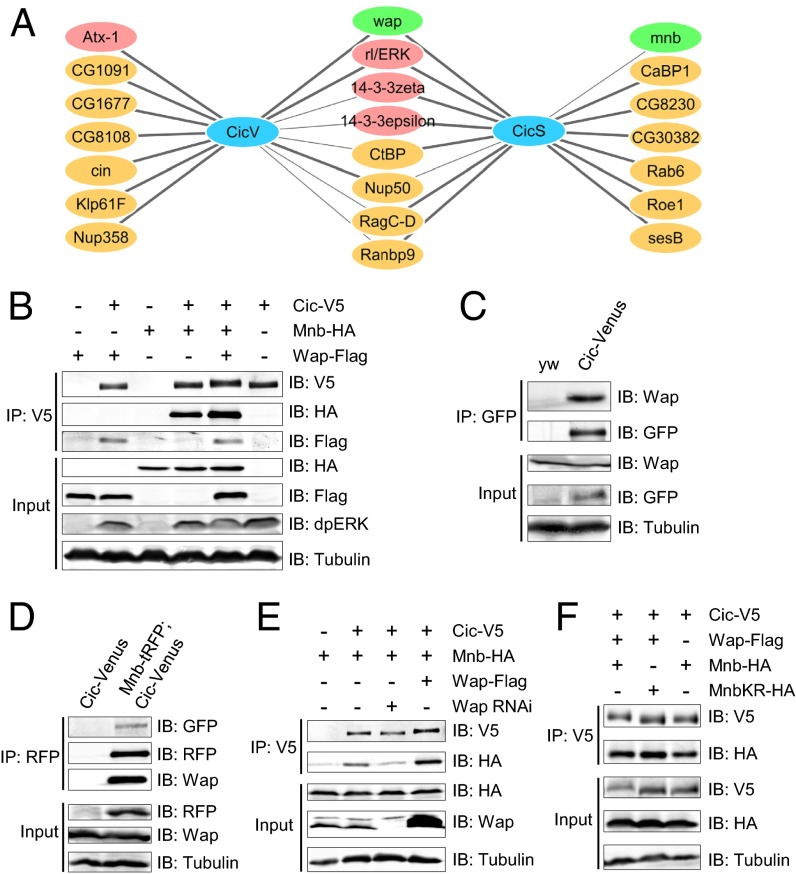
To identify Cic regulators, we used affinity purification/mass spectrometry (AP-MS) (22) to study the Cic protein interactome in Drosophila S2 cells and embryos. Embryonic cic-Venus was expressed at endogenous levels as part of a genomic rescue construct (13). We successfully recovered most of the known interactors of Cic, such as the Drosophila ERK ortholog Rolled (5), Ataxin-1 (9), and 14-3-3 proteins (16) (Fig. 1A). One of the prominent Cic interactors identified with high confidence in both cultured cells and embryos was Wap (also known as Riquiqui) (Fig. 1A and Dataset S1) (20, 21). Wap binds to the kinase Mnb (19, 20), and this interaction is conserved in mammals, because the Wap ortholog DDB1 and CUL4 associated factor 7 (DCAF7) forms a stable complex with the dual-specificity tyrosine phosphorylation-regulated kinase 1A (DYRK1A), which is an ortholog of Mnb (23). We have shown that Wap functions together with Mnb to regulate wing and leg tissue growth through the Hippo pathway (20). Our AP-MS experiments also identified four peptides of Mnb in the Cic-streptavidin binding peptide (SBP) pulldown in S2 cells (Fig. 1A and Dataset S1), suggesting that Wap, Mnb, and Cic form a protein complex. Coimmunoprecipitation in S2 cells using overexpressed proteins confirmed that Cic binds to both Wap and Mnb (Fig. 1B). To study the interactions between proteins expressed at endogenous levels in vivo, we generated tagged mnb-tagRFP-T (mnb-tRFP) and wap-Venus alleles by CRISPR/Cas9-mediated homologous recombination. Successful targeting was confirmed by RNAi (Fig. S1), and we found that both Mnb–tRFP and Wap–Venus were expressed throughout imaginal discs and the larval brain (Fig. S2). Endogenous Wap was detected in the Cic–Venus complex by using an anti-DCAF7 antibody (Fig. 1C), and both Wap and Cic–Venus were present in Mnb–tRFP complexes isolated from embryos in which Mnb–tRFP and Cic–Venus were coexpressed (Fig. 1D). Next, we asked whether Wap could serve as a bridge for the interaction between Mnb and Cic. RNAi depletion of wap in S2 cells led to a reduction in the binding of Cic to Mnb, whereas overexpression of Wap promoted the interaction (Fig. 1E). Collectively, these data suggest that Wap, Mnb, and Cic form a protein complex, with Wap likely serving as a bridging adaptor between Cic and Mnb.
Fig. 1.
Mnb and Wap physically interact with Cic, and Wap promotes the binding of Mnb to Cic. (A) The Cic protein interactome identified in Drosophila S2 cells (CicS) and embryos (CicV). Thick lines, highly significant interactions. A complete dataset is in Dataset S1. (B) Western blots showing coimmunoprecipitation of Cic, Mnb, and Wap in S2 cells. Endogenous dpERK is stabilized by Cic expression. (C and D) Coimmunoprecipitation of Cic, Mnb, and Wap in vivo using embryo lysates from yw (control), cic-Venus, or cic-Venus crossed with mnb-tRFP. (E) Wap is required and sufficient to bridge Cic and Mnb. (F) Cic mobility was changed when Cic was coexpressed with Wap and wild-type Mnb, but not kinase-dead Mnb (MnbKR).
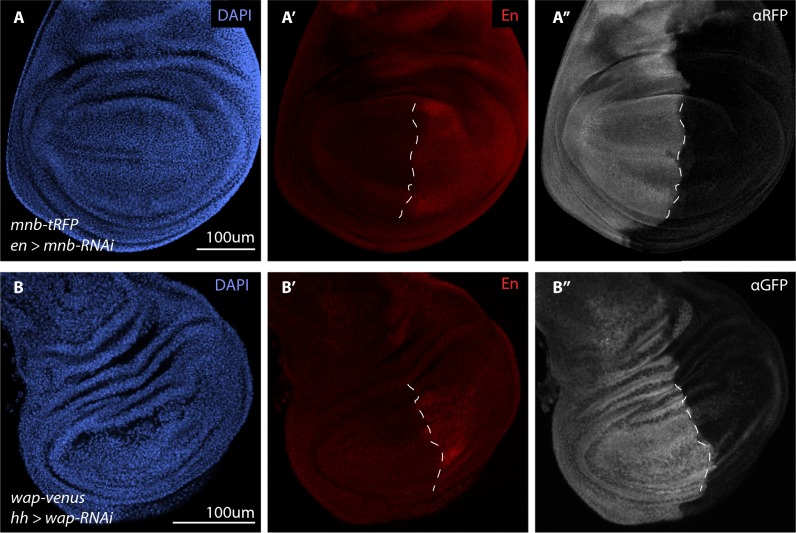
Fig. S1.
Depletion of fluorescently tagged endogenous Mnb and Wap confirms the on-target effect of mnb-RNAi and wap-RNAi. (A–B′′) Wing discs from third instar larvae of the indicated genotypes. The en-GAL4 and hh-GAL4 drivers are expressed in the posterior compartment of wing discs (red in A’ and B’) and were used to drive the expression of UAS-mnb RNAi (A–A′′) and UAS-wap RNAi (B–B′′) respectively. Cell nuclei were detected with DAPI (blue in A and B) whereas Mnb-tRFP (gray in A′′) and Wap-Venus (gray in B′′) were detected by anti-tRFP and anti-GFP antibodies, respectively. (Scale bars: 100 µm.) Dashed lines (A′ and B′) indicate the anterior-posterior compartment boundary of the wing pouch.
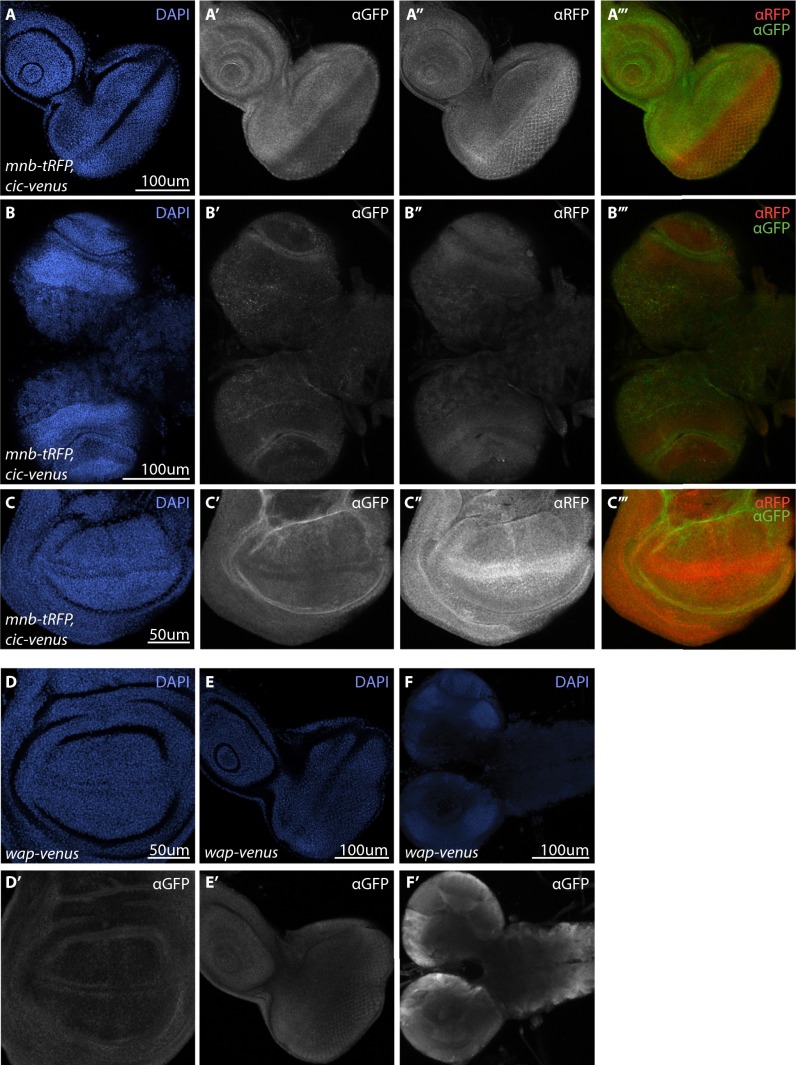
Fig. S2.
