Abstract
We present a method using a combination of enzymatic deconjugation and targeted LC-multiple reaction monitoring (MRM)-MS analysis for analyzing all common bile acids (BAs) in piglet urine, and in particular, for detecting conjugated BAs either in the absence of their standards, or when present in low concentrations. Initially, before enzymatic deconjugation, 19 unconjugated BAs (FBAs) were detected where the total concentration of the detected FBAs was 9.90 μmol/l. Sixty-seven conjugated BAs were identified by LC-MRM-MS analysis before and after enzymatic deconjugation. Four enzymatic assays were used to deconjugate the BA conjugates. FBAs in urine after cholylglycine hydrolase/sulfatase treatment were 33.40 μmol/l, indicating the urinary BAs were comprised of 29.75% FBAs and 70.25% conjugated BAs in single and multiple conjugated forms. For the conjugates in single form, released FBAs from cholylglycine hydrolase deconjugation indicated that the conjugates with amino acids were 14.54% of urinary BAs, 16.27% glycosidic conjugates were found by β-glucuronidase treatment, and sulfatase with glucuronidase inhibitor treatment liberated FBAs that constituted 16.67% of urinary BAs. Notably, chenodeoxycholic acid (CDCA) was initially detected only in trace amounts in urine, but was found at significant levels after the enzymatic assays above. These results support that CDCA is a precursor of γ-muricholic acid in BA biosynthesis in piglets.
Keywords: liquid chromatography-multiple reaction monitoring-mass spectrometry, enzymatic deconjugation, piglet urine, bile acid conjugates
Bile acids (BAs) have an important role in the control of lipid, glucose, and cholesterol homeostasis. Synthesis of BAs is the major pathway for the metabolism of cholesterol and for the excretion of excess cholesterol in mammals. BAs are biosynthesized from cholesterol in a complex process that utilizes at least 16 enzymes (1). BAs are remarkably diverse in structure and exist in many different forms in different species within mammals, but in the human, cholic acid (CA) and chenodeoxycholic acid (CDCA) are most common (2). As the prime experimental models for humans, the rat and mouse produce several additional BAs as primary BAs, which include ursodeoxycholic acid (UDCA), α-muricholic acid (MCA), and βMCA (2–4). The piglet is also a common model for human infants because of the similarities in gastrointestinal development between piglets and human infants (5). However, the domestic pig has been reported to be virtually unable to synthesize CA. As a substitute, γMCA (hyocholic acid), an isomer of CA, is synthesized in amounts equal to those of CA in humans. γMCA is considered to be a species-specific primary BA in the pig (6). Another species-specific primary BA is UDCA (an isomer of CDCA) in bears (7).
BAs constitute a large family of molecules composed of free BAs and their conjugated forms. Secondary free BAs are biosynthesized from primary BAs by various reactions, such as dehydroxylation and epimerization (8). Free BAs undergo further conjugation with two amino acids (glycine or taurine), sulfuric acid, glucoside (Glc), glucuronic acid (GlcUA), or N-acetylglucosamine (GlcNAc) to form conjugated BAs. BA composition can differ significantly between species, between gender in some species, between neonatal and adult periods of life, and also between healthy humans and patients with disorders, such as liver disease (9, 10). It has been reported that the inborn errors of BA formation involve isolated defects in the enzymes of the biosynthetic pathway (10–12), and that most inborn errors of the BA biosynthetic pathway involve the synthesis of excess intermediates and/or their metabolites that are excreted, in part, in urine. Also, insulin-resistant individuals had a blunted increase in blood glycochenodeoxycholic acid in oral glucose tolerance tests (13). Hence, the differentiated quantification of BAs in BA profiles may be an important tool for the diagnosis of disorders in the BA biosynthetic pathway. BA metabolites facilitate the absorption of dietary lipids and fat-soluble vitamins by formation of micelles, and diet has been reported to affect BA metabolism in infants (14). Thus, quantitative comparison of BAs from different synthetic pathways in the urine of piglets can be used to determine the effect of different diets on the BA metabolism and to further evaluate the nutritional value of different diets for infants.
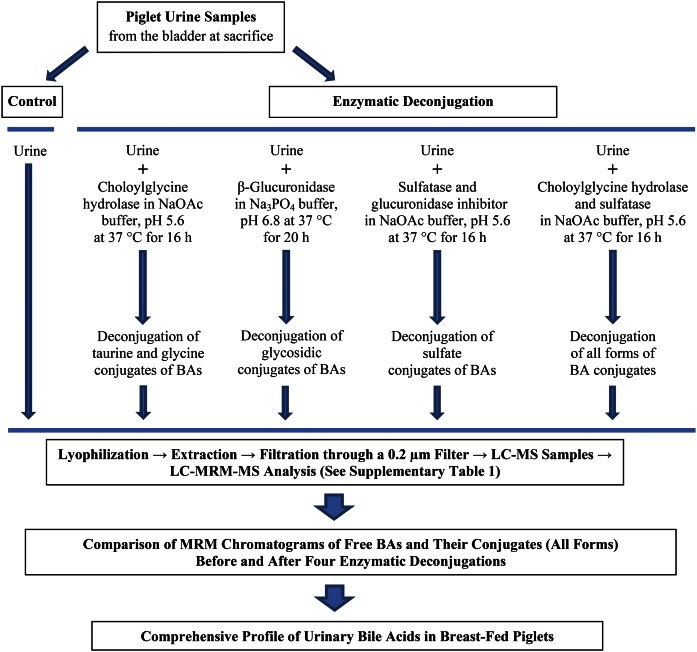
The purpose of this study was to establish a comprehensive profile of free and conjugated urinary BAs in piglets. Urine was collected from the bladder of 21-day-old breast-fed piglets at euthanization. A method using a combination of enzymatic deconjugation and targeted LC-multiple reaction monitoring (MRM)-MS analysis was developed: four enzymatic treatments were used to deconjugate BA conjugates, which included cholylglycine hydrolase (CGH) for taurine- and glycine-amidated conjugates, β-glucuronidase (GUS) for glycosidic conjugates, sulfatase with GUS inhibitor (S&GI) for sulfate conjugates, and CGH with sulfatase (CH&S) for all forms of conjugates. In the S&GI assay, sulfatase from Helix pomatia (type H-1) also has glucuronidase activity, and GUS inhibitor was added for inhibiting the glucuronidase activity. LC-MRM-MS was used for analysis of the BA conjugates, and the big advantage for using LC-MRM-MS scan is that each conjugated form has its own characteristic precursor-to-product ion transition for a specific ion pair. The qualitative and quantitative analysis of BA conjugates was determined from the comparison of selected ions [M-H]− chromatograms of free BAs and MRM chromatograms of their conjugates before and after four enzymatic deconjugations. Furthermore, this work will provide an analytical tool to study the conjugated metabolites of other chemical classes, which have not been well studied due to the absence of their standards commercially.
MATERIALS AND METHODS
Materials
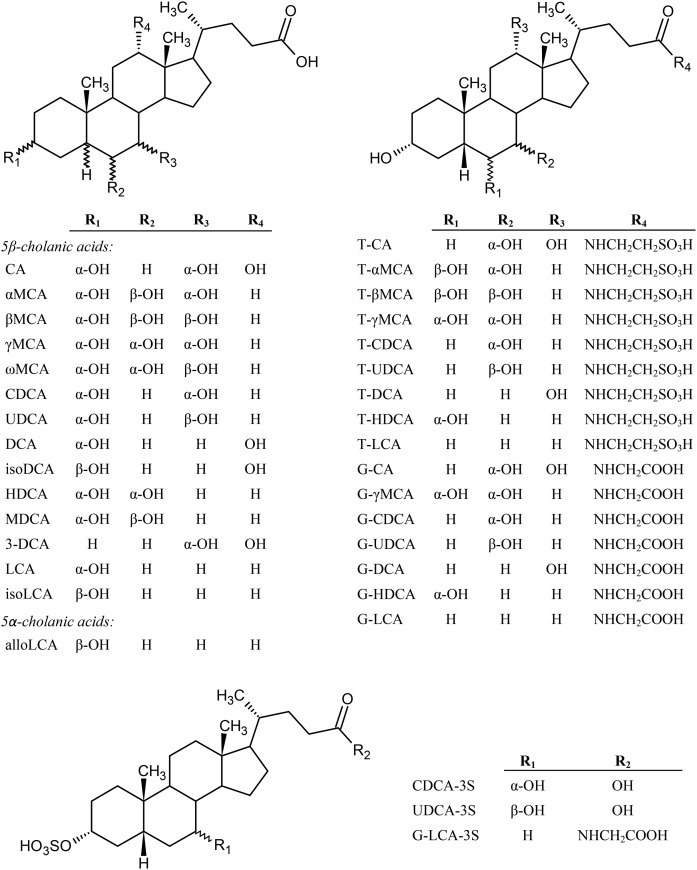
For the enzymatic assays (Fig. 1), CGH from Clostridium perfringens, sulfatase from Helix pomatia (type H-1), GUS from Escherichia coli (type VII-A), and d-saccharic acid 1,4-lactone monohydrate used as glucuronidase inhibitor, sodium acetate, sodium phosphate, sodium hydroxide, and hydrochloric acid were purchased from Sigma-Aldrich (St. Louis, MO). Thirty-four authentic standards of BAs were used in the study (Fig. 2). Twenty-four of the standards were purchased from Steraloids, Inc. (Newport, RI): αMCA, βMCA, γMCA (hyocholic acid), ωMCA, murideoxycholic acid (MDCA), 5β-cholanic acid-3β,12α-diol (isoDCA), 3-deoxycholic acid (3-DCA), isolithocholic acid (isoLCA), allolithocholic acid (alloLCA), taurocholic acid (T-CA), sodium tauro α-muricholic acid (T-αMCA), tauro β-muricholic acid (T-βMCA), sodium tauro γ-muricholic acid (T-γMCA sodium salt), taurohyodeoxycholic acid (T-HDCA), taurochenodeoxycholic acid (T-CDCA), tauroursodeoxycholic acid (T-UDCA), taurodeoxycholic acid (T-DCA), taurolithocholic acid (T-LCA), sodium glycohyocholic acid (G-γMCA sodium salt), glycohyodeoxycholic acid (G-HDCA), sodium glycochenodeoxycholate (G-CDCA sodium salt), glycoursodeoxycholic acid (G-UDCA), glycodeoxycholic acid (G-DCA), and glycolithocholic acid (G-LCA). Seven of the standards were obtained from Sigma-Aldrich: CA, CDCA, UDCA, deoxycholic acid (DCA), glycocholic acid (G-CA), hyodeoxycholic acid (HDCA), and lithocholic acid (LCA). Three sulfate conjugate standards, chenodeoxycholic acid 3-sulfate disodium salt (CDCA-3S disodium salt), ursodeoxycholic acid 3-sulfate disodium salt (UDCA-3S disodium salt), and glycolithocholic acid 3-sulfate disodium salt (G-LCA-3S disodium salt), were purchased from Alsachim (Strasbourg, France). Formic acid (for mass spectrometry, Fluka) and acetonitrile (Chromasolv®) were obtained from Sigma-Aldrich. Methanol (MeOH) (Spectranalyzed®) was purchased from Fisher Scientific (Fair Lawn, NJ). Water (H2O) was obtained from Milli-Q Integral Water Purification System (EMD Millipore Corporation, Billerica, MA).
Fig. 1.
Procedure for comprehensive profile of urinary BAs in breast-fed female piglets.
Fig. 2.
Chemical structures of BAs. Lipid Maps identification of BAs: CA, LMST04010001; αMCA, LMST04010066; βMCA, LMST04010067; γMCA, LMST04010064; ωMCA, LMST04010065; CDCA, LMST04010032; UDCA, LMST04010033; DCA, LMST04010040; isoDCA, LMST04010042; HDCA, LMST04010024; MDCA, LMST04010025; 3-DCA, LMST04010049; LCA, LMST04010003; isoLCA, LMST04010004; alloLCA, LMST04010005; T-CA, LMST05040001; T-βMCA, LMST05040012; T-γMCA, LMST05040010; T-CDCA, LMST05040005; T-UDCA, LMST05040015; T-DCA, LMST05040013; T-LCA, LMST05040018; G-CA, LMST05030001; G-CDCA, LMST05030008; G-UDCA, LMST05030016; G-DCA, LMST05030006; G-LCA, LMST05030009; CDCA-3S, LMST05020024; UDCA-3S, LMST05020033; and G-LCA-3S, LMST05030015.
