Abstract
Thraustochytrium, a unicellular marine protist, has been used as a commercial source of very long chain PUFAs (VLCPUFAs) such as DHA (22:6n-3). Our recent work indicates coexistence of a Δ4-desaturation-dependent pathway (aerobic) and a polyketide synthase-like PUFA synthase pathway (anaerobic) to synthesize the fatty acids in Thraustochytrium sp. 26185. Heterologous expression of the Thraustochytrium PUFA synthase along with a phosphopantetheinyl transferase in Escherichia coli showed the anaerobic pathway was highly active in the biosynthesis of VLCPUFAs. The amount of Δ4 desaturated VLCPUFAs produced reached about 18% of the total fatty acids in the transformant cells at day 6 in a time course of the induced expression. In Thraustochytrium, the expression level of the PUFA synthase gene was much higher than that of the Δ4 desaturase gene, and also highly correlated with the production of VLCPUFAs. On the other hand, Δ9 and Δ12 desaturations in the aerobic pathway were either ineffective or absent in the species, as evidenced by the genomic survey, heterologous expression of candidate genes, and in vivo feeding experiments. These results indicate that the anaerobic pathway is solely responsible for the biosynthesis for VLCPUFAs in Thraustochytrium.
Keywords: polyunsaturated fatty acid synthase, docosahexaenoic acid, docosapentaenoic acid, ω3-fatty acid, ω6-fatty acid, fatty acid biosynthesis
Very long chain PUFAs (VLCPUFAs), such as arachidonic acid (ARA, 20:4n-6) and DHA (22:6n-3), are essential components of cell membranes and precursors for biologically active signaling molecules in mammals. VLCPUFAs and their derived signaling molecules, such as eicosanoids and docosanoids, regulate the neurotransmission process in the brain, thereby affecting mood, cognition, and other neurological behaviors (1). In addition, VLCPUFAs and their derivatives can also regulate other physiological processes, such as blood circulation, metabolic pathways, and inflammatory status in mammals (2, 3). Imbalances of different types of VLCPUFAs and their derivatives in the body have been shown to implicate various pathogeneses in humans, such as neurological disorders, cardiovascular diseases, metabolic syndrome, and inflammatory conditions (4, 5). Appropriate dietary supplementation of these fatty acids is thus encouraged to provide protection against chronic diseases and improve performance of the brain, eyes, and immune system.
There are two distinct pathways in nature for the biosynthesis of VLCPUFAs (6, 7). The aerobic pathway follows an alternating desaturation and elongation process and occurs mainly in animals and eukaryotic microorganisms (8–11). The anaerobic pathway utilizes a polyketide synthase (PKS)-like PUFA synthase and takes place only in microorganisms (12–14). In the aerobic pathway, synthesis of VLCPUFAs, such as DHA in mammals, starts from α-linolenic acid (ALA, 18:3n-3) and goes through a retro-conversion process via a controlled β-oxidation step in the peroxisome for two carbon chain shortening (15, 16), while the synthesis of DHA in eukaryotic microorganisms starts from stearic acid (SA, 18:0) and ends with the final Δ4 desaturation step (17). In the anaerobic pathway, the VLCPUFA synthesis catalyzed by a PUFA synthase (12) differs from the aerobic pathway in that it does not require oxygen-dependent desaturation steps to introduce double bonds. Instead, double bonds are introduced during the process of fatty acid extension, as seen in the biosynthesis of unsaturated fatty acids in Escherichia coli (18). It is now known that de novo biosynthesis of VLCPUFAs occurs only in certain types of oceanic microorganisms, while animals and plants lack the PUFA synthase system and possess just part of the entire aerobic biosynthetic pathway, and thus are unable to completely synthesize these fatty acids. For those marine microbes that can de novo synthesize VLCPUFAs, the biosynthetic process goes through either an aerobic pathway employing desaturases and elongases to introduce double bonds and extend carbon chains of preexisting fatty acids for producing the final products or an anaerobic pathway employing a PUFA synthase to carry out all reactions required for conversion of initial acetyl-CoA to VLCPUFAs.
Thraustochytrium sp. 26185 is a unicellular marine protist that can produce more than 50% of its total fatty acids as VLCPUFAs in membrane and storage lipids. Our previous research indicates that there is an aerobic pathway in this species for the biosynthesis of VLCPUFAs and DHA can be synthesized through the final step of Δ4 desaturation (17). However, our recent genome sequencing revealed existence of an alternative pathway employing a PUFA synthase for the biosynthesis of VLCPUFAs in this species, as seen in Schizochytriun sp., a close relative of the Thraustochytrium sp. 26185 (12, 19, 20). The goal of this research was, thus, to interrogate the function and importance of the two pathways for the biosynthesis of VLCPUFAs in the species.
MATERIALS AND METHODS
Organisms and culture conditions
Thraustochytrium sp. 26185 was purchased from the American Type Culture Collection and cultured on BY+ medium (pH 6.0–6.5) consisting of 0.1% (w/v) yeast extract, 0.1% (w/v) peptone, 0.5% (w/v) D-glucose, and 4% (w/v) artificial sea salts (Sigma) at 25°C. The E. coli strain, BL21(star)DE3, was obtained from Novagen.
Cloning of putative open reading frames of the PUFA synthases from Thraustochytrium sp. 26185
By sequence analysis of the Thraustochytrium genome database, three putative genes encoding the PUFA synthase were identified (intronless genes). In order to clone the full-length genes, the genomic DNA isolated from Thraustochytrium was used as a template for PCR amplification using Q5, proofreading DNA polymerase (BioLabs). All specific primers were designed according to the sequence information obtained from the Thraustochytrium genome sequence, as listed in supplemental Table S1. The first open reading frame (ORF) of 8,439 bp encodes the PUFA synthase subunit-A, the second ORF of 6,150 bp codes for subunit-B, and the third ORF of 4,494 bp encodes subunit-C. All ORF sequences had both start and stop codons. To facilitate cloning and capacity of fidelity for DNA polymerase to synthesize a large DNA fragment, ORF-A was divided into three pieces according to internal restriction sites, PstI and AscI. Part I contained 3,254 bp including a start site with EcoRI restriction site and extended to a unique PstI site; part II contained 3,194 bp including PstI and AscI restriction sites; and part III contained 1,991 bp including AscI and a stop codon with HindIII restriction site. The ORF-B was divided into two pieces by restriction enzyme, BglII, first fragment (3,437 bp) contained a start site with EcoRI restriction site and extended to a unique BglII, and second fragment (2,713 bp) started from BglII until stop codon with HindIII restriction site. The ORF-C was directly amplified with primers from start until stop codons with EcoRI and HindIII, respectively, at both ends. All PCR fragments were cloned by TA cloning into pGEMT (Promega). All plasmids were sequenced to verify the amplification fidelity.
In order to get the full-length of ORF-A, part I, part II, and part III were combined by restriction cloning procedure. First, the PstI fragment was digested from part II-plasmid and cloned into part I-plasmid digested with PstI, producing plasmid-ORF-A-I+II. Second, the AscI and HindIII fragment was digested from part III-plasmid and cloned into plasmid-ORF-A-I+II digested with AscI and HindIII, producing plasmid-ORF-A-FL.
The same method was applied to clone the full-length of ORF-B where part I and II were ligated at BglII restriction site, producing plasmid-ORF-B-FL.
Cloning of phosphopantetheinyl transferase genes into pCOLADuet-1
Previous studies showed that E. coli endogenous phosphopantetheinyl transferase (PPTase) enzymes were unable to activate the apo-acyl carrier protein (ACP) into holo-ACP domains of PUFA synthases (19). A PPTase gene (HetI) from Nostoc sp. PCC7120 was chemically synthesized by Integrated DNA Technologies (USA) and subcloned into pGEM-T vector. The confirmed plasmids were digested by NdeI and KpnI and cloned into the second cloning site of ORF-A/pCOLADuet-1 digested with the same restriction enzymes.