Mnb-RFP, Cic-Venus, and Wap-Venus expression in third instar larval tissues. (A–F) Cic-Venus and Wap-Venus expression was detected by GFP antibody, Mnb-tRFP was detected by tRFP antibody and cell nuclei were detected by DAPI (blue). (A′–C′′′) Cic-Venus and Mnb-tRFP expression in eye imaginal discs (A–A’’’), brain optic lobes (B’–B’’’), and the pouch region of wing imaginal discs (C’–C’’’). For merged panels (A’’’–C’’’), Cic-Venus is in green and Mnb-tRFP is in red. (D–F’) Wap-Venus expression (gray) in wing imaginal discs (D’), eye imaginal discs (E’) and larval brain (F’). (Scale bars: A–B′′′, E, and F, 100 µm; C and D, 50 μm.)
Mnb Phosphorylates the Amino-Terminal Third of Cic in S2 Cells.
Given that Mnb/DYRK1A is a kinase (19, 20), we asked whether Mnb could phosphorylate Cic. Cic mobility on an SDS/PAGE was reduced when Cic, Mnb, and Wap were coexpressed (Fig. 1 B, E, and F). Notably, the levels of activated ERK (dpERK) did not increase in this condition (Fig. 1B). In contrast, a kinase-dead mutant of Mnb (MnbKR) (20) failed to reduce Cic mobility (Fig. 1F), suggesting that the kinase activity of Mnb is required to reduce Cic mobility and that this modification is likely to be phosphorylation.
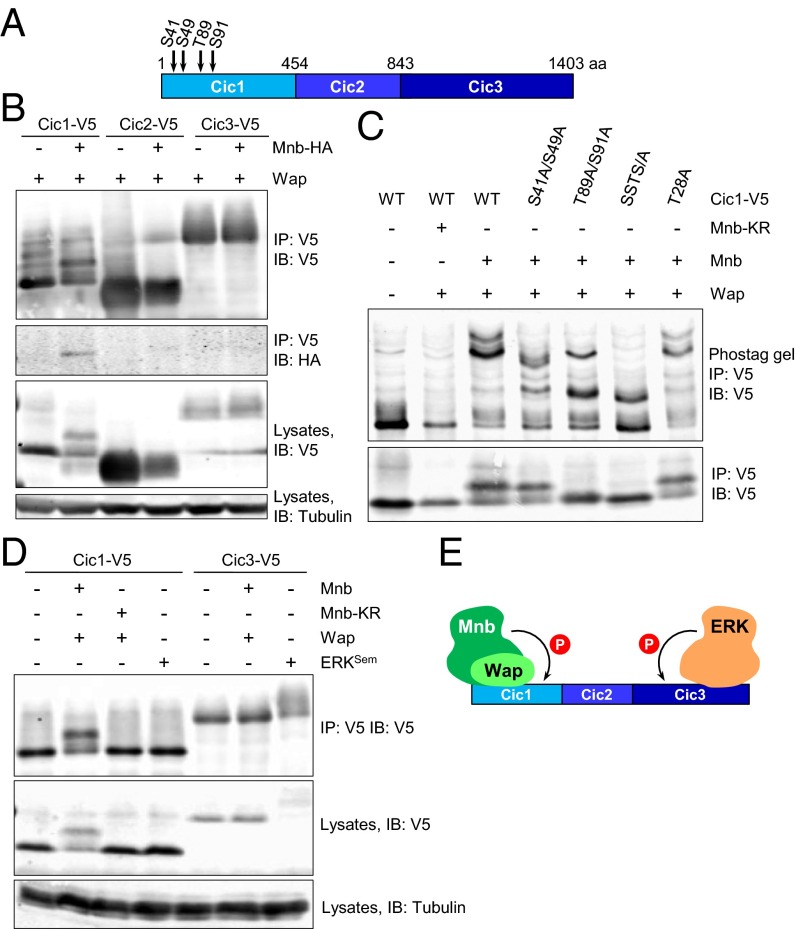
To determine which region of Cic was phosphorylated by Mnb, three Cic fragments (Cic1–3; Fig. 2A) were coexpressed with Wap in the presence or absence of Mnb in S2 cells. Only the amino-terminal fragment of Cic (Cic1, representing amino acids 1–453) was found to interact with Mnb (Fig. 2B). In addition, Mnb decreased the electrophoretic mobility of Cic1, but not Cic2 or Cic3 (Fig. 2B). Phos-tag gel analysis confirmed phosphorylation of Cic1 by wild-type but not kinase-dead Mnb (Fig. 2C). Next, we asked which residue(s) in Cic1 are phosphorylated by Mnb. Threonine 28 is part of a motif in Cic1 (RSATP) that closely matches the DYRK1A phosphorylation consensus RP(X)(S/T)P (24). Surprisingly, mutation of this residue (T28A) did not alter the phosphorylation pattern of Cic1 (Fig. 2C). To identify Cic residues that are phosphorylated by Mnb, Cic1 and Wap were coexpressed in S2 cells either with Mnb or MnbKR, Cic1 was purified, and its phosphorylation was analyzed by mass spectrometry. Four Cic1 residues (S41, S49, T89, and S91) were more highly phosphorylated by Mnb compared with MnbKR, with T89 and S91 phosphorylations found exclusively in the wild-type Mnb sample (Fig. 2A and Fig. S3). The S41 and S49 residues were also found to be phosphorylated in an unbiased global phosphoproteomic study in Drosophila embryos (25). Alanine substitutions of the four residues resulted in a reduction of phosphorylation of Cic1, with the most pronounced effect observed for a quadruple mutant, Cic-SSTS/A (Fig. 2C).
Fig. 2.
Mnb and ERK target different regions of Cic for phosphorylation. (A) Schematic diagram of the three Cic fragments (Cic1, Cic2, and Cic3) with locations of phosphorylation sites. (B) Mnb interacts with and phosphorylates only the amino-terminal Cic fragment, Cic1. (C) Phos-tag gel analysis of Cic1 phosphorylation. (C Bottom) Regular SDS/PAGE. (D) Mnb phosphorylates region Cic1, whereas activated ERK (ERKSem) phosphorylates region Cic3. (E) Summary of Cic binding and phosphorylation data.
Fig. S3.
Locations of Cic phosphorylation sites identified by mass spectrometry. A Cic1 region is shown (amino acids 1–453). The putative DYRK1A consensus is underlined, and the corresponding residue (T28) is highlighted in blue. T28 phosphorylation was not detected by mass spectrometry. S41 and S49 (green) were more highly phosphorylated in wild-type Mnb samples compared with MnbKR. T89 and S91 phosphorylations (red) were found exclusively in the wild-type Mnb samples.
Our previous studies showed that region Cic3 includes an ERK docking site and is subject to ERK-mediated phosphorylation (5, 14, 26). To compare the activities of Mnb and ERK, we coexpressed Cic1 or Cic3 with Mnb or a constitutively active Drosophila ERK, ERKSem (27). Mnb could only reduce the electrophoretic mobility of Cic1 but not Cic3, whereas ERKSem only reduced the electrophoretic mobility of Cic3 but not Cic1 (Fig. 2D). Collectively, these results suggest that Mnb and ERK target different regions of Cic for phosphorylation: Wap facilitates Mnb-dependent phosphorylation of the amino-terminal third of Cic, whereas ERK targets the carboxyl-terminal region (Fig. 2E).
Mnb and Wap Reduce Cic Repressor Activity.
