Abstract
Some symbiotic bacteria are capable of interfering with host reproduction in selfish ways. How such bacteria can manipulate host's sex-related mechanisms is of fundamental interest encompassing cell, developmental and evolutionary biology. Here, we uncover the molecular and cellular mechanisms underlying Spiroplasma-induced embryonic male lethality in Drosophila melanogaster. Transcriptomic analysis reveals that many genes related to DNA damage and apoptosis are up-regulated specifically in infected male embryos. Detailed genetic and cytological analyses demonstrate that male-killing Spiroplasma causes DNA damage on the male X chromosome interacting with the male-specific lethal (MSL) complex. The damaged male X chromosome exhibits a chromatin bridge during mitosis, and bridge breakage triggers sex-specific abnormal apoptosis via p53-dependent pathways. Notably, the MSL complex is not only necessary but also sufficient for this cytotoxic process. These results highlight symbiont's sophisticated strategy to target host's sex chromosome and recruit host's molecular cascades toward massive apoptosis in a sex-specific manner.
 Symbiotic bacteria are able to interfere with host reproduction in ways that are detrimental to the host organism. Here the authors show that Spiroplasma induces DNA damage on the male X chromosome in Drosophila, causing sex-specific apoptosis.
Symbiotic bacteria are able to interfere with host reproduction in ways that are detrimental to the host organism. Here the authors show that Spiroplasma induces DNA damage on the male X chromosome in Drosophila, causing sex-specific apoptosis.
The process, mechanism and origin of sex determination have been focal topics in genetics, cell biology, developmental biology and evolutionary biology1,2,3. Sex determination systems are strikingly diverse across animals, plants, fungi, protists and others, of which molecular mechanisms of sex determination and subsequent dosage compensation have been best documented for several model animals including fruit fly, nematode and mouse3.
In the fruit fly Drosophila melanogaster, a female-specific developmental switch gene, Sex lethal (Sxl), counts autosome/sex chromosome ratio in an early developmental stage to establish the choice between male and female alternative developmental pathways at the cellular level. Downstream of Sxl, a cascade of regulatory genes branches into several major pathways, which respectively control sexual differentiation of the soma and neural cells, development of the germ line, and dosage compensation for equalizing X chromosomal transcript levels between males with XY chromosomes and females with XX chromosomes4. Dosage compensation is mediated by a ribonucleoprotein complex, designated as the male-specific lethal (MSL) complex, consisting of at least five proteins (MSL1, MSL2, MSL3, MLE (Maleless) and MOF (Males absent on the first)) and two non-coding RNAs (roX1 and roX2), which concentrates on the single male X chromosome and up-regulates its transcriptional level approximately twofold5.
Diverse insects and other animals, including Drosophila species, are commonly associated with symbiotic bacteria6,7. These microbial associates substantially influence their host's biology in a variety of ways. Some symbionts like Wolbachia, Spiroplasma, Cardinium and Arsenophonus cause striking reproductive phenotypes such as cytoplasmic incompatibility, male-killing, parthenogenesis and feminization, whereby these symbionts drive their own infection to spread into their host populations in selfish ways8,9.
How these microbes interfere with host's reproduction and development is of fundamental interest, but the mechanisms have been poorly understood9,10. Previous studies provided some clues to the enigma in that symbionts are able to interact with a variety of eukaryotic molecular and cellular components including microtubules/centrosomes/mitotic spindles11,12,13,14,15,16, paternal chromosomes17,18,19,20 and somatic- and germline-stem cell niches21,22. As for male-killing Spiroplasma of Drosophila species, msl mutant hosts fail to express male-killing23 and the infection alters the localization of the MSL complex24, suggesting the involvement of the dosage compensation system, and infected male embryos suffer massive apoptosis25,26 and neural malformation26,27,28. In Ostrinia moths, male-killing Wolbachia was reported to suppress host's masculinizing gene expression, thereby disturbing dosage compensation in male embryos29. In the light of these previous works, however, the processes as to how the symbiont's interactions with the host's molecular and cellular components are causally connected to host's reproductive phenotypes are elusive.
In this study, by making use of ample genetic tools and resources available for Drosophila in combination with sophisticated cytological, molecular and genomic techniques, we demonstrate a number of previously unrecognized molecular and cellular aspects of Spiroplasma-induced male-killing, which provide an integrative understanding of mechanisms underlying the symbiont's reproductive manipulation at the molecular, chromosomal, cellular and organismal levels.
Results
Transcriptomic analysis of infected and uninfected embryos
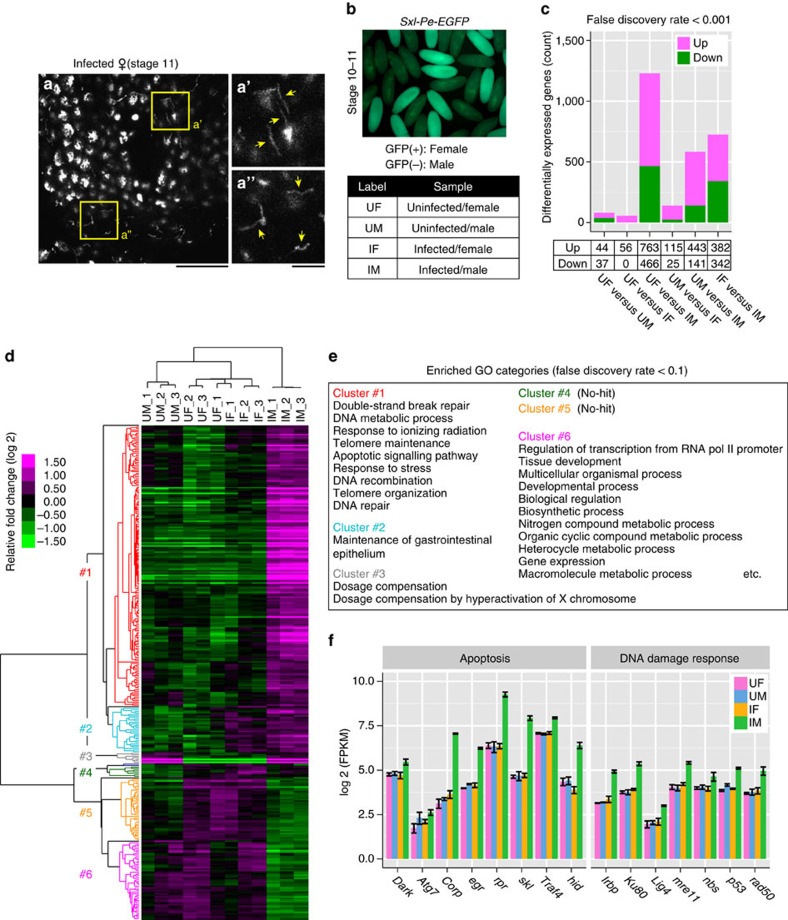
We collected Spiroplasma-infected and uninfected Drosophila embryos of both sexes at stage 10–11 when infection-associated male-specific abnormal apoptosis starts26 (Fig. 1a). For embryonic sexing, we used a transgenic strain with green fluorescence protein (GFP) reporter of Sxl gene, Sxl-Pe-EGFP, which expresses GFP only in females (Fig. 1b and Supplementary Fig. 1a). Four groups of pooled embryos (uninfected females, uninfected males, infected females and infected males; three replicates for each group) were subjected to RNA-sequencing (RNA-seq) analysis. Of all the genes annotated in the Drosophila genome, 8,387 genes were substantially expressed in the embryos (Supplementary Methods), of which we identified 1,430 differentially expressed genes by all pairwise comparisons between the groups (false discovery rate<0.001). Notably, more differentially expressed genes were associated with infected male embryos than other groups (Fig. 1c and Supplementary Fig. 1b). In infected male embryos, up-regulated genes were concentrated on the second and third chromosomes, whereas down-regulated genes were preferentially found on the X chromosome (Supplementary Fig. 1c). At a glance, this pattern may look like reflecting dosage compensation defects in infected male embryos. However, comparison with uninfected male embryos revealed only a small number of down-regulated X-encoded genes in infected male embryos (51 of 1,447 analyzed genes encoded on the X chromosome) (Supplementary Fig. 1c), suggesting that dosage compensation is still functioning in infected male embryos.
Figure 1. RNA-seq analysis of Spiroplasma-infected and uninfected embryos.
(a) Epithelial cells of an infected female embryo at stage 11. Three DNA-stained z-sections are projected to show the surface of the epithelium. Boxed regions in a are magnified in a′ and a′′ to highlight Spiroplasma cells (arrows). Scale bars, 20 μm (a) and 5 μm (a′,a′′). (b) Expression of Sxl-Pe-EGFP in uninfected embryos at stage 10–11. Only female embryos express GFP. Bottom panel shows abbreviations for RNA-seq samples. (c) The number of differentially expressed genes identified in pairwise comparisons between RNA-seq samples. In UF (uninfected female) versus UM (uninfected male), for example, numbers of differently expressed genes up- or down-regulated in UM compared with UF are plotted (the same applies hereafter). (d) A heat map of selected 320 differentially expressed genes. On the top is a clustering dendrogram of RNA-seq samples based on similarity of gene expression patterns across the samples. On the left is a clustering dendrogram of differentially expressed genes based on similarity of gene expression patterns across the genes, wherein gene clusters #1–#6 are depicted by different colours. (e) GO categories enriched in gene clusters #1–#6. (f) Expression levels (fragments per kilobase per million, FPKM) of major genes related to apoptosis (left) and DNA damage response (right) categorized to the gene cluster #1, represented as mean±s.d. of three independent experiments.