Cloning of PPTase genes from Thraustochytrium sp. 26185 and Schizochytrium limacinum ATCC MYA-1381
To identify genes encoding PPTase from Thraustochytrium and S. limacinum, HetI protein sequence from Nostoc PCC 7120 was used as a query to search Thraustochytrium sp. 26185 and S. limacinum genome databases. Homologous genes were identified from the two species. The full-length PPTase gene from Thraustochytrium was amplified by PCR using genomic DNA as template. The ORF was 867 bp encoding 288 amino acids with a molecular mass of 31.6 kDa. The full-length PPTase gene from Schizochytrium was 816 bp encoding a polypeptide of 271 amino acids and was chemically synthesized at Integrated DNA Technologies.
Relative expression of TcPUFA synthase and TcD4 by quantitative real-time PCR
A single colony of Thraustochytrium cells was grown in 10 ml of BY+ medium at 25°C overnight with 240 rpm. The cells were then diluted with the same medium and condition starting OD600 at 0.1. After two double times (8 h), cells were quickly harvested by centrifugation at 5,000 rpm and divided into two parts. The first part was used for total fatty acid analysis and the second part for the total RNA using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. Genomic DNA contamination was eliminated by on-column DNase I digestion with RNase-Free DNase (Qiagen). The first-strand cDNA was synthesized from 1 μg of the total RNA by qScript cDNA Supermix (Quanta Biosciences). Three genes, actin, GAPDH, and HSP90, were employed as internal references. Primers were designed using Primer3Plus online software (http://www.primer3plus.com/cgi-bin/dev/primer3plus.cgi). The real-time quantitative PCR was performed using SYBRgreen SuperMix (Quanta Biosciences) according to manufacturer’s recommendation. Reactions were carried out on a Bio-Rad CFX real-time PCR system (Bio-Rad, Mississauga, Ontario, Canada) with the following cycling reaction: 50°C for 2 min, 95°C for 2 min, 95°C for 15 s, 65°C for 30 s. Three biological replicates with three technical replicates were performed with each sample. Data obtained from the iCycler software (Bio-Rad) was used to analyze the expression of TcPUFAs and TcD4 compared with three internal standards. Their relative expression ratios were calculated using the comparative Ct method.
Functional expression of the PUFA synthase in E. coli
E. coli transformant cells containing pORF-C/ORF-B/ORF-A and either pHetI or pScPPTase or pTcPPTase were grown in LB or 765 medium supplemented with 10% glycerol, kanamycin (50 μg/ml), and spectinomycin (50 μg/ml) at 37°C. The overnight culture was inoculated to 50 vol of new medium containing the same supplementation. The bacteria were grown at 37°C to OD600 = 0.5–1.0. Induction was achieved by addition of 0.5 mM isopropyl-β-D-thiogalactopyranoside. The cells continued to grow at 20°C with 220 rpm and then were collected for lipid and total fatty acid analysis at the time course to check the expression level by centrifugation (3,000 rpm, 15 min).
Fatty acid analysis
Total fatty acids in E. coli were converted to fatty acid methyl esters (FAMEs) by adding 3 N methanolic HCl (Sigma-Aldrich) at 80°C for 2 h. After the transmethylation process, the sample was cooled down at room temperature before adding 1 ml of 0.9% NaCl and 2 ml of hexane. The sample was then mixed by vortex and centrifuged at 2,400 rpm for 5 min for phase separation. The hexane phase containing FAMEs was removed and dried under N2. After drying, the sample was resuspended in hexane and used for GC analysis. Two microliter samples of total FAME derivatives were analyzed on an Agilent 6890N gas chromatograph equipped with a DB-23 column (30 m × 0.25 mm) with 0.25 μm film thickness (J&W Scientific). The column temperature was maintained at 160°C for 1 min, and then raised to 240°C at a rate of 4°C/min. For mass spectrometry analysis, the mass selective detector was run under standard electron impact conditions (70 eV), scanning an effective m/z range of 40–700 at 2.26 scans/s.
RESULTS
Identification of genes involved in the biosynthesis of VLCPUFAs in the Thraustochytrium genome
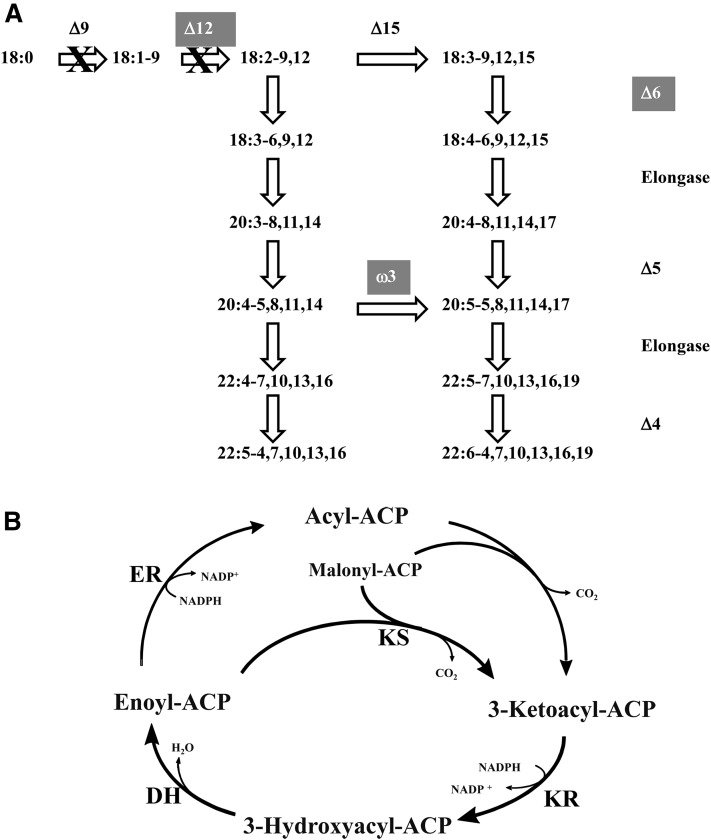
To further study the biosynthesis and assembly of VLCPUFAs in Thraustochytrium sp. 26185, we recently sequenced the whole genome of this species. From the genome sequence, we identified nearly all members in the aerobic pathway for the biosynthesis of DHA, except for acyl-CoA Δ9 desaturase, including five putative front-end desaturases (three Δ6 desaturases, one Δ5 desaturase, and one Δ4 desaturase) and three putative methyl-end desaturases for oxygenic desaturation, as well as at least three ELO type condensing enzymes for fatty acid elongations. In addition, a large type I FAS representing a fusion of fungus FASα and FASβ subunits for the synthesis of SA (18:0) and three large subunits of a type I PKS-like PUFA synthase, highly homologous to those of Schizochytrium for the biosynthesis of VLCPUFAs (19), were also identified. These data indicate that both aerobic and anaerobic pathways might coexist in the species (Fig. 1A, B).
Fig. 1.
Two pathways for the biosynthesis of VLCPUFAs in Thraustochytrium sp. 26185. A: Aerobic pathway for biosynthesis of VLCPUFAs. The biosynthesis of VLCPUFAs in the aerobic pathway starts with 18:0 and goes through alternating desaturation and elongation steps. Three highlighted desaturases (Δ12, Δ6, and ω3) were new enzymes identified and functionally characterized by this study. The rest of the enzymes in the pathway were cloned and characterized previously from the species. No Δ9 desaturase sequence was detected in the Thraustochytrium genome and feed experiments did not observe the activity in the species. One Δ12 desaturase sequence was detected in the genome, however little activity was observed when it was expressed in yeast. In addition, feed experiments did not observe any Δ12 desaturation activity in vivo. B: Anaerobic pathway for biosynthesis of VLCPUFAs. The biosynthesis of VLCPUFAs in the anaerobic pathway starts with acetate and proceeds with reiterative cycles of four reactions: condensation by KS, ketoreduction (KR), dehydration (DH), and enoylreduction (ER). However, the synthesis may omit the certain last step of a full cycle, causing a double bond to be retained in the acyl chain.