Animal experiments
Samples were derived from pig experiments previously described (15, 16). Briefly, White × Dutch Landrace × Duroc sows were artificially inseminated. Newborn male piglets (n = 5) were allowed to suckle with a sow for the duration of the experiment. All animals were housed in the animal facilities of the Arkansas Children’s Hospital Research Institute, an Association for the Assessment and Accreditation of Laboratory Animal Care-approved animal facility. Animal maintenance and experimental treatments were conducted in accordance with the ethical guidelines for animal research established and approved by the institutional Animal Care and Use Committee at the University of Arkansas for Medical Sciences. Twenty-one-day-old piglets were fasted 6–8 h following the last feeding and exsanguinated after anesthetization with isoflurane at 0800–1000 hours. Urine samples were obtained from the bladder at euthanization and stored at −70°C until analyzed. A pooled urine sample was made by combining urine from five piglets.
Enzymatic deconjugation and LC-MS/MS sample preparation
βMCA was added to urine (300 pmol/ml) as an internal standard (IS) and the urine containing IS was used for enzymatic assays and its control sample. For preparation of the control sample (Control), 0.5 ml urine with IS in a 1.5 ml graduated microcentrifuge tube was frozen at −80°C followed by lyophilization. The powdered urine in the tube was added with 0.5 ml 80% MeOH/H2O and vortexed vigorously for 2 min followed by sonication in ice-water for 5 min. The mixture was centrifuged at RCF (153,393 g) for 5 min and the supernatant solution was carefully drawn out from the tube. The extraction process was repeated two more times, the supernatants were combined and filtrated through a 17 mm 0.2 μm filter (National Scientific, Rockwood, TN), and adjusted to exactly 1.5 ml with 80% MeOH/H2O for LC-MS/MS analysis. The pellet was further extracted with pure MeOH (0.5 ml) and MeOH:methylene chloride (1:2) (0.5 ml). These two extracts were analyzed by LC-MS for examining the efficiency of three-time 80% MeOH/H2O extractions.
Four enzymatic assays (assays 1–4) were used to deconjugate BA conjugates, which included CGH, S&GI, CH&S, and GUS (Fig. 1). The urine with IS (250 μl) for each of assays 1–3 was added with sodium acetate buffer [100 mM (pH 5.6); 250 μl] containing different enzymes: 15 units of cholylglycine hydrolase (assay 1, CGH), 100 units of sulfatase with 200 mM d-saccharic 1,4-lactone glucuronidase inhibitor (assay 2, S&GI), and 15 units of cholylglycine hydrolase with 150 units of sulfatase (assay 3, CH&S). The solution in assays 1–3 was incubated at 37°C for 16 h. For assay 4 (GUS), sodium phosphate buffer [100 mM (pH 6.8), 250 μl] containing 1,000 units of GUS was added to urine with IS (250 μl), and the solution was incubated at 37°C for 20 h. Incubations (assays 1–4) were stopped with 500 μl of ice-cold MeOH, and the mixture was evaporated to dryness under an N2 stream, immediately. The same BA extraction procedure that was used for control sample preparation above was used here for extraction of BAs after enzymatic deconjugation. The hydrolysis rate of conjugates was determined by comparison of the peak areas between endogenous conjugates before and after enzymatic assays.
LC-MS/MS analysis
LC-MS/MS was performed using a 4000 Q TRAP system (Applied Biosystems, Foster City, CA) equipped with an Agilent 1100 series liquid chromatograph (Agilent Technologies, Wilmington, DE). The 4000 Q TRAP system included a hybrid triple quadrupole/linear ion trap mass spectrometer equipped with an ESI probe and Analyst® software. Data acquisition was performed using the AB Sciex Analyst 1.5.1 software in MRM.
To analyze the BAs in urine, a 5 μl aliquot of urine extract that was equivalent to 1.67 μl of starting sample urine was injected into the LC-MS/MS instrument. Two LC conditions with the same LC solvent flow rate of 0.5 ml/min were used in the LC-MS/MS analysis to confirm the peak yielded by its typical MRM ion pair. A 150 × 4.6 mm internal diameter 5 μm, Eclipse XDB-C18 column (Agilent Technologies) was used in LC-condition-I, and its LC gradient was 0.1% formic acid/acetonitrile (solvent B) in 0.1% formic acid/H2O (solvent A) as follows: 5% in 0–4 min; 0–30% from 4 to 5 min; 30–40% from 5 to 25 min; 40–85% from 25 to 29 min; held at 85% from 29 to 38 min; 85–100% from 38 to 40 min; held at 100% from 40 to 41 min, and finally back to 5% in 43 min, with 1 min as column re-equilibration in sequence analysis. For the LC-condition-II, a 150 mm × 4 mm internal diameter 5 μm ODS (2), SphereClone column (Phenomenex, Torrance, CA) was used, and its LC gradient was 0.1% formic acid/acetonitrile (solvent B) in 0.1% formic acid/H2O (solvent A) as follows: 38–38% in 15 min; 38–80% from 15 to 18 min; held at 80% from 18 to 21 min; 80–100% from 21 to 24 min; held at 100% from 24 to 25 min, and finally back to 38% in 26 min, with 1 min as column re-equilibration in sequence analysis.
Each BA component in the eluate was monitored by its typical MRM ion pair in LC-MS/MS with a negative ion mode (see supplemental Table S1). All parameters for ESI-MS/MS analysis of HPLC peaks were held constant at: CE, −50; DP, −125.0; EP, −10; and CXP, −10.0.
Validation of quantitative determination
βMCA was not detected in piglet urine in our preliminary experiment and has a chemical structure similar to that of endogenous free BAs, primary BA γMCA, and ωMCA. βMCA was used as IS for analysis of BAs in piglet urine. Individual stock solutions of BA standards were prepared at 3 μmol/ml in 80% MeOH/H2O containing 300 pmol/ml βMCA. Stock solutions were then diluted to the concentrations of 10,000, 3,000, 1,000, 300, 100, 30, 10, 3, and 1 pmol/ml using 80% MeOH/H2O containing 300 pmol/ml IS βMCA. Six concentrations were chosen according to the concentration of endogenous BAs in urine for establishment of six-point calibration curves. The peak-area ratio of each BA to the IS was used to construct calibration curves. Seventeen calibration curves were established for quantitative determination (ωMCA, γMCA, T-βMCA, T-γMCA, G- γMCA, MDCA, HDCA, CDCA, DCA, G-CDCA, G-HDCA, CDCA-3S disodium salt, alloLCA, LCA, T-LCA, G-LCA, and G-LCA-3S disodium salt). Two quantitative methods were used to calculate the concentration of endogenous BAs in the present study. The concentration of individual BA compounds was calculated using the peak-area ratio of each BA to the IS and its corresponding standard curve. When the standard of endogenous BA was not available, the standard curve of its structurally related BA standard was used (supplemental Table S2). For the concentration of each conjugate group, the liberated unconjugated BAs (FBAs) by enzymatic deconjugation of assays 1, 2, and 4 were used to determine the concentration of their corresponding conjugate groups in single conjugated form, such as liberated FBAs from CGH for amidated conjugates (taurine and glycine), liberated FBAs from GUS for glycosidic conjugates, and liberated FBAs from S&GI for sulfate conjugates. Liberated FBAs from assay 3 (CH&S) were from all forms of conjugates (Fig. 1). Data are expressed as mean ± SD (n = 3). The recovery (percent ± SD) of extraction was calculated as [IS recovery in urine sample/IS recovery in standard solution that used for standard curve] (n = 18).
RESULTS
Extraction of BAs in urine and recovery
The methods for analysis of BAs in piglet urine included exhaustive extraction of BAs from urine (using several methods) and LC-MRM-MS analysis was used to monitor the efficiency of BA extraction. In our preliminary experiment, several solvents, including MeOH, acetonitrile, and different percentages of aqueous MeOH or acetonitrile, were used to extract BAs from the lyophilized urine powder. The results indicated that 80% MeOH/H2O was more efficient for extraction of free and conjugated BAs from the lyophilized urine powder. Also, no significant amount of BAs could be extracted by 80% MeOH/H2O after three-time 80% MeOH/H2O extractions. In the present study, the process with three times of 80% MeOH/H2O extraction were used to extract BAs from the lyophilized urine powder, and this extraction process was considered nearly complete because there were no detectable ISs and endogenous BAs in the extracts from a fourth extraction with 100% MeOH and a fifth extraction with MeOH:methylene chloride (1:2). The recovery rate was 108.61 ± 3.15% supported the complete extraction of BAs. However, it is unusual that the recovery rate was more than 100%. The more than 100% recovery might be the result of matrix effects. It has been shown that the LC-MS methodology can encounter problems caused by matrix effects (17–20). In our previous study, the retention time (Rt.) and areas of LC-peaks of BAs in urine samples from piglets fed dissimilar diets differed significantly from each other under the same LC-MS conditions (21). Therefore, our 108.61% recovery rate may be partially explained by such effects.
Characterization of urinary BAs by LC-MRM-MS analysis and enzymatic deconjugations
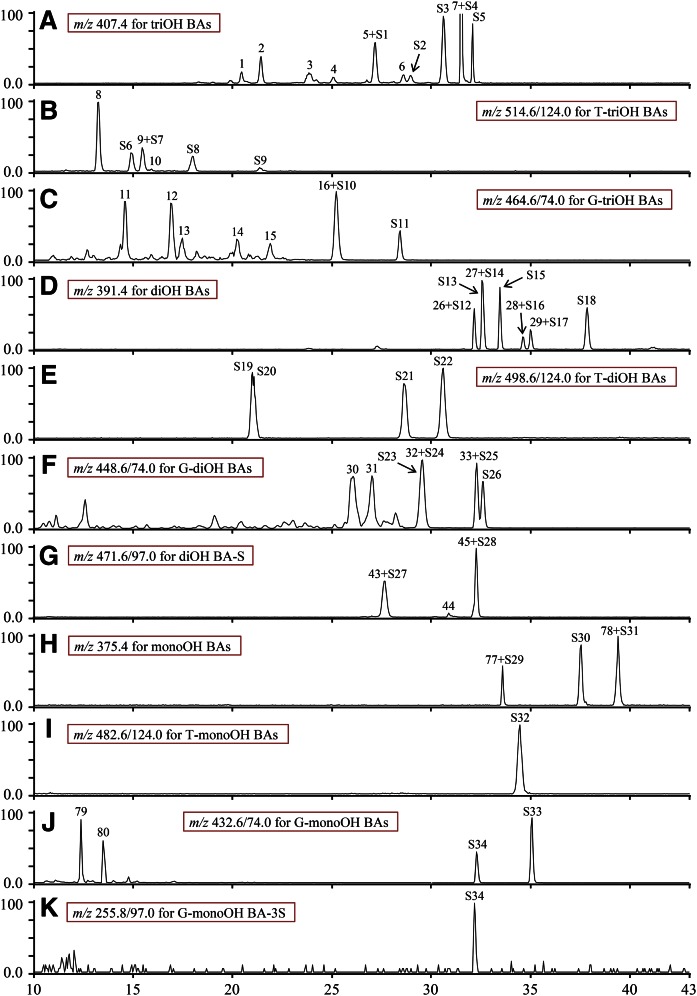
Conjugated BAs have a characteristic fragmentation pattern for each conjugated form and the typical MRM ion pairs from the characteristic fragmentation patterns were used to examine all common conjugated BAs in urine (supplemental Table S1). Unlike conjugated BAs, FBAs do not have a characteristic fragmentation pattern for the whole group in MS analysis. For example, CA and hyocholic acid have different typical MRM ion pairs, m/z 407.4/343.2 and m/z 407.4/389.4, respectively, and typical MRM ion pairs for unknown FBAs are not available. In order to examine all endogenous FBAs in urine, the selected ions [M-H]− were used to examine their corresponding FBAs (supplemental Table S1). Two LC conditions were used in the LC-MS/MS analysis to confirm the peaks of BAs. FBAs included primary BAs and their secondary BAs, which are biosynthesized from primary BAs by hydroxylation, dehydroxylation, and epimerization. The conjugates consisted of glycine- and taurine-amidated, sulfate, and glycosidic conjugates in single, double, and triple conjugate forms. It was notable that each conjugated BA group has its typical product ion, such as m/z 74 for glycine conjugates, m/z 124 for taurine conjugates, m/z 97 for sulfate conjugates, a neutral loss of 162 Da for Glc conjugates, a neutral loss of 176 Da for glucuronide conjugates, and a neutral loss of 203 Da for N-acetylglucosaminide conjugates. While the common Glcs of BAs are glucuronides, Glcs, and N-acetylglucosaminides, it has been reported that BA galactosides also exist in the urine (22). Therefore, it is also possible that the Glc conjugate, which is identified by a neutral loss of 162 Da in LC-MRM-MS analysis, is a galactoside. The typical ion pairs derived from characteristic fragmentation patterns of different BA conjugates can be used by targeted LC-MRM-MS analysis, and this targeted MS analysis enhances the lower detection limit for BA conjugates in biological samples. The enzymatic assays included CGH assay for taurine- and glycine-amidated conjugates, GUS assay for glycosidic conjugates, S&GI assay for sulfate conjugates, and CH&S assay for all forms of conjugates (Fig. 1). Deconjugation of conjugates by enzymatic assays was determined by MRM-MS analysis and used as evidence for identification of a conjugate. Thirty-four BA standards were used in this study and their chemical structures are shown in Fig. 2. We previously reported that matrix effects could change peak Rt. of BAs in LC-MS/MS analysis (21). In order to avoid matrix effects, the BA standards were mixed with urine samples and analyzed by LC-MRM-MS. Identification of endogenous BAs was achieved by overlap of standard and endogenous BA peaks (Fig. 3).
