Abstract
Juvenile neuronal ceroid lipofuscinosis (JNCL) is a childhood neurodegenerative disease with early-onset, severe central vision loss. Affected children develop seizures and CNS degeneration accompanied by severe motor and cognitive deficits. There is no cure for JNCL, and patients usually die during the second or third decade of life. In this study, independent lines of induced pluripotent stem cells (iPSCs) were generated from two patients with molecularly confirmed mutations in CLN3, the gene mutated in JNCL. Clinical-grade adeno-associated adenovirus serotype 2 (AAV2) carrying the full-length coding sequence of human CLN3 was generated in a U.S. Food and Drug Administration-registered cGMP facility. AAV2-CLN3 was efficacious in restoring full-length CLN3 transcript and protein in patient-specific fibroblasts and iPSC-derived retinal neurons. When injected into the subretinal space of wild-type mice, purified AAV2-CLN3 did not show any evidence of retinal toxicity. This study provides proof-of-principle for initiation of a clinical trial using AAV-mediated gene augmentation for the treatment of children with CLN3-associated retinal degeneration.
Introduction
The neuronal ceroid lipofuscinoses (NCLs) are a group of progressive neurodegenerative disorders that affect children and young adults.1 To date, 14 different genetic subtypes of NCL, with ages of onset varying from infantile to adult, have been described.2 The autosomal recessive juvenile form (JNCL), commonly known as Batten disease, is the most prevalent, presenting in roughly 1 in every 100,000 live births worldwide.3 Although 59 different mutations in the gene Ceroid Lipofuscinosis, Neuronal 3 (CLN3) have been reported to cause JNCL,3 approximately 85% of patients harbor a 1-kb genomic deletion, which induces a frameshift and insertion of a premature stop codon, in at least one allele.4 CLN3 encodes a 438-amino acid hydrophobic protein of the same name, which is trafficked to, and expressed on, lysosomal membranes.5,6 As mutations in CLN3 lead to a buildup of autofluorescent lipofuscin-like deposits within the lysosomes of affected neuronal cells,7 JNCL is often referred to as a “lysosomal storage” disorder.
CLN3-associated JNCL presents with rapidly progressive retinal degeneration that typically manifests between 4 and 7 years of age. Ophthalmoscopic findings include bull's eye maculopathy, optic nerve pallor, attenuated arterioles and peripheral retinal granularity. Within 2–5 years of diagnosis, patients usually experience profound vision loss that is due primarily to death of the light-sensing photoreceptor cells of the outer neural retina. In the ensuing decade, extensive death of CNS neurons leads to the onset of seizures, difficulty communicating, problems in walking, and ultimately death during the second or third decade of life. No U.S. Food and Drug Administration (FDA)-approved treatments currently exist for this fatal disease, but the fact that JNCL is a recessive disease, caused by variations in a gene expressed in the retina, raises the possibility of (1) lessening the risk of recurrence of the disease in affected families through a combination of genetic counseling and preimplantation genetic testing, and (2) preventing neural cell death and disease progression via gene addition.
Adeno-associated virus (AAV)-based gene augmentation for the treatment of the retinal degenerative disorder RPE65-associated Leber congenital amaurosis has been highly successful.8–10 A single subretinal injection of AAV2 (serotype 2) was shown to be well tolerated and sufficient to induce prolonged protein expression in humans.11 Likewise, improvement in visual acuity, pupillary light reflex, visual fields, and ambulatory vision was detected as early as 2 weeks postinjection and was closely correlated with the area of the retina that received treatment.11 No adverse immune responses or extraocular spread of AAV2-RPE65 was detected.11 These encouraging results, combined with the fact that AAV2 vectors have been shown to transduce both cone photoreceptor cells and inner retinal neurons,12 suggests that this vector could be ideal for delivery of full-length CLN3 as a means to mitigate disease progression and loss of vision in JNCL.
Advances in induced pluripotent stem cell (iPSC) technology have made it possible to generate patient-specific retinal photoreceptor cells in vitro.13–19 These cells can be used both to evaluate the pathophysiology of newly identified genetic variants and to determine the efficacy of novel gene-based therapeutics.20–22 In the case of rare diseases such as CLN3-associated JNCL, for which good animal models of retinal degeneration are lacking, the ability to generate patient-specific photoreceptor cells allows us to test whether AAV-based gene augmentation is capable of restoring wild-type transcript and protein without inducing overexpression toxicity in the patient for whom the treatment is intended. By combining this approach with in vivo toxicity studies in wild-type animals, one could significantly reduce the time between gene discovery and the first therapeutic trial in humans.
In this study, we generated independent iPSC lines from two patients with molecularly confirmed CLN3-associated JNCL: patient B775 (1.02-kb del/Leu101del3CTC) and patient B981 (1.02-kb del/c.1135_1138del). Clinical-grade AAV2 virus carrying full-length CLN3 was generated in an FDA-registered facility using current good manufacturing practices (cGMP). Delivery of full-length CLN3 via adeno-associated viral vector restored full-length CLN3 transcript and protein to patient-specific fibroblasts and iPSC-derived retinal cells in vitro. On injection into wild-type mice, this purified virus demonstrated a complete lack of retinal toxicity. This study provides proof-of-principle for a single-eye gene augmentation trial in children with JNCL.
Materials and Methods
Patients
This study was approved by the Institutional Review Board of the University of Iowa (project approval #199904167) and adhered to the tenets set forth in the Declaration of Helsinki. Patient reference numbers and corresponding molecularly confirmed CLN3 mutations are as follows: patient B775 (1.02-kb del/Leu101del3CTC) and patient B967 (1.02-kb del/1.02-kb del).
Isolation of fibroblasts from patient dermal biopsies
Skin biopsies (3 mm) were obtained from either the non–sun-exposed upper arm or lower abdomen and minced in Iowa xeno-free (i.e., free of animal-derived products) biopsy medium, IxMedia. IxMedia consists of 395 ml of minimal essential medium (MEM)-α (Cat. No. 12571-063; Gibco/Thermo Fisher Scientific, Grand Island, NY), 50 ml of KnockOut serum replacement (Cat. No. 10828-028; Gibco/Thermo Fisher Scientific), 50 ml of heat-inactivated human serum (Cat. No. IPLA-SERAB-HI; Innovative Research, Novi, MI), 5 ml of GlutaMAX supplement (Cat. No. 35050-061; Gibco/Thermo Fisher Scientific), 1 ml of Primocin (Cat. No. ant-pm-2; InvivoGen, San Diego, CA), and recombinant human fibroblast growth factor-2 (rhFGF2) cGMP grade (10 ng/ml) (Cat. No. rhFGF; Waisman Biomanufacturing, Madison, WI). After mincing, tissue fragments were allowed to adhere to 6-well tissue culture-treated plates via air drying, and then cultured in IxMedia with 5% CO2, 20% O2 at 37°C. These cultures were fed every other day with 2 ml of fresh IxMedia per well. For the first two feedings, 1% extracellular matrix (ECM) mixture (human type 1 and type 3 collagen–vitronectin–fibronectin [Advanced BioMatrix, Carlsbad, CA]: collagen type 1 [Cat. No. 5007-20ML], collagen type 3 [Cat. No. 5021-MG], vitronectin [Cat. No. 5051-0.1MG], and fibronectin [Cat. No. 5050-1MG]) was added to the medium. Once cells reached confluence, they were passaged with xeno-free TrypLE Express (Cat. No. 12604-013; Life Technologies/Thermo Fisher Scientific, Grand Island, NY) and used to generate patient-specific iPSCs.
