Significance
Much of our understanding of the chronology of human evolution relies on a fixed “molecular clock”; that is, a constant rate of substitutions per unit time. To evaluate the validity of this assumption, we analyze whole-genome sequences from 10 primate species. We find that there is substantial variation in the molecular clock between apes and monkeys and that rates even differ within hominines. Importantly, not all mutation types behave similarly; notably, transitions at CpG sites exhibit a more clocklike behavior than other substitutions, presumably because of their nonreplicative origin. Thus, the mutation spectra, and not just the overall substitution rates, are changing across primates. This finding suggests that events in primate evolution are most reliably dated using CpG transitions.
Keywords: molecular clock, mutation rate, primate evolution, CpG transition rate, human–ape divergence time
Abstract
Events in primate evolution are often dated by assuming a constant rate of substitution per unit time, but the validity of this assumption remains unclear. Among mammals, it is well known that there exists substantial variation in yearly substitution rates. Such variation is to be expected from differences in life history traits, suggesting it should also be found among primates. Motivated by these considerations, we analyze whole genomes from 10 primate species, including Old World Monkeys (OWMs), New World Monkeys (NWMs), and apes, focusing on putatively neutral autosomal sites and controlling for possible effects of biased gene conversion and methylation at CpG sites. We find that substitution rates are up to 64% higher in lineages leading from the hominoid–NWM ancestor to NWMs than to apes. Within apes, rates are ∼2% higher in chimpanzees and ∼7% higher in the gorilla than in humans. Substitution types subject to biased gene conversion show no more variation among species than those not subject to it. Not all mutation types behave similarly, however; in particular, transitions at CpG sites exhibit a more clocklike behavior than do other types, presumably because of their nonreplicative origin. Thus, not only the total rate, but also the mutational spectrum, varies among primates. This finding suggests that events in primate evolution are most reliably dated using CpG transitions. Taking this approach, we estimate the human and chimpanzee divergence time is 12.1 million years, and the human and gorilla divergence time is 15.1 million years.
Germline mutations are the ultimate source of genetic differences among individuals and species. They are thought to arise from a combination of errors in DNA replication (e.g., the chance misincorporation of a base pair) or damage that is unrepaired by the time of replication (e.g., the spontaneous deamination of methylated CpG sites) (1). If mutations are neutral (i.e., do not affect fitness), then the rate at which they arise will be equal to the substitution rate (2). A key consequence is that if mutation rates remain constant over time, substitution rates should likewise be constant.
This assumption of constancy of substitution rates plays a fundamental role in evolutionary genetics by providing a molecular clock with which to date events inferred from genetic data (3). Notably, important events in human evolution for which there is no fossil record (e.g., when humans and chimpanzees split, or when anatomically modern humans left Africa) are dated using a mutation rate obtained from contemporary pedigrees or phylogenetic analysis, assuming the per year rate has remained unchanged for millions of years (4).
However, we know from studies of mammalian phylogenies, as well as of other taxa, that there can be substantial variation in substitution rates per unit time (5–7). In particular, there is the well-known hypothesis of a “generation time effect” on substitution rates, based on the observation that species with shorter generation time (i.e., mean age of reproduction) have higher mutation rates (8). For instance, mice have a generation time on the order of months (∼10–12 mo) compared with ∼29 y in humans (9), and a two- to threefold higher substitution rate per year (8). More generally, a survey of 32 mammalian species found reproductive span to be the strongest predictor of substitution rate variation (5).
A generation time effect has also been suggested in humans, motivated by the observation that the yearly mutation rate estimated by sequencing human and chimpanzee pedigrees [∼0.4 × 10−9 per base pair per year (10, 11)] is approximately twofold lower than the mutation rate inferred from the number of substitutions observed between primates (1). Substitution-derived estimates of mutation rates are highly dependent on dating evolutionary lineages from the fossil record, and so are subject to considerable uncertainty. Nonetheless, one way to reconcile pedigree and substitution-derived estimates of the mutation rate would be to postulate that the generation time has increased toward the present, and led to a decrease in the yearly mutation rate (12).
Whether the association between generation time and substitution rates is causal remains unclear, however; correlated traits such as metabolic rate (13), body size (14), and sperm competition (15) may also affect substitution rates. For instance, the metabolic rate hypothesis posits that species with higher basal metabolic rates are subject to higher rates of oxidative stress, and hence have a higher mutation rate (13). Body mass has been shown to be negatively correlated to substitution rates, such that smaller animals tend to have higher substitution rates (13). Sexual selection on mating systems may also affect substitution rates, as more intense sperm competition leads to selection for higher sperm counts, leading to more cell divisions per unit time during spermatogenesis and a higher male mutation rate (15).
That said, an effect of life history traits such as generation time on the yearly mutation rate is expected from first principles, given our understanding of oogenesis and spermatogenesis (16, 17). In mammals, oogonial divisions are completed by the birth of the future mother, whereas the spermatogonial stem cells continue to divide postpuberty (16). Thus, the total number of replication-driven mutations inherited by a diploid offspring accrues in a piecewise linear manner with parental age, with the number depending on the number of cell divisions in each developmental stage, as well as the per cell division mutation rates (1, 17). These considerations indicate that changes in generation time, onset of puberty, and rate of spermatogenesis should all influence yearly mutation rates (1, 17).
Importantly, then, primates are well known to differ with regard to most of these traits. In addition to huge variation in body size and metabolic rates, generation time varies almost 10-fold, with the shortest generation time observed in prosimians [∼3 y in galago and mouse lemurs (18)] and the longest generation time observed in humans (∼29 y). Species also differ in the strength of sperm competition and rates of spermatogenesis: monkeys have a shorter spermatogenetic division, and thus consequently produce more sperm per unit time than do apes (19). Thus, even if the per cell division mutation rate remained constant, we should expect differences in yearly mutation rates among species.