Fig. 3.
Selected ion monitoring chromatograms (A, D, H) and MRM scan chromatograms (B, C, E–G, I–K) of BAs from co-injection of female piglet urine extract and standards. S1–S34 are BA standards. S1, ωMCA; S2, αMCA; S3, βMCA; S4, γMCA; S5, CA; S6, T-αMCA; S7, T-βMCA; S8, T-γMCA; S9, T-CA; S10, G-γMCA; S11, G-CA; S12, MDCA; S13, UDCA; S14, HDCA; S15, isoDCA; S16, CDCA; S17, DCA; S18, 3-DCA; S19, T-UDCA; S20, HDCA; S21, T-CDCA; S22, T-DCA; S23, G-UDCA; S24, G-HDCA; S25, G-CDCA; S26, G-DCA; S27, UDCA-3S; S28, CDCA-3S; S29, alloLCA; S30, isoLCA; S31, LCA; S32, T-LCA; S33, G-LCA; S34, G-LCA-3S.
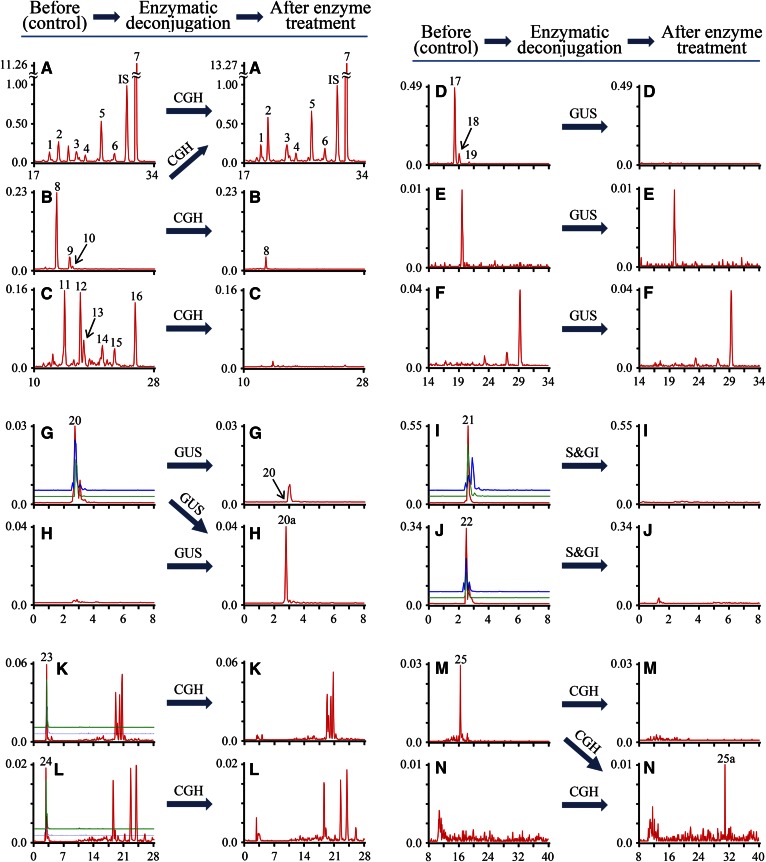
Identification of conjugates of trihydroxycholanoic acids
As shown in Fig. 4B, MRM ion pair m/z 514.6/124.0 yielded three peaks for possible taurine conjugates of triOH BAs in control samples. Hydrolysis by CGH treatment resulted in a 77.00 ± 0.77% reduction of peak area of the major peak (peak 8) and the elimination of the other two peaks (peaks 9, 10), permitting the identification of the three compounds of peaks 8–10 as T-trihydroxycholanoic acids (triOH BAs). Co-injection of pig urine and standards indicated that the compound of peak 9 was T-βMCA (Fig. 3B). Six LC-peaks (peaks 11–16) and several tiny peaks were generated by scan of MRM ion pair m/z 464.6/74.0 for G-triOH BAs and the peaks were eliminated after the CGH treatment (Fig. 4C). Therefore, the compounds of peaks 11–16 were identified as G-triOH BAs. Co-injection of urine samples and standards indicated that the major peak 16 was G-γMCA (Fig. 3C). Consistent with the deconjugation of taurine and glycine conjugates by CGH, the concomitant increase of the areas of peaks for free triOH BAs (peaks 1–7) from m/z 407.4 selected ion monitoring were observed after CGH deconjugation (Fig. 4A). The results from CGH deconjugation suggested that peaks 1–7 might be free triOH BAs and peaks 5 and 7 were identified as ωMCA and γMCA by their standards (Fig. 3A). Peaks for free tirOH BAs (peaks 1–7) were also generated by MRM m/z 407.4/389.4. On the basis of our experience from analyses of glycosidic conjugates and the literature reports, each type of glycosidic conjugate has a characteristic neutral loss: 162 Da for BA-Glc, 176 Da for BA-GlcUA, and 203 Da for BA-GlcNAc in LC-MS/MS with a negative ion mode (23–25). The product ion generated from the neutral losses of its precursor [M-H]− was used as product ion in its typical MRM ion pair to detect glycosidic conjugates (supplemental Table S1). Seven peaks for glycosidic conjugates were detected by their respective MRM ion pairs (supplemental Table S1). Three of them (peaks 17–19) from MRM m/z 583.7/407.4 were hydrolyzed by GUS (Fig. 4D) and identified as triOH-GlcUA. While one peak for triOH BA-Glc (Fig. 4E) and three peaks for triOH BA-GlcNAc (Fig. 4F) were suggested by MRM-MS scan, they were not affected by GUS treatment and were not considered as glycosidic conjugates. No sulfate conjugates of triOH BA, T-triOH BA, and G-triOH BA were detected in piglet urine by their respective MRM ion pairs (supplemental Table S1). However, a peak for T-triOH BA-S (peak 20a) was detected by MRM ion pair m/z 296.7[M-2H]2−/97.0 after GUS deconjugation (Fig. 4H), which suggested a glycosidic conjugate of T-triOH BA-S. T-triOH BA-S-Glc has three typical MRM ion pairs, m/z 377.9[M-2H]2−/97.0, 756.9/97.0, and 756.9/124.0, which generated the three peaks at the same Rt. (peak 20) and these peaks were eliminated in all four enzymatic assays. The results suggested that the compound of peak 20 is T-triOH BA-S-Glc (Fig. 4G). An unsaturated T-triOH BA-S (peak 21) was also detected by its typical MRM ion pairs, m/z 294.9[M-2H]2−/97.0, 590.7/97.0, and 590.7/124.0, and confirmed by enzymatic hydrolysis in treatment of S&GI (Fig. 4I) and CGH. Furthermore, MRM scan of m/z 396.4[M-2H]2−/97.0, 793.9/97.0, and 793.9/124.0 yielded three peaks at the same Rt. (peak 22) for GlcNAc conjugate of BA (peak 21) in the control sample and the disappearance of these peaks after hydrolysis by S&GI (Fig. 4J), CGH, and GUS, which suggested a structure, T-diOH BA-Δ-S-GlcNAc, for peak 22. Five peaks for T-triOH-S-GlcUA were yielded by MRM ion pair m/z 384.7[M-2H]2−/97.0 (Fig. 4K). However, only one of them (peak 23) was overlapped with peaks of m/z 770.8/97.0 and 770.8/124.0 and enzymatically hydrolyzed by CGH (Fig. 4K), GUS, and S&GI treatments. The results suggested that only peak 23 was a T-triOH-S-GlcUA. For the same interpretation of the results from LC-MRM-MS analysis and treatment of CGH (Fig. 4L), GUS, and S&GI, an unsaturated T-triOH-S-GlcUA (peak 24) was identified in piglet urine. One peak for unsaturated T-triOH BA (peak 25) was generated by MRM m/z 510.6/124.0 in the control sample and hydrolyzed by CGH treatment to liberate an unsaturated triOH BA (peak 25a) (Fig. 4M, N). Thus, the compound (peak 25) was identified as a taurine conjugate of unsaturated triOH BA. Because Δ4-3-oxo-steroid 5β-reductase is involved in the BA biosynthetic pathway (11), the compounds of peaks 21, 22, 24, 25, and 25a were tentatively identified as Δ4-3-oxo-BAs (Table 1). The glucuronidation is a conjugation pathway with preference for 6-hydroxylated BAs (26, 27). Therefore, we inferred that the compounds of peaks 17, 20, 22, 23, and 24 were BA-6-Glc (Table 1). To our knowledge, this is the first literature report of the triple conjugate of triOH BAs.
Fig. 4.
Comparison of selected ion monitoring chromatograms of triOH BAs and MRM scan chromatograms of their conjugates in urine of breast-fed female piglets before and after enzymatic deconjugation. The y-axis on the graph represents the peak-height ratio of endogenous BAs to the IS and the x-axis the Rt. in minutes. A: The chromatogram for free triOH BAs was generated by a selected ion m/z 407.4. MRM scan chromatograms generated by MRM ion pairs: m/z 514.6/124.0 for T-triOH BAs (B); m/z 464.6/74.0 for G-triOH BAs (C); m/z 583.7/407.4 for triOH BA-GlcUA (D); m/z 569.7/407.4 for TriOH BA-Glc (E); m/z 610.8/407.4 for TriOH BA-GlcNAc (F); m/z 377.9[M-2H]2−/97.0 (red line), 756.9/97.0 (blue line) and 756.9/124.0 (green line) for T-triOH BA-S-Glc (G); m/z 296.9[M-2H]2−/97.0 for T-triOH BA-S (H); m/z 294.9[M-2H]2−/97.0 (red line), 590.7/97.0 (blue line) and 590.7/124.0 (green line) for T-diOH BA-Δ-one-S (I); m/z 396.4[M-2H]2−/97.0 (red line), 793.9/97.0 (blue line) and 793.9/124.0 (green line) for T-diOH BA-Δ-one-S-GlcNAc (J); m/z 384.7[M-2H]2−/97.0 (red line), 770.8/97.0 (blue line) and 770.8/124.0 (green line) for T-triOH BA-S-GlcUA (K); m/z 382.7[M-2H]2−/97.0 (red line), 766.8/97.0 (blue line) and 766.8/124.0 (green line) for T-diOH BA-Δ-one-S-GlcUA (L); m/z 510.6/124.0 for T-dihydroxy-3-oxo-4-cholanoic acid (diOH BA-Δ4-3-one) (M); m/z 403.4/403.4 for free diOH BA-Δ4-3-one (N).
TABLE 1.