Patient-specific iPSC generation
Patient-specific dermal fibroblasts (250,000) were plated in 1 well of a 6-well culture dish with IxMedia. Twenty-four hours before transduction, medium was switched to xeno-free, serum-free IxMedia. The next day, cells were transduced with nonintegrating Sendai viral vectors16,17,19,23 driving expression of OCT4, SOX2, KLF4, and c-MYC at a multiplicity of infection (MOI) of 3 (CytoTune-iPS reprogramming kit, Cat. No. A16517; Invitrogen/Thermo Fisher Scientific, Waltham, MA) in viral transduction medium (serum-free IxMedia plus 50 μM Y-27632 ROCK inhibitor [Cat. No. 688000; EMD Millipore, Billerica, MA]). Transduction medium was removed 18 hr later and replaced daily with serum-free IxMedia. Five days after transduction, fibroblasts were passaged onto fresh xeno-free rLaminin-521 (human) (LN521)-coated 10-cm culture dishes (Cat. No. 354222; Corning Life Sciences, Tewksbury, MA) and fed with fresh serum-free IxMedia plus RevitaCell (Cat. No. A26445-01; Life Technologies/Thermo Fisher Scientific). On day 6, the medium was transitioned to xeno-free human Essential 6 (Cat. No. A1516401; Life Technologies/Thermo Fisher Scientific), using equal parts Essential 6 and serum-free IxMedia. From day 7 to day 21 cultures were fed daily with fresh Essential 6. On day 21, cultures were transitioned to human Essential 8 iPSC maintenance medium (Cat. No. A1517001; Life Technologies/Thermo Fisher Scientific) and fed daily. When iPSC colonies reached 1–2 mm in diameter they were manually isolated and passaged on fresh 12-well LN521-coated culture plates and clonally expanded in human Essential 8 medium. After expansion, cells were analyzed for pluripotency (i.e., expression of endogenous pluripotency factors) via rt-PCR and loss of transgene expression (i.e., lack of detectable expression of OCT4, SOX2, KLF4, or c-MYC), using a qPCR-based Scorecard assay.24,25
rt-PCR
Total RNA was isolated with an RNeasy mini kit (Cat. No. 74106; Qiagen, Germantown, MD) according to the manufacturer's instructions. One hundred nanograms of RNA template was amplified in one-step rt-PCRs, using the SuperScript III one-step RT-PCR system (Cat. No. 12574018; Life Technologies/Thermo Fisher Scientific) with primers hybridizing to human transcripts (listed in Table 1).
Table 1.
Primer sequences
| Human primer | Sequence |
|---|---|
| c-MYC-For | gctgcttagacgctggattt |
| c-MYC-Rev | agcagctcgaatttcttcca |
| KLF4-For | agaaggatctcggccaattt |
| KLF4-Rev | aagtcgcttcatgtgggaga |
| NANOG-For | ttcttccaccagtcccaaag |
| NANOG-Rev | ttgctccacattggaaggtt |
| OCT4-For | cgggttcaagtgattctcct |
| OCT4-Rev | agcttcctccacccacttct |
| SOX2-For | catcacccacagcaaatgac |
| SOX2-Rev | gcaaacttcctgcaaagctc |
| CLN3ex5-For | acatcctccccacactcgtcatc |
| CLN3ex11-Rev | acaatgtaccacagcagaccc |
| POLR2A-For | taacctgctgcggatgatctggaa |
| POLR2A-Rev | gatgttgaagagcagcgtggcatt |
TaqMan human pluripotent stem cell Scorecard panel
Pluripotency of patient-derived iPSCs was assessed with the TaqMan human pluripotent stem cell Scorecard panel (Cat. Nos. A15870 and A15872; Life Technologies/Thermo Fisher Scientific).24,25 Total RNA was isolated with an RNeasy mini kit (Qiagen) according to the manufacturer's instructions. One microgram of RNA was reverse transcribed with a SuperScript VILO cDNA synthesis kit (Cat. No. 11754050; Life Technologies/Thermo Fisher Scientific). cDNA was added to a human pluripotent stem cell (hPSC) Scorecard plate (Cat. No. A15870; Life Technologies/Thermo Fisher Scientific) and amplified with a QuantStudio 6 flex real-time PCR system with 384-well capability (Life Technologies/Thermo Fisher Scientific). Gene expression data were uploaded to the web-based hPSC Scorecard analysis software (Life Technologies/Thermo Fisher Scientific) for interpretation.
Two-dimensional differentiation of iPSC-derived photoreceptor precursor cells
Patient-derived iPSCs were cultured on ultralow-binding plates (Corning Life Sciences) in embryoid body formation medium (Dulbecco's modified Eagle's medium [DMEM] F-12 [Gibco/Thermo Fisher Scientific], 10% KnockOut serum replacement [Gibco/Thermo Fisher Scientific], 2% B27 supplement [Cat. No. 17504044; Gibco/Thermo Fisher Scientific], 1% N2 supplement [Cell Therapy Systems/Thermo Fisher Scientific], 1% l-glutamine [Cat. No. 25030081; Life Technologies/Thermo Fisher Scientific], 1 × nonessential amino acids [NEAA; Life Technologies], 0.2% Primocin [InvivoGen], 1-ng/ml Dkk-1 [Cat. No. 5439-DK-010; R&D Systems, Minneapolis, MN], 1-ng/ml insulin-like growth factor [IGF]-I [Cat. No. 291-G1-200; R&D Systems], 1-ng/ml Noggin [Cat. No. 6057-NG-100; R&D Systems], and 0.5-ng/ml basic fibroblast growth factor [bFGF] [Cat. No. 233-FB-01M; R&D Systems]) for 4–5 days. Embryoid bodies (200–300 per well) were plated on 6-well plates (Corning Life Sciences) coated with 25-μg/ml collagen (Cat. No. 354265; BD Biosciences, San Jose, CA), 50-μg/ml laminin (Life Technologies/Thermo Fisher Scientific), and 100-μg/ml fibronectin (Sigma-Aldrich, St. Louis, MO) and cultured in differentiation medium 1 (DMEM F-12 [Life Technologies], 2% B27 supplement [Gibco/Thermo Fisher Scientific], 1% N2 supplement [Life Technologies], 1% l-glutamine [Life Technologies/Thermo Fisher Scientific], 1 × NEAA [Life Technologies], 0.2% Primocin [InvivoGen], 10-ng/ml Dkk-1 [R&D Systems], 10-ng/ml IGF-I [R&D Systems], 10-ng/ml Noggin [R&D Systems], and 5-ng/ml bFGF [R&D Systems]). Embryoid bodies were differentiated for 10 days followed by 6 days in differentiation medium 2 (differentiation medium 1 plus 10 μM N-[N-(3,5-difluorophenacetyl-l-alanyl)]-S-phenylglycine t-butyl ester [DAPT; EMD Millipore]) and for an additional 12 days of culture in differentiation medium 3 (differentiation medium 2 plus 2-ng/ml acidic fibroblast growth factor [aFGF] [Cat. No. 232-FA-025; R&D Systems]). Human photoreceptor precursors were cultured for an additional 60 days in differentiation medium 4 (DMEM F-12 [Life Technologies], 2% B27 supplement [Life Technologies], 1% N2 supplement [Life Technologies], 1% l-glutamine [Life Technologies/Thermo Fisher Scientific], 1 × NEAA [Life Technologies], 0.2% Primocin [InvivoGen]).
Cloning of the transgene cassette plasmid carrying CLN3
AAV2, a member of the parvovirus family, is a nonpathogenic, helper-dependent single-stranded DNA virus. The expression cassette contains the minimal cytomegalovirus (CMV) promoter, the CMV immediate-early enhancer, and normal human full-length CLN3 coding sequence. CLN3 cDNA containing the coding sequence for human CLN3 (NM_001042432; IDT-DNA Technologies, Coralville, IA) was cloned into the pFBAAVmcsBgHpA AAV2 plasmid backbone (provided by the University of Iowa Viral Vector Core Facility).26 This plasmid backbone has the following characteristics: (1) it contains ampicillin and gentamicin resistance genes for selection and growth; (2) it contains a bacterial origin of replication (pUC); and (3) it contains inverted terminal repeats from AAV2. The final plasmid was sequence confirmed via bidirectional Sanger sequencing before packaging.
Treatment of patient fibroblasts and 2D iPSC-derived retinal precursor cells with AAV2-CMV-CLN3
Patient-specific iPSCs and imr90 control iPSCs were 2D differentiated17,23 for 30 days. Retinal precursor cells were infected for 16 hr with AAV2-CMV-CLN3 (viral titer, 1.63 × 1013) at an MOI of 10,000. Sixteen hours postinfection, the cells were washed with fresh differentiation medium and fed every other day for 10 days. Ten days postinfection, cells were harvested for Western blot analysis. This same procedure was also performed on patient-specific fibroblasts.