Categorization of differentially expressed genes
Of the 1,430 differentially expressed genes, 320 genes exhibiting at least twofold up- or down-regulation were selected and further analyzed. Hierarchical clustering grouped 314 genes into 6 clusters, whereas 6 genes were left ungrouped (Fig. 1d and Supplementary Data 1). Gene ontology (GO) analysis of these 6 clusters revealed that genes related to ‘apoptosis' and ‘DNA damage' were highly up-regulated in infected male embryos (cluster #1, 181/320=56.6%; Fig. 1e,f and Supplementary Data 1). These results are concordant with previous reports on the occurrence of abnormal apoptosis25,26, and notably, indicative of a high level of DNA damage in infected male embryos. On the other hand, genes related to ‘maintenance of gastrointestinal epithelium' were up-regulated in response to Spiroplasma infection irrespective of sex (cluster #2, 29/320=9.1%; Fig. 1e and Supplementary Data 1). Among them, unpaired 1 (upd1) and upd2 are ligands in the JAK-STAT (Janus kinase-signal transducers and activators of transcription) pathway, which are involved in host's survival upon intestinal-bacterial infection30,31,32. Furthermore, although not highlighted in the GO enrichment analysis, several genes related to ‘detoxification', ‘host defence' and ‘stress response' were also identified (GstE5, GstE9, Drsl5 and proPO45), which may reflect the general effects of Spiroplasma infection on host's physiology. Strikingly, no genes constituting the Toll and Imd (Immune deficiency) pathways were assigned to this cluster, which is in accordance with previous observations that Spiroplasma infection does not induce host's innate immune responses by evading host's recognition, presumably due to the absence of cell wall33,34,35. In male embryos irrespective of infection, ‘dosage compensation' related genes were up-regulated, though small in number (cluster #3, 7/320=2.2%; Fig. 1e and Supplementary Data 1), confirming that embryonic sexing by the Sxl-Pe-EGFP transgene worked well (Supplementary Fig. 1a). Spiroplasma infection in male embryos were also associated with down-regulation of miscellaneous genes such as transcription, development, metabolism and so on (cluster #6, 50/320=15.6%; Fig. 1e and Supplementary Data 1), likely reflecting systemic attenuation of gene expression in the infected male embryos that exhibit developmental arrest leading to death. In the remaining two clusters (cluster #4, 7/320=2.2%; cluster #5, 40/320=12.5%), no GO terms were enriched and we could not find any distinctive features (Fig. 1e and Supplementary Data 1).
DNA damage induces apoptosis in infected male embryos
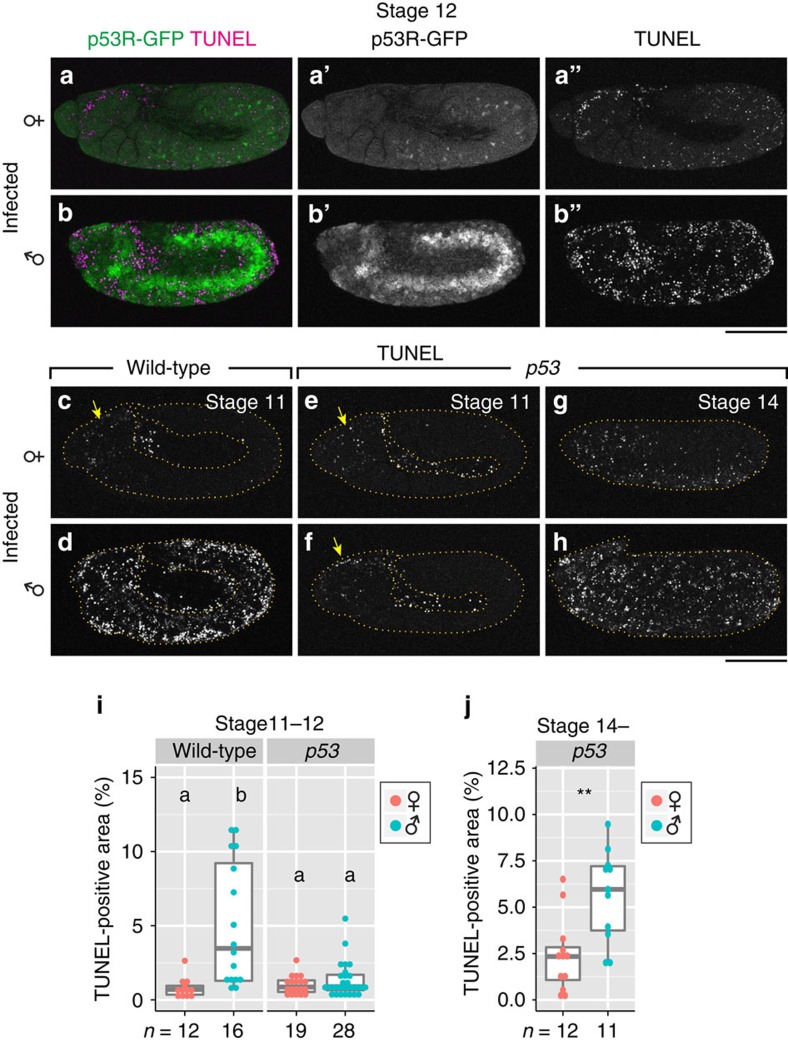
DNA damage is caused by a variety of genotoxic stresses including ionizing radiation, UV, chemicals, reactive oxygen species and replication stresses like stalling or delaying of replication fork progression. In response to DNA damage, a well-known tumour suppressor gene p53 is activated, which triggers an assemblage of p53-dependent pathways to control cell cycle, DNA repair and apoptosis36. Our analysis using a fly strain with p53-responsive GFP reporter (p53R-GFP) revealed that p53 was strongly activated in infected male embryos (Fig. 2a,b). The Drosophila genome encodes a single p53 family member, which is required for DNA damage-induced apoptosis37,38,39. Using a null allele of p53, we demonstrated that abnormal apoptosis in infected male embryos was significantly suppressed at stage 11–12 (Fig. 2c–f,i). On the other hand, developmental apoptosis prominent around the head region at these stages26,40 was not affected (Fig. 2c,e,f, arrows). Upon severe DNA damage such as double-strand breaks caused by ionizing radiation, apoptosis is induced in a time-delayed manner even when p53 is absent41,42. Concordantly, p53-independent apoptosis was observed in infected male embryos from stage 14 onward (Fig. 2g,h,j). Taken together, these results strongly suggest that cells of Spiroplasma-infected male embryos suffer DNA damage, and then abnormal apoptosis is triggered via p53-dependent pathways.
Figure 2. p53-dependent apoptosis in Spiroplasma-infected male embryos.
(a) p53R-GFP expression (green) and TUNEL staining (magenta) in an infected female embryo at stage 12 (n=12). Single-channel images are shown in a′ and a′′. (b) An image similar to a of an infected male embryo (n=10), wherein high p53 activity and massive apoptosis are seen. Single-channel images are shown in b′ and b′′. (c,d) TUNEL staining of infected female and male wild-type embryos at stage 11. The yellow arrow denotes developmental apoptosis in the head region. (e,f) Images similar to c,d of infected female and male embryos mutant for p53. (g,h) TUNEL staining of infected female and male embryos mutant for p53 at stage 14. In c–h, the edges of embryonic epidermis are depicted by dashed yellow lines. (i) Quantification of TUNEL-positive areas in infected female and male embryos, wild type and mutant for p53 at stage 11–12. Different letters (a,b) indicate statistically significant differences (P<0.01; Kruskal–Wallis test followed by Mann–Whitney U-tests). (j) Quantification of TUNEL-positive areas in infected female and male embryos mutant for p53 at stage 14 onward. Asterisks indicate a statistically significant difference (**, P<0.01; Mann–Whitney U-test). In i and j, box plots indicate the median (bold line), the 25th and 75th percentiles (box edges), and the range (whiskers). Sample sizes are shown at the bottom. Scale bars, 100 μm.