Although the factors discussed thus far apply to all sites, variation in substitution rates among species also depends on the type of mutation and the genomic context (i.e., flanking sequence) in which it occurs (6). For example, in mammals, CpG transitions show the least amount of variation in substitution rates among species (6). A plausible explanation is the source of mutations, as transitions at methylated CpG sites are thought to occur primarily through spontaneous deamination; if they arise at a constant rate and their repair is inefficient relative to the cell cycle length, as is thought to be the case, then their mutation rate should depend largely on absolute time, rather than the number of cell divisions (20–22).
In addition, even substitutions that have no effect on fitness may vary in their rate of accumulation among lineages because of biased gene conversion (BGC), the bias toward strong (S: G or C) rather than weak (W: A or T) bases that occurs in the repair of double-strand breaks (23). This phenomenon leads to the increased probability of fixation of S alleles (and loss of W alleles) in regions of higher recombination, and can therefore change substitution rates relative to mutation rates (23, 24). The strength of BGC is a function of the degree of bias, the local recombination rate, and the effective population size of the species (23). The latter varies by three- to fourfold among primates (25), and the fine-scale recombination landscape is also likely to differ substantially across species (26).
Empirically, the extent to which substitution rates vary among primate lineages remains unclear. Kim et al. (27) compared two hominoids (human and chimpanzee) and two Old World Monkeys (OWMs; baboon and rhesus macaque). Assuming that the average divergence time of the two pairs of species is identical, they reported that substitution rates at transitions at non-CpG sites differ by ∼31% between hominoids and OWMs, whereas rates of CpG transitions are almost identical (27). In turn, Elango et al. (28) found that the human branch is ∼2% shorter than that in chimpanzee (considering the rates from the human–chimpanzee ancestor), and ∼11% shorter than in gorilla (considering rates from the human–gorilla ancestor). Although these comparisons suggest that substitution rates are evolving across primates, they are based on limited data, make strong assumptions about divergence times, and rely on parsimony-based approaches that may underestimate substitution rates for divergent species, notably at CpG sites (29). We therefore revisit these questions using whole-genome sequence alignments of 10 primates, allowing for variable substitution rates along different lineages and explicitly modeling the context dependency of CpG substitutions.
Results
We first estimate the number of autosomal substitutions on 10 primate lineages by applying Phylofit (30) to the Multiz sequence alignment (excluding gorilla and gibbon because of concerns about incomplete lineage sorting; SI Appendix, Note S1). This method allows us to estimate branch lengths, accounting for uncertainty in the ancestral reconstruction, recurrent substitutions at a site, and context-dependent effects of neighboring nucleotides at CpG sites (30).
To focus on putatively neutral sites in the genome, in which substitutions more faithfully reflect mutation patterns, we exclude conserved elements, coding exons, and transposable elements (referred to as CET in what follows; SI Appendix, Note S1). After filtering CET sites and removing missing data, we obtain ∼562 Mb of whole-genome sequence alignment across 10 primates. In these filtered data, the total number of substitutions on the human lineage is similar to estimates in ancestral repeats (SI Appendix, Table S3), which are often considered a benchmark for strict neutral evolution (31), suggesting the substitutions we analyzed were largely neutral.
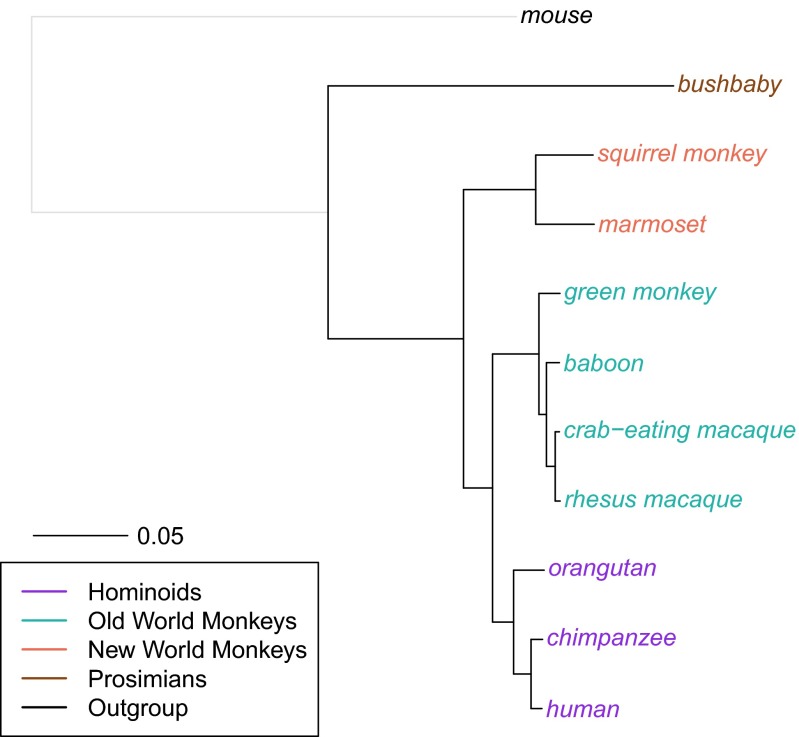
Across the 10 primate species, we find that the total substitution rates vary markedly (Fig. 1). For example, when we compare taxa pairwise, the substitution rates on lineages leading from the hominoid–OWM ancestor to hominoids are on average 2.68% (with a range of 2.63–2.74%), whereas rates on lineages leading to OWM are on average 3.57% (range: 3.55–3.59%), 1.33-fold higher. These findings are consistent with those of smaller studies (27). Similarly, when considering the distance from the hominoid–New World Monkey (NWM) ancestor, substitution rates leading to NWM are on average 6.92% (range: 6.89–6.94%), 1.64-fold higher than on the lineages leading to hominoids, which are on average 4.22% (range: 4.17–4.29%). Substitution rates are also 1.61-fold higher in lineages leading to bushbaby (a prosimian) compared with hominoids (Fig. 1). Because of challenges in accurately reconstructing the ancestral state for species that are closer to the outgroup, we believe this estimate to be less reliable, however, and hence do not consider bushbaby in further analyses.
