Abstract
Cholesterol 7α-hydroxylase (CYP7A1) plays a critical role in control of bile acid and cholesterol homeostasis. Bile acids activate farnesoid X receptor (FXR) and Takeda G protein-coupled receptor 5 (TGR5) to regulate lipid, glucose, and energy metabolism. However, the role of bile acids in hepatic inflammation and fibrosis remains unclear. In this study, we showed that adenovirus-mediated overexpression of Cyp7a1 ameliorated lipopolysaccharide (LPS)-induced inflammatory cell infiltration and pro-inflammatory cytokine production in WT and TGR5-deficient (Tgr5−/−) mice, but not in FXR-deficient (Fxr−/−) mice, suggesting that bile acid signaling through FXR protects against hepatic inflammation. Nuclear factor κ light-chain enhancer of activated B cells (NF-κB)-luciferase reporter assay showed that FXR agonists significantly inhibited TNF-α-induced NF-κB activity. Furthermore, chromatin immunoprecipitation and mammalian two-hybrid assays showed that ligand-activated FXR interacted with NF-κB and blocked recruitment of steroid receptor coactivator-1 to cytokine promoter and resulted in inhibition of NF-κB activity. Methionine/choline-deficient (MCD) diet increased hepatic inflammation, free cholesterol, oxidative stress, apoptosis, and fibrosis in CYP7A1-deficient (Cyp7a1−/−) mice compared with WT mice. Remarkably, adenovirus-mediated overexpression of Cyp7a1 effectively reduced hepatic free cholesterol and oxidative stress and reversed hepatic inflammation and fibrosis in MCD diet-fed Cyp7a1−/− mice. Current studies suggest that increased Cyp7a1 expression and bile acid synthesis ameliorate hepatic inflammation through activation of FXR, whereas reduced bile acid synthesis aggravates MCD diet-induced hepatic inflammation and fibrosis. Maintaining bile acid and cholesterol homeostasis is important for protecting against liver injury and nonalcoholic fatty liver disease.
Keywords: bile acid, nuclear receptor, farnesoid X receptor, Takeda G protein-coupled receptor 5
Bile acids are amphipathic molecules synthesized from cholesterol in the liver and secreted into the gastrointestinal track to facilitate digestion, absorption, and transport of lipids, nutrients, and vitamins (1). Bile acids are also recognized as signaling molecules that activate nuclear farnesoid X receptor (FXR) and membrane Takeda G protein-coupled receptor 5 [TGR5; also known as G protein-coupled bile acid receptor-1 (Gpbar1)] and cellular signaling pathways to regulate lipid, glucose, drug, and energy metabolism (2). The rate of bile acid synthesis is mainly controlled by transcriptional regulation of the gene encoding cholesterol 7α-hydroxylase (CYP7A1), the first and rate-limiting enzyme in the classic bile acid synthesis pathway, which synthesizes two primary bile acids, cholic acid (CA) and chenodeoxycholic acid (CDCA), and steroid 12α-hydroxylase (CYP8B1) is required for CA synthesis. In mice, CDCA is converted to α- and β-muricholic acids in the liver as primary bile acids. The alternative pathway is initiated by steroid 27-hydroxylase (CYP27A1) and followed by oxysterol 7α-hydroxylase (CYP7B1) to synthesize mainly CDCA. Bile acids are conjugated to taurine or glycine and are secreted into bile and stored in the gallbladder. After a meal, bile acids are released into the intestinal tract and are reabsorbed in the ileum and circulated back to the liver. This enterohepatic circulation of bile acids exerts a negative feedback mechanism to inhibit Cyp7a1 gene transcription and bile acid synthesis and maintains a constant bile acid pool. Alteration of bile acid homeostasis may contribute to inflammatory cholestatic liver diseases, diabetes, and obesity (1, 3).
The liver is the primary site of clearance of microbial products such as lipopolysaccharides (LPSs). LPS activates nuclear factor κ light-chain enhancer of activated B cells (NF-κB), which is a master regulator of inflammation and cell death and causes liver injury, fibrosis, and hepatocellular carcinoma (4). Accumulation of hydrophobic and toxic bile acids, such as CDCA, deoxycholic acid, and lithocholic acid can cause inflammation in the liver and intestine and contribute to inflammatory bowel diseases and cholestatic liver diseases. CDCA and deoxycholic acid have been shown to inhibit the LPS-induced pro-inflammatory cytokine production in macrophages and monocytes (5). However, several studies report that activation of FXR antagonizes NF-κB in the hepatic inflammatory response and regulates intestinal innate immunity and improves nonalcoholic fatty liver disease (6–9). Activation of TGR5 has been shown to inhibit atherosclerosis by reducing macrophage inflammation (10) and protect intestinal barrier function in mouse models of inflammatory bowel diseases (11). TGR5 is widely expressed in the gallbladder epithelium, ileum, and colon. In the liver, TGR5 is expressed in Kupffer cells and sinusoidal endothelial cells, but not in hepatocytes (12). Activation of TGR5 induces cAMP and stimulates glucagon-like peptide-1 (GLP-1) secretion from intestinal enteroendocrine L cells, which induces insulin secretion from β cells (13). It has been reported that bile acids activate the TGR5-cAMP pathway to inhibit LPS-induced cytokine production in Kupffer cells, and suggested that TGR5 in Kupffer cells may play a protective role against excessive cytokine production and liver injury in obstructive cholestasis (14), consistent with the finding that TGR5-deficient (Tgr5−/−) mice have more severe liver necrosis and inflammation compared with WT mice (15). On the contrary, several recent studies show that TGR5 induces cytokine production in Kupffer cells (16) and enhances LPS-induced inflammatory response in a human monocyte cell line (17). Thus, the role of TGR5 signaling in hepatic inflammation is controversial and requires further study.
Currently, it is thought that progression from simple steatosis to nonalcoholic steatohepatitis (NASH) involves “multiple hits,” such as endoplasmic reticulum stress, inflammatory cytokines, innate immunity, and gut microbiota and epigenetic factors (18–21). Increasing evidence shows that dietary cholesterol increases oxidative stress and exacerbates high-fat/high-cholesterol diet-induced hepatic steatosis and inflammation (22). Accumulation of free cholesterol in stellate cells and Kupffer cells aggravates liver fibrosis in mice (23–25). Our previous study reported that transgenic overexpression of CYP7A1 in the liver prevents high-fat/high-cholesterol diet-induced obesity and decreases inflammation in mice (26). CYP7A1 plays a key role in regulation of hepatic cholesterol homeostasis by converting excess cholesterol to bile acids, which facilitate biliary cholesterol secretion. The aims of this study are to investigate the roles and mechanisms of CYP7A1 and bile acid-activated receptors in anti-inflammation and fibrosis in the liver. Our data show that increasing bile acid synthesis by overexpression of CYP7A1 ameliorates hepatic inflammation through activation of FXR, but not TGR5. On the other hand, lack of Cyp7a1 aggravated methionine/choline-deficient (MCD) diet-induced liver fibrosis through accumulation of free cholesterol in hepatocytes. Our study suggests that CYP7A1 plays a critical role in control of hepatic inflammation and fibrosis by maintaining bile acid and cholesterol homeostasis.
MATERIALS AND METHODS
Animals
Male C57BL/6J mice, 6–8 weeks of age, were purchased from the Jackson Laboratory (Bar Harbor, ME). Tgr5−/− mice were obtained from Merck Research Laboratories (Kenilworth, NJ). FXR-deficient (Fxr−/−) mice were originally generated by Frank Gonzalez (National Cancer Institute, National Institutes of Health) and bred to C57BL/6J genetic background. CYP7A1-deficient (Cyp7a1−/−) mice in B6/129Sv background were obtained from Jackson Laboratory and backcrossed to C57BL/6J mice for seven generations to ∼99.6% C57BL6J background (27). All mice were maintained on a standard chow diet and water ad libitum and housed in a room with a 12 h light/dark cycle. The Institutional Animal Care and Use Committee of Northeast Ohio Medical University approved all animal protocols used in this study. To study the effect of overexpression of CYP7A1 on anti-inflammation in hepatocytes, WT, Fxr−/−, or Tgr5−/− mice were injected via the tail vein with 1 × 109 plaque-forming units (pfu) per mouse of adenovirus-Cyp7a1 (Ad-Cyp7a1), a generous gift from Dr. Michael Pandak (Virginia Commonwealth University, Richmond, VA). After 7 days, mice were treated with LPS (20 mg/kg) via ip injection and were euthanized 6 h later. To induce liver fibrosis, male WT and Cyp7a1−/− mice were fed a MCD (Harlan, catalog number TD90262) diet for 3 weeks and mice were euthanized for further analysis.
Cell culture
The human hepatocellular carcinoma cell line, HepG2, and human embryonic kidney cell line, HEK293T (American Type Culture Collection), were cultured in DMEM (Invitrogen, Carlsbad, CA) containing 10% fetal bovine serum (Gibco, Grand Island, NY), 100 U/ml penicillin, and 100 μg/ml streptomycin in 5% CO2 at 37°C. Primary human hepatocytes were obtained from the Liver Tissue Procurement and Distribution System (University of Pittsburgh, PA).
Luciferase reporter assay
For the luciferase reporter assays, HepG2 cells were plated in 24-well plates and transiently cotransfected with the pNL3.2-NF-κB-RE reporter plasmids, β-gal expressing plasmid (for normalization of transfection efficiency) and pcDNA3-hFXR (as indicated), using TransFast™ transfection reagent (Promega, Madison, WI). After 48 h, transfected HepG2 cells were treated with TNF-α for 6 h or transfected with pCMV4-p65 plasmid for 24 h to stimulate NF-κB reporter activity, and FXR agonist, GW4064 (1 μM; Sigma-Aldrich, St. Louis, MO) or obeticholic acid (OCA; 10 μM; Intercept Pharmaceuticals, Inc. New York, NY), was added to test their effect on reporter activity. Luciferase assays were performed and luciferase activity was normalized to β-gal activity as described previously (28).