Differential detection of apoptosis and DNA damage
Previous studies have established that, in response to DNA damage such as double-strand breaks and replication stress, a minor variant of histone H2A, called H2AX, is phosphorylated within the nucleus to form discrete foci, which are known as H2AX foci43. In Drosophila, an H2AX homologue, H2Av, has been reported to be phosphorylated following exposure to DNA damage44 (Supplementary Fig. 2a,b). When Spiroplasma-infected male embryos were stained with an antibody against phosphorylated form of H2Av (pH2Av), two types of signals were detected: strong signals covering the whole nucleus (Supplementary Fig. 2d, yellow arrows) and relatively small bright foci located within the nucleus (Supplementary Fig. 2d, light blue arrowheads). In mammalian cells, detailed immunocytochemical studies on the distribution of phosphorylated form of H2AX (pH2AX) have demonstrated that strong nuclear-wide pH2AX signals are associated with apoptosis whereas intra-nuclear focal pH2AX signals represent DNA damage foci45. Our data suggest that these criteria also apply to pH2Av signals in infected male embryos as follows: (i) many, if not all, embryonic cells with strong nuclear-wide pH2Av signals were also apoptotic with TUNEL (terminal deoxynucleotidyl transferase dUTP nick end labelling) signals (Supplementary Fig. 2g, arrows); (ii) strong nuclear-wide signals were preferentially found in the head region, where developmental apoptosis occurs (Supplementary Fig. 2c,e,f, arrows); (iii) in control embryos, only fewer and relatively obscure focal signals were detected (compare Supplementary Fig. 2c,d); and (iv) when abnormal apoptosis was suppressed in infected male embryos mutant for p53, nuclear-wide signals were reduced while focal signals were still prominent (compare Supplementary Fig. 2d,f). In subsequent experiments, we focused on focal pH2Av signals as cytological indicators of DNA damage.
DNA damage concentrated on the male X chromosome
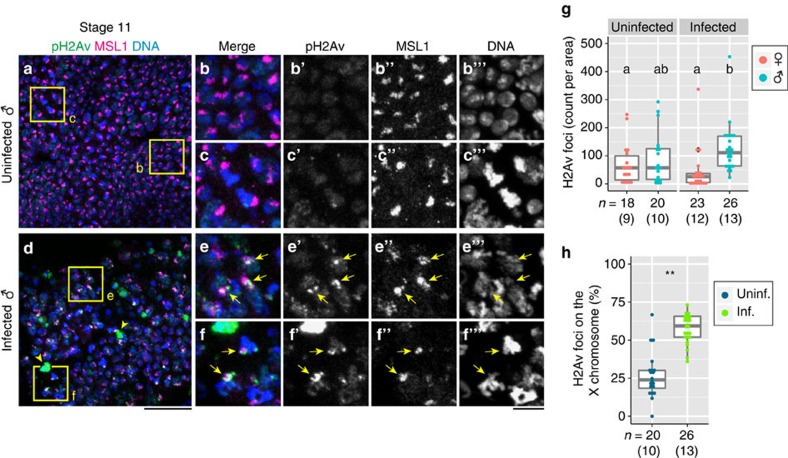
A previous study reported that maternal–zygotic Drosophila mutants for msl genes escape Spiroplasma-induced male-killing, suggesting that dosage compensation of the single male X chromosome is required for male-killing expression23. Hence, we hypothesized that the male X chromosome bound by the MSL complex may be the target of Spiroplasma-induced DNA damage, and tested the hypothesis by visualizing the male X chromosome and DNA-damage foci simultaneously using anti-MSL1 and anti-pH2Av antibodies in the embryonic epidermal cells where Spiroplasma-induced abnormal apoptosis predominantly occurs26. In infected male embryos, MSL1 signals and pH2Av signals were frequently overlapping, while such overlapped signals were infrequent in control embryos (Fig. 3a–f). Quantitative analysis of the co-localized MSL1 and pH2Av signals revealed that significantly more focal pH2Av signals were located on the X chromosome of infected male embryos in comparison with control embryos (Fig. 3g,h), indicating that DNA damage is specifically enriched on the X chromosome of infected male embryos. These results support the hypothesis that the male X chromosome is a major target of Spiroplasma-induced DNA damage, which plausibly underlies the p53-dependent apoptosis observed in infected male embryos.
Figure 3. DNA damage in the X chromosome of Spiroplasma-infected male embryos.
(a) Simultaneous detection of pH2Av (DNA damage foci or apoptotic nuclei; green), MSL1 (X chromosomes; magenta) and DNA (blue) in an uninfected male embryo at stage 11, wherein few green pH2Av signals are seen. (b,c) Magnified images of boxed regions in a. Single-channelled images of b and c are shown in b′–b′′′ and c′–c′′′, respectively. (d) An image similar to a of an infected male embryo, in which a number of green pH2Av signals are detected. (e,f) Magnified images of boxed regions in d. Single-channelled images of e and f are shown in e′–e′′′ and f′–f′′′, respectively. In d–f, arrowheads indicate large pH2AX signals representing apoptotic nuclei, whereas arrows depict focal pH2AX signals representing DNA damage foci. (g) Quantification of focal pH2AX signals in uninfected and infected embryos at stage 11. Different letters (a,b) indicate statistically significant differences (P<0.05; Kruskal–Wallis test followed by Mann–Whitney U-tests). (h) Quantification of focal pH2AX signals overlapping with MSL1-labelled X chromosomes in uninfected and infected male embryos at stage 11. Asterisks indicate a statistically significant difference (**, P<0.01; Pearson's χ2 test). Focal pH2Av signals obtained in g were used to calculate the enrichment on the X chromosome in h. In g and h, box plots are as in Fig. 2i,j. Sample sizes (numbers of images analyzed) are shown at the bottom. Numbers of embryos inspected are shown in parentheses. Scale bars, 20 μm (a,d) and 5 μm (b–c′′′,e–f′′′).
Bridge breakage of the X chromosome in male embryos
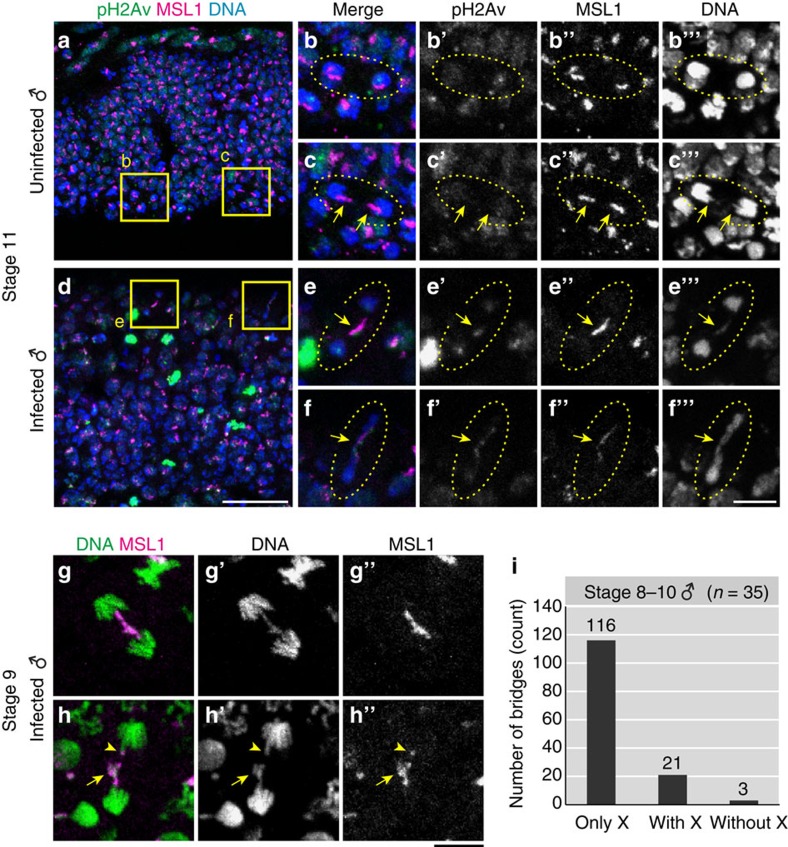
During the immunohistochemical experiments, we frequently observed inter-nuclear chromatin bridges in infected male embryos (Fig. 4d–f and Supplementary Fig. 3a). Notably, MSL1 signals frequently overlapped with chromatin bridges, suggesting that the male X chromosome may be involved in these abnormal structures (35/45 chromatin bridges observed in infected male embryos; Fig. 4e,f). To see more details, we analyzed 35 infected male embryos stained for both DNA and MSL1, and collected 140 mitotic cell images during anaphase, in which sister chromatids are about to separate and moving toward the opposite cell poles with a chromosomal bridge (Fig. 4g–i and Supplementary Fig. 3b–f). According to the extent of overlap between chromosomal bridges and MSL1 signals, we classified the images into three categories: only X, in which the chromosomal bridge and the MSL1 signal completely overlapped (116/140=83%; Fig. 4g–i and Supplementary Fig. 3c); with X, in which the chromosomal bridge and the MSL1 signal partially overlapped, or MSL1-labelled and unlabelled chromosomal bridges were observed simultaneously (21/140=15%; Supplementary Fig. 3d,e and Fig. 4i); and without X, in which the chromosomal bridge lacked the MSL1 signal (3/140=2%; Supplementary Fig. 3f and Fig. 4i). These results favour the idea that male X chromatids constitute chromosomal bridges. The chromosomal bridges were frequently associated with abnormally tangled DNA masses (95/140=68%; Fig. 4h and Supplementary Fig. 3c, arrows), suggesting compromised chromatin remodelling and/or condensation in male X chromatids. Notably, we observed that some X chromatids were asymmetrically segregated into two daughter cells (34/140=24%; Fig. 4h and Supplementary Fig. 3c,e, arrowheads), which presumably reflect the breakage of chromosomal bridges during cell division.
