Abstract
Spermatogonial stem cells (SSCs) propagate mammalian spermatogenesis throughout male reproductive life by continuously self-renewing and differentiating, ultimately, into sperm. SSCs can be cultured for long periods and restore spermatogenesis upon transplantation back into the native microenvironment in vivo. Conventionally, SSC research has been focused mainly on male infertility and, to a lesser extent, on cell reprogramming. With the advent of genome-wide sequencing technology, however, human studies have uncovered a wide range of pathogenic alleles that arise in the male germ line. A subset of de novo point mutations was shown to originate in SSCs and cause congenital disorders in children. This review describes both monogenic diseases (eg, Apert syndrome) and complex disorders that are either known or suspected to be driven by mutations in SSCs. We propose that SSC culture is a suitable model for studying the origin and mechanisms of these diseases. Lastly, we discuss strategies for future clinical implementation of SSC-based technology, from detecting mutation burden by sperm screening to gene correction in vitro.
Introduction
Stem cells play a pivotal role in maintaining homeostasis in adult tissues; they self-renew, differentiate, and regenerate tissues upon injury. Unlike other stem cells, spermatogonial stem cells (SSCs) have the unique job of propagating genetic information to subsequent generations. In the following sections, we focus on the properties and dynamics of mammalian SSCs in vivo and in vitro, with special emphasis on implications for human diseases caused by germ line mutations. We conclude by considering strategies for future clinical implementation of SSC-based technology.
In Vivo Biology of Mammalian SSCs
SSCs reside along the basement membrane of the seminiferous tubules in adults and are unipotent (ie, lineage restricted). Developmentally, these cells originate from the epiblast and undergo fate specification as primordial germ cells (PGCs) [1]. PGCs enter the nascent endoderm, spread along the hindgut epithelium, and begin to migrate toward the genital ridge, the future testis [2,3].
After colonization of the gonad, PGCs give rise to prospermatogonia, a change that occurs in the fetal/early neonatal period in rodents. During this phase, prospermatogonia become mitotically quiescent until proliferation eventually resumes within the first week after birth [4,5]. Prospermatogonia migrate to the basement membrane and the spermatogenesis phase begins [6]. The differentiation of PGCs into prospermatogonia in humans is not well understood, but there seem to be strong parallels between the two species [7]. Postnatally, prospermatogonia mature into SSCs.
Various theoretical models have been developed to explain how SSCs concomitantly maintain the stem cell population through self-renewal while providing sufficient numbers of differentiating progeny. In rodents, the pool of undifferentiated spermatogonia (Spg), which includes SSCs, comprises As (single), Apr (pair), and Aal (aligned) Spg. The definitions of A Spg subtypes were made according to their morphological arrangement on the basement membrane. The first model to describe stem cell renewal dates back to 1971 and is referred to as the As or static model [8]. It was proposed that As Spg comprise the only pool of true stem cells, whereas the balance of A Spg are irreversibly committed to differentiation.
An alternative model was presented recently by Yoshida's group, who demonstrated that these As, Apr, and Aal cells are extremely interchangeable with significant cell fate plasticity [9–11]. The authors developed a pulse-labeling system, in which a pulse of tamoxifen was given to mice to activate a Cre recombinase (creERT2) under control of the promoter either for Ngn3 (a marker of differentiating Spg [12]) or for Gfrα1 (a marker of undifferentiated Spg [9]). Upon labeling, the fates of the Spg were investigated in normal and challenged conditions. Moreover, using live-imaging analysis, the authors observed GFRα1+ cells that were actively and constantly moving along the basal compartment, revealing potential spatial heterogeneity within the SSC niche [9].
Compared with mice, primates show fewer spermatogonial subtypes, which can be divided into three distinct groups: Adark (Ad), Apale (Ap) (originally termed A1 and A2), and type B Spg [13,14]. Ad Spg, characterized by highly condensed chromatin, are thought to represent the reserve stem cells, whereas Ap Spg, which exhibit a more sparse chromatin distribution, may represent the active stem cells, even though definitive evidence is still lacking. Ad Spg, which are normally quiescent, became proliferative after irradiation but whether Ad Spg also contribute to steady-state spermatogenesis still remains to be addressed [15].
Some authors also proposed the existence of another type of A Spg, termed A transition Spg that display an intermediate phenotype between Ad and Ap [16–18]. Strikingly, this Ad/Ap model was proposed more than 50 years ago by Clermont, and, at present, very limited information is available about the kinetics of human Spg [19]. Nevertheless, substantial data exist for nonhuman primates, in which spermatogenesis has been extensively studied [14,17,20–23].
Derivation of SSC Cultures in Mice and Humans
Studies on SSCs have been hampered by the absence of specific markers, as well as the relative rarity of authentic stem cells present in the adult testis [11,24]. Yet, progress has been made in understanding the optimal culture conditions necessary to maintain SSCs in long-term culture, at least in rodents [25,26]. As with most other adult stem cells, SSCs require a specific niche to preserve their unique features.
In vivo, Sertoli and peritubular cells are the most prominent components of the niche, secreting factors that support SSC self-renewal and mitotic expansion [27,28], among which the most fundamental is glial cell line-derived neurotrophic factor (GDNF). The level of GDNF is critical for SSC homeostasis. Mouse Gdnf knock-out models exhibited impaired fertility [27,28], whereas Gdnf overexpression in the testis caused an initial abnormal expansion of undifferentiated Spg that resulted in sterility and tumor development [27]. As discussed hereunder, extensive efforts over the last 10 years have yielded a better understanding of how growth factors influence SSC renewal and differentiation.
A variety of strategies have enabled derivation of murine SSC lines with similar efficiency from both neonatal and adult mice [25,29]. Successful long-term culture conditions for SSCs were first established by the Shinohara group in 2003 [25]. The addition of GDNF, leukemia inhibitor factor, epithelial growth factor, and fibroblast growth factor-2 (FGF2) permitted maintenance of SSCs in vitro for up to five months. The colonizing potential of cultured SSCs was proven by injection into busulfan-treated mice and is discussed further hereunder. Another group reported the maintenance of SSCs in vitro for up to six months in serum-free conditions [26]. The outcome of SSC line derivation was also shown to be strain dependent [25,26].
In addition to SSC cultures from neonatal mice, long-term SSC cultures were derived from adult testis using modified culture conditions [29]. The studies already described have employed a variety of feeder cell types for maintenance of SSCs, including murine embryonic fibroblasts, testicular somatic cells (designated JK1), and SIM mouse embryo-derived thioguanine and ouabain-resistant (STO) cells [30]. Thus, knowledge of factors supporting SSC growth has gradually enabled development of robust in vitro models.
The story of human SSC (hSSC) culture is completely different. As already described, innumerable differences exist between humans and rodents. As such, the dearth of knowledge in identifying hSSCs and their niche has obstructed progress in the field. Current obstacles include the difficulties in accessing healthy human tissue samples, as well as isolation and expansion of highly purified hSSCs in culture. Of note, somatic cell overgrowth has been encountered as a major challenge to assessing hSSCs in culture [31].
Various markers that were previously studied in other organisms have been reported to be useful in enriching hSSCs, although most are controversial. One of the first reported markers was THY1 (or CD90) [32,33]. An obvious problem for employing this antigen is its robust expression by the mesenchymal stem cells [34], and later studies suggested that SSCs, in fact, do not express CD90 [35]. Another marker widely employed is CD49f [32,36]. As with CD90, CD49f is expressed by contaminating mesenchymal cells, as well as by testicular somatic cells, limiting its utility [37]. As a consequence of this overlap between putative hSSC markers and mesenchymal markers, most of the earlier reports that claimed the successful isolation of SSCs have interpretive constraints [38,39].
Our group studied whether several novel markers identified previously in mouse models could be useful for hSSCs [39,40]. For example, ID4 expression was recently found to mark a small population of bona fide SSCs in mice, making it an attractive candidate marker for other species [41]. GPR125 is an orphan adhesion-type G-protein-coupled receptor expressed by undifferentiated Spg and it, too, was first described in mice [42]. We employed a validated, human-specific anti-ID4 monoclonal antibody and found that ID4+ cells coexpress GPR125 and reside on the basal membrane of the seminiferous tubules [39,40]. Although He et al. succeeded in isolating a pure GPR125+ population from healthy donor testis, they were unable to maintain the SSCs in culture for long [39,40].
