Abstract
The rugged folding landscapes of functional proteins puts them at risk of misfolding and aggregation. Serine protease inhibitors, or serpins, are paradigms for this delicate balance between function and misfolding. Serpins exist in a metastable state that undergoes a major conformational change in order to inhibit proteases. However, conformational labiality of the native serpin fold renders them susceptible to misfolding, which underlies misfolding diseases such as α1-antitrypsin deficiency. To investigate how serpins balance function and folding, we used consensus design to create conserpin, a synthetic serpin that folds reversibly, is functional, thermostable, and polymerization resistant. Characterization of its structure, folding and dynamics suggest that consensus design has remodeled the folding landscape to reconcile competing requirements for stability and function. This approach may offer general benefits for engineering functional proteins that have risky folding landscapes, including the removal of aggregation-prone intermediates, and modifying scaffolds for use as protein therapeutics.
The rugged energy landscapes of functional proteins reflect the delicate balance between efficient folding and function1,2. For proteins to fold, the interactions of the native state must outweigh the non-native interactions, which result in a funnel-shaped energy landscape3,4,5. However, it is not obvious how the myriad of non-covalent interactions that stabilise the native state can do so selectively over the vastly larger number of non-native conformations. Effective protein engineering has typically focused on stabilising low energy configurations as observed in X-ray crystallography or nuclear magnetic resonance (NMR) spectroscopy6,7,8,9. However, engineering robust proteins with funnel shaped energy landscapes may require not only stabilisation of the native state (positive design)10,11,12, but also destabilisation of non-native states (negative design)12,13,14,15. This is especially true for engineering proteins with complex and rugged folding pathways, which often exhibit a delicate balance between function and misfolding1,2.
Such a balance is exemplified by members of the serine protease inhibitor, or serpin superfamily16,17,18,19. Inhibitory members fold to a metastable native state that undergoes a major conformational change in order to inhibit target proteases20. The inhibitory mechanism of serpins is structurally well understood20. Briefly, a target protease initially interacts with and cleaves the serpin reactive center loop (RCL) that protrudes from the main body of the molecule. Following RCL cleavage, but prior to the final hydrolysis of the acyl enzyme intermediate, the RCL inserts into the central β-sheet to form an extra strand20,21. Since the protease is still covalently linked to the serpin, the process of RCL insertion results in the translocation of the protease to the opposite end of the molecule. In the final complex, the protease active site is distorted and trapped as the acyl enzyme intermediate20,22. This remarkable conformational change is termed the stressed [S] to relaxed [R] transition and is accompanied by a major increase in stability of the serpin protein.
As a consequence of folding to a metastable active state, serpins are prone to misfolding. Without being cleaved by a protease, the serpin RCL can self-insert, either partially (delta), or fully (latent)16; or polymerize by insertion of the RCL of one serpin into the body of another serpin23,24,25. Both such RCL insertion events result in a more stable protein species that is no longer functional as a protease inhibitor. Misfolding of the archetypal serpin, α1-antitrypsin (α1-AT), results in a deficiency of active protein, inducing emphysema through uncontrolled protease activity, and the retention of α1-AT polymers in the liver that induce cell death18,26. Serpin misfolding and serpinopathies are a direct result of the ‘risky’ energy landscape required to fold the protein to a metastable state1,27. To investigate how the folding energy landscape of serpins balances the competing requirements for function and stability we used consensus design to build a synthetic serpin. Consensus design is based on the hypothesis that at a given position in a multiple sequence alignment (MSA) of homologous proteins, the respective consensus amino acid contributes more than average to the stability of the protein than non-consensus amino acids28,29,30,31. The efficacy of consensus design has been demonstrated to increase the stabilities of a wide range of proteins, usually by stabilising the native state29,31,32,33,34,35,36,37,38. However, its potential for altering folding landscapes has not been thoroughly explored29,31,32,33,34,35,36,37,38.
Hypothesizing that a serpin reflecting a highly conserved sequence may offer insight into the delicate balance between folding and function, we designed conserpin (consensus serpin). Characterization of its function, structure and folding reveal a serpin that is inhibitory, folds reversibly, is thermostable and resistant to polymerisation. Our results suggest that consensus design has smoothed the folding landscape, reducing the lifetime of aggregation-prone intermediates. This work provides insights into the serpin function-stability balance and emphasises the wider potential for consensus design to remodel the risky folding landscapes of functional proteins.
Results
Conserpin is an inhibitory serpin
To design conserpin we used the consensus approach and a previously reported MSA of 219 serpin sequences39. Conserpin (396 aa) shares the highest similarity with α1-AT (137 residue differences; 62% sequence identity). There is an overall loss of 10 residues located at the N-terminus of the D-helix and C-terminus of the protein. The RCL contains 7 residue differences compared to α1-AT, notably an arginine at P1 compared to the methionine of α1-AT, and the deletion of a residue at P2.
Purified conserpin inhibits trypsin with a stoichiometry of inhibition (SI) of 1.8 and a kassapp of 7.5 × 106 M−1 s−1 and hence a rate of association (kass) of 1.4 × 107 M−1 s−1 (Fig. S1A–C). Higher order complex formation of conserpin with trypsin was observed on SDS PAGE; however, it was atypical compared to α1-AT (Fig. S1D). This unusual behaviour and the increased SI of conserpin may be a consequence of shortening the RCL on the ‘prime’ side of the recognition sequence for trypsin, or due to other biophysical differences. The crystal structure of conserpin (Table S1), confirms that it adopts the archetypal native serpin fold (Fig. 1A,B). Taken together, we propose that inhibition by conserpin occurs via the classical serpin mechanism.
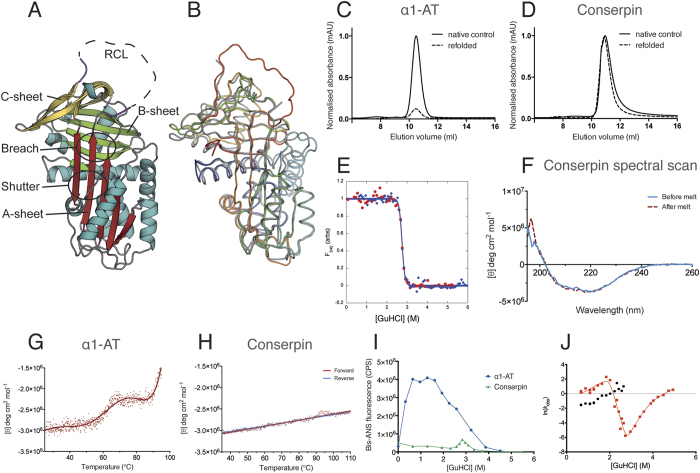
Figure 1. Conserpin conforms to the serpin fold and has superior biophysical properties compared with α1-AT.
(A) Cartoon representation of the 2.4 Å X-ray crystal structure of native conserpin, identifying the breach and shutter regions, the A, B and C sheets (colored in red, green and yellow respectively), and the RCL stumps (magenta). (B) Structural alignment of conserpin (grey) with α1-AT (PDB: 3NE4; spectrum, blue to red). Root mean square deviation (RMSD) = 0.91 Å across 296 backbone Cα atoms. Chemical refolding of (C) α1-AT and (D) conserpin shows that conserpin can refold to a monomer. Chromatograms from a Superdex 75 10/300 size exclusion column are shown. Final protein concentrations loaded onto column were 2 μM. Samples were unfolded in 5 M GuHCl and then diluted out to 0.5 M GuHCl (dotted line). Control samples of native protein are shown as the solid black line. (E) Intrinsic fluorescence equilibrium unfolding (red dots) and refolding (blue diamonds) curves of conserpin coincide, demonstrating reversible folding. (F) CD spectral scans of conserpin before (solid blue line) and after (dashed red line) heating to 110 °C. Variable temperature thermal melts of (G) α1-AT and (H) conserpin as measured by CD at 222 nm. (I) Conserpin shows a significant reduction of intermediate formation during bis-ANS fluorescent equilibrium unfolding of α1-AT (blue circles) and conserpin (green triangles). (J) Kinetic unfolding and refolding experiments. The plot shows the [GuHCl]-dependence of the natural logarithm of the rate constants for unfolding and refolding of conserpin (chevron plot). Two discernable refolding rates are observed (red squares, fast rate; black circles, slower folding rate). The positive slope in each refolding arm suggests the presence of intermediate species that have to partially unfold to reach the native state.