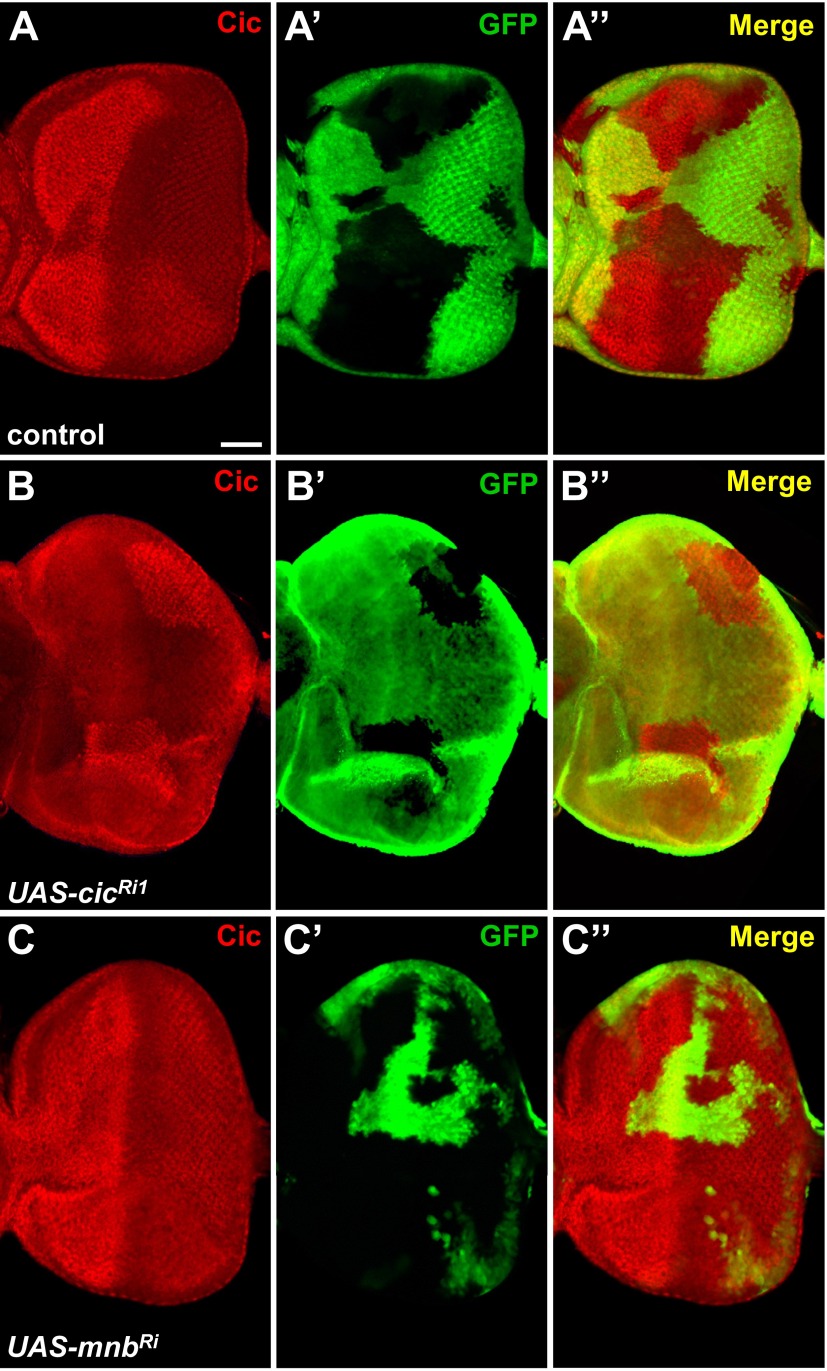
Previous studies have shown that phosphorylation of Cic by ERK can result in down-regulation of Cic by lowering its repressor activity, protein level, or nuclear localization (5, 6, 17). We hypothesized that Mnb may exert similar effects. First, we used the CoinFLP-GAL4 system (28) to generate RNAi-depletion clones in the eye imaginal discs (Fig. S4A). As expected, we observed reduced levels of Cic protein in UAS-cic-RNAi clones (Fig. S4B). However, no obvious increase in Cic protein level or change in subcellular localization was found in CoinFLP-generated UAS-mnb-RNAi clones (Fig. S4C). Therefore, Mnb is unlikely to control Cic at the level of protein turnover or nuclear access.
Fig. S4.
RNAi depletion of Mnb does not increase Cic levels in imaginal eye discs. (A–C′′) Mosaic eye discs of the indicated genotypes generated by using CoinFLP-GAL4 and stained with anti-Cic antibody (red). GAL4-positive cells are marked with UAS-GFP (green). (A–A′′) Control clones do not affect Cic protein levels. (B–B′′) Knockdown of cic reduces Cic protein levels. (C–C′′) Knockdown of mnb does not increase Cic protein level or change Cic subcellular localization. (Scale bar: 50 μm.)
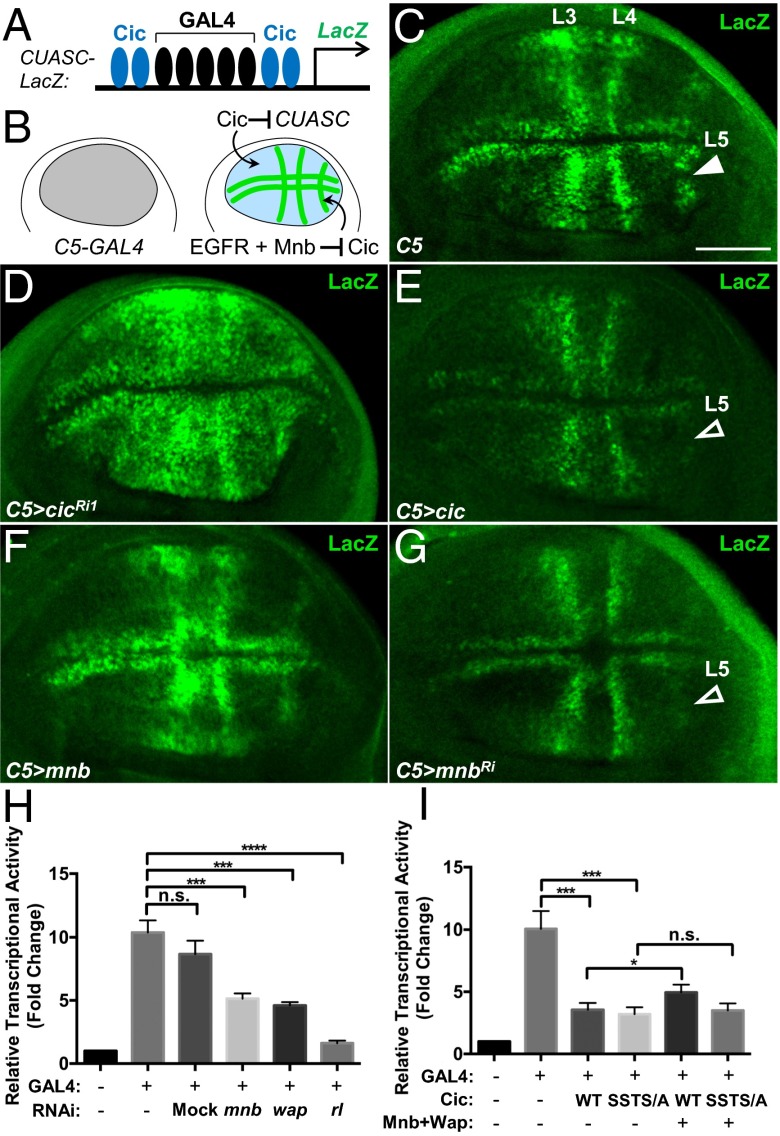
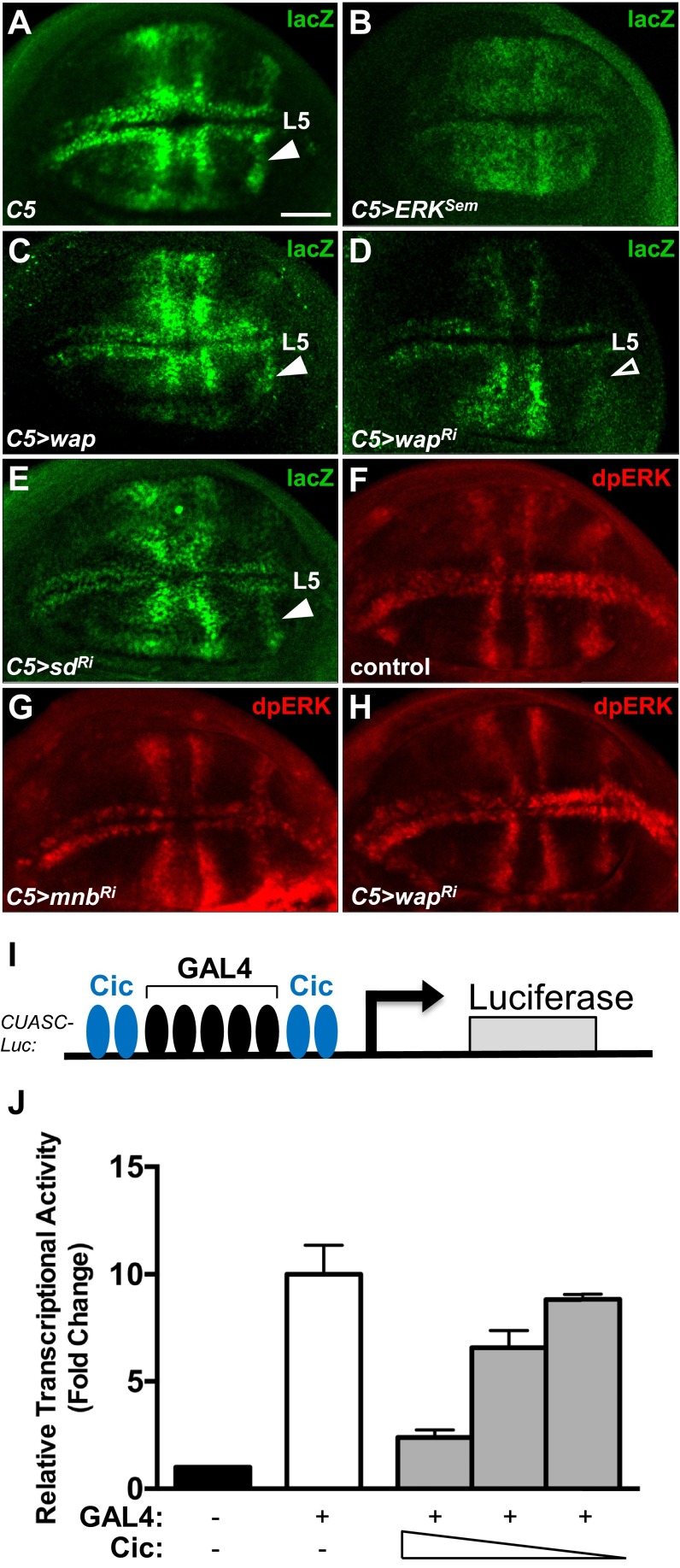
We have shown that the relief of Cic repressor function by ERK does not necessarily require reduction in Cic protein levels (17). To assess whether Mnb could similarly affect Cic repressor activity, we used a reporter, CUASC-lacZ, which contains five GAL4 binding sites flanked on either side by two Cic binding motifs (Fig. 3A) (6). This reporter is only responsive to GAL4 in areas where Cic activity is inhibited, e.g., by RTK signaling (Fig. 3B). Uniform induction of GAL4 expression in the wing pouch under the control of the C5-GAL4 driver (29) resulted in a localized activation of LacZ expression in prospective veins (Fig. 3C). This pattern results from epidermal growth factor receptor (EGFR)/ERK-mediated inactivation of Cic in these regions (Fig. 3B) (6). RNAi depletion of cic or overexpression of ERKSem throughout the wing pouch led to a much broader expression of LacZ (Fig. 3D and Fig. S5B), confirming that the normal restriction of the expression pattern of CUASC-lacZ to prospective veins is Cic-dependent. In contrast, overexpression of Cic resulted in the loss of LacZ expression in the vein L5 region (Fig. 3E, open arrowhead). Overexpression of Mnb or Wap induced a broader LacZ expression in the wing pouch (Fig. 3F and Fig. S5C). Conversely, RNAi depletion of mnb or wap under the control of C5-GAL4 resulted in reduced LacZ expression, particularly in vein L5 (Fig. 3G and Fig. S5D). These data suggest that Mnb and Wap limit Cic repressor function in the wing disc. This contribution likely complements the regulation by ERK, which appears to be insufficient on its own, at least for vein L5 (Fig. 3B).