Immunoblotting for CLN3
Western blots were performed as described previously.16,17,20 Briefly, cells were treated with 0.25% trypsin–EDTA (Life Technologies), homogenized in lysis buffer (50 mM Tris-HCl [pH 7.6], 150 mM NaCl, 10 mM CaCl2, 1% Triton X-100, 0.02% NaN3 [Sigma-Aldrich]), and centrifuged. Supernatant protein concentrations were determined via bicinchoninic acid (BCA) assay according to the manufacturer's instructions (Pierce, Rockford, IL). Forty micrograms each was subjected to SDS–PAGE (4–20% Tris–glycine), transferred to polyvinylidene difluoride (PVDF), and probed with anti-CLN3 (diluted 1:1000) (Cat. No. ab75959; Abcam, Cambridge, UK). Alexa Fluor-conjugated secondary antibodies (Life Technologies) were used and blots were visualized fluorescently on a VersaDoc imaging system (Bio-Rad, Hercules, CA). Anti-GAPDH (glyceraldehyde-3-phosphate dehydrogenase, diluted 1:2000) (Cat. No. ab9483; Abcam) antibody was used as an internal loading control.
Clinical-grade, cGMP AAV production
AAV2-CLN3 was manufactured under current Good Manufacturing Practices (cGMP) in the Steven W. Dezii Translational Vision Research Facility (DTVRF) within the Stephen A. Wynn Institute for Vision Research at the University of Iowa. This facility contains an independent high-efficiency particulate air (HEPA)-filtered equipment room, an ISO (International Organization for Standardization) class 7 (class 10,000) material storage and handling room, two ISO class 7 (class 10,000) gowning areas, two ISO class 6 (class 1000) processing rooms, and an unclassified service chase with dedicated access for equipment and facility maintenance. Each of the two processing rooms is equipped with custom BioSpherix Xvivo closed incubation systems, which are designed to exceed ISO class 5 (class 100) cleanliness requirements to ensure minimal biological contamination throughout standard operating procedures, while providing absolute optimal conditions for the long-term culturing of cells under defined atmospheric conditions.
AAV2-CLN3 was manufactured using a characterized human HEK293T master cell line. To package the CLN3 vector in AAV2 particles, HEK293T cells were transfected with a set of constructs encoding (1) normal human CLN3 (pAAV2-CMV-CLN3, which contains the expression cassette flanked by AAV2 ITRs) and (2) AAV (pXX2-R2C2, AAV2 packaging plasmid containing AAV2 rep and cap sequences) and helper virus-derived sequences (pHelper, containing E2A and E4 genes from adenovirus serotype 2), which are required for packaging. Each of these plasmids was sequence confirmed via bidirectional sequencing before packaging in the DTVRF. HEK293T cells taken from a qualified Master Cell Bank (DTVRF HEK293T) were plated and expanded in T600 multilayer tissue culture flasks (production batch scale of 20 flasks). On reaching confluence, cells were simultaneously transfected with pAAV2-CMV-CLN3, pXX2-R2C2, and pHelper. To purify AAV2-CLN3, the following steps were performed: (1) HEK293T cultures were passaged and centrifuged to remove cell culture reagents and low molecular weight impurities; (2) HEK293T cells were lysed to release the intracellular vector, and nuclease digestion was performed to remove nucleic acid impurities; (3) cell debris was removed from the lysate via filtration, using 0.45- and 0.2-μm filters; (4) density gradient ultracentrifugation was performed to separate empty capsids (a major impurity) from the vector product; (5) affinity chromatography was performed using an ÄKTA Pure 25 chromatography system (GE Healthcare Life Sciences, Pittsburgh, PA) to purify vector; (6) viral particles were subsequently concentrated via buffer exchange; and (7) working concentrations of AAV2-CLN3 were formulated in injection buffer (180 mM NaCl plus 10 mM Na3PO4 [trisodium phosphate] in water for injection at pH 7.2), filtered through a 0.2-μm filter, and vialed and labeled.
rt-PCR panel for virus contamination
Purified AAV2-CLN3 was assessed by rt-PCR for viral contaminants of human, simian, bovine, and porcine origin. Briefly, total RNA was isolated with an RNeasy mini kit (Qiagen) according to the manufacturer's instructions. One microgram of RNA was reverse transcribed, using a SuperScript VILO cDNA synthesis kit (Cat. No. 11754050; Life Technologies/Thermo Fisher Scientific). Standard rt-PCR was performed with primers targeting the following viruses or contaminants: Human panel—AAV (adeno-associated virus), EBV (Epstein-Barr virus), CMV (cytomegalovirus), HBV (hepatitis B virus), HHV8 (human herpesvirus 8), HHV6 (human herpesvirus 6), HHV7 (human herpesvirus 7), HTLV1 (human T-lymphotropic virus 1), HTLV2 (human T-lymphotropic virus 2), parvovirus B19, HAV (hepatitis A virus), HCV (hepatitis C virus), and HIV (human immunodeficiency virus)27–33; simian panel—SFV (simian foamy virus), STLV (simian T-cell leukemia virus), SV40 (simian vacuolating virus 40), and SRV (simian retrovirus)34–38; 9CFR panel—BVDV (bovine viral diarrhea virus), MRV1 (mammalian reovirus 1), MRV2 (mammalian reovirus 2), MRV3 (mammalian reovirus 3), and rabies virus39–41; bovine panel—BMaAd (bovine mastadenovirus), BAtAd (bovine atadenovirus), BPV (bovine parvovirus), BTV (bluetongue virus), and BRSV (bovine respiratory syncytial virus)42–44; porcine panel—PAd (porcine adenovirus), PPV (porcine parvovirus), PHE-CoV (porcine hemagglutinating encephalomyelitis coronavirus), and TGEV (transmissible gastroenteritis virus).45–48 gBlocks gene fragments (Integrated DNA Technologies, Coralville, IA) specific to each viral amplicon were designed to serve as positive controls.
Subretinal injection of AAV-CMV-CLN3
All animal procedures were approved by the University of Iowa's Animal Care and Use Committee. Ten C57BL/6J mice (Jackson Laboratory, Bar Harbor, ME) were anesthetized with ketamine–xylazine mixture and injected via the subretinal space as described previously49 at 2 months of age. Right eyes received purified and concentrated clinical-grade cGMP AAV2-CLN3 (109 vector genomes [VG]) in a volume of 1 μl and left eyes received the same dose of AAV2-GFP to serve as a contralateral control. Mice were sacrificed for histology 4 weeks after injection.
rt-PCR of vehicle- and AAV-CMV-CLN3-injected eyes
Ten C57BL/6J mice (Jackson Laboratory) were anesthetized with ketamine–xylazine mixture and injected via the subretinal space as described previously49 at 2 months of age. Right eyes received purified and concentrated clinical-grade cGMP AAV2-CLN3 (109 VG) in a volume of 1 μl and left eyes received sterile injection buffer (180 mM NaCl plus 10 mM Na3PO4 in water for injection at pH 7.2) to serve as a contralateral vehicle control. Mice were sacrificed for rt-PCR for human CLN3 expression 3 weeks postinjection. Briefly, eyes were enucleated and posterior eyecups dissected from the anterior chamber. Whole retinas were then harvested for RNA isolation. Total RNA was isolated with an RNeasy mini kit (Cat. No. 74106; Qiagen) according to the manufacturer's instructions. One hundred nanograms of RNA template was used to generate cDNA, using a SuperScript VILO cDNA synthesis kit (Cat. No. 11754050; Thermo Fisher Scientific). Two hundred nanograms of cDNA was then amplified with primers hybridizing to human CLN3 and POLR2A (listed in Table 1).