Fig. 1.
Phylogenetic tree for 10 primates. Autosomal neutral substitution rates for 10 primates and an outgroup (mouse, shown in gray) from the Multiz dataset were estimated using Phylofit (see SI Appendix, Note S1 for details of dataset and filtering). Branch lengths reflect the expected number of neutral substitutions per site along each lineage. R code to replicate this figure is available at: https://github.com/priyamoorjani/Molecular-clock_figures-and-data/blob/master/Figure1.R.
Using bootstrap resampling of 1-Mb regions of the genome suggests tiny SEs for the substitution rates (e.g., the SE on lineages leading from the hominoid–OWM ancestor to hominoids is 0.01%), as expected from such large datasets. These SEs are likely to be deceptively small, however, as the main source of uncertainty in our analysis is likely a result of systematic effects of varying sequence quality, mapping, and alignment artifacts among species. To evaluate the impact of these effects, we therefore repeat our analysis using a different sequence alignment of seven primates [the Enredo-Pecan-Ortheus (EPO) dataset (32)] and apply the same filters. When the species considered are matched between the two datasets, results are highly similar (SI Appendix, Note S3), and appear to be robust.
To evaluate how substitution patterns differ for mutations generated by distinct mechanisms, we distinguish between transitions at ancestrally CpG sites (referred to as CpG) outside of CpG islands (CGI), which are believed to occur mostly as a result of the spontaneous deamination of methylated cytosines, and transitions at ancestrally G or C sites outside of a CpG context (referred to as non-CpG G/C), which are thought to primarily occur as a result of replication errors. (Because CGI are often hypomethylated, we remove these regions from this analysis, and, unless specified otherwise, refer to transitions at CpG sites outside of CGIs as “CpG transitions.”) Mathematical modeling of different substitution mechanisms predicts that mutations that are nonreplicative in origin and highly inefficiently repaired should depend on absolute time, rather than on the number of cell divisions, and hence should be more clocklike among species (21). In contrast, mutations that arise from replication errors, or are nonreplicative in origin but well repaired, should depend on the generation time and other life history traits, and therefore their substitution rates could vary considerably across primates (21, 33). Thus, a priori, we expect CpG transitions outside CGI to be more clocklike than other types of substitutions (assuming similar rates of deamination across species).
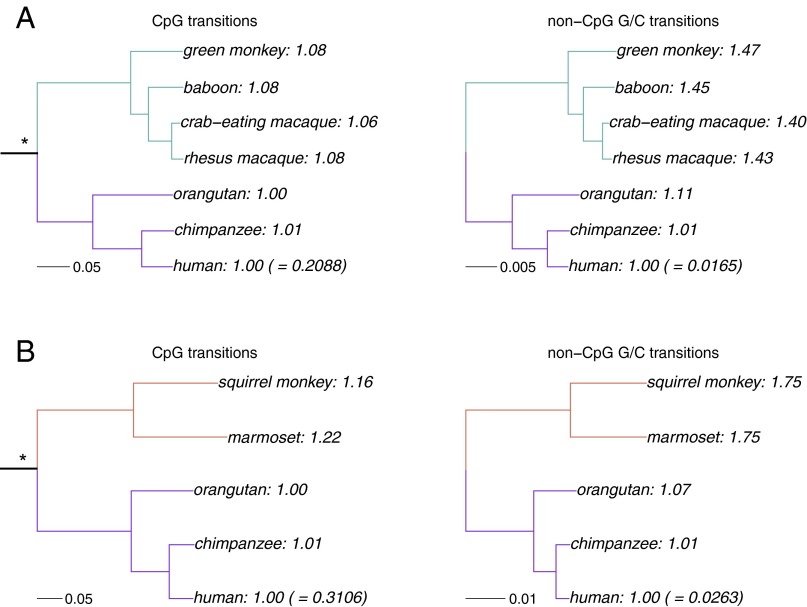
For our comparisons to not be confounded by biased gene conversion, we compare transitions at CpG sites with those occurring at non-CpG G/C sites. Because both types of mutations involve changes from G to A or C to T nucleotides, and both occur in regions with similar recombination rate profiles (SI Appendix, Fig. S2), they should, on average, be subject to similar strengths of biased gene conversion. Comparing hominoids and monkeys, the substitutions involving CpG transitions are on average 1.07-fold higher in lineages leading from the hominoid–OWM ancestor to OWM than they are in lineages leading to hominoids. Considering the hominoid–NWM ancestor, substitutions are 1.19-fold higher in lineages leading to NWM than to hominoids (Fig. 2). In contrast, when considering transitions at non-CpG G/C sites, there are on average 1.38-fold more substitutions from the hominoid–OWM ancestor to OWM lineages than to hominoid ones, and 1.71-fold more from the hominoid–NWM ancestor to NWM than to hominoid lineages (Fig. 2). Thus, CpG transition rates are more similar across species, as observed in comparisons of smaller datasets of primates and mammals (6, 27). These results are robust to the choice of species of OWM and hominoids used; e.g., using gorilla instead of chimpanzee or gibbon instead of orangutan yields similar findings (SI Appendix, Figs. S3 and S4).
Fig. 2.
Comparison of substitution rates in hominoids and monkeys. For transitions from CpG and non-CpG G/C sites, the total branch length is shown from either (A) the hominoid–OWM ancestor to each leaf, or (B) the hominoid–NWM ancestor to each leaf. The branch length from the root to the human tip was set to 1 (with the actual value in parenthesis), and other lineages normalized to the human branch length. Branches from root-hominoids are shown in purple, from root-OWM in green and from root-NWM in orange. *Hominoid–monkey (either OWM or NWM) ancestor used as root. R code to replicate this figure is available at: https://github.com/priyamoorjani/Molecular-clock_figures-and-data/blob/master/Figure2.R.