In 2009, the successful achievement of long-term culture of hSSCs was reported [38]. The study described the colonization of testicular tubules of nu/nu male mice by the hSSCs maintained in vitro. However, hSSC xenotransplantation has significant interpretative limitations (see “SSC Transplantation” hereunder), and, in addition, certain important data were not provided in the study. For example, although they showed a large relative increase in stem cell activity over time, the absolute number of stem cells expanded and the ability to freeze–thaw the long-term SSCs was not addressed. Finally, the genetic stability of hSSCs in culture remains unknown.
Improved colonization has been achieved by sorting testicular cells using SSEA-4 before injection into the testis, although the analysis was performed only one month post-transplantation [32]. The same year, another group reported the successful isolation and maintenance of hSSCs in culture using an exclusion approach, by plating the cells on collagen and further sorting the unattached cells for CD9 [36]. The authors demonstrated the differentiation of the isolated SSCs into spermatids, albeit at low efficiency.
Substantial further efforts have been made by several groups recently to characterize and enrich the hSSC population by using various combinations of markers [31,43–46]. However, the unwanted expansion of testicular somatic cells that are propagated in culture, irrespective of the method of derivation employed, remains an impediment. In conclusion, hSSC culture technology is not nearly as mature as that for mice and other species.
SSC Transplantation
As already discussed, experimental investigation of SSCs is complicated by their paucity in the testis and the inability to efficiently separate them from other unwanted cell types. Nevertheless, good tools are available to quantify the success of SSC isolation by observing their biological ability to produce and maintain spermatogenesis in a transplant model, first developed in 1994 by Brinster and Avarbock [47]. They showed how cells isolated from a donor testis were able to restore spermatogenesis once directly injected into the seminiferous tubules of infertile mice. The transplanted cells could produce de novo spermatogenesis regardless of the nature of the infertility, whether genetic [48] or experimentally induced by busulfan.
Today, this transplantation technique is routinely performed in both mice and rats [49] as a readout for the efficiency of SSC derivation. Given that only a true stem cell can give rise to a clone that will subsequently yield spermatogenic cells [50,51], the transplantation protocol represents a useful functional assay and constitutes the best way to identify SSCs. As an extension of the seminal Brinster study, which used freshly isolated testis cell suspensions, many studies currently employ this method to verify the presence of stem cells in culture [25,26]. As discussed later, the transplantation model has been successfully used recently to obtain experimental support for the paradigm of paternal age effect (PAE)-associated diseases, such as Apert syndrome [52].
In 2002, a similar transplantation strategy was developed for the study of hSSCs [53]. In this xenotransplant system, the authors employed busulfan-treated nu/nu mice as recipients for transplantation of donor hSSCs. Intriguingly, they were able to detect the presence of single hSSCs on the basement membrane of seminiferous tubules up to six months post-transplantation. Thus, xenografted human Spg colonize and undergo limited proliferation but do not differentiate in the mouse testis and gradually disappear. This indicates that although the human and mouse niches are somewhat compatible, the mouse testis does not provide a sufficient microenvironment to support human spermatogenesis. As such, the rigorous assays employed for rodent SSC activity remain unavailable for hSSCs.
Fertility Preservation
Preservation of fertility has historically been the most straightforward potential clinical application of SSCs. This issue is particularly urgent in the case of young boys, often with childhood leukemia, who undergo gonadotoxic chemotherapy as part of a curative regimen. The survival rate of such individuals has greatly improved in recent years [54]. This has resulted in an increased number of long-term survivors who may ultimately want to have families, raising the question of how to proceed if infertility occurs in the absence of banked sperm [55,56]. Theoretically, a way to overcome this problem is to cryopreserve testicular tissue and/or SSCs before chemotherapy and to transplant them back into the testis once the patient has been cured from cancer.
As already discussed, a similar strategy is routinely performed in animal models but has not yet been formally tested in humans. One obstacle to advancing such a strategy is the inability to efficiently expand hSSCs in culture, as the number of SSCs recovered from a small tissue sample is not typically enough to restore spermatogenesis.
Although success in propagating SSCs in vitro may be a prerequisite for autologous transplant, there are other concerns that need to be considered. First, the genetic and epigenetic stability of SSCs during in vitro expansion needs to be determined. It has been demonstrated that murine SSCs are genetically stable even after long-term expansion in culture, suggesting that they may possess a mechanism that prevents them from accumulating deleterious mutations [57].
At the same time, SSCs, when in culture, are not immune to telomere shortening and may not be considered immortal. Some data indicate that this genetic stability is shared by hSSCs, providing support for the possibility of employing SSCs to restore male fertility [58]. To this end, Radford et al. undertook a clinical trial in which testicular cells from Hodgkin lymphoma patients were cryopreserved and autotransplanted back to the testis after completion of chemotherapy, but the results of follow-up analyses remain to be published [59]. Finally, removal of contaminating cancer cells also needs to be taken into account.
Use of SSCs for the Study of Pluripotency
The ability of antenatal germ cells to acquire pluripotency in vitro was discovered many years ago (see Matsui et al. [60]), yet it was only in 2004 that a landmark article demonstrated that postnatal SSCs, too, retain such characteristics [61]. We subsequently showed that long-term wild-type adult SSC lines in vitro preserve the latent plasticity that enables reprogramming, which was ultimately confirmed by others [42,62,63]. These features of SSCs raised the possibility of exploiting SSCs for the study of cellular reprogramming and potentially for therapeutic use. Thus, a number of studies have produced differentiated cell types from pluripotent SSC-derived cell lines [42,64,65]. However, paternal methylation imprints persist in SSCs and their derivatives, which could ultimately affect their functionality and safety in the context of therapeutic use [61].
In conjunction with availability of alternative sources of pluripotent stem cells (ie, induced pluripotent stem cells), the enthusiasm for pursuing derivatives of SSCs in this realm is currently limited. Nonetheless, significant effort has been exerted toward developing pluripotent lines from hSSCs, but long-term, self-renewing hSSC cultures remain elusive. An initial high-profile report of pluripotent stem cells derived from human Spg was ultimately retracted [33]. A number of groups have since provided evidence that postnatal human male germ cells, like their murine counterparts, have developmental plasticity, but most studies demonstrated neither robust evidence of pluripotency nor clonal origins for the precursor to the reprogrammed cells [66–68]. As such, the most important current application of reprogrammed cells originating from SSCs is confined to the domain of basic research studies.
SSCs and PAEs
The advent of high throughput DNA sequencing has recently enabled the direct interrogation of a unique category of human genetic variation, comprising de novo point mutations (DNMs), defined here as single nucleotide variants that are present in offspring but absent in constitutional (ie, somatic cell) genomes of the parents and vanishingly rare in the general human population [69]. For the purposes of the subsequent sections, we focus on DNMs and ignore other important sources of genetic variation (eg, copy number variants, insertion/deletions, and other structural aberrations) and common variants (eg, single nucleotide polymorphisms), due to the significance of DNMs in known disorders related to paternal age.
Consistent with historical predictions, recent sequencing studies of healthy individuals revealed that humans are born with dozens of DNMs, and most of the variation in this number is attributable to paternal age at conception [69,70]. On average, two to three additional DNMs are apparent in children with each additional year of paternal age at conception [69,71]. Furthermore, the location of DNMs is significantly influenced by a variety of factors, including sequence context (eg, CpG site) and regional timing of replication [69,70,72]. Notably, mutation rates are higher in functional regions of the genome, such as exons [70,72]. Finally, genic regions (as opposed to intergenic regions) are disproportionately affected by paternal age, suggesting that such age-related mutations have greater potential to manifest phenotypically [70].
As a tractable alternative to studying humans, researchers have assessed the mutation frequency of germ line cells in an experimental mouse model [73,74]. This transgenic system allowed them to directly measure the mutation frequency of an insert (ie, a lacI mutation reporter), which revealed lower mutation frequencies of germ line cells than somatic cells [74,75]. These studies also revealed that spermatogenic cells from old mice have increased mutation frequencies and an increased prevalence of transversion mutations than those from young or middle-aged mice [74,76]. Together with the human data, these findings suggest that an age-related increase in point mutations is a general feature of male gametogenesis in mammals.
