Abstract
The human gastric pathogen, Helicobacter pylori, is becoming increasingly resistant to most available antibiotics. Peptidoglycan (PG) metabolism is essential to eubacteria, hence, an excellent target for the development of new therapeutic strategies. However, our knowledge on PG metabolism in H. pylori remains poor. We have further characterized an isogenic mutant of the amiA gene encoding a N-acetylmuramoyl-l-alanyl amidase. The amiA mutant displayed long chains of unseparated cells, an impaired motility despite the presence of intact flagella and a tolerance to amoxicillin. Interestingly, the amiA mutant was impaired in colonizing the mouse stomach suggesting that AmiA is a valid target in H. pylori for the development of new antibiotics. Using reverse phase high-pressure liquid chromatography, we analyzed the PG muropeptide composition and glycan chain length distribution of strain 26695 and its amiA mutant. The analysis showed that H. pylori lacked muropeptides with a degree of cross-linking higher than dimeric muropeptides. The amiA mutant was also characterized by a decrease of muropeptides carrying 1,6-anhydro-N-acetylmuramic acid residues, which represent the ends of the glycan chains. This correlated with an increase of very long glycan strands in the amiA mutant. It is suggested that these longer glycan strands are trademarks of the division site. Taken together, we show that the low redundancy on genes involved in PG maturation supports H. pylori as an actractive alternative model to study PG metabolism and cell shape regulation.
Introduction
Helicobacter pylori is the etiological agent of duodenal and gastric ulcers, of gastric adenocarcinoma and of mucosa-associated lymphoid tissue lymphoma. It colonizes around half of the human population. Despite its medical importance, we still have a fragmented knowledge of this human pathogen, in particular, regarding its physiology. The emergence of resistant strains to most available antibiotics active against H. pylori has stimulated the search for new therapeutic strategies against H. pylori. The peptidoglycan (PG or murein) is an essential macromolecule that surrounds the cytoplasmic membrane and functions as an exoskeleton. PG is structurally composed of glycan strands of repeating disaccharide units of N-acetyl-d-glucosamine-β(1,4)-N-acetylmuramic acid (GM) cross-linked via short stem peptides creating one single huge heteropolymer molecule surrounding each bacterial cell. This exoskeleton is required to withstand turgor pressure, to maintain cell shape and cell division. Therefore, during cell growth, the PG layer has to be enlarged to accompany cell enlargement, division, and daughter cell separation. The essential nature of the PG layer is evidenced by the wide success of antibiotics targeting bacterial cell wall synthesis such as β-lactams and glycopeptides. In this context, we were interested in studying PG metabolism in H. pylori for several reasons: (1) from the genome analysis, it appears that H. pylori has a restricted number of enzymes potentially involved in the PG metabolism in the periplasmic space. There are only three PG synthetases, penicillin-binding proteins (PBPs) 1 to 31–5; (2) a reduced number of PG maturation enzymes, two lytic transglycosylases, Slt and MltD,6 one N-acetylmuramoyl-l-alanyl amidase, AmiA,7 three M23-peptidases Csd1, Csd2, and Csd3/HdpA,8,9 one d,l-endopeptidase Csd4,10 and one l,d-endopeptidase Csd611; (3) H. pylori became a model organism to study the selective function of cell shape of bacteria.7–10,12 Hence, a better understanding of PG metabolism in H. pylori could in the long term lead to new therapeutic strategies. We continued addressing this issue by further characterizing the isogenic amiA mutant. We have combined physiological data with muropeptide composition analysis and glycan strand length distribution by reverse phase high-pressure liquid chromatography (HPLC) of the parental and amiA mutant. We provide evidence for enrichment of very long glycans at division sites that undergo maturation during cell daugther separation. In particular, we show that AmiA is required for daughter cell separation, correct motility, and full virulence of H. pylori.
Results
Modifications in PG composition of amiA mutant
Analysis of the muropeptide composition of the wild-type (WT) strain 26695 and the amiA mutant showed several modifications (Fig. 1A and Table 1). In exponentially growing bacteria, we observed an increase in proportion in muropeptides carrying pentapeptides and a decrease of the ones carrying tripeptides or dipeptides. The most striking difference concerned the proportion of the N-acetyl-d-glucosaminyl-β(1,4)-N-acetylmuramyl-l-Ala–d-Glu (GM-dipeptide) motif at different times of the growth curve. While the WT accumulated this motif in stationary phase (3.3% at 8 hr to 23.3% at 48 hr), the amiA mutant did it to a much lower extent (1.7% at 8 hr to 10.3% at 48 hr). In the contrary, the GM-tripeptide decreased over time in 26695 WT (from 16.8% at 8 hr to 4.96% at 48 hr) but stayed constant in the amiA mutant (around 14%). Those modifications were first described by Costa et al.13 and its biological relevance characterized by Chaput et al.7 Otherwise, another strong modification of the PG composition was that while the proportion of anhydromuropeptides increased over time in the 26695 WT strain (13% at 8 hr to 15.8% at 48 hr), the amiA mutant showed a drastic decrease (12.2% at 8 hr to 6.7% at 48 hr). Anhydromuropeptides consist of muropeptides carrying an N-acetyl-anhydromuramic acid residue (anhM), which is a signature for the end of glycan chains in Gram-negative bacteria. So, the relative amounts of anhydromuropeptides can be correlated to the length of glycan chain. This difference was mainly due to the decrease of dimeric GanhM-tetrapeptide-pentapeptide-GM (which contributed to 60% of the decrease of anhydromuropeptides in comparison with the WT), whereas the WT strain in the same period of time accumulated those motifs. During exponential growth, the amiA mutant had glycan chains of an average of 10.7 disaccharide units comparable to the WT strain (10.2). However, in stationary phase, the average increased to 18.7 disaccharide repeating units, compared to 8.3 disaccharide repeating units for the WT. Consequently, the amiA mutant appeared to have longer glycan chains than the parental strain 26695 in stationary phase. Even though, the major dimer GM-tetra-penta-GM increased in the amiA mutant (11.2% vs. 7.2%), the overall percentage of dimers was lower in the amiA mutant, particularly, in stationary phase (25.7% vs. 30.8%). Since in Escherichia coli when the endogenous amidases are mutated an increase of highly cross-linked muropeptides such as trimers or tetramers was observed, we were expecting that those motifs will appear in our amiA mutant even though WT H. pylori do not produce those motifs. Interestingly, no new muropeptides including highly cross-linked muropeptides were identified in the amiA mutant.