Figure 4. Bridge and breakage of the male X chromosome during mitosis.
(a) Epidermal cells of an uninfected male embryo at stage 11, in which pH2Av, MSL1 and DNA are visualized in green, magenta and blue as in Fig. 3a. (b,c) Magnified images of boxed regions in a, whose single-channelled images are shown in b′–b′′′ and c′–c′′′, respectively. Dotted circles highlight dividing cells in telophase. In b–b′′′, sister chromatids are normally segregating to daughter cells, whereas in c–c′′′, MSL1-labelled X chromatids seem to be segregating slightly slower than the other chromatids (arrows). (d) An image similar to a of a Spiroplasma-infected male embryo, exhibiting many pH2Av signals. (e,f) Magnified images of boxed regions in d, whose single-channelled images are shown in e′–e′′′ and f′–f′′′, respectively. Dotted circles highlight dividing cells in telophase, while arrows indicate inter-nuclear bridges overlapping with MSL1 and focal pH2Av signals, indicating that damaged male X chromatids constitute inter-nuclear bridges. (g,h) Two examples of anaphase chromatin bridges in infected male embryos at stage 9, wherein chromosomal DNA (green) and MSL1 representing X chromatids (magenta) are shown. Single-channelled images are shown in g′, g′′ and h′, h′′. Arrows and arrowheads in h–h′′ indicate an abnormally tangled DNA mass and asymmetrically segregated X chromatids, respectively. (i) Categorization of anaphase chromatin bridges in infected male embryos at stage 8–10. In total 140 anaphase bridges from 35 embryos were inspected. The categories ‘only X', ‘with X' and ‘without X' indicate complete, partial and no overlap between chromatin bridges and MSL1 signals. For more detail, see text. Scale bars, 20 μm (a,d) and 5 μm (b–c′′′, e–f′′′ and g–h′′).
The MSL complex is required for DNA damage and apoptosis
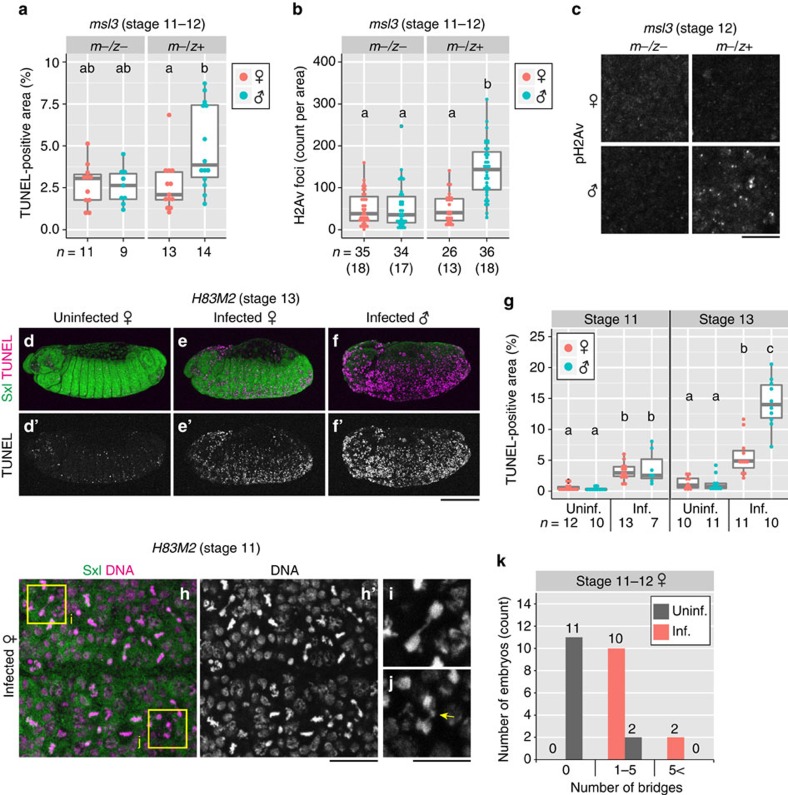
While MSL1 and MSL2 act as scaffold for MSL complex formation, MSL3, MOF and MLE are required for spreading the complex across the entire X chromosome5,46,47. Loss-of-function mutants of msl3, for example msl31, fail to form the complete MSL complex and exhibit male-specific larval lethality due to dosage compensation defects48,49,50. On account of the maternal and zygotic sources of msl3, we investigated a maternal–zygotic mutant (m–/z–; zygotic genotype msl31/msl31) with compromised MSL complex function in comparison with a maternal mutant (m–/z+; zygotic genotype msl31/TM3 ActGFP) with the functional MSL complex. When these fly strains were infected with Spiroplasma, DNA damage and abnormal apoptosis in male embryos were attenuated under the msl3-deficient maternal–zygotic mutant genotype (Fig. 5a–c), indicating that the MSL complex is necessary for Spiroplasma-induced DNA damage and abnormal apoptosis.
Figure 5. The MSL complex is necessary and sufficient for Spiroplasma-induced DNA damage and abnormal apoptosis.
(a–c) Apoptosis (a) and DNA damage (b,c) in Spiroplasma-infected male and female embryos of msl3 maternal–zygotic mutant (m−/z−; zygotic genotype msl31/msl31) and maternal mutant (m−/z+; zygotic genotype msl31/TM3 ActGFP). (a) Quantification of TUNEL-positive areas at stage 11–12. Different letters (a,b) indicate statistically significant differences (P<0.05; Kruskal–Wallis test followed by Mann–Whitney U-tests). (b) Quantification of focal pH2Av signals at stage 11–12. Different letters (a,b) indicate statistically significant differences (P<0.01; Kruskal–Wallis test followed by Mann–Whitney U-tests). (c) Focal pH2Av signals in msl3 mutant embryos at stage 12. (d–k) Ectopic MSL complex formation by the H83M2 transgene. (d) An uninfected H83M2 female embryo exhibiting little abnormal apoptosis. (e,f) Infected H83M2 embryos showing abnormal apoptosis (e, female; f, male). In d–f, stage 13 embryos are stained for Sxl (green) and TUNEL (magenta), whereas single-channelled TUNEL images are shown in d′–f′. (g) Quantification of TUNEL-positive areas in uninfected and infected H83M2 embryos at stage 11 (left) and 13 (right). Different letters (a–c) indicate statistically significant differences (P<0.01; Kruskal–Wallis test followed by Mann–Whitney U-tests). (h) Epidermal cells of an infected H83M2 female embryo at stage 11, stained for Sxl (green) and DNA (magenta), whereas single-channelled DNA image is shown in h′. (i,j) Enlarged images of dividing cells with a chromatin bridge (i) and an abnormally tangled DNA mass (j, arrow), representing boxed regions in h. (k) Quantification of chromatin bridges in the epidermal cells of uninfected and infected H83M2 female embryos at stage 11–12. The number of chromatin bridges per × 63 objective view are categorized into three classes: no bridge (0); 1 to 5 bridges (1–5); and 6 or more bridges (5<). In a, b and g, box plots are as in Fig. 2i,j. and sample sizes are indicated at the bottom. In b, numbers of embryos observed are shown in parentheses. Scale bars, 10 μm (c, i and j), 100 μm (d–f′) and 20 μm (h,h′).
Ectopic MSL complex induces male-killing phenotypes
In females of Drosophila, Sxl directly inhibits translation of MSL2 to prevent the formation of the functional MSL complex5. A previous study showed that the H83M2 transgene, which encodes a suppression-resistant form of msl2 mRNA, induces inappropriate dosage compensation of female X chromosomes (Supplementary Fig. 4a–d), thereby causing reduced viability and developmental delay with a few escaper adult females51 (Supplementary Fig. 4e, purple bars). When H83M2 females were infected with Spiroplasma, no adult escapers were obtained (Supplementary Fig. 4f), suggesting occurrence of ectopic male-killing in infected females. It has been shown that msl1 gene exhibits an allelic dosage effect in H83M2 females, where even heterozygosity (msl1/+) suppresses toxicity of this transgene due to reduced amounts of ectopic MSL complex51. We observed that both the deleterious effects and the ectopic male-killing were suppressed in msl1L60/+ heterozygotes (Supplementary Fig. 4e,f, red bars), supporting the notion that ectopic MSL complex formation is causative of these phenotypes. During the development of Spiroplasma-infected H83M2 female embryos, abnormal apoptosis was observed throughout the body (Fig. 5e,g) and chromatin bridges were frequently found (Fig. 5h–k). Without Spiroplasma infection, by contrast, H83M2 females did not show these abnormal phenotypes (Fig. 5d,g,k), confirming that these abnormal phenotypes are associated with the Spiroplasma infection and are not ascribed to secondary effects of the ectopic MSL complex formation. Taken together, these results indicate that ectopic expression of the MSL complex can reproduce male-killing and associated cytological defects, including DNA damage, chromatin bridge formation and abnormal apoptosis in Spiroplasma-infected female embryos, which is in agreement with a recent report24.