Urinary BAs and standard BAs
| Number | In Figure | Rt. (min) | Negative Mode | Structures | Concentration in Urine (μmol/l) (mean ± SD) | |
| Q1 Ion (m/z) | Q3 Ion (m/z) | |||||
| TriOH cholanic acids and their conjugates: | ||||||
| 1 | 3A, 4A | 20.41 | 407.4a | TriOH BA | 0.31 ± 0.01 | |
| 2 | 3A, 4A | 21.36 | 407.4 | TriOH BA | 0.17 ± 0.01 | |
| 3 | 3A, 4A | 23.76 | 407.4 | TriOH BA | 0.18 ± 0.01 | |
| 4 | 3A, 4A | 24.98 | 407.4 | TriOH BA | 0.09 ± 0.00 | |
| 5 + S1 | 3A, 4A | 27.02 | 407.4 | ωMCA | 0.57 ± 0.02 | |
| 6 | 3A, 4A | 28.47 | 407.4 | TriOH BA | 0.15 ± 0.01 | |
| S2 | 3A | 28.87 | 407.4 | αMCA | n.d. | |
| S3 | 3A | 30.61 | 407.4 | βMCA | n.d. | |
| 7 + S4 | 3A, 4A | 31.58 | 407.4 | γMCA | 8.11 ± 0.22 | |
| S5 | 3A | 32.16 | 407.4 | CA | n.d. | |
| 8 | 3B, 4B | 13.18 | 514.6 | 124.0 | T-triOH BA | 11.24 ± 0.37 |
| S6 | 3B | 14.85 | 514.6 | 124.0 | T-αMCA | n.d. |
| 9 + S7 | 3B, 4B | 15.42 | 514.6 | 124.0 | T-βMCA | 2.30 ± 0.10 |
| 10 | 3B, 4B | 15.87 | 514.6 | 124.0 | T-triOH BA | 0.42 ± 0.08 |
| S8 | 3B | 17.96 | 514.6 | 124.0 | T-γMCA | n.d. |
| S9 | 3B | 21.34 | 514.6 | 124.0 | T-CA | n.d. |
| 11 | 4C | 14.48 | 464.6 | 74.0 | G-triOH BA | 0.15 ± 0.01 |
| 12 | 4C | 16.89 | 464.6 | 74.0 | G-triOH BA | 0.18 ± 0.01 |
| 13 | 4C | 17.23 | 464.6 | 74.0 | G-triOH BA | 0.04 ± 0.02 |
| 14 | 4C | 20.29 | 464.6 | 74.0 | G-triOH BA | 0.04 ± 0.00 |
| 15 | 4C | 21.74 | 464.6 | 74.0 | G-triOH BA | 0.04 ± 0.00 |
| 16 + S10 | 3C, 4C | 25.15 | 464.6 | 74.0 | G-γMCA | 0.12 ± 0.01 |
| S11 | 3C | 28.37 | 464.6 | 74.0 | G-CA | n.d. |
| 17 | 4D | 18.32 | 583.7 | 407.4 | ωMCA-6-GlcUA | 0.51 ± 0.01 |
| 18 | 4D | 19.08 | 583.7 | 407.4 | βMCA-6-GlcUA | 0.06 ± 0.00 |
| 19 | 4D | 20.76 | 583.7 | 407.4 | TriOH BA-GlcUA | 0.01 ± 0.00 |
| 20 | 4G | 2.89 | 377.9 | 97.0 | T-triOH BA-S-6-Glc | 1.83 ± 0.35 |
| 756.9 | 97.0 | |||||
| 756.9 | 124.0 | |||||
| 20a | 4H | 2.82 | 296.9 | 97.0 | T-triOH BA-S | 1.61 ± 0.04 |
| 21 | 4I | 2.67 | 294.9 | 97.0 | T-diOH BA-Δ4-3-one-S | 11.55 ± 0.29 |
| 590.7 | 97.0 | |||||
| 590.7 | 124.0 | |||||
| 22 | 4J | 2.58 | 396.4 | 97.0 | T-diOH BA-Δ4-3-one-S-GlcNAc | 6.10 ± 0.16 |
| 793.9 | 97.0 | |||||
| 793.9 | 124.0 | |||||
| 23 | 4K | 2.90 | 384.9 | 97.0 | T-triOH BA-S-6-GlcUA | 2.70 ± 0.90 |
| 770.8 | 97.0 | |||||
| 770.8 | 124.0 | |||||
| 24 | 4L | 2.83 | 382.9 | 97.0 | T-diOH BA-Δ4-3-one-S-6-GlcUA | 1.27 ± 0.25 |
| 766.8 | 97.0 | |||||
| 766.8 | 124.0 | |||||
| 25 | 4M | 16.03 | 510.6 | 124.0 | T-diOH-Δ4-3-one | 0.99 ± 0.11 |
| 25a | 4N | 31.25 | 403.4 | 403.4 | DiOH-Δ4-3-one | 0.39 ± 0.09 |
| DiOH cholanic acids and their conjugates: | ||||||
| 26 + S12 | 3D,5A | 32.15 | 391.4a | MDCA | Traceb | |
| S13 | 3D | 32.55 | 391.4 | UDCA | n.d. | |
| 27 + S14 | 3D,5A | 32.55 | 391.4 | HDCA | Trace | |
| S15 | 3D | 33.44 | 391.4 | isoDCA | n.d. | |
| 28 + S16 | 3D,5A | 34.61 | 391.4 | CDCA | Trace | |
| 29 + S17 | 3D,5A | 34.99 | 391.4 | DCA | Trace | |
| S18 | 3D | 37.83 | 391.4 | 3-DCA | n.d. | |
| S19 | 3E | 21.10 | 498.6 | 124.0 | T-UDCA | n.d. |
| S20 | 3E | 21.17 | 498.6 | 124.0 | T-HDCA | n.d. |
| S21 | 3E | 28.84 | 498.6 | 124.0 | T-CDCA | n.d. |
| S22 | 3E | 30.83 | 498.6 | 124.0 | T-DCA | n.d. |
| 30 | 3F,5B | 26.17 | 448.6 | 74.0 | G-diOH BA | 0.01 ± 0.00 |
| 31 | 3F,5B | 27.13 | 448.6 | 74.0 | G-diOH BA | 0.01 ± 0.00 |
| S23 | 3F | 29.59 | 448.6 | 74.0 | G-UDCA | n.d. |
| 32 + S24 | 3F,5B | 29.59 | 448.6 | 74.0 | G-HDCA | 0.01 ± 0.00 |
| 33 + S25 | 3F,5B | 32.23 | 448.6 | 74.0 | G-CDCA | Trace |
| S26 | 3F,5B | 32.65 | 448.6 | 74.0 | G-DCA | n.d. |
| 34 | 5C | 23.21 | 567.7 | 391.4 | DiOH BA-GlcUA | Trace |
| 35 | 5C | 24.81 | 567.7 | 391.4 | DiOH BA-GlcUA | Trace |
| 36 | 5C | 29.18 | 567.7 | 391.4 | MDCA-6-GlcUA | 0.05 ± 0.01 |
| 37 | 5C | 31.01 | 567.7 | 391.4 | HDCA-6-GlcUA | 0.23 ± 0.04 |
| 38 | 5C | 31.35 | 567.7 | 391.4 | CDCA-6-GlcUA | 0.02 ± 0.00 |
| 39 | 5C | 32.11 | 567.7 | 391.4 | DCA-6-GlcUA | Trace |
| 40 | 5D | 22.40 | 594.8 | 391.4 | DiOH BA-GlcNAc | Trace |
| 41 | 5D | 23.57 | 594.8 | 391.4 | DiOH BA-GlcNAc | 0.01 ± 0.00 |
| 42 | 5E | 11.03 | 624.7624.7 | 74.0391.4 | G-diOH BA-GlcNAc | 0.04 ± 0.00 |
| 42a | 5B | 28.60 | 448.6 | 74.0 | G-diOH BA | 0.03 ± 0.00 |
| S27 | 3G | 27.67 | 471.6 | 97.0 | UDCA-3S | n.d. |
| 43 | 3G,5F | 27.67 | 471.6 | 97.0 | HDCA-3S | Trace |
| 44 | 3G,5F | 30.91 | 471.6 | 97.0 | DiOH BA-3S | 0.01 ± 0.00 |
| 45 + S28 | 3G,5F | 32.29 | 471.6 | 97.0 | CDCA-3S | 0.03 ± 0.00 |
| 46 | 5G | 2.97 | 288.9 | 97.0 | T-HDCA-3S | 0.05 ± 0.01 |
| 578.8 | 97.0 | |||||
| 578.8 | 124.0 | |||||
| 47 | 5G | 11.90 | 288.9 | 97.0 | T-diOH BA-3S | 0.02 ± 0.00 |
| 48 | 5G | 13.26 | 288.9 | 97.0 | T-CDCA-3S | 0.08 ± 0.00 |
| 49 | 5H | 2.81 | 286.9 | 97.0 | T-monoOH BA-Δ4-3-one-S | 0.14 ± 0.03 |
| 574.7 | 97.0 | |||||
| 574.7 | 124.0 | |||||
| 50 | 5I | 2.66 | 390.4 | 97.0 | T-diOH-S-GlcNAc | 0.17 ± 0.03 |
| 781.9 | 97.0 | |||||
| 781.9 | 124.0 | |||||
| 51 | 5I | 2.82 | 390.4 | 97.0 | T-diOH-S-GlcNAc | 3.10 ± 0.61 |
| 781.9 | 97.0 | |||||
| 781.9 | 124.0 | |||||
| TetraOH cholanic acids and their conjugates: | ||||||
| 52 | 6A | 16.17 | 423.4a | TetraOH BA | 0.05 ± 0.00 | |
| 53 | 6A | 19.18 | 423.4 | TetraOH BA | 0.06 ± 0.00 | |
| 54 | 6A | 19.97 | 423.4 | TetraOH BA | 0.06 ± 0.01 | |
| 55 | 6A | 21.08 | 423.4 | TetraOH BA | 0.02 ± 0.00 | |
| 56 | 6A | 24.77 | 423.4 | TetraOH BA | 0.01 ± 0.00 | |
| 57 | 6A | 27.79 | 423.4 | TetraOH BA | 0.06 ± 0.00 | |
| 58 | 6B | 15.51 | 530.6 | 124.1 | T-tetraOH BA | 1.30 ± 0.08 |
| 59 | 6C | 14.50 | 480.6 | 74.0 | G-tetraOH BA | 0.13 ± 0.00 |
| 60 | 6C | 16.58 | 480.6 | 74.0 | G-tetraOH BA | 0.04 ± 0.00 |
| 61 | 6C | 18.68 | 480.6 | 74.0 | G-tetraOH BA | 0.12 ± 0.01 |
| 62 | 6D | 2.58 | 679.8 | 97.0 | TetraOH BA-S-GlcUA | 0.03 ± 0.00 |
| 63 | 6D | 2.73 | 679.8 | 97.0 | TetraOH BA-S-GlcUA | 0.02 ± 0.00 |
| 64 | 6E | 2.51 | 665.8 | 97.0 | TetraOH BA-S-Glc | 0.01 ± 0.00 |
| 65 | 6E | 2.66 | 665.8 | 97.0 | TetraOH BA-S-Glc | 0.05 ± 0.00 |
| 66 | 6F | 2.76 | 304.9 | 97.0 | T-tetraOH-S | 0.04 ± 0.00 |
| 610.8 | 97.0 | |||||
| 610.8 | 124.0 | |||||
| 67 | 6F | 2.88 | 304.9 | 97.0 | T-tetraOH-S | 0.06 ± 0.00 |
| 610.8 | 97.0 | |||||
| 610.8 | 124.0 | |||||
| 68 | 6G | 2.59 | 392.9 | 97.0 | T-tetraOH BA-S-GlcUA | 0.13 ± 0.01 |
| 786.8 | 97.0 | |||||
| 786.8 | 124.0 | |||||
| 69 | 6G | 2.73 | 392.9 | 97.0 | T-tetraOH BA-S-GlcUA | 0.21 ± 0.01 |
| 786.8 | 97.0 | |||||
| 786.8 | 124.0 | |||||
| 70 | 6H | 2.82 | 390.9 | 97.0 | T-triOH BA-Δ4-3-one-S-GlcUA | 6.41 ± 0.04 |
| 782.8 | 97.0 | |||||
| 782.8 | 124.0 | |||||
| 71 | 6I | 2.61 | 406.4 | 97.0 | T-tetraOH BA-S-GlcNAc | 0.24 ± 0.03 |
| 813.9 | 97.0 | |||||
| 813.9 | 124.0 | |||||
| 72 | 6I | 2.91 | 406.4 | 97.0 | T-tetraOH BA-S-GlcNAc | 0.04 ± 0.01 |
| 813.9 | 97.0 | |||||
| 813.9 | 124.0 | |||||
| 73 | 6J | 2.60 | 404.4 | 97.0 | T-triOH BA-Δ4-3-one-S-GlcNAC | 0.01 ± 0.00 |
| 809.8 | 97.0 | |||||
| 809.8 | 124.0 | |||||
| 74 | 6J | 2.75 | 404.4 | 97.0 | T-triOH BA-Δ4-3-one-S-GlcNAC | 0.04 ± 0.01 |
| 809.8 | 97.0 | |||||
| 809.8 | 124.0 | |||||
| 75 | 6K | 2.92 | 385.9 | 97.0 | T-tetraOH BA-S-Glc | 0.15 ± 0.01 |
| 772.9 | 97.0 | |||||
| 772.9 | 124.0 | |||||
| 76 | 6L | 2.69 | 383.9 | 97.0 | T-triOH BA-Δ4-3-one-S-Glc | 0.09 ± 0.01 |
| 768.8 | 97.0 | |||||
| 768.8 | 124.0 | |||||
| MonoOH cholanic acids and their conjugates: | ||||||
| 77+S29 | 3H,7A | 33.54 | 375.4a | alloLCA | 0.05 ± 0.00 | |
| S30 | 3H | 37.54 | 375.4 | isoLCA | n.d. | |
| 78+S31 | 3H,7A | 39.09 | 375.4 | LCA | Trace | |
| S32 | 3I | 34.43 | 482.6 | 124.0 | T-LCA | n.d. |
| 79 | 3J,7B | 11.62 | 432.6 | 74.0 | G-monoOH BA | 0.03 ± 0.00 |
| 80 | 3J,7B | 12.75 | 432.6 | 74.0 | G-monoOH BA | 0.03 ± 0.00 |
| S33 | 3J | 35.01 | 432.6 | 74.0 | G-LCA | n.d. |
| S34 | 3J,3K | 32.21 | 255.8 | 97.0 | G-LCA-3S | n.d. |
| 432.6 | 74.0 | |||||
| 81 | 7C | 2.83 | 455.6 | 97.0 | MonoOH BA-S | 0.84 ± 0.13 |
| 82 | 7D | 15.42 | 551.7 | 375.4 | MonoOH BA-GlcUA | 1.76 ± 0.05 |
| 83 | 7D | 21.04 | 551.7 | 375.4 | MonoOH BA-GlcUA | 1.51 ± 0.53 |
| 84 | 7D | 25.15 | 551.7 | 375.4 | MonoOH BA-GlcUA | 0.85 ± 0.18 |
| 85 | 7E | 2.75 | 658.8 | 124.0 | T-monoOH BA-GlcUA | 2.14 ± 0.11 |
| 86 | 7E | 2.83 | 658.8 | 124.0 | T-monoOH BA-GlcUA | 2.10 ± 0.20 |
triOH BA-Δ4-3-one, trihydroxy-3-oxo-4-cholanoic acid; diOH BA-Δ4-3-one, dihydroxy-3-oxo-4-cholanoic acid; n.d., not detected.