Incomplete aerobic pathway for the biosynthesis of VLCPUFAs in Thraustochytrium
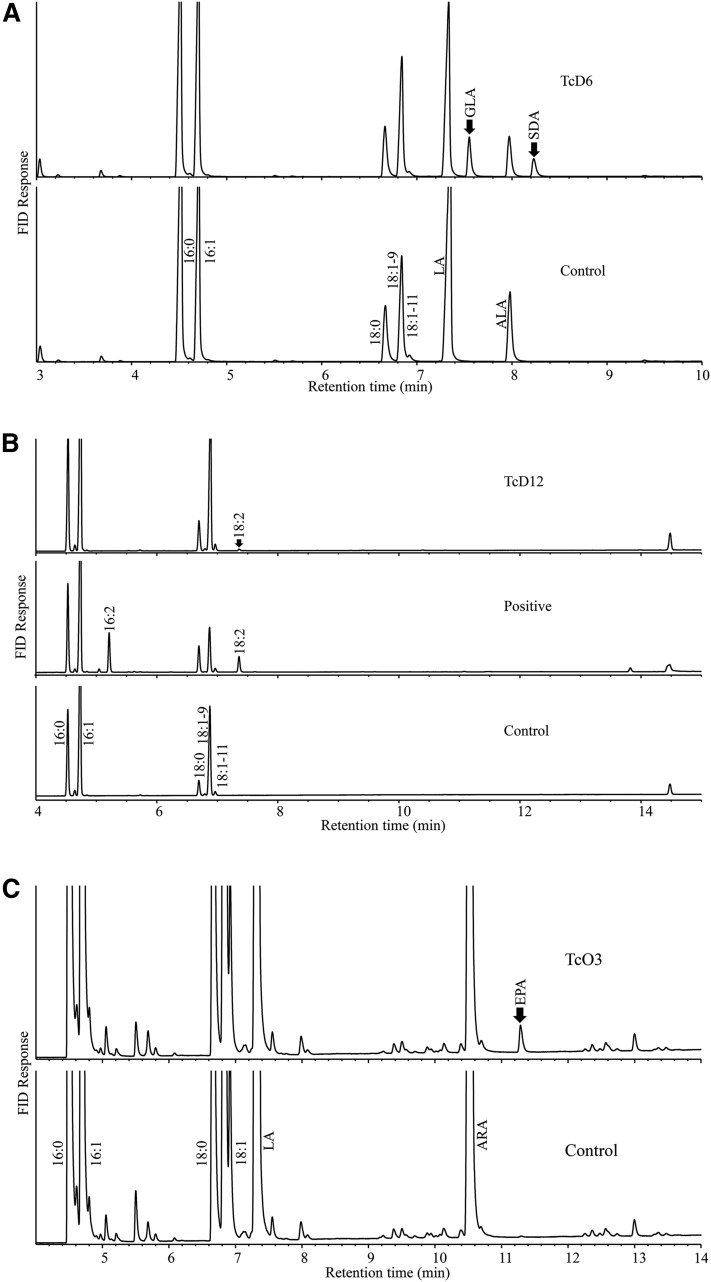
To interrogate whether both pathways were functional for the VLCPUFA biosynthesis, we attempted to clone all putative genes in the two pathways that were not biochemically characterized previously, and then expressed them in yeast or E. coli for the functional analysis. In the aerobic pathway, several genes encoding front-end desaturases and elongases had been previously cloned and functionally characterized from the species. For instance, one Δ5 desaturase and one Δ4 desaturase could introduce a double bond into eicosatetraenoic acid (20:4n-3) and docosapentaenoic acid (DPA, 22:5n-3), giving EPA (20:5n-3) and DHA, respectively (17). One functional elongase capable of elongating both 18C and 20C PUFAs was characterized and used for stepwise engineering of DHA in an oilseed crop (21). In addition to these genes, however, three putative front-end and three putative methyl-end desaturase genes in the aerobic pathway identified from the genome were previously uncharacterized. To functionally analyze these genes, pairs of specific primers targeting outside the individual ORFs were designed and used for reverse transcriptase-PCR amplifications with the total RNA as the template. The amplified coding regions of these putative desaturase genes were individually cloned into a yeast vector and expressed in yeast. The results showed that one of three putative Δ6 desaturase genes indeed encoded an effective front-end desaturase introducing a Δ6 double bond into linoleic acid (LA, 18:2-9,12) and ALA (18:3-9,12,15), giving γ-linolenic acid (GLA, 18:3-6,9,12) and stearidonic acid (18:4-6,9,12,15), respectively (Fig. 2A). The rest did not possess any fatty acid desaturation activity. One of three putative methyl-end desaturase genes, when expressed in yeast, showed little Δ12 desaturase activity on oleic acid (18:1-9) to give product LA (18:2-9,12) at the level of less than 0.1% of the total fatty acids, as compared with the positive control (the yeast transformant with a Δ12 desaturase from fungus Claviceps purpurea) where a substantial amount of two Δ12 desaturated products (16:2-9,12 and 18:2-9,12) on two substrates (16:1-9 and 18:1-9) was observed (22) (Fig. 2B). Another putative methyl-end desaturase gene conferred ω3 desaturation activity converting 20C ω6 fatty acids, such as ARA (20:4-5,8,11,14) and dihomo-GLA (20:3-8,11,14), into their corresponding ω3 fatty acids, such as EPA and eicosatetraenoic acid (20:4-8,11,14,17) (Fig. 2C, supplemental Fig. S1). However, the third one did not show any desaturase activity when expressed in yeast. Collectively, these data indicate that all catalytic steps, except for two ineffective ones (Δ9 and Δ12) from SA (18:0) to LA, in the aerobic pathway might be functional in this species.
Fig. 2.
Functional analysis of three new Thraustochytium desaturases identified in the aerobic pathway in yeast. A: GC analysis of FAMEs prepared from yeast transformant with Thraustochytium Δ6 desaturase (TcD6) fed with LA and ALA. B: GC analysis of FAMEs prepared from yeast transformant with Thraustochytium Δ12 desaturase (TcD12). The positive control (Positive) was Δ12 desaturase from Claviceps purpurea. The negative control (Control) is yeast with an empty vector. C: GC analysis of FAMEs prepared from yeast transformant with Thraustochytium ω3 desaturase (TcO3) fed with LA and ARA.