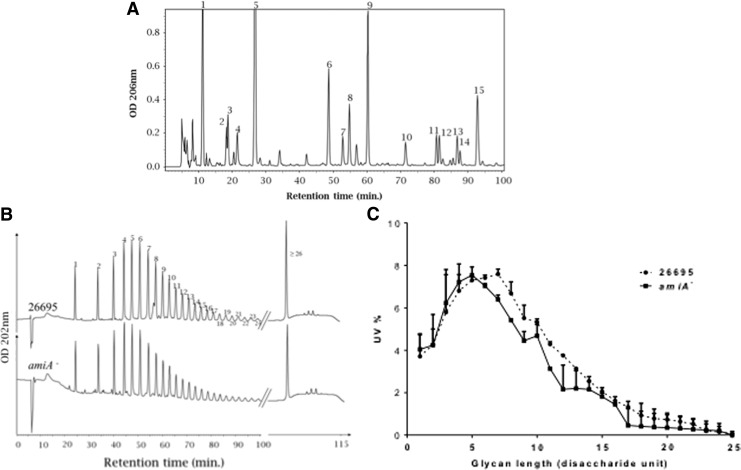
FIG. 1.
Representative HPLC chromatograms of Helicobacter pylori muropeptide composition of 26695 (A) and distribution of glycan chain length of 26695 and isogenic amiA mutant (B, C). Muropeptide peaks (from 1 to 15) correspond to the nomenclature in Table 1. Glycan strand peaks (from 1 to 25 and >26) correspond to the nomenclature in Table 2. From the Glycan chain chromatograms (B), UV percentage was calculated taking into account the total glycan strand UV absorbing material separated by HPLC. The UV% of the glycan chain was plotted for the chain from 1 to 25 disaccharide units for 26695 and the amiA mutant (C). Comparative analysis of strains 26695 and 26695 amiA is presented in Tables 1 and 2 for the muropeptide and glycan chain distribution, respectively. HPLC, high-pressure liquid chromatography.
Table 1.
Peptidoglycan Muropeptide Composition of Helicobacter pylori 26695 and amiA Mutant
| 26695 | 26695 amiA− | ||||||
|---|---|---|---|---|---|---|---|
| Peaks | 8 hr | 24 hr | 48 hr | 8 hr | 24 hr | 48 hr | |
| Monomers | |||||||
| 1 | GM-Tri | 16.8% ± 0.9% | 13.7% ± 0.2% | 4.9% ± 0.1% | 13.5% ± 1.0% | 17.7% ± 2.2% | 14.6% ± 1.5% |
| 2 | GM-Tetra | 5.2% ± 1.6% | 3.7% ± 0.2% | 2.6% ± 0.1% | 6.7% ± 1.0% | 4.7% ± 0.8% | 3.8% ± 0.7% |
| 3 | GM-Tetra-Gly | 4.0% ± 1.4% | 4.8% ± 0.2% | 5.0% ± 0.0% | 5.0% ± 1.2% | 4.0% ± 1.0% | 5.6% ± 0.4% |
| 4 | GM-Di | 3.3% ± 1.0% | 10.9% ± 0.2% | 23.3% ± 0.4% | 1.7% ± 0.7% | 3.8% ± 1.0% | 10.3% ± 1.0% |
| 5 | GM-Penta | 37.6% ± 2.4% | 31.9% ± 0.7% | 31.6% ± 0.3% | 41.2% ± 1.6% | 39.8% ± 2.6% | 38.6% ± 3.6% |
| Dimers | |||||||
| 6 | GM-Tetra-Tri-GM | 5.1% ± 0.8% | 5.6% ± 0.0% | 4.6% ± 0.1% | 3.5% ± 0.4% | 4.5% ± 0.4% | 4.1% ± 0.4% |
| 7 | GM-Tetra-TetraGly-GM | 2.0% ± 0.8% | 1.7% ± 0.1% | 1.4% ± 0.2% | 1.9% ± 0.4% | 2.0% ± 0.5% | 2.0% ± 0.2% |
| 8 | GM-Tetra-Tetra-GM | 3.6% ± 0.2% | 3.8% ± 0.1% | 3.7% ± 0.4% | 3.0% ± 0.3% | 2.8% ± 0.5% | 3.1% ± 0.1% |
| 9 | GM-Tetra-Penta-GM | 9.4% ± 0.5% | 8.4% ± 0.0% | 7.2% ± 0.0% | 11.3% ± 1.0% | 10.0% ± 1.2% | 11.2% ± 0.5% |
| Anhydromuropeptides | |||||||
| 10 | GanhM-Penta | 2.6% ± 0.5% | 2.3% ± 0.2% | 1.8% ± 0.0% | 2.7% ± 0.6% | 1.6% ± 0.6% | 1.4% ± 0.1% |
| 11 | GanhM-Tri-Tetra-GM | 1.9% ± 0.4% | 1.8% ± 0.0% | 1.9% ± 0.0% | 1.5% ± 0.3% | 2.0% ± 0.4% | 1.6% ± 0.5% |
| 12 | GanhM-Tetra-Tri-GM | 1.4% ± 0.4% | 2.6% ± 0.1% | 2.7% ± 0.0% | 1.2% ± 0.2% | 1.7% ± 0.4% | 1.5% ± 0.2% |
| 13 | GanhM-Tetra-Tetra-GM | 1.4% ± 0.5% | 1.8% ± 0.1% | 2.0% ± 0.0% | 1.3% ± 0.2% | 1.2% ± 0.2% | 1.2% ± 0.0% |
| 14 | GanhM-Tetra-Tetra-GM | 1.0% ± 0.3% | 1.4% ± 0.1% | 1.5% ± 0.1% | 0.6% ± 0.2% | 0.7% ± 0.2% | 0.6% ± 0.1% |
| 15 | GanhM-Tetra-Penta-GM | 4.8% ± 0.1% | 5.7% ± 0.3% | 5.9% ± 0.2% | 5.0% ± 1.2% | 3.3% ± 2.0% | 0.4% ± 0.5% |
| 4 | Dipeptides | 3.3% ± 1.0% | 10.9% ± 0.2% | 23.3% ± 0.4% | 1.7% ± 0.7% | 3.8% ± 1.0% | 10.3% ± 1.0% |
| 1, 6, 11, 12 | Tripeptides | 25.2% ± 1.4% | 23.6% ± 0.3% | 14.1% ± 0.1% | 19.7% ± 0.7% | 25.9% ± 1.9% | 21.8% ± 1.5% |
| 2, 3, 7–9, 11–15 | Tetrapeptides | 41.8% ± 1.2% | 43.3% ± 0.6% | 40.6% ± 1.1% | 40.9% ± 2.7% | 37.7% ± 2.3% | 34.4% ± 0.5% |
| 3, 7 | Tetrapeptides-Glycin | 6.0% ± 1.1% | 6.5% ± 0.1% | 6.4% ± 0.2% | 6.9% ± 1.0% | 6.1% ± 1.1% | 7.6% ± 0.6% |
| 5, 9, 10, 15 | Pentapeptides | 54.4% ± 1.8% | 48.3% ± 0.7% | 46.4% ± 0.5% | 60.2% ± 1.7% | 54.7% ± 1.6% | 51.6% ± 3.5% |
| 1–5, 10 | Monomers | 69.4% ± 1.2% | 67.4% ± 0.5% | 69.2% ± 0.5% | 70.7% ± 3.0% | 71.7% ± 2.2% | 74.3% ± 0.0% |
| 6–9, 11–15 | Dimers | 30.6% ± 1.2% | 32.6% ± 0.5% | 30.8% ± 0.5% | 29.3% ± 3.0% | 28.3% ± 2.2% | 25.7% ± 0.0% |
| 10–15 | Anhydromuropeptides | 13.0% ± 0.9% | 15.5% ± 0.6% | 15.8% ± 0.2% | 12.2% ± 1.9% | 10.6% ± 2.3% | 6.7% ± 0.2% |
| Average glycan chains length | 10.2 ± 0.8 | 8.5 ± 0.3 | 8.3 ± 0.1 | 10.7 ± 2.1 | 12.5 ± 2.3 | 18.7 ± 0.7 | |
Each peak numbering are illustrated in Fig. 1A and corresponds to the nomenclature described by Costa et al.13 Each muropeptide structure was confirmed by MALDI-MS. Muropeptide abundance is expressed as molar percentage and was calculated as desbribed by Glauner.21 Average glycan chain length was calculated as described by Harz.23
MALDI-MS, matrix-assisted laser desorption ionization mass spectrometry.