Histology of AAV2-CLN3-injected eyes
Mouse eyes were enucleated and fixed in 4% paraformaldehyde (PFA) for 2 hr and then embedded in paraffin. Paraffin-embedded eyes were sectioned, deparaffinized and counterstained with hematoxylin and eosin according to standard procedures.16,17,20 Slides were masked as to whether they were from an AAV2-CLN3- or AAV2-GFP-injected eye. Slides were imaged with an EVOS XL Core microscope (Cat. No. AMEX1000; Life Technologies/Thermo Fisher Scientific) to assess retinas for AAV2-CLN3-mediated retinal toxicity. Retinal toxicity was assessed by measurement of the thickness of the outer nuclear layer, using ImageJ64 software (National Institutes of Health, Bethesda, MA). Ten random images (one from each of 10 different sections) were obtained by a person masked to the treatment groups in each of 10 AAV2-CLN3- or AAV2-GFP-injected eyes, for a total of 100 images per group. Outer nuclear layer thickness was measured in three different areas of each image and averaged for each treatment group. To assess uninjected wild-type retinas, outer nuclear layer thickness was measured in a section from the center of the globe from each of 10 eyes from 10 different C57BL/6J mice. A paired, two-tailed Student t test was performed to determine significance, using Prism 6 (Graphpad Software, La Jolla, CA). p < 0.05 was considered statistically significant.
Results
Generation of patient-specific CLN3-associated JNCL iPSCs
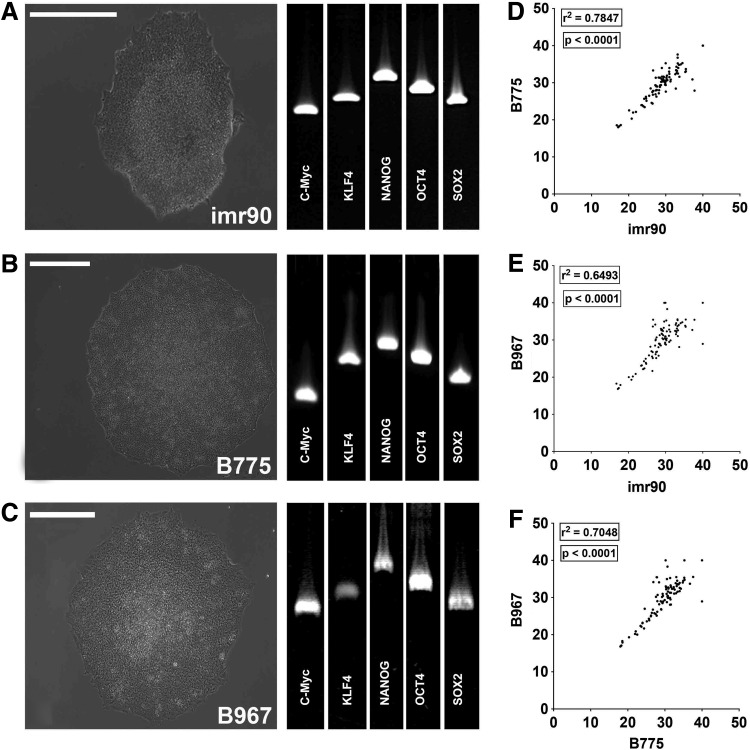
To test the efficacy of virus-mediated correction of CLN3 disease in vitro, we first generated patient-derived iPSCs and iPSC-derived neuronal retinal precursor cells. Skin biopsies were acquired from two patients with molecularly confirmed mutations in CLN3: patient B775 (compound heterozygote; 1.02-kb del/Leu101del3CTC) and patient B967 (homozygote; 1.02-kb del/1.02-kb del). The imr90 (WiCell Research Institute, Madison, WI) human iPSC line was used as a nondisease control. Fibroblasts were isolated, cultured, and reprogrammed as induced pluripotent stem cells (iPSCs) via viral transduction of the transcription factors OCT4, SOX2, KLF4, and c-MYC as previously described.16,17,19,23 Approximately 3 weeks after transduction, densely packed colonies of cells with a high nucleus-to-cytoplasm ratio (Fig. 1A–C) were isolated and clonally expanded. After 10 passages expression of the pluripotency transcripts c-MYC, KLF4, NANOG, OCT4, and SOX2 was confirmed via rt-PCR (Fig. 1A–C). Pluripotency was further assessed with the newly available TaqMan human pluripotent stem cell Scorecard panel.24,25 This assay uses real-time PCR to compare transcript expression levels required for self-renewal, ectoderm, mesoderm, or endoderm formation in lines of interest to an undifferentiated iPSC reference gene set. Pair-wise comparison of all Ct values for genes tested showed that imr90, B775, and B967 were pluripotent and transcriptionally similar to one another as demonstrated by scatter plots and correlation coefficients (Fig. 1D–F; imr90 vs. B775 = 0.7847, p < 0.0001; imr90 vs. B967 = 0.6493, p < 0.0001; B775 vs. B967 = 0.7048, p < 0.0001). Taken together, these data show that we had successfully generated two independent, iPSC lines from patients with CLN3 mutations and a human control iPSC line.
Figure 1.
Generation and characterization of induced pluripotent stem cells (iPSCs) from patients with CLN3-associated juvenile neuronal ceroid lipofuscinosis (JNCL). (A–C) Representative light micrographs of iPSC colonies and rt-PCR for standard pluripotency markers from control iPSC line imr90 (A) and two CLN3 patient lines, B775 (B) and B967 (C). (D and E) Pair-wise comparison of all Ct values for genes tested in each line. Scatter plots are shown comparing each line with one another, with corresponding correlation coefficients and p values: imr90 versus B775 (D; r2 = 0.7847; p < 0.0001), imr90 versus B967 (E; r2 = 0.6493; p < 0.0001), and B775 versus B967 (F; r2 = 0.7048; p < 0.0001). Scale bars: 400 μm.
AAV2-CLN3 restores full-length CLN3 protein in patient-specific iPSC-derived retinal cells
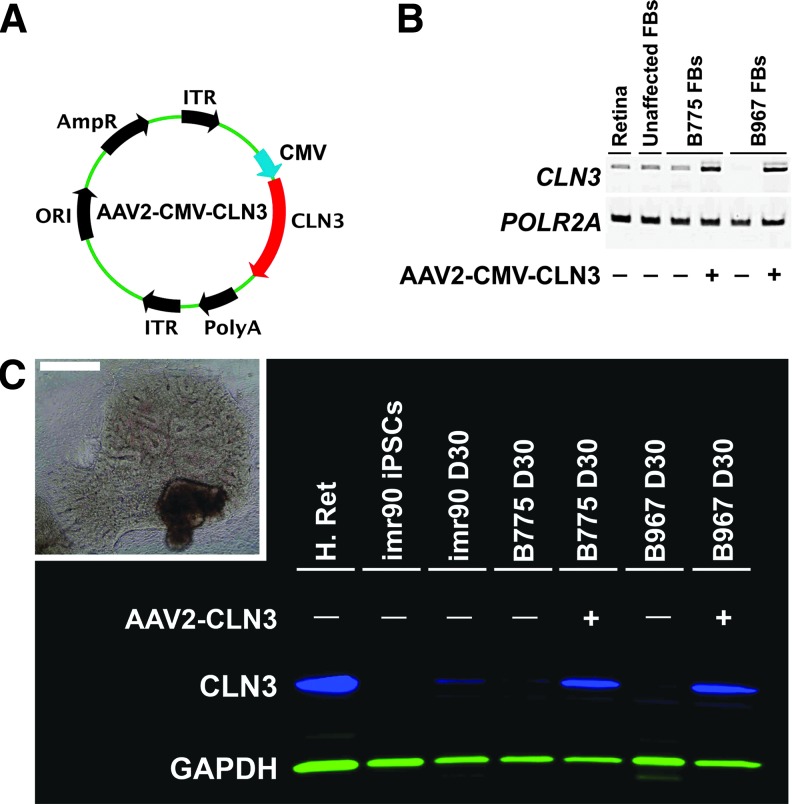
After successfully generating patient-specific iPSCs, we differentiated these lines to develop iPSC-derived retinal cells to test the ability of adeno-associated viral particles carrying wild-type, full-length CLN3 (Table 1) to restore expression of CLN3 protein. For these experiments, we used a two-dimensional differentiation protocol that we have previously demonstrated for the development of patient-specific retinal cells (Fig. 2C, inset).16,17 The AAV we chose for these studies was the serotype 2 AAV vector designed with a CMV promoter driving CLN3 gene expression (Fig. 2A). This AAV vector is the same serotype that has been successfully used in clinical human gene augmentation trials to treat RPE65-associated Leber congenital amaurosis.8,11,50 Initially, to test the ability of this vector to drive CLN3 transcript, we transduced CLN3 patient fibroblasts and performed rt-PCR using primers specific to human CLN3. Little (B775) or no (B967) CLN3 transcript was detected in fibroblasts derived from either patient with JNCL before transduction (Fig. 2B). AAV2-CLN3 was able to drive and restore robust expression of CLN3 transcript in the cells of both patients (Fig. 2B). To test the efficacy of AAV2-CLN3 in restoring full-length CLN3 protein in patient-specific retinal neurons, we differentiated patient iPSCs for 30 days, the time point at which neuronal rosettes are clearly visible in differentiating cultures (Fig. 2C, inset). To assess CLN3 protein levels we produced an immunoblot, using a human anti-CLN3 antibody. CLN3 was robustly expressed in whole retinal protein lysate isolated from a human donor eye, as expected (Fig. 2C, H. Ret). Neurons from an unaffected control patient, imr90, at 30 days postdifferentiation expressed CLN3, whereas undifferentiated iPSCs did not (Fig. 2C, imr90 iPSCs vs. imr90 D30). AAV2-CLN3 restored full-length CLN3 protein to retinal neurons derived from both CLN3 patients, B775 and B967, compared with uninfected differentiated neurons (Fig. 2C). These data demonstrate that we successfully cloned and packaged an AAV vector driving CLN3 and that this vector is efficacious in restoring full-length CLN3 transcript and protein to patient-specific neural progenitor cells.