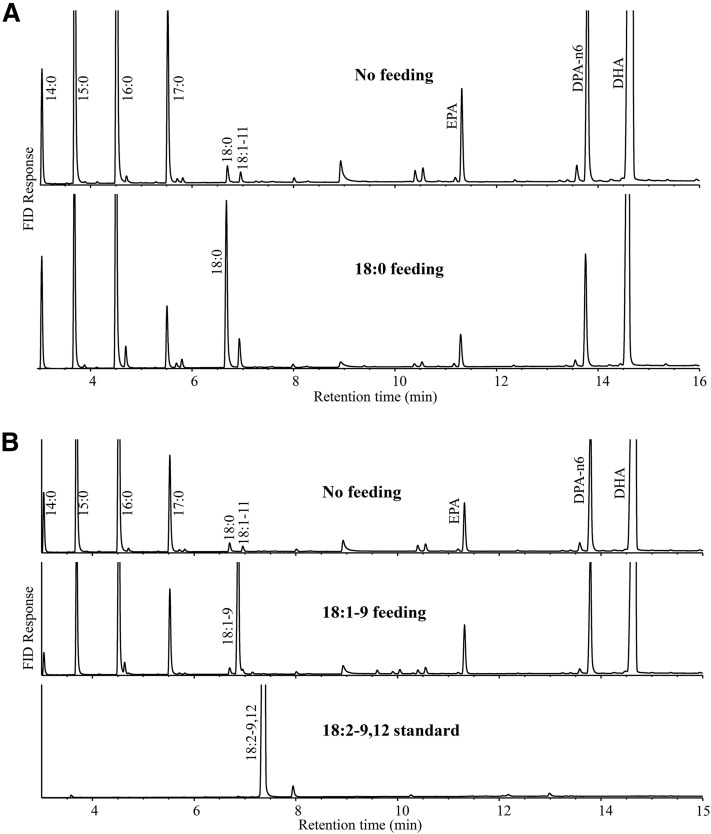
To confirm that the aerobic pathway might be incomplete by missing the two critical desaturation steps in the species, we fed Thraustochytrium with several key fatty acid intermediates in the aerobic pathway to test the desaturation and elongation activities in vivo. The results showed that Thraustochytrium could effectively elongate both an 18C Δ6 desaturated fatty acid, such as GLA (18:3-6,9,12), and a 20C Δ5 desaturated fatty acid, such as ARA (20:4-5,8,11,14), with particularly highly efficient elongation on 18C PUFAs. The elongation efficiency from 18:3-6,9,12 to 20:3-8,11,14 reached more than 50% (Table 1). The front-end and ω3 desaturation activities were also observed in the species on the fed substrates. However, no Δ9 desaturation on fed 18:0 and no Δ12 desaturation on fed 18:1-9 were observed. In Thraustochytrium, a very small amount of 18:0 (∼1% of the total fatty acids) was produced. When fed with a large amount of 18:0, it still did not produce any 18:1-9 from the fed substrate (Fig. 3A). Thraustochytrium did not produce any 18:1-9. When fed with this fatty acid, it did not produce any detectable 18:2-9,12 from the fed substrate (Fig. 3B, Table 1). These data unequivocally confirmed that the aerobic pathway was incomplete in Thraustochytium sp. 26185 where two critical desaturation activities (Δ9 and Δ12) were either ineffective or missing from the pathway.
TABLE 1.
In vivo desaturation and elongation activities on fatty acids fed to Thraustochytrium sp. 26185
| Fatty Acid Substrate | Desaturated or Elongated Product | Conversion Efficiency |
| 18:0 | 18:1-9 | 0.0 ± 0.00 |
| 18:1-9 | 18:2-9,12 (Δ12 desaturation) | 0.0 ± 0.00 |
| 18:2-9,12 | 18:3-9,12,15 (Δ15 desaturation) | 2.7 ± 0.11 |
| 18:3-6,9,12 (Δ6 desaturation) | 5.7 ± 0.10 | |
| 18:3-6,9,12 | 20:3-8,11,14 (Δ6 elongation) | 56.8 ± 2.49 |
| 18:4-6,9,12,15 (Δ15 desaturation) | 0.0 ± 0.00 | |
| 20:3-8,11,14 | 20:4-5,8,11,14 (Δ5 desaturation) | 16.4 ± 0.22 |
| 20:4-8,11,14,17 (Δ17 desaturation) | 29.6 ± 2.95 | |
| 20:4-5,8,11,14 | 22:4-7,10,13,16 (Δ5 elongation) | 34.2 ± 2.29 |
| 20:5-5,8,11,14,17 (Δ17 desaturation) | 27.0 ± 3.79 |
Conversion efficiency was calculated by [products/(substrate plus products)] × 100. Values are mean ± SD of four replicates.
Fig. 3.
Fatty acid analysis of Thraustochytrium fed with different fatty acids. A: GC analysis of Thraustochytrium fed with 18:0. B: GC analysis of Thraustochytrium fed with 18:1-9.
Functional analysis of the PUFA synthase in E. coli
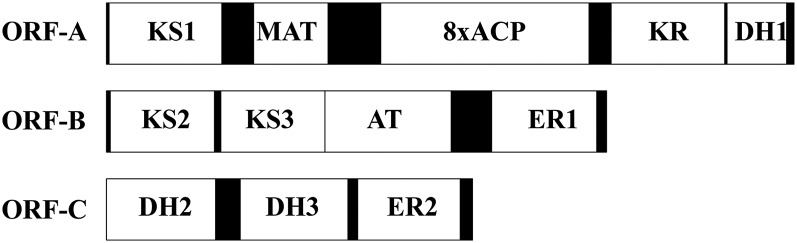
Next, we looked into the anaerobic pathway for the VLCPUFA biosynthesis in Thraustochytium. From the genome sequence, three large ORFs (∼8.5 kb, 6.2 kb, and 4.5 kb) encoding three subunits (ORF-A, 2,813 amino acids; ORF-B, 2,049 amino acids; and ORF-C, 1,497 amino acids) of the mega-enzyme that was highly homologous to a PUFA synthase of Schizochytrium (12, 19) were identified. Most catalytic domains in the three subunits were predicted based on the presence of characteristic active sites (23). Subunit I comprised one β-ketoacyl-ACP synthase (KS1) domain, one malonyl-CoA:ACP acyltransferase domain, eight ACP domains, one ketoacyl reductase domain, and one dehydratase (DH1) domain. Subunit II comprised two KS domains (KS2 and KS3), one acyltransferase domain, and one enoyl-ACP reductase (ER1) domain. Subunit III consisted of two DH domains (DH2 and DH3) and one ER domain (ER2) (Fig. 4). Furthermore, a putative gene encoding PPTase required for attaching a phosphopantetheine prosthetic group to the ACP domains (19) was also identified from the genome.
Fig. 4.
Diagrammatic representation of the structure of the PUFA synthase of Thraustochytrium sp. 26185. It is comprised of three OFRs with ORF-A of 8,439 bp for 2,812 amino acids, ORF-B of 6,150 bp for 2,049 amino acids, and ORF-C of 4494 bp for 1,497 amino acids.
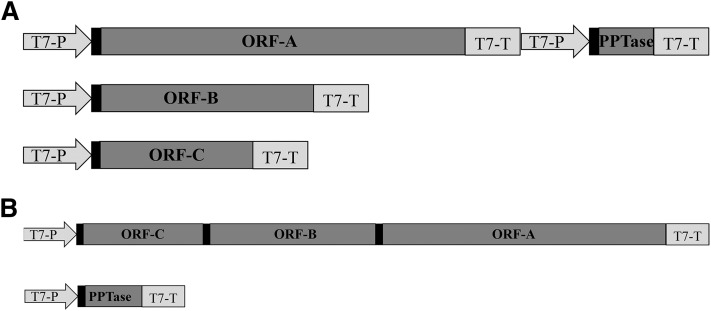
To functionally characterize the putative PUFA synthase, the three ORFs were first individually cloned into three E. coli vectors, each behind a T7 promoter (pCOLADuet-1 for ORF-A, pCDFDuet-1 for ORF-B, and pETDuet-1 for ORF-C). In addition, a PPTase (HetI) from Nostoc PCC7120 (24) was also included in pCOLADuet-1 beside ORF-A for the expression analysis (Fig. 5A). As the ORFs were large, cloning them into the expression vectors was challenging. To facilitate the cloning process, ORF-A was divided into three pieces, and ORF-B was divided into two pieces according to the internal restriction sites for sequential cloning. The final constructs were built by assembling these pieces together according to their order using a restriction-cloning procedure.