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation (ChIP) assays were performed with a ChIP assay kit (Millipore, Bedford, MA) according to the manufacturer’s instructions. Briefly, liver tissues were homogenized and lysates were cross-linked in 1% formaldehyde and sonicated to break the DNA into 0.2–2.0 kb fragments. Sonicated cell lysates were incubated with either anti-p65 (#8242, Cell Signaling Technology) or anti-FXR (#13063, Santa Cruz Biotechnology) antibody to precipitate protein/chromatin complex. Rabbit IgG (#sc-2027, Santa Cruz Biotechnology) was used as a negative control. Protein A/G-agarose beads were added to pull down protein/DNA complexes. DNA fragments were amplified by PCR using the primers designed to probe the NF-κB binding sites on the Tnf-α and interleukin (Il)-1β gene promoters. SYBR Green primers for real-time PCR analysis were as follows: mouse IL-1β (forward primer, −319CCCATTTCCACCACGATGA; reverse primer, −216GAGGCTATTGCTACCCTGAAATA); mouse TNF-α (forward primer, −800CCACTTCCTCCAAGAACTCAA; reverse primer, −600TCTGAAAG CTGGGTGCATAA).
Bile acid analysis
For total bile acid content analysis, bile acids in liver, intestine (whole with its content), and gallbladder were extracted in 95% ethanol overnight, in 80% ethanol for 2 h, and methanol:chloroform (2:1) for 2 h at 50°C. Bile acids were determined with a bile acid assay kit (Genzyme Diagnostic, Framingham, MA). Bile acid composition was analyzed by ultra-high performance liquid chromatography/quadrupole time-of-flight mass spectrometry (by Dr. Feng Li, Baylor College of Medicine, Huston, TX) as described previously (27).
ELISA assay of pro-inflammatory cytokines
After LPS injection for 6 h, blood was collected from mice and centrifuged at 3,000 g for 10 min. The levels of IL-1β, IL-6, and TNF-α in the serum were measured using ELISA kits (Qiagen, Valencia, CA) according to the manufacturer’s instructions.
Mammalian two-hybrid assay
For mammalian two-hybrid assays, HepG2 cells were cotransfected with luciferase reporter plasmid, pG5luc, and Gal4-fusion protein plasmids and VP-16-p65 plasmids for 48 h. Luciferase reporter activity was determined by dual luciferase reporter assay system (Promega) according to the manufacturer’s instructions. GAL4-FXR-FULL, GAL4-FXR-ligand binding domain (LBD), GAL4-FXR-DNA binding domain (DBD), and GAL4-FXR-HINGE plasmids were generous gifts from Dr. Frederick J. Suchy (University of Colorado Health Sciences Center, Aurora, CO).
Histochemical and immunohistochemical staining
Liver tissues from mice were fixed in 10% formalin and embedded in paraffin. Tissue sections were then subjected to hematoxylin and eosin (H&E). For F4/80 (#6640, Abcam) and α-smooth muscle actin (α-SMA) (#5694, Abcam) immunohistochemistry, livers were fixed with paraformaldehyde and stained by using an avidin-biotin complex staining kit (Santa Cruz Biotechnology, Santa Cruz, CA) according to the manufacturer’s instructions. The images were visualized by microscopy (IX73; Olympus, Japan).
Sirius red and Masson’s trichrome staining
Livers were fixed in 10% neutral-buffered formalin and embedded in paraffin. For Sirius red staining, liver sections were incubated with 0.1% Sirius red in saturated picric acid for 90 min. For Masson’s trichrome staining, liver sections were stained by a Masson’s trichrome staining kit (Sigma-Aldrich). The images were visualized by microscopy (IX73; Olympus).
Filipin staining of free cholesterol
Liver sections were fixed with 4% paraformaldehyde (fresh) for 1 h and stained with 0.05 mg/ml filipin working solution for 2 h at room temperature. The images were visualized by fluorescence microscopy using a UV filter set (340–380 nm excitation, 430 nm emission).
Hepatic hydroxyproline assay
Livers were homogenized in water and hydrolyzed in concentrated hydrochloric acid at 120°C for 3 h. Hepatic hydroxyproline contents were determined by the reaction of oxidized hydroxyproline with 4-(dimethylamino) benzaldehyde by a colorimetric kit (Cell Biolabs, San Diego, CA).
RNA isolation and quantitative real-time PCR
Total RNA was isolated with Trizol reagent (Invitrogen). Reverse transcription reactions and real-time PCR were performed as described previously (29). All primer/probe sets for real-time PCR were TaqMan gene expression assays (Applied Biosystems, Foster City, CA). Amplification of GAPDH was used as an internal control. Relative mRNA expression was quantified using the comparative CT (Ct) method and expressed as 2^ (−ΔΔCt).
Western blot
Liver tissues were homogenized in RIPA buffer containing protease and phosphatase inhibitor cocktails (Cell Signaling Technology, Danvers, MA). The protein contents were determined by a BCATM protein assay kit (Pierce, Rockford, IL). The protein lysates were separated by 10% SDS-PAGE and transferred onto a polyvinylidene difluoride membrane (Millipore, Billerica, MA). After blocking with 5% nonfat milk for 1 h at room temperature, the membrane was incubated with primary antibody overnight and conjugated secondary antibody for 1 h. Protein bands were visualized using a Western blotting detection system according to the manufacturer’s instructions.
Quantification of tissue and plasma lipids
Liver tissues were homogenized and lipids were extracted in a mixture of chloroform and methanol (2:1), dried, and dissolved in 5% Triton X-100 in isopropanol. Hepatic and plasma triglyceride and cholesterol levels were then quantified using Infinity reagents from Thermo Scientific (Waltham, MA). Hepatic total free cholesterol was quantified using a kit from BioVision (Milpitas, CA). Hepatic and plasma free fatty acid were quantified using a kit from Wako Pure Chemical Industries, Ltd. (Osaka, Japan).
Thiobarbituric acid reactive substance, superoxide dismutase, and terminal deoxynucleotidyl transferase dUTP nick end labeling assays
Liver tissues were homogenized and hepatic malondialdehyde and superoxide dismutase (SOD) levels were determined by using a thiobarbituric acid reactive substance (TBARS) assay kit and SOD assay kit, respectively (Cayman Chemical, Ann Arbor, MI). A terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assay kit was used to assay apoptotic cells in liver tissues (R&D, Minneapolis, MN).
Statistical analysis
All experimental data are presented as mean ± SE. Statistical analysis was performed either by Student’s t-test for analysis of two variants or ANOVA for analysis of four variants. P < 0.05 was considered statistically significant.
RESULTS
Adenovirus-mediated expression of CYP7A1 ameliorated LPS-induced hepatic inflammation
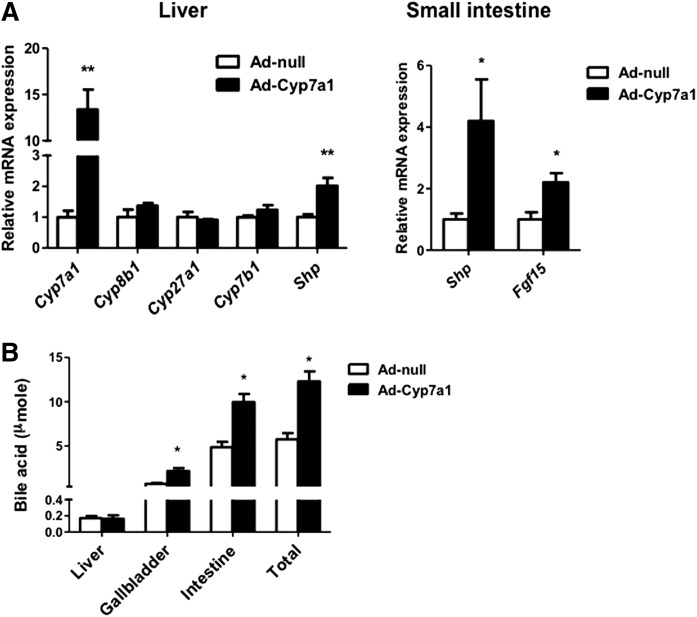
Adenovirus-mediated Cyp7a1 gene transduction (Ad-Cyp7a1) has been used to increase CYP7A1 activity and bile acid synthesis in rat primary hepatocytes (30). We used Ad-Cyp7a1 to overexpress CYP7A1 in mice. Figure 1A shows that Ad-Cyp7a1 increased hepatic Cyp7a1 mRNA levels by 12-fold compared with adenovirus-null (Ad-null) mice, but did not affect CYP8B1, CYP27A1, and CYP7B1 mRNA levels. The mRNA levels of a FXR target gene in the liver, small heterodimer partner (SHP), and intestine FXR target genes, SHP and fibroblast growth factor 15 (FGF15), were increased in Ad-Cyp7a1 mice compared with Ad-null mice (Fig. 1A). The tissue bile acid contents increased significantly in the small intestine and gallbladder, but not in the liver, of Ad-Cyp7a1 mice (Fig. 1B). The total bile acid pool increased 2-fold in Ad-Cyp7a1 mice compared with Ad-null mice. Analysis of gallbladder bile acids shows that TCA content was reduced and tauro-muricholic acids increased in Ad-Cyp7a1 mice compared with Ad-null mice, indicating stimulation of the alternative bile acid synthesis pathway to change bile acid composition (supplemental Fig. S1).
Fig. 1.
Adenovirus-mediated Cyp7a1 gene transduction activated FXR signaling in liver and intestine and increased bile acid pool size. Mice were injected in the tail vein with 1 × 109 pfu per mouse of either Ad-null (control) or Ad-Cyp7a1 for 7 days. A: mRNAs were isolated from liver (Left) and small intestine (Right) and mRNA levels of bile acid synthesis genes (Cyp7a1, Cyp8b1, Cyp27a1, and Cyp7b1) and FXR-induced genes [Shp and intestine fibroblast growth factor 15 (Fgf15)] were determined by qPCR. Amplification of Gapdh was used as a control. B: Bile acid contents in liver, intestine, and gallbladder of Ad-null and Ad-Cyp7a1 mice were determined, and total bile acid pool sizes were calculated. Results are shown as mean ± SE. *P < 0.05, **P < 0.01. Studentʼs t-test was used for statistical analysis (n = 12 male mice per group).
