Significance
Establishment of the progenitor of chloroplasts by the host plant cell during endosymbiosis required the integration of two sets of biological membranes, the endoplasmic reticulum and the chloroplast envelopes, participating in the synthesis of galactolipid precursors for the photosynthetic membranes. Galactolipid synthesis is unequally distributed between the two envelope membranes, necessitating lipid transfer between the envelopes and toward the thylakoids. Here we show that the N-terminal sequence of digalactosyldiacylglycerol synthase 1 is essential for the integration of the chloroplast galactolipid synthesis machinery into the host cell. This N-terminal sequence was invented at the time the endosymbiotic organelle was established, providing a basic glycosyltransferase with a neofunction essential for lipid mobilization between organelles and endomembrane systems in plants.
Keywords: galactolipid, chloroplast, envelope, lipid transfer
Abstract
Galactolipids [monogalactosyldiacylglycerol (MGDG) and digalactosyldiacylglycerol (DGDG)] are the hallmark lipids of photosynthetic membranes. The galactolipid synthases MGD1 and DGD1 catalyze consecutive galactosyltransfer reactions but localize to the inner and outer chloroplast envelopes, respectively, necessitating intermembrane lipid transfer. Here we show that the N-terminal sequence of DGD1 (NDGD1) is required for galactolipid transfer between the envelopes. Different diglycosyllipid synthases (DGD1, DGD2, and Chloroflexus glucosyltransferase) were introduced into the dgd1-1 mutant of Arabidopsis in fusion with N-terminal extensions (NDGD1 and NDGD2) targeting to the outer envelope. Reconstruction of DGDG synthesis in the outer envelope membrane was observed only with diglycosyllipid synthase fusion proteins carrying NDGD1, indicating that NDGD1 enables galactolipid translocation between envelopes. NDGD1 binds to phosphatidic acid (PA) in membranes and mediates PA-dependent membrane fusion in vitro. These findings provide a mechanism for the sorting and selective channeling of lipid precursors between the galactolipid pools of the two envelope membranes.
The two galactolipids, monogalactosyldiacylglycerol (MGDG) and digalactosyldiacylglycerol (DGDG), are most abundant in land plants, green algae, and cyanobacteria (1). MGDG and DGDG are predominant in thylakoid membranes of chloroplasts, where they are integral components of photosystems I and II and of the light-harvesting complex II (2–4), and are essential for photosynthesis and growth (5, 6). Galactolipids are synthesized in the envelope membranes of chloroplasts (7). In tobacco and Arabidopsis, the MGDG synthase MGD1 localizes to the inner envelope where it produces the major proportion of MGDG (8, 9). The outer chloroplast envelope of Arabidopsis harbors two DGDG synthases, DGD1 and DGD2 (10, 11). DGD1 synthesizes the predominant proportion of DGDG, whereas DGD2 is active during growth under phosphate limitation. Phosphate deprivation results in the accumulation of glycolipids, including DGDG, at the expense of phospholipids and the redirection of phosphate to other cellular processes (12). DGDG synthesized by DGD1 and DGD2 is transported to thylakoid and extraplastidial membranes, respectively (11, 12).
Transport processes are required to channel lipid molecules between organelles and across and between different membranes (13). An ATP-binding cassette (ABC) transporter composed of three different subunits [trigalactosyldiacylglycerol1 (TGD1), -2 (TGD2), and -3 (TGD3)] is involved in the transfer of lipid precursors from the endoplasmic reticulum (ER) to the chloroplast where they are used for galactolipid synthesis. Galactolipid molecules derived from imported precursors can be distinguished from galactolipids directly synthesized in the chloroplast by the acyl composition at the sn2 position of the glycerol, with molecules containing sn2-16C acyl groups being chloroplast-derived (prokaryotic) and molecules containing sn2-18C acyl groups being ER-derived (eukaryotic). In Arabidopsis, MGDG consists of similar proportions of eukaryotic (sn1-18:3/sn2-18:3-MGDG) and prokaryotic molecules (18:3/16:3-MGDG), but DGDG is mostly eukaryotic (18:3/18:3-DGDG) (14).
DGD1 carries a long N-terminal extension (NDGD1), which is required for insertion into the outer envelope (10). The presence of this NDGD1 domain is unique to DGD1 among the proteins involved in galactolipid synthesis; the other proteins (MGD1, MGD2, MGD3, and DGD2) carry shorter, cleavable N-terminal sequences with targeting information to the chloroplast.
In the present study we address the role of NDGD1 in galactolipid synthesis and transfer. Expression of fusion proteins of plant DGDG synthases and of a bacterial glucosylgalactosyldiacylglycerol (GlcGalDG) synthase (GlcT) with different N-terminal extensions in Escherichia coli and Arabidopsis demonstrated that NDGD1 is not required for DGDG synthesis per se but is essential for enabling transfer of galactolipids between envelope membranes and therefore for DGDG accumulation in thylakoid membranes.
Results and Discussion
DGDG Synthases from Plants and Eukaryotic Algae.
DGD1 carries a unique N-terminal extension (amino acids 1–338, NDGD1) required for insertion into the outer envelope membrane (oEM), in addition to its glycosyltransferase domain (amino acids 339–808, CDGD1) (10, 15). CDGD1/DGD2-like sequences are found in all Streptophyta and in some Chlorophyta and Rhodophyta (SI Appendix, Figs. S1 and S2) (16). Spermatophyta contain two sequences, DGD1 and DGD2, with only DGD1 encompassing the NDGD1 extension (SI Appendix, Fig. S2). The genomes of Selaginella, Physcomitrella, and Klebsormidium contain DGD1 with the NDGD1 extension but are devoid of DGD2-related genes (SI Appendix, Fig. S1). Only one DGD1-like gene with a long N-terminal extension is found in some Chlorophyta (Chlamydomonas, Volvox, and Ostreococcus), whereas Coccomyxa and Bathycoccus contain two genes, one is a DGD1-type and the other a DGD2-type sequence. Other Chlorophyta (Chlorella, Auxenochlorella, and Micromonas) contain only a single DGD2 gene without the extension. Within Rhodophyta, Chondrus contains a single DGD2-like gene (16), and Porphyridium contains two genes, a DGD1-like sequence and a DGD2 sequence (SI Appendix, Figs. S1 and S2). The Cyanidiales (Rhodophyta) harbor DGDG synthases distinct from those in plants; the Cyanidiales sequences are related to cyanobacterial DgdA (17, 18). Database searches with PSI BLAST using NDGD1 from Arabidopsis resulted in the retrieval of NDGD1 sequences in all Streptophyta, including Klebsormidium, but not in other organisms. The long N-terminal extensions of DGD1 proteins in Chlorophyta and Rhodophyta show only low similarity with Streptophyta NDGD1 sequences. Therefore, it is possible that in some Chlorophyta and Rhodophyta a polypeptide with low sequence similarity to NDGD1 but with a similar function was established in Streptophyta and has evolved further to or has been replaced by NDGD1. NDGD1 sequences of Streptophyta are only found in translational fusions in DGD1 proteins, raising an intriguing question: whether the origin of this domain coincided with chloroplast endosymbiosis and perhaps was a necessary neofunctionalization enabling the integration of chloroplast and host-cell lipid metabolisms.