a A selected ion [M-H]− was used to monitor FBAs.
b Trace: calculated value was less than 0.01 μmol/l in urine.
Identification of conjugates of dihydroxycholanoic acids
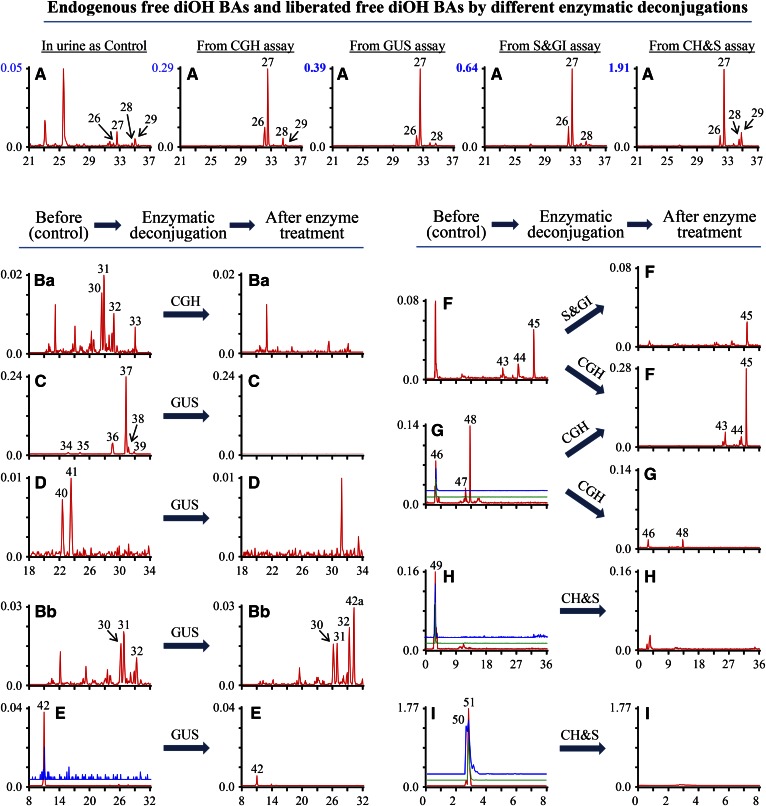
Four peaks (peaks 30–33) for G-dihydroxycholanoic acids (diOH BAs) were found in urine by their typical MRM ion pair, m/z 448.6/74.0, and eliminated by CGH treatment (Fig. 5Ba). Comparison with the standards indicated that conjugates (peaks 32 and 33) were G-HDCA and G-CDCA, respectively (Fig. 3F). As expected, a selected ion m/z 391.4 monitoring yielded several tiny peaks in the urine sample and four of them (peaks 26–29) increased in the urine sample after CGH treatment (Fig. 5A). Analysis of urine samples spiked with standards indicated that peaks 26–29 were MDCA, HDCA, CDCA, and DCA, respectively (Fig. 3D). HDCA and UDCA were shown in one peak in this LC condition (LC-condition-I). Using LC-condition-II, LC-peaks for HDCA and UDCA were separated, and HDCA had a longer Rt. than UDCA. GUS enzymatic deconjugation resulted in the substantial increase of peak intensity for free diOH BAs of peaks 26–29 (Fig. 5A, from GUS assay), which corresponded with the elimination of six peaks (peaks 34–39) for diOH BA-GlcUA (Fig. 5C). Furthermore, the chromatogram profiles (Rt. and peak intensity) of peaks 36–39 for glucuronide conjugates (Fig. 5C) and their released free diOH BAs (peaks 26–29) (Fig. 5A from GUS or CH&S assay) were similar. Because released free diOH BAs (peaks 26–29) were MDCA, HDCA, CDCA, and DCA, their corresponding peaks (peaks 36–39) were identified as MDCA-GlcUA, HDCA-GlcUA, CDCA-GlcUA, and DCA-GlcUA, respectively. Two diOH BA-GlcNAc (peaks 40 and 41) were detected by their typical MRM m/z 594.8/391.4 and GUS assay (Fig. 5D). Two typical MRM ion pairs m/z 624.3/74.0 and 624.3/391.4 for glucuronide conjugates of G-diOH BA yielded two peaks at the same Rt. (peak 42) suggesting that the compound of peak 42 might be a G-diOH BA-GlcUA (Fig. 5E). GUS treatment caused an 87.75 ± 1.69% decrease of the peak area and added an extra peak (peak 42a) in the profile of a G-diOH BA (Fig. 5Bb). The GUS assay confirmed the structural assignment of G-diOH BA-GlcUA to peak 42. Study of diOH BA-Glc and -GlcUA in human urine indicated that the sugar moiety of Glcs and glucuronide of CDCA, HDCA, and DCA was attached at a ring position and not at C-24 (28), which suggested that the sites of glycosidic conjugations in diOH BA-GlcUA of peaks 40–42 were at a ring position. Three sulfate conjugates of diOH BA (peaks 43–45) were detected in piglet urine by MRM m/z 471.3/97.0 and confirmed by S&GI assay (Fig. 5F). Peak 45 was overlapped with the peaks of CDCA-3S (Fig. 3G) and identified as CDCA-3S. Peak 43 was a tiny peak (Fig. 5F) and overlapped with that of standard UDCA-3S (Fig. 3G). G-conjugates of UDCA and HDCA (Fig. 3F) had similar Rt., and HDCA and its G-conjugate were found in urine. Thus, the compound of peak 43 was tentatively identified as HDCA-3S even though peak 43 was overlapped with UDCA-3S peak. Though the peak at Rt. 2.70 min was hydrolyzed by S&GI, it was also hydrolyzed by CGH (Fig. 5F). Moreover the 2.70 min Rt. in RP C18 LC condition was too short for a glucuronide conjugate of diOH BA in comparison with the Rt. of peaks 43–45, and therefore the peak at Rt. 2.70 min in Fig. 5F was not considered as a glucuronide conjugate of diOH BA. MRM m/z 288.7[M-2H]2−/97.0 scan yielded three peaks (peaks 46–48) for T-diOH BA-S and another two typical MRM ion pairs for T-diOH BA-S, m/z 578.8/97.0 and 578.8/124.0, did not generate the peak for peaks 47and 48 (Fig. 5G). However, the enzymatic deconjugation of compounds (peaks 47 and 48) strongly suggested that these compounds were T-diOH BA-S. As shown in Fig. 5F, G, CGH deconjugation caused a nearly 10-fold decrease of peak intensity of T-diOH BA-3S (peaks 46–48) and a more than 2-fold increase of the peak intensity of diOH BA-S (peaks 43–45), which led to identification of peaks 46–48 as T-diOH BA-S. Comparison of the profiles of T-diOH-S in the control and diOH-S after CGH deconjugation makes it reasonable to assume that peaks 46 and 48 are the taurine conjugates of UDCA-3S (peak 43) and CDCA-3S (peak 45), respectively. An unsaturated T-diOH BA-S (peak 49) and two GlcNAc conjugates of T-diOH BA-S (peaks 50 and 51) were also detected by their multiple typical MRM ion pairs scan and enzyme assays (Fig. 5H, I). A total of 25 diOH BAs were detected in piglet urine, which included four FBAs, twenty conjugated BAs, and one unsaturated BA.
Fig. 5.
Comparison of selected ion monitoring chromatograms of diOH BAs and MRM scan chromatograms of their conjugates in urine of breast-fed female piglets before and after enzymatic deconjugation. Y-axis on the graph represents the peak-height ratio of endogenous BAs to the IS and the X axis the Rt. in minutes. A: Chromatogram for free diOH BAs was generated by a selected ion m/z 391.4. MRM scan chromatograms generated by MRM ion pairs: m/z 448.6/74.0 for G-diOH BAs (Ba) and (Bb); m/z 567.7/391.4 for diOH BA-GlcUA (C); m/z 594.8/391.4 for diOH BA-GlcNAc (D); m/z 624.7/97.0 (red line), 624.7/391.4 (blue line) for G-diOH BA-GlcUA (E); m/z 471.6/97.0 for diOH BA-GlcNAc (F); m/z 288.9[M-2H]2−/97.0 (red line), 578.8/97.0 (blue line) and 578.8/124.0 (green line) for T-diOH BA-S (G); m/z 286.9[M-2H]2−/97.0 (red line), 574.7/97.0 (blue line) and 574.7/124.0 (green line) for T-monoOH BA-Δ4−3-one-S (H); m/z 390.4[M-2H]2−/97.0 (red line), 781.9/97.0 (blue line) and 781.9/124.0 (green line) for T-diOH BA-S-GlcNAc (I).