Glycan chain length distribution
Since a major feature of the amiA mutant was the decrease of anhydromuropeptides, we analyzed the glycan chain length of the WT and the amiA mutant at 8 hr of growth (Fig. 1B and Table 2). Generation of glycan chains was obtained using the human serum amidase, which has a specificity for stem peptides with 3 or more amino acids but is unable to cleave the GM-dipeptide.14 Hence, we were unable to compare the glycan chain length at 24 and 48 hr because the WT strain accumulates the GM-dipeptide motif. As expected, glycan strand analysis did not require prior amino sugar reduction for HPLC separation of the different peaks confirming that the glycan strands end exclusively by 1,6-anhydro-N-acetylmuramic acid residues (Fig. 1B). In the 1 to 24 disaccharide repetition unit range, the distribution of the glycan chain length in the amiA mutant showed a shift toward shorter glycan chains (Table 2 and Fig. 1C). The highest proportion of glycan is at 5 disaccharide units for amiA mutant versus 7 for the WT strain. While the proportion of short glycan chains (≤5 disaccharide repeating units) increased, glycan chains between 6 and 16 disaccharide repeating units decreased. However, the overall average glycan chain length of strands up to 25 disaccharide units decreased moderately from 5.3 to 4.9 disaccharide repeating units. Inversely, the proportion of very long glycan chains (≥26 disaccharide repeating units) increased substantially from 16.9% for WT to 25% for the amiA mutant.
Table 2.
Glycan Strand Length Distribution Analysis of H. pylori
| % UV | % molar | |||
|---|---|---|---|---|
| Disaccharide units | 26695 | amiA− | 26695 | amiA− |
| 1 | 3.71% ± 1.0% | 4.04% ± 0.7% | 3.71% ± 1.0% | 4.04% ± 0.7% |
| 2 | 4.26% ± 1.4% | 4.24% ± 0.8% | 2.13% ± 0.7% | 2.12% ± 0.4% |
| 3 | 5.79% ± 2.0% | 6.23% ± 1.3% | 1.93% ± 0.7% | 2.08% ± 0.4% |
| 4 | 6.80% ± 0.8% | 7.22% ± 0.8% | 1.70% ± 0.2% | 1.81% ± 0.2% |
| 5 | 7.30% ± 0.2% | 7.54% ± 0.4% | 1.46% ± 0.0% | 1.51% ± 0.1% |
| 6 | 7.42% ± 0.1% | 7.03% ± 0.1% | 1.24% ± 0.0% | 1.17% ± 0.0% |
| 7 | 7.60% ± 0.2% | 6.39% ± 0.2% | 1.09% ± 0.0% | 0.91% ± 0.0% |
| 8 | 6.68% ± 0.5% | 5.42% ± 0.0% | 0.84% ± 0.1% | 0.68% ± 0.0% |
| 9 | 5.52% ± 0.6% | 4.45% ± 0.4% | 0.61% ± 0.1% | 0.49% ± 0.0% |
| 10 | 5.32% ± 0.2% | 4.69% ± 0.5% | 0.53% ± 0.0% | 0.47% ± 0.1% |
| 11 | 4.29% ± 0.1% | 3.13% ± 0.1% | 0.39% ± 0.0% | 0.28% ± 0.0% |
| 12 | 3.75% ± 0.1% | 2.16% ± 1.1% | 0.31% ± 0.0% | 0.18% ± 0.1% |
| 13 | 3.14% ± 0.1% | 2.20% ± 0.8% | 0.24% ± 0.0% | 0.17% ± 0.1% |
| 14 | 2.55% ± 0.2% | 2.15% ± 0.4% | 0.18% ± 0.0% | 0.15% ± 0.0% |
| 15 | 2.06% ± 0.2% | 1.81% ± 0.2% | 0.14% ± 0.0% | 0.12% ± 0.0% |
| 16 | 1.62% ± 0.2% | 1.44% ± 0.1% | 0.10% ± 0.0% | 0.09% ± 0.0% |
| 17 | 1.32% ± 0.3% | 0.46% ± 0.8% | 0.08% ± 0.0% | 0.03% ± 0.0% |
| 18 | 0.93% ± 0.6% | 0.41% ± 0.7% | 0.05% ± 0.0% | 0.02% ± 0.0% |
| 19 | 0.78% ± 0.5% | 0.37% ± 0.6% | 0.04% ± 0.0% | 0.02% ± 0.0% |
| 20 | 0.73% ± 0.3% | 0.35% ± 0.6% | 0.04% ± 0.0% | 0.02% ± 0.0% |
| 21 | 0.64% ± 0.3% | 0.31% ± 0.5% | 0.03% ± 0.0% | 0.01% ± 0.0% |
| 22 | 0.52% ± 0.3% | 0.27% ± 0.5% | 0.02% ± 0.0% | 0.01% ± 0.0% |
| 23 | 0.19% ± 0.3% | 0.24% ± 0.4% | 0.01% ± 0.0% | 0.01% ± 0.0% |
| 24 | 0.15% ± 0.3% | 0.20% ± 0.3% | 0.01% ± 0.0% | 0.01% ± 0.0% |
| 25 | 0.06% ± 0.1% | 0.00% ± 0.0% | 0.00% ± 0.0% | 0.00% ± 0.0% |
| ≥26 | 16.86% ± 2.1% | 24.97% ± 4.6% | 0.50% ± 0.1% | 0.81% ± 0.2% |
Each glycan strand species corresponds to the different peaks in Fig. 1B. The nomenclature of each peak refers to the number of disaccharide repeating units per glycan strand specie. The UV percentage takes into account the total glycan strand UV absorbing material separated by HPLC (Fig. 1B). The molar percentage can be calculated for the 25 first peaks by dividing the UV percentage by the number of disaccharide units of each glycan species. The final glycan strand peak is a mixture of different species for which the relative proportions are unknown. Therefore, we estimated the average glycan strand length of the very long chains to have a gross estimate of their molar proportion. To determine the average chain length for glycans ≥26 disaccharide units, we used the following formula = (average length − UV% [peaks 1–25] × average length [peaks 1–25])/UV% [peaks ≥26]). The average glycan strand length was calculated in Table 1 (10.2 and 10.7 for 26695 and 26695 amiA, respectively). The average length for the glycan chains up to 25 disaccharide units were calculated as described by Harz.23 We obtained an average of 5.3 and 4.9 for 26695 and 26695 amiA, respectively. The average length of glycans with more than 26 disaccharide units is 33.4 and 30.7 disaccharide repeating units for 26695 and 26695 amiA, respectively. HPLC, high-pressure liquid chromatography.