The N-Terminal Extension NDGD1 Is Dispensable for DGDG Synthesis by Recombinant DGD1.
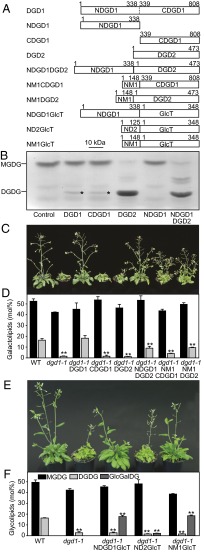
To study the biochemical and molecular function of NDGD1, different DGDG synthase constructs were introduced into E. coli coexpressing cucumber MGD1 to provide MGDG. DGDG accumulation was observed by TLC. DGD1, CDGD1, and DGD2 converted MGDG into DGDG, but NDGD1 was enzymatically inactive, indicating that CDGD1 was necessary and sufficient for DGDG synthesis (Fig. 1). The amount of DGDG synthesized by NDGD1DGD2 (the fusion of NDGD1 to DGD2 to make it DGD1-like) was similar to that synthesized by DGD2. Introduction of NDGD1-containing constructs (DGD1, NDGD1, and NDGD1DGD2) into E. coli affected growth, indicating that NDGD1 production is detrimental to the bacterial cells (SI Appendix, Fig. S3). Production of CDGD1 and DGD2 as monitored by immunoblot analysis was strong and comparable, whereas the production of NDGD1-containing proteins was very low (SI Appendix, Fig. S3).
Fig. 1.
DGDG and GlcGalDG formation in E. coli and Arabidopsis after expression of plant and bacterial diglycosyllipid synthases. (A) Constructs for DGDG synthases from Arabidopsis and GlcT from Chloroflexus. Numbers indicate amino acid positions. (B) Thin-layer chromatogram stained for sugars on which lipid extracts from E. coli expressing MGD1 and various DGDG synthases are separated. Small amounts of DGDG are marked with an asterisk. The bands between MGDG and DGDG comigrate with lyso-MGDG. (C and E) Complementation of diglycosyllipid and growth deficiency of dgd1-1. Transformed dgd1-1 plants expressing DGD1, DGD2, or GlcT fusion constructs were grown on soil for 35 d. (D and F) Galactolipid and GlcGalDG contents of dgd1-1 plants expressing DGD1, DGD2, or GlcT. Values (mean ± SD, measurements of three plants) significantly different from WT are indicated (**P < 0.01, Student's t test).
Mutations in the NDGD1 or CDGD1 Part of DGD1 Affect in Vivo DGDG Synthesis Activity.
The Arabidopsis dgd1-1 mutant carries a premature stop codon in the CDGD1 part of the DGD1 gene resulting in decreased DGDG content and plant growth (SI Appendix, Fig. S4) (5, 15). Immunoblot analysis with anti-NDGD1 antibodies revealed the presence of similar amounts of a 91-kDa DGD1 protein in WT Arabidopsis and a 64-kDa protein (a truncated NDGD1-containing polypeptide, amino acids 1–563) in the dgd1-1 mutant. If NDGD1 is functionally relevant in dgd1-1, a more severe phenotype would be expected for dgd1 alleles deficient in NDGD1. Two additional mutant plants (dgd1-2 and dgd1-3) carrying insertions in the first exon and third intron, respectively, were obtained (SI Appendix, Fig. S4). Immunoblot analysis revealed that no residual DGD1 polypeptide was observed in dgd1-2 (M1–A189, calculated size, 21 kDa), presumably because it was degraded, whereas a 31-kDa polypeptide (M1–D273) was detected in dgd1-3 (SI Appendix, Fig. S4). Growth and galactolipid contents of dgd1-2 and dgd1-3 were indistinguishable from dgd1-1. Therefore, all lines carry DGD1-null mutations. Furthermore the production of the truncated NDGD1 polypeptide in dgd1-1 per se does not contribute to galactolipid synthesis in planta. It is possible that additional truncated versions of DGD1 are expressed in Arabidopsis. Indeed, one splice variant (At3g11670.2) is annotated in The Arabidopsis Information Resource (TAIR) database (www.arabidopsis.org), in addition to the correctly spliced DGD1 mRNA (At3g11670.1). This variant is derived from mis-splicing of intron 6 resulting in a truncated ORF that encodes a polypeptide encompassing the entire NDGD1 sequence but only part of the glycosyltransferase domain (SI Appendix, Fig. S5). Compared with At3g11670.1, the expression of At3g11670.2 under normal or phosphate deficient conditions was not detectable or was extremely low, indicating that it presumably has no physiological function (SI Appendix, Fig. S5).
NDGD1 Is Essential for DGDG Mobilization from the oEM to the Inner Envelope of Chloroplasts.
Next, the relevance of NDGD1 for glycolipid production in planta was studied by introducing NDGD1 constructs or chloroplast envelope-targeting sequences of MGD1 (NM1, targeting the inner envelope) or DGD2 (ND2, a short N-terminal sequence targeting the oEM), fused to CDGD1, DGD2, or a glucosyltransferase, GlcT, from Chloroflexus aurantiacus, into dgd1-1. GlcT adds a glucose to carbon 6 of the galactose moiety of MGDG, thereby producing GlcGalDG (19). The synthesis of DGDG or GlcGalDG was monitored by TLC separation and quantification by GC (Fig. 1). Transfer of glycosyltransferases mistargeted to the inner envelope membrane (iEM) by fusion with the N terminus of MGD1 (NM1CDGD1, NM1DGD2, or NM1GlcT) into dgd1-1 complemented diglycosyllipid deficiency and growth, confirming that all domains are functional and indicating that diglycosyllipid production can be translocated to the iEM. Introduction of DGD1, NDGD1DGD2, or NDGD1GlcT (the fusion of NDGD1 with GlcT) into dgd1-1 again resulted in complementation of diglycosyllipid deficiency and growth, whereas the expression of DGD2 or ND2GlcT (the fusion of ND2 with GlcT) in dgd1-1 resulted in the synthesis of low amounts of DGDG/GlcGalDG insufficient for complementation. In summary, only constructs targeting the diglycosyllipid synthase to the iEM or in fusion with NDGD1 were able to complement dgd1-1 lipid deficiency and growth retardation. Therefore, NDGD1 is essential for diglycosyllipid production in the oEM and must allow galactolipid transfer between envelopes. This conclusion is corroborated by the analysis of Arabidopsis dgd1-1 plants during phosphate deprivation, when DGD2 is produced. DGDG increases to 9.0% in dgd1-1 (11, 12); however, this pool of extra DGDG cannot complement dgd1-1 growth deficiency, indicating that in the absence of NDGD1 it is not transferred to the inner envelope and to the thylakoids (12, 20). The NDGD1 polypeptide is still produced in dgd1-1 plants (SI Appendix, Fig. S4), but its expression in trans in the genome together with CDGD1, DGD2, or ND2GlcT is insufficient to complement lipid and growth deficiency. Therefore NDGD1 and diglycosyllipid synthases cannot form functional complexes in the oEM, but the two sequences must be present in a translational fusion.
