Abstract
LPL contains two principal domains: an amino-terminal catalytic domain (residues 1–297) and a carboxyl-terminal domain (residues 298–448) that is important for binding lipids and binding glycosylphosphatidylinositol-anchored high density lipoprotein binding protein 1 (GPIHBP1) (an endothelial cell protein that shuttles LPL to the capillary lumen). The LPL sequences required for GPIHBP1 binding have not been examined in detail, but one study suggested that sequences near LPL’s carboxyl terminus (residues ∼403–438) were crucial. Here, we tested the ability of LPL-specific monoclonal antibodies (mAbs) to block the binding of LPL to GPIHBP1. One antibody, 88B8, abolished LPL binding to GPIHBP1. Consistent with those results, antibody 88B8 could not bind to GPIHBP1-bound LPL on cultured cells. Antibody 88B8 bound poorly to LPL proteins with amino acid substitutions that interfered with GPIHBP1 binding (e.g., C418Y, E421K). However, the sequences near LPL’s carboxyl terminus (residues ∼403–438) were not sufficient for 88B8 binding; upstream sequences (residues 298–400) were also required. Additional studies showed that these same sequences are required for LPL binding to GPIHBP1. In conclusion, we identified an LPL mAb that binds to LPL’s GPIHBP1-binding domain. The binding of both antibody 88B8 and GPIHBP1 to LPL depends on large segments of LPL’s carboxyl-terminal domain.
Keywords: chylomicrons, endothelial cells, lipids/chemistry, lipolysis and fatty acid metabolism, triglycerides, lipoprotein lipase, glycosylphosphatidylinositol-anchored high density lipoprotein binding protein 1
For more than 50 years, it has been known that LPL, a triglyceride hydrolase secreted by myocytes and adipocytes, is crucial for the intravascular processing of triglyceride-rich lipoproteins (TRLs) (1–3). For most of that time, it was assumed that LPL was attached to the heparan-sulfate proteoglycans along the lumen of blood vessels (4), but how LPL reached the lumen of blood vessels was a stubborn mystery. Within the past few years, that mystery has been solved (5, 6). Glycosylphosphatidylinositol-anchored high density lipoprotein binding protein 1 (GPIHBP1), a GPI-anchored protein of capillary endothelial cells, picks up freshly secreted LPL within the interstitial spaces and shuttles it across endothelial cells to the capillary lumen (7, 8). In the absence of GPIHBP1, LPL remains in the interstitial spaces and never reaches the capillary lumen, resulting in an accumulation of plasma TRLs and extremely high plasma triglyceride levels (“chylomicronemia”) (8). Recent studies showed that GPIHBP1 (and GPIHBP1-bound LPL) are also crucial for the margination of TRLs along the capillary lumen, allowing triglyceride hydrolysis to proceed (9).
GPIHBP1 has two main structural features—an amino-terminal acidic domain and a cysteine-rich three-fingered “LU domain” (7, 10). Recent studies have shown that the LU domain is primarily responsible for high-affinity binding of LPL, while the acidic domain augments the interaction and promotes an initial interaction complex between LPL and GPIHBP1 (6, 11). A variety of missense mutations in GPIHBP1’s LU domain have been identified in patients with chylomicronemia (12–22), and all of those abolish the ability of GPIHBP1 to bind LPL (6). Most of these mutations interfere with the formation of disulfide bonds in the LU domain, leading to disulfide-linked dimers and multimers (23). Alanine-scanning mutagenesis studies showed that the highly conserved second finger of the three-fingered LU domain is particularly important for binding LPL (24). Mutagenizing W109 in finger 2 abolishes LPL binding without promoting the formation of GPIHBP1 dimers/multimers, suggesting that W109 participates directly in binding LPL (23).
In contrast to the situation with GPIHBP1, our understanding of the LPL sequences required for binding to GPIHBP1 is meager, but the relevant sequences appear to be located in LPL’s carboxyl-terminal “lipid-binding” domain (residues 298–448) rather than in LPL’s catalytic domain (residues 1–297). A pair of LPL mutations (C418Y, E421K), first identified in patients with hypertriglyceridemia (25, 26), interfere with the binding of LPL to GPIHBP1. Mutation of nearby LPL sequences (residues 403–438) also impaired LPL binding to GPIHBP1 (27). Those studies were interpreted as showing that sequences near the carboxyl terminus of LPL are singularly important for mediating LPL binding to GPIHBP1.