Adenovirus-mediated expression of Cyp7a1 ameliorated LPS-induced hepatic inflammation via FXR, but not TGR5
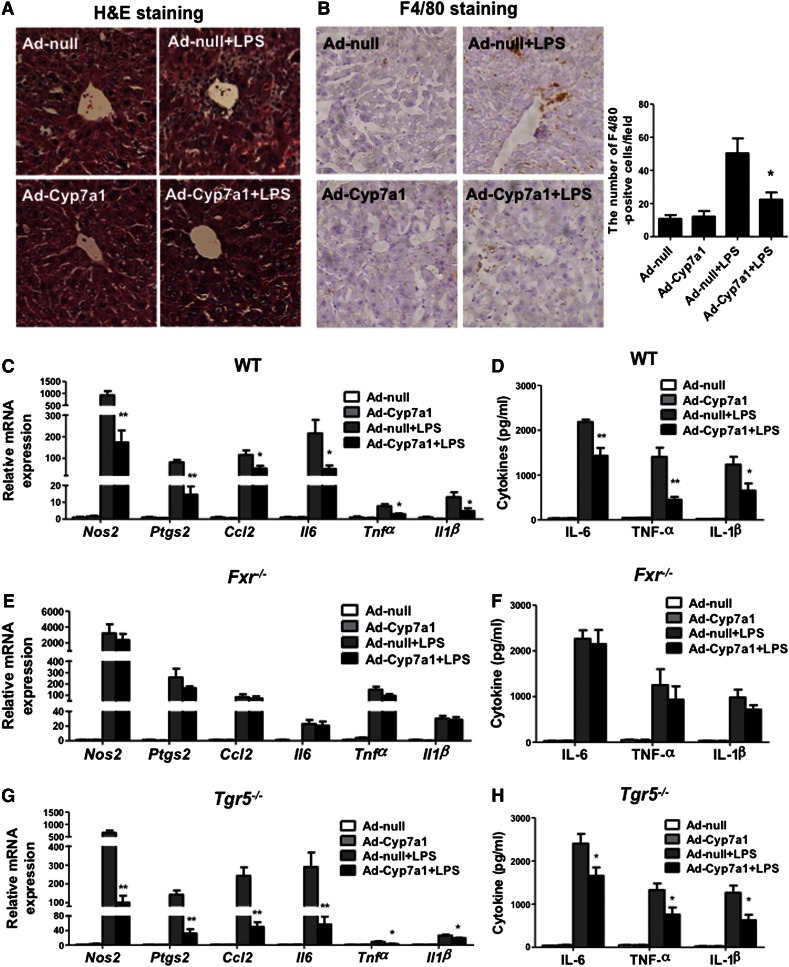
To study the roles of bile acid-activated receptors FXR and TGR5 in hepatic inflammation, WT, Fxr−/−, and Tgr5−/− mice were injected with LPS to induce acute liver inflammation and to test the effect of Ad-Cyp7a1 on hepatic inflammation. H&E staining shows that LPS induced massive inflammation in WT mouse livers of Ad-null mice (Control), but not Ad-Cyp7a1 mice (Fig. 2A). Immunohistochemistry staining of WT mouse livers shows that LPS treatment increased a macrophage marker, F4/80, in Ad-null mice, but much less in Ad-Cyp7a1 mice (Fig. 2B). In addition, Ad-Cyp7a1 reduced mRNA expression of LPS-induced pro-inflammatory gene mRNA expression, such as nitric oxide synthase 2 (Nos2), prostaglandin-endoperoxide synthase 2 (Ptgs2), chemokine (C-C motif) ligand 2 (Ccl2), IL-6, TNF-α, and IL-1β in WT mouse livers (Fig. 2C). Serum IL-1β, IL-6, and TNF-α protein levels were also significantly decreased in Ad-Cyp7a1-treated WT mice compared with Ad-null mice (Fig. 2D). It has been reported that hepatocyte lipoprotein receptors, scavenger receptor-B1 (Scarb1) (31) and LDL receptor (Ldlr) (32), are involved in LPS clearance. Data in supplemental Fig. S2 shows that Ad-Cyp7a1 did not induce Scarb1 and Ldlr mRNA expression in WT mouse livers and there was no change of serum endotoxin levels in LPS-treated Ad-null and Ad-Cyp7a1 mice, suggesting that Scarb1 and Ldlr are not involved in LPS-induced endotoxin clearance in Ad-Cyp7a1 mice.
Fig. 2.
Ad-Cyp7a1 ameliorated LPS-induced hepatic inflammation via FXR, but not TGR5. WT (C57BL/6J), Fxr−/−, and Tgr5−/− mice were injected in the tail vein with 1 × 109 pfu per mouse of either Ad-null or Ad-Cyp7a1. After 7 days, mice were injected ip with 20 mg/kg LPS or PBS. After 6 h, liver tissues and sera were collected for analysis. A: Ad-Cyp7a1 reduced LPS-induced hepatic inflammation. Fresh WT mouse liver tissues were kept in 10% formalin solution and 5 μM sections were stained with H&E. B: Ad-Cyp7a1 reduced macrophage infiltration in LPS-induced hepatic inflammation. Representative immunohistochemistry staining for F4/80 from WT mouse liver sections. The number of F4/80-positive cells in five microscopic fields was counted and statistically analyzed. C: Ad-Cyp7a1 reduced LPS-induced pro-inflammatory cytokines and chemokine mRNA expression in WT mice. Total RNAs from liver tissues were isolated and hepatic mRNA levels of inflammatory genes were determined by qPCR. Amplification of Gapdh was used as a control. D: Ad-Cyp7a1 reduced serum cytokines induced by LPS in WT mice. The levels of cytokines in the serum were measured by ELISA assay. Results are shown as mean ± SE. *P < 0.05, **P < 0.01 versus mice injected with Ad-null and treated with LPS. E: Ad-Cyp7a1 did not affect LPS-induced inflammatory gene mRNA expression in Fxr−/− mice. Total RNA from livers of Fxr−/− mice was isolated and hepatic mRNA levels of inflammatory genes were determined by qPCR. Amplification of Gapdh was used as control. F: Ad-Cyp7a1 did not affect serum cytokine protein levels in Fxr−/− mice. The levels of cytokines in the sera of Fxr−/− mice were measured by ELISA assay. G: Ad-Cyp7a1 reduced LPS-induced inflammatory cytokine mRNA levels in Tgr5−/− mouse liver. Total RNA from livers of Tgr5−/− mice was isolated and hepatic mRNA levels of inflammatory genes were determined by qPCR. Amplification of Gapdh was used as control. H: Ad-Cyp7a1 reduced serum cytokines in Tgr5−/− mice. The levels of cytokines in the sera of Tgr5−/− mice were measured by ELISA assay. Results are shown as mean ± SE. *P < 0.05, **P < 0.01 versus mice injected with Ad-null and treated with LPS. ANOVA was used for statistical analysis (n = 8 male mice per group).
Several studies reported that activation of FXR and TGR5 protects the liver from inflammation (6–9). However, TGR5 is not expressed in hepatocytes and the role of TGR5 signaling in liver inflammation is controversial (14, 16). Ad-Cyp7a1 did not affect LPS-induced inflammatory cell infiltration (supplemental Fig. S3A) or the population of F4/80-positive cells in Fxr−/− mice (supplemental Fig. S3B), but did reduce hepatic inflammation in Tgr5−/− mice (supplemental Fig. S4A, B). Furthermore, Ad-Cyp7a1 increased bile acid pool size by about 2.5-fold in both WT and Fxr−/− mice (supplemental Fig. S5A). It is surprising to notice that the total bile acid pool size in Ad-null Fxr−/− mice was not significantly increased compared with Ad-null WT mice. Quantitative (q)PCR analysis shows that in Fxr−/− mice, Cyp7a1 and Cyp8b1 mRNA levels were increased, but Cyp27a1, Cyp7b1, and Shp mRNA levels were reduced (supplemental Fig. S5B), indicating that bile acid synthesis via the classic pathway may be increased, but bile acid synthesis via the alternative pathway may be reduced and results in no significant change in bile acid pool size in Fxr−/− mice.
Real-time PCR analysis and ELISA assay showed that Ad-Cyp7a1 did not significantly affect LPS-induced pro-inflammatory cytokine and chemokine mRNAs (Fig. 2E) and serum cytokine levels (Fig. 2F) in Fxr−/− mice compared with Ad-null mice. However, Ad-Cyp7a1 ameliorated LPS-induced hepatic cytokine and chemokine mRNAs (Fig. 2G) and serum cytokine levels (Fig. 2H) in Tgr5−/− mice compared with Ad-null mice. These data indicate that activation of FXR, but not TGR5, ameliorates LPS-induced hepatic inflammation.
Activation of FXR inhibited NF-κB activity
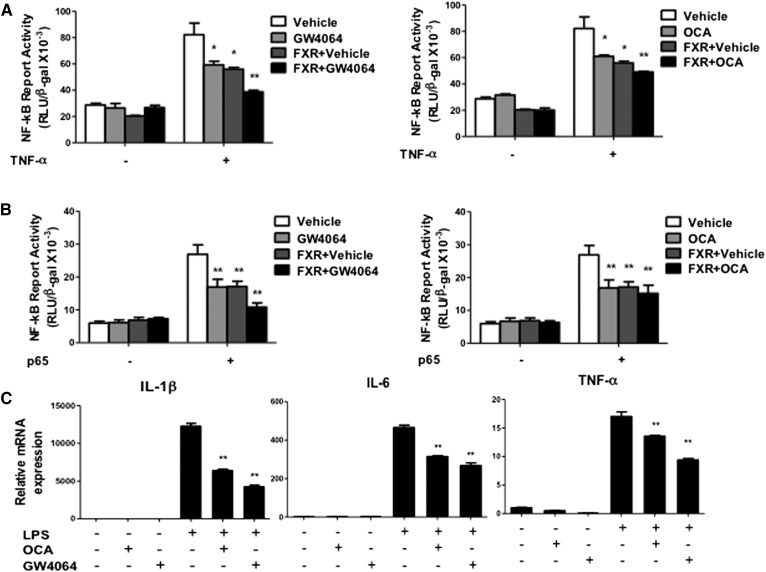
To further study the role of FXR in anti-inflammation, we used NF-κB/luciferase reporter assay to test whether activation of FXR reduces NF-κB/luciferase reporter activity. The NF-κB-luciferase reporter contains a canonical NF-κB response sequence. Figure 3A shows that both FXR selective agonists, GW4064 (1 μM) and OCA (10 μM), reduced the TNF-α-activated NF-κB reporter activity, and cotransfection with a FXR expression plasmid exacerbated the inhibitory effect of FXR agonists on the NF-κB reporter activity. Furthermore, GW4064 or OCA also inhibited the NF-κB reporter activity stimulated by p65, a component of the NF-κB complex (Fig. 3B). In addition, activation of FXR by GW4064 or OCA inhibited LPS-induced pro-inflammatory cytokine mRNA expression in human primary hepatocytes (Fig. 3C). These results demonstrated that activation of FXR inhibited NF-κB activity and pro-inflammatory cytokine production in hepatocytes.