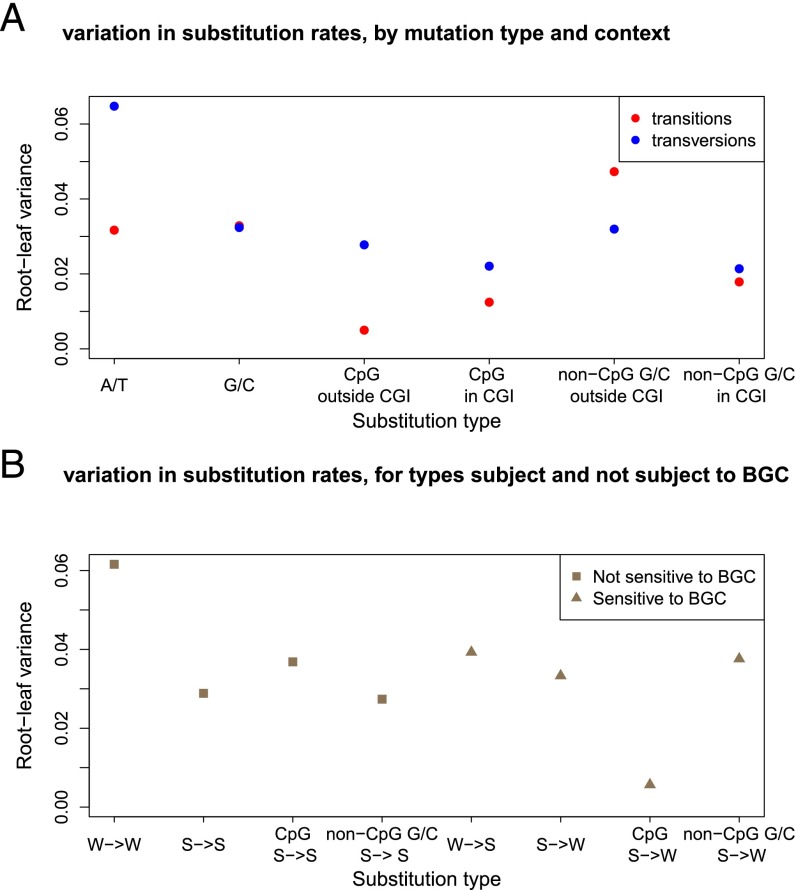
We then consider different substitution types in more detail, focusing on eight types: transitions and transversions occurring at either ancestrally A or T (referred to as A/T), ancestrally G or C (G/C), and CpG and non-CpG G/C, again excluding CGI. As a measure of variation among species, we use the variance of the normalized root-to-leaf distances across all remaining nine species (SI Appendix, Note S1), which is expected to be 0 if substitution rates are all identical. In general, transversions are more variable than transitions, with the largest variance (0.065) observed at A/T transversions (Fig. 3A). In turn, the variance is lowest for CpG transitions outside of annotated CGI (0.005), as observed previously in comparisons of smaller datasets of 19 mammals (1.7 Mb) (6) and 9 primates (0.15 Mb) (34). Interestingly, transitions at CpG sites inside CGI have a greater variance in substitution rates and behave similar to G/C transitions (Fig. 3A). The difference in behavior of CpGs inside and outside CGI is again consistent with the notion that when the source of mutation is primarily nonreplicative, mutations may depend more on absolute time than numbers of cell divisions, whereas when they have sources that are dependent on the numbers of cell divisions, they will be more variable among species. If this interpretation is correct, an interesting implication is that germline methylation levels and spontaneous deamination rates have remained very similar across primate species.
Fig. 3.
Variance among lineages for different substitution types. (A) For each ancestral state and each context shown on the x axis, we estimate the total branch length from the root to each terminal leaf in the Multiz dataset as the inferred number of substitutions per site. We then calculate the variance in the normalized root-to-leaf distance across nine primate species (human, chimpanzee, orangutan, rhesus macaque, crab-eating macaque, baboon, green monkey, squirrel monkey, and marmoset). (B) For each substitution type [S (G/C) and W (A/T)] in different substitution contexts shown on the x axis, we estimate the total branch length from the root to each terminal leaf in the Multiz dataset and calculate the variance in the root-to-leaf distance across the nine primates used in A. Using only one species from each taxon yields similar results (not shown). R code to replicate this figure is available at: https://github.com/priyamoorjani/Molecular-clock_figures-and-data/blob/master/Figure3.R.
Patterns of substitutions may also vary across species as a result of the effects of biased gene conversion, notably because of differences in effective population sizes (23). To examine this possibility, we compare the variance of the normalized root-to-leaf distances for substitutions that are subject to BGC (such as W→S or S→W) and those that should not be affected by BGC (such as W→W and S→S). If the strength of BGC varies across primates, we expect larger variance across species at W→S and S→W substitutions. Instead, there is no significant difference in the estimates of variances across the two classes of substitutions (Fig. 3B; P = 0.3, based on a permutation test). Although this finding seems puzzling, given the three- to fourfold difference in effective population size of these species (25), it is consistent with results of Do et al., who found no significant difference in the extent of biased gene conversion across diverse groups of West African and non-African human populations that differ up to twofold in their effective population sizes (35). If the strength of biased gene conversion at a site is typically very weak, both findings could reflect lack of power.
Given the importance of a steady molecular clock for dating events in human evolution, we next focus specifically on hominines (human, chimpanzee, and gorilla). In these comparisons, subtle differences in sequence quality, coverage, or the extent of mapping artifacts can lead to misleading evidence for variation in substitution rates across species. To minimize these effects, we generated pairwise sequence alignments for high-coverage (∼30×) genomes of human and chimpanzee, and human and gorilla. These pairs of genomes were mapped to the orangutan reference genome (which should be equidistant to all three species, assuming no differences in substitution rates among species), matching the alignment and variant calling pipeline for all three species (SI Appendix, Note S1). After removing missing data, nonneutral sites, and CGI, we obtain ∼1.03 Gb of sequence for human–chimpanzee and ∼1.02 Gb of sequence for human–gorilla whole-genome sequence alignments.