Figure 2.
Transduction of patient-specific fibroblasts and two-dimensional (2D) iPSC-derived retinal neurons restores expression of CLN3. (A) Vector diagram of AAV2-CLN3 used to transduce patient fibroblasts and iPSC-derived retinal neurons. (B) rt-PCR panel comparing transcripts from human retina (Retina), unaffected human foreskin-derived fibroblasts (Unaffected FBs), and skin fibroblasts from two patients with CLN3-associated disease, B775 (B775 FBs) and B967 (B967 FBs). Results are shown for CLN3 (primers flanking exons 5–12) and the housekeeping gene, POLR2A. Transduction of patient fibroblasts with AAV2-CMV-CLN3 restores CLN3 transcript compared with untransduced fibroblasts. (C) Western blot comparing unaffected human donor retinal lysate (H. Ret), undifferentiated imr90 iPSCs, imr90 cells differentiated for 30 days (imr90 D30), and B775 and B967 patient cells differentiated for 30 days with and without addition of AAV2-CLN3. Transduction of iPSC-derived retinal neurons with AAV2-CLN3 rescued full-length CLN3 protein (blue) in both patient lines. Anti-GAPDH (green) was used as an internal loading control. Inset: A typical neural retina-like structure with retinal pigment epithelium (RPE) pigmentation and numerous neural rosettes. Scale bar: 400 μm. ORI, origin of replication; AmpR, ampicillin resistance cassette; ITR, inverted terminal repeats; CMV, cytomegalovirus promoter; CLN3, full-length CLN3 cDNA; PolyA, polyadenylation signal.
Production of clinical-grade cGMP AAV2-CLN3
After demonstrating that AAV2-CLN3 successfully restores full-length transcript and protein to patient-derived cells, we next sought to produce this vector in a clinical-grade manner suitable for use in clinical trials in children with CLN3 JNCL. To achieve this, we produced AAV2-CLN3 under cGMP conditions in an onsite, FDA-registered cGMP facility. This facility exceeds ISO class 5 (class 100) cleanliness requirements to ensure minimal biological contamination throughout standard operating procedures, while providing optimal conditions for the long-term culturing of cells under defined atmospheric conditions.
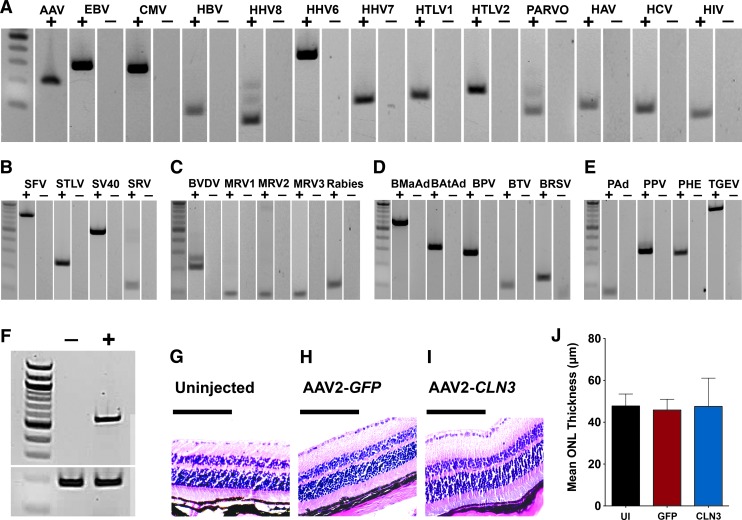
AAV2-CLN3 was manufactured with a characterized human HEK293T master cell line. HEK293T cells were transfected with a transgene cassette plasmid AAV2-CMV-CLN3, an AAV2 packaging plasmid containing AAV2 rep and cap sequences, and a plasmid carrying helper virus-derived sequences (pHelper, containing E2A and E4 genes from adenovirus serotype 2). To purify AAV2-CLN3, HEK293T cultures were passaged and centrifuged to remove cell culture reagents and low molecular weight impurities and lysed and digested to release the intracellular vector and remove nucleic acid impurities. Cellular debris was removed from the lysate via filtration followed by density gradient ultracentrifugation to separate empty capsids (a major impurity) from the vector product. Finally, affinity chromatography was performed to purify the vector and AAV2-CLN3 was subsequently concentrated via buffer exchange. To demonstrate the purity of AAV2-CLN3, we performed a series of rt-PCRs to assess the presence of viruses and vector contaminants. As demonstrated in Fig. 3, AAV2-CLN3 was completely devoid of viral DNA of human (Fig. 3A), simian (Fig. 3B), bovine (Fig. 3D), and porcine origin (Fig. 3E). AAV2-CLN3 also lacked expression of all viruses specified by the code of federal regulations (Fig. 3C).
Figure 3.
Purified AAV2-CLN3 displays a lack of retinal toxicity in wild-type mice. (A–E) rt-PCR for virus panels specific for human (A), simian (B), required code of federal regulations (C), bovine (D), and porcine (E) viruses and contaminants in purified AAV2-CLN3. (F) rt-PCR for human CLN3 (top band) and POLR2A (bottom band) in mouse eyes injected with sterile buffer only (−) or AAV2-CLN3 (+) at 3 weeks postinjection. (G–I) Representative hematoxylin- and eosin-stained paraffin sections from uninjected (G), AAV2-GFP-injected (H), and AAV2-CLN3-injected (I) eyes. (J) Histogram comparing the mean thickness of the outer nuclear layer (ONL) between uninjected (n = 10 eyes), AAV2-GFP-injected (n = 10 eyes), and AAV2-CLN3-injected (n = 10 eyes) mice (AAV2-GFP vs. AAV2-CLN3, p = 0.72). Scale bars: 100 μm.
To demonstrate that subretinal injection of AAV2-CLN3 transduced neural retina, we injected wild-type C57BL/6J mice with either sterile injection buffer (n = 10) or 109 VG of AAV2-CLN3 (n = 10). At 3 weeks postinjection rt-PCR, using primers specific for human CLN3, showed robust expression of CLN3 transcript in AAV2-CLN3-injected retinas as compared with vehicle-injected controls (Fig. 3F). With a highly pure and concentrated AAV2-CLN3 vector in hand, we next asked whether delivery of CLN3, at a viral titer exceeding that which would likely be tested in humans, would be toxic when injected into wild-type murine retinas. We injected wild-type C57BL/6J mice with either 109 VG of AAV2-GFP (n = 10 right eyes) or 109 VG of AAV2-CLN3 (n = 10 left eyes) and assessed retinal histology 2 months postinjection. Uninjected wild-type eyes were used as an additional control to ensure there was no green fluorescent protein (GFP)-induced retinal toxicity (Fig. 3G). Vector-induced toxicity was assessed by measuring and comparing the mean thickness of the outer nuclear layer between treatment groups. Mean outer nuclear layer thickness did not differ between AAV2-GFP- and AAV2-CLN3-injected eyes (Fig. 3J; p = 0.72). Likewise, hematoxylin- and eosin-stained sections were morphologically identical between uninjected (Fig. 3G), AAV2-GFP-injected (Fig. 3H), and AAV2-CLN3-injected eyes (Fig. 3I). Together, these data demonstrate the production of a highly pure, potent, and safe cGMP-compliant clinical-grade AAV2-CLN3 vector.