Here, we sought to better define LPL sequences that are important for GPIHBP1 binding. As part of these efforts, we tested the capacity of three LPL-specific monoclonal antibodies (mAbs) (5D2, 88B8, 57A5) (28–30) to block the binding of LPL to GPIHBP1. We reasoned that if we were to identify a “blocking antibody,” then efforts to define the epitope would lead to new insights into LPL sequences that are important for LPL binding to GPIHBP1.
MATERIALS AND METHODS
Monoclonal antibodies
We examined three LPL-specific mouse mAbs (5D2, 57A5, 88B8) (28–30). The epitope for 5D2 has been studied in detail and is located between residues 380 and 410 in LPL’s carboxyl-terminal domain (29, 30). mAbs 57A5 and 88B8 were generated against human LPL (hLPL) and have been used previously in LPL immunoassays (28), but data on the epitopes for these antibodies have never been reported. Fab′ fragments were prepared with immobilized papain and Fc fragments removed with Protein A–Sepharose. mAb 5D2 was a gift from Dr. John Brunzell, and mAbs 57A5 and 88B8 were acquired from Immuno-Biological Laboratories (Gunma, Japan).
LPL–GPIHBP1 binding assays
Cell-free assay.
Secreted versions of wild-type human GPIHBP1 and GPIHBP1-W109S were stably expressed in Drosophila S2 cells. Both GPIHBP1 proteins contain a uPAR epitope tag (23) as well as the epitope for mAb 11A12 (31). For the assay, the conditioned medium from Drosophila S2 cells expressing soluble human GPIHBP1 was incubated for 1 h at 4°C with V5-tagged hLPL (32), with or without mAbs (20 μg/ml final), and agarose beads coated with mAb 11A12 (33). After washing the beads, GPIHBP1 and any GPIHBP1-bound LPL were eluted from the antibody-coated beads by heating the beads in SDS sample buffer for 5 min at 90°C. The amounts of GPIHBP1 and LPL in the starting material, unbound fractions, wash fractions, and elution fractions were assessed by Western blotting. Proteins were separated on a 12% NuPAGE SDS-PAGE gel with MES buffer, followed by transfer to a sheet of nitrocellulose membrane. The membrane was then incubated with IRdye680-conjugated antibody 11A12 and an IRdye800-conjugated V5-antibody.
Cell-based assay.
CHO pgsA-745 cells (2 × 106) were electroporated with 2 μg plasmids encoding either an S-protein–tagged wild-type human GPIHBP1 or an S-protein–tagged mutant human GPIHBP1 (W109S) and then plated on coverslips in 24-well plates. After 24 h, the cells were incubated with either V5-tagged hLPL alone, or V5-tagged hLPL with one of the three mAbs (5D2, 57A5, 88B8) (20 μg/ml) for 1 h at 4°C. The cells were then washed and processed for Western blots or immunofluorescence microscopy. For the Western blots, cell lysates were collected by incubating cells with M-PER mammalian protein extraction reagent (ThermoFisher Scientific) with EDTA-free complete protease inhibitor cocktail (Roche) for 5 min at 4°C, followed by centrifugation at 14,000 g for 10 min to remove insoluble material. The nitrocellulose membrane was then blocked for 1 h at room temperature with Odyssey blocking buffer (Li-Cor), and then incubated with an IRdye800–conjugated mouse mAb against the V5 tag (ThermoFisher Scientific; 2.32 μg/ml) and a goat polyclonal antibody against the S-protein tag (Abcam; 1:1,000) followed by an IRdye680–conjugated donkey anti-goat IgG (Li-Cor). Signals were visualized and quantified with a Li-Cor Odyssey scanner. For the immunofluorescence microscopy assay, cells were fixed in 3% paraformaldehyde for 15 min and blocked with 10% donkey serum in PBS/Mg/Ca. Cells were then incubated overnight at 4°C with an Alexa 647–conjugated mouse mAb against the V5 tag (ThermoFisher Scientific; 11.6 μg/ml) and a goat polyclonal antibody against the S-protein tag (Abcam; 1:800), followed by a 30-min incubation with an Alexa 568–conjugated donkey anti-goat IgG (ThermoFisher Scientific; 1:800). After washing, the cells were fixed with 3% paraformaldehyde for 15 min and stained with 4′,6-diamidino-2-phenylindole (DAPI) to visualize DNA. Images were recorded with an Axiovert 200M microscope and processed with Zen 2010 software (all from Zeiss). Within each experiment, the exposure conditions for each construct were identical.