Recent studies have found that a distinct subset of human genetic diseases originates from DNMs in the male germ line. Historically, these disorders, including Apert syndrome (FGFR2 mutations), achondroplasia (FGFR3), Noonan syndrome (mutations in PTPN11 and other genes), and Costello syndrome (mutations in HRAS), have been observed frequently in children from older fathers irrespective of mothers’ age [77–79]. In fact, one of the earliest clues to the existence of such disorders was the observation by Weinberg in the early 20th century of a birth order effect on the incidence of achondroplasia [80]. The mutations in these disorders are paternally derived, yet the fathers do not harbor the mutations in their somatic tissues, indicating the occurrence of de novo male germ line mutations [81–83]. These disorders are now commonly referred to as PAE disorders (see Goriely and Wilkie [84] for a detailed review).
The genes mutated in PAE disorders often belong to the receptor tyrosine kinase/RAS/MAPK signaling pathway and almost always comprise gain-of-function mutations. For example, in Apert syndrome, the mutation of FGFR2 at Ser252Trp manifests increased affinity of the mutant receptor for FGF ligand, which is likely to lead to activation of signaling even when availability of ligand is reduced [85]. Of note, PAE mutations generally exhibit milder gains-of-function than somatic mutations in cancer cells. Specific PAE mutations have been identified in sperm donors and increase with advanced donor age [86–89]. The increase in mutations is exponential, which is significant, because such an amplification could not be driven simply by the linear increase expected in the cumulative number of cell divisions in the normal aging testis.
Furthermore, testes obtained from normal older males have more mutations in PAE genes, and the cells bearing mutations are arranged in a spatially clustered manner [90–93]. The clustering phenomenon is reminiscent of tumor expansion in somatic tissues and cannot be explained in statistical models by a local increase in hot-spot mutations. Thus, the simplest explanation is positive selection of PAE mutations in SSCs. That is, a PAE mutation may arise in a single SSC that acquires a selective growth advantage as compared with its wild-type neighbors (Fig. 1). Goriely and Wilkie termed this phenomenon “selfish spermatogonial selection” [84].
FIG. 1.
Schematic representation of the paternal age effect hypothesis. De novo point mutations (DNMs) may occur in isolated spermatogonial stem cells (SSCs, red dots on the second testis) conferring an advantage to the mutant cells over the wild type cells. During the course of the years, the mutant cells subsequently take over the wild type cells leading to a clonal expansion and colonization of the testis tubules (red fragment in the third and fourth testes), increasing the possibility to pass those mutations to the offspring (red spermatozoa).
Although several attempts have been made to prove the mechanism of this phenomenon using computer modeling, selfish spermatogonial selection, in the absence of prospective, experimental data, is still largely hypothetical. One caveat of such statistical models is that many assumptions are required, including, for example, the mitotic rate of hSSCs, which is unknown and has only been inferred through a single, ethically dubious study in which a handful of prisoners were first administered H3-thymidine and then subjected to testicular biopsies [94].
De Novo Mutations and Complex Disorders
As discussed in the previous paragraphs, DNMs arise in the germ cells of older fathers, giving rise to various defects. In contrast to monogenic disorders, complex developmental disorders are genetically quite heterogeneous. Thus, it is typically more difficult to establish a direct association between specific DNMs and the disease phenotype. Also, unlike the monogenic disorders already described, the DNMs described to date in complex disorders tend to result in loss of function of the corresponding genes. Several studies detailed hereunder have highlighted a major contribution of DNMs in developmentally important genes. These mutations are believed to originate in parental germ cells before fertilization, and they tend to increase with the father's age [95].
Compared with whole-genome sequencing, which confers a greater sensitivity in detecting pathogenic mutations, whole-exome sequencing (WES) is more efficient in identifying novel disease genes [96]. One limitation of both approaches is the relatively small number of families typically analyzed and the lack of a systematic analysis of the male germ lines, to determine the extent of accumulating mutations [71].
Neuropsychiatric Disorders, Congenital Heart Disease, and Cancer
The autism spectrum disorders (ASDs) comprise an array of developmental syndromes defined by a variety of symptoms and degrees of disability. Although a strong genetic component is well recognized, the underlying causes are still largely unknown, despite rapid progress. Recently, various independent WES studies demonstrated how disruptive (ie, nonsense, splice site, and frame shift) DNMs are highly recurrent in ASDs [91,97–99].
In particular, two of these studies carried out exome analyses not on family trios but rather on “quads” (two healthy parents, an affected child, and an unaffected sibling) [91,98]. Strikingly, the studies reached a common conclusion that the affected children exhibit an excess of potentially pathogenic mutations. These occurred in genes commonly expressed in the brain [91], were associated with the fragile X protein, FMRP [98], or involved β-catenin/chromatin remodeling protein networks, often associated with neuronal development [99]. Furthermore, both Neale et al. and O'Roak et al. observed a positive correlation between CHD8 mutations and ASDs, suggesting the CHD8 mutant phenotype as a novel syndromic form of autism [97,99]. Remarkably, a strong bias toward paternal rather than maternal origin for the culprit DNMs was also noted [98,99].
Similarly, a different study, using the same general strategy (ie, trio WES), showed a significant excess of DNMs in congenital heart disease with rare variants in genes expressed in the developing heart [100]. In particular, they found that DNMs in congenital heart disease tend to be associated with the methylation of lysine 4 on histone 3 (H3K4) [100]. In the same vein, earlier studies had indicated a strong effect of paternal age on the incidence of schizophrenia, another common genetically complex and heterogeneous disorder [101]. However, a very recent study employed five different models and concluded that, although age-related DNMs may account for 10%–20% of the increased risk, DNMs do not fully explain the predisposition to schizophrenia in the offspring of older fathers [102].
It has also been proposed that DNMs in tumor-associated genes may have a role in cancer risk in children. Proving this assumption is again complicated by the heterogeneous, multistep nature of the disease. There are a number of studies that describe an association between early onset cancer and DNMs in father's gametes [103]. Recently a comprehensive study analyzed numerous genes known to be recurrently mutated in cancer, looking for evidence of germ line mosaicism [104]. Interestingly, they found that 8.5% of the patients analyzed harbored germ line mutations in genes related to cancer predisposition. Unfortunately, they did not assess the presence of these mutations in the parents of the affected children, which precludes addressing the origin of DNMs. Thus, further efforts are required to unravel a possible link between DNMs and cancer.
Development of Systems to Study SSC-Derived Mutations
Although de novo paternally derived mutations are likely to occur in many species, the disease phenotypes attributable to such mutations, thus far, appear to be a uniquely human phenomenon. As such, it would be ideal to employ hSSCs as a model system for studying paternal age-associated mutations. Unfortunately, the relatively primitive technology available for functional studies of hSSCs obviates substantial mechanistic studies. For example, if clonally derived, cryopreserved hSSC lines were readily available to investigators, significant in vitro studies could be combined with xenotransplantation assays to address the significance of key genes and pathways. However, as already noted, it would be important to first determine the genetic stability of hSSCs in culture.
Although such studies are not yet possible, important insights have come from static observations of archived human testis samples. The identification and marking of apparently clonal populations of mutant germ cells in the human testis were pioneered by the Wilkie and Goriely groups. In a series of unique studies, these investigators first predicted that certain immunohistochemical features would be unique to mutant clones present in the adult testis. Based on their earlier studies of spermatocytic seminoma [105], the investigators found two populations of tubules in the normal testis with differential levels of anti-FGFR3 staining: one with both normal signal intensity and morphology and a second with stronger FGFR3 expression and intense coexpression of MAGEA4 and phospho-AKT [106]. The latter (termed “immunopositive tubules”) also frequently exhibited an unusual double layer of Spg.
In a recent subsequent study, the authors used laser capture microdissection, exon capture, and high-throughput sequencing to show that immunopositive tubules, as defined previously, were strongly enriched for mutations in growth factor signaling-related genes that could be implicated in PAE disorders [107]. Critically, adjacent nonimmunopositive regions selected as negative controls from the same testis did not have any of the tested mutations, indicating that their detection system was highly specific. Although the sample size was relatively small, it was somewhat unexpected that they detected only one of the common Apert syndrome-associated substitutions in FGFR2 (P253R) but not the other (S252W), nor any mutations in RET, another well-studied gene implicated in PAE.
Also of note, the individual immunopositive regions were surprisingly large (ie, up to millimeters in length of the tubule) but were relatively deficient in spermatogenesis compared with adjacent areas. These findings provide evidence for discrete clusters of mutant, clonal premeiotic germ cells, bearing DNMs in signaling-related genes, on a background of otherwise normal human germ cells, but suggest that many such putative clones do not actually contribute to disease in offspring.