Fig. 5.
Diagrammatic representation of the PUFA synthase constructs for E. coli expression. Black squares represent ribosome binding sites, T7-P represents T7 promoter, and T7-T represents T7 terminator. A: Three different plasmids were used to express three ORFs of the PUFA synthase ORFs in E. coli. B: One plasmid was used to express three ORFs of the PUFA synthase as one operon in E. coli.
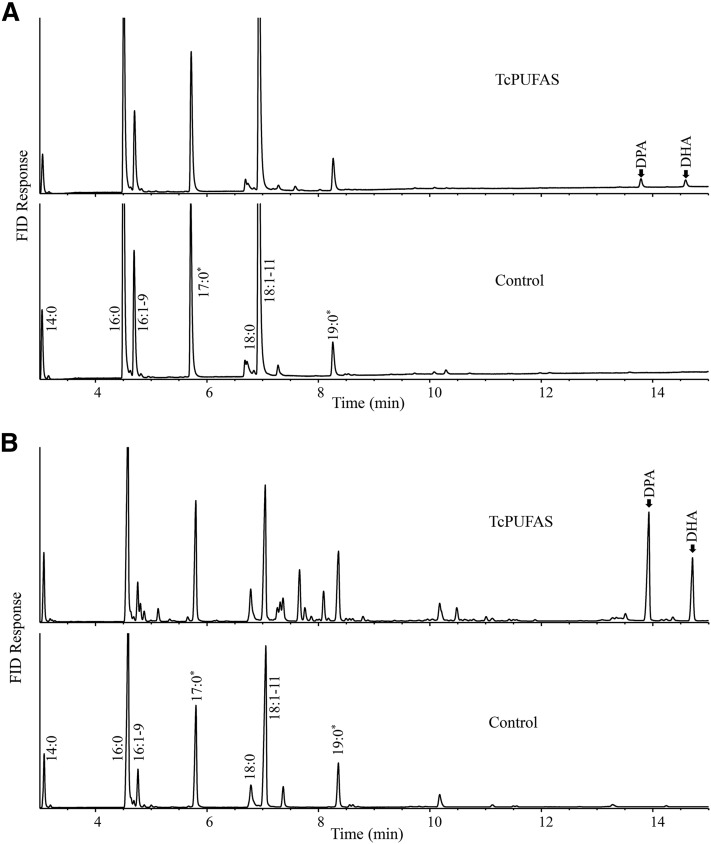
To reconstitute the anaerobic pathway heterologously, all three recombinant plasmids were simultaneously transformed into the E. coli strain BL21(star)DE3. The functional analysis showed that the transformant produced two new fatty acids, DHA and DPA (22:5n-6), compared with the strain transformed with empty vectors. Although the amount of the two Δ4 desaturated VLCPUFAs produced was at only about 1–2% of the total fatty acids in the transformant (Fig. 6A), the result confirmed that the PUFA synthase from Thraustochytrium was functional for the biosynthesis of VLCPUFAs.
Fig. 6.
GC analysis of E. coli transformants expressing the PUFA synthase genes. TcPUFAS, the transformant expressing the Thraustochytrium PUFA synthase; Control, the transformant with vector control. *Cyclopropane fatty acid. A: Expressing three ORFs of the PUFA synthase in three different plasmids in E. coli. B: Expressing three ORFs of the PUFA synthase as one operon in a single plasmid in E. coli.
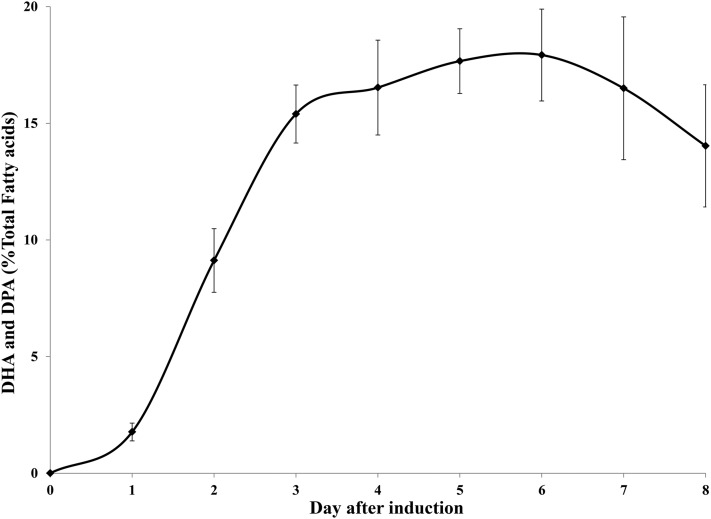
To improve production of VLCPUFAs in E. coli, we further attempted to coordinate expressions of three ORFs of the PUFA synthase in an operon by constructing one recombinant plasmid comprising the three ORFs behind a single promoter with a ribosome binding site motif upfront of each ORF. The total length of the operon reached ∼20 kb, making construction of the expression plasmid highly challenging. The whole operon was divided into more than 10 pieces and a sequential cloning strategy was used to assemble all these pieces together into a single recombinant plasmid. Functional analysis of this recombinant plasmid showed that the transformant with the operon indeed improved the production of DHA and DPA substantially (Fig. 6B). Surprisingly however, DPA was higher than the DHA produced in the E. coli transformant, which was a striking contrast to that in the native Thraustochytrium. Furthermore, in a time course of the induced expression, the amount of DHA and DPA in the transformant was almost linearly increased from day 1 to day 3 and reached the peak at day 6 at about 18% of the total fatty acids. After that, the amount of VLCPUFAs was gradually reduced in the reconstituted strain (Fig. 7).
Fig. 7.
Production of two VLCPUFAs, DPA and DHA, in the E. coli transformant expressing the PUFA synthase along with a PPTase from Nostoc PCC7120 (HetI) during a time course of the induction. Values are the mean of three replicates with SD.