Testing the binding of hepatic lipase–LPL chimeras to GPIHBP1 on the surface of cultured cells
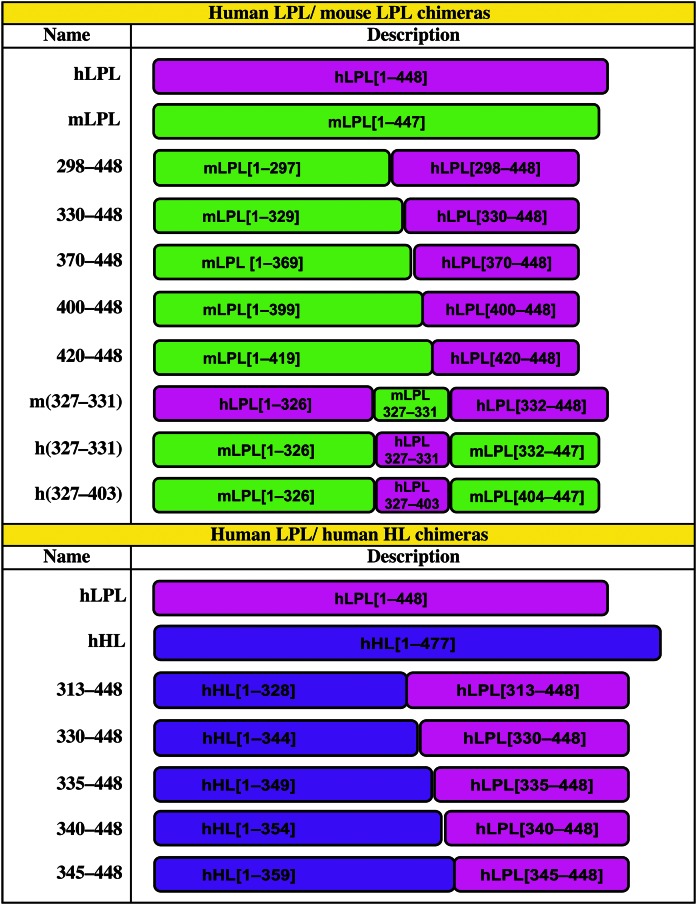
Earlier studies by Wong et al. (34) showed that it was possible to express catalytically active, dimeric HL–LPL chimeras by exchanging sequences in HL with the corresponding sequences in LPL. Here, we used that approach to create HL–LPL chimeras containing hLPL residues 313–448, 330–448, 335–448, 340–448, or 345–448 (Fig. 1). To test the ability of HL, LPL, and the HL–LPL chimeras to bind to GPIHBP1, we used a “co-plating assay” described previously by Beigneux et al. (24). CHO-K1 cells (1 × 106) were electroporated with 1.0 mg of either an S-protein–tagged human GPIHBP1 construct or an expression vector for one of the V5-tagged lipases. The independently transfected cells were then mixed together and plated on coverslips in 24-well plates. Twenty-four hours later, the cells were fixed in 3% paraformaldehyde. When indicated, the cells were permeabilized with 0.2% Triton X-100. Cells were blocked with 10% donkey serum in PBS/Mg/Ca and then incubated for 1 h in the blocking buffer containing a goat antibody against the S-protein tag (Abcam; 1:500) and a mouse mAb against the V5 tag (Invitrogen; 1:100), followed by a 30-min incubation with an Alexa 568–conjugated donkey anti-goat IgG (Invitrogen; 1:800) and an Alexa 488–conjugated donkey anti-mouse IgG (Invitrogen; 1:800). After washing, the cells were stained with DAPI to visualize DNA. Images were recorded with the Axiovert 200M microscope. The exposure conditions for each construct were identical. In this system, cells that expressed wild-type GPIHBP1 captured LPL that was secreted by the LPL-transfected cells, thus, GPIHBP1 and LPL signals colocalized on the merged image (24). HL does not bind to GPIHBP1 (31); consequently, there was no colocalization of HL and GPIHBP1 signals.