In contrast to humans, animal models afford the opportunity to address SSC competition in a prospective, experimental manner. In fact, two historical studies incidentally revealed evidence of adult SSC selection. The first direct evidence for germ line selection in mammals may have come from Mintz. In the course of analyzing chimeric animals comprising cells from two different mouse strains, she cleverly noted a progressive shift in the proportion of offspring of each strain that appeared in subsequent litters from chimeric fathers [108]. Later, Jaenisch made a similar observation using a male mouse that was genetically mosaic for integrated Moloney leukemia provirus [109].
SSC competition has also been interrogated using transplantation assays in mice. The Shinohara group demonstrated that transplanted SSCs compete with endogenous stem cells for colonization [110]. In another series of relevant studies, the Brinster group assessed effects of aging and the niche microenvironment on SSC self-renewal [111,112]. They demonstrated that SSCs from old mice consecutively transplanted into young testes maintained self-renewal capacity and spermatogenesis for long past the normal life span of mice [111]. Although cell competition was not specifically interrogated, the data suggest that normal mouse SSCs do not acquire gross genetic or epigenetic abnormalities over time. Nonetheless, niche failure or deterioration could facilitate cell competition, particularly in humans.
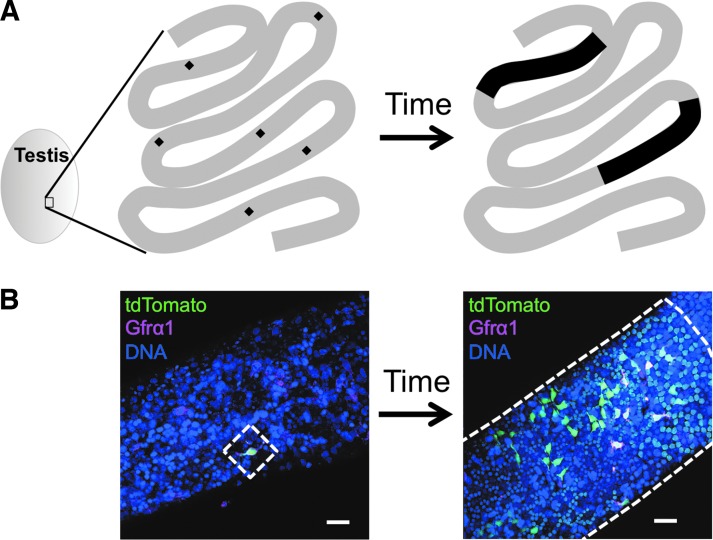
To assess homeostatic behavior of SSCs in vivo, Nakagawa et al. first described life-long kinetics of mouse SSCs using genetic lineage tracing, as already mentioned [11]. In this study, it was demonstrated that stem cells are occasionally lost and compensated for by other stem cells that form larger sized clones over time. Indeed, using a similar lineage tracing setup in the mouse testis, we observed that certain SSC clones survive for more than 1 year, expanding their territories, but the remaining clones disappear (unpublished data) (Fig. 2). By analyzing SSC clonal dynamics using mathematical models, Klein and Simons proposed that neutral competition confers all stem cells with equal proliferation potential, but some stem cells are lost in a stochastic manner and are replaced by others [113].
FIG. 2.
Lineage tracing reveals the long-term kinetics of labeled SSCs. (A) An example of lineage tracing. In this segment of seminiferous tubules, a single SSC (black diamond; six SSCs are shown in the segment) is being labeled after induction. Certain clones (eg, only two clones out of six in this case) self-renew and expand to occupy the long segment (black area), but the rest disappear over time. By applying mathematical models to this phenomenon, Klein and Simons proposed a neutral competition model, in which all stem cells have equal proliferation potential, but some stem cells are lost in a stochastic manner [113]. (B) Actual images of clonal expansion in the lineage tracing system using the Sox2-creERT2 driver. Tamoxifen was administered to a Sox2-creERT2; Rosa-tdTomato mouse at 7 weeks of age. Right after the induction, Sox2-creERT2 driver labeled Asingle SSCs (left). After a long period of chase, SSC progeny clonally expand to form a large colony (right). Some of the undifferentiated spermatogonia retained Gfrα1 expression, which is known to mark SSCs. Scale bars: 40 μm.
Drosophila melanogaster has provided excellent models for cell competition, enabling comparisons of pairs of clones with different genotypes (ie, non-neutral competition; see Stine and Matunis [114] for a review). However, few non-neutral competition models in mammalian germ line systems have been described. One study used a competitive transplantation assay in mice, in which mutant stem cells were mixed with wild-type SSCs for transplantation into recipient testes. These data showed that most of offspring from transplanted SSCs, consisting of mixed populations of wild-type cells and those with loss-of-function for p21 cyclin-dependent kinase inhibitor, were derived from the p21-deficient SSCs and not from wild-type SSCs [115]. This suggests that the mutant SSCs successfully compete with wild-type cells to support long-term repopulation.
Using adult mouse SSCs, our group developed an experimental approach for interrogating PAE disorders [52]. In this study, SSCs with an ectopically expressed, Apert syndrome-associated FGFR2 mutation (S252W) exhibited significantly greater stem cell activity than those with ectopic wild-type FGFR2 in transplantation assays. Importantly, the S252W SSCs did not show any proliferative advantage when cultured alone in vitro in standard media, but exhibited increased competitiveness in the presence of wild-type SSCs. At the cellular level, S252W SSCs showed enhanced sensitivity to FGF, as evidenced by enhanced MAPK signaling with low dose FGF, suggesting an increased self-renewal capability of S252W SSCs, when growth factors are scarce. This experimental model supports the central hypothesis of positive selection propagation of SSCs in mammals.
Common genome editing techniques such as TALEN and CRISPR enable simple, rapid gene modification [116]. These new approaches will likely enhance the development of SSC culture systems designed to directly interrogate both the mechanisms of positive selection for PAE mutations and also the functions of other mutations arising in germ line stem cells. However, studies of SSC cultures are dependent on the post-transplantation reconstitution of spermatogenesis, a milieu that does not fully reflect the physiological behavior of SSC in vivo. To understand in vivo SSC competition in a normal environment, one would require a system to induce mutations in situ (ie, a genetic mosaic) and follow mutated cell fates over long intervals.
Significance of SSCs in the Setting of Disease Prevention and Diagnosis
The genetic contribution to PAE disorders is driven by DNMs. Although most of the best understood such disorders are individually rare and monogenic, it is likely that a substantial fraction of the risk also of common, genetically complex disorders (eg, autism and early onset cancers) is also driven by paternally derived DNMs. In fact, a recent preliminary report by the Developmental Disorders Study consortium estimates that the aggregate risk of a child being born with a developmental disorder (of any kind) attributable to DNMs is very large (∼1:450 to 1:200 live births) [96]. Since the majority of such DNMs are of paternal origin, it is extremely important to understand the detailed pathogenic process that occurs upstream of transmission in the male germ line.
As techniques to efficiently derive and propagate hSSCs are being developed, new clinical potential applications of SSCs are coming into focus. With the advent of such lines, a number of strategies may emerge that could enable the prevention of genetic paternal age-related disorders. First, however, it will be necessary to determine which individually rare disorders are, in fact, exclusively of paternal origin. This information can sometimes be obtained using existing WES data by means of adjacent polymorphisms unique to the father or mother. However, informative polymorphisms are not always available, necessitating more in-depth genetic analysis.
For pathogenic DNMs that are exclusively of paternal origin, we can envision the development of genetic screening panels that could be applied to sperm samples of prospective older fathers, analogous to the diagnostic gene panels that are currently available for genotyping of somatic cancers. If evidence is found for a significant degree of risk, this would allow the prospective parents to make informed decisions about whether to proceed.
A series of further applications of hSSCs can be imagined, but each step in implementation entails serious concerns related to both safety and ethics. For men with insufficient healthy sperm (ie, those lacking known pathogenic DNMs), clonal SSC lines could be derived from a testis biopsy, expanded, and retested. If healthy lines are found, these could theoretically be employed in several ways. The first is autotransplantation back into the testis. Notably, proof-of-principle for autotransplantation has been already demonstrated in animal models [117–120]. Second, given recent improvements in experimental derivation of gametes in vitro from stem cell precursors, it is likely that production of functional sperm from hSSCs will eventually become feasible. For example, functional male haploid gametes were recently generated in vitro from rodent stem cells and were successfully used to fertilize oocytes and produce offspring [121]. Also, chemical treatment (ie, retinoic acid exposure) has been used to efficiently induce meiosis in cultured mouse SSCs [121]. If such technologies could be adapted to hSSCs, these could be used for intracytoplasmic sperm injection to generate embryos for implantation.