Because the acyl compositions of MGDG and DGDG in the iEM and oEM are similar, it has been suggested that galactolipid transfer between the envelopes is not selective (21). This notion is corroborated here, because DGDG in transgenic dgd1-1 lines transformed with DGD1 (oEM) or NM1CDGD1 (iEM) was mostly eukaryotic (18:3/18:3), because it contained only 2–6 mol% 16:3, similar to WT (SI Appendix, Tables S1 and S2). On the other hand, DGDG or GlcGalDG in dgd1-1 lines with DGD2 or GlcT transgenes contained high 16:3 content (12–19 mol%), suggesting that prokaryotic MGDG (18/16) was also used for diglycosyllipid synthesis.
Secondary Structure of NDGD1.
The structures of DGD2 and CDGD1 can be predicted based on sequence similarities to glycosyltransferases of the CAZY family GT-4 (22). However, no 3D structure was available for the Arabidopsis NDGD1 sequence. Secondary structure prediction of NDGD1 using the I-TASSER algorithm revealed that it presumably harbors coiled–coil domains and α-helices without β-sheets (23). The top threading templates used by I-TASSER are 4jioA [BCK1-like resistance to osmotic shock protein 1, V domain (Bro1V)], 4wj1A [ESX-1 secretion-associated protein B (EspB)], 4bm5A [translocon at the inner chloroplast envelope membrane protein 110 (TIC110)], and 4mu6A [Legionella pneumophila effector protein C3 (LegC3)] (SI Appendix, Fig. S6). The same threading templates were obtained with NDGD1 sequences from rice, Physcomitrella, Selaginella, Klebsormidium, and other plants, indicating that these templates represent relevant models. The four proteins used for threading of NDGD1 are rich in α-helices and are associated with membranes. TIC110 is a component of the chloroplast protein import complex (24). EspB from Mycobacterium tuberculosis and LegC3 from Legionella pneumophila are bacterial effector proteins causing phagosome rupture or preventing phagosome fusion with lysosomes, respectively, after uptake into the host cell (25, 26). The Bro1V domain is part of the yeast Bro1 protein which is homologous to the human Alix (ALG-2–interacting protein X) protein. Alix binds to the unusual acidic lipid lysobisphosphatidic acid and is recruited to late endosomal membranes (27, 28). The interaction between lysobisphosphatidic acid and Alix is crucial for the formation of multivesicular liposomes (28). The structures of the four proteins used for threading of NDGD1 resemble those of SNARE (soluble N-ethylmaleimide-sensitive-factor attachment receptor) proteins, which also harbor α-helices and coiled–coil domains and are involved in tethering and fusion events between vesicles and target membranes (29). Therefore NDGD1 is predicted to harbor coiled–coil domains and α-helices and might interact with membranes and possibly bind to acidic phospholipids.
NDGD1 Binds to Phosphatidic Acid.
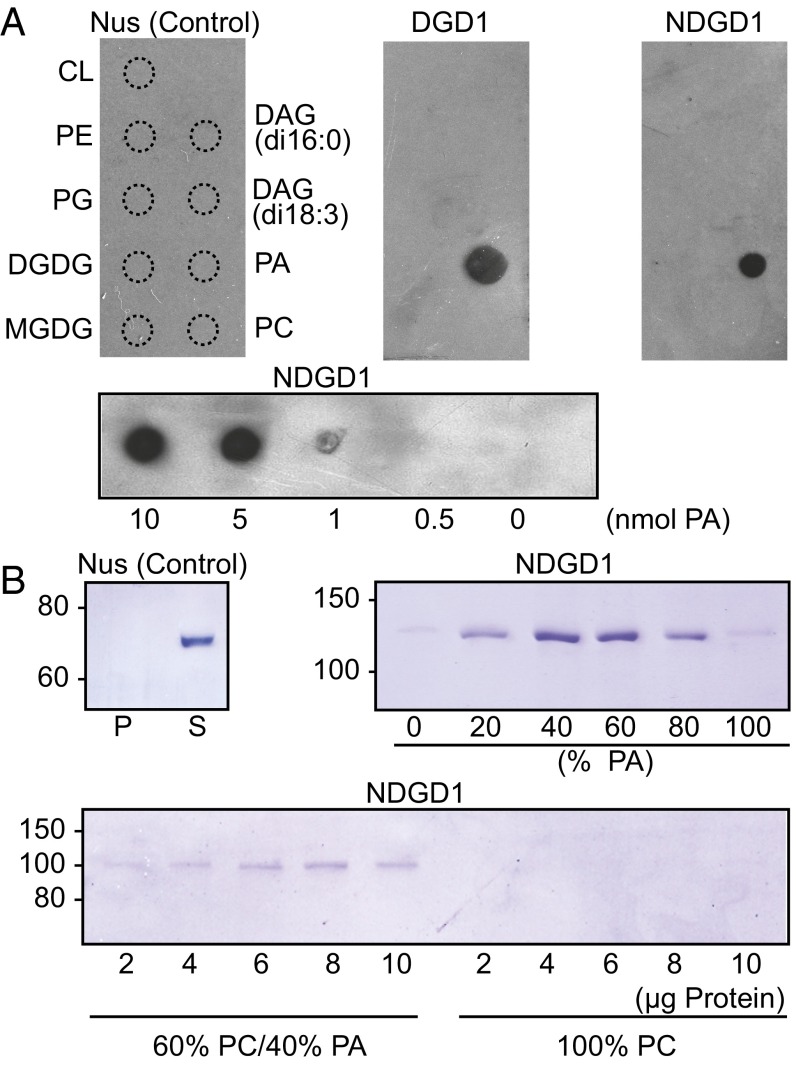
To determine if DGD1 and NDGD1 interact directly with membranes, lipid binding was investigated by incubating recombinant proteins with lipid-nitrocellulose strips. Strong binding of DGD1 and NDGD1 to the acidic phospholipid phosphatidic acid (PA), but not to other membrane lipids, was observed (Fig. 2). The lowest amount of PA sufficient for NDGD1 binding was 1 nmol (Fig. 2). Liposomes composed of phosphatidylcholine (PC) and PA were incubated with NDGD1, and liposome-associated proteins were harvested by centrifugation and analyzed in protein gels (Fig. 2). NDGD1 bound to liposomes containing PA but not to PA-free liposomes. NDGD1 binding was stronger with increasing PA content up to 60%, but binding with liposomes of 100% PA was compromised, probably because PA does not form bilayers and affects liposome stability (30). The lowest amount of NDGD1 detectable in liposome binding assays was 2 µg.