Fig. 3.
Activation of FXR inhibited TNF-α and p65 activated NF-κB activity. A: NF-κB reporter assay showed that activation of FXR by GW4064 and OCA reduced TNF-α-induced NF-κB activity. B: NF-κB reporter assay showed that activation of FXR by GW4064 and OCA reduced p65-induced NF-κB activity. For reporter assay, HepG2 cells were cotransfected with NF-κB/luciferase reporter plasmid (pNL3.2-NF-κB-RE) and pCMV-β-gal (for normalization of β-galactosidase activity), and FXR expression plasmid (pcDNA3-hFXR), p65 expression plasmid (pCMV-p65), or pcDNA3 plasmid (control) as indicated. After 48 h, cells were treated with TNF-α (20 ng/ml), GW4064 (1 μM), or OCA (10 μM), as indicated for 6 h. Luciferase activities were measured by luciferase assay kit and luciferase activity was normalized to β-gal activity and expressed as relative luciferase units (RLU)/β-gal activity. Results shown are mean ± SE. *P < 0.05, **P < 0.01 versus vehicle (DMSO). C: Primary human hepatocytes were treated with GW4064 (1 μM) or OCA (10 μM) in the absence or presence of LPS (100 ng/ml) for 6 h. Total RNA was isolated and mRNA levels of inflammatory genes were determined by qPCR. Amplification of Gapdh was used as control. Results shown are mean ± SE. *P < 0.05, **P < 0.01 versus LPS treatment. ANOVA was used for statistical analysis.
Ad-Cyp7a1 inhibited NF-κB binding to cytokine gene promoters via a ligand-dependent interaction between FXR and p65 and steroid receptor coactivator-1
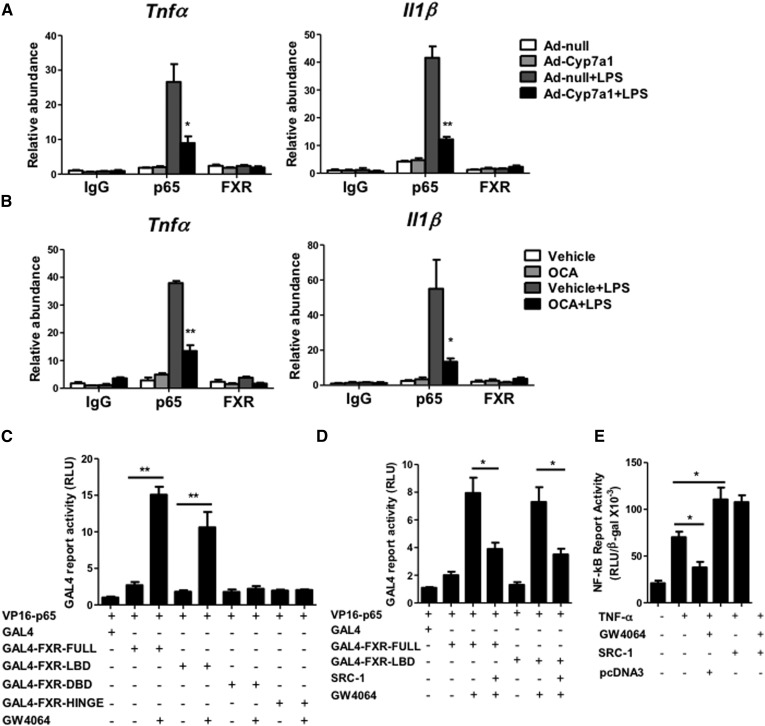
To further investigate the mechanisms of CYP7A1 and FXR in anti-inflammation in hepatocytes, we performed ChIP assay to evaluate the effect of CYP7A1 on NF-κB binding to the Tnf-α and Il-1β gene promoters, which contain the NF-κB binding sites. Liver lysates were isolated from mice treated with LPS for ChIP assay. An antibody against p65 was used to immunoprecipitate chromatin, and DNA fragments were PCR amplified with the primers designed to probe the NF-κB binding sites on the Tnf-α and Il-1β gene promoters. Figure 4A shows that p65 occupancy on the TNF-α and IL-1β gene promoters was enhanced by LPS (Ad-null + LPS), and that Ad-Cyp7a1 significantly reduced p65 occupancy on the TNF-α and IL-1β gene promoters (Ad-Cyp7a1 + LPS). An antibody against FXR was used for ChIP assay and showed that FXR did not bind to the NF-κB binding sites on the Tnf-α and Il-1β promoters under the same treatment. Next, we studied whether activation of FXR affects p65 binding to the Tnf-α and Il-1β gene promoters. Figure 4B shows that OCA had a similar effect as Ad-Cyp7a1 treatment to significantly inhibit p65 binding to the Tnf-α and Il-1β gene promoters. These data suggest that activation of FXR by bile acids and FXR agonist blocked p65 binding to the Tnf-α and Il-1β gene promoters.
Fig. 4.
Ad-Cyp7a1 inhibited LPS-induced NF-κB binding to cytokine gene promoters through inducing a ligand-dependent interaction between FXR and p65 and coactivator. A: ChIP assay of the NF-κB binding site showed that Ad-Cyp7a1 ameliorated LPS-induced p65 occupancy to the NF-κB binding site on Tnf-α and Il-1β gene promoters. B: ChIP assay showed that activation of FXR by OCA ameliorated LPS-induced p65 binding to the NF-κB binding site on Tnf-α and Il-1β gene promoters. For ChIP assay, WT mice were injected in the tail vein with 1 × 109 pfu per mouse of either Ad-null or Ad-Cyp7a1, or treated with 30 mg/kg OCA by gavage once per day. After 7 days of adenovirus or OCA treatment, mice were injected ip with 20 mg/kg LPS or PBS (control). After 6 h, liver tissues were homogenized and the chromatin protein and DNA were cross-linked in 1% formaldehyde and sonicated to 0.2–2.0 kb fragments and mixed with either anti-p65 or anti-FXR antibody to precipitate protein/chromatin complexes. Rabbit IgG was used as negative control. DNA fragments were quantified by amplification with the primers designed to probe the NF-κB binding sites on the Tnf-α and Il-1β gene promoters. C: Mammalian two-hybrid assay showed that GW4064 stimulated FXR interaction with p65 through the FXR LBD. D: Mammalian two-hybrid assay showed that SRC-1 reduced GW4064-activated FXR and p65 interaction by competition for interaction with FXR and p65. For two-hybrid assay, HEK293T cells were cotransfected with GAL4-luciferase reporter plasmid pG5luc (or Gal4 empty as control), VP16 fusion protein plasmid, VP16-p65 and GAL4-fusion protein plasmid, GAL-FXR (full length), GAL-FXR-LBD, GAL-FXR-DBD, GAL-FXR-HINGE, and SRC-1 plasmid in the absence or presence of GW4064 (1 μM) as indicated for 48 h. Luciferase reporter activity was assayed using a luciferase assay kit. Relative luciferase activity (RLU) was measured and normalized to Renilla luciferase activity. E: NF-κB reporter assay showed that coactivator SRC-1 stimulated TNF-α-induced NF-κB activity. HepG2 cells were cotransfected with pNL3.2-NF-κB reporter plasmid and pCMV-β-gal plasmid, and pcDNA3-SRC-1 or pcDNA3 (control). After 48 h, cells were treated with GW4064 (1 μM) in the absence or presence of TNF-α (20 ng/ml) for 6 h. NF-κB reporter activity was measured by luciferase assay kit. Results shown are mean ± SE. *P < 0.05, **P < 0.01. ANOVA was used for statistical analysis.
We then performed mammalian two-hybrid assays to study whether FXR interacts with p65 to prevent p65 from binding to the NF-κB binding site as a mechanism for the anti-inflammatory effects of FXR. In mammalian two-hybrid assay, a VP16-p65 fusion protein plasmid was cotransfected with GAL4-FXR fusion plasmids to stimulate GAL4-Luc reporter activity. As shown in Fig. 4C, GW4064 significantly increased the interaction between FXR and p65 through the LBD of FXR, because the DBD and the hinge domain of FXR do not interact with p65. Ligand-activated FXR is known to recruit coactivators such as steroid receptor coactivator-1 (SRC-1) to activate target gene transcription (33). SRC-1 is also a coactivator of NF-κB (34). Two-hybrid assays show that SRC-1 significantly inhibited the interaction between FXR and p65 (Fig. 4D), likely by interaction with the LBD of FXR to reduce FXR and p65 interaction. Furthermore, NF-κB reporter assay shows that GW4064 significantly inhibited TNF-α-induced NF-κB activity, whereas SRC-1 stimulated TNF-α-induced NF-κB activity and prevented GW4064 inhibition of reporter activity (Fig. 4E). These data suggest that activation of FXR promotes FXR interaction with p65 to prevent p65 binding to the NF-κB response elements on the TNF-α and IL-1β gene promoters. In addition, FXR and p65 may compete for interaction with SRC-1 and result in inhibiting TNF-α-induced NF-κB activation.