Finally, given recent technical advances in genome editing, it may be possible to revert mutant alleles in SSCs back to the wild-type state before use of sperm. Each of the procedures already outlined would require extremely extensive, potentially prohibitive safety testing, first in primates and then in humans.
Although we do not attempt to delve into the ethical intricacies of assisted reproduction, several major issues bear brief a note. The following concerns are not entirely unique to the technical strategies already described, but are potentially more significant (eg, relative to simple in vitro fertilization [IVF]), given the greater extent of artificial manipulation of the cells involved. First, the genomes and epigenomes of gametes derived from long-term cultured cells could be altered in unforeseen ways. In contrast, short-term culture of human embryos is already employed in standard IVF protocols, suggesting that at least some period of culture is compatible with normal development. Second, many future generations could be affected by genomic or epigenomic alterations due to long-term culture. From our current vantage point, these barriers are certainly formidable and may even appear prohibitive.
However, it is difficult to predict how novel technology may be able to successfully overcome such hurdles. Therefore, it is incumbent upon the research community to consider the risks and benefits far in advance of each stage of clinical testing and to proceed with extreme caution. In fact, recent guidelines from the International Society for Stem Cell Research, although supportive of research studies, prohibit clinical application of gametes that have undergone modification of the nuclear genome [122]. The growing interest around this new area is highlighted by the fact that human germ line editing is a major focus of a comprehensive study recently undertaken by the National Academies to address the clinical, ethical, legal, and social implications of the use of gene-editing technologies [123].
Conclusion
Myriad reasons exist to be optimistic about the future for using SSCs to understand and ameliorate human disease. The spectrum of human disorders for which the strategies already outlined are relevant is only likely to expand, as the ability to gather and interpret genomic sequencing data increases. Although impressive technological advances have already occurred in the field of SSC biology, many are limited to other organisms and have not been successfully applied to hSSCs. Yet, given the lag period for translating basic findings with other stem cell types (eg, embryonic stem cells) from model systems to humans, the question is not whether this will happen, but rather when will it happen. In the interim, the research and clinical communities will have ample time to consider the weighty issues that surround any manipulation of human gametes for therapeutic purposes.
Acknowledgments
This study was supported, in part, by the Eunice Kennedy Shriver National Institute Of Child Health & Human Development of National Institutes of Health grant 1DP2HD080352-01, New York State Stem Cell Science Board grant C029156, and a fellowship (to M.Y.) from the Tri-Institutional Stem Cell Initiative.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Lawson KA. and Hage WJ. (1994). Clonal analysis of the origin of primordial germ cells in the mouse. Ciba Found Symp 182:68–84; discussion 84–91. [DOI] [PubMed] [Google Scholar]
- 2.Chiquoine AD. (1954). The identification, origin, and migration of the primordial germ cells in the mouse embryo. Anat Rec 118:135–146 [DOI] [PubMed] [Google Scholar]
- 3.Molyneaux KA, Stallock J, Schaible K. and Wylie C. (2001). Time-lapse analysis of living mouse germ cell migration. Dev Biol 240:488–498 [DOI] [PubMed] [Google Scholar]
- 4.Hilscher B, Hilscher W, Bulthoff-Ohnolz B, Kramer U, Birke A, Pelzer H. and Gauss G. (1974). Kinetics of gametogenesis. I. Comparative histological and autoradiographic studies of oocytes and transitional prospermatogonia during oogenesis and prespermatogenesis. Cell Tissue Res 154:443–470 [DOI] [PubMed] [Google Scholar]
- 5.Wartenberg H. (1976). Comparative cytomorphologic aspects of the male germ cells, especially of the “Gonia”. Andrologia 8:117–130 [DOI] [PubMed] [Google Scholar]
- 6.Clermont Y. and Perey B. (1957). Quantitative study of the cell population of the seminiferous tubules in immature rats. Am J Anat 100:241–267 [DOI] [PubMed] [Google Scholar]
- 7.De Felici M. (2013) Origin, migration, and proliferation of human primordial germ cells. In: Oogenesis. Coticchio G, Albertini DF, DeSantis L, eds. Springer Verlag, London; New York, pp. 19–37 [Google Scholar]
- 8.Oakberg EF. (1971). Spermatogonial stem-cell renewal in the mouse. Anat Rec 169:515–531 [DOI] [PubMed] [Google Scholar]
- 9.Hara K, Nakagawa T, Enomoto H, Suzuki M, Yamamoto M, Simons BD. and Yoshida S. (2014). Mouse spermatogenic stem cells continually interconvert between equipotent singly isolated and syncytial states. Cell Stem Cell 14:658–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakagawa T, Sharma M, Nabeshima Y, Braun RE. and Yoshida S. (2010). Functional hierarchy and reversibility within the murine spermatogenic stem cell compartment. Science 328:62–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakagawa T, Nabeshima Y. and Yoshida S. (2007). Functional identification of the actual and potential stem cell compartments in mouse spermatogenesis. Dev Cell 12:195–206 [DOI] [PubMed] [Google Scholar]
- 12.Yoshida S, Takakura A, Ohbo K, Abe K, Wakabayashi J, Yamamoto M, Suda T. and Nabeshima Y. (2004). Neurogenin3 delineates the earliest stages of spermatogenesis in the mouse testis. Dev Biol 269:447–458 [DOI] [PubMed] [Google Scholar]
- 13.Clermont Y. (1966). Renewal of spermatogonia in man. Am J Anat 118:509–524 [DOI] [PubMed] [Google Scholar]
- 14.Clermont Y. and Leblond CP. (1959). Differentiation and renewal of spermatogonia in the monkey, Macacus rhesus. Am J Anat 104:237–273 [DOI] [PubMed] [Google Scholar]
- 15.van Alphen MM, van de Kant HJ. and de Rooij DG. (1988). Repopulation of the seminiferous epithelium of the rhesus monkey after X irradiation. Radiat Res 113:487–500 [PubMed] [Google Scholar]
- 16.Ehmcke J. and Schlatt S. (2006). A revised model for spermatogonial expansion in man: lessons from non-human primates. Reproduction 132:673–680 [DOI] [PubMed] [Google Scholar]
- 17.Fouquet JP. and Dadoune JP. (1986). Renewal of spermatogonia in the monkey (Macaca fascicularis). Biol Reprod 35:199–207 [DOI] [PubMed] [Google Scholar]
- 18.van Alphen MM, van de Kant HJ. and de Rooij DG. (1988). Depletion of the spermatogonia from the seminiferous epithelium of the rhesus monkey after X irradiation. Radiat Res 113:473–486 [PubMed] [Google Scholar]
- 19.Clermont Y. (1966). Spermatogenesis in man. A study of the spermatogonial population. Fertil Steril 17:705–721 [PubMed] [Google Scholar]
- 20.Clermont Y. (1969). Two classes of spermatogonial stem cells in the monkey (Cercopithecus aethiops). Am J Anat 126:57–71 [DOI] [PubMed] [Google Scholar]
- 21.Clermont Y. and Antar M. (1973). Duration of the cycle of the seminiferous epithelium and the spermatogonial renewal in the monkey Macaca arctoides. Am J Anat 136:153–165 [DOI] [PubMed] [Google Scholar]
- 22.Kluin PM, Kramer MF. and de Rooij DG. (1983). Testicular development in Macaca irus after birth. Int J Androl 6:25–43 [DOI] [PubMed] [Google Scholar]
- 23.Ehmcke J, Luetjens CM. and Schlatt S. (2005). Clonal organization of proliferating spermatogonial stem cells in adult males of two species of non-human primates, Macaca mulatta and Callithrix jacchus. Biol Reprod 72:293–300 [DOI] [PubMed] [Google Scholar]
- 24.Tegelenbosch RA. and de Rooij DG. (1993). A quantitative study of spermatogonial multiplication and stem cell renewal in the C3H/101 F1 hybrid mouse. Mutat Res 290:193–200 [DOI] [PubMed] [Google Scholar]