Fig. 3.
Mnb reduces Cic repressor activity. (A) Diagram of the CUASC-lacZ reporter. (B) Summary diagram of expression patterns. (C–G) LacZ expression pattern resulting from C5-GAL4–directed activation of CUASC-lacZ in wing discs from control (C), UAS-cicRNAi1 (D), UAS-cic (E), UAS-mnb (F), and UAS-mnbRNAi (G) larvae. (Scale bar: 50 μm.) (H and I) Luciferase assays using CUASC-Luc reporter in S2 cells. (H) mnb, wap, and rl (ERK) are required to limit the activity of Cic. (I) Mnb and Wap reduce transcriptional repressor activity of wild-type Cic, but not of the phosphorylation site mutant, Cic-SSTS/A. n.s., not significant, *P < 0.05, **P < 0.01, ***P < 0.001, statistical significance was analyzed by using unpaired Student’s t test. Error bars represent SD.
Fig. S5.
Modulation of Cic repressor activity by Mnb and Wap does not require Sd and does not affect activity of ERK. (A–E) LacZ expression pattern resulting from C5-GAL4–directed activation of CUASC-lacZ in wing imaginal discs from control (A), UAS-ERKSem (B), UAS-wap (C), UAS-wapRNAi (D), and UAS-sdRNAi (E) larvae. Expression of ERKSem and Wap led to a broader activation of the reporter (B and C), whereas knockdown of wap resulted in lower LacZ expession, particularly in presumptive vein L5 (D). Knockdown of sd did not alter the normal pattern of expression in presumptive veins. (F–H) dpERK expression pattern in wing discs from control (F), C5-GAL4 > UAS-mnbRNAi (G), and C5-GAL4 > UAS-wapRNAi (H) larvae. (I) Schematic diagram of the CUASC-Luc reporter construct. (J) Dose-dependent repression of CUASC-Luc expression by Cic. S2 cells were cotransfected with the reporter CUASC-Luc, pMT-GAL4, and decreasing amounts of Cic-expressing plasmid (500 ng, 250 ng, 125 ng). The values shown are fold changes over the negative control set at 1. (Scale bar: 50 μm.)
Mnb and Wap have been shown to phosphorylate and inhibit Warts, which results in elevated Yki activity (20). To test whether CUASC-lacZ expression was affected by Hippo signaling, we depleted the levels of the Yki-interacting transcription factor Scalloped (Sd), which is required for the activation of Yki targets (30). We observed that knockdown of sd using RNAi had no obvious effect on the expression of CUASC-lacZ (Fig. S5E), suggesting that Hippo signaling is not involved in the regulation of Cic repressor activity in this context. To further assess whether Mnb and Wap engage RTK/ERK signaling to control Cic, we analyzed dpERK levels in wing pouches expressing mnb-RNAi or wap-RNAi. We found that RNAi depletion of wap or mnb did not alter the dpERK pattern in wing discs (Fig. S5 F–H). This result is in agreement with the observation that overexpression of Mnb did not increase dpERK levels in S2 cells (Fig. 1B). We conclude that Mnb and Wap down-regulates Cic repressor activity independently from the RTK/ERK and the Hippo pathways.
To directly address how Mnb and Wap affect Cic function as a transcriptional repressor, we studied the activity of a reporter, CUASC-Luc, which is controlled by GAL4 and Cic, in S2 cells (Fig. S5I). Transfection of GAL4 activated this reporter ∼10-fold, and this activation was repressed by coexpression of Cic in a dose-dependent manner (Fig. S5J). Depletion of endogenous mnb, wap, or rl (ERK) by RNAi resulted in a reduction of reporter activity (Fig. 3H), suggesting that Mnb, Wap and ERK are required to limit the activity of Cic. We next tested whether Mnb and Wap could reduce the capacity of Cic to repress CUASC-Luc expression, and found that cotransfection of Cic with Mnb and Wap partially relieved Cic-mediated repression of this reporter (Fig. 3I). Whereas the Cic-SSTS/A mutant repressed the reporter gene expression to a similar level as wild-type Cic, coexpression of this mutant with Mnb and Wap did not affect its ability to repress CUASC-Luc (Fig. 3I). Collectively, these results indicate that Mnb and Wap reduce the activity of Cic as a transcriptional repressor, likely via Mnb-mediated phosphorylation of residues located in the amino terminus of the Cic protein.
Mnb Opposes Cic Function in Controlling Wing and Eye Growth.
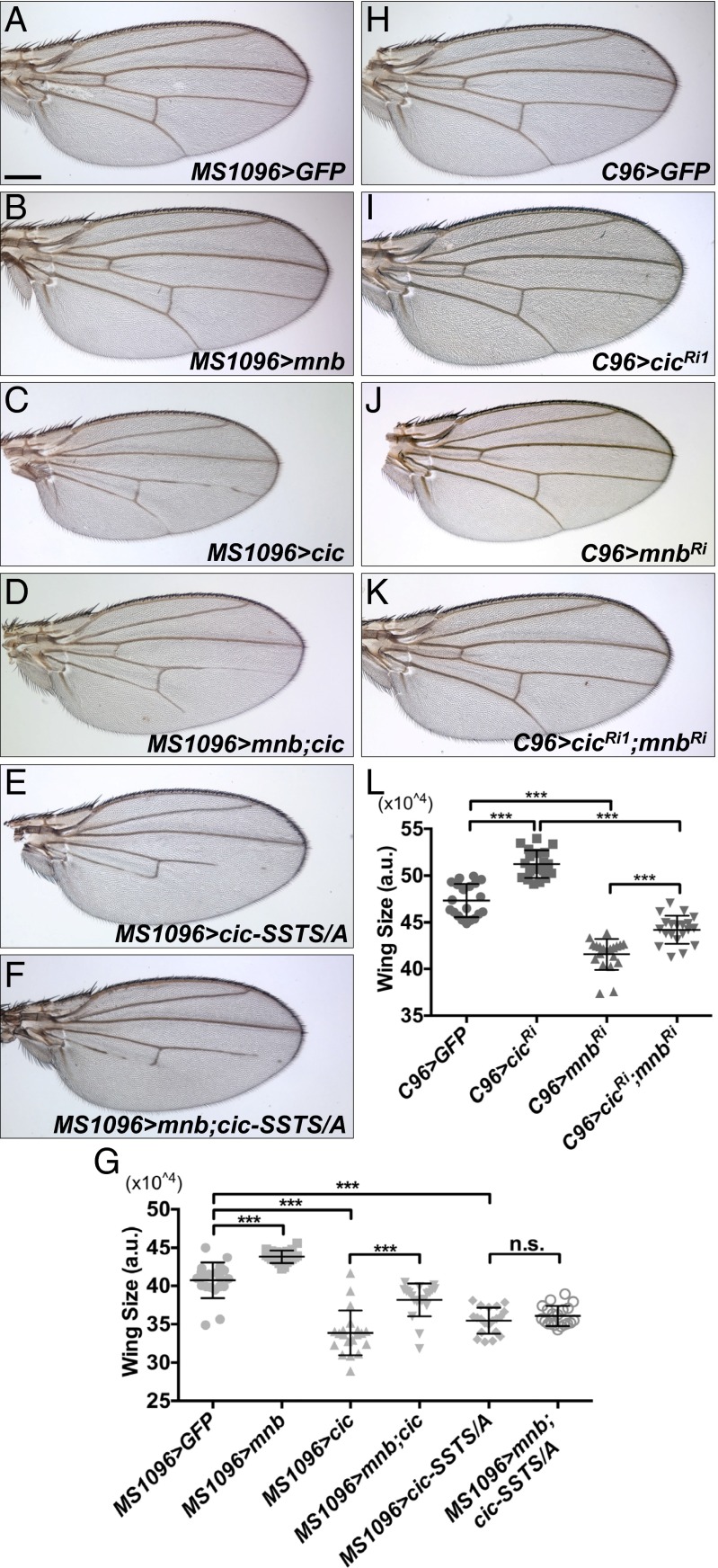
We next investigated whether the inhibitory effects of Mnb/Wap on Cic were involved in the control of organ growth. Overexpression of Mnb using the wing pouch MS1096-GAL4 driver (31) promoted wing growth (Fig. 4 B and G). Conversely, overexpression of Cic or the Cic-SSTS/A mutant resulted in a reduction of wing size (Fig. 4 C, E, and G). Whereas coexpression of Mnb with Cic suppressed the smaller wing size associated with Cic overexpression (Fig. 4 C, D, and G), coexpression of Mnb with Cic-SSTS/A did not modify this phenotype (Fig. 4 E–G). These data suggest that Mnb regulates Cic function at least in part through the phosphorylation of the SSTS residues. We also asked whether mnb and cic would display opposing effects on growth in a reduction-of-function context. RNAi depletion of cic using the MS1096-GAL4 driver caused a severe defect in wing development. We thus used a weaker driver, C96-GAL4, which is expressed primarily around the wing margin (32), to study the effects of reduced levels of mnb and cic. Knockdown of cic caused wing overgrowth (Fig. 4 I and L), and RNAi depletion of mnb resulted in an opposite effect (Fig. 4 J and L). Importantly, RNAi depletion of cic partially rescued the small wing phenotype induced by expression of mnb-RNAi (Fig. 4 K and J). Mutually antagonistic effects of Mnb and Cic on growth were also observed in the eye (Fig. S6). Collectively, we conclude that Mnb and Wap promote wing and eye growth by antagonizing the growth-restricting function of Cic.
Fig. 4.