Genetically dissecting effects of bridge breakage
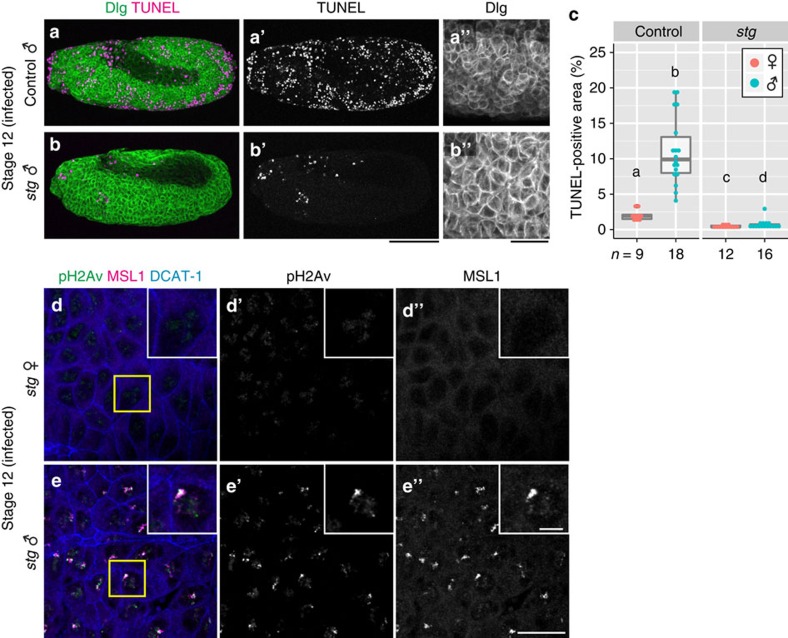
In an attempt to gain further insight into the relationship between DNA damage, chromosomal breakage and abnormal apoptosis, we genetically blocked cell division during embryogenesis. String (Stg), a CDC25 homologue of Drosophila, is essential for the initiation of G2/M transition in the cell cycle52. In zygotic mutants of strong alleles of stg, embryonic cells initially undergo normal cleavage cycles by using maternal transcripts during mitoses 1–13, and after cellularization when zygotically regulated cell division starts (from mitosis 14 onward), cells are arrested at G2 phase during the rest of embryogenesis, thereby resulting in embryos with fewer and larger cells52 (Fig. 6a,b). Considering that the recruitment of the MSL complex to the male X chromosome is first detected in cellularized embryos at mitosis 14 (refs 53, 54), embryonic cells mutant for stg do not undergo cell division after the formation of the MSL complex, which is required for Spiroplasma-induced DNA damage. Therefore, using stg mutant embryos, we can genetically dissect whether bridge breakage in the male X chromosome has a causative role for the chromosome-specific DNA damage induced by Spiroplasma.
Figure 6. DNA damage and apoptosis in Spiroplasma-infected embryos mutant for stg.
(a,b) Spiroplasma-infected control male embryo (genotype stgAR2/TM3 or stg7B/TM3) and stg mutant male embryo (genotype stgAR2/stg7B) at stage 12. Cell membranes and apoptotic cells are visualized by anti-Discs large (Dlg; green) and TUNEL (magenta) staining, respectively. Single-channel images of TUNEL staining and high magnification images of Dlg staining are shown in a′–b′ and a′′–b′′, respectively. (c) Quantification of TUNEL-positive areas in infected control and stg mutant embryos. Box plots are as in Fig. 2i,j. Different letters (a–d) indicate statistically significant differences (P<0.05 for stg females versus males, P<0.01 for the others; Kruskal–Wallis test followed by Mann–Whitney U-tests). Sample sizes are indicated at the bottom. (d,e) Infected stg mutant female (n=7) and male (n=11) embryos at stage 12, in which DNA damage (pH2Av; green), the X chromosome (MSL1; magenta), and cell membrane (DCAT-1; blue) are visualized. Single-channel images of pH2Av signals and MSL1 signals are shown in d′–e′ and d′′–e′′, respectively. Boxed regions in d and e are magnified in insets of d–d′′ and e–e′′. Scale bars, 100 μm (a–b′), 25 μm (a′′,b′′), 20 μm (d–e′′) and 5 μm (insets in d–e′′).
Chromosome-specific DNA damage precedes bridge breakage
In Spiroplasma-infected male embryos mutant for stg, abnormal apoptosis was significantly suppressed in comparison with control embryos (Fig. 6a–c), indicating that cell division is required for the expression of abnormal apoptosis. On the ground that chromosomal bridge-breakage occurs during mitosis, abnormal apoptosis is likely attributable to the DNA damage response activated by the bridge breakage in the X chromosome. On the other hand, even in Spiroplasma-infected male embryos mutant for stg, remaining apoptosis was observed around the head region (Fig. 6b), indicating that the blockage of cell division mainly suppresses p53-dependent abnormal apoptosis rather than developmental apoptosis. Despite the suppression of abnormal apoptosis, focal pH2Av signals on the male X chromosome were still prominent in Spiroplasma-infected male embryos mutant for stg (Fig. 6d,e), indicating that the male X chromosome has been damaged even in the absence of bridge breakage.
Partially attenuated neural disorder in p53 mutant embryos
In addition to abnormal apoptosis, disordered neurogenesis is among the most prominent defective phenotypes of Spiroplasma-infected male embryos; while highly organized central and peripheral nervous systems develop in control embryos, whole nervous systems are severely disorganized in Spiroplasma-infected male embryos26,27,28 (Supplementary Fig. 5c,d). We examined whether and how DNA damage in the male X chromosome and subsequent activation of p53-dependent signalling pathways are relevant to Spiroplasma-induced neural defects. In neural precursor cells called neuroblasts, focal pH2Av signals were overlapping with MSL1 signals in Spiroplasma-infected male embryos (Supplementary Fig. 5a,b), indicating that DNA damage certainly occurs in the male X chromosome of neuroblasts as in the epidermal cells where p53-dependent abnormal apoptosis occurs (Figs 2 and 3). When differentiated neural cells were visualized with an antibody against a specific marker protein Elav (embryonic lethal abnormal vision)55, neural organization was severely disordered in Spiroplasma-infected control male embryos (Supplementary Fig. 5c,d), and notably, the neural disorder was partially recovered in Spiroplasma-infected male embryos mutant for p53: the overall morphology of the ventral nerve cord was restored considerably, but each neural cluster was still disorganized (Supplementary Fig. 5e,f).
Apoptosis suppression similarly attenuates neural disorder
Abnormal apoptosis in Spiroplasma-infected male embryos is concentrated on epidermal cells and scarcely associated with neural cells26,28. Considering that the massive apoptosis is suppressed in p53 mutant embryos, the recovery of overall neural morphology may be attributable to suppression of the extensive cell death in surrounding non-neural tissues. To test this hypothesis, we analyzed the homozygous H99 mutant in which pro-apoptotic genes are deleted and apoptosis is almost completely blocked during embryogenesis56. When we examined Spiroplasma-infected male embryos deficient for apoptosis, the entire structure of the ventral nerve cord was considerably restored (Supplementary Fig. 5g,h), in comparison with Spiroplasma-infected control embryos (Supplementary Fig. 5c,d), but the structure of each neural cluster remained abnormal (Supplementary Fig. 5g,h), which was a reminiscent of the p53 mutant phenotype (Supplementary Fig. 5e,f). These results suggest that the neural malformation in Spiroplasma-infected male embryos is, at least partly, a secondary effect of p53-dependent massive apoptosis, whereas the possibility that some apoptosis-independent pathway(s) may underlie Spiroplasma-induced neural defects cannot be excluded.