Susceptibility to different antibiotics
As shown above, AmiA has a major role in the structure and composition of H. pylori PG. Thus, we were interested in characterizing the resistance phenotype of the amiA mutant to several classes of antibiotics, in particular, β-lactam antibiotics. The minimal inhibitory concentration (MIC) and minimum bactericidal concentration (MBC) values of amoxicillin were both 0.06 μg/ml for the parental strain 26695 (Table 3). So, MBC/MIC ratio was 1 for strain 26695. The amiA mutant showed MIC value 0.06 μg/ml of amoxicillin identical to the parental strain. But MBC value for the mutant was superior than the maximum amoxicillin concentration tested (32 μg/ml). Therefore, the amiA mutant showed a MBC/MIC ratio >256 and could be considered as tolerant to amoxicillin. The complemented amiA mutant had similar MIC value than parental strain and the mutant. It had a MBC/MIC ratio of 2, similar to the parental strain value. These results showed that AmiA is needed for the bactericidal activity of amoxicillin. Finally, we tested the resistance phenotype to several other classes of antibiotics. The amiA mutant had the same pattern of antibiotic resistance as the parental strain (Table 3). This indicates that contrary to E. coli,15 inactivation of the single amidase of H. pylori does not affect the overall outer membrane architecture but rather only PG metabolism.
Table 3.
Minimum Bactericidal and Inhibitor Concentration of Amoxicillin and Other Antibiotics for H. pylori Strain 26695, Mutant amiA, and Complemented Mutant
| MIC (μg/ml) and MBC (μg/ml) | 26695a | 26695 amiA− | 26695 amiA−complemented |
|---|---|---|---|
| Amoxicillin | 0.06 (0.06) | 0.06 (>32) | 0.125 (0.25) |
| Streptomycin | 1 (10) | 1 (10) | 1 (10) |
| Bacitracin | >1000 | >1000 | >1000 |
| Nalidixic acid | 30 (1000) | 30 (1000) | 30 (1000) |
| Metronidazole | 1 (1) | 1 (1) | 1 (1) |
| Vancomycin | >1000 | >1000 | >1000 |
| Trimethroprim | >100 | >100 | >100 |
MBC are indicated in parenthesis for those antibiotics tested.
MBC, minimum bactericidal concentration; MIC, minimal inhibitory concentration.
Morphological analysis of the amiA mutant
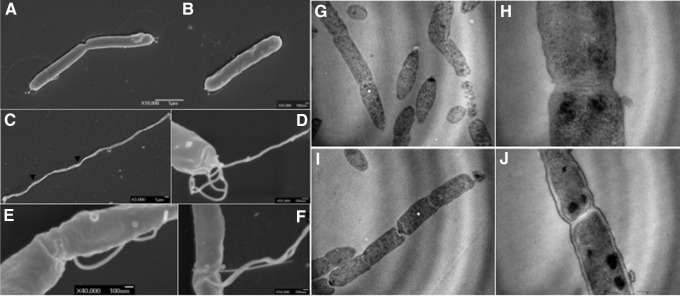
Next, we were interested in analyzing the general morphological phenotype of the amiA mutant since amidases have been implicated in daughter cell separation both in Gram-positive and Gram-negative bacteria. As observed for several other bacteria, the inactivation of the amiA gene resulted in a chaining phenotype (Fig. 2). Also, H. pylori is known for undergoing a morphological transition from spiral to coccoid form during entry in stationary phase. We observed that associated with the chaining phenotype, the amiA mutant failed to undergo morphological transition as previously described.7 Since the sequenced strain 26695 lacks flagella, we also constructed several independent clones in other H. pylori backgrounds. Interestingly, when the amiA gene was inactivated in strains that were motile such as X47-2AL (Fig. 2C–F) and B128 (data not shown), the mutants were still able to synthesize at the poles (Fig. 2D) and some division sites intact flagella (Fig. 2E, F). Although some bacterial chains were motile under the optical microscope, the vast majority was not. Using a soft agar mobility assay, all the amiA independent mutant clones were unable to migrate from the site of inoculation in contrast to the WT strain (data not shown).
FIG. 2.
Electron microscopy of WT H. pylori strain X47-2AL (A, B) and its isogenic amiA mutant (C–F). (C) Shows the chaining phenotype of the amiA mutants. Arrows heads highlight flagella located in the middle of a bacterial chain. Examples of higher magnifications of flagella of the amiA mutant are illustrated in (D–F). (D) Shows polar flagella and (E, F) illustrate flagella at division sites. (G–J) Show transmission electron microscopy of the amiA mutant showing evenly spaced septa that failed to separate daugther cells. WT, wild-type.