Conserpin folds reversibly, is thermostable and resistant to polymerization
The majority of serpins unfold through an aggregation-prone intermediate ensemble and do not completely refold after chemical and/or thermal denaturation40,41,42,43,44,45,46,47. This is exemplified by α1-AT, which shows a very small amount of refolded monomer via chemical denaturation, rapid dilution and gel filtration (Fig. 1C). In contrast, conserpin refolds to a monomeric state (Fig. 1D). Equilibrium chemical unfolding and refolding curves overlay well, revealing a midpoint of denaturation, [D]50, of 2.75 ± 0.10 M, an equilibrium m-value, mD-N, of 8.45 ± 0.65 kcal mol−1 M−1, and hence a stability, ∆GD-N, of −23.2 ± 2.0 kcal mol−1 (Fig. 1E). The correlation of unfolding and refolding curves, the single unfolding transition, and the steep m-value all suggest minimal formation of an intermediate ensemble. Refolded conserpin retained inhibitory activity, resulting in no significant change in SI (increased from 1.8 to 2.3, Fig. S1E), confirming that conserpin refolds to the native state after chemical denaturation.
Variable temperature circular dichroism (CD) thermal melt analysis at 222 nm reveals a highly thermostable protein with no defined unfolding transition up to a temperature of 110 °C (Fig. 1H). Far-UV spectral scans before and after the thermal melt showed no change in signal, indicating no detectable heat-induced structural changes (Fig. 1F). This contrasts with α1-AT, which upon heating undergoes a three-state transition with an initial midpoint temperature (Tm) of 61.8 °C and an incomplete transition that starts at 90 °C (consistent with other reports48,49; Fig. 1G). Upon cooling of α1-AT, we observed a white precipitate in the cuvette, consistent with irreversible aggregation. Refolding transverse urea gradient (TUG) gels further demonstrate that conserpin is more resistant to polymerization than α1-AT, which mostly formed polymers on refolding, with no formation of native protein (Fig. S2).
To test if conserpin undergoes a transition to the more stable latent state upon heating, we assessed its inhibitory activity and structure after heating at 80 °C for 20 minutes. Heating caused a complete loss in inhibitory activity (Fig. S1F), suggesting formation of the latent state, which was then confirmed by native PAGE (Fig. S3A) and crystal structure determination (Table S1 & Fig. S3B).
Conserpin avoids polymerization by minimizing formation of folding intermediates
Although equilibrium unfolding/refolding data using intrinsic fluorescence indicated minimal formation of intermediates in the folding pathway of conserpin (Fig. 1E), this method is dependent on the difference in solvation of tryptophan residues during unfolding/refolding. In order to more thoroughly interrogate folding intermediates, we repeated the equilibrium unfolding experiments in the presence of bis-ANS (Fig. 1I). In native conditions, both folded conserpin and α1-AT show similar levels of fluorescence, however, by ~ 1 M GuHCl, a high intensity fluorescent peak indicated the presence of a folding intermediate(s) for α1-AT (Fig. 1I), consistent with previous reports47. In contrast, the unfolding profile of conserpin in bis-ANS shows a small, sharp peak at approximately 3 M GuHCl (Fig. 1I). This is consistent with our [D]50 measurement by intrinsic fluorescence (Fig. 1E) and confirms our hypothesis that conserpin has reduced intermediate ensemble formation.
To observe the kinetics of the folding intermediate, we used rapid mixing techniques during unfolding and refolding (Fig. 1J). As expected, the unfolding traces fitted well to a single exponential. When the protein was refolded from an equilibrated denatured solution (single-jump), the resulting traces could not be fitted to fewer than three exponentials (Fig. S1G) and showed inconsistencies between repeats (likely due to aggregate from previous runs). However, unfolding native conserpin followed by refolding (double-jump) resulted in more consistent refolding traces that fitted to a double exponential (SI methods; Fig. S1H). Both single and double jump refolding identified two rates that were independent of the delay time (Figs S1I,J and 1J). Plausible explanations for the presence of two refolding rates are: two denatured states folding on different timescales (e.g. folding limited by proline isomerisation); a fast rate of refolding to an intermediate, followed by a slow rate of refolding from that intermediate; or two fluorophores reporting on independent folding events (e.g. two independently nucleating subdomains). In our data, it is most likely that we are detecting folding from two similarly structured ground states. If we were observing a fast rate, followed by a slow rate, we should expect the fast rate to become kinetically invisible when the two rates cross (~2 M GuHCl), which it does not. Similarly, if there are two independent folding events, then the relative amplitudes of each rate should be consistent, which they are not. Most interestingly, the refolding m-values are positive at low concentrations of denaturant (<2 M), suggesting that the two populated ground states are more structured than the subsequent folding transition state(s). Therefore, the starting states cannot be denatured states, and must be structured intermediates (I1 and I2) that fold on different time-scales (Figs 1J and S1I,J). The fast folding rate (red squares) matches up with the unfolding rate at the expected [D]50%, (2.75 M), verifying that this rate shows folding over the major transition state. The “rollover” in this rate demonstrates that the first intermediate I1 is in rapid pre-equilibrium with the denatured state (D) and there is a switch in ground state from I1 to D when the two species are of equal stability (2 M GuHCl, red squares in Fig. 1J). The second intermediate I2 (Fig. 1J, black circles) shows an almost identical folding m-value and, assuming this also folds over the major transition state, is likely to be very similar in structure to I1. However, I2 is more stable than I1 and persists until the denaturant midpoint (2.75 M). As such, we propose that I1 is likely to be the previously observed polymerogenic folding branch point19,40,47,50,51,52,53. As this species is highly aggregation prone in other serpins, it is possible that the second intermediate (I2) is a multimer of the first intermediate.
Global structural features of native conserpin are not typical for a thermostable protein
Our data imply that the folding reversibility and low polymerization propensity of conserpin is due to alteration of the folding landscape, resulting in minimal formation of a folding intermediate. Comparison of native and latent state conserpin structures with available native, latent and cleaved structures of α1-AT54,55,56, plasminogen activator inhibitor 1 (PAI-1)57,58, α1-antichymotrypsin (ACH)59, neuroserpin60,61, antithrombin62 and the thermostable serpins, thermopin63 and tengpin64 reveal that despite having the highest thermostability, native conserpin has the fewest H-bonds and salt bridges (Table S2). Further, native state conserpin has the largest accessible surface area and largest solvent inaccessible cavity volume of all assessed serpins. These characteristics are unusual for thermostable proteins, which typically feature more interactions and optimized packing compared to their mesophilic counterparts37,65,66,67,68,69,70,71,72,73. Comparison of the electrostatic surface potential of conserpin with that of α1-AT reveals minor differences on the surface-exposed face of the A-sheet, whilst the opposite face of the molecule is substantially more positively charged (Fig. 2A), consistent with the reported aggregation resistance of proteins featuring increased electrostatic surface potential74,75. In contrast to the majority of mesophilic proteins and their thermophilic homologues, a correlation between overall number of H-bonds/salt bridges and thermostability is not apparent for serpins, which must balance the relative stabilities of native and RCL-inserted states to enable unique conformational plasticity underpinning inhibitory function63,64 (Table S2). This reasoning suggests that more subtle, context-dependent structural and dynamical features play a more dominant role in conserpin, which we explore next.
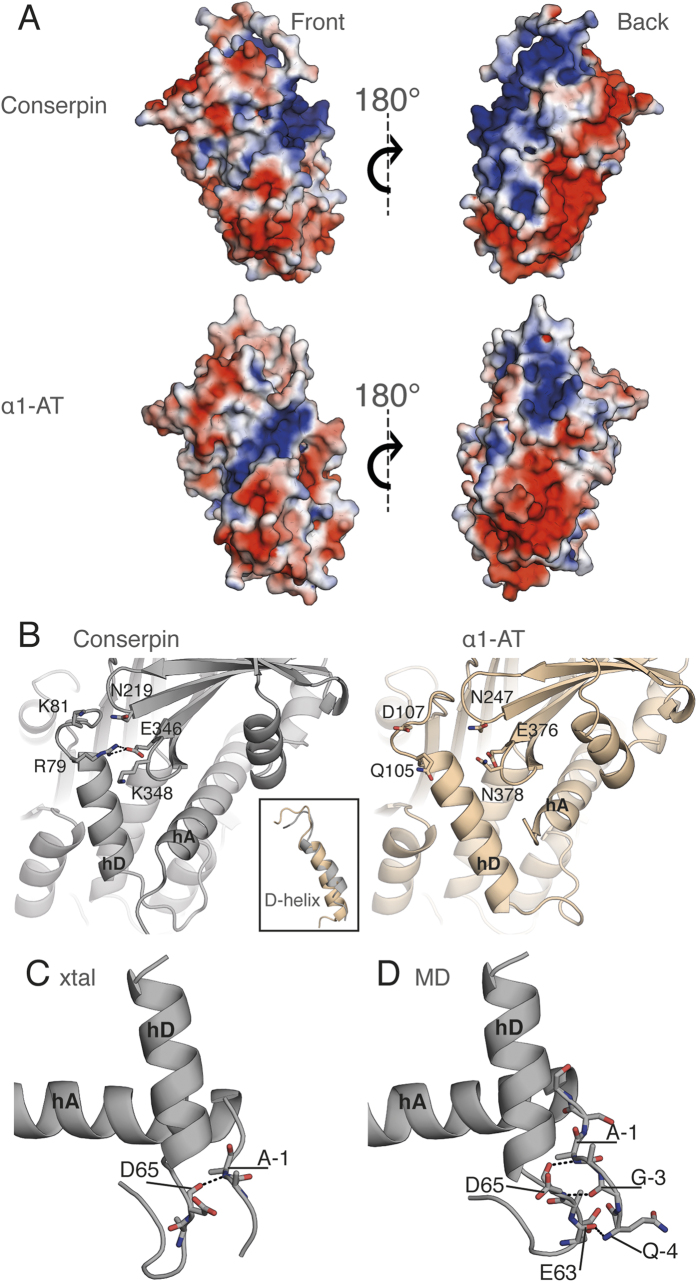
Figure 2. Structural analysis reveals alterations of the electrostatic surface and stabilization of the D-helix in conserpin.