Fig. 2.
NDGD1 interactions with lipids and membranes. (A, Upper) NDGD1 binds to PA as revealed after incubation of nitrocellulose strips containing different glycerolipids with recombinant Nus (control), DGD1, or NDGD1 protein. Binding was visualized by immunodetection. (Lower) The blot shows NDGD1 binding to different amounts of PA (0.1–10 nmol). CL, cardiolipin; DAG, diacylglycerol; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PG, phosphatidylglycerol. (B) NDGD1 binding to liposomes is PA-dependent. Liposomes with different proportions of PA and PC were incubated with recombinant proteins. Bound proteins were detected in polyacrylamide gels after centrifugation of liposomes. (Upper Left) Control (Nus) protein; P, pellet, S, supernatant. (Upper Right) NDGD1 binding to liposomes composed of PA and PC. Composition is expressed as percent PA. (Lower) Binding of NDGD1 (2–10 µg) to liposomes consisting of 60% PC/40% PA or 100% PC.
NDGD1 Causes Liposome Fusions in a PA-Dependent Manner.
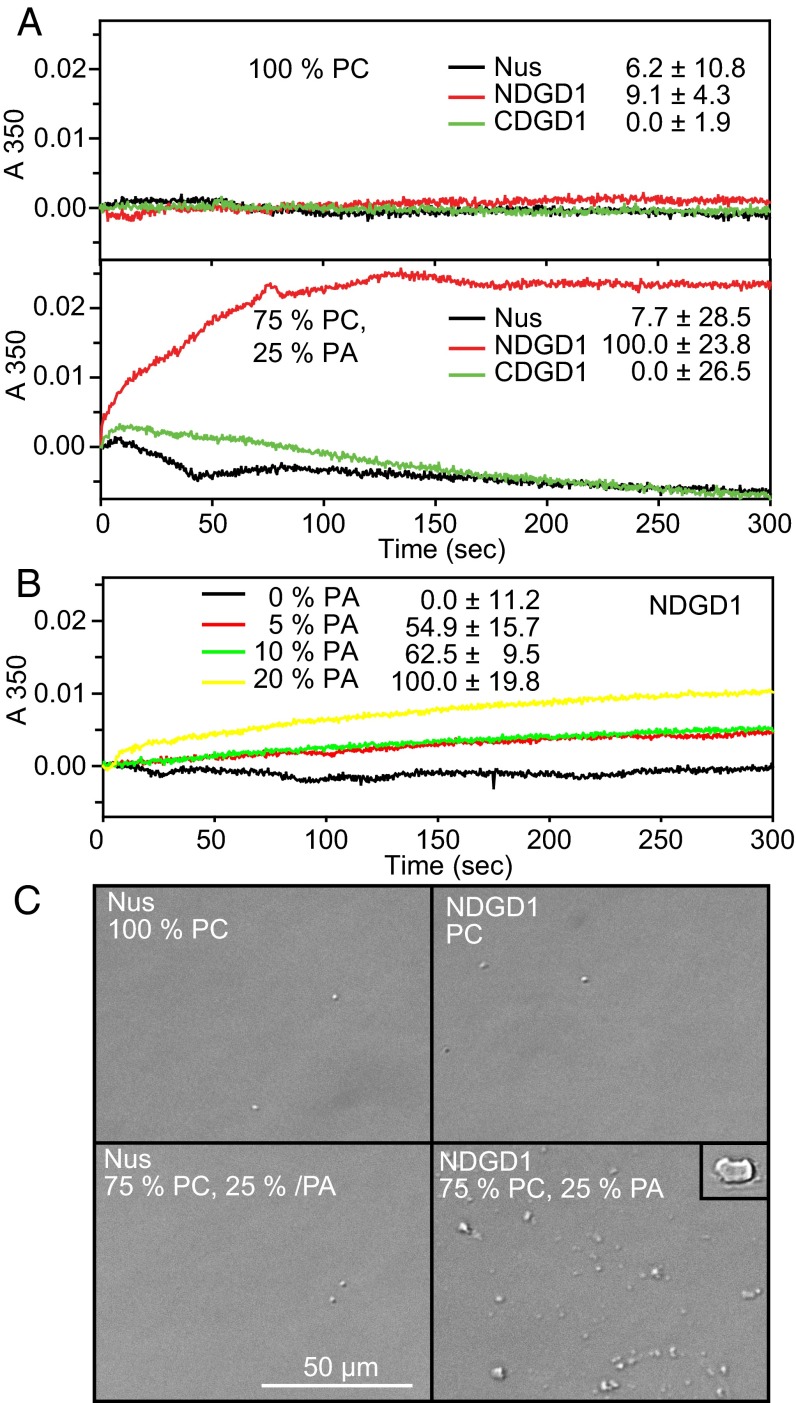
Protein–lipid interactions with unilamellar vesicles can cause membrane fusion or liposome aggregation resulting in giant liposomes or multi-liposome complexes, which can be detected by increased turbidity (31). In contrast to CDGD1, the addition of NDGD1 to unilamellar vesicles composed of 75% PC/25% PA resulted in increased turbidity, (Fig. 3A). No turbidity changes were observed with control protein or with vesicles lacking PA. NDGD1-dependent liposome fusion/aggregation could be observed with vesicles containing as little as 5% PA, a concentration close to that found in chloroplast membranes (32), indicating that this effect also can occur in vivo (Fig. 3B).
Fig. 3.