Identification and functional analysis of PPTase from Thraustochytrium
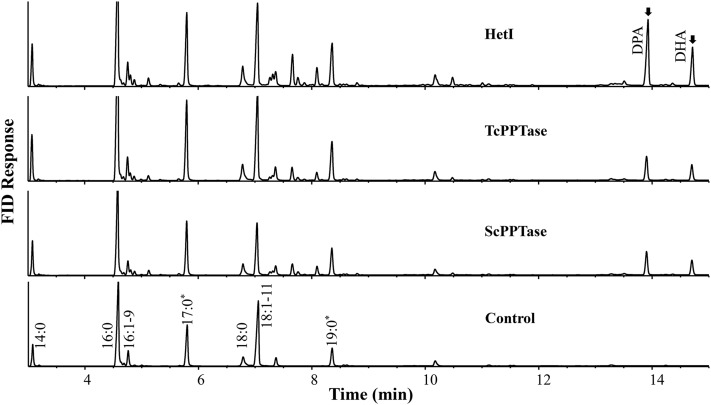
PPTase was essential for attaching phosphopantetheine to ACP domains of a PUFA synthase converting inactive apo-synthase to active holo-synthase. This function could not be fulfilled by E. coli PPTase, an AcpS-type PPTase (19). Based on the structure, PPTases were classified into three groups, AcpS-type, Sfp-type, and type I FAS PPTase. Among the three, the second group had broad substrates, including PKS, some type II synthase, PUFA synthase, and nonribosomal polypeptide synthase. PPTases of this group from prokaryotes for the PUFA synthase were previously characterized. However, no such PPTase has ever been functionally analyzed from eukaryotes. To identify the PPTase, a previously characterized PPTase from bacteria (HetI) for a PKS was used as a query to search genome databases of Thraustochytrium sp. 26185 and Schizochytrium sp. ATCC MYA-1381, a closely related species of Thraustochytrium. One homolog was identified from each species (TcPPTase from the Thraustochytrium and ScPPTase from the Schizochytrium). Sequence analysis indicated that they encoded polypeptides of 288 and 271 amino acids, respectively, with 52% amino acid sequence identity with each other. However, both proteins displayed low sequence similarity to PPTases for PUFA synthases from bacterial Moritella and Schewanella (in a range of 17–22% amino acid sequence identity). As the Sfp-type PPTases, TcPPTase and ScPPTase, also comprised two conserved motifs [(V/I/L)G(V/I/L/T)D(V/I/L/A)] and [(F/W)(A/S/C/T)XKE(A/S)Z(Z/S)K(A/G)] (X is any amino acid, Z is a hydrophobic amino acid). However, unlike bacterial Sfp-type PPTases where two motifs were separated by 38–41 amino acids, the two PPTases possessed more than 50 amino acids between the two conserved motifs (supplemental Fig. S2). To functionally characterize TcPPTase and ScPPTase, each gene was then expressed along with Thraustochytrium PUFA synthase in E. coli. The result showed that the native TcPPTase did not produce any VLCPUFAs, and the native ScPPTase also produced only a small amount of DPA and DHA in the E. coli transformants. This result showed that these two PPTases either had no activity or had very weak activity in E. coli. When we looked at the sequences in detail, it was found that both TcPPTase and ScPPTase possessed high GC content, especially TcPPTase where the entire coding region had 70% GC content with the first 100 bp from the start codon at 91%. To address the potential issue, both TcPPTase and ScPPTase were then codon-optimized for the E. coli expression. As expected, the codon optimized versions of TcPPTase and ScPPTase improved the activity substantially in E. coli for the production of the two VLCPUFAs. However, despite the improvement, the two eukaryotic PPTases were still less active as compared with the prokaryotic PPTase for producing VLCPUFAs in E. coli. The amounts of the two VLCPUFAs produced with a PPTase from bacterial Nostoc PCC7120 (HetI) were 42.9 and 67.1% higher than those with PPTases from Thraustochytrium and Schizochytrium, respectively (Fig. 8).
Fig. 8.
GC analysis of FAMEs prepared from E. coli transformants containing Thraustochytrium PUFA synthase genes along with empty vector (Control), PPTase from Nostoc PCC7210 (HetI), PPTase from Schizochytrium (ScPPTase), and Thraustochytrium (TcPPTase). *Cyclopropane fatty acid.
Comparison of gene expressions in the aerobic and anaerobic pathways
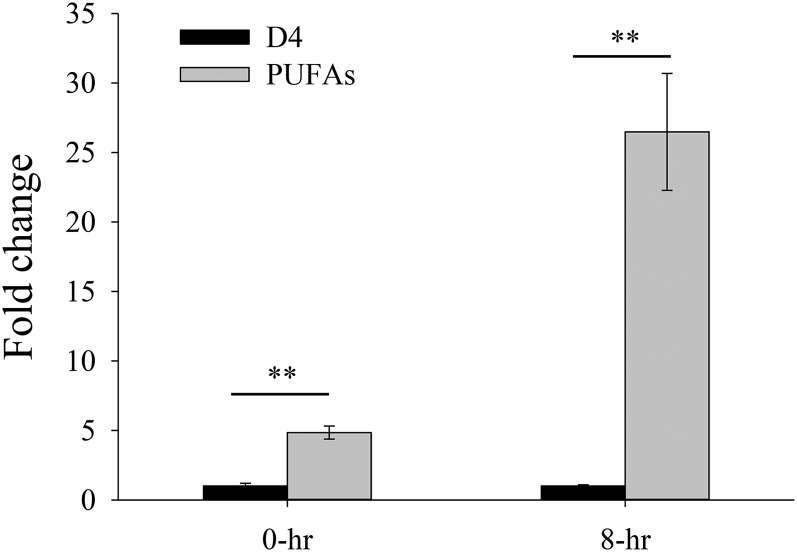
To investigate the relationship between the expression level of the genes in the two pathways and production of VLCPUFAs in the Thraustochytrium, TcD4 encoding Δ4 desaturase catalyzing the final step of DHA biosynthesis in the aerobic pathway and the 3′-end of ORF-C of the PUFA synthase in the anaerobic pathway were selected for quantitative real-time PCR analysis. The result showed that production of DHA was highly correlated with the expression level of the PUFA synthase gene, but not with that of the Δ4 desaturase gene in the cell. At the starting time point (0 h), the amount of DHA produced in Thraustochytrium accounted for about 33% of total fatty acids, and the amount of saturated fatty acids was about 55%. However, at the 8 h time point of culturing (at the mid-log phase), the amount of DHA reached up to 53% of the total fatty acids in the cell and the amount of saturated fatty acids dropped to 35%. In other words, the amount of DHA produced at 8 h was increased by about 60% from that at 0 h (Table 2). Concurrently, the expression level of the PUFA synthase gene at 0 h was about five times that of the Δ4 desaturase gene in the cell, while at 8 h the expression level of the PUFA synthase gene was about 26 times that of the Δ4 desaturase gene in the cell (Fig. 9). Alternatively, when the expressed level of the same gene at both time points was considered, the expression level of the Δ4 desaturase gene at 8 h was only half of that at 0 h, while the expression level of the PUFA synthase gene at 8 h was two and one-half times more than that at 0 h. These results provided further evidence that the PUFA synthase pathway was responsible for the VLCPUFA biosynthesis in Thraustochytrium sp. 26185.
TABLE 2.
Fatty acids composition of Thraustochytrim sp. 26185 cultured at the 0 h and 8 h
| Time | 14:0 | 15:0 | 16:0 | 17:0 | 18:0 | 18:1-11 | ARA | EPA | DPA-n6 | DHA |
| 0 h | 7.5 ± 2.34 | 18.1 ± 2.76 | 24.3 ± 3.92 | 4.2 ± 1.13 | 0.6 ± 0.13 | 1.0 ± 0.17 | 0.3 ± 0.04 | 1.7 ± 0.29 | 8.0 ± 0.48 | 33.1 ± 2.44 |
| 8 h | 3.4 ± 1.22 | 9.9 ± 0.78 | 18.2 ± 1.45 | 3.1 ± 0.23 | 0.4 ± 0.05 | 1.0 ± 0.28 | 0.3 ± 0.01 | 2.2 ± 0.28 | 7.7 ± 0.31 | 53.3 ± 3.52 |
Numbers were calculated from the peak area of FAMEs. Values are the mean of three replicates with SD.
Fig. 9.
Comparison of the expressions of two genes in aerobic (TcD4 for Δ4 desaturase) and anaerobic (TcPUFAs for PUFA synthase) pathways in Thraustochytrium. Values are the mean ± SD of six biological replicates. **Statistically significant at P < 0.01.