(A) The electrostatic potential surface of conserpin and α1-AT models (blue = +ve, red = −ve), in the same orientation as Fig. 1A (front) and a 180° rotation reveals an overall increase in positive charge on the back face of conserpin. (B) The introduced salt bridge in hD of conserpin with residues Q105R79 and E376346. There is no comparable interaction present in α1-AT. Inset shows the shortened D-helix in conserpin. (C) H-bonding between A-1 of the extended N-terminus and D65 of hD, as seen in the conserpin crystal structure. (D) Persistent hydrogen bonding between Q-4, G-3 and A-1 of the extended N-terminus and E63 and D65 of hD in conserpin as seen in MD simulation.
Favorable interactions and reduced dynamics surrounding the D-helix
Given the conformational plasticity required for serpin function, we next performed molecular dynamics (MD) simulations for 0.5 μs at 300 K in triplicate for both conserpin and α1-AT. Both systems reach equilibrium by 150 ns (Fig. S4A). Although the increased mobility of the RCL and the C-terminus of hA of conserpin leads to a higher overall RMSD, inspection of root mean square fluctuations (RMSFs) shows conserpin to exhibit an overall reduction in dynamics in the majority of regions, specifically the extended N-terminus of hA, hC/hD loop, hD, hE, hF, hG, hH (Fig. S4B,C). This is further supported by a large reduction in conformational sampling as shown by principle component analysis (Fig. S4D). The most notable reduction in dynamics is in the D-helix (hD; RMSD of 0.58 vs. 1.65 Å; Figs 2B and S4B,C). The D-helix of α1-AT has been implicated in stability; notably two mutations (T114Fα1-AT and G117Fα1-AT) stabilize the D-helix and rescue the polymerogenic Z-variant76,77. The D-helix of conserpin is shortened by the deletion of five residues, four at the N-terminus (L84, E86, I87 and P88 in α1-AT) and one at the C-terminus (Q109 in α1-AT; Figs 2B & S5A). The deletion of L84α1-AT and I87α1-AT reduces overall hydrophobicity without affecting the packing of hD against the core of conserpin (Fig. S5A). Residue numbering will adhere to the following convention unless explicitly stated: Q105α1-AT or R79conserpin or Q105R79, where Q105 from α1-AT has been mutated to an R, which is residue number 79 in conserpin.
The rigidity of hD in conserpin is probably due to a salt bridge between Q105R79 of hD and E376346 and interactions of the N-terminus with hD. The salt bridge between the B-sheet and hD is present throughout the MD simulation and possibly stabilizes the top of the D-helix (Fig. 2B). In contrast, there are no similar salt bridges in the α1-AT crystal structure or during MD (Fig. 2B). Rather, hD in α1-AT undergoes conformational rearrangement and loss of secondary structure in one of the replicates (Fig. S5B). This is consistent with other reports, which indicates that minor changes to hD may accelerate or reduce polymer formation76,77. The N-terminus in conserpin is extended by the addition of a purification tag. Four residues of the extension were resolved in the crystal structure and a single H-bond is observed between the backbone of residue A-1conserpin and the N-terminus of hD (D65conserpin; Fig. 2C). This H-bond is persistent throughout MD and extends to form a small β-sheet (Fig. 2D). Therefore, the extended N-terminus may impart stability to hD and may reflect similar interactions seen in the naturally extended N-termini of thermophilic serpins63,64. Taken together, our observations suggest that optimized interactions in and around hD increase the stability of the native state.
The electrostatic network of the serpin breach region is extended in conserpin
The breach region, consisting of a highly conserved electrostatic network between residues E342α1-AT, K290α1-AT and D341α1-AT at the top of the A-sheet is important for controlling the conformational change that drives protease inhibition39,49,79. This network is significantly extended in conserpin, compared to α1-AT (Fig. 3A). Specifically, the mutations of T339E310 and S292K264 contribute to a salt bridge network spanning s3A, s5A and s6A with K191163. T294E266 also forms a new salt bridge with K335306 between s6A and s5A, whilst D341N312 mediates an unfavorably charged cluster of E310conserpin, E313conserpin and E314conserpin that is not present in α1-AT (Fig. 3A). These observations are interesting in the context of serpin polymerization, which involves insertion of the RCL and/or s5A from one molecule into the flexible A-sheet of another19,23,24,40,51,78. In particular, the disease-causing Z-variant, E342Kα1-AT induces repulsion with K290α1-AT, which either retards the formation of the A-sheet during folding, increasing the lifetime of the polymerogenic intermediate ensemble, or destabilizes the structure and increases the dynamics of the native state, allowing for s5A and s6A to separate, and reduce the energy barrier for polymerization19,40,80,81,82.
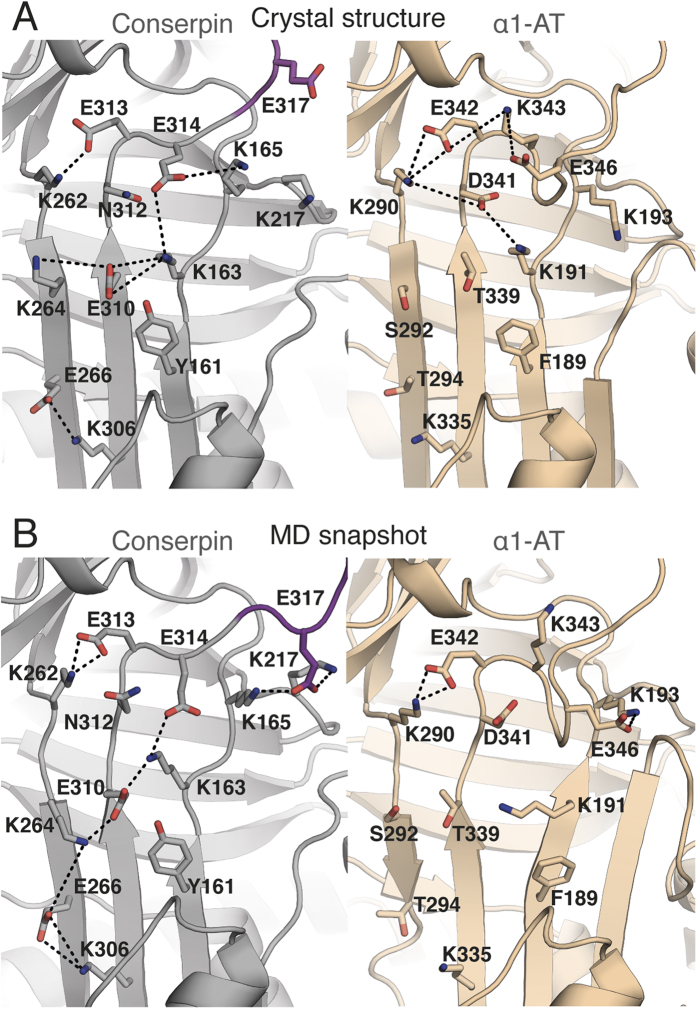
Figure 3. The electrostatic network of the breach region is extended in conserpin.
(A) A-sheet salt bridge interactions (dashed lines) in the crystal structures of conserpin (carbon atoms in grey) and α1-AT (carbon atoms in wheat; PDB: 3NE4). (B) A simulation snapshot taken at 500 ns, showing A-sheet salt bridge interactions as described above. The modeled RCL of conserpin is colored magenta.
It is difficult to ascertain the effect of the extended salt bridge network on A-sheet dynamics during folding of conserpin as MD simulations only describe the dynamics of the native state. Nevertheless, simulations of α1-AT reveal its A-sheet salt bridge network to be weaker over time (Fig. 3B), allowing for the transient separation of strands s5A and s3A (Fig. S6). In contrast, the extensive network in conserpin is present throughout the majority of the simulation, with additional interactions being formed, notably an alternate conformation of K264conserpin mediating interactions between E310conserpin, and E266conserpin (Fig. 3B). Furthermore, E317conserpin in the RCL of conserpin is able to adopt a stable conformation, mediating the salt-bridge between K165conserpin and K217conserpin, with K165conserpin forming transient interactions to E314conserpin. Equilibrium and kinetic folding studies of α1-AT provide compelling evidence for the late folding of s5A during transition through the polymerogenic intermediate state19,40. Taken together, our observations suggest an improved energetically stable conserpin native state with possible increases to the folding cooperativity in this region, which may also be augmented by the hydrophobic core behind the A-sheet.