Mnb opposes Cic function in controlling wing growth. (A–F) Wings from adult female flies expressing UAS-GFP as a control (A), UAS-mnb (B), UAS-cic (C), UAS-mnb together with UAS-cic (D), UAS-cic-SSTS/A (E), and UAS-mnb together with UAS-cic-SSTS/A (F) using the MS1096-GAL4 driver. (G) Quantification of the wing areas for the genotypes shown in A–F (n = 20 for each genotype). (H–K) Wings from adult female flies expressing UAS-GFP as a control (H), UAS-cicRNAi1 (I), UAS-mnbRNAi (J), and UAS-cicRNAi1 together with UAS-mnbRNAi (K) using the C96-GAL4 driver. (L) Quantification of the wing areas for the genotypes shown in H–K (n = 20 for each genotype). *P < 0.05, **P < 0.01, ***P < 0.001. Statistical significance was analyzed by using Student’s t test. Error bars represent SD. (Scale bar: 200 μm.)
Fig. S6.
Mnb opposes Cic function in controlling eye growth. Adult female flies expressing UAS-GFP (A), UAS-cicRNAi1 (B), UAS-mnb-RNAi (C), and UAS-mnb-RNAi together with UAS-cicRNAi1 (D) under the control of the da-GAL4 driver. Knockdown of mnb results in a smaller eye (C), which is reversed by a concomitant knockdown of cic (D). (Scale bar: 100 μm.)
Reduction of cic Level Restores Adult Brain Size in mnb Mutants.
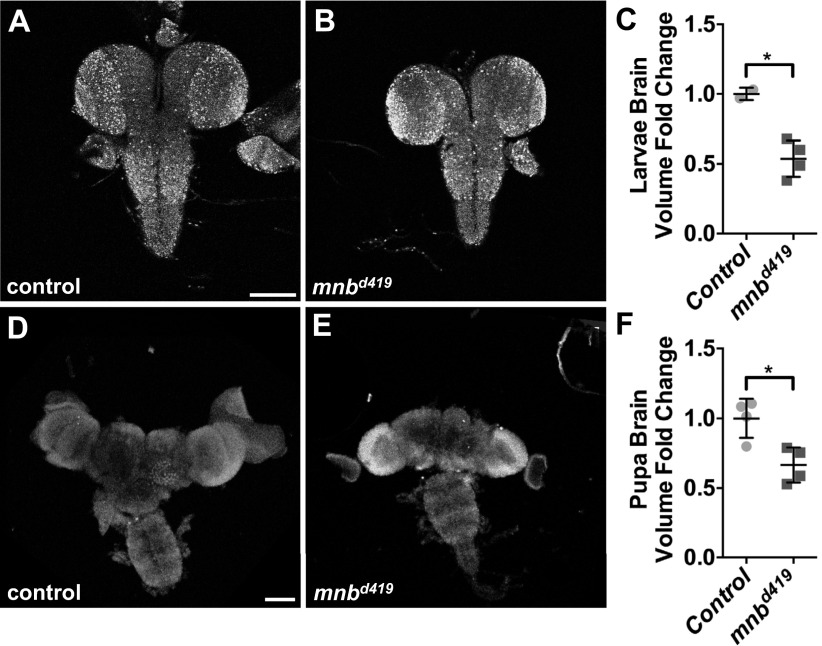
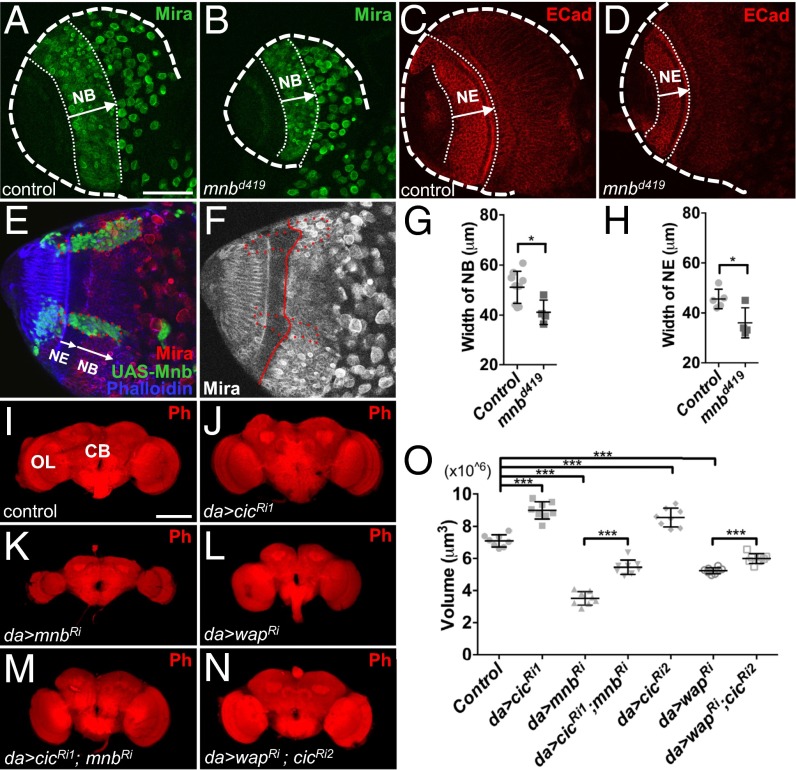
Mnb was originally identified in a genetic screen for mutants with altered brain structure (19). Mutant mnb adult animals have smaller brains, with the optic lobes (OLs) most significantly affected (19). In Drosophila development, the size of the central brain (CB) is determined by the proliferative ability of the neuroblasts (NBs) that are of embryonic origin, whereas the OLs are generated by the neuroepithelium (NE), which gives rise to the OL NBs during the larval stages (33). To identify the tissue origins of the reduction in adult brain size, we studied the larval and pupal brains from the wild-type and mnbd419 animals (mnbd419 is a null allele; ref. 34). The volumes of the larval and pupal brains in the mnbd419 mutants were significantly smaller than controls (Fig. S7), suggesting that the effects of loss of mnb can be traced to these developmental stages. We asked whether the smaller OLs in mnb mutants could result from altered proliferation in the NE and/or NB regions during the larval stages. The widths of both the NB and NE regions in the larval brains from mnbd419 animals were significantly reduced, compared with controls (Fig. 5 A–D, G, and H). Conversely, overexpression of Mnb in MARCM clones resulted in an increase of the width of NE specifically in the clone area (Fig. 5 E and F). These results suggest that Mnb is required for the proper growth of both the NE and NB regions in the OL. Additionally, Mnb may be involved in controlling the timing of NE to NB differentiation (35).
Fig. S7.
Loss of mnb leads to smaller brain size in larvae and pupae. (A and B) Third instar larval brains from control (w1118) (A) and mnbd419 (B) animals. (C) Quantification of the larval brain volumes in A and B (n = 2, 4). (D and E) Pupal brains from control (w1118) (D) and mnbd419 (E) animals. (F) Quantification of the pupal brain volumes in D and E (n = 4, 4). *P < 0.05. Statistical significance was analyzed by using Student’s t test. (Scale bars: 100 μm.)
Fig. 5.
Reduction in cic level restores adult brain size in mnb mutants. (A and B) Neuroblast (NB) regions (Mira-positive cells) in larval CNS from control (w1118) (A) and mnbd419 animals (B). (C and D) Neuroepithelium (NE) regions (E-cad positive cells) in larval CNS from control (w1118) (C) and mnbd419 animals (D). (E and F) NE region is expanded cell-autonomously in UAS-mnb overexpression clones (marked in green in E). Dotted red line, clone areas; solid red line, boundary between NB and NE. (G) Quantification of results in A and B (n = 9, 4). (H) Quantification of results in C and D (n = 5, 4). (I–N) Brains from adult female flies with the indicated genotypes. da-GAL4 driver was used to drive the expression of UAS-GFP (I), UAS-cicRNAi1 (J), UAS-mnbRNAi (K), UAS-wapRNAi (L), UAS-cicRNAi1 together with UAS-mnbRNAi (M), or UAS-cicRNAi2 together with UAS-wapRNAi (N). Ph, phalloidin stain. (O) Quantification of brain volumes for the genotypes shown in I–N (n = 8 for each genotype). (Scale bars: A–H, 50 μm; I–N, 100 μm.) *P < 0.05, **P < 0.01, ***P < 0.001. Error bars represent SD. Statistical significance was analyzed by using Student’s t test.
We next asked whether interactions among cic, mnb, and wap were involved in the control of adult brain size. RNAi knockdown of mnb or wap with a ubiquitous da-GAL4 driver (36) resulted in a smaller adult brain, especially in the optic lobes (Fig. 5 K, L, and O). This result suggests that both Mnb and Wap are required for normal brain growth. Knockdown of cic resulted in an increased adult brain size (Fig. 5 J and O). Strikingly, depletion of cic strongly suppressed the small brain phenotype caused by the knockdown of mnb (Fig. 5 K, M, and O), suggesting that Mnb promotes brain growth via down-regulation of Cic. Similarly, RNAi depletion of cic rescued the smaller brain phenotype of wap-RNAi (Fig. 5 L, N, and O). Overall, our results implicate Cic, Mnb, and Wap in a common pathway controlling organ growth and suggest that at least some of the growth-promoting functions of Mnb and Wap are mediated via their inhibition of Cic activity.
Mnb and ERK Have Additive Effects on Cic Activity.