Colonization of mice stomachs
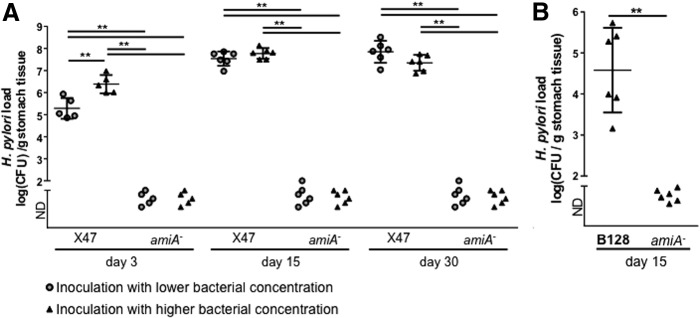
Since the amiA mutant had two major cell morphological defects, impaired daughter cell separation and motility, we investigated the impact on the amiA inactivation on H. pylori capability to colonize mice stomachs. We infected C57/BL6J mice with two parental and fully motile strains, X47-2AL (Fig. 3A) and B128 (Fig. 3B), and their isogenic amiA mutants. We then analyzed their ability to colonize the mouse gastric mucosa at different time points (3, 15, and 30 days after infection; see Fig. 3). Note that the infections were done with an even mixture of three independent clones of amiA mutants in each background. Clearly, the amiA mutant was unable to colonize the stomach of C57/BL6J mice under any conditions tested, indicating that the AmiA protein is required for efficient colonization of the stomach.
FIG. 3.
Mice colonization with WT X47-2AL and its isogenic amiA mutants after 3, 15, and 30 days of infections (A), and with WT B128 and its isogenic amiA mutants 15 days after infection (B). For each experiment, we used an even mixture of three independent clones of the amiA mutants. Since the amiA mutant chains, we considered it was plausible that we were not able to detect colonization of the mutant using a low infectious dose (represented with gray circles). Therefore, a higher dose was also tested (represented with dark triangles). The amiA mutant was still unable to colonize C57/BL6J mice (ND = non detectable). The data were submitted to a Mann–Whitney test (**p < 0.01).
Discussion
In H. pylori the single amiA gene appears to fulfill the same role in daughter cell separation as that played collectively by the three amidases of E. coli. The amiA mutants constructed in different genetic backgrounds (26695, X47-2AL and B128) present long bacterial chains with up to 30–40 bacteria in which the division site was completely formed but without daughter cell separation (Fig. 2). This observation underlines the major role played by amidases in daughter cell separation both in Gram-negative and Gram-positive bacteria. Interestingly, despite impaired daughter cell separation, amiA mutants derived from parental flagellated H. pylori were still able to assemble intact flagella at the site of cell division. We can thus assume that these new division sites are fully functional for flagella assembly, although these flagella appeared to be “paralyzed.” Therefore, whatever are the structural modification of the PG layer at the new cell poles in the amiA mutant, these do not appear to hinder flagella assembly but only flagella function. As it is well known that fully motile bacteria are essential for H. pylori colonization of the stomach,16 our observation that amiA-deficient strains do not colonize is consistent with their “paralyzed” phenotype. This result is reminiscent of the phenotype of lytic transglycosylase mutants of H. pylori, E. coli and Salmonella typhimirium.17 Lytic transglycolases are the enzymes generating anhydromuropeptides and have been reported to be co-regulated, potentially via protein complex, with amidase(s) to orchestrate their activities.18 The abundance of those muropeptides decreased in the amiA mutant (Table 1), suggesting a partial defect on lytic transglycolase activity hypothetically due to destabilization or dysregulation of the protein complex. This defect could lead to “paralyzed” phenotype of the amiA mutant. The amiA mutant also failed to accumulate the GM-dipeptide (Table 1 and Chaput et al.7). Generation of the GM-dipeptide depends on a novel PG maturation enzyme Csd4.10 In accordance to the current model of PG hydrolase regulation by protein complex formation,18 our data suggest that AmiA could participate in a protein complex for septal PG maturation that includes AmiA, lytic transglycosylases, and Csd4, and, in the absence of AmiA, these other activities are impaired. These results make AmiA an attractive new target against H. pylori. H. pylori is one of the few bacteria against which a specific antibiotherapy that does not affect the commensal flora is recommended due to its high world prevalence. Interfering with normal H. pylori AmiA function would probably fit such a strategy. The C-terminus of H. pylori AmiA where the active domain is located has 32% identity with AmiC from E. coli. Nevertheless, phylogenitically H. pylori AmiA is closer to the amidases CwlU and CwlV from Paenibacillus polymyxa and an amidase from Deinococcus radiodurans, two environnemental Gram-positive bacteria. Hence, specific inhibitors of AmiA function would probably not affect amidases from other commensal bacteria. It should be noted that multiple attempts to produce active recombinant AmiA have failed and we have never been able to detect any enzymatic activity in vitro. It is possible that despite the sequence homology with other amidases, the AmiA might have lost its enzymatic activity and function exclusively as a scaffolding protein for other enzymes. Alternatively, AmiA might require a protein partner for activity such as described for E. coli amidases.19 These are activated by LytM-domain containing proteins EnvC and NlpD that lost the endopeptidase activity. H. pylori has four LytM-like proteins.8,9 We have tested their ability to activate AmiA without succes. We are pursuing this work to find potential partners of AmiA that could activate its amidase domain.
Amidases have also been involved in the mechanism of β-lactam induced lysis and death. Interestingly, the H. pylori amiA mutant became tolerant to amoxicillin similar to the lytA mutant of Streptococcus pneumoniae.20 The ratio of MBC over MIC was higher than 256, while complementation of the amiA mutant restored a WT ratio (ratio of 2). As for other bacteria, in H. pylori, AmiA plays a major role in the mechanism of β-lactam induced death. However, we have shown that β-lactam antibiotics do not induce lysis of H. pylori7 but only cell rounding (or coccoid formation). Exposure of the amiA mutant to 100 times its MIC to amoxicillin still induced coccoid formation.7 Hence, the cell rounding can be dissociated from cell death since the amiA mutant is tolerant to amoxicillin. The mechanism of cell death in WT bacteria and tolerance of the amiA mutant remain a mystery. However, we can reasonably assume that it is directly related to the three-dimensional modifications of the PG layer that occurs at the division site.