Biophysical and structural analysis of Z-conserpin
Intrigued by the stabilizing electrostatic interactions in the breach region of conserpin, we assessed the effect of introducing the disease-causing Z-mutation, E342K313, into conserpin. Mutation of E342α1-AT to a lysine results in an increased propensity of α1-AT to polymerize in the endoplasmic reticulum of hepatocytes, leading to a lack of secretion into the circulation26. Studying the effects of the Z-variant in α1-AT is difficult due to expression as insoluble aggregate76,83. The most likely mechanism of Z-variant polymerization involves perturbation of the folding energy landscape, thus increasing the lifetime of the polymerogenic intermediate ensemble24,40,81,83. Although there is evidence to suggest that the Z-mutation also results in structural and dynamic changes to the native state80,82,84, a recent crystal structure of Z α1-AT shows minimal perturbation in comparison to wild-type83. In order to investigate the effects of a highly destabilizing mutation on conserpin, we introduced E342K313 into conserpin to produce Z-conserpin.
Z-conserpin expressed well as a soluble monomer in E. coli, which has not been possible with Z α1-AT76,83. Z-conserpin showed a highly similar inhibitory profile to conserpin, with an SI of 2.3 and a kass of 2.1 × 107 M−1 s−1 (Fig. S7A,B). Z-conserpin exhibits reversible, two-state folding upon chemical denaturation (Fig. S7C,D). The equilibrium unfolding and refolding curves overlay almost perfectly, revealing a midpoint of denaturation, [D]50 to be 2.51 ± 0.01 M, an equilibrium m-value, mD-N, of 5.18 kcal mol−1 M−1, and a stability ∆GD-N, of −12.8 kcal mol−1 (a loss of −10.04 kcal mol−1; Fig. S7D). As with conserpin, equilibrium data did not reveal the presence of an intermediate species. We therefore repeated equilibrium unfolding using bis-ANS fluorescence, detecting a fluorescent peak at ~2.5 M GuHCl, that is slightly broader and more intense than observed in conserpin, indicating an increase in the intermediate ensemble population, but still smaller than in α1-AT (Fig. S7E). Variable-temperature far-UV CD melting curves in 2 M GuHCl gave a Tm of 60.7 °C (conserpin Tm = 72.5 °C; (Fig. S7F)). Native PAGE shows conserpin to remain monomeric except when heated to 90 °C for 10 minutes, whilst Z-conserpin has a complete loss of monomer at 80 °C and forms a slightly higher molecular weight species when heated to 70 °C for 10 minutes (Fig. S7G). The crystal structure of native Z-conserpin (Table S1) reveals almost no structural differences upon mutation (backbone RMSD = 0.23 Å); the sole differences surrounding E342K313 are small side-chain shifts of K342313 and K290262, most likely as a result of electrostatic repulsion (Fig. S7H). A caveat is one local residue difference, K343E314, in conserpin that may partially negate the effects of E342K313, due to its salt bridge with K165conserpin. As such, future studies of the double mutant E342K313/E314Kconserpin would be insightful. Regardless, the structure of Z-conserpin reveals essentially no structural changes to the native state which disagrees with reports of structural perturbations within the native state80,82,84; therefore favoring the mechanism of Z-variant polymerization via a folding intermediate83. However, the intermediate versus native state polymerization mechanisms may be reconciled if the intermediate ensemble is native-like in structure, consistent with our kinetic (un)folding data for conserpin. Considering the evidence in support of this for a wide range of proteins85, our data is therefore consistent with the Z-mutation altering the folding energy landscape, possibly by lowering the kinetic barrier of the unfolding transition to the polymerogenic intermediate ensemble81,83.
Importance of A-sheet/F-helix hydrophobic core packing
The hydrophobic core buried by the A-sheet is important for serpin stability48,86,87. Amongst 19 mutations designed to probe the stability of α1-AT, seven mutations in the hydrophobic core were found to be stabilizing86. Four of these mutations are found in conserpin (T59S37, T68A46, A70G48 and M374I244). In the remaining three mutations, the local environment adapts to improve packing and local interactions (Fig. S8).
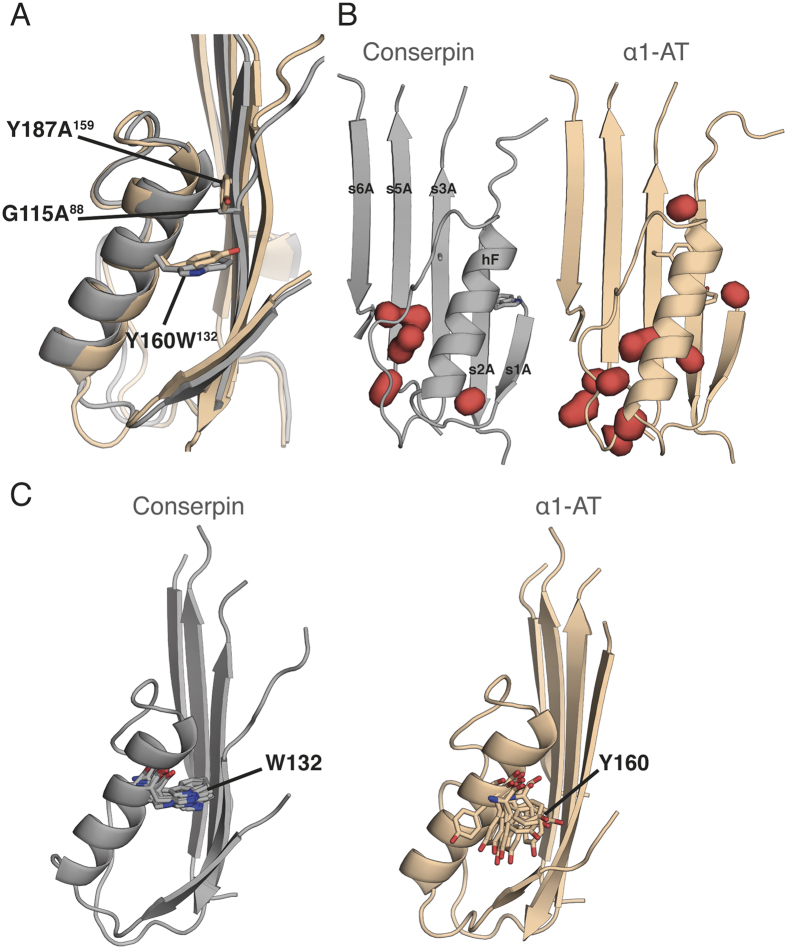
Packing between hF and the A-sheet also stabilizes the native serpin state, with hF acting as a physical barrier for RCL insertion into the A-sheet during protease inhibition and polymerization19,88,89,90,91. Conserpin contains three mutations in this region (Fig. 4A); Y187A159 and G115A88, which allow s2A to more tightly pack against hF, and Y160W132, which further improves the packing density (Fig. 4B). This is consistent with mutagenesis studies of α1-AT, where Y160A resulted in a 5 °C decrease in Tm and was attributed to the loss of a hydrogen bond and formation of a cavity88. In contrast, Y160W raised the Tm of α1-AT to 65 °C, and slowed the rate of polymerization88. MD reveals hF of conserpin to be slightly less flexible than that of α1-AT, with W160132 remaining conformationally locked compared to Y160 of α1-AT, which frequently flips in and out of the hydrophobic pocket (Fig. 4C). Interactions within the “clasp” motif at the F-helix are structurally conserved in conserpin and maintained throughout simulation, consistent with its proposed role in regulating conformational change92. Taken together, these changes likely contribute to the stability of the native state.
Figure 4. W160 stabilizes hF in conserpin.
(A) A structural overlay of hF in conserpin (grey) and α1-AT (wheat), highlighting the positions of Y160W132, Y187A159 and G115A88. (B) Solvent inaccessible cavities (red blobs) surrounding hF of conserpin and α1-AT. Y160W132 reduces cavity volumes from 233.8 to 120.9 Å3. (C) MD simulation frames (every 50 ns), highlighting the dynamic differences of W132 in conserpin and Y160 in α1-AT.