- 25.Kanatsu-Shinohara M, Ogonuki N, Inoue K, Miki H, Ogura A, Toyokuni S. and Shinohara T. (2003). Long-term proliferation in culture and germline transmission of mouse male germline stem cells. Biol Reprod 69:612–616 [DOI] [PubMed] [Google Scholar]
- 26.Kubota H, Avarbock MR. and Brinster RL. (2004). Growth factors essential for self-renewal and expansion of mouse spermatogonial stem cells. Proc Natl Acad Sci U S A 101:16489–16494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meng X, Lindahl M, Hyvonen ME, Parvinen M, de Rooij DG, Hess MW, Raatikainen-Ahokas A, Sainio K, Rauvala H, et al. (2000). Regulation of cell fate decision of undifferentiated spermatogonia by GDNF. Science 287:1489–1493 [DOI] [PubMed] [Google Scholar]
- 28.Chen LY, Willis WD. and Eddy EM. (2016). Targeting the Gdnf Gene in peritubular myoid cells disrupts undifferentiated spermatogonial cell development. Proc Natl Acad Sci U S A 113:1829–1834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin LA. and Seandel M. (2013). Serial enrichment of spermatogonial stem and progenitor cells (SSCs) in culture for derivation of long-term adult mouse SSC lines. J Vis Exp e50017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim J, Seandel M, Falciatori I, Wen D. and Rafii S. (2008). CD34+ testicular stromal cells support long-term expansion of embryonic and adult stem and progenitor cells. Stem Cells 26:2516–2522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zheng Y, Thomas A, Schmidt CM. and Dann CT. (2014). Quantitative detection of human spermatogonia for optimization of spermatogonial stem cell culture. Hum Reprod 29:2497–2511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Izadyar F, Wong J, Maki C, Pacchiarotti J, Ramos T, Howerton K, Yuen C, Greilach S, Zhao HH, et al. (2011). Identification and characterization of repopulating spermatogonial stem cells from the adult human testis. Hum Reprod 26:1296–1306 [DOI] [PubMed] [Google Scholar]
- 33.(2014). Retraction: generation of pluripotent stem cells from adult human testis. Nature 512:338. [DOI] [PubMed] [Google Scholar]
- 34.Maleki M, Ghanbarvand F, Reza Behvarz M, Ejtemaei M. and Ghadirkhomi E. (2014). Comparison of mesenchymal stem cell markers in multiple human adult stem cells. Int J Stem Cells 7:118–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith JF, Yango P, Altman E, Choudhry S, Poelzl A, Zamah AM, Rosen M, Klatsky PC. and Tran ND. (2014). Testicular niche required for human spermatogonial stem cell expansion. Stem Cells Transl Med 3:1043–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lim JJ, Sung SY, Kim HJ, Song SH, Hong JY, Yoon TK, Kim JK, Kim KS. and Lee DR. (2010). Long-term proliferation and characterization of human spermatogonial stem cells obtained from obstructive and non-obstructive azoospermia under exogenous feeder-free culture conditions. Cell Prolif 43:405–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chikhovskaya JV, van Daalen SK, Korver CM, Repping S. and van Pelt AM. (2014). Mesenchymal origin of multipotent human testis-derived stem cells in human testicular cell cultures. Mol Hum Reprod 20:155–167 [DOI] [PubMed] [Google Scholar]
- 38.Sadri-Ardekani H, Mizrak SC, van Daalen SK, Korver CM, Roepers-Gajadien HL, Koruji M, Hovingh S, de Reijke TM, de la Rosette JJ, et al. (2009). Propagation of human spermatogonial stem cells in vitro. JAMA 302:2127–2134 [DOI] [PubMed] [Google Scholar]
- 39.He Z, Kokkinaki M, Jiang J, Dobrinski I. and Dym M. (2010). Isolation, characterization, and culture of human spermatogonia. Biol Reprod 82:363–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sachs C, Robinson BD, Andres Martin L, Webster T, Gilbert M, Lo HY, Rafii S, Ng CK. and Seandel M. (2014). Evaluation of candidate spermatogonial markers ID4 and GPR125 in testes of adult human cadaveric organ donors. Andrology 2:607–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oatley MJ, Kaucher AV, Racicot KE. and Oatley JM. (2011). Inhibitor of DNA binding 4 is expressed selectively by single spermatogonia in the male germline and regulates the self-renewal of spermatogonial stem cells in mice. Biol Reprod 85:347–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seandel M, James D, Shmelkov SV, Falciatori I, Kim J, Chavala S, Scherr DS, Zhang F, Torres R, et al. (2007). Generation of functional multipotent adult stem cells from GPR125+ germline progenitors. Nature 449:346–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nickkholgh B, Mizrak SC, Korver CM, van Daalen SK, Meissner A, Repping S. and van Pelt AM. (2014). Enrichment of spermatogonial stem cells from long-term cultured human testicular cells. Fertil Steril 102:558–565.e5. [DOI] [PubMed] [Google Scholar]
- 44.Kossack N, Terwort N, Wistuba J, Ehmcke J, Schlatt S, Scholer H, Kliesch S. and Gromoll J. (2013). A combined approach facilitates the reliable detection of human spermatogonia in vitro. Hum Reprod 28:3012–3025 [DOI] [PubMed] [Google Scholar]
- 45.Valli H, Sukhwani M, Dovey SL, Peters KA, Donohue J, Castro CA, Chu T, Marshall GR. and Orwig KE. (2014). Fluorescence- and magnetic-activated cell sorting strategies to isolate and enrich human spermatogonial stem cells. Fertil Steril 102:566–580.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.von Kopylow K, Schulze W, Salzbrunn A. and Spiess AN. (2016). Isolation and gene expression analysis of single potential human spermatogonial stem cells. Mol Hum Reprod 22:229–239 [DOI] [PubMed] [Google Scholar]
- 47.Brinster RL. and Avarbock MR. (1994). Germline transmission of donor haplotype following spermatogonial transplantation. Proc Natl Acad Sci U S A 91:11303–11307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Koshimizu U, Watanabe D, Tajima Y. and Nishimune Y. (1992). Effects of W (c-kit) gene mutation on gametogenesis in male mice: agametic tubular segments in Wf/Wf testes. Development 114:861–867 [DOI] [PubMed] [Google Scholar]
- 49.Ryu BY, Orwig KE, Oatley JM, Lin CC, Chang LJ, Avarbock MR. and Brinster RL. (2007). Efficient generation of transgenic rats through the male germline using lentiviral transduction and transplantation of spermatogonial stem cells. J Androl 28:353–360 [DOI] [PubMed] [Google Scholar]
- 50.Dobrinski I, Ogawa T, Avarbock MR. and Brinster RL. (1999). Computer assisted image analysis to assess colonization of recipient seminiferous tubules by spermatogonial stem cells from transgenic donor mice. Mol Reprod Dev 53:142–148 [DOI] [PubMed] [Google Scholar]
- 51.Kanatsu-Shinohara M, Inoue K, Miki H, Ogonuki N, Takehashi M, Morimoto T, Ogura A. and Shinohara T. (2006). Clonal origin of germ cell colonies after spermatogonial transplantation in mice. Biol Reprod 75:68–74 [DOI] [PubMed] [Google Scholar]
- 52.Martin LA, Assif N, Gilbert M, Wijewarnasuriya D. and Seandel M. (2014). Enhanced fitness of adult spermatogonial stem cells bearing a paternal age-associated FGFR2 mutation. Stem Cell Reports 3:219–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nagano M, Patrizio P. and Brinster RL. (2002). Long-term survival of human spermatogonial stem cells in mouse testes. Fertil Steril 78:1225–1233 [DOI] [PubMed] [Google Scholar]
- 54.Smith MA, Altekruse SF, Adamson PC, Reaman GH. and Seibel NL. (2014). Declining childhood and adolescent cancer mortality. Cancer 120:2497–2506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Picton HM, Wyns C, Anderson RA, Goossens E, Jahnukainen K, Kliesch S, Mitchell RT, Pennings G, Rives N, et al. (2015). A European perspective on testicular tissue cryopreservation for fertility preservation in prepubertal and adolescent boys. Hum Reprod 30:2463–2475 [DOI] [PubMed] [Google Scholar]
- 56.Moss JL, Choi AW, Fitzgerald Keeter MK. and Brannigan RE. (2016). Male adolescent fertility preservation. Fertil Steril 105:267–273 [DOI] [PubMed] [Google Scholar]