Discussion
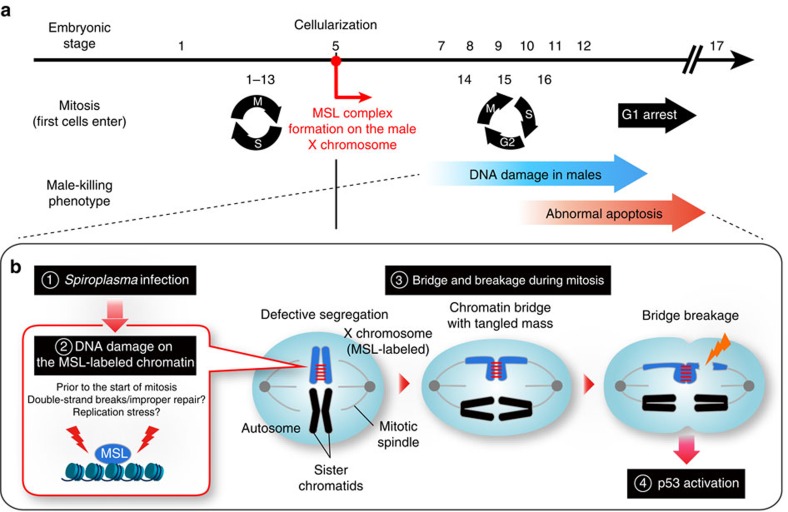
In this study, we uncovered a number of previously unrecognized molecular and cellular aspects underlying Spiroplasma-induced male-killing during Drosophila's embryogenesis, which include: (i) a large number of genes related to DNA damage and apoptosis are up-regulated specifically in Spiroplasma-infected male embryos (Fig. 1; Supplementary Fig. 1; Supplementary Data 1); (ii) Spiroplasma causes DNA damage on the male X chromosome interacting with the functional MSL complex (Fig. 3 and Supplementary Fig. 2); (iii) the damaged male X chromosome exhibits chromosomal bridge and breakage during cell division (Fig. 4 and Supplementary Fig. 3); (iv) the functional MSL complex is not only necessary but also sufficient for triggering Spiroplasma-induced DNA damage, chromatin bridge and apoptosis (Fig. 5 and Supplementary Fig. 4); (v) bridge breakage in the male X chromosome is responsible for abnormal apoptosis via p53-dependent pathways (Fig. 2); and (vi) the mitosis-associated chromatin bridge-breakage is preceded by the induction of chromosome-specific DNA damage (Fig. 6). On the basis of these results, we propose a hypothetical model as to what molecular and cellular mechanisms are operating in the developmental events of Spiroplasma-infected male embryos, which finally result in massive apoptosis and associated developmental abnormalities leading to male-specific embryonic lethality (Fig. 7). In conclusion, Spiroplasma targets the dosage-compensated male X chromosome with the clue of the functional MSL complex and somehow introduces DNA damage on it, thereby causing male-specific chromosomal segregation defects and recruiting host's p53-dependent pathways to induce apoptosis.
Figure 7. Model for the mechanism of Spiroplasma-induced male-killing in Drosophila.
(a) The time line of male-killing phenotypes during embryogenesis. Mitotic cycles are shown below the line. First 13 cleavage cycles are rapid and synchronous, consisting of only S and M phases. After cellularization at stage 5, cells obtain G2 phase and undergo three rounds of mitosis with specific pattern and timing (cycle 14–16), followed by G1 arrest. (b) The cytological model of Spiroplasma-induced male-killing. See the text for details.
It remains unknown how Spiroplasma damages the MSL-bound male X chromosome. On the grounds that (i) Spiroplasma is enriched extracellularly in Drosophila hosts57, (ii) the X chromosome located within the nucleus is damaged in Spiroplasma-infected male embryos (this study) and (iii) mosaic and gynandromorph analyses reveal specific killing of male cells even when male cells and female cells coexist in the same embryos26,58, it is conceivable, although speculative, that Spiroplasma-produced factors, so-called effectors or toxins, may be involved in the process. Some bacterial toxins, such as colibactin of Escherichia coli, typhoid toxin of Salmonella typhi and cytolethal distending toxins of various Gram-negative bacteria, are known to cause DNA crosslinking and induce double-strand breaks in eukaryotic cells, though probably not specific to sex chromosomes59,60. In this context, it may be notable that the Spiroplasma genome encodes specific prophages61 and a plenty of phage particles are found in Spiroplasma-infected Drosophila hosts62,63. It has been reported that a bacterial endosymbiont Hamiltonella defensa produces a phage-encoded toxin, thereby protecting its aphid host against parasitoid wasps64. Similar symbiont-mediated defence against natural enemies has been found in several Spiroplasma-associated Drosophila species65,66, wherein a symbiont-derived ribosome-inactivating protein was identified as a defensive factor67. Meanwhile, the possibility cannot be excluded that Spiroplasma may act on the host cells to induce some eukaryotic factors that interact with and damage the MSL-bound X chromosome. Future studies should focus on these possibilities.
Our finding that the functional MSL complex is necessary as well as sufficient for triggering Spiroplasma-induced male-killing (Fig. 5 and Supplementary Fig. 4) implies that the Spiroplasma-induced damage on the male X chromosome depends on the functional MSL complex either directly or indirectly. A simple scenario is that the protein complex itself serves as a molecular target. In this context, it may be relevant that MSL proteins evolve rapidly under strong positive selection in Drosophila, suggesting the possibility of evolutionary arms race between the host's dosage compensation system and the symbiont's selfish reproductive manipulation68. Recently, it was reported that Spiroplasma infection alters the localization of the MSL complex in male embryos, suggesting that Spiroplasma directly targets the dosage compensation machinery to induce genome-wide disruption of gene expression24. On the other hand, considering that the functional MSL complex is associated with various types of histone modifications and subsequent structural/transcriptional changes5, Spiroplasma may influence these modifications rather than target the MSL complex itself. Recent studies revealed that some bacteria can affect chromatin structure and transcriptional activity of host cells by modulating diverse epigenetic factors such as histone modifications, DNA methylation and chromatin-associated complexes69. It was reported that acetylation of histone H4K16, one of the major chromatin modifications mediated by the MSL complex, weakens nucleosome packing, thereby making chromatins more accessible for DNA binding factors70. It is possible that the MSL-bound X chromosome similarly becomes susceptible to Spiroplasma-induced DNA damage.
In theory, any male-specific essential molecular, cellular and/or structural aspects of host organisms can potentially be exploited by symbiotic microorganisms to induce male-killing9. Probably reflecting this, symbiont-induced male-killing has evolved repeatedly in diverse bacterial lineages including Wolbachia, Spiroplasma, Arsenophonus, Rickettsia and others, where the symbiotic bacteria interact with a variety of host's molecular and cellular components8,9: the male-killing Spiroplasma damages the dosage-compensated male X chromosome bound by the MSL complex in Drosophila melanogaster (this study); a male-killing Wolbachia suppresses host's masculinizing gene expression and thereby disturbs dosage compensation of the male Z chromosome in Ostrinia moths29; another male-killing Wolbachia induces defective chromatin remodelling and subsequent abnormal mitotic spindle formation in the male embryos of Drosophila bifasciata16; and a male-killing Arsenophonus inhibits formation of maternal centrosomes required for early male development in Nasonia vitripennis14. In the light of the diversity and commonality of male-killing mechanisms, the Ostrinia's Wolbachia is of particular interest in comparison with the Drosophila's Spiroplasma in that: (i) the entirely different symbiotic bacteria, Wolbachia (α-Proteobacteria) and Spiroplasma (Mollicutes), cause similar male-killing phenotypes in the entirely different insect hosts, Drosophila (Diptera; male heterogametic with XY chromosomes) and Ostrinia (Lepidoptera; female heterogametic with ZW chromosomes); (ii) both symbiotic bacteria interact with host's dosage compensation mechanisms for inducing male-killing; (iii) however, while Wolbachia disturbs the dosage compensation of the male Z chromosome in Ostrinia, Spiroplasma scarcely affects the dosage compensation of the male X chromosome in Drosophila; and (iv) Wolbachia's male-killing in Ostrinia is due to dosage compensation defects, whereas Spiroplasma's male-killing in Drosophila is caused by bridge breakage of the male X chromosome and subsequent p53-mediated massive apoptosis.
In this study, we provide an integrative picture as to what mechanisms underlie Spiroplasma-induced male-killing, which encompass molecular, chromosomal, cellular and organismal levels. By accumulating such in-depth knowledge for different host–symbiont systems, we will be able to gain insights into the diversity and commonality of symbiont's strategies for interfering with host's sex-related cellular mechanisms, which should lead to a promising avenue for broadening the frontier of cell biology towards the realm of evolutionary biology and ecology.
Methods
Fly stocks used in this study were obtained from the Bloomington Drosophila Stock Center (Indiana University), the Drosophila Genetic Resource Center (Kyoto Institute of Technology) and several Drosophila researchers. RNA-seq libraries of Spiroplasma-infected and uninfected embryos were constructed by TruSeq RNA Sample Preparation Kit (Illumina) and sequenced by HiSeq 2000/2500 (Illumina). Short reads were aligned to the reference genome sequence of D. melanogaster provided by University of California, Santa Cruz (dm3, Berkeley Drosophila Genome Project Release 5) (Supplementary Data 2). Of all genes annotated in the Drosophila genome, 8,387 genes achieved at least one CPM (counts per million reads) for at least three libraries were subjected to identification of differentially expressed genes (Supplementary Data 3). Immunofluorescence staining and other cytological procedures were as described26, which were subjected to imaging analyses using custom R scripts with the EBImage package. Further details of the methods can be found in the Supplementary Methods.
Data availability
Nucleotide sequence data that support the findings of this study have been deposited in the DNA Data Bank of Japan (DDBJ: http://www.ddbj.nig.ac.jp) Sequence Read Archive with the accession numbers PRJDB4469/DRA004268/SAMD00044983-SAMD00044986 (Supplementary Data 2). All other relevant data supporting the findings of this study are included within the article and its Supplementary Information files or available on request.
Additional information
Accession codes: Nucleotide sequence data have been deposited in the DNA Data Bank of Japan (DDBJ: http://www.ddbj.nig.ac.jp) Sequence Read Archive with the accession numbers PRJDB4469/DRA004268/SAMD00044983-SAMD00044986 (Supplementary Data 2).