NDGD1 and PA-dependent liposome fusion. (A) NDGD1-mediated fusion/aggregation of PA-containing unilamellar vesicles. Unilamellar vesicles containing 100% PC or 75% PC/25% PA were incubated with Nus (control), NDGD1, or CDGD1, and the increase in turbidity at 350 nm was measured. (B) Fusion/aggregation of vesicles containing PC and different amounts of PA after the addition of NDGD1. Numbers in A and B indicate changes in absorption after 300 s (mean and SD of three experiments). (C) Vesicles after incubation with Nus (control) or NDGD1 visualized by DIC microscopy. The original vesicles produced by extrusion cannot be observed because of their small diameter (0.1 µm). Incubation of PC/PA vesicles with NDGD1 results in the formation of numerous large (1–5 µm) liposomes, which are barely detectable in the control experiments. (Inset) A very large liposome observed only with PC/PA vesicles incubated with NDGD1. The experiments were repeated three times with fresh liposomes with comparable results.
To visualize liposome fusion or aggregation directly, the vesicles were observed by differential interference contrast (DIC) microscopy after the addition of NDGD1. The unilamellar vesicles used for the experiment have a diameter of ∼0.1 µm and therefore cannot be observed by DIC microscopy (Fig. 3C). The addition of control [N-utilization substance protein A (Nus)] or NDGD1 protein to PC-containing vesicles or of Nus protein to PC/PA vesicles resulted in the occurrence of very few large vesicles (1–5 µm in diameter). However, after the addition of NDGD1 to PC/PA vesicles, numerous large vesicles were observed. In addition, some giant (∼10 µm) vesicles were found, which were absent from the controls. This result demonstrates that NDGD1 causes PA-dependent liposome fusion in vitro.
Overexpression of NDGD1 in Transgenic Arabidopsis Plants Affects Galactolipid Accumulation and Growth.
Because NDGD1 cannot function unless fused with a glycosyltransferase domain but still binds PA and causes liposome fusion in vitro, it was hypothesized that NDGD1 would compete with endogenous DGD1 for PA binding and would affect galactolipid production if overexpressed in vivo. Two independent Arabidopsis NDGD1 overexpression lines (WT-NDGD1#45 and WT-NDGD1#40) were selected by Northern hybridization (SI Appendix, Fig. S7). Immunoblot analysis revealed the presence of two bands at 38 and 36 kDa (NDGD1, M1–E338, and a degradation product/polypeptide derived from an alternative start codon, respectively) in transgenic lines in addition to the 91-kDa DGD1 band (SI Appendix, Fig. S7). The presence of full-length DGD1 mRNA and protein bands with intensities similar to those in WT Arabidopsis indicated that DGD1 expression was not compromised by cosuppression. The NDGD1 plants were bushy and smaller than WT plants. Chlorophyll content was reduced, but photosynthetic quantum yield at different light intensities was not changed, indicating that photosynthesis was not affected. No obvious differences in chloroplast envelope structures were observed by electron microscopy (SI Appendix, Fig. S8). Absolute amounts of galactolipids lipids were reduced by one third, but the acyl composition remained unchanged (SI Appendix, Fig. S7). It is possible that PA binding to NDGD1 affects galactolipid transfer between envelope membranes and MGDG synthesis by MGD1, which is known to require PA for optimal activity (33).
Galactolipid Transfer Between Envelope Membranes.
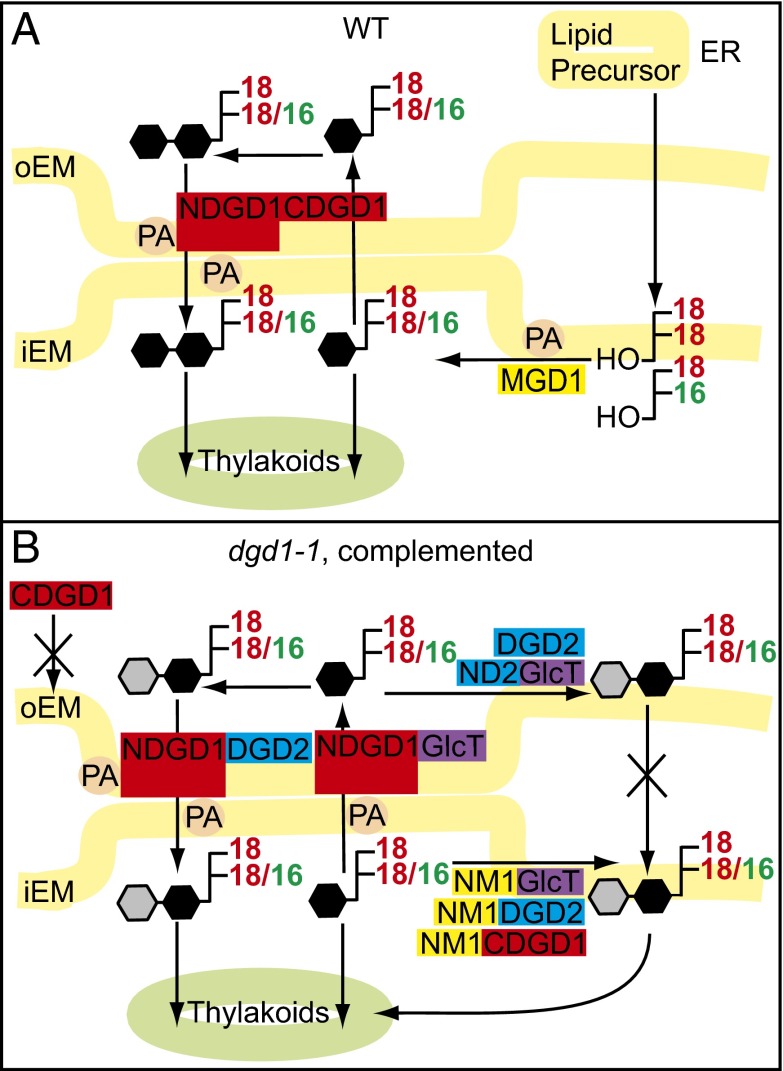
NDGD1 harbors several hydrophobic domains, behaves as an integral oEM protein, and causes PA-dependent liposome fusion (10). It remains unclear whether membrane fusion mediated by NDGD1 encompasses the entire lipid bilayer or is restricted to the outer leaflets, resulting in membrane hemifusions, as suggested for membrane interactions between the oEM and the ER (34). As a nonbilayer-forming, hexagonal phase II (HII) lipid, arrangements of PA can form membrane protrusions and contribute to membrane fusions (35). Therefore, NDGD1 binding to PA might result in the local aggregation of PA, resulting in the local formation of HII phases that cause fusions of neighboring membranes (35). In the proposed model, membrane fusions/associations between the iEM and oEM mediated by PA–NDGD1 interaction (Fig. 4) can enable the transfer of lipid precursors and of galactolipids (MGDG and DGDG) between the two envelopes. The importance of PA as a central lipid metabolite is demonstrated by the use of PA for phospholipid synthesis and by diacylglycerol derived from PA dephosphorylation serving as substrate for galactolipid synthesis. PA binds to TGD2, a subunit of the iEM ABC lipid transporter, and stimulates liposome aggregation/membrane fusion by TGD2 (31). PA is also bound to TGD4 in the oEM, and PA stimulates MGDG synthesis by binding to MGD1 (33). PA binding of proteins involved in galactolipid metabolism in the iEM and oEM might lead to self-organized aggregation (Fig. 4), possibly including other proteins involved in lipid or protein transport through the envelope, including TGD1/TGD2/TGD3, TGD4, and MGD1 (36), resulting in the establishment of protein/membrane microdomains between the iEM and oEM. The evolutionary origin of NDGD1 remains obscure, because it is absent from prokaryotic genomes. DGDG synthases with long N-terminal extensions can be observed first in Chlorophyta and in some Rhodophyta (Porphyridium) (SI Appendix, Fig. S1), and NDGD1 sequences occur in Streptophyta (SI Appendix, Fig. S1) (16–18). Cyanobacteria are surrounded by two envelope membranes, the presumed progenitors for the iEM and the oEM of the chloroplast (37). Although DgdA in cyanobacteria is localized to the inner membrane (38), replacement of DgdA with plant DGD1 in the chloroplasts resulted in the relocation of DGDG synthesis to the oEM. Therefore, NDGD1 might have evolved along with the establishment of the chloroplast endosymbiont to neofunctionalize DGD1, enabling integration of lipid metabolism between the endosymbiont and the host (39). NDGD1 accumulation by itself in cells is detrimental (SI Appendix, Figs. S3 and S7) and hence is strongly selected against. Thus we speculate that the fusion of NDGD1 to the DGDG synthase might represent a crucial event during plant evolution, enabling galactolipid exchange between the iEM and oEM membranes; without this exchange a functionalized and permanent integration of the endosymbiont into the metabolic fabric of the host cell would not have been possible.