Lack of Cyp7a1 aggravated MCD diet-induced liver steatosis and fibrosis
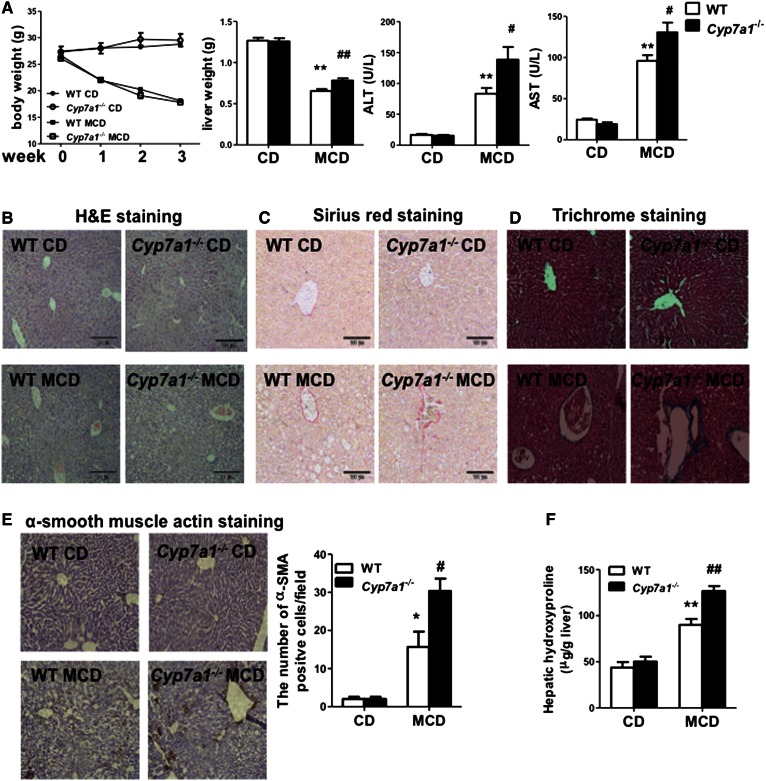
Hepatic inflammation plays a critical role in fibrosis progression. MCD diet induces steatohepatitis and fibrosis associated with elevated hepatic triglyceride and cholesterol contents (23). We therefore used a MCD diet model to study the role of CYP7A1 in hepatic inflammation and steatosis and fibrosis. After feeding MCD diet for three weeks, both WT and Cyp7a1−/− mice had significantly reduced body weights, but increased liver weight, hepatic steatosis, and serum aspartate aminotransferase and alanine aminotransferase levels, more in Cyp7a1−/− mice than WT mice (Fig. 5A). MCD diet feeding increased hepatic inflammation indicated by H&E staining (Fig. 5B), and increased collagen fibers by Sirius red (Fig. 5C), Masson’s trichrome (Fig. 5D), and α-SMA staining (Fig. 5E), more in Cyp7a1−/− mice than in WT mice. MCD diet also increased hydroxyproline collagen content more in Cyp7a1−/− mice than in WT mice. However, MCD diet feeding significantly reduced plasma cholesterol, triglycerides, and free fatty acid content in both WT and Cyp7a1−/− mice (supplemental Fig. S6A). Oil-Red-O staining of neutral lipids further confirmed that chow diet-fed Cyp7a1−/− mice had more hepatic steatosis than WT mice, and MCD diet feeding increased hepatic steatosis more in Cyp7a1−/− mice than in WT mice (supplemental Fig. S6B). These data suggested that lack of Cyp7a1 aggravated MCD diet-induced hepatic inflammation and fibrosis in mouse liver.
Fig. 5.
Lack of Cyp7a1 aggravated MCD diet-induced liver fibrosis. WT and Cyp7a1−/− mice were fed with chow diet or MCD diet for 3 weeks. A: MCD diet affected body weight, liver weight, and serum aspartate aminotransferase and alanine aminotransferase levels of WT and Cyp7a1−/− mice. B–E: MCD diet increased hepatic inflammation and collagen fibrosis in Cyp7a1−/− mouse livers. Fresh liver samples fixed in 10% formalin solution and 5 μM sections were stained with H&E (B), Sirius red (C), Masson’s trichrome stain (aniline blue solution) (D), and α-SMA (E). Representative immunohistochemistry stains of liver sections are shown. The number of α-SMA-positive cells in five microscopic fields was counted and statistically analyzed. F: MCD diet increased collagen hydroxyproline contents in Cyp7a1−/− mouse livers. Hydroxyproline contents in liver collagen were determined by the reaction of oxidized hydroxyproline with 4-(dimethylamino) benzaldehyde using a hydroxyproline assay kit. Results shown are mean ± SE. *P < 0.05, **P < 0.01 versus WT mice fed with chow diet. #P < 0.05, ##P < 0.01 versus WT mice fed with MCD diet. ANOVA was used for statistical analysis (n = 7–9 male mice per group).
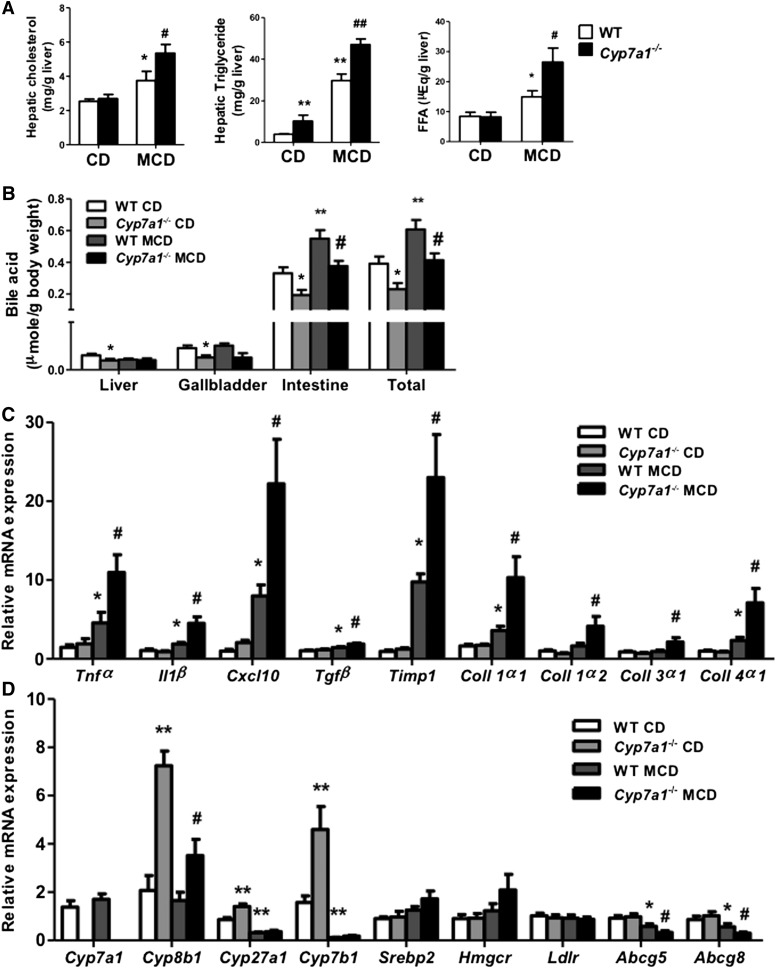
Figure 6A shows that only hepatic triglycerides were higher in chow diet-fed Cyp7a1−/− mice than WT mice, but MCD diet significantly increased hepatic cholesterol, triglyceride, and free fatty acid contents more in Cyp7a1−/− mice than in WT mice. Moreover, MCD diet did not affect total bile acid pool size in either WT or Cyp7a1−/− mice, the latter of which have a bile acid pool size about 60% of WT mice due to decreased bile acid contents in the liver, gallbladder, and intestine (Fig. 6B), as we reported previously (27). MCD diet significantly increased inflammatory genes, Tnf-α, Il-1β, and C-X-C motif chemokine ligand 10 (Cxcl-10), and fibrosis genes transforming growth factor β (Tgfβ), tissue inhibitor of metalloproteinase (Timp1), and collagen-1α1, -1α2, -3α1, and -4α1 mRNA expression in Cyp7a1−/− mice compared with WT mice (Fig. 6C). Cyp8b1, Cyp27a1, and Cyp7b1 mRNA levels were significantly increased in Cyp7a1−/− mice compared with WT mice (Fig. 6D) (27). The increase of Cyp8b1 mRNA may be due to reduced bile acid synthesis and pool size. Increased Cyp7b1 may increase bile acid synthesis via the alternative bile acid synthesis pathway to partially compensate for reduced bile acid synthesis via the classic pathway (27). MCD diet strongly reduced Cyp8b1, Cyp7b1, and Cyp27a1 mRNA levels in Cyp7a1−/− mice (Fig. 6D), consistent with changing bile acid composition to that of WT mice (supplemental Fig. S7). There was no significant difference in mRNA levels of cholesterol metabolism genes Srebp2, Hmgcr, and Ldlr. Interestingly, mRNA levels of cholesterol transporters Abcg5 and Abcg8 in the liver were significantly decreased in WT mice, and even more in MCD diet-fed Cyp7a1−/− mice, consistent with reduced cholesterol secretion and accumulation of hepatic cholesterol in MCD diet-fed Cyp7a1−/− mice. Hepatic Abcg5 and Abcg8 genes are induced by both LXR (35) and FXR (36). Reduced Abcg5/g8 expression may be due to reduced bile acid/FXR signaling in MCD diet-fed mice. It is not known whether oxysterol-activated LXR is activated. It is also possible that increased hepatic cytokines may decrease Abcg5/g8 expression (37).
Fig. 6.