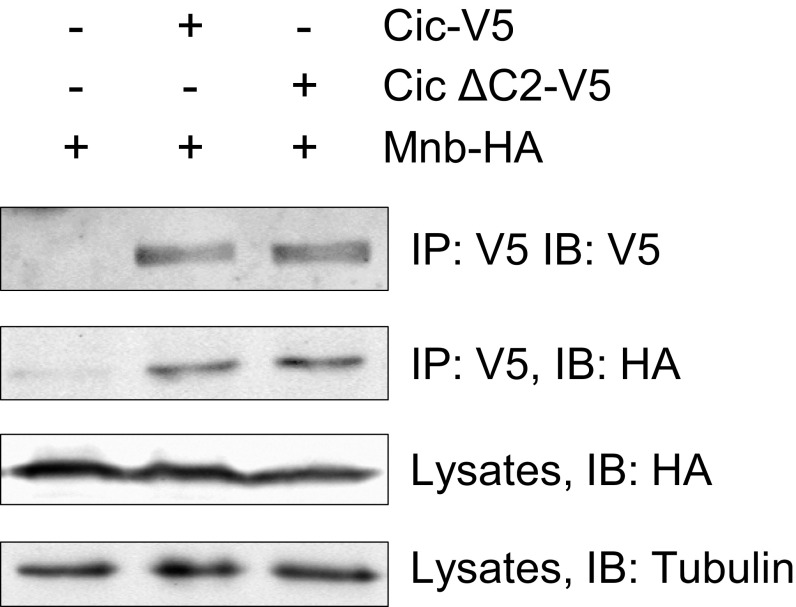
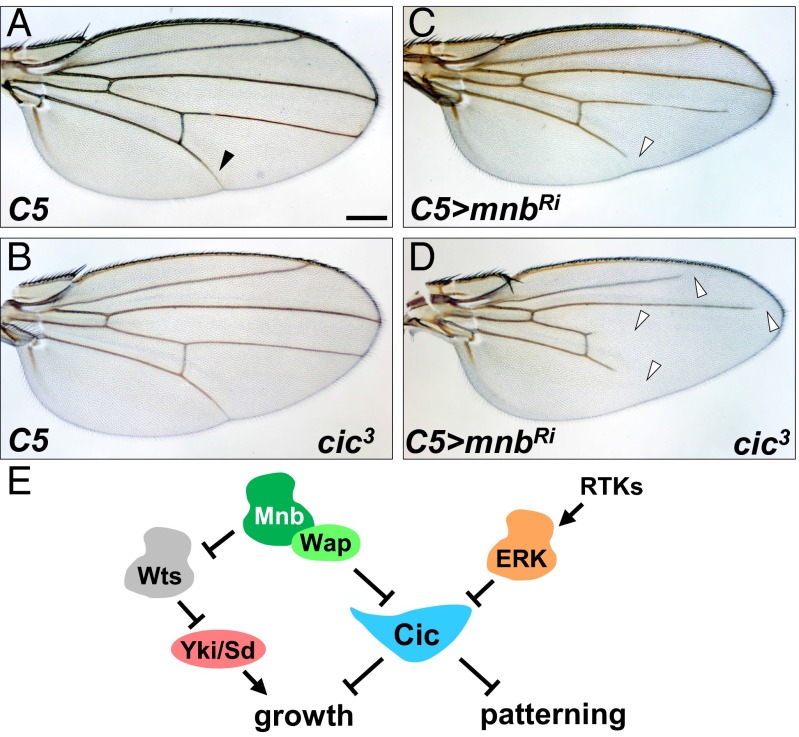
Our results so far have shown that Mnb is required for inhibiting Cic activity in various tissue contexts, which is also how ERK transmits signals from RTKs to control growth and patterning. We next asked whether the effects of ERK and Mnb on Cic are additive by first individually and then simultaneously reducing their ability to inhibit Cic. ERK-mediated down-regulation of Cic depends on the conserved C2 motif located in region Cic3, which serves as the ERK docking site (5). Deletion of the C2 motif abrogated Cic–ERK interaction (5), and a single amino acid substitution, F1054A, in the C2 motif (QQFILAPTPAQLG) reduced the binding of Cic to ERK (26). Importantly, deletion of the C2 domain did not affect the binding of Cic to Mnb (Fig. S8). Using CRISPR/Cas9-mediated mutagenesis, we generated a cic allele (cic3) lacking residue F1054, which is predicted to specifically disrupt the interaction of Cic with ERK. Most of the cic3 mutant animals showed normal wing vein pattern (Fig. 6 A and B); however, a partial loss of vein L5 was observed in ∼30% of adult flies, indicating that this is a gain-of-function mutation. RNAi depletion of mnb using C5-GAL4 resulted in a partial loss of veins L4 and L5 (Fig. 6C, arrowheads), which is in agreement with our observation that CUASC-lacZ expression was lost in the L5 region in this background (Fig. 3G). We reasoned that if Mnb and ERK had additive effects on Cic activity, reduction of mnb level in the cic3 background would cause a more severe vein loss phenotype compared with depletion of mnb alone. Indeed, we observed not only a more severe loss of veins L4 and L5, but also partial loss of veins L2 and L3 in the C5 > mnbRi; cic3 animals (Fig. 6D). We conclude that Mnb and ERK function additively to regulate wing tissue patterning via inhibition of Cic activity.
Fig. S8.
Deletion of the C2 motif does not affect the binding between Cic and Mnb. Protein lysates from S2 cells transfected with the indicated plasmids were incubated with anti-V5 beads, and immunocomplexes were analyzed on Western blots probed with anti-V5, anti-HA, and anti-tubulin antibodies.
Fig. 6.
Mnb and ERK function additively to inhibit Cic. Wings from adult female flies of the following genotypes: C5-GAL4 (A), C5-GAL4 cic3/+ (B), C5-GAL4/UAS-mnbRNAi (C), C5-GAL4 cic3/UAS-mnbRNAi (D). (Scale bar: 200 μm.) (E) Cic integrates upstream signals to control organ growth and tissue patterning.
Discussion
Our knowledge of upstream signals controlling Cic activity has been largely limited to its regulation by the RTK/ERK pathway (2). This study identifies a previously unknown mechanism for the regulation of Cic by the kinase Mnb and its adaptor Wap. Wap facilitates Mnb-dependent phosphorylation of Cic in the amino-terminal region, which is necessary for down-regulation of Cic activity. We found that the primary mechanism of Cic down-regulation by Mnb is through the relief of Cic-dependent transcriptional repression. Given that the DYRK family kinases autoactivate themselves soon after translation (37), it is likely that the effects of Mnb and Wap on Cic are constitutive.
Inhibition of Cic activity by Mnb/Wap has two developmentally important consequences (Fig. 6E). First, this regulation is important for the proper growth of several organs, such as the wings, eyes, and the brain. Second, down-regulation of Cic activity by Mnb/Wap is required for proper tissue patterning. Given the broad expression patterns of Cic, Mnb, and Wap (Fig. S2), the inhibitory mechanism we describe appears to operate in most, if not all, cells. In relation to ERK, the contribution from Mnb and Wap to Cic down-regulation depends on the tissue context and includes three possible scenarios: In some cells (e.g., developing vein L5 in the wing), both pathways are required for complete inhibition of Cic and operate additively. In other cells, ERK is the primary inhibitory signal, whereas the contribution of Mnb/Wap is less prominent (e.g., veins L2 and L3). Finally, in yet other cells in which ERK is not active, the function of Mnb and Wap to limit Cic activity would be dominant.
In addition to the RTK/ERK pathway, Cic was also shown to be regulated by Hippo signaling (18), and we have previously implicated Mnb and Wap as Hippo pathway regulators downstream of Dachsous (20). In this study, we found that knockdown of sd, a required component of Hippo signaling, did not affect the pattern of expression of CUASC-LacZ, and that knockdown of mnb or wap did not alter the pattern of ERK activation (Fig. S5), suggesting that Mnb and Wap control Cic activity independently from ERK and Hippo signaling. Altogether, current evidence suggests that Cic functions as an integrator of upstream developmental signals that converge on Cic to limit its activity, which is necessary for the proper execution of developmental programs responsible for tissue patterning and organ growth (Fig. 6E). Cic controls these developmental programs by direct binding to the enhancers of the genes encoding regulators of tissue patterning and cell proliferation in Drosophila and mammals (6, 8, 38, 39).
Interactions between Mnb/Wap and Cic in the brain have interesting parallels in human biology. The majority (>70%) of oligodendrogliomas, which are aggressive brain tumors, have been recently shown to harbor loss-of-function mutations in CIC, suggesting that it functions as a tumor suppressor (11, 12). Higher expression of DYRK1A was also found in a subset of oligodendroglioma patient samples (40), raising a possibility that DYRK1A may suppress Cic activity in human cells, much like Mnb does in Drosophila. A connection between DYRK1A and Cic in controlling brain development may extend even deeper, because both proteins have been implicated in neurodegenerative diseases (9, 41–43).
Materials and Methods
Experimental procedures are provided in SI Materials and Methods. They include a description of Drosophila stocks; information on antibodies, expression plasmids, and cell culture; procedures for luciferase reporter assays and mass spectrometry; and methods used for quantification of wing and brain size.
SI Materials and Methods
Drosophila melanogaster Stocks.
All Drosophila stocks were maintained on standard yeast-cornmeal-agar medium at 25 °C or 18 °C as indicated. MS1096-GAL4, da-GAL4, C96-GAL4, en-GAL4, hh-GAL4 UAS-GFP, ey-FLP UAS-dcr2 (#58757), and CoinFLP UAS-GFP (#58751) were from the Bloomington Drosophila Stock Center. UAS-wap RNAi (KK 107076), UAS-mnb RNAi (GD 28628), UAS-cic RNAi1 (KK 103012), UAS-cic RNAi2 (GD 40867), and UAS-sd RNAi (KK 101497) were from the Vienna Drosophila Resource Center (VDRC). UAS-cic, UAS-mnb, and UAS-wap were made by using standard cloning methods, and transgenic lines were generated by Genetic Services. Cic-Venus is a genomic Cic rescue construct from ref. 13. Other stocks were C5-GAL4 and CUASC-lacZ (6), mnbd419 (34). mnb-tagRFP-T and wap-Venus were generated by CRISPR/Cas9-mediated targeted transgene integration as described (44). The targeting vectors comprised the tagRFP-T (45) or Venus gene, the 3xP3-RFP gene for transformant selection and 1-kb homology arms. The vectors were designed such that the fluorescent protein gene is inserted immediately in front of the stop codon of the target gene and expressed as a fusion protein. The cic3 allele was obtained by CRISPR/Cas9-mediated mutagenesis. A guide RNA (gRNA) sequence (5′-GTGGGTGCCAAAATAAACTGC-3′) targeting the C2 coding sequence was subcloned in vector pCFD3 (46) and inserted at the attP40 genomic site via PhiC31-based integration. Transgenic gRNA males were crossed to nanos-cas9 females, and the resulting founder males were then crossed to TM3-bearing females for recovery of mutations. Induced alleles were identified by sequencing PCR products amplified from candidate flies.