Fig. 4.
Galactolipid synthesis in chloroplast envelope membranes. (A) In Arabidopsis, eukaryotic, ER-derived lipid precursors are transported to the chloroplast. Prokaryotic chloroplast-derived and imported eukaryotic diacylglycerols are used for MGDG synthesis by MGD1 in the iEM. In the oEM, MGDG is converted into DGDG by DGD1 (NDGD1CDGD1). NDGD1 binding to PA mediates the association of the iEM and oEM, thereby facilitating the transfer of MGDG from the iEM to the oEM for further galactosylation and of DGDG from the oEM to the iEM. (B) DGDG or GlcGalDG accumulates in dgd1-1 plants complemented with diglycosyllipid synthases (CDGD1, DGD2, or GlcT). Transformation with NM1 fusion constructs (targeting the iEM) relocates diglycosyllipid synthesis to the iEM, resulting in complementation. Transformation with ND2 fusion constructs (targeting the oEM) results in the synthesis of low amounts of DGDG or GlcGalDG without complementation because of the lack of MGDG, DGDG, or GlcGalDG transfer between envelopes. Transformation with DGD2 or GlcT in fusion with NDGD1 results in complementation mediated by PA-dependent aggregations between the iEM and oEM, enabling glycolipid transfer for efficient diglycosyllipid synthesis. Black hexagons indicate galactose; gray hexagons indicate glucose or galactose.
Materials and Methods
Mutant Lines and Growth Conditions.
Arabidopsis plants were grown at 150 µmol⋅m−2⋅s−1 with 16 h light/d. The dgd1-1 mutant was derived from chemical mutagenesis (5, 15), and the transfer DNA (T-DNA) insertion lines dgd1-2 (518_A01.b.1a.Lb3Fa) and dgd1-3 (73_B08.b.1a.Lb3Fa) were from the Syngenta Arabidopsis Insertion Library (SAIL) collection (Syngenta) (40). Resequencing of the flanking regions revealed that the T-DNAs in dgd1-2 and dgd1-3 are inserted into exon 1 after amino acid A189 and into intron 3 N-terminal to D282, respectively.
Northern Blot and Immunoblot.
Total RNA was isolated from leaves, separated by agarose gel electrophoresis, and transferred to nylon membranes. Northern blots were hybridized to an NDGD1 probe, and bands were visualized by autoradiography.
Polyclonal antiserum was raised in rabbits against the synthetic polypeptide V159LEMSRLRRRRNSD172 derived from NDGD1 and was immunopurified (BioGenes). Proteins from Arabidopsis leaves were separated by SDS-PAGE and, after blotting to nitrocellulose, were immunodetected with anti-DGD1 antibodies and alkaline phosphatase-coupled goat anti-rabbit antibodies (Kirkegaard & Perry Laboratories). His-tagged proteins were detected using the HisDetector Nickel-HRP kit (Kirkegaard & Perry Laboratories).
Measurement of Chlorophyll, Lipids, Chlorophyll Fluorescence, and Electron Microscopy.
Chlorophyll was measured photometrically. Chlorophyll fluorescence was recorded using a JUNIOR-PAM pulse amplitude modulation fluorometer (Heinz Walz) after exposure to different light intensities (0, 500, or 1,000 µmol⋅m−2⋅s−1) for 30 min. The quantum yield was calculated according to ref. 41. Transmission electron microscopy of leaf ultrathin sections was performed as described (42).
Lipids were isolated from leaves and separated by TLC (5). Fatty acid methyl esters were transmethylated and quantified by GC using pentadecanoic acid (15:0) as an internal standard (43). Galactolipids and phospholipids were quantified by direct infusion mass spectrometry (44).
Expression of DGDG Synthases in E. coli and Arabidopsis.
The cDNAs from Arabidopsis DGD1, DGD2, and Chloroflexus GlcT (5, 8, 15, 19, 20, 45, 46) were cloned into E. coli expression vectors for lipid measurements and lipid binding or liposome aggregation experiments (31, 47) or into binary vectors (48) for Arabidopsis transformation (SI Appendix, SI Material and Methods). The Arabidopsis dgd1-1 mutant was transformed using the Agrobacterium floral-dip method (49). A minimum of 15 independent transgenic dgd1-1 plants were screened for lipid accumulation by TLC for each experiment, and one line with highest DGDG amount was selected for further analysis.
Phylogenetic Analysis and Structure Prediction.
Amino acid sequences of DGDG synthases were obtained from GenBank, from genome.microbedb.jp/klebsormidium (Klebsormidium), or from the cyanophora database cyanophora.rutgers.edu/porphyridium (Porphyridium). Phylogenetic analyses and alignments were done with MEGA 6 (50) and with the ClustalW algorithm. Unrooted phylogenetic trees were constructed using the neighbor-joining method, and the bootstrap values were derived from 1,000 replicates. The I-TASSER structural analysis (zhanglab.ccmb.med.umich.edu/I-TASSER/) (23) was used with NDGD1 sequences from Arabidopsis and other Streptophyta.