How to cite this article: Harumoto, T. et al. Male-killing symbiont damages host's dosage-compensated sex chromosome to induce embryonic apoptosis. Nat. Commun. 7:12781 doi: 10.1038/ncomms12781 (2016).
Supplementary Material
Supplementary Figures 1-5, Supplementary Methods and Supplementary References
Cluster analysis and gene annotation of selected differentially expressed genes. Gene sets identified in the cluster analysis (Fig. 1d) are summarized with their Flybase IDs and gene annotations (sheet 1). In sheets 2-7, red color highlights remarkably enriched GO terms (false discovery rate < 0.1).
Summary of RNA-seq datasets and mapping results. The number of total reads obtained from each RNA-seq library. Mapped reads, aligned pairs to the D. melanogaster reference genome sequence (dm3, BDGP Release 5) and their percentages are also summarized with their accession numbers in the DNA Data Bank of Japan (DDBJ).
Transcriptional changes in Spiroplasma-infected and uninfected embryos. Summary of edgeR differential expression analysis. In sheet 1, gene names and normalized read count (counts per million, CPM) are indicated. Results of significance tests are also shown (significance, yes/no). In sheet 2, gene names and their locus (chromosome) are indicated with log2-transformed fold-change (logFC), average of log2-transformed counts per million reads (logCPM), P-value, and false discovery rate (FDR).
Acknowledgments
The Drosophila Genetic Resource Center at Kyoto Institute of Technology, Japan and the Bloomington Stock Center, USA, provided fly stocks. The Developmental Studies Hybridoma Bank at the University of Iowa, USA, provided antibodies. We thank Takehide Murata, John Abrams, Mitzi Kuroda, Bruce Edgar and John Jaenike for providing fly strains, John Lucchesi for providing antibodies, Yoichi Kamagata and Takafumi Mizuno for confocal microscopy, Shuji Shigenobu and Tomoko Shibata for RNA-seq data acquisition, Takashi Kiuchi and Lemaitre lab members for comments on the manuscript, and Junko Makino and Wakana Kikuchi for technical and secretarial assistance. This work was supported by Japan Society for the Promotion of Science (JSPS) KAKENHI Grant Number 12J05307 to TH. T.H. was also supported by JSPS Fellowship for Young Scientists. RNA-seq analysis was supported by KAKENHI Grant Number 22128001. Part of this work, including the efforts of T.H. and B.L., was funded by the European Research Council (ERC) Advanced Grant 339970 and the Swiss National Science Foundation (SNSF) Sinergia grant no. CRSII3_154396.
Footnotes
Author contributions T.H. and T.F. conceived the study. T.H. performed most of the experiments. H.A. carried out some of the genetic experiments. T.H, B.L and T.F wrote the paper. All authors edited and commented on the paper.
References
- Maynard Smith J. The Evolution of Sex Cambridge University Press (1978). [Google Scholar]
- Bull J. J. Evolution of Sex Determining Mechanisms Benjamin/Cummings Pub. Co. (1983). [Google Scholar]
- Beukeboom L. & Perrin N. The Evolution of Sex Determination Oxford University Press (2014). [Google Scholar]
- Salz H. K. & Erickson J. W. Sex determination in Drosophila: the view from the top. Fly (Austin) 4, 60–70 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchesi J. C. & Kuroda M. I. Dosage compensation in Drosophila. Cold Spring Harb. Perspect. Biol. 7, a019398 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran N. A., McCutcheon J. P. & Nakabachi A. Genomics and evolution of heritable bacterial symbionts. Annu. Rev. Genet. 42, 165–190 (2008). [DOI] [PubMed] [Google Scholar]
- McFall-Ngai M. et al. Animals in a bacterial world, a new imperative for the life sciences. Proc. Natl Acad. Sci. USA 110, 3229–3236 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werren J. H., Baldo L. & Clark M. E. Wolbachia: master manipulators of invertebrate biology. Nat. Rev. Microbiol. 6, 741–751 (2008). [DOI] [PubMed] [Google Scholar]
- Hurst G. D. D. & Frost C. L. Reproductive parasitism: maternally inherited symbionts in a biparental world. Cold Spring Harb. Perspect. Biol. 7, a017699 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serbus L. R., Casper-Lindley C., Landmann F. & Sullivan W. The genetics and cell biology of Wolbachia-host interactions. Annu. Rev. Genet. 42, 683–707 (2008). [DOI] [PubMed] [Google Scholar]
- Counce S. J. & Poulson D. F. Developmental effects of the sex-ratio agent in embryos of Drosophila willistoni. J. Exp. Zool. 151, 17–31 (1962). [DOI] [PubMed] [Google Scholar]
- Ferree P. M. et al. Wolbachia utilizes host microtubules and Dynein for anterior localization in the Drosophila oocyte. PLoS Pathog. 1, e14 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serbus L. R. & Sullivan W. A cellular basis for Wolbachia recruitment to the host germline. PLoS Pathog. 3, e190 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferree P. M., Avery A., Azpurua J., Wilkes T. & Werren J. H. A bacterium targets maternally inherited centrosomes to kill males in Nasonia. Curr. Biol. 18, 1409–1414 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertson R., Casper-Lindley C., Cao J., Tram U. & Sullivan W. Symmetric and asymmetric mitotic segregation patterns influence Wolbachia distribution in host somatic tissue. J. Cell. Sci. 122, 4570–4583 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riparbelli M. G., Giordano R., Ueyama M. & Callaini G. Wolbachia-mediated male killing is associated with defective chromatin remodeling. PLoS ONE 7, e30045 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaini G., Dallai R. & Riparbelli M. G. Wolbachia-induced delay of paternal chromatin condensation does not prevent maternal chromosomes from entering anaphase in incompatible crosses of Drosophila simulans. J. Cell. Sci. 110, (Pt 2): 271–280 (1997). [DOI] [PubMed] [Google Scholar]
- Tram U. & Sullivan W. Role of delayed nuclear envelope breakdown and mitosis in Wolbachia-induced cytoplasmic incompatibility. Science 296, 1124–1126 (2002). [DOI] [PubMed] [Google Scholar]
- Tram U., Fredrick K., Werren J. H. & Sullivan W. Paternal chromosome segregation during the first mitotic division determines Wolbachia-induced cytoplasmic incompatibility phenotype. J. Cell. Sci. 119, 3655–3663 (2006). [DOI] [PubMed] [Google Scholar]
- Landmann F., Orsi G. A., Loppin B. & Sullivan W. Wolbachia-mediated cytoplasmic incompatibility is associated with impaired histone deposition in the male pronucleus. PLoS Pathog. 5, e1000343 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frydman H. M., Li J. M., Robson D. N. & Wieschaus E. Somatic stem cell niche tropism in Wolbachia. Nature 441, 509–512 (2006). [DOI] [PubMed] [Google Scholar]
- Fast E. M. et al. Wolbachia enhance Drosophila stem cell proliferation and target the germline stem cell niche. Science 334, 990–992 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veneti Z., Bentley J. K., Koana T., Braig H. R. & Hurst G. D. D. A functional dosage compensation complex required for male killing in Drosophila. Science 307, 1461–1463 (2005). [DOI] [PubMed] [Google Scholar]
- Cheng B., Kuppanda N., Aldrich J. C., Akbari O. S. & Ferree P. M. Male-killing Spiroplasma alters behavior of the dosage compensation complex during Drosophila melanogaster embryogenesis. Curr. Biol. 26, 1339–1345 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley J. K., Veneti Z., Heraty J. & Hurst G. D. D. The pathology of embryo death caused by the male-killing Spiroplasma bacterium in Drosophila nebulosa. BMC Biol. 5, 9 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harumoto T., Anbutsu H. & Fukatsu T. Male-killing Spiroplasma induces sex-specific cell death via host apoptotic pathway. PLoS Pathog. 10, e1003956 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiyama-Omura S., Sakaguchi B., Koga K. & Poulson D. F. Morphological features of embryogenesis in Drosophila melanogaster infected with a male-killing Spiroplasma. Zoolog. Sci. 5, 375–383 (1988). [Google Scholar]
- Martin J., Chong T. & Ferree P. M. Male killing Spiroplasma preferentially disrupts neural development in the Drosophila melanogaster embryo. PLoS ONE 8, e79368 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui T. et al. The endosymbiotic bacterium Wolbachia selectively kills male hosts by targeting the masculinizing gene. PLoS Pathog. 11, e1005048 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronin S. J. F. et al. Genome-wide RNAi screen identifies genes involved in intestinal pathogenic bacterial infection. Science 325, 340–343 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H. et al. Cytokine/Jak/Stat signaling mediates regeneration and homeostasis in the Drosophila midgut. Cell 137, 1343–1355 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman D. et al. Autocrine and paracrine unpaired signaling regulate intestinal stem cell maintenance and division. J. Cell. Sci. 125, 5944–5949 (2012). [DOI] [PubMed] [Google Scholar]
- Hurst G. D. D., Anbutsu H., Kutsukake M. & Fukatsu T. Hidden from the host: Spiroplasma bacteria infecting Drosophila do not cause an immune response, but are suppressed by ectopic immune activation. Insect Mol. Biol. 12, 93–97 (2003). [DOI] [PubMed] [Google Scholar]