MCD diet increased hepatic lipids and decreased mRNA expression of alternative bile acid synthesis pathway genes in Cyp7a1−/− mice. WT and Cyp7a1−/− mice were fed with chow diet or MCD diet for 3 weeks. A: MCD diet increased cholesterol, triglyceride, and free fatty acid contents in Cyp7a1−/− mouse livers. Hepatic lipids were detected using lipid analysis kits. B: MCD diet did not alter bile acid pool size in Cyp7a1−/− mice. Bile acids were analyzed using a bile acid analysis kit. Total bile acid pool sizes were determined as the total amount of bile acids in liver, intestine, and gallbladder. C: MCD diet increased inflammatory cytokines and collagen gene expression in Cyp7a1−/− mouse livers. Total RNAs from liver tissues were isolated and hepatic mRNA levels were determined by qPCR. Amplification of Gapdh was used as control. D: MCD diet decreased mRNA levels of alternative bile acid synthesis pathway genes in Cyp7a1−/− mouse livers. Quantitative real-time PCR was used to analyze mRNA expression levels of bile acid synthesis and cholesterol synthesis and transport genes. Total RNAs from liver tissues were isolated and hepatic mRNA levels were determined by qPCR. Amplification of Gapdh was used as control. Results shown are mean ± SE. *P < 0.05, **P < 0.01 versus WT mice fed with chow diet. #P < 0.05, ##P < 0.01 versus WT mice fed with MCD diet. ANOVA was used for statistical analysis (n = 7–9 male mice per group).
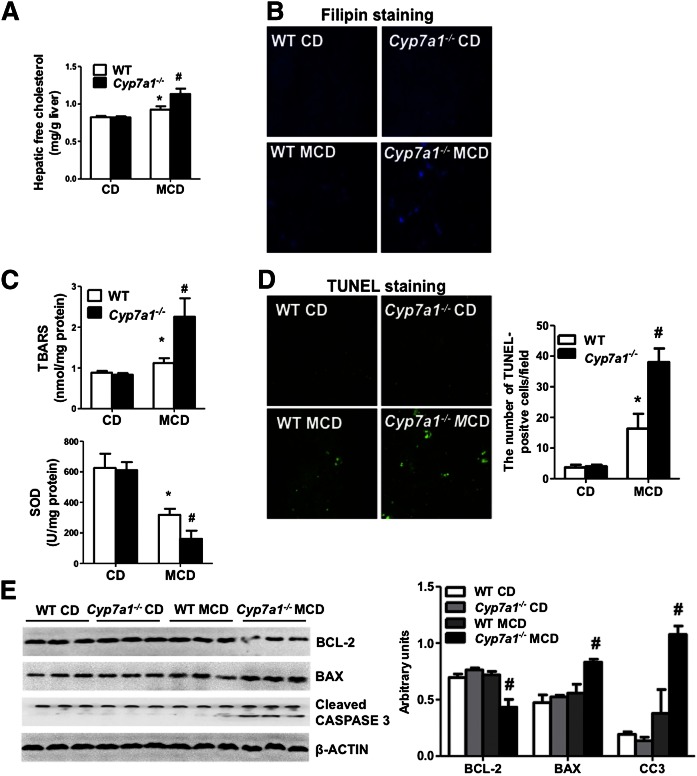
MCD diet increased free cholesterol, oxidative stress, and apoptosis, and aggravated hepatic fibrosis in Cyp7a1−/− mice
It has been reported that free cholesterol increases oxidative stress and aggravates liver fibrosis (24). We thus determined free cholesterol content in the liver of Cyp7a1−/− mice. Figure 7A shows that hepatic free cholesterol levels increased in MCD diet-fed Cyp7a1−/− mice compared with WT mice. In addition, filipin staining of free cholesterol in liver tissue further confirmed increased free cholesterol in MCD diet-fed Cyp7a1−/− mice compared with WT mice (Fig. 7B). TBARS assay of malondialdehyde, a product of oxidative stress, was significantly increased in MCD diet-fed Cyp7a1−/− mice compared with WT mice. SOD, the key enzyme for reducing reactive oxygen species, was much lower in MCD diet-fed Cyp7a1−/− mice than WT mice (Fig. 7C). Further, TUNEL assay for apoptosis revealed that the numbers of cell death were markedly increased in MCD diet-fed Cyp7a1−/− mice compared with WT mice (Fig. 7D). Immunoblot analysis showed a significant decrease of anti-apoptotic B cell lymphoma-2 (Bcl-2) and an increase of apoptotic Bcl-2-associated X (Bax) protein and cleaved-caspase 3 (CC3) protein in liver lysates of MCD diet-fed Cyp7a1−/− mice compared with WT mice (Fig. 7E). These data suggest that lack of Cyp7a1 increases hepatic free cholesterol, which induces oxidative stress and apoptosis in hepatocytes and aggravates hepatic fibrosis in MCD diet-fed Cyp7a1−/− mice.
Fig. 7.
MCD diet increased free cholesterol, oxidative stress, and apoptosis, and aggravated hepatic fibrosis in Cyp7a1−/− mice. WT and Cyp7a1−/− mice were fed with chow diet or MCD diet for 3 weeks. A: MCD diet increased free cholesterol in Cyp7a1−/− mouse livers. Free cholesterol contents in the liver were detected using a cholesterol assay kit. B: MCD diet increased free cholesterol in Cyp7a1−/− mouse livers. Fresh liver tissues were kept in 10% formalin solution and 5 μM sections were stained with filipin and determined by fluorescence microscope. Fluorescence density was analyzed by Image J software. C: MCD diet increased oxidative stress in Cyp7a1−/− mouse livers. Liver tissues were homogenized and oxidative stress was assayed using a TBARS assay kit. SOD activity was determined by a SOD kit. D: MCD diet increased hepatic apoptosis in Cyp7a1−/− mouse livers. Representative TUNEL apoptosis staining of liver sections from WT and Cyp7a1−/− mice are shown. The number of TUNEL-positive cells in five microscopic fields was counted and statistically analyzed. E: MCD diet increased apoptosis proteins in Cyp7a1−/− mouse livers. Liver tissues were homogenized and cell death-related proteins were determined by Western blot. β-Actin was used as control. Results shown are mean ± SE. *P < 0.05, versus WT mice fed with chow diet. #P < 0.05, versus WT mice fed with MCD diet. ANOVA was used for statistical analysis (n = 7–9 male mice per group).
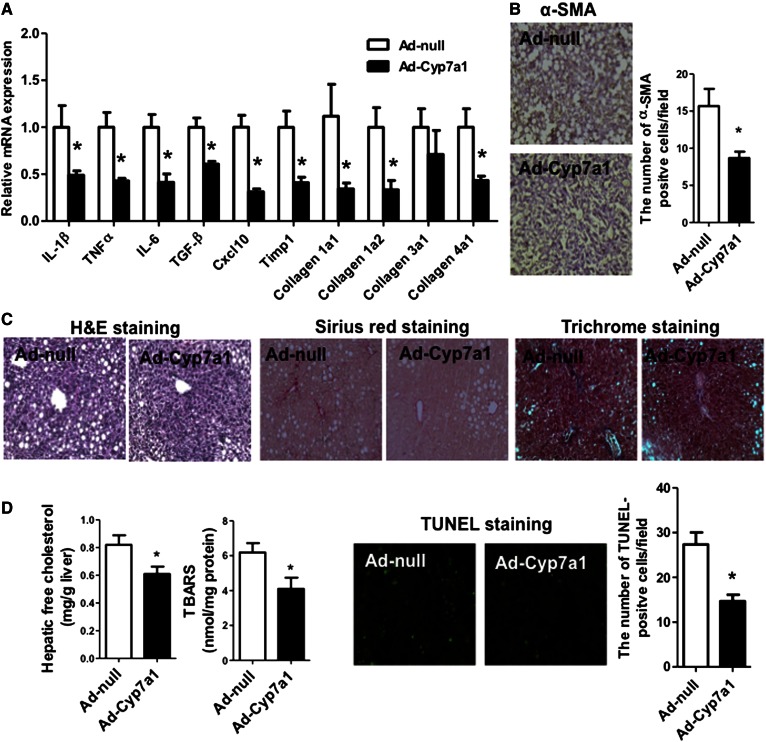
Ad-Cyp7a1 reversed MCD diet-induced hepatic steatosis and fibrosis in Cyp7a1−/− mice
In previous studies, we found that transgenic expression of CYP7A1 in the liver prevents high-fat diet-induced obesity and insulin resistance in mice and ameliorates LPS-induced hepatic inflammation (26). We further investigated to determine whether overexpression of CYP7A1 could reverse hepatic steatosis and fibrosis in MCD diet-fed Cyp7a1−/− mice. Cyp7a1−/− mice were fed with MCD diet for 3 weeks. On week 2, mice were injected with 1 × 109 pfu per mouse of either Ad-null (control) or Ad-Cyp7a1 and were euthanized 1 week later. Figure 8A shows that Ad-Cyp7a1 reduced MCD diet-induced hepatic fibrosis and inflammatory genes by 50% in Cyp7a1−/− mice compared with Ad-null Cyp7a1−/− mice. Ad-Cyp7a1 also reduced hepatic α-SMA of Cyp7a1−/− mice by ∼50% compared with Ad-null Cyp7a1−/− mice (Fig. 8B). H&E staining showed that Ad-Cyp7a1 reduced MCD diet-induced hepatic steatosis in Cyp7a1−/− mice compared with Ad-null Cyp7a1−/− mice. Sirius red and Masson’s trichrome staining showed that Ad-Cyp7a1 reduced MCD diet-induced collagen level compared with Ad-null Cyp7a1−/− mice (Fig. 8C). Furthermore, Ad-Cyp7a1 reduced MCD diet-induced hepatic free cholesterol content, TBARS level, and apoptosis (Fig. 8D). In addition, it was found that Ad-Cyp7a1 reduced total hepatic cholesterol and triglyceride contents, but not free fatty acid levels in Cyp7a1−/− mice (supplemental Fig. S8). These data suggested that overexpression of Cyp7a1 prevented MCD-diet-induced NASH in Cyp7a1−/− mice.
Fig. 8.