Discussion
In this study we describe the generation of clinical-grade, cGMP-compliant patient-specific iPSCs from two individuals with CLN3-associated retinal degeneration. We demonstrate the usefulness of these cells for testing the efficacy of a clinical-grade AAV-mediated gene augmentation strategy. Furthermore, we show that AAV2-CLN3 is nontoxic to mouse retinas when delivered in vivo. We propose that these data provide proof-of-concept for the design and initiation of a clinical trial in children with CLN3 JNCL disease.
Nonintegrating AAV2 has been successfully used as a clinical gene transfer vehicle to treat human retinal degenerative disease.10,11,50–53 It has been established that AAV2 is safe, well tolerated, and restricted to the eye after subretinal delivery.11,50,51,53–55 For example, AAV2 carrying the RPE65 gene has been reproducibly demonstrated to be safe and beneficial for the treatment of RPE65-associated Leber congenital amaurosis.10,11,51–54 The safety and efficacy profile of AAV2, coupled with the fact that this vector is capable of carrying full-length CLN3, and has been shown to be capable of transducing both cone photoreceptor and inner retinal neurons,12 makes it a logical choice for the treatment of JNCL.
The retina is a highly specialized extension of the brain derived from the neural ectoderm.56 Like the cerebral cortex, it is an organized, laminated structure, consisting of a diverse mixture of neurons and glia. Similarities between the neural retina and the cerebral cortex suggest that success of a gene augmentation-based approach in the eye could serve as proof-of-principle for a similar treatment in the brain. In vivo animal studies assessing both the efficacy and safety of AAV-mediated gene delivery to the brain have been demonstrated for two forms of NCL: infantile NCL (caused by mutations in the gene PPT1) and late infantile NCL (caused by mutations in the gene TPP1). AAV delivery of TPP1 to the brain has been demonstrated in mice,57–59 rats,60 nonhuman primates,60 and canines.61 Likewise, one group has demonstrated the efficacy of AAV-mediated delivery of PPT1 in murine eyes62 before showing that CNS delivery ameliorated functional deficits in the same Ppt1−/− mouse.63 Importantly, AAV2-mediated treatment of 10 children with TPP1-associated late infantile NCL, administered throughout the CNS, slowed disease progression.64 These studies have laid the groundwork for similar treatment of children with CLN3 disease.
If AAV-mediated therapy is found to be safe and effective in the eye, then delivery of AAV2-CLN3 to the brain would be the next logical step. Although observing efficacy in the eyes of these children would be a great achievement, and would significantly reduce the suffering from this disease, blindness is only one of the problems faced by patients with JNCLs. Development of a treatment that could be used to slow or prevent the cognitive decline, onset of seizures, and loss of motor function associated with JNCL is the ultimate goal. Unlike the genes TPP1 and PPT1, both of which transcribe soluble enzymes,58,63,65–67 CLN3 encodes an intracellular protein that is localized to the membrane of lysosomes. Unfortunately, this means that compared with TPP1- or PPT1-associated NCL, which can be effectively treated by transducing a relatively small number of cells and relying on the secreted gene product to induce a widespread rescue effect, for CLN3 gene augmentation to be effective it is likely that extensive CNS transduction will be necessary. Although AAV2-based transduction of cone photoreceptor cells and inner retinal neurons may be sufficient to prevent vision loss, it is likely that a serotype such as AAV9, which has been demonstrated to be significantly better at transducing CNS neurons,68,69 will be required. Even then, to treat the entire CNS, one would need to deliver a large quantity of viral vector by some means, perhaps by repeated intrathecal injection. Whether a patient could tolerate such treatments, and whether they could ameliorate the condition sufficiently to make it worthwhile, are completely unknown at the present time.
An alternative approach would be to identify systemically active compounds, perhaps even orally deliverable, that would act to slow or halt disease progression. As JNCL has been demonstrated to have an autoimmune component, several groups have attempted to use various immunosuppressive agents for this purpose.70 For instance, Seehafer and colleagues demonstrated that the immunomodulatory compound mycophenolate mofetil (trade name CellCept), when given to Cln3−/− mice, decreased immunoglobulin G deposition, promoted cell survival, and improved motor function.71 A phase 2 clinical trial designed to evaluate the usefulness of this compound in children with CLN3 JNCL is currently underway (clinical trial NCT01399047).70 As promising as these findings are, immunosuppressive agents are less than ideal. They decrease the patient's ability to respond to infections,72 increase the chance of developing cancer,73 and can cause injury to vital organs.74 Although animal models of disease are valuable for testing therapeutic efficacy, large-scale screening for compounds that may be capable of altering disease progression will require in vitro models such as patient-specific iPSCs that recapitulate key disease phenotypes such as autofluorescent lysosomal storage granules.
Many patients with JNCL have retinal disease that has progressed beyond the point at which gene replacement therapy would be beneficial. For these patients, transplantation of iPSC-derived photoreceptor precursors may eventually be able to restore some of their vision. For autologous cell replacement therapy, one would first need to correct the CLN3 mutations to prevent the newly transplanted cells from suffering the same fate as the original ones. This is particularly important for JNCL because of its early onset and aggressive photoreceptor death. Fortunately, advances in CRISPR/Cas9-mediated genome editing now allow the highly specific correction of disease-causing alleles in patient-specific stem cells. As CLN3-associated JNCL is inherited in an autosomal recessive fashion, correction of either of a patient's disease-causing alleles should be sufficient. It is advantageous that the most common mutation in CLN3, a 1-kb genomic deletion, occurs in approximately 85% of patients.4 For example, the two patients investigated in this study each harbor this mutation: one heterozygously and the other homozygously. Designing a strategy to correct this variant would allow for the production of healthy, immunologically matched photoreceptor precursor cells for use in cell replacement therapy.
In conclusion, we have demonstrated the development of a clinical-grade AAV2 vector that is capable of restoring full-length CLN3 transcript and protein to retinal cells derived from two patients with CLN3-associated JNCL. Furthermore, AAV2-CLN3 is safe in the eyes of wild-type mice, displaying a complete lack of vector-induced overexpression toxicity. This study provides preclinical data for the use of AAV2-CLN3 to treat retinal degenerative blindness caused by mutations in the gene CLN3.
Acknowledgments
The authors thank the patients for their participation in this research study. This work was supported by grants from the Stephen A. Wynn Foundation, the Elmer and Sylvia Sramek Charitable Foundation, the National Institutes of Health (1-DP2-OD007483-01, EY-024605, and EY-024588), Research to Prevent Blindness, and the Foundation Fighting Blindness.
Author Disclosure
No competing financial interests exist.