Immunohistochemistry and Immunoblotting.
Primary antibodies were as follows: mouse anti-β-galactosidase (LacZ) 1:100 (Promega), mouse anti-dpERK 1:100 (Sigma), rabbit anti-GFP 1:1,000 (Abcam), mouse anti-V5 1:1,000 (Sigma), rabbit anti-Flag 1:1,000 (Sigma), rabbit anti-HA 1:1,000 (Sigma), guinea pig anti-Cic 1:100 (gift from Iswar Hariharan, University of California, Berkeley, CA), rabbit anti-DCAF7 1:100 (Novus Biologicals), rabbit anti-tRFP 1:1,000 (Evrogen), mouse anti-Mira 1:100 (gift from Alex Gould, The Francis Crick Institute, London), mouse anti-Ecad 1:50 (Developmental Studies Hybridoma Bank), and mouse anti-Engrailed 1:5 (Developmental Studies Hybridoma Bank). Secondary antibodies used were as follows: Alexa Donkey anti Rabbit-647 (Life Technologies), Alexa Donkey anti-Mouse 488 (Abcam), Alexa Donkey anti-Rabbit 555 (Life Technologies), IRDye 800CW Donkey anti-Rabbit IgG (LI-COR), IRDye 680CW Donkey anti-Guinea pig IgG (LI-COR), and IRDye 680CW Donkey anti-Mouse IgG (LI-COR). Dissected imaginal discs were stained as in ref. 20. Stained tissues were mounted with Prolong Gold anti-fade mounting reagent with DAPI (Life Technologies), and images were acquired with Zeiss LSM 510 or Zeiss LSM 880 confocal microscopes. Tissues from the Mnb-tRFP, Wap-Venus, and Cic-Venus stocks were mounted in 90% (vol/vol) glycerol, and images were acquired with the Nikon C2 confocal microscope and processed in Fiji.
Quantification of Wing and Brain Size.
Wing area from 20 flies was quantified by using Adobe Photoshop. For quantification of brain size, flies were raised at 18 °C. Brains from eight 10-day-old female flies were dissected in cold 1× PBS, fixed with 4% formaldehyde/PBS, and stained with Alexa Fluor 594 Phalloidin (Life Technologies) overnight. Stained brains were mounted with Prolong Gold anti-fade mounting reagent with DAPI (Life Technologies). Brain volume measurements were done from 3D reconstructions of 2.5 µm-spaced confocal Z stacks acquired with a Zeiss LSM 880 or Leica SP5 confocal microscopes, using a 3D Viewer plugin in Fiji or Volocity software. Significance was calculated with a Student’s t test. In all figures, the indications used are the following: *P < 0.05, **P < 0.01, ***P < 0.001.
Expression Plasmids and Cell Culture.
For establishing a stable S2 cell line, full-length Cic ORF was tagged with SBP at the C terminus and cloned into pMK33 vector (47). Full-length ORF and fragments of Cic were cloned into pMT/V5-His A vector (Invitrogen). Full-length Wap was tagged with the Flag tag and cloned into the pMT vector. pMT-Mnb-HA and pMT-MnbKR-HA plasmids were from ref. 20. The pGL2-CUASC-Luc (“CUASC-Luc”) reporter was generated by PCR amplification of the promoter regions of pC4PLZ-CUASC vector (6) with primers CUASC-XhoI-UP1 (5′-ATCGCTCGAGGAATTCCCAGTTTATG-3′) and CUASC-XhoI-DN1 (5′-GCTACTCGAGTTATCACCCACGGCTCTGCTC-3′), which was subcloned into pGL2 basic (Promega). Full-length GAL4 was cloned into pMT/V5-His A vector (Invitrogen) to generate pMT-GAL4. pIE4-lacZ was described (48).
Drosophila S2 cells were maintained in standard Schneider’s S2 medium with FBS (Gibco) at 25 °C, and transfections were performed by using Effectene transfection reagent (Qiagen). In some instances, cells were treated with 30 µg of dsRNA specific for wap, mnb, or rl (ERK) for 96 h. After 96 h, cells were transfected with indicated plasmids. CuSO4 was added to culture media at a final concentration of 0.35 mM for inducing expression. Cells were lysed by using default lysis buffer (50 mM Tris pH 7.5, 125 mM NaCl, 5% glycerol, 0.2% IGEPAL, 1.5 mM MgCl2, 1 mM DTT, 25 mM NaF, 1 mM Na3VO4, 1 mM EDTA and 2× Complete protease inhibitor, Roche). Clear cell lysates were incubated with anti-V5 beads (Sigma), streptavidin beads (Pierce), GFP-Trap, or RFP-Trap resin (ChromoTek) for 2 h at 4 °C. Beads were washed three times with lysis buffer, and protein complexes were eluted with SDS buffer. To visualize differences in Cic1 mobility, 6% Tris-Glycine gels were used with 50 µM Phos-tag (Wako Laboratory Chemicals) and 100 µM MnCl2.
Luciferase Reporter Assays.
Cells were cotransfected with the luciferase reporter vector pGL2-CUASC-Luc and with the effector plasmids. pIE4-LacZ was used to normalize transfection efficiencies. Each transfection point was assayed in triplicate, and each experiment was repeated three times. Luciferase and β-galactosidase activities were measured in S2 cell lysates by Luc-Screen Extended-Glow Luciferase Reporter Gene Assay System (Thermo Fisher) and Galacto-Star One-Step β-Galactosidase Reporter Gene Assay System (Thermo Fisher), respectively. Luminescence signals were acquired on POLARstar Omega multifunction microplate reader (BMG Labtech).
Mass Spectrometry.
Cic-interacting proteins were purified from Drosophila S2 cells (Cic-SBP) essentially as described in ref. 47, using a modified single-step procedure for SBP-tagged Cic. For expression in S2 cells, pMK33-Cic-SBP construct was transfected by using Effectene transfection reagent (Qiagen), and stable cell lines were selected in the presence of 300 µg/mL hygromycin (Sigma). Cic-SBP purifications were performed in two biological replicates. To analyze Cic complexes in vivo, 0- to 16-h Cic-Venus embryos were collected and the proteins were extracted and purified on GFP-Trap resin (ChromoTek) as described in ref. 49. The Cic-Venus construct uses genomic regulatory sequences of the cic gene and, therefore, expresses the Cic-Venus protein at endogenous levels (13). Functionality of this construct was previously confirmed in a rescue assay (13). Cic-Venus purifications were performed in three biological replicates. Protein complexes were analyzed by nanoLC-MS/MS as described in ref. 47 at the Taplin Mass Spectrometry Facility at Harvard Medical School. Identified Cic-interacting proteins were analyzed by the SAINT program (50). A complete mass spectrometry dataset is shown in Dataset S1. Interactions with SAINT scores >0.8 were considered significant.
To identify Cic residues that are phosphorylated by Mnb, Cic1-V5 and Wap-Flag were coexpressed in S2 cells either with Mnb-HA or MnbKR-HA, Cic1-V5 was purified on anti-V5 agarose resin, and its phosphorylation was analyzed by nanoLC-MS/MS.
Supplementary Material
Acknowledgments
We thank Iswar Hariharan for the Cic antibody; the Bloomington Drosophila Stock Center, Vienna Drosophila Resource Centre, and Developmental Studies Hybridoma Bank for their services; Walter Flores for help with preparation of Cic mutant constructs; and Jonathan Celli for help with brain size quantification. Mass spectrometry was performed at the Taplin Mass Spectrometry Facility at Harvard Medical School. This work was supported by the NIH Grants GM105813 (to A.V. and Ken Moberg) and HD085870 (to A.V. and S.Y.S.), L.Y. was supported by the University of Massachusetts Boston Sanofi Genzyme Doctoral Fellowship, S.Y.S. was supported by the NIH Grant GM086537, and G.J. is supported by Spanish Government Grant BFU2014-52863-P and Fundació La Marató de TV3 Grant 20131730.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1609417113/-/DCSupplemental.