- 57.Kanatsu-Shinohara M, Ogonuki N, Iwano T, Lee J, Kazuki Y, Inoue K, Miki H, Takehashi M, Toyokuni S, et al. (2005). Genetic and epigenetic properties of mouse male germline stem cells during long-term culture. Development 132:4155–4163 [DOI] [PubMed] [Google Scholar]
- 58.Nickkholgh B, Mizrak SC, van Daalen SK, Korver CM, Sadri-Ardekani H, Repping S. and van Pelt AM. (2014). Genetic and epigenetic stability of human spermatogonial stem cells during long-term culture. Fertil Steril 102:1700–1707.e1. [DOI] [PubMed] [Google Scholar]
- 59.Radford J, Shalet S. and Lieberman B. (1999). Fertility after treatment for cancer. Questions remain over ways of preserving ovarian and testicular tissue. BMJ 319:935–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Matsui Y, Zsebo K. and Hogan BL. (1992). Derivation of pluripotential embryonic stem cells from murine primordial germ cells in culture. Cell 70:841–847 [DOI] [PubMed] [Google Scholar]
- 61.Kanatsu-Shinohara M, Inoue K, Lee J, Yoshimoto M, Ogonuki N, Miki H, Baba S, Kato T, Kazuki Y, et al. (2004). Generation of pluripotent stem cells from neonatal mouse testis. Cell 119:1001–1012 [DOI] [PubMed] [Google Scholar]
- 62.Izadyar F, Pau F, Marh J, Slepko N, Wang T, Gonzalez R, Ramos T, Howerton K, Sayre C. and Silva F. (2008). Generation of multipotent cell lines from a distinct population of male germ line stem cells. Reproduction 135:771–784 [DOI] [PubMed] [Google Scholar]
- 63.Ko K, Tapia N, Wu G, Kim JB, Bravo MJ, Sasse P, Glaser T, Ruau D, Han DW, et al. (2009). Induction of pluripotency in adult unipotent germline stem cells. Cell Stem Cell 5:87–96 [DOI] [PubMed] [Google Scholar]
- 64.Baba S, Heike T, Umeda K, Iwasa T, Kaichi S, Hiraumi Y, Doi H, Yoshimoto M, Kanatsu-Shinohara M, Shinohara T. and Nakahata T. (2007). Generation of cardiac and endothelial cells from neonatal mouse testis-derived multipotent germline stem cells. Stem Cells 25:1375–1383 [DOI] [PubMed] [Google Scholar]
- 65.Glaser T, Opitz T, Kischlat T, Konang R, Sasse P, Fleischmann BK, Engel W, Nayernia K. and Brustle O. (2008). Adult germ line stem cells as a source of functional neurons and glia. Stem Cells 26:2434–2443 [DOI] [PubMed] [Google Scholar]
- 66.Kossack N, Meneses J, Shefi S, Nguyen HN, Chavez S, Nicholas C, Gromoll J, Turek PJ. and Reijo-Pera RA. (2009). Isolation and characterization of pluripotent human spermatogonial stem cell-derived cells. Stem Cells 27:138–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mizrak SC, Chikhovskaya JV, Sadri-Ardekani H, van Daalen S, Korver CM, Hovingh SE, Roepers-Gajadien HL, Raya A, Fluiter K, et al. (2010). Embryonic stem cell-like cells derived from adult human testis. Hum Reprod 25:158–167 [DOI] [PubMed] [Google Scholar]
- 68.Golestaneh N, Kokkinaki M, Pant D, Jiang J, DeStefano D, Fernandez-Bueno C, Rone JD, Haddad BR, Gallicano GI. and Dym M. (2009). Pluripotent stem cells derived from adult human testes. Stem Cells Dev 18:1115–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kong A, Frigge ML, Masson G, Besenbacher S, Sulem P, Magnusson G, Gudjonsson SA, Sigurdsson A, Jonasdottir A, et al. (2012). Rate of de novo mutations and the importance of father's age to disease risk. Nature 488:471–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Francioli LC, Polak PP, Koren A, Menelaou A, Chun S, Renkens I, Genome of the Netherlands Consortium, van Duijn CM, Swertz M, et al. (2015). Genome-wide patterns and properties of de novo mutations in humans. Nat Genet 47:822–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rahbari R, Wuster A, Lindsay SJ, Hardwick RJ, Alexandrov LB, Al Turki S, Dominiczak A, Morris A, Porteous D, et al. (2016). Timing, rates and spectra of human germline mutation. Nat Genet 48:126–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Michaelson JJ, Shi Y, Gujral M, Zheng H, Malhotra D, Jin X, Jian M, Liu G, Greer D, et al. (2012). Whole-genome sequencing in autism identifies hot spots for de novo germline mutation. Cell 151:1431–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kohler SW, Provost GS, Fieck A, Kretz PL, Bullock WO, Sorge JA, Putman DL. and Short JM. (1991). Spectra of spontaneous and mutagen-induced mutations in the lacI gene in transgenic mice. Proc Natl Acad Sci U S A 88:7958–7962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Walter CA, Intano GW, McCarrey JR, McMahan CA. and Walter RB. (1998). Mutation frequency declines during spermatogenesis in young mice but increases in old mice. Proc Natl Acad Sci U S A 95:10015–10019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Murphey P, McLean DJ, McMahan CA, Walter CA. and McCarrey JR. (2013). Enhanced genetic integrity in mouse germ cells. Biol Reprod 88:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Walter CA, Intano GW, McMahan CA, Kelner K, McCarrey JR. and Walter RB. (2004). Mutation spectral changes in spermatogenic cells obtained from old mice. DNA Repair (Amst) 3:495–504 [DOI] [PubMed] [Google Scholar]
- 77.Jones KL, Smith DW, Harvey MA, Hall BD. and Quan L. (1975). Older paternal age and fresh gene mutation: data on additional disorders. J Pediatr 86:84–88 [DOI] [PubMed] [Google Scholar]
- 78.Risch N, Reich EW, Wishnick MM. and McCarthy JG. (1987). Spontaneous mutation and parental age in humans. Am J Hum Genet 41:218–248 [PMC free article] [PubMed] [Google Scholar]
- 79.Tolarova MM, Harris JA, Ordway DE. and Vargervik K. (1997). Birth prevalence, mutation rate, sex ratio, parents’ age, and ethnicity in Apert syndrome. Am J Med Genet 72:394–398 [DOI] [PubMed] [Google Scholar]
- 80.Weinberg W. (1912). On inheritance of dwarfism. Arch Rassen Ges Biol 9:710–718 [Google Scholar]
- 81.Tartaglia M, Cordeddu V, Chang H, Shaw A, Kalidas K, Crosby A, Patton MA, Sorcini M, van der Burgt I, Jeffery S. and Gelb BD. (2004). Paternal germline origin and sex-ratio distortion in transmission of PTPN11 mutations in Noonan syndrome. Am J Hum Genet 75:492–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sol-Church K, Stabley DL, Nicholson L, Gonzalez IL. and Gripp KW. (2006). Paternal bias in parental origin of HRAS mutations in Costello syndrome. Hum Mutat 27:736–741 [DOI] [PubMed] [Google Scholar]
- 83.Wilkin DJ, Szabo JK, Cameron R, Henderson S, Bellus GA, Mack ML, Kaitila I, Loughlin J, Munnich A, et al. (1998). Mutations in fibroblast growth-factor receptor 3 in sporadic cases of achondroplasia occur exclusively on the paternally derived chromosome. Am J Hum Genet 63:711–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Goriely A. and Wilkie AO. (2012). Paternal age effect mutations and selfish spermatogonial selection: causes and consequences for human disease. Am J Hum Genet 90:175–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Anderson J, Burns HD, Enriquez-Harris P, Wilkie AO. and Heath JK. (1998). Apert syndrome mutations in fibroblast growth factor receptor 2 exhibit increased affinity for FGF ligand. Hum Mol Genet 7:1475–1483 [DOI] [PubMed] [Google Scholar]
- 86.Goriely A, McVean GA, Rojmyr M, Ingemarsson B. and Wilkie AO. (2003). Evidence for selective advantage of pathogenic FGFR2 mutations in the male germ line. Science 301:643–646 [DOI] [PubMed] [Google Scholar]