References
- 1.Mole SE, Cotman SL. Genetics of the neuronal ceroid lipofuscinoses (Batten disease). Biochim Biophys Acta 2015;1852:2237–2241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kmoch S, Stránecký V, Emes RD, et al. Bioinformatic perspectives in the neuronal ceroid lipofuscinoses. Biochim Biophys Acta 2013;1832:1831–1841 [DOI] [PubMed] [Google Scholar]
- 3.Drack AV, Miller JN, Pearce DA. A Novel c.1135_1138delCTGT mutation in CLN3 leads to juvenile neuronal ceroid lipofuscinosis. J Child Neurol 2013;28:1112–1116 [DOI] [PubMed] [Google Scholar]
- 4.de los Reyes E, Dyken PR, Phillips P, et al. Profound infantile neuroretinal dysfunction in a heterozygote for the CLN3 genetic defect. J Child Neurol 2004;19:42–46 [DOI] [PubMed] [Google Scholar]
- 5.Margraf LR, Boriack RL, Routheut AA, et al. Tissue expression and subcellular localization of CLN3, the Batten disease protein. Mol Genet Metab 1999;66:283–289 [DOI] [PubMed] [Google Scholar]
- 6.Mao Q, Xia H, Davidson BL. Intracellular trafficking of CLN3, the protein underlying the childhood neurodegenerative disease, Batten disease. FEBS Lett 2003;555:351–357 [DOI] [PubMed] [Google Scholar]
- 7.Gardiner M, Sandford A, Deadman M, et al. Batten disease (Spielmeyer-Vogt disease, juvenile onset neuronal ceroid-lipofuscinosis) gene (CLN3) maps to human chromosome 16. Genomics 1990;8:387–390 [DOI] [PubMed] [Google Scholar]
- 8.Maguire AM, High KA, Auricchio A, et al. Age-dependent effects of RPE65 gene therapy for Leber's congenital amaurosis: a phase 1 dose-escalation trial. Lancet 2009;374:1597–1605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stein L, Roy K, Lei L, et al. Clinical gene therapy for the treatment of RPE65-associated Leber congenital amaurosis. Expert Opin Biol Ther 2011;11:429–439 [DOI] [PubMed] [Google Scholar]
- 10.Jacobson SG, Cideciyan AV, Ratnakaram R, et al. Gene therapy for Leber congenital amaurosis caused by RPE65 mutations: safety and efficacy in 15 children and adults followed up to 3 years. Arch Ophthalmol 2012;130:9–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maguire AM, Simonelli F, Pierce EA, et al. Safety and efficacy of gene transfer for Leber's congenital amaurosis. N Engl J Med 2008;358:2240–2248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Watanabe S, Sanuki R, Ueno S, et al. Tropisms of AAV for subretinal delivery to the neonatal mouse retina and its application for in vivo rescue of developmental photoreceptor disorders. PLoS One 2013;8:e54146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jin Z-B, Okamoto S, Osakada F, et al. Modeling retinal degeneration using patient-specific induced pluripotent stem cells. PLoS One 2011;6:e17084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Singh R, Shen W, Kuai D, et al. iPS cell modeling of Best disease: insights into the pathophysiology of an inherited macular degeneration. Hum Mol Genet 2013;22:593–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Phillips MJ, Perez ET, Martin JM, et al. Modeling human retinal development with patient-specific induced pluripotent stem cells reveals multiple roles for visual system homeobox 2. Stem Cells 2014;32:1480–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tucker BA, Scheetz TE, Mullins RF, et al. Exome sequencing and analysis of induced pluripotent stem cells identify the cilia-related gene male germ cell-associated kinase (MAK) as a cause of retinitis pigmentosa. Proc Natl Acad Sci USA 2011;108:E569–E576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tucker BA, Mullins RF, Streb LM, et al. Patient-specific iPSC-derived photoreceptor precursor cells as a means to investigate retinitis pigmentosa. Elife 2013;2:e00824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tucker BA, Cranston C, Anfinson KR, et al. Using patient specific iPSCs to interrogate the pathogenicity of a novel RPE65 cryptic splice site mutation and confirm eligibility for enrollment into a clinical gene augmentation trial. Transl Res 2015;166:740–749.e1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Small KW, DeLuca AP, Whitmore SS, et al. North Carolina macular dystrophy is caused by dysregulation of the retinal transcription factor PRDM13. Ophthalmology 2015;123:9–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burnight ER, Wiley LA, Drack AV, et al. CEP290 gene transfer rescues Leber congenital amaurosis cellular phenotype. Gene Ther 2014;21:662–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cereso N, Pequignot MO, Robert L, et al. Proof of concept for AAV2/5-mediated gene therapy in iPSC-derived retinal pigment epithelium of a choroideremia patient. Mol Ther Methods Clin Dev 2014;1:14011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vasireddy V, Mills JA, Gaddameedi R, et al. AAV-mediated gene therapy for choroideremia: preclinical studies in personalized models. PLoS One 2013;8:e61396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tucker BA, Anfinson KR, Mullins RF, et al. Use of a synthetic xeno-free culture substrate for induced pluripotent stem cell induction and retinal differentiation. Stem Cells Transl Med 2013;2:16–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsankov AM, Akopian V, Pop R, et al. A qPCR ScoreCard quantifies the differentiation potential of human pluripotent stem cells. Nat Biotechnol 2015;33:1182–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fergus J, Quintanilla R, Lakshmipathy U. Characterizing pluripotent stem cells using the TaqMan hPSC Scorecard panel. Methods Mol Biol 2016;1307:25–37 [DOI] [PubMed] [Google Scholar]
- 26.Urabe M, Ding C, Kotin RM. Insect cells as a factory to produce adeno-associated virus type 2 vectors. Hum Gene Ther 2002;13:1935–1943 [DOI] [PubMed] [Google Scholar]
- 27.Engelmann I, Petzold DR, Kosinska A, et al. Rapid quantitative PCR assays for the simultaneous detection of herpes simplex virus, varicella zoster virus, cytomegalovirus, Epstein-Barr virus, and human herpesvirus 6 DNA in blood and other clinical specimens. J Med Virol 2008;80:467–477 [DOI] [PubMed] [Google Scholar]
- 28.Kodani M, Mixson-Hayden T, Drobeniuc J, et al. Rapid and sensitive approach to simultaneous detection of genomes of hepatitis A, B, C, D and E viruses. J Clin Virol 2014;61:260–264 [DOI] [PubMed] [Google Scholar]
- 29.Watzinger F, Suda M, Preuner S, et al. Real-time quantitative PCR assays for detection and monitoring of pathogenic human viruses in immunosuppressed pediatric patients. J Clin Microbiol 2004;42:5189–5198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lawrence DC, Stover CC, Noznitsky J, et al. Structure of the intact stem and bulge of HIV-1 Psi-RNA stem-loop SL1. J Mol Biol 2003;326:529–542 [DOI] [PubMed] [Google Scholar]
- 31.Styer LM, Miller TT, Parker MM. Validation and clinical use of a sensitive HIV-2 viral load assay that uses a whole virus internal control. J Clin Virol 2013;58(Suppl 1):e127–e133 [DOI] [PubMed] [Google Scholar]
- 32.Pripuzova N, Wang R, Tsai S, et al. Development of real-time PCR array for simultaneous detection of eight human blood-borne viral pathogens. PLoS One 2012;7:e43246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Costa C, Terlizzi ME, Solidoro P, et al. Detection of parvovirus B19 in the lower respiratory tract. J Clin Virol 2009;46:150–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Switzer WM, Bhullar V, Shanmugam V, et al. Frequent simian foamy virus infection in persons occupationally exposed to nonhuman primates. J Virol 2004;78:2780–2789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schweizer M, Neumann-Haefelin D. Phylogenetic analysis of primate foamy viruses by comparison of pol sequences. Virology 1995;207:577–582 [DOI] [PubMed] [Google Scholar]
- 36.White JA, Todd PA, Rosenthal AN, et al. Development of a generic real-time PCR assay for simultaneous detection of proviral DNA of simian betaretrovirus serotypes 1, 2, 3, 4 and 5 and secondary uniplex assays for specific serotype identification. J Virol Methods 2009;162:148–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van Dooren S, Shanmugam V, Bhullar V, et al. Identification in gelada baboons (Theropithecus gelada) of a distinct simian T-cell lymphotropic virus type 3 with a broad range of Western blot reactivity. J Gen Virol 2004;85:507–519 [DOI] [PubMed] [Google Scholar]