Despite the differences in generation time and onset of puberty among extant chimpanzees and humans, rates of evolution on the two lineages are very similar, at 0.621% and 0.633%, respectively. This difference of 1.9% is, however, highly statistically significant, under the assumption of no systematic errors (P < 10−20; SI Appendix, Note S1). When we consider the substitution rates at different mutation types, there are somewhat more pronounced differences for some types of substitutions, in inconsistent directions. For example, when comparing chimpanzee with human branches for substitutions involving transversions from CpG sites, the difference is 0.91-fold, whereas it is 1.07-fold for transversions at A/T sites (SI Appendix, Fig. S9). Comparing human and gorilla lineages, differences in substitution rates are more pronounced: the gorilla branch (0.824%) is longer than the human (0.773%) branch by, on average, 6.6% (P < 10−20; SI Appendix, Note S1). Again, different types of substitutions show distinct patterns, ranging between 0.96-fold at CpG transversions on the gorilla versus human branch to 1.10-fold for A/T transitions (SI Appendix, Fig. S10).
To check the reliability of these inferences, we also estimate the substitution rates using a second method based on a maximum-likelihood approach (36) (SI Appendix, Note S1). Although the absolute values of the substitution rates differ between the two methods, possibly as a result of methodological differences in calling ancestral states and assumptions about stationarity, the ratios of substitution rates between humans and chimpanzees (SI Appendix, Figs. S9 and S11) and between humans and gorillas (SI Appendix, Figs. S10 and S12) are almost identical. Although these results for the human–chimpanzee comparison match those obtained by Elango et al. (28), based on 75 Mb of data, our estimate of 1.07 for human–gorilla sequence difference is lower than the previous estimate of 1.11, based on 2 Mb of sequence data (28). Because our study is able to take advantage of a much larger dataset, accounts for differences in coverage and mapping among reference genomes, and considers only putatively neutral sites, we surmise that the earlier estimate of the extent of substitution rate variation among human and gorilla was slightly too high.
Importantly, these observations imply that the mutation spectra, and not just the yearly mutation rate, are changing across primates. Notably, although the rate of substitutions involving CpG transitions is relatively stable across species, the proportion of substitutions involving CpG transitions varies across species. Beyond that, the substitution rates for other mutation types also vary considerably (SI Appendix, Fig. S13). More fundamentally, our findings underscore that the mutation spectrum appears to have changed over the course of primate evolution. In this regard, it mirrors observations from even shorter time scales; for example, the recent report that transitions from 5′-TCC-3′→5′-TTC-3′ occurred at a proportionally higher rate in Europeans compared with Asians and Africans since these populations split (37).
Discussion
Evolutionary rates are faster in NWMs compared with OWMs, and in turn, rates in OWMs are faster than in humans and apes. These findings support the hominoid rate slowdown hypothesis (38, 39), indicating that since the split of hominoids and monkeys, per year mutation rates have decreased considerably. Moreover, the ordering of substitution rates is consistent with the generation time hypothesis, in that NWMs have a substantially shorter generation time (g = ∼6 y) than OWMs (g = ∼11 y), who in turn reproduce at younger ages than apes (g = ∼25 y; SI Appendix, Table S2). Within hominines, gorillas (g = ∼19 y) have a faster yearly rate than humans (g = ∼29 y) and chimpanzees (g = ∼25 y; SI Appendix, Table S2). To investigate whether the association between generation time and substitution rates is significant after controlling for the underlying phylogeny, we perform the phylogenetically independent contrast analysis (40) (SI Appendix, Note S1). Specifically, we assume the underlying phylogeny based on CpG transition rates (effectively assuming these are strictly clocklike) and then estimate the correlation between generation times and non-CpG substitution rates, controlling for the shared phylogenetic history. Using the nine species available for the analysis, the association is not significant (r = 0.17; P = 0.7), so the causal relationship remains to be established for primates.
An alternative approach is to ask whether differences in generation times and other life history traits can plausibly explain the variation in substitution rates. To this end, we use a model introduced by Amster and Sella (33) to describe mutations that are replicative in origin, which should also apply to mutations that are nonreplicative but well repaired (21). This model relates substitution rates to sex-specific life history and reproductive traits, and thus predicts how substitution rates are expected to differ among species. In applying the model, one option would be to examine the effect of one trait at a time. However, across primates, the average time between puberty and reproduction is positively correlated with age of onset of puberty in males (r = 0.74; P = 0.01, using Spearman’s correlation test), and the rate of spermatogenesis [measured by estimating the seminiferous epithelium cycle length (SECL)] is positively correlated with generation time (r = 0.90; P = 0.002) (SI Appendix, Table S4). We therefore vary the generation time, age of onset of puberty, and SECL for each lineage, relying on values estimated for extant humans, chimpanzees, and OWMs and mutation parameters estimated from human pedigree studies (SI Appendix, Note S1 and Table S2). On that basis, we predict that yearly mutation rates in humans and chimpanzees should differ by ∼19%, and hominoids (using humans and chimpanzees here as not all parameter values are available for orangutans) and OWMs should differ by ∼86%. Thus, if anything, differences in life history traits in extant species predict even more variation in substitution rates than is observed (33).
The use of life history traits in extant species will exacerbate the expected differences in substitution rates if closely related species have had similar life histories throughout much of their evolutionary past. Fossil evidence suggests the age of puberty on the human lineage may have only recently increased, for example, and was lower in Homo erectus and Neanderthals (41, 42). Similar changes are likely to have occurred on the chimpanzee lineage as well. If we change the age of onset of puberty in humans to 9 y, the difference in rates between humans and chimpanzees is only expected to be ∼5%. One implication, then, of finding such similar substitution rates in humans and chimpanzees is that their life histories may have been fairly similar for much of their evolutionary history.