The H. pylori amiA mutant appears to have longer glycan chains. Despite inhibition of PG synthesis by amoxicillin, a localized resistance to the action of endogenous hydrolases at the poles could also account for the observed tolerance of the amiA mutant, as we observed in the present article and previously.7 The remaining phenotypes diverged substantially from the E. coli example. One of the major observations supporting the 3-for-1 model is the presence of trimeric muropeptides in the PG of E. coli (and a variety of other Gram-negative bacteria).21,22 Interestingly, H. pylori appeared to be an exception since it lacked trimeric muropeptides or muropeptides with a higher degree of cross-linking (Table 1 and Ref.13) at detectable levels by HPLC analysis and UV detection. Since inactivation of the three amidases of E. coli resulted in an increase of trimeric and tetrameric muropeptides and consequently an increase in the degree of cross-linking, we reasoned that the amiA mutant of H. pylori should exhibit the presence of such structures in the PG layer of H. pylori. To our surprise, we did not observe any detectable amounts of trimeric muropeptides (Fig. 1 and Table 1). Other major differences in muropeptide composition between the parental and amiA strain were observed when bacteria entered stationary phase (24 and 48 hr of growth). The WT strain showed an increase of the anhydromuropeptides from exponential to stationary phase. Those muropeptides represent the glycan chain ends,23 and their proportion gives an estimate of the average length of the glycan chains. The same is valid for H. pylori as shown by the glycan chain analysis by HPLC. Exponentially growing and stationary phase bacteria had glycan chains with an average of 10.2 and 8.3 disaccharides units, respectively. The amiA had the same average as the WT during exponential growth (10.7 disaccharide repeating units). However, in stationary phase the average increased drastically to 18.7 disaccharide repeating units. Furthermore, the degree of cross-linking in the amiA mutant decreased. This is in sharp contrast with the triple amidase mutant of E. coli, for which not only the degree of cross-linking was increased but where the average glycan chain length decreased.15 We can postulate that when the degree of cross-linking decreases one expects to have a looser network. Therefore, increasing the glycan chain length could increase the chances of two distinct glycan chains to be connected by a cross-bridge. These changes in glycan strand structure were confirmed by a more precise analysis of the glycan chain length distribution by HPLC. Comparison of the HPLC profiles of the WT and the amiA mutant (Fig. 1) revealed that the overall distribution of the different glycan species was distinct. The amiA mutant showed enrichment in very short and very long glycans, while glycans with intermediate length decrease (Table 2 and Fig. 1C). From the microscopy observation of the amiA mutant, the only morphological distinct difference concerned the impaired daughter cell division (Fig. 2G–J). This observation taken together with a net increase of the glycan chain length in stationary phase for the amiA mutant when the bacterial chains increased the most, strongly suggests that the septum PG is composed primarily of very long glycan chains while the lateral wall PG would be of very short ones. Interestingly, an E. coli ftsZ84 thermosensitive mutant grown at permissive temperature fails to initiate cell division and filaments. Consequently, the ftsZ84 mutant synthesizes exclusively lateral wall PG. Glycan chain length distribution of the ftsZ84 mutant showed an enrichment of very short glycan chains and a substantial decrease of very long chains again.24 Unfortunately, we could not corroborate the phenotype in H. pylori since ftsZ (hp0979) is an essential gene and attempts to generate a conditional ftsZ mutant have failed.
All together, our work validates H. pylori as a suitable alternative model to study PG metabolism and cell shape regulation. It also shows that AmiA metabolises the PG layer together with lytic transglycosylases and d,l-endopeptidase and that targeting its activity would impact multiple activites simultaneously. This is supported by the inability of a amiA mutant to colonize the mouse model (Fig. 3) presumably due to its impaired motility and role in escaping the host innate immune system.24
Experimental Procedures
Bacteria, cells and growth conditions
Escherichia coli MC106125 and DH5α were used as hosts for the construction and preparation of plasmids. They were cultivated in Luria Bertani solid or liquid media supplemented as appropriate with spectinomycin (100 μg·ml−1) or kanamycin (40 μg·ml−1) or both. H. pylori strain 26695,5 X47-2AL26 and B12827 were used to construct mutants. PG was extracted from strain 26695. H. pylori was grown microaerobically at 37°C on blood agar plates or in liquid medium consisting of brain-heart infusion (Oxoid) with 0.2% β-cyclodextrin (Sigma) supplemented with antibiotic-antifungic mix.28 H. pylori mutants were selected on 20 μg·ml−1 kanamycin.
Construction of mutants and complementation
Genes were disrupted as described previously.29 H. pylori mutants were constructed by allelic exchange after transformation with suicide plasmids or PCR products carrying the gene of interest interrupted by a nonpolar cassette aphA-329 and selected on kanamycin. PCRs were used to confirm that correct allelic exchange occurred. Gene constructions were sequenced to ensure sequence fidelity. All reagents, enzymes, and kits were used according to manufacturers' recommendations. Midiprep (HiSpeed Plasmid Midi Kit) and DNA extraction kits (QIAamp DNA extraction kit) were purchased from QIAGEN. The plasmid, pILL2000, was used to construct the amiA mutant. pILL570 carrying ORF hp0772 (amiA gene) was used as the template for an Expand High Fidelity PCR (Amersham) with oligonucleotides 772-1 (5′-GAUGAUGAUGGTACCAAGGATTTTAACTTCATAAGTC-3′ in which the underlined sequence corresponds to a KpnI site) and 772-2 (5′-AUCAUCAUCGGATCCAACACGCAGCGATTGATCGTCTCTAAC-3′ the underlined sequence corresponds to a BamHI site). PCR products were digested with BamHI (Amersham) and KpnI (Amersham) and ligated (T4 DNA ligase; Amersham) with the aphA-3 nonpolar cassette digested with the same endonucleases. For complementation, the promorterless WT amiA gene was introduced in the rdxA gene carried by plasmid pILL570. The plasmid was used as the template for an Expand High Fidelity PCR (Amersham) with oligonucleotides 954-2KpnI (5′-CGGGGTACCTACATGCAAAATCTCTATCCG-3′ in which the underlined sequence corresponds to a KpnI site) and 954-1BamHI (5′-CGCGGATCCGTGTGGTAACAACTCGCTGGG-3′ the underlined sequence corresponds to a BamHI site). The amiA gene was amplified using the following primers: 772-compl-1Bis (5′-GGGGATCCGAGGGTTAATTTGTAGTGCTTGTGAGGTTAGGGG-3′ in which the underlined sequence corresponds to a BamHI site) and 772-compl-2Bis (5′-CGGGTACCCTAATCATTCTTGCTGAAAAACTATCAATGCC-3′ the underlined sequence corresponds to a KpnI site). PCR products were digested with BamHI (Amersham) and KpnI (Amersham) and ligated (T4 DNA ligase; Amersham).