- 87.Goriely A, McVean GA, van Pelt AM, O'Rourke AW, Wall SA, de Rooij DG. and Wilkie AO. (2005). Gain-of-function amino acid substitutions drive positive selection of FGFR2 mutations in human spermatogonia. Proc Natl Acad Sci U S A 102:6051–6056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yoon SR, Qin J, Glaser RL, Jabs EW, Wexler NS, Sokol R, Arnheim N. and Calabrese P. (2009). The ups and downs of mutation frequencies during aging can account for the Apert syndrome paternal age effect. PLoS Genet 5:e1000558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Giannoulatou E, McVean G, Taylor IB, McGowan SJ, Maher GJ, Iqbal Z, Pfeifer SP, Turner I, Burkitt Wright EM, et al. (2013). Contributions of intrinsic mutation rate and selfish selection to levels of de novo HRAS mutations in the paternal germline. Proc Natl Acad Sci U S A 110:20152–20157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Choi SK, Yoon SR, Calabrese P. and Arnheim N. (2008). A germ-line-selective advantage rather than an increased mutation rate can explain some unexpectedly common human disease mutations. Proc Natl Acad Sci U S A 105:10143–10148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sanders SJ, Murtha MT, Gupta AR, Murdoch JD, Raubeson MJ, Willsey AJ, Ercan-Sencicek AG, DiLullo NM, Parikshak NN, et al. (2012). De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature 485:237–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Shinde DN, Elmer DP, Calabrese P, Boulanger J, Arnheim N. and Tiemann-Boege I. (2013). New evidence for positive selection helps explain the paternal age effect observed in achondroplasia. Hum Mol Genet 22:4117–4126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yoon SR, Choi SK, Eboreime J, Gelb BD, Calabrese P. and Arnheim N. (2013). Age-dependent germline mosaicism of the most common noonan syndrome mutation shows the signature of germline selection. Am J Hum Genet 92:917–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Heller CH. and Clermont Y. (1964). Kinetics of the germinal epithelium in man. Recent Prog Horm Res 20:545–575 [PubMed] [Google Scholar]
- 95.Deciphering Developmental Disorders Study. (2015). Large-scale discovery of novel genetic causes of developmental disorders. Nature 519:223–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.McRae JF, Clayton S, Fitzgerald TW, Kaplanis J, Prigmore E, Rajan D, Sifrim A, Aitken S, Akawi N, et al. (2016). Prevalence, phenotype and architecture of developmental disorders caused by de novo mutation. bioRxiv. http://dx.doi.org/10.1101/049056 [Google Scholar]
- 97.Neale BM, Kou Y, Liu L, Ma'ayan A, Samocha KE, Sabo A, Lin CF, Stevens C, Wang LS, et al. (2012). Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature 485:242–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Iossifov I, Ronemus M, Levy D, Wang Z, Hakker I, Rosenbaum J, Yamrom B, Lee YH, Narzisi G, et al. (2012). De novo gene disruptions in children on the autistic spectrum. Neuron 74:285–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.O'Roak BJ, Vives L, Girirajan S, Karakoc E, Krumm N, Coe BP, Levy R, Ko A, Lee C, et al. (2012). Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature 485:246–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zaidi S, Choi M, Wakimoto H, Ma L, Jiang J, Overton JD, Romano-Adesman A, Bjornson RD, Breitbart RE, et al. (2013). De novo mutations in histone-modifying genes in congenital heart disease. Nature 498:220–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Malaspina D, Harlap S, Fennig S, Heiman D, Nahon D, Feldman D. and Susser ES. (2001). Advancing paternal age and the risk of schizophrenia. Arch Gen Psychiatry 58:361–367 [DOI] [PubMed] [Google Scholar]
- 102.Gratten J, Wray NR, Peyrot WJ, McGrath JJ, Visscher PM. and Goddard ME. (2016). Risk of psychiatric illness from advanced paternal age is not predominantly from de novo mutations. Nat Genet 48:718–724 [DOI] [PubMed] [Google Scholar]
- 103.Stadler ZK, Esposito D, Shah S, Vijai J, Yamrom B, Levy D, Lee YH, Kendall J, Leotta A, et al. (2012). Rare de novo germline copy-number variation in testicular cancer. Am J Hum Genet 91:379–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhang J, Nichols KE. and Downing JR. (2016). Germline mutations in predisposition genes in pediatric cancer. N Engl J Med 374:1391. [DOI] [PubMed] [Google Scholar]
- 105.Goriely A, Hansen RM, Taylor IB, Olesen IA, Jacobsen GK, McGowan SJ, Pfeifer SP, McVean GA, Rajpert-De Meyts E. and Wilkie AO. (2009). Activating mutations in FGFR3 and HRAS reveal a shared genetic origin for congenital disorders and testicular tumors. Nat Genet 41:1247–1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lim J, Maher GJ, Turner GD, Dudka-Ruszkowska W, Taylor S, Rajpert-De Meyts E, Goriely A. and Wilkie AO. (2012). Selfish spermatogonial selection: evidence from an immunohistochemical screen in testes of elderly men. PLoS One 7:e42382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Maher GJ, McGowan SJ, Giannoulatou E, Verrill C, Goriely A. and Wilkie AO. (2016). Visualizing the origins of selfish de novo mutations in individual seminiferous tubules of human testes. Proc Natl Acad Sci U S A 113:2454–2459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Mintz B. (1968). Hermaphroditism, sex chromosomal mosaicism and germ cell selection in allophenic mice. J Anim Sci 27 Suppl 1:51–60 [PubMed] [Google Scholar]
- 109.Jaenisch R. (1976). Germ line integration and Mendelian transmission of the exogenous Moloney leukemia virus. Proc Natl Acad Sci U S A 73:1260–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Shinohara T, Orwig KE, Avarbock MR. and Brinster RL. (2002). Germ line stem cell competition in postnatal mouse testes. Biol Reprod 66:1491–1497 [DOI] [PubMed] [Google Scholar]
- 111.Ryu BY, Orwig KE, Oatley JM, Avarbock MR. and Brinster RL. (2006). Effects of aging and niche microenvironment on spermatogonial stem cell self-renewal. Stem Cells 24:1505–1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Schmidt JA, Abramowitz LK, Kubota H, Wu X, Niu Z, Avarbock MR, Tobias JW, Bartolomei MS. and Brinster RL. (2011). In vivo and in vitro aging is detrimental to mouse spermatogonial stem cell function. Biol Reprod 84:698–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Klein AM, Nakagawa T, Ichikawa R, Yoshida S. and Simons BD. (2010). Mouse germ line stem cells undergo rapid and stochastic turnover. Cell Stem Cell 7:214–224 [DOI] [PubMed] [Google Scholar]
- 114.Stine RR. and Matunis EL. (2013). Stem cell competition: finding balance in the niche. Trends Cell Biol 23:357–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kanatsu-Shinohara M, Takashima S. and Shinohara T. (2010). Transmission distortion by loss of p21 or p27 cyclin-dependent kinase inhibitors following competitive spermatogonial transplantation. Proc Natl Acad Sci U S A 107:6210–6215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gaj T, Gersbach CA. and Barbas CF., 3rd. (2013). ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol 31:397–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Izadyar F, Den Ouden K, Stout TA, Stout J, Coret J, Lankveld DP, Spoormakers TJ, Colenbrander B, Oldenbroek JK, et al. (2003). Autologous and homologous transplantation of bovine spermatogonial stem cells. Reproduction 126:765–774 [PubMed] [Google Scholar]
- 118.Schlatt S, Foppiani L, Rolf C, Weinbauer GF. and Nieschlag E. (2002). Germ cell transplantation into X-irradiated monkey testes. Hum Reprod 17:55–62 [DOI] [PubMed] [Google Scholar]
- 119.Jahnukainen K, Ehmcke J, Quader MA, Saiful Huq M, Epperly MW, Hergenrother S, Nurmio M. and Schlatt S. (2011). Testicular recovery after irradiation differs in prepubertal and pubertal non-human primates, and can be enhanced by autologous germ cell transplantation. Hum Reprod 26:1945–1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hermann BP, Sukhwani M, Winkler F, Pascarella JN, Peters KA, Sheng Y, Valli H, Rodriguez M, Ezzelarab M, et al. (2012). Spermatogonial stem cell transplantation into rhesus testes regenerates spermatogenesis producing functional sperm. Cell Stem Cell 11:715–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zhou Q, Wang M, Yuan Y, Wang X, Fu R, Wan H, Xie M, Liu M, Guo X, et al. (2016). Complete meiosis from embryonic stem cell-derived germ cells in vitro. Cell Stem Cell 18:330–340 [DOI] [PubMed] [Google Scholar]
- 122.International Society for Stem Cell Research. (2016). Guidelines for Stem Cell Research and Clinical Translation. www.isscr.org/home/publications/2016-guidelines
- 123.The National Academies of Sciences, Engineering, and Medicine. (2016). Human Gene-Editing Initiative Consensus Study. www.nationalacademies.org/gene-editing/consensus-study/index.htm