That substitution rates should and do vary with life history traits highlights the challenges of using the molecular clock for dating evolutionary events, even within hominines. One way to overcome this difficulty is to explicitly model the changes in life history traits within species and over the course of primate evolution (33). Taking this approach, Amster and Sella (33) show that accounting for variation in generation time, age of onset of puberty, and rate of spermatogenesis in extant apes helps to reconcile the split times estimated on the basis of molecular and fossil evidence (33). Their method, however, requires knowledge of life history traits in both extant and ancestral populations.
An alternative is to focus on mutation types that are much less sensitive to life history traits, such as CpG transitions outside CGIs. Even for this mutation type, the variance in substitution rates across species is nonzero, possibly because a subset of these mutations occurs due to replication errors, or because repair is not completely inefficient, or mutations do not accumulate in strict proportion to parental ages (21). Nonetheless, CpG transitions appear to be least affected by life history differences across species, accumulating in a quasi-clocklike manner. Moreover, in humans, they contribute almost a fifth of all de novo mutations, and so provide enough data for precise estimation (10).
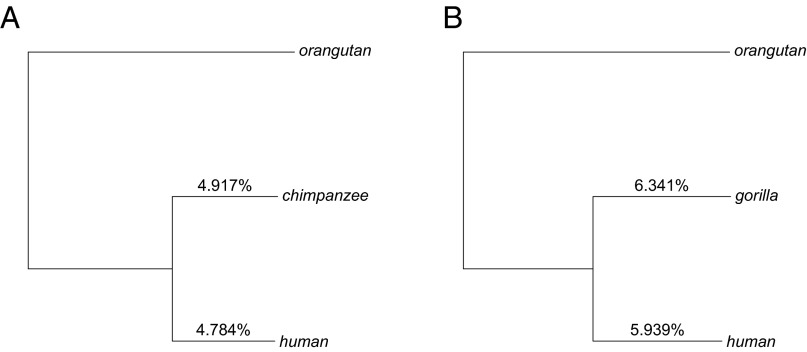
With these considerations in mind, we reestimate the divergence and split times of humans, chimpanzees, and gorillas, using substitution rates estimated only at CpG transitions. Assuming the per year mutation rate for CpG transitions obtained in ref. 10 (SI Appendix, Note S1), we estimate that humans diverged from chimpanzees ∼12.1 Mya and from gorillas ∼15.1 Mya (Fig. 4). Assuming further that the effective population size of the human–ape ancestor was five times the current population size (as estimated by refs. 43, 44), the human–chimpanzee split time is ∼7.9 Mya, and the human–gorilla split time is 10.8 Mya. We note that there is substantial uncertainty in estimates of ancestral population size of apes, with previous estimates ranging between 50,000 and 100,000 (43–45). Accounting for this uncertainty provides estimates of human–chimpanzee split time in the range of 6.5–9.3 Mya, and human–gorilla split time in the range of 9.4–12.2 Mya. Reassuringly, these estimates are similar to those obtained by explicitly modeling the dependence of replicative mutations on life history traits in hominines (33). Moreover, they are in broad agreement with evidence from the fossil record, which suggests a human–chimpanzee split time of 6–10 Mya and a human–gorilla split time of 7–12 Mya (46–51). Thus, within hominines, there is no obvious discrepancy between phylogenetic and pedigree-based estimates of mutation rates, once the effect of life history traits on mutation rates is taken into account (33).
Fig. 4.
Revised divergence time for hominines. We estimate the autosomal substitution rates for transitions at CpG sites by applying Phylofit to the high-coverage pairwise alignment of (A) human and chimpanzee and (B) human and gorilla. All hominines were were mapped to the orangutan reference genome. To infer divergence times, we use germline mutation rates for CpG transitions estimated from sequencing human pedigrees (see SI Appendix, Note S1 for details). We estimate average human and chimpanzee divergence time as 12.1 Mya, and average human and gorilla divergence time as 15.1 Mya. R code to replicate this figure is available at https://github.com/priyamoorjani/Molecular-clock_figures-and-data/blob/master/Figure4.R.
Materials and Methods
We used Phylofit (30) to estimate autosomal substitution rates for different mutation types, using the following three datasets: a 12-primate whole-genome sequence alignment, with mouse as an outgroup, that is part of a 100-way mammalian phylogeny, mapped using Multiz (52) (referred to as the Multiz dataset); a seven-primate whole-genome alignment, mapped using the Enredo-Pecan-Ortheus pipeline (32) (referred to the EPO dataset); and high coverage genomes for a human (of European descent) that we sequenced (SI Appendix, Note S2), a chimpanzee (Ind-D from ref. 11), and a gorilla [Delphi from ref. 44; data kindly provided by Tomas Marques-Bonet, Institut Biologia Evolutiva, Universitat Pompeu Fabra/Spanish National Research Council (CSIC) (referred to as the high-coverage hominoid dataset)]. These genomes were mapped to the orangutan reference genome (ponAbe2) (53), which should be equidistant to humans and extant African great apes (assuming no variation in substitution rates). We matched these species for coverage, alignment, and mapping pipelines to minimize the effects of technical artifacts. For details, see SI Appendix, Note S1.
Supplementary Material
Acknowledgments
We thank David Pilbeam, Guy Amster, Guy Sella, Jenny Tung, Minyoung Wyman, Susan Alberts, and Ziyue Gao for helpful discussions. We thank Nick Patterson and Heng Li for technical advice for mapping and alignment of high-coverage genomes and Melissa Hubisz and Adam Siepel for advice on running Phylofit. P.M. was supported by the National Institutes of Health under Ruth L. Kirschstein National Research Service Award F32 GM115006-01. C.E.G.A. was supported by a Science Without Borders fellowship from Conselho Nacional de Desenvolvimento Científico e Tecnológico Brazil (PDE 201145/2015-4). The computing in this project was supported by two National Institutes of Health instrumentation grants (S10OD012351 and S10OD021764) received by the Department of Systems Biology at Columbia University.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1600374113/-/DCSupplemental.