Identification of conjugates of tetrahydroxycholanoic acids
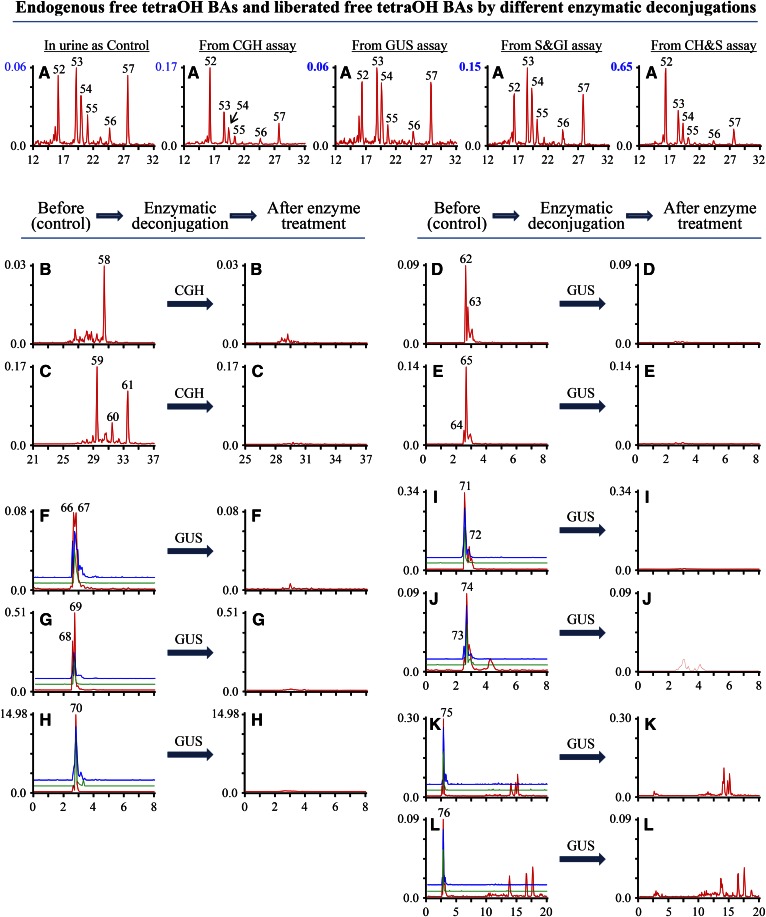
Tetrahydroxycholanoic acid (TetraOH BA) was examined by its selected ion [M-H]−, and T-tetraOH BA and G-tetraOH BA were examined by their respective MRM ion pairs shown in supplemental Table S1. A selected ion m/z 423.4 for tetraOH BAs yielded six peaks in control samples and all of them (peaks 52–57) had the peak area increase after CGH, S&GI, and CH&S treatments (Fig. 6A), which suggested that compounds (peaks 52–57) were tetraOH BAs. One peak for T-tetraOH BA (peak 58) and three peaks for G-tetraOH BAs (peaks 59–61) were detected in the control sample and eliminated by CGH deconjugation (Fig. 6B, C), which was in line with a more than 2-fold increase of the intensity of peak 52 and mild increase of peak 53 (Fig. 6A, from CGH assay). The results suggested that hydrolysis by CGH cleaved the amide bond of T-tetraOH BA (peak 58) and G-tetraOH BAs (peaks 59–61), producing two FBAs (peaks 52 and 53). On the basis of the Rt. and intensity increase of peaks for two released FBAs (peaks 52 and 53), it is reasonable to assume that the compounds (peaks 59 and 61) might be glycine conjugates of peaks 52 and 53, respectively. However, the final structural identification of peaks 52, 53, 59, and 61 could not be completed here because their standards were unavailable. No glycosidic conjugates of tetraOH BA were detected by their typical MRM ion pairs (supplemental Table S1), and the conclusions were confirmed by unaffected free tetraOH BA in GUS assay (Fig. 6A, from GUS assay). Three peaks for glycosidic conjugates of T-tetraOH were yielded by their typical MRM ion pairs: m/z 706.4/124.0 for T-tetraOH BA-GlcUA, m/z 733.4/124.0 for T-tetraOH BA-GlcNAc, and m/z 692.3/124.0 for T-tetraOH BA-Glc in the control sample, and were eliminated by GUS or CGH/sulfatase treatments (chromatograms not shown). A combination of product ions and neutral losses has been reported to be the typical MRM ion pairs for glycosidic conjugates of BAs (24, 25). However, the typical MRM ion pairs for glycosidic conjugates of tetraOH BA ([M-1]−/530.6 or [M-1]−/423.4) did not yield any peak. Therefore, the three peaks generated by their typical MRM ion pairs ([M-1]−/124) would not be identified as glycosidic conjugates of T-tetraOH BA. Three typical MRM ion pairs for T-tetraOH BA-S, including m/z 304.9[M-2H]2−/97.0, 610.8/97.0, and 610.8/124.0, yielded peaks at the same Rt. (peaks 66 and 67), and the peaks were enzymatically hydrolyzed by CGH and GUS (Fig. 6F). The results led to the identification of these two compounds (peaks 66 and 67) as T-tetraOH BA-S. Also, T-tetraOH BA-S’s GlcUA conjugates (peaks 68 and 69), GlcNAc conjugates (peaks 71 and 72), and Glc conjugate (peak 75) were identified by their multiple typical MRM ion pairs and enzymatic deconjugation in CGH, GUS, S&GI, and CH&S (Fig. 6G, I, K). The glycosidic conjugates of T-tetraOH-S (peaks 68, 69, 71, 72, and 75) were in triple conjugate form and had high levels in the control (Fig. 6G, I, K). CH&S treatment deconjugated these conjugates of triple conjugate form and led a more than 7-fold increase of free tetraOH BAs (Fig. 6A, from CH&S), which confirmed the structural assignments for peaks 68, 69, 71, 72 and 75 (Table 1). Furthermore, the same procedure identified their corresponding unsaturated BAs, such as T-tetraOH BA-Δ-S-GlcUA (peak 70), T-tetraOH BA-Δ-S-GlcNAc (peaks 73 and 74), and T-tetraOH BA-Δ-S-GlcUA (peak 76) (Fig. 6H, J, L). The unsaturated BAs (peaks 70, 73, 74, and 76) were identified as Δ4-3-oxo BAs due to the Δ4-3-oxo-steroid 5β-reductase in a complex biochemical pathway of BAs. For the tetraOH BAs, six free tetraOH BAs, eighteen conjugated BAs, and four unsaturated BAs were characterized in piglet urine (Table 1).
Fig. 6.
Comparison of selected ion monitoring chromatograms of tetraOH BAs and MRM scan chromatograms of their conjugates in urine of breast-fed female piglets before and after enzymatic deconjugation. The y-axis on the graph represents the peak-height ratio of endogenous BAs to the IS and the x-axis the Rt. in minutes. A: The chromatogram for free tetraOH BAs was generated by a selected ion m/z 423.4. MRM scan chromatograms generated by MRM ion pairs: m/z 530.6/124.0 for T-tetraOH BAs (B); m/z 480.6/74.0 for G-tetraOH BAs (C); m/z 679.8/97.0 for tetraOH BA-S-GlcUA (D); m/z 665.8/97.0 for tetraOH BA-S-Glc (E); m/z 304.9[M-2H]2−/97.0 (red line), 610.8/97.0 (blue line) and 610.8/124.0 (green line) for T-tetraOH BA-S (F); m/z 392.9[M-2H]2−/97.0 (red line), 786.8/97.0 (blue line) and 786.8/124.0 (green line) for T-tetraOH BA-S-GlcUA (G); m/z 390.9[M-2H]2−/97.0 (red line), 782.8/97.0 (blue line) and 782.8/124.0 (green line) for T-trihydroxy-3-oxo-4-cholanoic acid (triOH BA-Δ4-3-one)-S-GlcUA (H); m/z 406.4[M-2H]2−/97.0 (red line), 813.9/97.0 (blue line) and 813.9/124.0 (green line) for T-tetraOH BA-S-GlcNAc (I); m/z 404.4[M-2H]2−/97.0 (red line), 809.8/97.0 (blue line) and 809.8/124.0 (green line) for T-triOH BA-Δ4-3-one-S-GlcNAc (J); m/z 385.9[M-2H]2−/97.0 (red line), 772.9/97.0 (blue line) and 772.9.9/124.0 (green line) for T-tetraOH BA-S-Glc (K); m/z 383.9[M-2H]2−/97.0 (red line), 768.8/97.0 (blue line) and 768.8/124.0 (green line) for T-triOH BA-Δ4-3-one-S-Glc (L).
Identification of conjugates of monohydroxycholanoic acids
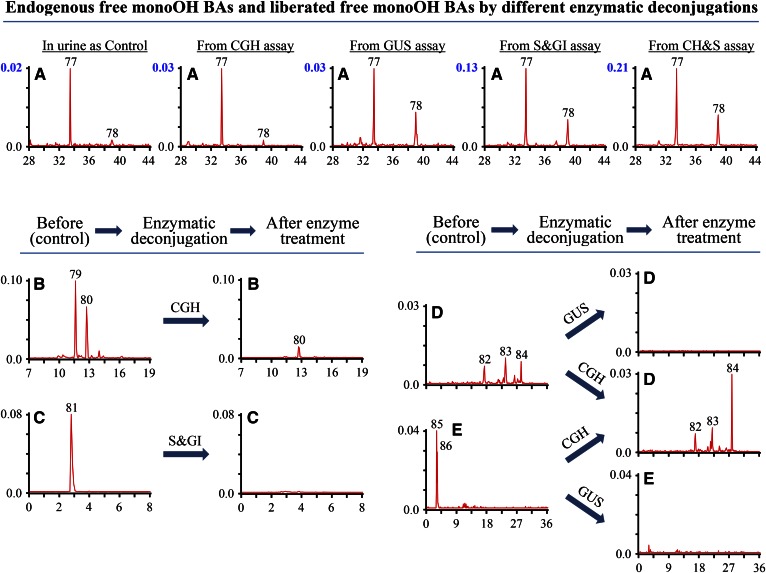
As shown in Fig. 7A, B, the selected ion [M-H]− monitoring detected two peaks (peaks 77 and 78) for monohydroxycholanoic acids (monoOH BAs) in the control sample, and compounds of peaks 77 and 78 were identified as alloLCA and LCA by co-injection of sample and standard in LC-MS analysis (Fig. 3H). Also, two peaks (peaks 79 and 80) were detected for glycine conjugates of monoOH BAs. Enzymatic deconjugation using CGH resulted in the hydrolysis of these two compounds (Fig. 7B) and the increase of peaks 77 and 78 (Fig. 7A, from CGH assay). However, the increase was too small (Fig. 7A, from CGH) for the hydrolyzed conjugates of peaks 79 and 80 (Fig. 7B). Also, standard comparison indicated that the compounds of peaks 79 and 80 were not G-LCA (Fig. 3J). Thus, the compounds of peaks 79 and 80 were tentatively identified as glycine conjugates of monoOH BAs. As neither NMR data of these minor compounds nor their standards were available, identification of these compounds could not be completed by LC-MRM-MS analysis in this study. For the sulfate conjugate of monoOH BAs, one good peak (peak 81) at Rt. 2.83 min for monoOH BA-3S was detected in the control by its typical MRM ion pair, m/z 455.6/97.0. In comparison with the Rt. of diOH BA-3S (Rt. 29.58 min for UDCA-3S and 32.36 min for CDCA-3S), the peak at 2.83 min should not be a monoOH BA-S. However, the elimination of the peak at 2.83 min (Fig. 7C) and increase of free monoOH BAs (Fig. 7A, from S&GI) by S&GI assay suggested that peak 81 might be a monoOH BA-S. No amidated (T- and G-) conjugates of monoOH BA-S were detected by their corresponding MRM ion pairs (supplemental Table S1). Three peaks (peaks 82–84) were generated by MRM m/z 551.3/375.2 for monoOH BA-GlcUA and two peaks (peaks 85 and 86) by MRM m/z 658.3/124.0 for T-monoOH BA-GlcUA (Fig. 7D, E). Elimination of all five peaks (peaks 82–86) after GUS deconjugation suggested that they were GlcUA conjugates. Furthermore, CGH treatment totally hydrolyzed the compounds of peaks 85 and 86. In agreement with the hydrolysis of T-monoOH BA-GlcUA of peaks 85 and 86 by CGH treatment, CGH treatment caused a more than 2-fold increase of monoOH BA-GlcUA peak 84 (Fig. 7D, E), which indicated that peaks 85 and 86 were T-monoOH BA-GlcUA. G-monoOH BA-GlcUA was not detected in the piglet urine and in the samples after CGH and GUS enzymatic deconjugation. Monohydroxylated BA has been reported to have two potential glucuronidation sites: the 3-hydroxy group and the side chain carboxyl group (29), and compounds (peaks 82–86) should be 3-0- or carboxyl-attached glucuronides of LCA. However, their particular glucuronidation sites could not be determined without their authentic standards in the present study.