References
- 1.Jiménez G, Guichet A, Ephrussi A, Casanova J. Relief of gene repression by torso RTK signaling: Role of capicua in Drosophila terminal and dorsoventral patterning. Genes Dev. 2000;14(2):224–231. [PMC free article] [PubMed] [Google Scholar]
- 2.Jiménez G, Shvartsman SY, Paroush Z. The Capicua repressor--a general sensor of RTK signaling in development and disease. J Cell Sci. 2012;125(Pt 6):1383–1391. doi: 10.1242/jcs.092965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goff DJ, Nilson LA, Morisato D. Establishment of dorsal-ventral polarity of the Drosophila egg requires capicua action in ovarian follicle cells. Development. 2001;128(22):4553–4562. doi: 10.1242/dev.128.22.4553. [DOI] [PubMed] [Google Scholar]
- 4.Roch F, Jiménez G, Casanova J. EGFR signalling inhibits Capicua-dependent repression during specification of Drosophila wing veins. Development. 2002;129(4):993–1002. doi: 10.1242/dev.129.4.993. [DOI] [PubMed] [Google Scholar]
- 5.Astigarraga S, et al. A MAPK docking site is critical for downregulation of Capicua by Torso and EGFR RTK signaling. EMBO J. 2007;26(3):668–677. doi: 10.1038/sj.emboj.7601532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ajuria L, et al. Capicua DNA-binding sites are general response elements for RTK signaling in Drosophila. Development. 2011;138(5):915–924. doi: 10.1242/dev.057729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tseng AS, et al. Capicua regulates cell proliferation downstream of the receptor tyrosine kinase/ras signaling pathway. Curr Biol. 2007;17(8):728–733. doi: 10.1016/j.cub.2007.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jin Y, et al. EGFR/Ras signaling controls Drosophila intestinal stem cell proliferation via Capicua-regulated genes. PLoS Genet. 2015;11(12):e1005634. doi: 10.1371/journal.pgen.1005634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lam YC, et al. ATAXIN-1 interacts with the repressor Capicua in its native complex to cause SCA1 neuropathology. Cell. 2006;127(7):1335–1347. doi: 10.1016/j.cell.2006.11.038. [DOI] [PubMed] [Google Scholar]
- 10.Bettegowda C, et al. Mutations in CIC and FUBP1 contribute to human oligodendroglioma. Science. 2011;333(6048):1453–1455. doi: 10.1126/science.1210557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sahm F, et al. CIC and FUBP1 mutations in oligodendrogliomas, oligoastrocytomas and astrocytomas. Acta Neuropathol. 2012;123(6):853–860. doi: 10.1007/s00401-012-0993-5. [DOI] [PubMed] [Google Scholar]
- 12.Wesseling P, van den Bent M, Perry A. Oligodendroglioma: Pathology, molecular mechanisms and markers. Acta Neuropathol. 2015;129(6):809–827. doi: 10.1007/s00401-015-1424-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grimm O, et al. Torso RTK controls Capicua degradation by changing its subcellular localization. Development. 2012;139(21):3962–3968. doi: 10.1242/dev.084327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim Y, et al. MAPK substrate competition integrates patterning signals in the Drosophila embryo. Curr Biol. 2010;20(5):446–451. doi: 10.1016/j.cub.2010.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim Y, et al. Substrate-dependent control of MAPK phosphorylation in vivo. Mol Syst Biol. 2011;7(7):467. doi: 10.1038/msb.2010.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dissanayake K, et al. ERK/p90(RSK)/14-3-3 signalling has an impact on expression of PEA3 Ets transcription factors via the transcriptional repressor capicúa. Biochem J. 2011;433(3):515–525. doi: 10.1042/BJ20101562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lim B, et al. Kinetics of gene derepression by ERK signaling. Proc Natl Acad Sci USA. 2013;110(25):10330–10335. doi: 10.1073/pnas.1303635110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herranz H, Hong X, Cohen SM. Mutual repression by bantam miRNA and Capicua links the EGFR/MAPK and Hippo pathways in growth control. Curr Biol. 2012;22(8):651–657. doi: 10.1016/j.cub.2012.02.050. [DOI] [PubMed] [Google Scholar]
- 19.Tejedor F, et al. minibrain: A new protein kinase family involved in postembryonic neurogenesis in Drosophila. Neuron. 1995;14(2):287–301. doi: 10.1016/0896-6273(95)90286-4. [DOI] [PubMed] [Google Scholar]
- 20.Degoutin JL, et al. Riquiqui and minibrain are regulators of the hippo pathway downstream of Dachsous. Nat Cell Biol. 2013;15(10):1176–1185. doi: 10.1038/ncb2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morriss GR, et al. The Drosophila wings apart gene anchors a novel, evolutionarily conserved pathway of neuromuscular development. Genetics. 2013;195(3):927–940. doi: 10.1534/genetics.113.154211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Veraksa A. Regulation of developmental processes: Insights from mass spectrometry-based proteomics. Wiley Interdiscip Rev Dev Biol. 2013;2(5):723–734. doi: 10.1002/wdev.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Skurat AV, Dietrich AD. Phosphorylation of Ser640 in muscle glycogen synthase by DYRK family protein kinases. J Biol Chem. 2004;279(4):2490–2498. doi: 10.1074/jbc.M301769200. [DOI] [PubMed] [Google Scholar]
- 24.Himpel S, et al. Specificity determinants of substrate recognition by the protein kinase DYRK1A. J Biol Chem. 2000;275(4):2431–2438. doi: 10.1074/jbc.275.4.2431. [DOI] [PubMed] [Google Scholar]
- 25.Zhai B, Villén J, Beausoleil SA, Mintseris J, Gygi SP. Phosphoproteome analysis of Drosophila melanogaster embryos. J Proteome Res. 2008;7(4):1675–1682. doi: 10.1021/pr700696a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Futran AS, Kyin S, Shvartsman SY, Link AJ. Mapping the binding interface of ERK and transcriptional repressor Capicua using photocrosslinking. Proc Natl Acad Sci USA. 2015;112(28):8590–8595. doi: 10.1073/pnas.1501373112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brunner D, et al. A gain-of-function mutation in Drosophila MAP kinase activates multiple receptor tyrosine kinase signaling pathways. Cell. 1994;76(5):875–888. doi: 10.1016/0092-8674(94)90362-x. [DOI] [PubMed] [Google Scholar]
- 28.Bosch JA, Tran NH, Hariharan IK. CoinFLP: A system for efficient mosaic screening and for visualizing clonal boundaries in Drosophila. Development. 2015;142(3):597–606. doi: 10.1242/dev.114603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yeh E, Gustafson K, Boulianne GL. Green fluorescent protein as a vital marker and reporter of gene expression in Drosophila. Proc Natl Acad Sci USA. 1995;92(15):7036–7040. doi: 10.1073/pnas.92.15.7036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang L, et al. The TEAD/TEF family of transcription factor Scalloped mediates Hippo signaling in organ size control. Dev Cell. 2008;14(3):377–387. doi: 10.1016/j.devcel.2008.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Capdevila J, Guerrero I. Targeted expression of the signaling molecule decapentaplegic induces pattern duplications and growth alterations in Drosophila wings. EMBO J. 1994;13(19):4459–4468. doi: 10.1002/j.1460-2075.1994.tb06768.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Helms W, et al. Engineered truncations in the Drosophila mastermind protein disrupt Notch pathway function. Dev Biol. 1999;215(2):358–374. doi: 10.1006/dbio.1999.9477. [DOI] [PubMed] [Google Scholar]
- 33.Sousa-Nunes R, Cheng LY, Gould AP. Regulating neural proliferation in the Drosophila CNS. Curr Opin Neurobiol. 2010;20(1):50–57. doi: 10.1016/j.conb.2009.12.005. [DOI] [PubMed] [Google Scholar]
- 34.Hong SH, et al. Minibrain/Dyrk1a regulates food intake through the Sir2-FOXO-sNPF/NPY pathway in Drosophila and mammals. PLoS Genet. 2012;8(8):e1002857. doi: 10.1371/journal.pgen.1002857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reddy BV, Rauskolb C, Irvine KD. Influence of fat-hippo and notch signaling on the proliferation and differentiation of Drosophila optic neuroepithelia. Development. 2010;137(14):2397–2408. doi: 10.1242/dev.050013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wodarz A, Hinz U, Engelbert M, Knust E. Expression of crumbs confers apical character on plasma membrane domains of ectodermal epithelia of Drosophila. Cell. 1995;82(1):67–76. doi: 10.1016/0092-8674(95)90053-5. [DOI] [PubMed] [Google Scholar]
- 37.Becker W, Sippl W. Activation, regulation, and inhibition of DYRK1A. FEBS J. 2011;278(2):246–256. doi: 10.1111/j.1742-4658.2010.07956.x. [DOI] [PubMed] [Google Scholar]
- 38.Kawamura-Saito M, et al. Fusion between CIC and DUX4 up-regulates PEA3 family genes in Ewing-like sarcomas with t(4;19)(q35;q13) translocation. Hum Mol Genet. 2006;15(13):2125–2137. doi: 10.1093/hmg/ddl136. [DOI] [PubMed] [Google Scholar]
- 39.Forés M, et al. Origins of context-dependent gene repression by capicua. PLoS Genet. 2015;11(1):e1004902. doi: 10.1371/journal.pgen.1004902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pozo N, et al. Inhibition of DYRK1A destabilizes EGFR and reduces EGFR-dependent glioblastoma growth. J Clin Invest. 2013;123(6):2475–2487. doi: 10.1172/JCI63623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guimerá J, et al. A human homologue of Drosophila minibrain (MNB) is expressed in the neuronal regions affected in Down syndrome and maps to the critical region. Hum Mol Genet. 1996;5(9):1305–1310. doi: 10.1093/hmg/5.9.1305. [DOI] [PubMed] [Google Scholar]
- 42.Møller RS, et al. Truncation of the Down syndrome candidate gene DYRK1A in two unrelated patients with microcephaly. Am J Hum Genet. 2008;82(5):1165–1170. doi: 10.1016/j.ajhg.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tejedor FJ, Hämmerle B. MNB/DYRK1A as a multiple regulator of neuronal development. FEBS J. 2011;278(2):223–235. doi: 10.1111/j.1742-4658.2010.07954.x. [DOI] [PubMed] [Google Scholar]
- 44.Poon CL, Mitchell KA, Kondo S, Cheng LY, Harvey KF. The Hippo pathway regulates neuroblasts and brain size in Drosophila melanogaster. Curr Biol. 2016;26(8):1034–1042. doi: 10.1016/j.cub.2016.02.009. [DOI] [PubMed] [Google Scholar]
- 45.Shaner NC, et al. Improving the photostability of bright monomeric orange and red fluorescent proteins. Nat Methods. 2008;5(6):545–551. doi: 10.1038/nmeth.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Port F, Chen HM, Lee T, Bullock SL. Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proc Natl Acad Sci USA. 2014;111(29):E2967–E2976. doi: 10.1073/pnas.1405500111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kyriakakis P, Tipping M, Abed L, Veraksa A. Tandem affinity purification in Drosophila: The advantages of the GS-TAP system. Fly (Austin) 2008;2(4):229–235. doi: 10.4161/fly.6669. [DOI] [PubMed] [Google Scholar]
- 48.Frolov MV, et al. Functional antagonism between E2F family members. Genes Dev. 2001;15(16):2146–2160. doi: 10.1101/gad.903901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Neumüller RA, et al. Stringent analysis of gene function and protein-protein interactions using fluorescently tagged genes. Genetics. 2012;190(3):931–940. doi: 10.1534/genetics.111.136465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Choi H, et al. SAINT: Probabilistic scoring of affinity purification-mass spectrometry data. Nat Methods. 2011;8(1):70–73. doi: 10.1038/nmeth.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.