Ad-Cyp7a1 reduced MCD diet-induced hepatic steatosis and fibrosis in Cyp7a1−/− mice. Cyp7a1−/− mice were fed with MCD diet for 3 weeks. On week 2, mice were injected in the tail vein with 1 × 109 pfu per mouse of either Ad-null or Ad-Cyp7a1. One week later, mice were euthanized and liver tissues were collected for analysis. A: Ad-Cyp7a1 reduced MCD diet-induced inflammatory cytokines and collagen gene mRNA expression in Cyp7a1−/− mouse livers. Total RNAs from liver tissues were isolated and mRNA levels were determined by qPCR. B: Ad-Cyp7a1 reduced α-SMA in Cyp7a1−/− mice. C: Ad-Cyp7a1 reduced collagen and fibrosis in Cyp7a1−/− mouse livers. Representative H&E, Sirius red, and Masson’s trichrome stains are shown. D: Ad-Cyp7a1 reduced free cholesterol contents, TBARS levels, and apoptosis in Cyp7a1−/− mouse livers. All assays were performed as described in the Fig. 7 legend. Results a shown are mean ± SE. *P < 0.05 versus Ad-null Cyp7a1−/− mice fed with MCD diet. Studentʼs t-test was used for statistical analysis (n = 5–7 male mice per group).
DISCUSSION
In this study, we used gain-of-function and loss-of-function mouse models to investigate the roles of CYP7A1, FXR, and TGR5 in hepatic steatosis, inflammation, and fibrosis. Results from this study suggest that overexpression of CYP7A1 ameliorated LPS-induced inflammatory cell infiltration and pro-inflammatory cytokine production, and that FXR, but not TGR5, mediates the anti-inflammatory function of bile acids in hepatocytes. Conversely, genetic ablation of the Cyp7a1 gene exacerbated MCD diet-induced hepatic steatosis and fibrosis in mice that was likely caused by accumulation of free cholesterol, which increases oxidative stress and aggravates hepatic fibrosis and apoptosis. It has been reported recently that a high-cholesterol diet exacerbates liver fibrosis in mice and free cholesterol is accumulated in hepatic stellate cells (23, 24) and Kupffer cells (25), and accelerates liver fibrosis and NASH progression. Increasing free cholesterol accelerated oxidative stress via upregulation of the enzymes involved in the generation of reactive oxygen species and the downregulation of antioxidant enzymes (38). The mitochondrial overloading of free cholesterol, but not triglycerides and free fatty acids, decreases mitochondrial glutathione and sensitizes it to the TNF-α-mediated apoptosis of hepatocytes (39). The accumulation of lipids in the cytoplasm of hepatocytes is considered the first step in the development of liver fibrosis. Interestingly, we also found that triglyceride and free fatty acid levels were higher in Cyp7a1−/− mice than WT mice fed with MCD diet. On the other hand, overexpression of CYP7A1 stimulates bile acid synthesis to reduce hepatic free cholesterol and hepatic inflammation.
Our current study suggests a mechanism in which ligand-activated FXR directly interacts with p65 and blocks p65 from binding to the NF-κB binding sites on the cytokine promoters or competes for coactivator SRC-1 and results in inhibiting cytokine production in hepatocytes. A previous study reports that sumoylation of FXR stimulates FXR interaction with p65 to recruit corepressors to inhibit NF-κB-activated pro-inflammatory cytokine production (40). Another study reports that TGR5 antagonizes NF-κB activity to regulate inflammatory response in hepatocytes (15), but TGR5 induced Il-1β and Tnf-α expression in Kupffer cells (16). Our findings that LPS stimulates inflammatory cytokine production in Tgr5−/− mice and that overexpression of CYP7A1 in WT or Tgr5−/− mice reduces LPS-induced pro-inflammatory cytokine production. These findings suggest that TGR5 signaling may not play a major role in anti-inflammation in the liver.
Bile acid synthesis plays a critical role in maintaining hepatic cholesterol homeostasis, which is maintained by the balance of uptake of dietary cholesterol and de novo cholesterol synthesis, and catabolism of cholesterol to bile acids, biliary cholesterol secretion, and steroid hormone synthesis. It should be noted that bile acid synthesis accounts for catabolism of about 50% cholesterol and disposal of 45% cholesterol as acidic steroids (41). Thus bile acid synthesis plays a major role in maintaining whole-body cholesterol homeostasis. Stimulating CYP7A1 activity and bile acid synthesis is known to activate liver FXR signaling and intestine TGR5 signaling to reduce hepatic inflammation, increase insulin sensitivity, improve glycemic control, and improve obesity (7, 11, 42); however, the underlying mechanism is not completely understood (42, 43). In Cyp7a1 transgenic mice, bile acid pool size is doubled but TCA content is markedly reduced and these mice are resistant to Western diet-induced obesity and diabetes (26). CA is most efficacious among all bile acids in absorption of dietary cholesterol and fats, and has been linked to obesity and diabetes in human patients (44). We reported recently that Cyp7a1−/− mice, with a smaller bile acid pool and lower TCA, have improved glucose and insulin tolerance (27). Interestingly, dietary supplementation of CA to Cyp7a1−/− mice reverses glucose and insulin tolerance phenotypes. This current study also suggests that increased CA content in the bile acid pool of MCD diet-treated Cyp7a1−/− mice may increase dietary fat and cholesterol absorption and contribute to increased hepatic cholesterol and oxidative stress and hepatic fibrosis. Importantly, this study shows that overexpression of Cyp7a1 in MCD diet-fed Cyp7a1−/− mice could reduce hepatic free cholesterol and reverse hepatic steatosis, inflammation, and fibrosis. These data further prove our hypothesis that Cyp7a1 plays a critical role in protection of the liver from inflammation and fibrosis by maintaining cholesterol homeostasis.
In summary, our current study suggests that increased Cyp7a1 expression and bile acid synthesis ameliorates hepatic inflammation through activation of FXR, whereas reduced bile acid synthesis in Cyp7a1−/− mice aggravates MCD diet-induced hepatic inflammation and fibrosis. Maintaining bile acid and cholesterol homeostasis is important for protecting the liver from injury and nonalcoholic fatty liver disease.
Supplementary Material
Footnotes
Abbreviations:
- Ad-Cyp7a1
- adenovirus-Cyp7a1
- Ad-null
- adenovirus-null
- CA
- cholic acid
- CDCA
- chenodeoxycholic acid
- ChIP
- chromatin immunoprecipitation
- CYP7A1
- cholesterol 7α-hydroxylase
- Cyp7a1−/−
- CYP7A1-deficient
- CYP7B1
- oxysterol 7α-hydroxylase
- CYP8B1
- steroid 12α-hydroxylase
- CYP27A1
- steroid 27-hydroxylase
- DBD
- DNA binding domain
- FXR
- farnesoid X receptor
- Fxr−/−
- FXR-deficient
- H&E
- hematoxylin and eosin
- IL
- interleukin
- LBD
- ligand binding domain
- Ldlr
- LDL receptor
- LPS
- lipopolysaccharide
- MCD
- methionine/choline-deficient
- NASH
- nonalcoholic steatohepatitis
- NF-κB
- nuclear factor κ light-chain enhancer of activated B cells
- OCA
- obeticholic acid
- pfu
- plaque-forming units
- Scarb1
- scavenger receptor-B1
- SHP
- small heterodimer partner
- α-SMA
- α-smooth muscle actin
- SOD
- superoxide dismutase
- SRC-1
- steroid receptor coactivator-1
- TBARS
- thiobarbituric acid reactive substance
- TGR5
- Takeda G protein-coupled receptor 5
- Tgr5−/−
- TGR5-deficient
- TUNEL
- terminal deoxynucleotidyl transferase dUTP nick end labeling
This study was supported by National Institute of Diabetes and Digestive and Kidney Disease Grants DK44442 and DK58379. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors report no conflicts of interest.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Li T., and Chiang J. Y.. 2014. Bile acid signaling in metabolic disease and drug therapy. Pharmacol. Rev. 66: 948–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chiang J. Y. 2009. Bile acids: regulation of synthesis. J. Lipid Res. 50: 1955–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Trauner M., Fickert P., and Stauber R. E.. 1999. Inflammation-induced cholestasis. J. Gastroenterol. Hepatol. 14: 946–959. [DOI] [PubMed] [Google Scholar]
- 4.Luedde T., and Schwabe R. F.. 2011. NF-kappaB in the liver-linking injury, fibrosis and hepatocellular carcinoma. Nat. Rev. Gastroenterol. Hepatol. 8: 108–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Greve J. W., Gouma D. J., and Buurman W. A.. 1989. Bile acids inhibit endotoxin-induced release of tumor necrosis factor by monocytes: an in vitro study. Hepatology. 10: 454–458. [DOI] [PubMed] [Google Scholar]
- 6.Zhang S., Wang J., Liu Q., and Harnish D. C.. 2009. Farnesoid X receptor agonist WAY-362450 attenuates liver inflammation and fibrosis in murine model of non-alcoholic steatohepatitis. J. Hepatol. 51: 380–388. [DOI] [PubMed] [Google Scholar]
- 7.Vavassori P., Mencarelli A., Renga B., Distrutti E., and Fiorucci S.. 2009. The bile acid receptor FXR is a modulator of intestinal innate immunity. J. Immunol. 183: 6251–6261. [DOI] [PubMed] [Google Scholar]
- 8.Gadaleta R. M., van Erpecum K. J., Oldenburg B., Willemsen E. C., Renooij W., Murzilli S., Klomp L. W., Siersema P. D., Schipper M. E., Danese S., et al. 2011. Farnesoid X receptor activation inhibits inflammation and preserves the intestinal barrier in inflammatory bowel disease. Gut. 60: 463–472. [DOI] [PubMed] [Google Scholar]