DISCUSSION
Thraustochytrium sp. 26185 produces a high level of VLCPUFAs, mainly DHA and DPA, in the membrane and storage lipids. The first Δ4 desaturase in the biosynthesis of VLCPUFAs is identified from the species, implying that DHA can be synthesized by the aerobic pathway (17). However, recent studies on microbial VLCPUFA producers show that an anaerobic pathway employing a PUFA synthase can also be used to synthesize VLCPUFAs (12, 19, 20, 25). These results prompted us to investigate whether a similar pathway also exists in Thraustochytrium, and if so, which pathway is more important for the biosynthesis of VLCPUFAs in the species. To answer these questions, we first carried out a genomic survey of genes involved in the biosynthesis of VLCPUFAs in the species. From the genome sequence, nearly all members in both pathways, except for the Δ9 desaturase gene in the aerobic pathway, in the biosynthesis of VLCPUFAs were identified. To interrogate which pathway was functional and important in vivo, we first attempted to generate mutants deficient in a gene in the two pathways through homologous recombination by introducing disruption constructs into the species. However, due to unknown reasons, we were unable to generate any knockout mutants in both pathways by using this approach in the recalcitrant species. Afterwards, we turned to the use of in vivo feeding experiments and functional analysis of all the important genes in both pathways in heterologous systems. All of the results point to the notion that the anaerobic pathway is responsible for the VLCPUFA biosynthesis in Thraustochytrium. First, the genomic survey indicated that the Δ9 desaturase gene was not found in the genome. Heterologous expression of the Δ12 desaturase candidate in yeast showed little Δ12 desaturase activity. This result indicates that two critical steps in the aerobic pathway are ineffective for the biosynthesis of VLCPUFAs. Second, the fatty acid feeding experiments confirmed that no Δ9 and Δ12 desaturation was observed on the culture supplemented with 18:0 and 18:1-9, respectively. Third, heterologous expression of three ORFs of the PUFA synthase along with its PPTase from Thraustochytrium indicated that the anaerobic pathway was highly active for the biosynthesis of VLCPUFAs in E. coli. Fourth, quantitative RT-PCR analysis of the genes in the two pathways showed that the expression level of the PUFA synthase genes was much higher than that of the Δ4 desaturase gene, particularly in the cells at the mid-log phase (at the 8 h culturing) where the expression level of the PUFA synthase gene was about 26 times more than that of the Δ4 desaturase gene. Moreover, the expression pattern was highly correlated with the amount of DHA produced in the cells at the two culturing time points. Therefore, although both aerobic and anaerobic pathways for DHA biosynthesis coexist in Thraustochytrium, the capacity in the aerobic pathway is void due to two ineffective catalytic steps and the PUFA synthase is solely responsible for the biosynthesis of VLCPUFAs in this species. The phenomenon that both aerobic and anaerobic pathways coexist in a species has been observed in other protists. However, the biosynthetic machinery of VLCPUFAs in those species is very different. In Thraustochytrium aureum, another species of the same genus of Thraustochytrium, both pathways are functionally involved in the biosynthesis of VLCPUFAs (26). In Schizochytrium sp., a member of the same family Thraustochytriaceae, the aerobic pathway is not complete, however, with missing different components (27). In Thraustochytrium sp. 26185, both Δ9 desaturation and Δ12 desaturation activities are ineffective, leading to inability to synthesize both 18:1-9 and 18:2-9,12 in the aerobic pathway in the species. These results suggest that the progenitor aerobic pathway might become relic in the species as the newly acquired anaerobic pathway is more efficient. As such, during evolution, different components in the aerobic pathway could be lost or retained in different species.
This study has reconstituted the anaerobic pathway in E. coli with a PUFA synthase from Thraustochytrium along with a PPTase, which results in the production of a high level of two VLCPUFAs, DHA and DPA, in the reconstituted strain (∼18% of the total fatty acids). However, surprisingly, unlike the native species where the ratio of DHA and DPA is about 4:1 (Table 2), the reconstituted strain produces a higher level of DPA (22:5n-6) than DHA (22:6n-3). The reason for this is currently unknown. E. coli utilizes a type II FAS with discrete enzymes for fatty acid synthesis. It is possible that some of these enzymes might be able to interact with the Thraustochytrium PUFA synthase to slightly modify the catalytic activity, resulting in the difference in retaining double bonds during the VLCPUFA synthesis. PPTase is essential for attaching a phosphopantetheine group to the ACP domains of a PUFA synthase converting inactive apo-PUFA synthase to active holo-PUFA synthase. Although PPTases for a PUFA synthase from prokaryotics were cloned and biochemically characterized, no such enzyme has been identified from eukaryotes. This study identifies two new Sfp-type PPTases from Thraustochytrium and Schizochytrium for activating the apo-PUFA synthase by attaching phosphopantetheine to the ACP domains, as this function could not be fulfilled by the AcpS-type PPTase of E. coli. However, coexpression of the Thraustochytrium PPTase along with the Thraustochytrium PUFA synthase, even after codon optimization, produces VLCPUFAs at a level that is incomparable to that of the prokaryotic PPTase along with the PUFA synthase in E. coli. The reason for this remains elusive. It might be relevant to the system used for the expression of these genes. Although TcPPTase derives from the same species as the PUFA synthase, they are eukaryotic proteins. The prokaryotic PPTase HetI from Nostoc PCC7120 might have advantage to express in E. coli, resulting in being more effective in coordinating the expression with a eukaryotic PUFA synthase.
PUFA synthase has recently been found in a number of microorganisms, including bacteria, algae, and fungi, for the biosynthesis of VLCPUFAs (12–14, 28, 29). The advantage of the system to synthesize VLCPUFAs over the desaturation/elongation pathway is the relatively pure end products, which makes production of VLCPUFAs by the system very attractive (16). Similar to type I PKS and type I FAS, the PUFA synthase can comprise multiple subunits, each with several catalytic domains. The fatty acid synthesis uses malonate as a substrate and ACP as the covalent attachment for the chain extension, proceeding with reiterative cycles. A full biosynthetic cycle also comprises four chemical reactions: condensation of an acyl-ACP with a malonyl-ACP to produce a ketoacyl-ACP, ketoreduction to convert ketoacyl-ACP to hydroxyacyl-ACP, dehydration to remove a water molecule from hydroxyacyl-ACP resulting in an unsaturated enoyl-ACP, and reduction of enoyl-ACP to a saturated acyl chain. However, unlike type I fatty acid synthesis, the synthesis of VLCPUFAs by a PUFA synthase may omit the last step of a full cycle, causing a double bond to be retained in the acyl chain (1, 7). However, nothing is known about how a PUFA synthase positions multiple cis-double bonds in VLCPUFAs. Further molecular analysis of the Thraustochytrium PUFA synthase would not only contribute to understanding of the mechanisms underlying the biosynthesis of VLCPUFAs in the species, but also be instrumental in metabolic engineering of VLCPUFAs using this new system in heterologous systems, particularly in oilseed crops (30).
Supplementary Material
Acknowledgments
The authors would like to thank Yan Chen for technical assistance for fatty acid analysis, Jin Wang for statistical analysis, and Dr. Mark Smith for critical reading of the manuscript.