Remodeling the B/C barrel, a folding nucleus
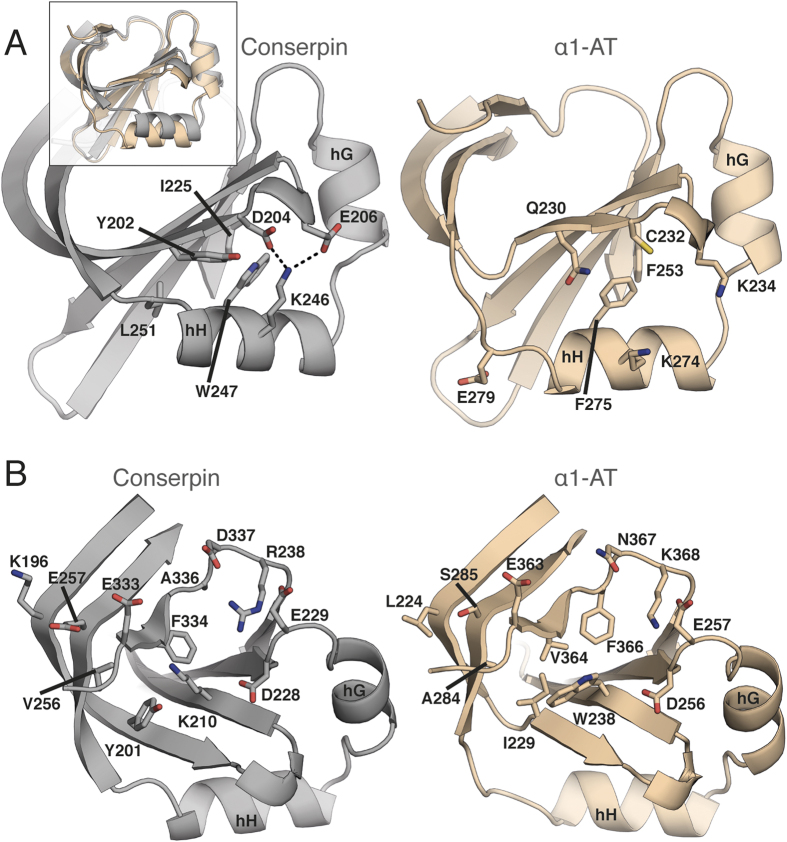
Formation of the B/C barrel is thought to occur early in the folding pathway of α1-AT, preceding formation of the A-sheet and acting as a “kinetic trap” that captures the RCL and prevents folding to other more stable states19,22,40,55,93. Conserpin contains several mutations in the B/C barrel that improve hydrophobic packing and form favorable interactions within the native state (Fig. 5A). Specifically, F275W247 and E279L251 allow tighter packing of hH. The introduction of a small salt-bridge network between K274246, C232D204 and K234E206 in hH may further stabilise the hydrophobic core of the B/C barrel (Fig. 5A). Conserpin harbors two potentially destabilizing mutations, but surrounding mutations have compensatory effects: the known destabilizing mutation F366A336 40,55, which in isolation would create a destabilizing cavity, is compensated by the mutation V364F334 and the introduction of a coordinated salt-bridge network between D256228, E257229, K368R338 and N367D337 (Fig. 5B); the potentially destabilizing mutation W238K210, which would likely weaken hydrophobic packing and introduce a large cavity, is offset by backbone polar contacts with E363333, and together with I229Y201 and A284V256 may function as a solvent barrier that shields the hydrophobic core (Fig. 5B). MD simulation also indicates a transient salt bridge between W238K210 and D256228. Conserpin also contains L224K196 and S285E257, which staples s2C and s3C together, further stabilizing the native state (Fig. 5B). Finally, L241E213 and N228Y200 are close to the B-sheet hydrophobic core and the region in which citrate was found to bind and stabilize α1-AT, thus potentially providing extra stability93. Taken together, these features may contribute to core nucleation rates during early protein folding, as well as native state resistance to unfolding, consistent with our unfolding and refolding data (Fig. 1).
Figure 5. Structural analysis of the B/C barrel in conserpin (grey) and α1-AT (wheat).
(A) Stabilizing hydrophobic mutations surrounding F275W247. (B) Remodeling of the inner barrel surrounding W238K210.
Conserpin is less frustrated than α1-AT
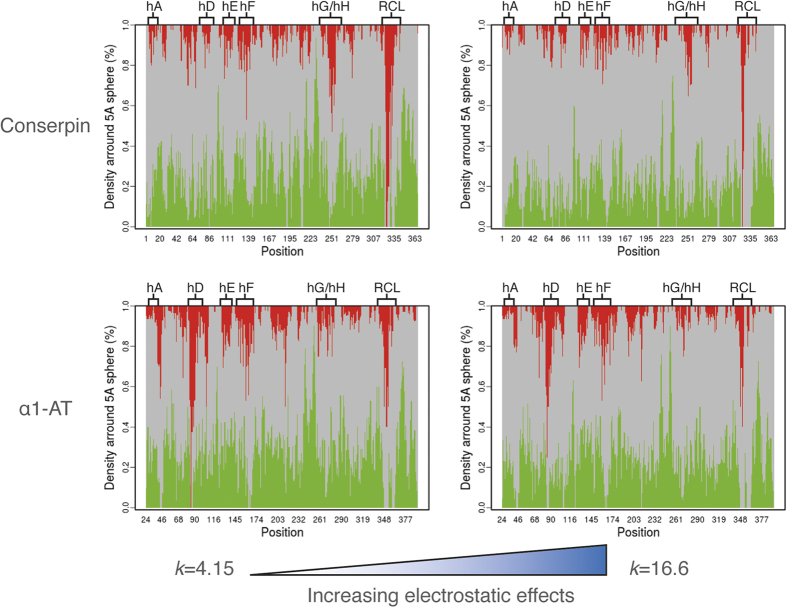
We next investigated the distribution of energetic frustration within the structures of conserpin and α1-AT using the frustratometer webserver94,95. As proteins are thought to be minimally frustrated polymers with rugged energy landscapes, the degree of energetic frustration is related to the description of the proteins energy landscape2,27,94; that is, a high level of frustration implies flexibility and a more rugged energy landscape. We therefore used configurational frustration analysis which describes interactions with respect to structural decoys that may be encountered during the folding process94,95. Overall, conserpin is less frustrated than α1-AT in several regions, with the exception of the RCL, which is shown to be more dynamic during MD simulation (Fig. 6 and S4B,C). In combination with MD simulation, these results show an inverse correlation between the degree of frustration and degree of dynamics, which is mediated by electrostatic effects (Figs 6 and S4B,C). By modulating the electrostatic constant (k) from 4.15 to 16.6, it becomes apparent that long-range interactions are essential to the reduced frustration of conserpin, but are not as significant in α1-AT (Fig. 6). This is particularly noticeable for helix D and F, which have fewer highly frustrated contacts in conserpin (Fig. 6). The improved folding properties and increased conserpin stability may therefore be related to the higher number of charged residues that stabilise local contacts and introduce repulsion between patches enriched in like charges, which in turn must be correctly oriented in unfolded forms to avoid aggregation96.
Figure 6. Configurational frustration analysis for conserpin and α1-AT.
Minimal, neutral and highly frustrated contacts are represented in green, gray and red respectively. Calculations were performed with different electrostatic strengths by varying the electrostatic constant (k). According to ref 95, larger k values are related to stronger effects of the Debye–Hückel term.
Discussion
The puzzle of how the folding polypeptide chain of serpins achieves a metastable native state has proven challenging to solve. Their unusual and complicated mechanism of protease inhibition challenges the characterization of their folding pathway42. It has been established that α1-AT, ACH and PAI-1 all unfold from their native states via an aggregation-prone intermediate ensemble19,22,40,42,43,47,53,55,89,97. More recent studies of α1-AT revealed relatively fast folding of the core B/C sheet β-barrel followed by much slower formation of the central A β-sheet40. These observations were consistent with models of how off-pathway α1-AT polymers form due to a folding “race” between the core barrel and the central β-sheet19,24. However, despite two decades of effort, the aggregation-prone nature and poor refolding properties of serpins have prevented a full, atomic level characterization of their folding landscape.
Our study reports for the first time, the successful engineering of a reversibly folding serpin that is highly resistant to polymerization and aggregation, even after the introduction of the polymerogenic, disease-causing Z-mutation. Structural analysis reveals the presence of many context dependant and stabilising interactions in regions that are known to be important for folding. These include stabilizing interactions around the D-helix, a salt bridge network in the A-sheet that may resist aberrant RCL insertion, optimization of A-sheet hydrophobic core packing, stabilising mutations in the F-helix that may raise the energy barrier for RCL insertion, and improved packing in the B/C barrel. Although some single mutations in these same regions have been reported to stabilise α1-AT, we found that many mutations within conserpin act together cooperatively. The relatively large accessible surface area and solvent inaccessible cavity volume compared to all other serpins are unusual for a thermostable protein, suggesting that stabilization is achieved by highly context-specific interactions. Although structure and dynamics suggest stabilisation of the native state in key regions, this represents a conundrum: how can function be maintained, which requires metastability, flexibility and conformational change, in combination with a high degree of stability?