- 9.McMahan R. H., Wang X. X., Cheng L. L., Krisko T., Smith M., El Kasmi K., Pruzanski M., Adorini L., Golden-Mason L., Levi M., et al. 2013. Bile acid receptor activation modulates hepatic monocyte activity and improves nonalcoholic fatty liver disease. J. Biol. Chem. 288: 11761–11770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pols T. W., Nomura M., Harach T., Lo Sasso G., Oosterveer M. H., Thomas C., Rizzo G., Gioiello A., Adorini L., Pellicciari R., et al. 2011. TGR5 activation inhibits atherosclerosis by reducing macrophage inflammation and lipid loading. Cell Metab. 14: 747–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cipriani S., Mencarelli A., Chini M. G., Distrutti E., Renga B., Bifulco G., Baldelli F., Donini A., and Fiorucci S.. 2011. The bile acid receptor GPBAR-1 (TGR5) modulates integrity of intestinal barrier and immune response to experimental colitis. PLoS One. 6: e25637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keitel V., Reinehr R., Gatsios P., Rupprecht C., Gorg B., Selbach O., Haussinger D., and Kubitz R.. 2007. The G-protein coupled bile salt receptor TGR5 is expressed in liver sinusoidal endothelial cells. Hepatology. 45: 695–704. [DOI] [PubMed] [Google Scholar]
- 13.Katsuma S., Hirasawa A., and Tsujimoto G.. 2005. Bile acids promote glucagon-like peptide-1 secretion through TGR5 in a murine enteroendocrine cell line STC-1. Biochem. Biophys. Res. Commun. 329: 386–390. [DOI] [PubMed] [Google Scholar]
- 14.Keitel V., Donner M., Winandy S., Kubitz R., and Haussinger D.. 2008. Expression and function of the bile acid receptor TGR5 in Kupffer cells. Biochem. Biophys. Res. Commun. 372: 78–84. [DOI] [PubMed] [Google Scholar]
- 15.Wang Y. D., Chen W. D., Yu D., Forman B. M., and Huang W.. 2011. The G-protein-coupled bile acid receptor, Gpbar1 (TGR5), negatively regulates hepatic inflammatory response through antagonizing nuclear factor kappa light-chain enhancer of activated B cells (NF-kappaB) in mice. Hepatology. 54: 1421–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lou G., Ma X., Fu X., Meng Z., Zhang W., Wang Y. D., Van Ness C., Yu D., Xu R., and Huang W.. 2014. GPBAR1/TGR5 mediates bile acid-induced cytokine expression in murine Kupffer cells. PLoS One. 9: e93567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mobraten K., Haugbro T., Karlstrom E., Kleiveland C. R., and Lea T.. 2015. Activation of the bile acid receptor TGR5 enhances LPS-induced inflammatory responses in a human monocytic cell line. J. Recept. Signal Transduct. Res. 35: 402–409. [DOI] [PubMed] [Google Scholar]
- 18.Tilg H., and Moschen A. R.. 2010. Evolution of inflammation in nonalcoholic fatty liver disease: the multiple parallel hits hypothesis. Hepatology. 52: 1836–1846. [DOI] [PubMed] [Google Scholar]
- 19.Farrell G. C., and Larter C. Z.. 2006. Nonalcoholic fatty liver disease: from steatosis to cirrhosis. Hepatology. 43: S99–S112. [DOI] [PubMed] [Google Scholar]
- 20.Buzzetti E., Pinzani M., and Tsochatzis E. A.. 2016. The multiple-hit pathogenesis of non-alcoholic fatty liver disease (NAFLD). Metabolism. 65: 1038–1048. [DOI] [PubMed] [Google Scholar]
- 21.Woo Baidal J. A., and Lavine J. E.. 2016. The intersection of nonalcoholic fatty liver disease and obesity. Sci. Transl. Med. 8: 323rv1. [DOI] [PubMed] [Google Scholar]
- 22.Wouters K., van Gorp P. J., Bieghs V., Gijbels M. J., Duimel H., Lutjohann D., Kerksiek A., van Kruchten R., Maeda N., Staels B., et al. 2008. Dietary cholesterol, rather than liver steatosis, leads to hepatic inflammation in hyperlipidemic mouse models of nonalcoholic steatohepatitis. Hepatology. 48: 474–486. [DOI] [PubMed] [Google Scholar]
- 23.Tomita K., Teratani T., Suzuki T., Shimizu M., Sato H., Narimatsu K., Okada Y., Kurihara C., Irie R., Yokoyama H., et al. 2014. Free cholesterol accumulation in hepatic stellate cells: mechanism of liver fibrosis aggravation in nonalcoholic steatohepatitis in mice. Hepatology. 59: 154–169. [DOI] [PubMed] [Google Scholar]
- 24.Teratani T., Tomita K., Suzuki T., Oshikawa T., Yokoyama H., Shimamura K., Tominaga S., Hiroi S., Irie R., Okada Y. , et al. 2012. A high-cholesterol diet exacerbates liver fibrosis in mice via accumulation of free cholesterol in hepatic stellate cells. Gastroenterology. 142: 152–164.e10. [DOI] [PubMed] [Google Scholar]
- 25.Ioannou G. N., Haigh W. G., Thorning D., and Savard C.. 2013. Hepatic cholesterol crystals and crown-like structures distinguish NASH from simple steatosis. J. Lipid Res. 54: 1326–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li T., Owsley E., Matozel M., Hsu P., Novak C. M., and Chiang J. Y.. 2010. Transgenic expression of cholesterol 7alpha-hydroxylase in the liver prevents high-fat diet-induced obesity and insulin resistance in mice. Hepatology. 52: 678–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ferrell J. M., Boehme S., Li F., and Chiang J. Y.. 2016. Cholesterol 7{alpha}-hydroxylase-deficient mice are protected from high fat/high cholesterol diet-induced metabolic disorders. J. Lipid Res. 57: 1144–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pathak P., Li T., and Chiang J. Y.. 2013. Retinoic acid-related orphan receptor alpha regulates diurnal rhythm and fasting induction of sterol 12alpha-hydroxylase in bile acid synthesis. J. Biol. Chem. 288: 37154–37165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li T., Francl J. M., Boehme S., and Chiang J. Y.. 2013. Regulation of cholesterol and bile acid homeostasis by the cholesterol 7alpha-hydroxylase/steroid response element-binding protein 2/microRNA-33a axis in mice. Hepatology. 58: 1111–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pandak W. M., Schwarz C., Hylemon P. B., Mallonee D., Valerie K., Heuman D. M., Fisher R. A., Redford K., and Vlahcevic Z. R.. 2001. Effects of CYP7A1 overexpression on cholesterol and bile acid homeostasis. Am. J. Physiol. Gastrointest. Liver Physiol. 281: G878–G889. [DOI] [PubMed] [Google Scholar]
- 31.Guo L., Zheng Z., Ai J., Huang B., and Li X. A.. 2014. Hepatic scavenger receptor BI protects against polymicrobial-induced sepsis through promoting LPS clearance in mice. J. Biol. Chem. 289: 14666–14673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Topchiy E., Cirstea M., Kong H. J., Boyd J. H., Wang Y., Russell J. A., and Walley K. R.. 2016. Lipopolysaccharide is cleared from the circulation by hepatocytes via the low density lipoprotein receptor. PLoS One. 11: e0155030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bramlett K. S., Yao S., and Burris T. P.. 2000. Correlation of farnesoid X receptor coactivator recruitment and cholesterol 7alpha-hydroxylase gene repression by bile acids. Mol. Genet. Metab. 71: 609–615. [DOI] [PubMed] [Google Scholar]
- 34.Gao Z., Chiao P., Zhang X., Zhang X., Lazar M. A., Seto E., Young H. A., and Ye J.. 2005. Coactivators and corepressors of NF-kappaB in IkappaB alpha gene promoter. J. Biol. Chem. 280: 21091–21098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Repa J. J., Berge K. E., Pomajzl C., Richardson J. A., Hobbs H., and Mangelsdorf D. J.. 2002. Regulation of ATP-binding cassette sterol transporters ABCG5 and ABCG8 by the liver X receptors alpha and beta. J. Biol. Chem. 277: 18793–18800. [DOI] [PubMed] [Google Scholar]
- 36.Li T., Matozel M., Boehme S., Kong B., Nilsson L. M., Guo G., Ellis E., and Chiang J. Y.. 2011. Overexpression of cholesterol 7alpha-hydroxylase promotes hepatic bile acid synthesis and secretion and maintains cholesterol homeostasis. Hepatology. 53: 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Khovidhunkit W., Moser A. H., Shigenaga J. K., Grunfeld C., and Feingold K. R.. 2003. Endotoxin down-regulates ABCG5 and ABCG8 in mouse liver and ABCA1 and ABCG1 in J774 murine macrophages: differential role of LXR. J. Lipid Res. 44: 1728–1736. [DOI] [PubMed] [Google Scholar]
- 38.Matsuzawa N., Takamura T., Kurita S., Misu H., Ota T., Ando H., Yokoyama M., Honda M., Zen Y., Nakanuma Y., et al. 2007. Lipid-induced oxidative stress causes steatohepatitis in mice fed an atherogenic diet. Hepatology. 46: 1392–1403. [DOI] [PubMed] [Google Scholar]
- 39.Marí M., Caballero F., Colell A., Morales A., Caballeria J., Fernandez A., Enrich C., Fernandez-Checa J. C., and Garcia-Ruiz C.. 2006. Mitochondrial free cholesterol loading sensitizes to TNF- and Fas-mediated steatohepatitis. Cell Metab. 4: 185–198. [DOI] [PubMed] [Google Scholar]
- 40.Kim D. H., Xiao Z., Kwon S., Sun X., Ryerson D., Tkac D., Ma P., Wu S. Y., Chiang C. M., Zhou E., et al. 2015. A dysregulated acetyl/SUMO switch of FXR promotes hepatic inflammation in obesity. EMBO J. 34: 184–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dietschy J. M., and Turley S. D.. 2002. Control of cholesterol turnover in the mouse. J. Biol. Chem. 277: 3801–3804. [DOI] [PubMed] [Google Scholar]
- 42.Thomas C., Gioiello A., Noriega L., Strehle A., Oury J., Rizzo G., Macchiarulo A., Yamamoto H., Mataki C., Pruzanski M., et al. 2009. TGR5-mediated bile acid sensing controls glucose homeostasis. Cell Metab. 10: 167–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lefebvre P., Cariou B., Lien F., Kuipers F., and Staels B.. 2009. Role of bile acids and bile acid receptors in metabolic regulation. Physiol. Rev. 89: 147–191. [DOI] [PubMed] [Google Scholar]
- 44.Haeusler R. A., Astiarraga B., Camastra S., Accili D., and Ferrannini E.. 2013. Human insulin resistance is associated with increased plasma levels of 12alpha-hydroxylated bile acids. Diabetes. 62: 4184–4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.