Peptidoglycan extraction and analysis
Liquid cultures of H. pylori parental strain and isogenic mutant strains were stopped after various times of growth and chilled in an ice-ethanol bath. The crude murein sacculus was immediately extracted in boiling sodium dodecyl sulfate (4% final). Purification steps and HPLC analyses were done as described previously.30 Mutanolysin (Sigma) digested samples were analyzed by HPLC on a Hypersil ODS18 reverse phase column (250 by 4.6 mm, 3 μm particle size) with a methanol (Fischer; HPLC grade) gradient from 0% to 15% in sodium phosphate buffer pH 4.3 to 5.0. Chromatograms were obtained by monitoring at 206 nm. Each peak was collected, desalted, and identified by matrix-assisted laser desorption ionization mass spectrometry (MALDI-MS) as described previously.31 Glycan chain analysis was done as previously described.23,32 Briefly, H. pylori PG was digested with purified human serum amidase kindly provided by Waldemar Vollmer. The digestion was done in 50 mM Tris–HCl pH 7.9, 5 mM MgCl2, 0.02% NaN3. Soluble material was first purified on a MonoS (HR5/5) column (Amersham Pharmacia) using a 10 mM sodium phosphate buffer pH 2. Glycans eluted with the flow-through and were collected. Free peptides were eluted by one step using 10 mM sodium phosphate buffer pH 2, 1 M NaCl. The runs performed at room temperature using a flow of 1 ml/min. Purified glycans were analyzed by reverse phase HPLC using a 5 μm Nucleosil 300 C18 column (250 × 4.6 mm) at 50°C. A convex gradient from 0% to 10.5% acetonitrile (−4 curve of the Shimadzu CLASS-VP software) in 100 mM sodium phosphate buffer pH 2 was used over 90 min at a flow rate of 0.5 ml/min. Unresolved glycan material was eluted after the convex gradient in a single step with 30% acetonitrile in 100 mM sodium phosphate buffer pH 2. Glycan material was detected at 202 nm.
Electronic microscopy
For scanning electron microscopy (SEM), samples were washed in phosphate-buffered saline, prefixed in 2.5% glutaraldehyde in 0.1 M cacodylate buffer for 30 min, and then rinsed in 0.2 M cacodylate buffer. Postfixation in 1% osmium tetraoxide (in 0.2 M cacodylate buffer), bacteria were dehydrated in a series of ethanol concentrations. Specimens were critical point dried using carbon dioxide, coated with gold, and examined with a JEOL JSM-6700F SEM.
Minimal inhibitory concentration
To determine the MIC of different antibiotics, suspensions of H. pylori estimated to contain 108 bacteria/ml (OD600nm of 0.1) were serially diluted and grown on plates containing various concentrations of amoxicillin. The MIC was defined as the minimal concentration leading to a decrease of 3 log of CFU/ml as compared to growth without antibiotic. MBC for amoxicillin was done as follows. Bacteria were grown in increasing concentrations of amoxicillin in liquid culture and OD600nm was monitored. After 18 hr, CFU/ml counts were determined for each amoxicillin concentration. MBC was defined as the concentration leading to a 3 log decrease of CFU/ml as compared to growth without amoxicillin.
Mice experiments
Five-week-old female C57/BL6J mice (Charles River) were intragastrically infected with around 106–5 × 106 (low dose) and 5 × 107–108 (high dose) cfu/mouse as previously described.33,34 The presence of H. pylori infection in mice was determined by quantitative culture of gastric tissue fragments containing both the antrum and corpus, from mice sacrificed at day 3, 15, and 30 postinfection.33
Acknowledgments
We would like to thank Waldemar Vollmer for providing purified human serum amidase and Marie-Christine Prévost for access to the electron scanning microscope. C.C. was supported by fellowship from the French Ministry (MENRT) and from La Fondation pour la Recherche Médicale (FRM). This research was supported by a Génopole Grant (Institut Pasteur), by a “Programme Transversal de Recherche” Grant 153 (Institut Pasteur), an ACI (Action Concertée Incitative Microbiology, INSERM No. MIC 0321) grant from the Ministère chargé de la Recherche and an ERC starting grant (202283 PGNfromSHAPEtoVIR).
Disclosure Statement
No competing financial interests exist.
References
- 1.Alm R.A., Ling L.S., Moir D.T., King B.L., Brown E.D., Doig P.C., Smith D.R., Noonan B., Guild B.C., deJonge B.L., et al. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 397:176–180 [DOI] [PubMed] [Google Scholar]
- 2.Boneca I.G., de Reuse H., Epinat J.C., Pupin M., Labigne A., and Moszer I. 2003. A revised annotation and comparative analysis of Helicobacter pylori genomes. Nucleic Acids Res. 31:1704–1714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boneca I.G., Ecobichon C., Chaput C., Mathieu A., Guadagnini S., Prevost M.C., Colland F., Labigne A., and de Reuse H. 2008. Development of inducible systems to engineer conditional mutants of essential genes of Helicobacter pylori. Appl Environ Microbiol. 74:2095–2102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.El Ghachi M., Mattei P.J., Ecobichon C., Martins A., Hoos S., Schmitt C., Colland F., Ebel C., Prevost M.C., Gabel F., et al. 2011. Characterization of the elongasome core PBP2: MreC complex of Helicobacter pylori. Mol Microbiol. 82:68–86 [DOI] [PubMed] [Google Scholar]
- 5.Tomb J.F., White O., Kerlavage A.R., Clayton R.A., Sutton G.G., Fleischmann R.D., Ketchum K.A., Klenk H.P., Gill S., Dougherty B.A., et al. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 388:539–547 [DOI] [PubMed] [Google Scholar]
- 6.Chaput C., Labigne A., and Boneca I.G. 2007. Characterization of Helicobacter pylori lytic transglycosylases Slt and MltD. J Bacteriol. 189:422–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chaput C., Ecobichon C., Cayet N., Girardin S.E., Werts C., Guadagnini S., Prevost M.C., Mengin-Lecreulx D., Labigne A., and Boneca I.G. 2006. Role of AmiA in the morphological transition of Helicobacter pylori and in immune escape. PLoS Pathog. 2:e97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bonis M., Ecobichon C., Guadagnini S., Prevost M.C., and Boneca I.G. 2010. A M23B family metallopeptidase of Helicobacter pylori required for cell shape, pole formation and virulence. Mol Microbiol. 78:809–819 [DOI] [PubMed] [Google Scholar]