Biophysical and structural analysis paints a complex picture. Although native conserpin features fewer overall number of polar contacts compared to other metastable serpins, new, specific interactions stabilise a rigidified native state that is less frustrated than α1-AT. The inhibitory activity of conserpin confirms that its native state retains metastability required for function. The slightly increased SI of conserpin is most likely due its sub-optimal RCL sequence hindering association with the target protease. However, functional impairment might also be caused by a slowing of the rate of insertion of its RCL into the central A β-sheet, which may also contribute to its aggregation resistance. The latent state structure reveals an overall increase in H-bonds and salt bridges. These observations underline the functional importance of maintaining the relative stabilities of the native versus RCL-inserted state (latent/cleaved) irrespective of the specific stabilising features, as observed previously for thermostable serpins63,64. However, the structural data do not completely explain the remarkable stability of conserpin. Indeed, the most interesting properties are seen during folding and on exposure to heat; conserpin folds in a concerted fashion, with a relatively minimised population of the aggregation prone intermediate ensemble, and avoids aggregation on heating, with preferential transition to the latent state. Taken together, these results reveal two distinct effects from chemical denaturation and thermal treatment that uniquely provides conserpin with two-state reversible folding, a high degree of thermostability and aggregation resistance. Furthermore, our findings suggest that the robustness of conserpin folding is due to remodelling of its energy landscape, specifically the smoothing of rugged features that trap aggregation-prone intermediates.
Remodelling of the energy landscape is fascinating from an evolutionary and protein engineering perspective. Consensus design typically accumulates residues important to native state stability28,29,31,32,34,98, but could equally alter the folding landscape37,38,99. As such, conserved features of the energy landscape would be solidified or even amplified, whilst non-conserved features would be minimised. For conserpin, this mechanism implies that aggregation and off-pathway folding events are not conserved across the serpin family, possibly because the functional and regulatory requirements of divergently evolved serpin clades sculpted rugged landscapes as an unfortunate consequence, as may be the case for other functional proteins1,2. Therefore, consensus design can potentially smooth the “risky”, rugged folding landscapes of functional proteins. This may offer several benefits for protein engineering in general, including the removal of aggregation-prone intermediates and modifying protein scaffolds for use as protein therapeutics and diagnostic reagents. In the case of serpins, our structural and folding data for conserpin and Z-conserpin demonstrate the potential of this engineered scaffold as a model system for studying pathological disease mutations. Finally, the fragile nature of serpin folding has thus far hindered residue-level kinetic characterization of all species on the folding pathway, for example using phi-value analysis100; the robustness of conserpin may finally provide the basis for such characterization.
Materials and Methods
To design conserpin we used a previously reported MSA of 219 serpin sequences39 to generate a consensus sequence as described previously37. After filtering to remove incomplete sequences and the application of redundancy reduction, we aligned 212 sequences and generated a new protein sequence by selecting the most frequently observed residue at each column of the MSA (the ‘consensus method’) (Dataset S1). Protein expression and purification, SI measurement and spectroscopic analysis was performed as described previously89. Protein Crystallography was performed at the MX1 and MX2 beamlines at the Australian Synchrotron101. All experimental and computational methods are described in detail in SI Methods.
Additional Information
Accession Numbers: The coordinates and structure factors have been deposited in the Protein Data Bank under accession codes 5CDX, 5CDZ and 5CE0.
How to cite this article: Porebski, B. T. et al. Smoothing a rugged protein folding landscape by sequence-based redesign. Sci. Rep. 6, 33958; doi: 10.1038/srep33958 (2016).
Supplementary Material
Acknowledgments
We thank Jane Clarke, Andrew Ellisdon, and Gordon Lloyd for helpful discussions and advice. BTP is a Medical Research Council Career Development Fellow. AAN and JJH are supported by the Wellcome Trust (grant number WT 095195). SM acknowledges fellowship support from the Australian Research Council (FT100100960). NAB is an Australian Research Council Future Fellow (110100223). GIW is an Australian Research Council Discovery Outstanding Researcher Award Fellow (DP140100087). AMB is a National Health and Medical Research Senior Research Fellow (1022688). JCW is an NHMRC Senior Principal Research fellow and also acknowledges the support of an ARC Federation Fellowship. We thank the Australian Synchrotron for beam-time and technical assistance. This work was supported by the Multi-modal Australian ScienceS Imaging and Visualisation Environment (MASSIVE) (www.massive.org.au). We acknowledge the Monash Protein Production Unit and Monash Macromolecular Crystallization Facility.
Footnotes
Author Contributions B.T.P., J.I., S.M., S.P.B. and A.M.B. designed the study. J.A.I., J.C.W., L.Z., S.P.B. and G.I.W. performed the protein design. S.K., E.M.M., M.A.P., W.D. and B.T.P. performed the protein expression, purification and CD thermal melt experiments. B.T.P., N.A.B., S.M. and S.K. performed the crystallography. B.T.P. performed molecular dynamics simulations and analysis with contributions from IK. D.E.H. assisted in biophysical data analysis. M.G.S.C. performed the frustration analysis, structural analysis and contributed to discussion of data. S.K., J.J.H. and A.A.N. performed the folding kinetics and equilibrium measurement experiments. B.T.P. and S.K. generated figures. B.T.P., S.M., A.A.N. and A.M.B. wrote the manuscript.
References
- Gershenson A., Gierasch L. M., Pastore A. & Radford S. E. Energy landscapes of functional proteins are inherently risky. Nat. Chem. Biol. 10, 884–891 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gianni S. et al. Understanding the frustration arising from the competition between function, misfolding, and aggregation in a globular protein. Proc. Natl. Acad. Sci. USA 111, 14141–14146 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leopold P. E., Montal M. & Onuchic J. N. Protein folding funnels: a kinetic approach to the sequence-structure relationship. Proceedings of the National Academy of Sciences 89, 8721–8725 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onuchic J. N., Wolynes P. G., Luthey-Schulten Z. & Socci N. D. Toward an outline of the topography of a realistic protein-folding funnel. Proceedings of the National Academy of Sciences 92, 3626–3630 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill K. A. & Chan H. S. From Levinthal to pathways to funnels. Nat. Struct. Biol. 4, 10–19 (1997). [DOI] [PubMed] [Google Scholar]
- Hilvert D. Design of protein catalysts. Annu. Rev. Biochem. 82, 447–470 (2013). [DOI] [PubMed] [Google Scholar]
- Loladze V. V., Ibarra-Molero B., Sanchez-Ruiz J. M. & Makhatadze G. I. Engineering a thermostable protein via optimization of charge-charge interactions on the protein surface. Biochemistry 38, 16419–16423 (1999). [DOI] [PubMed] [Google Scholar]
- Schreiber G., Buckle A. M. & Fersht A. R. Stability and function: two constraints in the evolution of barstar and other proteins. Structure 2, 945–951 (1994). [DOI] [PubMed] [Google Scholar]
- Russell R. J. & Taylor G. L. Engineering thermostability: lessons from thermophilic proteins. Current Opinion in Biotechnology 6, 370–374 (1995). [DOI] [PubMed] [Google Scholar]
- Dantas G., Kuhlman B., Callender D., Wong M. & Baker D. A Large Scale Test of Computational Protein Design: Folding and Stability of Nine Completely Redesigned Globular Proteins. Journal of Molecular Biology 332, 449–460 (2003). [DOI] [PubMed] [Google Scholar]
- Kuhlman B. Design of a Novel Globular Protein Fold with Atomic-Level Accuracy. Science 302, 1364–1368 (2003). [DOI] [PubMed] [Google Scholar]
- Koga N. et al. Principles for designing ideal protein structures. Nature 491, 222–227 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson J. S. & Richardson D. C. Natural beta-sheet proteins use negative design to avoid edge-to-edge aggregation. Proc. Natl. Acad. Sci. USA 99, 2754–2759 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht M. H., Richardson J. S., Richardson D. C. & Ogden R. C. De novo design, expression, and characterization of Felix: a four-helix bundle protein of native-like sequence. Science 249, 884–891 (1990). [DOI] [PubMed] [Google Scholar]