Fig. 7.
Comparison of selected ion monitoring chromatograms of monoOH BAs and MRM scan chromatograms of their conjugates in urine of breast-fed female piglets before and after enzymatic deconjugation. The y-axis on the graph represents the peak-height ratio of endogenous BAs to the IS and the x-axis the Rt. in minutes. A: The chromatogram for free monoOH BAs was generated by a selected ion m/z 375.4. MRM scan chromatograms generated by MRM ion pairs: m/z 432.6/74.0 for G-tetraOH BAs (B); m/z 455.6/97.0 for MonoOH BA-S (C); m/z 551.7/375.4 for MonoOH BA-GlcUA (D); m/z 658.8/124.0 for T-monoOH BA-GlcUA (E).
Quantitative determination
In this study, two quantitative methods were used to calculate the concentrations of endogenous BAs. In the first method, the concentration of each individual BA was calculated using the ratio of its peak-area to the IS and its corresponding standard curve (supplemental Table S2). The concentrations of the individual BAs are presented in Table 1. BAs are remarkably diverse in structure and exist in the piglet urine, and BA standards were not available for all detected BAs, in which case the concentration of a detected BA was calculated using the standard curve of a structurally related BA standard. However, the calculated concentration may differ depending on which structurally related standard was chosen. For example, given the peak area of an analyte, the concentration of the analyte calculated using the T-γMCA standard curve would be 43 times higher than that calculated using the G-γMCA standard curve (supplemental Table S2) even though both T-γMCA and G-γMCA are in the amidated form with the only difference between them being that they are amidated with different amino acids. Thus, the concentration of an analyte calculated using the standard curve of a structurally related standard might be different from the analyte’s real concentration in urine due to the difference in sensitivity between the analyte and the structurally related standard in LC-MRM-MS analysis (supplemental Table S2).
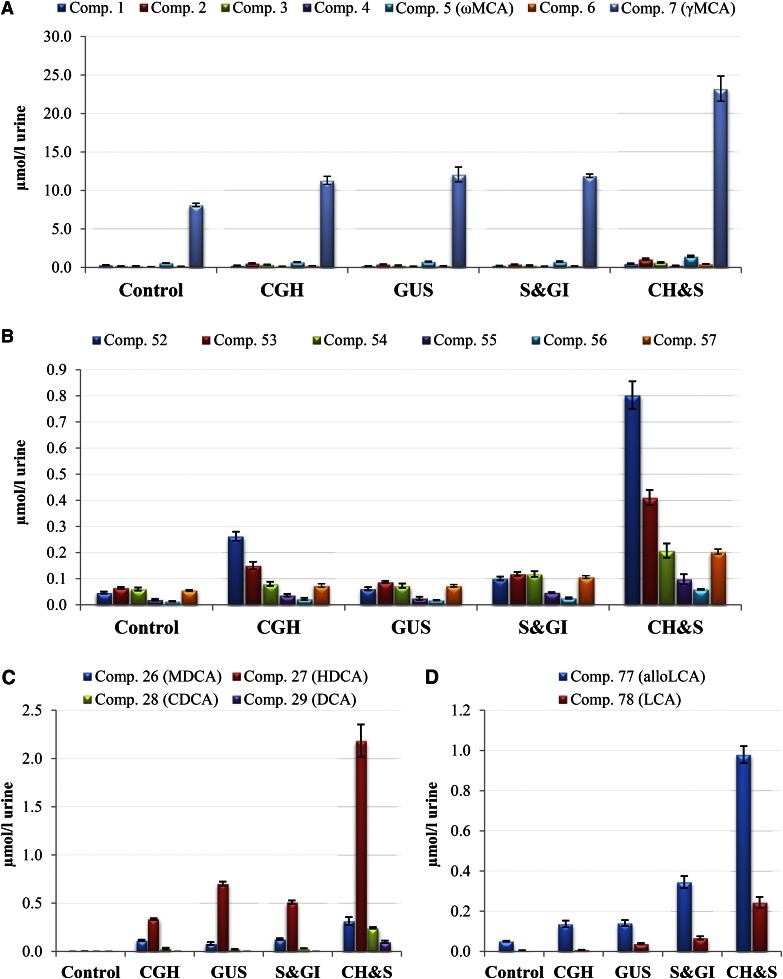
In the second method, the concentration of each conjugate group was determined by the FBAs liberated from enzymatic deconjugation. Conjugates were hydrolyzed by four enzymatic assays: CGH for amidated conjugates (taurine and glycine), GUS for glycosidic conjugates, S&GI for sulfate conjugates, and CH&S for all forms of conjugates (Fig. 1). The results are shown in Fig. 8. The total concentration of FBAs was 9.90 ± 0.20 μmol/l in urine control and increased to 33.40 ± 2.31 μmol/l in urine after CH&S treatment, indicating the urine was comprised of 29.75 ± 2.44% FBAs and 70.25 ± 2.44% conjugated BAs in single and multiple conjugated forms. CGH hydrolyzed the conjugates with two amino acids (glycine or taurine) and released 4.87 ± 0.64 μmol/l of FBAs, which was 14.54 ± 1.08% of total urinary BAs. Glycosidic conjugates at 5.43 ± 0.92 μmol/l were found by GUS treatment and represented 16.27 ± 2.65% of urinary BAs. S&GI treatment liberated 5.56 ± 0.33 μmol/l of FBAs, which constituted 16.67 ± 0.42% of urinary BAs. The released FBAs in CGH, GUS, and S&GI treatments only represented the concentration of single conjugates because the conjugated BAs in multiple conjugated forms could not liberate the FBAs during their enzymatic deconjugation by CGH, GUS, and S&GI treatments. For example, T-triOH BA-S-Glc (peak 20 in Fig. 4G) was enzymatically hydrolyzed by GUS to release T-triOH BA-S (peak 20a in Fig. 4H). γMCA was found to be a dominant FBA and comprised 81.94 ± 0.70% of FBAs (8.11 ± 0.22 μmol/l) in female piglet urine. The total concentration of triOH BAs and their conjugates was 27.54 ± 1.99 μmol/l and 82.47 ± 0.34% of total urinary BAs. DiOH BAs were initially detected only in trace amounts in urine, but were found in significant levels after the enzymatic assays above (Fig. 8C). The most abundant diOH BA released by CH&S treatment was HDCA (2.19 ± 0.17 μmol/l), and the concentration of released CDCA was 0.24 ± 0.01 μmol/l.
Fig. 8.
Endogenous free diOH BAs and liberated free diOH BAs in urine of breast-fed female piglets by different enzymatic deconjugations. Control, urine sample without enzymatic treatment; CGH, urine sample after CGH treatment; GUS, urine sample after GUS treatment; S&GI, urine sample after sulfatase with glucuronidase inhibitor treatment; CH&S, after choloylglycine hydrolase and sulfatase treatment. Free triOH BAs (A), Free tetraOH BAs (B), Free diOH BAs (C), Free monoOH BAs (D).
DISCUSSION
The purpose of the present work was to obtain a comprehensive profile of urinary BAs in piglets, an animal model increasingly common in studies of postnatal development and nutrition. Because conjugated BAs have characteristic fragmentation patterns in MS analysis, and most endogenous conjugated BAs are present in low concentrations in biological samples, targeted LC-MRM-MS analysis is generally accepted as the preferred technique for detecting and quantitating conjugated BAs in a biological matrix. However, in the absence of a corresponding standard for a particular BA, a non-BA compound in the biological sample would be mistakenly identified as the BA if the MS of the non-BA compound contained the same molecular weight and product ion as that of a typical MRM ion pair for the BA. Therefore, herein we present a method for identifying and quantitating BAs, especially in the absence of their corresponding standards. This method identifies conjugated BAs using a two-step process: 1) the typical MRM ion pairs (supplemental Table S1) were used to scan the BA conjugates in urine and yielded the peaks of potential BA conjugates; and 2) four enzymatic assays were used to deconjugate the conjugated BAs and examine the changes of possible BA peaks by LC-MRM-MS analysis. The MRM peaks for conjugated BAs in urine that were diminished or eliminated and liberated new BA peaks by enzymatic treatment were considered as conjugated BAs. For example, as shown in Fig. 4E, F, two good peaks for Glc and GlcNAc conjugates of triOH BAs were generated by their typical MRM ion pairs, respectively. These two peaks were not considered to be glycosidic conjugates because they were not affected by GUS. On the other hand, peaks 17–19 were identified as GlcUA conjugates of triOH BAs because compounds (peaks 17–19) were hydrolyzed (Fig. 4D) and concomitantly released the free triOH BAs (Fig. 8A) by GUS treatment. In the present study, enzymatic deconjugation was used as additional evidence for confirming the identification of BAs in urine. Also, multiple typical MRM ion pairs were used for the identification of BAs in multiple conjugated form in this study (Fig. 4G, I–L). Five peaks for T-triOH-S-GlcUA were generated by the MRM ion pair m/z 384.7[M-2H]2−/97.0. However, four of these peaks did not have the peak at the same Rt. by the other two typical MRM ion pairs (m/z 770.8/97.0 and 770.8/124.0) for T-triOH-S-GlcUA (Fig. 4K). Furthermore, these four peaks were not affected by CGH treatment. This finding clearly indicates that the four peaks were not those of T-triOH-S-GlcUA, despite being generated by the typical MRM ion pair m/z 384.7[M-2H]2−/97.0 of T-triOH-S-GlcUA.
The primary BAs in most mammals are CA and CDCA, and the CYP8B subfamily is required for introduction of a 12α-hydroxyl group to CDCA in CA biosynthesis. However, it is known that the domestic pig does not synthesize CA (6). The enzyme (CYP4A21) catalyzing the 6α-hydroxylation in γMCA biosynthesis, an atypical member of the CYP4A subfamily, was found in pig (6, 30). An isomer of CA, 3α,6α,7α-trihydroxy-5β-cholanoic acid (γMCA), is the major BA constituent of porcine bile and considered to be a species-specific primary BA in the pig. Hence, the identification of γMCA as a dominant urinary BA component (81.94 ± 0.70% of FBAs in urine) in the present study is consistent with the literature (6). CDCA is considered to be a precursor of γMCA biosynthesis using 6α-hydroxylase (CYP4A21), and it is expected that the ratio between γMCA and CDCA in pigs is comparable to that of CA and CDCA in most other mammals (6). In the present study, free diOH BAs, including CDCA, were only detected in trace amounts in urine by a selected ion m/z 391.4, and the ratio between γMCA and CDCA in pigs is not comparable to that of CA and CDCA in most other mammals. However, 22 conjugates of diOH BAs were identified in urine (Table 1) and a significant level of CDCA was found after enzymatic deconjugation (Figs. 5A, 8C), which supported a role of CDCA in biosynthesis of BAs and, more specifically, in the biosynthesis of γMCA via the action of 6α-hydroxylase (CYP4A21). Also, CYP3A4 has been reported to be involved in 6α-hydroxylation of both taurochenodeoxycholic acid and lithocholic acid (31). For the secondary free BAs, six triOH BAs, three diOH BAs, six tetraOH BAs, and two monoOH BAs were detected, which may be biosynthesized from primary BA γMCA and CDCA by epimerization, hydroxylation, and dehydroxylation, respectively.
Biosynthesis of BAs from cholesterol results in accumulation of BAs, and BAs in high concentration can be toxic in the liver. Metabolism of BAs, including various conjugation reactions, protects the liver by both converting hepatic toxic BAs into polar BA conjugates and increasing their urinary elimination. In humans, taurine and glycine conjugates of primary BAs, CA and CDCA, are a major metabolic pathway of BAs. Sulfation and three types of glycosidic conjugation are also known metabolic pathways of BA synthesis (27), which can be synthesized from nonamidated BAs and their glycine and taurine conjugates as excretion products in urine. Sixty-seven conjugated BAs, including 38 in single conjugated form, 14 in double conjugated form, and 15 in triple conjugated form, were detected in female piglet urine by the method of a combination of enzymatic deconjugation and targeted LC-MRM-MS analysis. The qualitative and quantitative results of conjugated BAs in female piglet urine are summarized in Table 1 and Fig. 8. Liberated FBAs from urinary BA conjugates by CH&S treatment was 23.50 ± 2.46 μmol/l, which represented 70.25 ± 2.44% of urinary BAs. The conjugated BAs in triple conjugated forms are the most hydrophilic BAs and constitute a major portion of the urinary BAs in piglets (Table 1). Identification of glycine and taurine conjugates of unsaturated BAs has been reported in urine of children with defects in the gene or promoter region of Δ4-3-oxo-steroid 5β-reductase (gene AKR1D1) (11). Nine conjugates of unsaturated BAs were detected in this study and suggested an involvement of Δ4-3-oxo-steroid 5β-reductase in biosynthesis of BAs in piglets.