Footnotes
Abbreviations:
- ACP
- acyl carrier protein
- ALA
- α-linolenic acid
- ARA
- arachidonic acid
- DH
- dehydratase
- DPA
- docosapentaenoic acid
- ER
- enoyl-ACP reductase
- FAME
- fatty acid methyl ester
- GLA
- γ-linolenic acid
- KS
- ketoacyl-ACP synthase
- LA
- linoleic acid
- ORF
- open reading frame
- PKS
- polyketide synthase
- PPTase
- phosphopantetheinyl transferase
- SA
- stearic acid
- VLCPUFA
- very long chain PUFA
This work was supported by the Canadian Network for Research and Innovation in Machining Technology, Natural Sciences and Engineering Research Council of Canada.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Bazinet R. P., and Laye S.. 2014. Polyunsaturated fatty acids and their metabolites in brain function and disease. Nat. Rev. Neurosci. 15: 771–785. [DOI] [PubMed] [Google Scholar]
- 2.Guichardant M., Calzada C., Bernoud-Hubac N., Lagarde M., and Vericel E.. 2015. Omega-3 polyunsaturated fatty acids and oxygenated metabolism in atherothrombosis. Biochim. Biophys. Acta. 1851: 485–495. [DOI] [PubMed] [Google Scholar]
- 3.Wang W., Zhu J., Lyu F., Panigrahy D., Ferrara K. W., Hammock B., and Zhang G.. 2014. omega-3 polyunsaturated fatty acids-derived lipid metabolites on angiogenesis, inflammation and cancer. Prostaglandins Other Lipid Mediat. 113–115: 13–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Robinson L. E., and Mazurak V. C.. 2013. N-3 polyunsaturated fatty acids: relationship to inflammation in healthy adults and adults exhibiting features of metabolic syndrome. Lipids. 48: 319–332. [DOI] [PubMed] [Google Scholar]
- 5.Janssen C. I., and Kiliaan A. J.. 2014. Long-chain polyunsaturated fatty acids (LCPUFA) from genesis to senescence: the influence of LCPUFA on neural development, aging, and neurodegeneration. Prog. Lipid Res. 53: 1–17. [DOI] [PubMed] [Google Scholar]
- 6.Qiu X. 2003. Biosynthesis of docosahexaenoic acid (DHA, 22:6-4,7,10,13,16,19): two distinct pathways. Prostaglandins Leukot. Essent. Fatty Acids. 68: 181–186. [DOI] [PubMed] [Google Scholar]
- 7.Khozin-Goldberg I., Iskandarov U., and Cohen Z.. 2011. LC-PUFA from photosynthetic microalgae: occurrence, biosynthesis, and prospects in biotechnology. Appl. Microbiol. Biotechnol. 91: 905–915. [DOI] [PubMed] [Google Scholar]
- 8.Sprecher H., Luthria D. L., Mohammed B. S., and Baykousheva S. P.. 1995. Reevaluation of the pathways for the biosynthesis of polyunsaturated fatty acids. J. Lipid Res. 36: 2471–2477. [PubMed] [Google Scholar]
- 9.Harwood J. L., and Guschina I. A.. 2009. The versatility of algae and their lipid metabolism. Biochimie. 91: 679–684. [DOI] [PubMed] [Google Scholar]
- 10.Meesapyodsuk D., and Qiu X.. 2012. The front-end desaturase: structure, function, evolution and biotechnological use. Lipids. 47: 227–237. [DOI] [PubMed] [Google Scholar]
- 11.Graham I. A., Larson T., and Napier J. A.. 2007. Rational metabolic engineering of transgenic plants for biosynthesis of omega-3 polyunsaturates. Curr. Opin. Biotechnol. 18: 142–147. [DOI] [PubMed] [Google Scholar]
- 12.Metz J. G., Roessler P., Facciotti D., Levering C., Dittrich F., Lassner M., Valentine R., Lardizabal K., Domergue F., Yamada A., et al. . 2001. Production of polyunsaturated fatty acids by polyketide synthases in both prokaryotes and eukaryotes. Science. 293: 290–293. [DOI] [PubMed] [Google Scholar]
- 13.Okuyama H., Orikasa Y., Nishida T., Watanabe K., and Morita N.. 2007. Bacterial genes responsible for the biosynthesis of eicosapentaenoic and docosahexaenoic acids and their heterologous expression. Appl. Environ. Microbiol. 73: 665–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morita N., Tanaka M., and Okuyama H.. 2000. Biosynthesis of fatty acids in the docosahexaenoic acid-producing bacterium Moritella marina strain MP-1. Biochem. Soc. Trans. 28: 943–945. [PubMed] [Google Scholar]
- 15.Voss A., Reinhart M., Sankarappa S., and Sprecher H.. 1991. The metabolism of 7,10,13,16,19-docosapentaenoic acid to 4,7,10,13,16,19-docosahexaenoic acid in rat liver is independent of a 4-desaturase. J. Biol. Chem. 266: 19995–20000. [PubMed] [Google Scholar]
- 16.Sprecher H., Chen Q., and Yin F. Q.. 1999. Regulation of the biosynthesis of 22:5n-6 and 22:6n-3: a complex intracellular process. Lipids. 34(Suppl): S153–S156. [DOI] [PubMed] [Google Scholar]
- 17.Qiu X., Hong H., and MacKenzie S. L.. 2001. Identification of a Delta 4 fatty acid desaturase from Thraustochytrium sp. involved in the biosynthesis of docosahexanoic acid by heterologous expression in Saccharomyces cerevisiae and Brassica juncea. J. Biol. Chem. 276: 31561–31566. [DOI] [PubMed] [Google Scholar]
- 18.Cronan J. E., and Thomas J.. 2009. Bacterial fatty acid synthesis and its relationships with polyketide synthetic pathways. Methods Enzymol. 459: 395–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hauvermale A., Kuner J., Rosenzweig B., Guerra D., Diltz S., and Metz J. G.. 2006. Fatty acid production in Schizochytrium sp.: Involvement of a polyunsaturated fatty acid synthase and a type I fatty acid synthase. Lipids. 41: 739–747. [DOI] [PubMed] [Google Scholar]
- 20.Metz J. G., Kuner J., Rosenzweig B., Lippmeier J. C., Roessler P., and Zirkle R.. 2009. Biochemical characterization of polyunsaturated fatty acid synthesis in Schizochytrium: release of the products as free fatty acids. Plant Physiol. Biochem. 47: 472–478. [DOI] [PubMed] [Google Scholar]
- 21.Wu G., Truksa M., Datla N., Vrinten P., Bauer J., Zank T., Cirpus P., Heinz E., and Qiu X.. 2005. Stepwise engineering to produce high yields of very long-chain polyunsaturated fatty acids in plants. Nat. Biotechnol. 23: 1013–1017. [DOI] [PubMed] [Google Scholar]
- 22.Meesapyodsuk D., Reed D. W., Covello P. S., and Qiu X.. 2007. Primary structure, regioselectivity, and evolution of the membrane-bound fatty acid desaturases of Claviceps purpurea. J. Biol. Chem. 282: 20191–20199. [DOI] [PubMed] [Google Scholar]
- 23.Keatinge-Clay A. T. 2012. The structures of type I polyketide synthases. Nat. Prod. Rep. 29: 1050–1073. [DOI] [PubMed] [Google Scholar]
- 24.Black T. A., and Wolk C. P.. 1994. Analysis of a Het- mutation in Anabaena sp. strain PCC 7120 implicates a secondary metabolite in the regulation of heterocyst spacing. J. Bacteriol. 176: 2282–2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yazawa K. 1996. Production of eicosapentaenoic acid from marine bacteria. Lipids. 31(Suppl): S297–S300. [DOI] [PubMed] [Google Scholar]
- 26.Matsuda T., Sakaguchi K., Hamaguchi R., Kobayashi T., Abe E., Hama Y., Hayashi M., Honda D., Okita Y., Sugimoto S., et al. . 2012. Analysis of Delta12-fatty acid desaturase function revealed that two distinct pathways are active for the synthesis of PUFAs in T. aureum ATCC 34304. J. Lipid Res. 53: 1210–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lippmeier J. C., Crawford K. S., Owen C. B., Rivas A. A., Metz J. G., and Apt K. E.. 2009. Characterization of both polyunsaturated fatty acid biosynthetic pathways in Schizochytrium sp. Lipids. 44: 621–630. [DOI] [PubMed] [Google Scholar]
- 28.Allen E. E., and Bartlett D. H.. 2002. Structure and regulation of the omega-3 polyunsaturated fatty acid synthase genes from the deep-sea bacterium Photobacterium profundum strain SS9. Microbiology. 148: 1903–1913. [DOI] [PubMed] [Google Scholar]
- 29.Shulse C. N., and Allen E. E.. 2011. Widespread occurrence of secondary lipid biosynthesis potential in microbial lineages. PLoS One. 6: e20146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ruiz-Lopez N., Usher S., Sayanova O. V., Napier J. A., and Haslam R. P.. 2015. Modifying the lipid content and composition of plant seeds: engineering the production of LC-PUFA. Appl. Microbiol. Biotechnol. 99: 143–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.