- Jin W., Kambara O., Sasakawa H., Tamura A. & Takada S. De novo design of foldable proteins with smooth folding funnel: automated negative design and experimental verification. Structure/Folding and Design 11, 581–590 (2003). [DOI] [PubMed] [Google Scholar]
- Gettins P. G. W. Serpin Structure, Mechanism, and Function. Chem. Rev. 102, 4751–4804 (2002). [DOI] [PubMed] [Google Scholar]
- Lomas D. A. & Carrell R. W. Serpinopathies and the conformational dementias. Nat Rev Genet 3, 759–768 (2002). [DOI] [PubMed] [Google Scholar]
- Law R. H. P. et al. An overview of the serpin superfamily. Genome Biol. 7, 216 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan B. & Gierasch L. M. Dynamic local unfolding in the serpin α-1 antitrypsin provides a mechanism for loop insertion and polymerization. Nature Structural & Molecular Biology 18, 222–226 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntington J. A., Read R. J. & Carrell R. W. Structure of a serpin-protease complex shows inhibition by deformation. Nature 407, 923–926 (2000). [DOI] [PubMed] [Google Scholar]
- Stratikos E. & Gettins P. G. Major proteinase movement upon stable serpin-proteinase complex formation. Proceedings of the National Academy of Sciences 94, 453–458 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tew D. J. & Bottomley S. P. Probing the equilibrium denaturation of the serpin α1-antitrypsin with single tryptophan mutants; evidence for structure in the urea unfolded state. Journal of Molecular Biology 313, 1161–1169 (2001). [DOI] [PubMed] [Google Scholar]
- Yamasaki M., Li W., Johnson D. J. D. & Huntington J. A. Crystal structure of a stable dimer reveals the molecular basis of serpin polymerization. Nature 455, 1255–1258 (2008). [DOI] [PubMed] [Google Scholar]
- Yamasaki M., Sendall T. J., Pearce M. C., Whisstock J. C. & Huntington J. A. Molecular basis of α1-antitrypsin deficiency revealed by the structure of a domain-swapped trimer. EMBO Rep. 12, 1011–1017 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekeowa U. I. et al. Defining the mechanism of polymerization in the serpinopathies. Proc. Natl. Acad. Sci. USA 107, 17146–17151 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomas D. A., Evans D. L., Finch J. T. & Carrell R. W. The mechanism of Z alpha 1-antitrypsin accumulation in the liver. Nature 357, 605–607 (1992). [DOI] [PubMed] [Google Scholar]
- Ferreiro D. U., Komives E. A. & Wolynes P. G. Frustration in biomolecules. Q. Rev. Biophys. 47, 285–363 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann M. & Wyss M. Engineering proteins for thermostability: the use of sequence alignments versus rational design and directed evolution. Current Opinion in Biotechnology 12, 371–375 (2001). [DOI] [PubMed] [Google Scholar]
- Steipe B., Schiller B., Plückthun A. & Steinbacher S. Sequence statistics reliably predict stabilizing mutations in a protein domain. Journal of Molecular Biology 240, 188–192 (1994). [DOI] [PubMed] [Google Scholar]
- Steipe B. Evolutionary approaches to protein engineering. Curr. Top. Microbiol. Immunol. 243, 55–86 (1999). [DOI] [PubMed] [Google Scholar]
- Lehmann M. et al. The consensus concept for thermostability engineering of proteins: further proof of concept. Protein Eng. 15, 403–411 (2002). [DOI] [PubMed] [Google Scholar]
- Wang Q., Buckle A. M., Foster N. W., Johnson C. M. & Fersht A. R. Design of highly stable functional GroEL minichaperones. Protein Sci. 8, 2186–21193 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell K. L. & Davidson A. R. Mutagenesis of a Buried Polar Interaction in an SH3 Domain: Sequence Conservation Provides the Best Prediction of Stability Effects †. Biochemistry 37, 16172–16182 (1998). [DOI] [PubMed] [Google Scholar]
- Nikolova P. V., Henckel J., Lane D. P. & Fersht A. R. Semirational design of active tumor suppressor p53 DNA binding domain with enhanced stability. Proc. Natl. Acad. Sci. USA 95, 14675–14680 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai M. et al. The creation of a novel fluorescent protein by guided consensus engineering. Protein Eng. Des. Sel. 20, 69–79 (2007). [DOI] [PubMed] [Google Scholar]
- Jacobs S. A. et al. Design of novel FN3 domains with high stability by a consensus sequence approach. Protein Eng. Des. Sel. 25, 107–117 (2012). [DOI] [PubMed] [Google Scholar]
- Porebski B. T. et al. Structural and dynamic properties that govern the stability of an engineered fibronectin type III domain. Protein Eng. Des. Sel. 28, 67–78 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porebski B. T. & Buckle A. M. Consensus protein design. Protein Eng. Des. Sel. 29, 245–251 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irving J. A., Pike R. N., Lesk A. M. & Whisstock J. C. Phylogeny of the serpin superfamily: implications of patterns of amino acid conservation for structure and function. Genome Research 10, 1845–1864 (2000). [DOI] [PubMed] [Google Scholar]
- Tsutsui Y., Cruz, Dela R. & Wintrode P. L. Folding mechanism of the metastable serpin α1-antitrypsin. Proc. Natl. Acad. Sci. USA 109, 4467–4472 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon K. S., Lee S. & Yu M. H. Refolding of alpha 1-antitrypsin expressed as inclusion bodies in Escherichia coli: characterization of aggregation. Biochim. Biophys. Acta 1247, 179–184 (1995). [DOI] [PubMed] [Google Scholar]
- Whisstock J. C. & Bottomley S. P. Molecular gymnastics: serpin structure, folding and misfolding. Current Opinion in Structural Biology 16, 761–768 (2006). [DOI] [PubMed] [Google Scholar]
- Wang Z., Mottonen J. & Goldsmith E. J. Kinetically controlled folding of the serpin plasminogen activator inhibitor 1. Biochemistry 35, 16443–16448 (1996). [DOI] [PubMed] [Google Scholar]
- Shirai N., Tani F., Higasa T. & Yasumoto K. Linear polymerization caused by the defective folding of a non-inhibitory serpin ovalbumin. J. Biochem. 121, 787–797 (1997). [DOI] [PubMed] [Google Scholar]
- Takehara S. et al. Refolding and polymerization pathways of neuroserpin. Journal of Molecular Biology 403, 751–762 (2010). [DOI] [PubMed] [Google Scholar]
- Onda M. & Hirose M. Refolding mechanism of ovalbumin: investigation by using a starting urea-denatured disulfide isomer with mispaired CYS367-CYS382. J. Biol. Chem. 278, 23600–23609 (2003). [DOI] [PubMed] [Google Scholar]
- Pearce M. C., Rubin H. & Bottomley S. P. Conformational change and intermediates in the unfolding of alpha 1-antichymotrypsin. J. Biol. Chem. 275, 28513–28518 (2000). [DOI] [PubMed] [Google Scholar]
- Kwon K. S., Kim J., Shin H. S. & Yu M. H. Single amino acid substitutions of alpha 1-antitrypsin that confer enhancement in thermal stability. J. Biol. Chem. 269, 9627–9631 (1994). [PubMed] [Google Scholar]
- Dafforn T. R., Mahadeva R., Elliott P. R., Sivasothy P. & Lomas D. A. A kinetic mechanism for the polymerization of alpha1-antitrypsin. J. Biol. Chem. 274, 9548–9555 (1999). [DOI] [PubMed] [Google Scholar]
- James E. L., Whisstock J. C., Gore M. G. & Bottomley S. P. Probing the unfolding pathway of alpha1-antitrypsin. J. Biol. Chem. 274, 9482–9488 (1999). [DOI] [PubMed] [Google Scholar]
- Knaupp A. S. et al. The Roles of Helix I and Strand 5A in the Folding, Function and Misfolding of α1-Antitrypsin. PLoS ONE 8, e54766 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran S. T. & Shrake A. The folding of alpha-1-proteinase inhibitor: kinetic vs equilibrium control. Archives of Biochemistry and Biophysics 385, 322–331 (2001). [DOI] [PubMed] [Google Scholar]
- Kim D. & Yu M. H. Folding pathway of human alpha 1-antitrypsin: characterization of an intermediate that is active but prone to aggregation. Biochem. Biophys. Res. Commun. 226, 378–384 (1996). [DOI] [PubMed] [Google Scholar]
- Patschull A. O. M. et al. Therapeutic target-site variability in α1-antitrypsin characterized at high resolution. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 67, 1492–1497 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im H. Interactions Causing the Kinetic Trap in Serpin Protein Folding. J. Biol. Chem. 277, 46347–46354 (2002). [DOI] [PubMed] [Google Scholar]
- Dunstone M. A. et al. Cleaved antitrypsin polymers at atomic resolution. Protein Sci. 9, 417–420 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou A., Huntington J. A., Pannu N. S., Carrell R. W. & Read R. J. How vitronectin binds PAI-1 to modulate fibrinolysis and cell migration. Nat. Struct. Biol. 10, 541–544 (2003). [DOI] [PubMed] [Google Scholar]
- Stout T. J., Graham H., Buckley D. I. & Matthews D. J. Structures of active and latent PAI-1: a possible stabilizing role for chloride ions. Biochemistry 39, 8460–8469 (2000). [DOI] [PubMed] [Google Scholar]