In summary, a method using a combination of enzymatic deconjugation and targeted LC-MRM-MS analysis was developed for analyzing BA conjugates in piglet urine. Four enzymatic assays were used to deconjugate BA conjugates: CGH for taurine- and glycine-amidated conjugates, GUS for glycosidic conjugates, S&GI for sulfate conjugates, and CH&S for all forms of conjugates (Fig. 1). γMCA, an isomer of CA, was identified as a predominant FBA in female piglet urine, which agrees with the previous report that γMCA is a species-specific primary BA in the pig (6). CDCA was detected only in trace amounts in urine before the enzymatic assays, but was found in a significant amount after the enzymatic assays, which supports that CDCA is a precursor of γMCA in BA biosynthesis in piglets. A high concentration of conjugated BAs in double and triple conjugated forms in urine suggests that multiple conjugated forms constitute one of the major pathways for the excretion of excess cholesterol. Furthermore, the developed method also can be used in detection of other classes of metabolites in single and multiple conjugated forms where their standards are unavailable.
Supplementary Material
Acknowledgments
The authors wish to thank Phaedra Yount for assistance with figure preparation.
Footnotes
Abbreviations:
- alloLCA
- allolithocholic acid
- BA
- bile acid
- CA
- cholic acid
- CDCA
- chenodeoxycholic acid
- CGH
- choloylglycine hydrolase
- CH&S
- choloylglycine hydrolase and sulfatase
- DCA
- deoxycholic acid
- 3-DCA
- 3-deoxycholic acid
- diOH BA
- dihydroxycholanoic acid
- FBA
- unconjugated bile acid
- G-
- glyco- (glycine)
- Glc
- glucoside
- GlcNAc
- N-acetylglucosamine
- GlcUA
- glucuronic acid
- GUS
- β-glucuronidase
- HDCA
- hyodeoxycholic acid
- H2O
- water
- IS
- internal standard
- isoDCA
- 5β-cholanic acid-3β,12α-diol
- isoLCA
- isolithocholic acid
- LCA
- lithocholic acid
- MCA
- muricholic acid
- MDCA
- murideoxycholic acid
- MeOH
- methanol
- monoOH BA
- monohydroxycholanoic acid
- MRM
- multiple reaction monitoring
- Rt.
- retention time
- S
- sulfate
- S&GI
- sulfatase with glucuronidase inhibitor
- T-
- tauro- (taurine)
- T-CA
- taurocholic acid
- tetraOH BA
- tetrahydroxycholanoic acid
- triOH BA
- trihydroxycholanoic acid
- UDCA
- ursodeoxycholic acid
These studies were supported by the U.S. Department of Agriculture, Agriculture Research Service Project 6026-51000-010-05S. The content is solely the responsibility of the authors and does not necessarily represent the official views or policies of the US Department of Agriculture.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Russell D. W. 2009. Fifty years of advances in bile acid synthesis and metabolism. J. Lipid Res. 50: S120–S125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.García-Cañaveras J. C., Donato M. T., Castell J. V., and Lahoz A.. 2012. Targeted profiling of circulating and hepatic bile acids in human, mouse, and rat using a UPLC-MRM-MS-validated method. J. Lipid Res. 53: 2231–2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sayin S. I., Wahlstrom A., Felin J., Jantti S., Marschall H., Bamberg K., Angelin B., Hyotylainen T., Oresic M., and Backhed F.. 2013. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 17: 225–235. [DOI] [PubMed] [Google Scholar]
- 4.Li-Hawkins J., Gåfvels M., Olin M., Lund E. G., Andersson U., Schuster G., Björkhem I., Russell D. W., and Eggertsen G.. 2002. Cholic acid mediates negative feedback regulation of bile acid synthesis in mice. J. Clin. Invest. 110: 1191–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oosterloo B. C., Premkumarb M., Stolla B., Olutoyec O., Thymanne T., Sangilde P. T., and Burrin D. G.. 2014. Dual purpose use of preterm piglets as a model of pediatricGI disease. Vet. Immunol. Immunopathol. 159: 156–165. [DOI] [PubMed] [Google Scholar]
- 6.Lundell K., Hansson R., and Wikvall K.. 2001. Cloning and expression of a pig liver taurochenodeoxycholic acid 6a-hydroxylase (CYP4A21). J. Biol. Chem. 276: 9606–9612. [DOI] [PubMed] [Google Scholar]
- 7.Thomas C., Pellicciari R., Pruzanski M., Auwerx J., and Schoonjans K.. 2008. Targeting bile-acid signalling for metabolic diseases. Nat. Rev. Drug Discov. 7: 678–693. [DOI] [PubMed] [Google Scholar]
- 8.Eyssen H. J., De Pauw G., and Van Eldere J.. 1999. Formation of hyodeoxycholic acid from muricholic acid and hyocholic acid by an unidentified gram-positive rod termed HDCA-1 isolated from rat intestinal microflora. Appl. Environ. Microbiol. 65: 3158–3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Griffiths W. J., and Sjövall J.. 2010. Bile acids: analysis in biological fluids and tissues. J. Lipid Res. 51: 23–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clayton P. T. 2011. Disorders of bile acid synthesis. J. Inherit. Metab. Dis. 34: 593–604. [DOI] [PubMed] [Google Scholar]
- 11.Muto A., Takei H., Unno A., Murai T., Kurosawa T., Ogawa S., Iida T., Ikegawa S., Mori J., Ohtake A., et al. . 2012. Detection of Δ4-3-oxo-steroid 5β-reductase deficiency by LC–ESI-MS/MS measurement of urinary bile acids. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 900: 24–31. [DOI] [PubMed] [Google Scholar]
- 12.Hanson R. F., Isenberg J. N., Williams G. C., Hachey D., Szczepanik P., Klein P. D., and Sharp H. L.. 1975. The metabolism of 3α, 7α, 12α-trihydorxy-5β-cholestan-26-oic acid in two siblings with cholestasis due to intrahepatic bile duct anomalies. An apparent inborn error of cholic acid synthesis. J. Clin. Invest. 56: 577–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shaham O., Wei R., Wang T. J., Ricciardi C., Lewis G. D., Vasan R. S., Carr S. A., Thadhani R., Gerszten R. E., and Mootha V. K.. 2008. Metabolic profiling of the human response to a glucose challenge reveals distinct axes of insulin sensitivity. Mol. Syst. Biol. 4: 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wahlén E., and Strandvik B.. 1994. Effects of different formula feeds on the developmental pattern of urinary bile acid excretion in infants. J. Pediatr. Gastroenterol. Nutr. 18: 9–19. [DOI] [PubMed] [Google Scholar]
- 15.Chen J. R., Lazarenko O. P., Blackburn M. L., Badeaux J. V., Badger T. M., and Ronis M. J.. 2009. Infant formula promotes bone growth in neonatal piglets by enhancing osteoblastogenesis through bone morphogenic protein signaling. J. Nutr. 139: 1839–1847. [DOI] [PubMed] [Google Scholar]
- 16.Ronis M. J., Chen Y., Shankar K., Gomez-Acevedo H., Cleves M. A., Badeaux J., Blackburn M. L., and Badger T. M.. 2011. Formula feeding alters hepatic gene expression signature, iron and cholesterol homeostasis in the neonatal pig. Physiol. Genomics. 43: 1281–1293. [DOI] [PubMed] [Google Scholar]
- 17.Matuszewski B. K., Constanzer M. L., and Chavez-Eng C. M.. 2003. Strategies for the assessment of matrix effect in quantitative bioanalytical methods based on HPLC-MS/MS. Anal. Chem. 75: 3019–3030. [DOI] [PubMed] [Google Scholar]
- 18.Fu I., Woolf E. J., and Matuszewski B. K.. 1998. Effect of the sample matrix on the determination of indinavir in human urine by HPLC with turbo ion spray tandem mass spectrometric detection. J. Pharm. Biomed. Anal. 18: 347–357. [DOI] [PubMed] [Google Scholar]
- 19.Buhrman D. L., Price P. I., and Rudewicz P. J.. 1996. Quantitation of SR 27417 in human plasma using electrospray liquid chromatography-tandem mass spectrometry: a study of ion suppression. J. Am. Soc. Mass Spectrom. 7: 1099–1105. [DOI] [PubMed] [Google Scholar]
- 20.Hall T. G., Smukste I., Bresciano K. R., Wang Y., McKearn D., and Savage R. E.. 2012. Identifying and overcoming matrix effects in drug discovery and development. In Tandem Mass Spectrometry-Applications and Principles. J. K. Prasain, editor. InTech, Rijeka, Croatia. 389–421. [Google Scholar]
- 21.Fang N., Yu S., Ronis M. J., and Badger T. M.. 2015. Matrix effects break the LC behavior rule for analytes in LC-MS/MS analysis of biological samples. Exp. Biol. Med. (Maywood). 240: 488–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goto T., Shibata A., Sasaki D., Suzuki N., Hishinuma T., Kakiyama G., Iida T., Manoa N., and Goto J.. 2005. Identification of a novel conjugate in human urine: bile acid acyl galactosides. Steroids. 70: 185–192. [DOI] [PubMed] [Google Scholar]
- 23.Fang N., Yu S., and Badger T. M.. 2002. Characterization of isoflavones and their conjugates in female rat urine using LC/MS/MS. J. Agric. Food Chem. 50: 2700–2707. [DOI] [PubMed] [Google Scholar]
- 24.Ellis E., Roeb E., and Marschall H-U.. 2001. Primary cultures of human hepatocytes but not HepG2 hepatoblastoma cells are suitable for the study of glycosidic conjugation of bile acids. Biochim. Biophys. Acta. 1530: 155–161. [DOI] [PubMed] [Google Scholar]
- 25.Maekawa M., Shimada M., Iida T., Goto J., and Mano N.. 2014. Tandem mass spectrometric characterization of bile acids and sterokd confugates based on low-energy collision-induced dissociation. Steroids. 80: 80–91. [DOI] [PubMed] [Google Scholar]
- 26.Marschall H-U., Matem H., Wietholtz H., Egestad B., Matem S., and Sjovall J.. 1992. Bile acid N-acetylglucosaminidation in vivo and in vitro evidence for a selective conjugation reaction of 7β-Hydroxylated bile acids in humans. J. Clin. Invest. 89: 1981–1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Radomińska-Pyrek A., Zimniak P., Irshaid Y. M., Lester R., Tephly T. R., and Pyrek J. S.. 1987. Glucuronidation of 6a-hydroxy bile acids by human liver microsomes. J. Clin. Invest. 80: 234–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marschall H-U., Egestad B., Matem H., Matem S., and Sjovall J.. 1987. Evidence for bile acid glucosides as normal constituents in human urine. FEBS Lett. 213: 411–414. [DOI] [PubMed] [Google Scholar]
- 29.Radomińska-Pyrek A., Zimniak P., Chari M., Golunski E., Lester R., and Pyrek J. S.. 1986. Glucuronides of monohydroxylated bile acids: specificity of microsomal glucuronyltransferase for the glucuronidation site, C-3 configuration, and side chain length. J. Lipid Res. 27: 89–101. [PubMed] [Google Scholar]
- 30.Lundell K., and Wikvall K.. 2008. Species-specific and age-dependent bile acid composition: aspects on CYP8B and CYP4A subfamilies in bile acid biosynthesis. Curr. Drug Metab. 9: 323–331. [DOI] [PubMed] [Google Scholar]
- 31.Araya Z., and Wikvall K.. 1999. 6K-hydroxylation of taurochenodeoxycholic acid and lithocholic acid by CYP3A4 in human liver microsomes. Biochim. Biophys. Acta. 1438: 47–54. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.