- 38.Fagrouch Z, Karremans K, Deuzing I, et al. Molecular analysis of a novel simian virus 40 (SV40) type in rhesus macaques and evidence for double infections with the classical SV40 type. J Clin Microbiol 2011;49:1280–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wernike K, Hoffmann B, Beer M. Simultaneous detection of five notifiable viral diseases of cattle by single-tube multiplex real-time RT-PCR. J Virol Methods 2015;217:28–35 [DOI] [PubMed] [Google Scholar]
- 40.Gallagher EM, Margolin AB. Development of an integrated cell culture—real-time RT-PCR assay for detection of reovirus in biosolids. J Virol Methods 2007;139:195–202 [DOI] [PubMed] [Google Scholar]
- 41.Wacharapluesadee S, Tepsumethanon V, Supavonwong P, et al. Detection of rabies viral RNA by TaqMan real-time RT-PCR using non-neural specimens from dogs infected with rabies virus. J Virol Methods 2012;184:109–112 [DOI] [PubMed] [Google Scholar]
- 42.Sibley SD, Goldberg TL, Pedersen JA. Detection of known and novel adenoviruses in cattle wastes via broad-spectrum primers. Appl Environ Microbiol 2011;77:5001–5008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bae JE, Kim IS. Multiplex PCR for rapid detection of minute virus of mice, bovine parvovirus, and bovine herpesvirus during the manufacture of cell culture-derived biopharmaceuticals. Biotechnol Bioproc Eng 2011;15:1031–1037 [Google Scholar]
- 44.Thonur L, Maley M, Gilray J, et al. One-step multiplex real time RT-PCR for the detection of bovine respiratory syncytial virus, bovine herpesvirus 1 and bovine parainfluenza virus 3. BMC Vet Res 2012;8:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hundesa A, Maluquer de Motes C, Albinana-Gimenez N, et al. Development of a qPCR assay for the quantification of porcine adenoviruses as an MST tool for swine fecal contamination in the environment. J Virol Methods 2009;158:130–135 [DOI] [PubMed] [Google Scholar]
- 46.Zeng Z, Liu Z, Wang W, et al. Establishment and application of a multiplex PCR for rapid and simultaneous detection of six viruses in swine. J Virol Methods 2014;208:102–106 [DOI] [PubMed] [Google Scholar]
- 47.Zhao J, Shi B-J, Huang X-G, et al. A multiplex RT-PCR assay for rapid and differential diagnosis of four porcine diarrhea associated viruses in field samples from pig farms in East China from 2010 to 2012. J Virol Methods 2013;194:107–112 [DOI] [PubMed] [Google Scholar]
- 48.Quiroga MA, Cappuccio J, Piñeyro P, et al. Hemagglutinating encephalomyelitis coronavirus infection in pigs, Argentina. Emerging Infect Dis 2008;14:484–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Seo S, Mullins RF, Dumitrescu AV, et al. Subretinal gene therapy of mice with bardet-biedl syndrome type 1. Invest Ophthalmol Vis Sci 2013;54:6118–6132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jacobson SG, Acland GM, Aguirre GD, et al. Safety of recombinant adeno-associated virus type 2-RPE65 vector delivered by ocular subretinal injection. Mol Ther 2006;13:1074–1084 [DOI] [PubMed] [Google Scholar]
- 51.Simonelli F, Maguire AM, Testa F, et al. Gene therapy for Leber's congenital amaurosis is safe and effective through 1.5 years after vector administration. Mol Ther 2010;18:643–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bennett J, Ashtari M, Wellman J, et al. AAV2 gene therapy readministration in three adults with congenital blindness. Sci Transl Med 2012;4:120ra15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Testa F, Maguire AM, Rossi S, et al. Three-year follow-up after unilateral subretinal delivery of adeno-associated virus in patients with Leber congenital amaurosis type 2. Ophthalmology 2013;120:1283–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jacobson SG, Boye SL, Aleman TS, et al. Safety in nonhuman primates of ocular AAV2-RPE65, a candidate treatment for blindness in Leber congenital amaurosis. Hum Gene Ther 2009;17:845–858 [DOI] [PubMed] [Google Scholar]
- 55.Barker SE, Broderick CA, Robbie SJ, et al. Subretinal delivery of adeno-associated virus serotype 2 results in minimal immune responses that allow repeat vector administration in immunocompetent mice. J Gene Med 2009;11:486–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Heavner W, Pevny L. Eye development and retinogenesis. Cold Spring Harb Perspect Biol 2012;4:a008391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Haskell RE, Hughes SM, Chiorini JA, et al. Viral-mediated delivery of the late-infantile neuronal ceroid lipofuscinosis gene, TPP-I to the mouse central nervous system. Gene Ther 2003;10:34–42 [DOI] [PubMed] [Google Scholar]
- 58.Passini MA, Dodge JC, Bu J, et al. Intracranial delivery of CLN2 reduces brain pathology in a mouse model of classical late infantile neuronal ceroid lipofuscinosis. J Neurosci 2006;26:1334–1342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sondhi D, Hackett NR, Peterson DA, et al. Enhanced survival of the LINCL mouse following CLN2 gene transfer using the rh.10 rhesus macaque-derived adeno-associated virus vector. Mol Ther 2007;15:481–491 [DOI] [PubMed] [Google Scholar]
- 60.Crystal RG, Sondhi D, Hackett NR, et al. Clinical protocol. Administration of a replication-deficient adeno-associated virus gene transfer vector expressing the human CLN2 cDNA to the brain of children with late infantile neuronal ceroid lipofuscinosis. Hum Gene Ther 2004;15:1131–1154 [DOI] [PubMed] [Google Scholar]
- 61.Katz ML, Tecedor L, Chen Y, et al. AAV gene transfer delays disease onset in a TPP1-deficient canine model of the late infantile form of Batten disease. Sci Transl Med 2015;7:313ra180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Griffey M, Macauley SL, Ogilvie JM, et al. AAV2-mediated ocular gene therapy for infantile neuronal ceroid lipofuscinosis. Mol Ther 2005;12:413–421 [DOI] [PubMed] [Google Scholar]
- 63.Griffey MA, Wozniak D, Wong M, et al. CNS-directed AAV2-mediated gene therapy ameliorates functional deficits in a murine model of infantile neuronal ceroid lipofuscinosis. Mol Ther 2006;13:538–547 [DOI] [PubMed] [Google Scholar]
- 64.Worgall S, Sondhi D, Hackett NR, et al. Treatment of late infantile neuronal ceroid lipofuscinosis by CNS administration of a serotype 2 adeno-associated virus expressing CLN2 cDNA. Hum Gene Ther 2008;19:463–474 [DOI] [PubMed] [Google Scholar]
- 65.Chang M, Cooper JD, Sleat DE, et al. Intraventricular enzyme replacement improves disease phenotypes in a mouse model of late infantile neuronal ceroid lipofuscinosis. Mol Ther 2008;16:649–656 [DOI] [PubMed] [Google Scholar]
- 66.Lu J-Y, Hu J, Hofmann SL. Human recombinant palmitoyl-protein thioesterase-1 (PPT1) for preclinical evaluation of enzyme replacement therapy for infantile neuronal ceroid lipofuscinosis. Mol Genet Metab 2010;99:374–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Griffey M, Bible E, Vogler C, et al. Adeno-associated virus 2-mediated gene therapy decreases autofluorescent storage material and increases brain mass in a murine model of infantile neuronal ceroid lipofuscinosis. Neurobiol Dis 2004;16:360–369 [DOI] [PubMed] [Google Scholar]
- 68.Jakovcevski M, Guo Y, Su Q, et al. rAAV9—a human-derived adeno-associated virus vector for efficient transgene expression in mouse cingulate cortex. Cold Spring Harb Protoc 2010;4.:pdb.prot5417. [DOI] [PubMed] [Google Scholar]
- 69.Gray SJ, Nagabhushan Kalburgi S, McCow TJ, et al. Global CNS gene delivery and evasion of anti-AAV-neutralizing antibodies by intrathecal AAV administration in non-human primates. Gene Ther 2013;20:450–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Drack AV, Mullins RF, Pfeifer WL, et al. Immunosuppressive treatment for retinal degeneration in juvenile neuronal ceroid lipofuscinosis (juvenile Batten disease). Ophthalmic Genet 2015;36:359–364 [DOI] [PubMed] [Google Scholar]
- 71.Seehafer SS, Ramirez-Montealegre D, Wong AM, et al. Immunosuppression alters disease severity in juvenile Batten disease mice. J Neuroimmunol 2011;230:169–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Simon DM, Levin S. Infectious complications of solid organ transplantations. Infect Dis Clin North Am 2001;15:521–549 [DOI] [PubMed] [Google Scholar]
- 73.Kasiske BL, Snyder JJ, Gilbertson DT, et al. Cancer after kidney transplantation in the United States. Am J Transplant 2004;4:905–913 [DOI] [PubMed] [Google Scholar]
- 74.Briggs JD. Causes of death after renal transplantation. Nephrol Dial Transplant 2001;16:1545–1549 [DOI] [PubMed] [Google Scholar]