- Anbutsu H. & Fukatsu T. Evasion, suppression and tolerance of Drosophila innate immunity by a male-killing Spiroplasma endosymbiont. Insect Mol. Biol. 19, 481–488 (2010). [DOI] [PubMed] [Google Scholar]
- Herren J. K. & Lemaitre B. Spiroplasma and host immunity: activation of humoral immune responses increases endosymbiont load and susceptibility to certain Gram-negative bacterial pathogens in Drosophila melanogaster. Cell. Microbiol. 13, 1385–1396 (2011). [DOI] [PubMed] [Google Scholar]
- Ciccia A. & Elledge S. J. The DNA damage response: making it safe to play with knives. Mol. Cell 40, 179–204 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky M. H. et al. Drosophila p53 binds a damage response element at the reaper locus. Cell 101, 103–113 (2000). [DOI] [PubMed] [Google Scholar]
- Jin S. et al. Identification and characterization of a p53 homologue in Drosophila melanogaster. Proc. Natl Acad. Sci. USA 97, 7301–7306 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollmann M. et al. Drosophila p53 is a structural and functional homologue of the tumor suppressor p53. Cell 101, 91–101 (2000). [DOI] [PubMed] [Google Scholar]
- Abrams J. M., White K., Fessler L. I. & Steller H. Programmed cell death during Drosophila embryogenesis. Development 117, 29–43 (1993). [DOI] [PubMed] [Google Scholar]
- Wichmann A., Jaklevic B. & Su T. T. Ionizing radiation induces caspase-dependent but Chk2- and p53-independent cell death in Drosophila melanogaster. Proc. Natl Acad. Sci. USA 103, 9952–9957 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamee L. M. & Brodsky M. H. p53-independent apoptosis limits DNA damage-induced aneuploidy. Genetics 182, 423–435 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinner A., Wu W., Staudt C. & Iliakis G. γ-H2AX in recognition and signaling of DNA double-strand breaks in the context of chromatin. Nucleic Acids Res. 36, 5678–5694 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madigan J. P., Chotkowski H. L. & Glaser R. L. DNA double-strand break-induced phosphorylation of Drosophila histone variant H2Av helps prevent radiation-induced apoptosis. Nucleic Acids Res. 30, 3698–3705 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleaver J. E. γH2Ax: biomarker of damage or functional participant in DNA repair ‘all that glitters is not gold!'. Photochem. Photobiol. 87, 1230–1239 (2011). [DOI] [PubMed] [Google Scholar]
- Conrad T. & Akhtar A. Dosage compensation in Drosophila melanogaster: epigenetic fine-tuning of chromosome-wide transcription. Nat. Rev. Genet. 13, 123–134 (2011). [DOI] [PubMed] [Google Scholar]
- Straub T. & Becker P. B. Transcription modulation chromosome-wide: universal features and principles of dosage compensation in worms and flies. Curr. Opin. Genet. Dev. 21, 147–153 (2011). [DOI] [PubMed] [Google Scholar]
- Okuno T., Satou T. & Oishi K. Studies on the sex-specific lethals of Drosophila melanogaster. VII. Sex-specific lethals that do not affect dosage compensation. Jpn. J. Genet. 59, 237 (1984). [Google Scholar]
- Palmer M. J., Richman R., Richter L. & Kuroda M. I. Sex-specific regulation of the male-specific lethal-1 dosage compensation gene in Drosophila. Genes Dev. 8, 698–706 (1994). [DOI] [PubMed] [Google Scholar]
- Gorman M., Franke A. & Baker B. S. Molecular characterization of the male-specific lethal-3 gene and investigations of the regulation of dosage compensation in Drosophila. Development 121, 463–475 (1995). [DOI] [PubMed] [Google Scholar]
- Kelley R. L. et al. Expression of Msl-2 causes assembly of dosage compensation regulators on the X chromosomes and female lethality in Drosophila. Cell 81, 867–877 (1995). [DOI] [PubMed] [Google Scholar]
- Edgar B. A. & O'Farrell P. H. Genetic control of cell division patterns in the Drosophila embryo. Cell 57, 177–187 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastelli L., Richman R. & Kuroda M. I. The dosage compensation regulators MLE, MSL-1 and MSL-2 are interdependent since early embryogenesis in Drosophila. Mech. Dev. 53, 223–233 (1995). [DOI] [PubMed] [Google Scholar]
- Franke A., Dernburg A., Bashaw G. J. & Baker B. S. Evidence that MSL-mediated dosage compensation in Drosophila begins at blastoderm. Development 122, 2751–2760 (1996). [DOI] [PubMed] [Google Scholar]
- Robinow S. & White K. Characterization and spatial distribution of the ELAV protein during Drosophila melanogaster development. J. Neurobiol. 22, 443–461 (1991). [DOI] [PubMed] [Google Scholar]
- White K. et al. Genetic control of programmed cell death in Drosophila. Science 264, 677–683 (1994). [DOI] [PubMed] [Google Scholar]
- Anbutsu H. & Fukatsu T. Tissue-specific infection dynamics of male-killing and nonmale-killing spiroplasmas in Drosophila melanogaster. FEMS Microbiol. Ecol. 57, 40–46 (2006). [DOI] [PubMed] [Google Scholar]
- Tsuchiyama S., Sakaguchi B. & Oishi K. Analysis of gynandromorph survivals in Drosophila melanogaster infected with the male-killing SR organisms. Genetics 89, 711–721 (1978). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasso F. & Frisan T. Bacterial genotoxins: merging the DNA damage response into infection biology. Biomolecules 5, 1762–1782 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcaino M. I. & Crawford J. M. The colibactin warhead crosslinks DNA. Nat. Chem. 7, 411–417 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredes J. C. et al. Genome sequence of the Drosophila melanogaster male-killing Spiroplasma strain MSRO endosymbiont. MBio 6, e02437–14 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oishi K. & Poulson D. F. A virus associated with SR-spirochetes of Drosophila nebulosa. Proc. Natl Acad. Sci. USA 67, 1565–1572 (1970). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson D. L., Oishi K. & Poulson D. F. in The Atlas of Insect and Plant Viruses ed. Maramorosch K. 465–472Academic Press (1977). [Google Scholar]
- Oliver K. M., Degnan P. H., Hunter M. S. & Moran N. A. Bacteriophages encode factors required for protection in a symbiotic mutualism. Science 325, 992–994 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenike J., Unckless R., Cockburn S. N., Boelio L. M. & Perlman S. J. Adaptation via symbiosis: recent spread of a Drosophila defensive symbiont. Science 329, 212–215 (2010). [DOI] [PubMed] [Google Scholar]
- Xie J., Vilchez I. & Mateos M. Spiroplasma bacteria enhance survival of Drosophila hydei attacked by the parasitic wasp Leptopilina heterotoma. PLoS ONE 5, e12149 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton P. T., Peng F., Boulanger M. J. & Perlman S. J. A ribosome-inactivating protein in a Drosophila defensive symbiont. Proc. Natl Acad. Sci. USA 113, 350–355 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez M. A., Vermaak D., Bayes J. J. & Malik H. S. Species-specific positive selection of the male-specific lethal complex that participates in dosage compensation in Drosophila. Proc. Natl Acad. Sci. USA 104, 15412–15417 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierne H., Hamon M. & Cossart P. Epigenetics and bacterial infections. Cold Spring Harb. Perspect. Med. 2, a010272 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell O. et al. Accessibility of the Drosophila genome discriminates PcG repression, H4K16 acetylation and replication timing. Nat. Struct. Mol. Biol. 17, 894–900 (2010). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures 1-5, Supplementary Methods and Supplementary References
Cluster analysis and gene annotation of selected differentially expressed genes. Gene sets identified in the cluster analysis (Fig. 1d) are summarized with their Flybase IDs and gene annotations (sheet 1). In sheets 2-7, red color highlights remarkably enriched GO terms (false discovery rate < 0.1).
Summary of RNA-seq datasets and mapping results. The number of total reads obtained from each RNA-seq library. Mapped reads, aligned pairs to the D. melanogaster reference genome sequence (dm3, BDGP Release 5) and their percentages are also summarized with their accession numbers in the DNA Data Bank of Japan (DDBJ).
Transcriptional changes in Spiroplasma-infected and uninfected embryos. Summary of edgeR differential expression analysis. In sheet 1, gene names and normalized read count (counts per million, CPM) are indicated. Results of significance tests are also shown (significance, yes/no). In sheet 2, gene names and their locus (chromosome) are indicated with log2-transformed fold-change (logFC), average of log2-transformed counts per million reads (logCPM), P-value, and false discovery rate (FDR).
Data Availability Statement
Nucleotide sequence data that support the findings of this study have been deposited in the DNA Data Bank of Japan (DDBJ: http://www.ddbj.nig.ac.jp) Sequence Read Archive with the accession numbers PRJDB4469/DRA004268/SAMD00044983-SAMD00044986 (Supplementary Data 2). All other relevant data supporting the findings of this study are included within the article and its Supplementary Information files or available on request.