- 9.Sycuro L.K., Pincus Z., Gutierrez K.D., Biboy J., Stern C.A., Vollmer W., and Salama N.R. 2010. Peptidoglycan crosslinking relaxation promotes Helicobacter pylori's helical shape and stomach colonization. Cell. 141:822–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sycuro L.K., Wyckoff T.J., Biboy J., Born P., Pincus Z., Vollmer W., and Salama N.R. 2012. Multiple peptidoglycan modification networks modulate Helicobacter pylori's cell shape, motility, and colonization potential. PLoS Pathog. 8:e1002603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sycuro L.K., Rule C.S., Petersen T.W., Wyckoff T.J., Sessler T., Nagarkar D.B., Khalid F., Pincus Z., Biboy J., Vollmer W., et al. 2013. Flow cytometry-based enrichment for cell shape mutants identifies multiple genes that influence Helicobacter pylori morphology. Mol Microbiol. 90:869–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martinez L.E., Hardcastle J.M., Wang J., Pincus Z., Tsang J., Hoover T.R., Bansil R., and Salama N.R. 2016. Helicobacter pylori strains vary cell shape and flagellum number to maintain robust motility in viscous environments. Mol Microbiol. 99:88–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Costa K., Bacher G., Allmaier G., Dominguez-Bello M.G., Engstrand L., Falk P., de Pedro M.A., and Garcia-del Portillo F. 1999. The morphological transition of Helicobacter pylori cells from spiral to coccoid is preceded by a substantial modification of the cell wall. J Bacteriol. 181:3710–3715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Z.M., Li X., Cocklin R.R., Wang M., Wang M., Fukase K., Inamura S., Kusumoto S., Gupta D., and Dziarski R. 2003. Human peptidoglycan recognition protein-L is an N-acetylmuramoyl-L-alanine amidase. J Biol Chem. 278:49044–49052 [DOI] [PubMed] [Google Scholar]
- 15.Heidrich C., Templin M.F., Ursinus A., Merdanovic M., Berger J., Schwarz H., de Pedro M.A., and Holtje J.V. 2001. Involvement of N-acetylmuramyl-L-alanine amidases in cell separation and antibiotic-induced autolysis of Escherichia coli. Mol Microbiol. 41:167–178 [DOI] [PubMed] [Google Scholar]
- 16.Ottemann K.M., and Lowenthal A.C. 2002. Helicobacter pylori uses motility for initial colonization and to attain robust infection. Infect Immun. 70:1984–1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roure S., Bonis M., Chaput C., Ecobichon C., Mattox A., Barriere C., Geldmacher N., Guadagnini S., Schmitt C., Prevost M.C., et al. 2012. Peptidoglycan maturation enzymes affect flagellar functionality in bacteria. Mol Microbiol. 86:845–856 [DOI] [PubMed] [Google Scholar]
- 18.Typas A., Banzhaf M., Gross C.A., and Vollmer W. 2012. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat Rev Microbiol. 10:123–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Uehara T., Parzych K.R., Dinh T., and Bernhardt T.G. 2010. Daughter cell separation is controlled by cytokinetic ring-activated cell wall hydrolysis. EMBO J. 29:1412–1422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tomasz A., Albino A., and Zanati E. 1970. Multiple antibiotic resistance in a bacterium with suppressed autolytic system. Nature. 227:138–140 [DOI] [PubMed] [Google Scholar]
- 21.Glauner B., Holtje J.V., and Schwarz U. 1988. The composition of the murein of Escherichia coli. J Biol Chem. 263:10088–10095 [PubMed] [Google Scholar]
- 22.Quintela J.C., Caparros M., and de Pedro M.A. 1995. Variability of peptidoglycan structural parameters in gram-negative bacteria. FEMS Microbiol Lett. 125:95–100 [DOI] [PubMed] [Google Scholar]
- 23.Harz H., Burgdorf K., and Holtje J.V. 1990. Isolation and separation of the glycan strands from murein of Escherichia coli by reversed-phase high-performance liquid chromatography. Anal Biochem. 190:120–128 [DOI] [PubMed] [Google Scholar]
- 24.Ishidate K., Ursinus A., Holtje J.V., and Rothfield L. 1998. Analysis of the length distribution of murein glycan strands in ftsZ and ftsI mutants of E. coli. FEMS Microbiol Lett. 168:71–75 [DOI] [PubMed] [Google Scholar]
- 25.Casadaban M.J., and Cohen S.N. 1980. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 138:179–207 [DOI] [PubMed] [Google Scholar]
- 26.Londono-Arcila P., Freeman D., Kleanthous H., O'Dowd A.M., Lewis S., Turner A.K., Rees E.L., Tibbitts T.J., Greenwood J., Monath T.P., et al. 2002. Attenuated Salmonella enterica serovar Typhi expressing urease effectively immunizes mice against Helicobacter pylori challenge as part of a heterologous mucosal priming-parenteral boosting vaccination regimen. Infect Immun. 70:5096–5106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Israel D.A., Salama N., Arnold C.N., Moss S.F., Ando T., Wirth H.P., Tham K.T., Camorlinga M., Blaser M.J., Falkow S., et al. 2001. Helicobacter pylori strain-specific differences in genetic content, identified by microarray, influence host inflammatory responses. J Clin Invest. 107:611–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bury-Mone S., Thiberge J.M., Contreras M., Maitournam A., Labigne A., and De Reuse H. 2004. Responsiveness to acidity via metal ion regulators mediates virulence in the gastric pathogen Helicobacter pylori. Mol Microbiol. 53:623–638 [DOI] [PubMed] [Google Scholar]
- 29.Skouloubris S., Thiberge J.M., Labigne A., and De Reuse H. 1998. The Helicobacter pylori UreI protein is not involved in urease activity but is essential for bacterial survival in vivo. Infect Immun. 66:4517–4521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Glauner B. 1988. Separation and quantification of muropeptides with high-performance liquid chromatography. Anal Biochem. 172:451–464 [DOI] [PubMed] [Google Scholar]
- 31.Antignac A., Boneca I.G., Rousselle J.C., Namane A., Carlier J.P., Vazquez J.A., Fox A., Alonso J.M., and Taha M.K. 2003. Correlation between alterations of the penicillin-binding protein 2 and modifications of the peptidoglycan structure in Neisseria meningitidis with reduced susceptibility to penicillin G. J Biol Chem. 278:31529–31535 [DOI] [PubMed] [Google Scholar]
- 32.Boneca I.G., Huang Z.H., Gage D.A., and Tomasz A. 2000. Characterization of Staphylococcus aureus cell wall glycan strands, evidence for a new beta-N-acetylglucosaminidase activity. J Biol Chem. 275:9910–9918 [DOI] [PubMed] [Google Scholar]
- 33.Ferrero R.L., Thiberge J.M., Huerre M., and Labigne A. 1998. Immune responses of specific-pathogen-free mice to chronic Helicobacter pylori (strain SS1) infection. Infect Immun. 66:1349–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ferrero R.L., Thiberge J.M., Kansau I., Wuscher N., Huerre M., and Labigne A. 1995. The GroES homolog of Helicobacter pylori confers protective immunity against mucosal infection in mice. Proc Natl Acad Sci U S A. 92:6499–6503 [DOI] [PMC free article] [PubMed] [Google Scholar]