Supplementary Material
Acknowledgments
We thank R. Wendenburg and H. Peisker for help with plant work and lipid analyses. This work was funded in part by Deutsche Forschungsgemeinschaft Grants SFB429 and Do520/10 (to P.D.); the Wenner Gren Foundation (A.A.K.); and US Department of Energy, Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences Grant DE-FG02-98ER20305 (to C.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1609184113/-/DCSupplemental.
References
- 1.Douce R, Joyard J. Plant galactolipids. In: Stumpf PK, editor. The Biochemistry of Plants. Lipids: Structure and Function. Academic; New York: 1980. pp. 321–362. [Google Scholar]
- 2.Liu Z, et al. Crystal structure of spinach major light-harvesting complex at 2.72 Å resolution. Nature. 2004;428(6980):287–292. doi: 10.1038/nature02373. [DOI] [PubMed] [Google Scholar]
- 3.Jordan P, et al. Three-dimensional structure of cyanobacterial photosystem I at 2.5 Å resolution. Nature. 2001;411(6840):909–917. doi: 10.1038/35082000. [DOI] [PubMed] [Google Scholar]
- 4.Umena Y, Kawakami K, Shen JR, Kamiya N. Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9 Å. Nature. 2011;473(7345):55–60. doi: 10.1038/nature09913. [DOI] [PubMed] [Google Scholar]
- 5.Dörmann P, Hoffmann-Benning S, Balbo I, Benning C. Isolation and characterization of an Arabidopsis mutant deficient in the thylakoid lipid digalactosyl diacylglycerol. Plant Cell. 1995;7(11):1801–1810. doi: 10.1105/tpc.7.11.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kobayashi K, Kondo M, Fukuda H, Nishimura M, Ohta H. Galactolipid synthesis in chloroplast inner envelope is essential for proper thylakoid biogenesis, photosynthesis, and embryogenesis. Proc Natl Acad Sci USA. 2007;104(43):17216–17221. doi: 10.1073/pnas.0704680104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Douce R. Site of biosynthesis of galactolipids in spinach chloroplasts. Science. 1974;183(4127):852–853. doi: 10.1126/science.183.4127.852. [DOI] [PubMed] [Google Scholar]
- 8.Miège C, et al. Biochemical and topological properties of type A MGDG synthase, a spinach chloroplast envelope enzyme catalyzing the synthesis of both prokaryotic and eukaryotic MGDG. Eur J Biochem. 1999;265(3):990–1001. doi: 10.1046/j.1432-1327.1999.00801.x. [DOI] [PubMed] [Google Scholar]
- 9.Awai K, et al. Two types of MGDG synthase genes, found widely in both 16:3 and 18:3 plants, differentially mediate galactolipid syntheses in photosynthetic and nonphotosynthetic tissues in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2001;98(19):10960–10965. doi: 10.1073/pnas.181331498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Froehlich JE, Benning C, Dörmann P. The digalactosyldiacylglycerol (DGDG) synthase DGD1 is inserted into the outer envelope membrane of chloroplasts in a manner independent of the general import pathway and does not depend on direct interaction with monogalactosyldiacylglycerol synthase for DGDG biosynthesis. J Biol Chem. 2001;276(34):31806–31812. doi: 10.1074/jbc.M104652200. [DOI] [PubMed] [Google Scholar]
- 11.Kelly AA, Froehlich JE, Dörmann P. Disruption of the two digalactosyldiacylglycerol synthase genes DGD1 and DGD2 in Arabidopsis reveals the existence of an additional enzyme of galactolipid synthesis. Plant Cell. 2003;15(11):2694–2706. doi: 10.1105/tpc.016675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Härtel H, Dörmann P, Benning C. DGD1-independent biosynthesis of extraplastidic galactolipids after phosphate deprivation in Arabidopsis. Proc Natl Acad Sci USA. 2000;97(19):10649–10654. doi: 10.1073/pnas.180320497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hurlock AK, Roston RL, Wang K, Benning C. Lipid trafficking in plant cells. Traffic. 2014;15(9):915–932. doi: 10.1111/tra.12187. [DOI] [PubMed] [Google Scholar]
- 14.Browse J, Warwick N, Somerville CR, Slack CR. Fluxes through the prokaryotic and eukaryotic pathways of lipid synthesis in the ‘16:3’ plant Arabidopsis thaliana. Biochem J. 1986;235(1):25–31. doi: 10.1042/bj2350025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dörmann P, Balbo I, Benning C. Arabidopsis galactolipid biosynthesis and lipid trafficking mediated by DGD1. Science. 1999;284(5423):2181–2184. doi: 10.1126/science.284.5423.2181. [DOI] [PubMed] [Google Scholar]
- 16.Petroutsos D, et al. Evolution of galactoglycerolipid biosynthetic pathways--from cyanobacteria to primary plastids and from primary to secondary plastids. Prog Lipid Res. 2014;54:68–85. doi: 10.1016/j.plipres.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 17.Awai K, Watanabe H, Benning C, Nishida I. Digalactosyldiacylglycerol is required for better photosynthetic growth of Synechocystis sp. PCC6803 under phosphate limitation. Plant Cell Physiol. 2007;48(11):1517–1523. doi: 10.1093/pcp/pcm134. [DOI] [PubMed] [Google Scholar]
- 18.Sakurai I, Mizusawa N, Wada H, Sato N. Digalactosyldiacylglycerol is required for stabilization of the oxygen-evolving complex in photosystem II. Plant Physiol. 2007;145(4):1361–1370. doi: 10.1104/pp.107.106781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hölzl G, et al. Functional differences between galactolipids and glucolipids revealed in photosynthesis of higher plants. Proc Natl Acad Sci USA. 2006;103(19):7512–7517. doi: 10.1073/pnas.0600525103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Härtel H, Dörmann P, Benning C. Galactolipids not associated with the photosynthetic apparatus in phosphate-deprived plants. J Photochem Photobiol B. 2001;61(1-2):46–51. doi: 10.1016/s1011-1344(01)00144-0. [DOI] [PubMed] [Google Scholar]
- 21.Block MA, Dorne AJ, Joyard J, Douce R. Preparation and characterization of membrane fractions enriched in outer and inner envelope membranes from spinach chloroplasts. II. Biochemical characterization. J Biol Chem. 1983;258(21):13281–13286. [PubMed] [Google Scholar]
- 22.Henrissat B, Coutinho PM, Davies GJ. A census of carbohydrate-active enzymes in the genome of Arabidopsis thaliana. Plant Mol Biol. 2001;47(1-2):55–72. [PubMed] [Google Scholar]