References
- 1.Ségurel L, Wyman MJ, Przeworski M. Determinants of mutation rate variation in the human germline. Annu Rev Genomics Hum Genet. 2014;15:47–70. doi: 10.1146/annurev-genom-031714-125740. [DOI] [PubMed] [Google Scholar]
- 2.Kimura M. The Neutral Theory of Molecular Evolution. Cambridge Univ Press; New York: 1984. [Google Scholar]
- 3.Zuckerkandl E, Pauling L. 1965. Evolutionary divergence and convergence in proteins. Evolving Genes and Proteins, eds Bryson V, Vogel HJ (Academic, New York), pp 97–166. [DOI] [PubMed]
- 4.Kumar S. Molecular clocks: Four decades of evolution. Nat Rev Genet. 2005;6(8):654–662. doi: 10.1038/nrg1659. [DOI] [PubMed] [Google Scholar]
- 5.Wilson Sayres MA, Venditti C, Pagel M, Makova KD. Do variations in substitution rates and male mutation bias correlate with life-history traits? A study of 32 mammalian genomes. Evolution. 2011;65(10):2800–2815. doi: 10.1111/j.1558-5646.2011.01337.x. [DOI] [PubMed] [Google Scholar]
- 6.Hwang DG, Green P. Bayesian Markov chain Monte Carlo sequence analysis reveals varying neutral substitution patterns in mammalian evolution. Proc Natl Acad Sci USA. 2004;101(39):13994–14001. doi: 10.1073/pnas.0404142101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Britten RJ. Rates of DNA sequence evolution differ between taxonomic groups. Science. 1986;231(4744):1393–1398. doi: 10.1126/science.3082006. [DOI] [PubMed] [Google Scholar]
- 8.Wu C-I, Li W-H. Evidence for higher rates of nucleotide substitution in rodents than in man. Proc Natl Acad Sci USA. 1985;82(6):1741–1745. doi: 10.1073/pnas.82.6.1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fenner JN. Cross-cultural estimation of the human generation interval for use in genetics-based population divergence studies. Am J Phys Anthropol. 2005;128(2):415–423. doi: 10.1002/ajpa.20188. [DOI] [PubMed] [Google Scholar]
- 10.Kong A, et al. Rate of de novo mutations and the importance of father’s age to disease risk. Nature. 2012;488(7412):471–475. doi: 10.1038/nature11396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Venn O, et al. Nonhuman genetics. Strong male bias drives germline mutation in chimpanzees. Science. 2014;344(6189):1272–1275. doi: 10.1126/science.344.6189.1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scally A, Durbin R. Revising the human mutation rate: Implications for understanding human evolution. Nat Rev Genet. 2012;13(10):745–753. doi: 10.1038/nrg3295. [DOI] [PubMed] [Google Scholar]
- 13.Martin AP, Palumbi SR. Body size, metabolic rate, generation time, and the molecular clock. Proc Natl Acad Sci USA. 1993;90(9):4087–4091. doi: 10.1073/pnas.90.9.4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fontanillas E, Welch JJ, Thomas JA, Bromham L. The influence of body size and net diversification rate on molecular evolution during the radiation of animal phyla. BMC Evol Biol. 2007;7(1):95. doi: 10.1186/1471-2148-7-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wong A. Covariance between testes size and substitution rates in primates. Mol Biol Evol. 2014;31(6):1432–1436. doi: 10.1093/molbev/msu091. [DOI] [PubMed] [Google Scholar]
- 16.Drost JB, Lee WR. Biological basis of germline mutation: Comparisons of spontaneous germline mutation rates among drosophila, mouse, and human. Environ Mol Mutagen. 1995;25(Suppl 26):48–64. doi: 10.1002/em.2850250609. [DOI] [PubMed] [Google Scholar]
- 17.Crow JF. The origins, patterns and implications of human spontaneous mutation. Nat Rev Genet. 2000;1(1):40–47. doi: 10.1038/35049558. [DOI] [PubMed] [Google Scholar]
- 18.Gage TB. The comparative demography of primates: With some comments on the evolution of life histories. Annu Rev Anthropol. 1998;27:197–221. doi: 10.1146/annurev.anthro.27.1.197. [DOI] [PubMed] [Google Scholar]
- 19.Ramm SA, Stockley P. Sperm competition and sperm length influence the rate of mammalian spermatogenesis. Biol Lett. 2009;6(2):219–221. doi: 10.1098/rsbl.2009.0635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duncan BK, Miller JH. Mutagenic deamination of cytosine residues in DNA. Nature. 1980;287(5782):560–561. doi: 10.1038/287560a0. [DOI] [PubMed] [Google Scholar]
- 21.Gao Z, Wyman MJ, Sella G, Przeworski M. Interpreting the dependence of mutation rates on age and time. PLoS Biol. 2016;14(1):e1002355. doi: 10.1371/journal.pbio.1002355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coulondre C, Miller JH, Farabaugh PJ, Gilbert W. Molecular basis of base substitution hotspots in Escherichia coli. Nature. 1978;274(5673):775–780. doi: 10.1038/274775a0. [DOI] [PubMed] [Google Scholar]
- 23.Duret L, Galtier N. Biased gene conversion and the evolution of mammalian genomic landscapes. Annu Rev Genomics Hum Genet. 2009;10:285–311. doi: 10.1146/annurev-genom-082908-150001. [DOI] [PubMed] [Google Scholar]