- Baumann U. et al. Crystal structure of cleaved human alpha 1-antichymotrypsin at 2.7 A resolution and its comparison with other serpins. Journal of Molecular Biology 218, 595–606 (1991). [DOI] [PubMed] [Google Scholar]
- Takehara S. et al. The 2.1-A crystal structure of native neuroserpin reveals unique structural elements that contribute to conformational instability. Journal of Molecular Biology 388, 11–20 (2009). [DOI] [PubMed] [Google Scholar]
- Ricagno S., Caccia S., Sorrentino G., Antonini G. & Bolognesi M. Human neuroserpin: structure and time-dependent inhibition. Journal of Molecular Biology 388, 109–121 (2009). [DOI] [PubMed] [Google Scholar]
- Skinner R. et al. The 2.6 A structure of antithrombin indicates a conformational change at the heparin binding site. Journal of Molecular Biology 266, 601–609 (1997). [DOI] [PubMed] [Google Scholar]
- Fulton K. F. et al. The high resolution crystal structure of a native thermostable serpin reveals the complex mechanism underpinning the stressed to relaxed transition. J. Biol. Chem. 280, 8435–8442 (2005). [DOI] [PubMed] [Google Scholar]
- Zhang Q. et al. The N terminus of the serpin, tengpin, functions to trap the metastable native state. EMBO Rep. 8, 658–663 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horovitz A., Serrano L., Avron B., Bycroft M. & Fersht A. R. Strength and co-operativity of contributions of surface salt bridges to protein stability. Journal of Molecular Biology 216, 1031–1044 (1990). [DOI] [PubMed] [Google Scholar]
- Serrano L., Horovitz A., Avron B., Bycroft M. & Fersht A. R. Estimating the contribution of engineered surface electrostatic interactions to protein stability by using double-mutant cycles. Biochemistry 29, 9343–9352 (1990). [DOI] [PubMed] [Google Scholar]
- Fersht A. R. & Serrano L. Principles of protein stability derived from protein engineering experiments. Current Opinion in Structural Biology 3, 75–83 (1993). [Google Scholar]
- Tokuriki N., Stricher F., Serrano L. & Tawfik D. S. How protein stability and new functions trade off. PLoS Comput Biol 4, e1000002 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpusas M., Baase W. A., Matsumura M. & Matthews B. W. Hydrophobic packing in T4 lysozyme probed by cavity-filling mutants. Proc. Natl. Acad. Sci. USA 86, 8237–8241 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chothia C. & Finkelstein A. V. The classification and origins of protein folding patterns. Annu. Rev. Biochem. 59, 1007–1039 (1990). [DOI] [PubMed] [Google Scholar]
- DeDecker B. S. et al. The crystal structure of a hyperthermophilic archaeal TATA-box binding protein. Journal of Molecular Biology 264, 1072–1084 (1996). [DOI] [PubMed] [Google Scholar]
- Levitt M., Gerstein M., Huang E., Subbiah S. & Tsai J. Protein folding: the endgame. Annu. Rev. Biochem. 66, 549–579 (1997). [DOI] [PubMed] [Google Scholar]
- Kellis J. T., Nyberg K., Sali D. & Fersht A. R. Contribution of hydrophobic interactions to protein stability. Nature 333, 784–786 (1988). [DOI] [PubMed] [Google Scholar]
- Lawrence M. S., Phillips K. J. & Liu D. R. Supercharging proteins can impart unusual resilience. J. Am. Chem. Soc. 129, 10110–10112 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miklos A. E. et al. Structure-based design of supercharged, highly thermoresistant antibodies. Chemistry & Biology 19, 449–455 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parfrey H. et al. Targeting a Surface Cavity of 1-Antitrypsin to Prevent Conformational Disease. J. Biol. Chem. 278, 33060–33066 (2003). [DOI] [PubMed] [Google Scholar]
- Gooptu B. et al. Crystallographic and Cellular Characterisation of Two Mechanisms Stabilising the Native Fold of α1-Antitrypsin: Implications for Disease and Drug Design. Journal of Molecular Biology 387, 857–868 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivasothy P., Dafforn T. R., Gettins P. G. W. & Lomas D. A. Pathogenic 1-Antitrypsin Polymers Are Formed by Reactive Loop–Sheet A Linkage. J. Biol. Chem. 275, 33663–33668 (2000). [DOI] [PubMed] [Google Scholar]
- James E. L. & Bottomley S. P. The mechanism of alpha 1-antitrypsin polymerization probed by fluorescence spectroscopy. Archives of Biochemistry and Biophysics 356, 296–300 (1998). [DOI] [PubMed] [Google Scholar]
- Kass I., Knaupp A. S., Bottomley S. P. & Buckle A. M. Conformational properties of the disease-causing Z variant of α1-antitrypsin revealed by theory and experiment. Biophys. J. 102, 2856–2865 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knaupp A. S., Levina V., Robertson A. L., Pearce M. C. & Bottomley S. P. Kinetic instability of the serpin Z alpha1-antitrypsin promotes aggregation. Journal of Molecular Biology 396, 375–383 (2010). [DOI] [PubMed] [Google Scholar]
- Hughes V. A., Meklemburg R., Bottomley S. P. & Wintrode P. L. The Z mutation alters the global structural dynamics of α1-antitrypsin. PLoS ONE 9, e102617 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X. et al. Molecular Mechanism of Z α1-Antitrypsin Deficiency. J. Biol. Chem. 291, 15674–15686 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knaupp A. S. & Bottomley S. P. Structural change in β-sheet A of Z α(1)-antitrypsin is responsible for accelerated polymerization and disease. Journal of Molecular Biology 413, 888–898 (2011). [DOI] [PubMed] [Google Scholar]
- Best R. B., Hummer G. & Eaton W. A. Native contacts determine protein folding mechanisms in atomistic simulations. Proc. Natl. Acad. Sci. USA 110, 17874–17879 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K. N., Park S. D. & Yu M. H. Probing the native strain in alpha1-antitrypsin. Nat. Struct. Biol. 3, 497–500 (1996). [DOI] [PubMed] [Google Scholar]
- Kim J., Lee K. N., Yi G. S. & Yu M. H. A thermostable mutation located at the hydrophobic core of alpha 1-antitrypsin suppresses the folding defect of the Z-type variant. J. Biol. Chem. 270, 8597–8601 (1995). [DOI] [PubMed] [Google Scholar]
- Cabrita L. D., Whisstock J. C. & Bottomley S. P. Probing the Role of the F-Helix in Serpin Stability through a Single Tryptophan Substitution †. Biochemistry 41, 4575–4581 (2002). [DOI] [PubMed] [Google Scholar]
- Cabrita L. D., Dai W. & Bottomley S. P. Different Conformational Changes within the F-Helix Occur during Serpin Folding, Polymerization, and Proteinase Inhibition †. Biochemistry 43, 9834–9839 (2004). [DOI] [PubMed] [Google Scholar]
- Gettins P. G. W. The F-helix of serpins plays an essential, active role in the proteinase inhibition mechanism. FEBS Letters 523, 2–6 (2002). [DOI] [PubMed] [Google Scholar]
- Gooptu B. et al. Inactive conformation of the serpin alpha(1)-antichymotrypsin indicates two-stage insertion of the reactive loop: implications for inhibitory function and conformational disease. Proc. Natl. Acad. Sci. USA 97, 67–72 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyon M. P. et al. Structural dynamics associated with intermediate formation in an archetypal conformational disease. Structure 20, 504–512 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce M. C. et al. Preventing serpin aggregation: the molecular mechanism of citrate action upon antitrypsin unfolding. Protein Sci. 17, 2127–21133 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreiro D. U., Hegler J. A., Komives E. A. & Wolynes P. G. Localizing frustration in native proteins and protein assemblies. Proc. Natl. Acad. Sci. USA 104, 19819–19824 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra R. G. et al. Protein Frustratometer 2: a tool to localize energetic frustration in protein molecules, now with electrostatics. Nucleic Acids Res. 44, W356–W360 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai M.-Y. et al. Electrostatics, structure prediction, and the energy landscapes for protein folding and binding. Protein Sci. 25, 255–269 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce M. C., Cabrita L. D., Rubin H., Gore M. G. & Bottomley S. P. Identification of residual structure within denatured antichymotrypsin: implications for serpin folding and misfolding. Biochem. Biophys. Res. Commun. 324, 729–735 (2004). [DOI] [PubMed] [Google Scholar]
- Lehmann M. et al. From DNA sequence to improved functionality: using protein sequence comparisons to rapidly design a thermostable consensus phytase. Protein Eng. 13, 49–57 (2000). [DOI] [PubMed] [Google Scholar]
- Barrick D., Ferreiro D. U. & Komives E. A. Folding landscapes of ankyrin repeat proteins: experiments meet theory. Current Opinion in Structural Biology 18, 27–34 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fersht A. R. Characterizing transition states in protein folding: an essential step in the puzzle. Current Opinion in Structural Biology 5, 79–84 (1995). [DOI] [PubMed] [Google Scholar]
- Cowieson N. P. et al. MX1: a bending-magnet crystallography beamline serving both chemical and macromolecular crystallography communities at the Australian Synchrotron. Journal of Synchrotron Radiation 22, 187–190 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.