- 23.Yang J, et al. The I-TASSER Suite: Protein structure and function prediction. Nat Methods. 2015;12(1):7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsai JY, et al. Structural characterizations of the chloroplast translocon protein Tic110. Plant J. 2013;75(5):847–857. doi: 10.1111/tpj.12249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Solomonson M, et al. Structure of EspB from the ESX-1 type VII secretion system and insights into its export mechanism. Structure. 2015;23(3):571–583. doi: 10.1016/j.str.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 26.Yao D, Cherney M, Cygler M. Structure of the N-terminal domain of the effector protein LegC3 from Legionella pneumophila. Acta Crystallogr D Biol Crystallogr. 2014;70(Pt 2):436–441. doi: 10.1107/S139900471302991X. [DOI] [PubMed] [Google Scholar]
- 27.Bissig C, Gruenberg J. ALIX and the multivesicular endosome: ALIX in Wonderland. Trends Cell Biol. 2014;24(1):19–25. doi: 10.1016/j.tcb.2013.10.009. [DOI] [PubMed] [Google Scholar]
- 28.Matsuo H, et al. Role of LBPA and Alix in multivesicular liposome formation and endosome organization. Science. 2004;303(5657):531–534. doi: 10.1126/science.1092425. [DOI] [PubMed] [Google Scholar]
- 29.Zhou Q, et al. Architecture of the synaptotagmin-SNARE machinery for neuronal exocytosis. Nature. 2015;525(7567):62–67. doi: 10.1038/nature14975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aguilar L, et al. Phospholipid membranes form specific nonbilayer molecular arrangements that are antigenic. J Biol Chem. 1999;274(36):25193–25196. doi: 10.1074/jbc.274.36.25193. [DOI] [PubMed] [Google Scholar]
- 31.Roston R, Gao J, Xu C, Benning C. Arabidopsis chloroplast lipid transport protein TGD2 disrupts membranes and is part of a large complex. Plant J. 2011;66(5):759–769. doi: 10.1111/j.1365-313X.2011.04536.x. [DOI] [PubMed] [Google Scholar]
- 32.Uemura M, Steponkus PL. Effect of cold acclimation on the lipid composition of the inner and outer membrane of the chloroplast envelope isolated from rye leaves. Plant Physiol. 1997;114(4):1493–1500. doi: 10.1104/pp.114.4.1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dubots E, et al. Activation of the chloroplast monogalactosyldiacylglycerol synthase MGD1 by phosphatidic acid and phosphatidylglycerol. J Biol Chem. 2010;285(9):6003–6011. doi: 10.1074/jbc.M109.071928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mehrshahi P, et al. Transorganellar complementation redefines the biochemical continuity of endoplasmic reticulum and chloroplasts. Proc Natl Acad Sci USA. 2013;110(29):12126–12131. doi: 10.1073/pnas.1306331110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wong-Baeza C, et al. Molecular organization of the non-bilayer phospholipid arrangements that induce an autoimmune disease resembling human lupus in mice. Mol Membr Biol. 2012;29(2):52–67. doi: 10.3109/09687688.2012.667577. [DOI] [PubMed] [Google Scholar]
- 36.Roston RL, Gao J, Murcha MW, Whelan J, Benning C. TGD1, -2, and -3 proteins involved in lipid trafficking form ATP-binding cassette (ABC) transporter with multiple substrate-binding proteins. J Biol Chem. 2012;287(25):21406–21415. doi: 10.1074/jbc.M112.370213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Inoue K. Emerging roles of the chloroplast outer envelope membrane. Trends Plant Sci. 2011;16(10):550–557. doi: 10.1016/j.tplants.2011.06.005. [DOI] [PubMed] [Google Scholar]
- 38.Selão TT, Zhang L, Ariöz C, Wieslander Å, Norling B. Subcellular localization of monoglucosyldiacylglycerol synthase in Synechocystis sp. PCC6803 and its unique regulation by lipid environment. PLoS One. 2014;9(2):e88153. doi: 10.1371/journal.pone.0088153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chothia C, Gough J, Vogel C, Teichmann SA. Evolution of the protein repertoire. Science. 2003;300(5626):1701–1703. doi: 10.1126/science.1085371. [DOI] [PubMed] [Google Scholar]
- 40.Sessions A, et al. A high-throughput Arabidopsis reverse genetics system. Plant Cell. 2002;14(12):2985–2994. doi: 10.1105/tpc.004630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schreiber U, Schliwa U, Bilger W. Continuous recording of photochemical and non-photochemical chlorophyll fluorescence quenching with a new type of modulation fluorometer. Photosynth Res. 1986;10(1-2):51–62. doi: 10.1007/BF00024185. [DOI] [PubMed] [Google Scholar]
- 42.Hölzl G, et al. The role of diglycosyl lipids in photosynthesis and membrane lipid homeostasis in Arabidopsis. Plant Physiol. 2009;150(3):1147–1159. doi: 10.1104/pp.109.139758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Browse J, McCourt PJ, Somerville CR. Fatty acid composition of leaf lipids determined after combined digestion and fatty acid methyl ester formation from fresh tissue. Anal Biochem. 1986;152(1):141–145. doi: 10.1016/0003-2697(86)90132-6. [DOI] [PubMed] [Google Scholar]
- 44.Gasulla F, et al. The role of lipid metabolism in the acquisition of desiccation tolerance in Craterostigma plantagineum: A comparative approach. Plant J. 2013;75(5):726–741. doi: 10.1111/tpj.12241. [DOI] [PubMed] [Google Scholar]
- 45.Kelly AA, Dörmann P. DGD2, an Arabidopsis gene encoding a UDP-galactose-dependent digalactosyldiacylglycerol synthase is expressed during growth under phosphate-limiting conditions. J Biol Chem. 2002;277(2):1166–1173. doi: 10.1074/jbc.M110066200. [DOI] [PubMed] [Google Scholar]
- 46.Shimojima M, et al. Cloning of the gene for monogalactosyldiacylglycerol synthase and its evolutionary origin. Proc Natl Acad Sci USA. 1997;94(1):333–337. doi: 10.1073/pnas.94.1.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lu B, Benning C. A 25-amino acid sequence of the Arabidopsis TGD2 protein is sufficient for specific binding of phosphatidic acid. J Biol Chem. 2009;284(26):17420–17427. doi: 10.1074/jbc.M109.016014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Höfgen R, Willmitzer L. Biochemical and genetic analysis of different patatin isoforms expressed in various organs of potato (Solanum tuberosum) Plant Sci. 1990;66(2):221–230. [Google Scholar]
- 49.Clough SJ, Bent AF. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16(6):735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 50.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.