- 24.Capra JA, Hubisz MJ, Kostka D, Pollard KS, Siepel A. A model-based analysis of GC-biased gene conversion in the human and chimpanzee genomes. PLoS Genet. 2013;9(8):e1003684. doi: 10.1371/journal.pgen.1003684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leffler EM, et al. Revisiting an old riddle: What determines genetic diversity levels within species? PLoS Biol. 2012;10(9):e1001388. doi: 10.1371/journal.pbio.1001388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stevison LS, et al. Great Ape Genome Project The Time Scale of Recombination Rate Evolution in Great Apes. Mol Biol Evol. 2016;33(4):928–945. doi: 10.1093/molbev/msv331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim S-H, Elango N, Warden C, Vigoda E, Yi SV. Heterogeneous genomic molecular clocks in primates. PLoS Genet. 2006;2(10):e163. doi: 10.1371/journal.pgen.0020163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Elango N, Thomas JW, Yi SV. NISC Comparative Sequencing Program Variable molecular clocks in hominoids. Proc Natl Acad Sci USA. 2006;103(5):1370–1375. doi: 10.1073/pnas.0510716103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patterson N, Richter DJ, Gnerre S, Lander ES, Reich D. Genetic evidence for complex speciation of humans and chimpanzees. Nature. 2006;441(7097):1103–1108. doi: 10.1038/nature04789. [DOI] [PubMed] [Google Scholar]
- 30.Siepel A, Haussler D. Phylogenetic estimation of context-dependent substitution rates by maximum likelihood. Mol Biol Evol. 2004;21(3):468–488. doi: 10.1093/molbev/msh039. [DOI] [PubMed] [Google Scholar]
- 31.Ananda G, Chiaromonte F, Makova KD. A genome-wide view of mutation rate co-variation using multivariate analyses. Genome Biol. 2011;12(3):R27. doi: 10.1186/gb-2011-12-3-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paten B, Herrero J, Beal K, Fitzgerald S, Birney E. Enredo and Pecan: Genome-wide mammalian consistency-based multiple alignment with paralogs. Genome Res. 2008;18(11):1814–1828. doi: 10.1101/gr.076554.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Amster G, Sella G. Life history effects on the molecular clock of autosomes and sex chromosomes. Proc Natl Acad Sci USA. 2016;113(6):1588–1593. doi: 10.1073/pnas.1515798113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee H-J, Rodrigue N, Thorne JL. Relaxing the Molecular Clock to Different Degrees for Different Substitution Types. Mol Biol Evol. 2015;32(8):1948–1961. doi: 10.1093/molbev/msv099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Do R, et al. No evidence that selection has been less effective at removing deleterious mutations in Europeans than in Africans. Nat Genet. 2015;47(2):126–131. doi: 10.1038/ng.3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Duret L, Arndt PF. The impact of recombination on nucleotide substitutions in the human genome. PLoS Genet. 2008;4(5):e1000071. doi: 10.1371/journal.pgen.1000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harris K. Evidence for recent, population-specific evolution of the human mutation rate. Proc Natl Acad Sci USA. 2015;112(11):3439–3444. doi: 10.1073/pnas.1418652112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goodman M. Serological analysis of the systematics of recent hominoids. Hum Biol. 1963;35:377–436. [PubMed] [Google Scholar]
- 39.Yi SV. Morris Goodman’s hominoid rate slowdown: The importance of being neutral. Mol Phylogenet Evol. 2013;66(2):569–574. doi: 10.1016/j.ympev.2012.07.031. [DOI] [PubMed] [Google Scholar]
- 40.Felsenstein J. Phylogenies and the comparative method. Am Nat. 1985;125(1):1–15. [Google Scholar]
- 41.Graves RR, Lupo AC, McCarthy RC, Wescott DJ, Cunningham DL. Just how strapping was KNM-WT 15000? J Hum Evol. 2010;59(5):542–554. doi: 10.1016/j.jhevol.2010.06.007. [DOI] [PubMed] [Google Scholar]
- 42.Gluckman PD, Hanson MA. Changing times: The evolution of puberty. Mol Cell Endocrinol. 2006;254-255:26–31. doi: 10.1016/j.mce.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 43.Wall JD. Estimating ancestral population sizes and divergence times. Genetics. 2003;163(1):395–404. doi: 10.1093/genetics/163.1.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Prado-Martinez J, et al. Great ape genetic diversity and population history. Nature. 2013;499(7459):471–475. doi: 10.1038/nature12228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scally A, et al. Insights into hominid evolution from the gorilla genome sequence. Nature. 2012;483(7388):169–175. doi: 10.1038/nature10842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wilkinson RD, et al. Dating primate divergences through an integrated analysis of palaeontological and molecular data. Syst Biol. 2011;60(1):16–31. doi: 10.1093/sysbio/syq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Begun DR. 2015. Fossil record of Miocene hominoids. Handbook of Paleoanthropology, eds Henke W, Tattersall I (Springer, Berlin), pp 1261–1332.
- 48.Brunet M, et al. A new hominid from the Upper Miocene of Chad, Central Africa. Nature. 2002;418(6894):145–151. doi: 10.1038/nature00879. [DOI] [PubMed] [Google Scholar]
- 49.Suwa G, Kono RT, Katoh S, Asfaw B, Beyene Y. A new species of great ape from the late Miocene epoch in Ethiopia. Nature. 2007;448(7156):921–924. doi: 10.1038/nature06113. [DOI] [PubMed] [Google Scholar]
- 50.White TD, et al. Ardipihtecus ramidus and the paleobiology of early hominids. Science. 2009;326(5949):64–86. [PubMed] [Google Scholar]
- 51.Wood B, Harrison T. The evolutionary context of the first hominins. Nature. 2011;470:347–352. doi: 10.1038/nature09709. [DOI] [PubMed] [Google Scholar]
- 52.Karolchik D, et al. The UCSC Genome Browser database: 2014 update. Nucleic Acids Res. 2014;42(Database issue) D1:D764–D770. doi: 10.1093/nar/gkt1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Locke DP, et al. Comparative and demographic analysis of orang-utan genomes. Nature. 2011;469(7331):529–533. doi: 10.1038/nature09687. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.