Fig. 1.
List of the mLPL-hLPL chimeras, and of the human HL-hLPL chimeras generated for the studies. Numbers in brackets correspond to amino acid residues.
Lipase expression vectors
hLPL–mouse LPL (mLPL) chimeras, and HL–LPL chimeras (Fig. 1) were created with a PCR-based method (In-Fusion HD cloning kit, Clontech). Point mutations were introduced with the QuikChange Lightning kit (Agilent Technologies).
Testing the ability of the mAbs to bind to GPIHBP1-bound LPL
CHO pgsA-745 cells (2 × 106) were electroporated with 2 μg of a plasmid encoding an S-protein–tagged wild-type human GPIHBP1 and plated on coverslips in 24-well plates. Twenty-four hours later, cells were incubated with a V5-tagged hLPL or buffer for 1 h at 4°C. Cells were then washed and incubated with either buffer or Alexa 568–conjugated mAbs (5D2, 57A5, or 88B8) (20 μg/ml) for 1 h at 4°C. The cells were then washed and processed for immunocytochemistry. Cells on coverslips were fixed in 3% paraformaldehyde for 15 min, blocked with 10% donkey serum in PBS/Mg/Ca, and then incubated overnight at 4°C with an Alexa 647–conjugated mouse mAb against the V5 tag (ThermoFisher Scientific; 11.6 μg/ml) and a goat polyclonal antibody against the S-protein tag (Abcam; 1:800), followed by a 30-min incubation with an Alexa 488–conjugated donkey anti-goat IgG (ThermoFisher Scientific; 1:800). After washing, the cells were fixed with 3% paraformaldehyde for 15 min and stained with DAPI to visualize DNA. Microscopy was performed as described earlier.
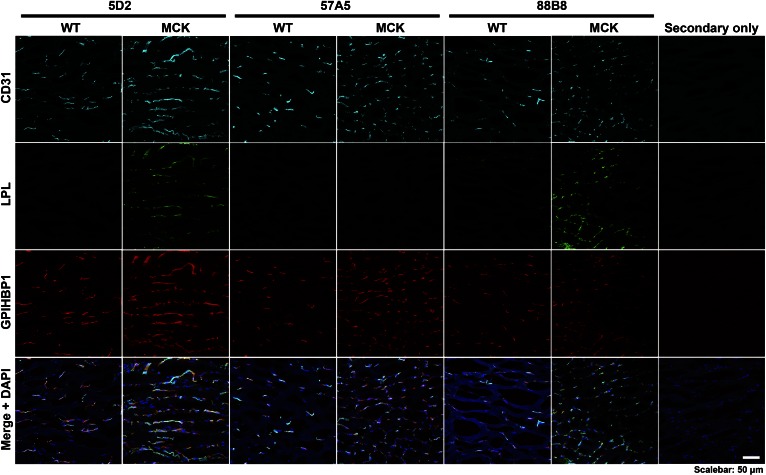
Testing the ability of the mAbs to bind to LPL in capillaries of tissue sections
Wild-type and Lpl−/−MCK-hLPL mice (35) were perfused with PBS followed by 3% paraformaldehyde; quadriceps muscle was harvested and embedded in OCT on dry ice. Tissue sections (7 μm) were fixed in methanol at −20°C for 10 min, permeabilized with 0.2% Triton X-100 for 5 min, and blocked with 5% donkey serum, 10% FBS, and 0.2% BSA in PBS/Mg/Ca. Tissues were incubated overnight at 4°C with Alexa 568–conjugated mouse mAbs (5D2, 57A5, or 88B8) (8 μg/ml) and a rabbit polyclonal antibody against mouse CD31 (Abcam; 1:50), followed by a 45-min incubation with Alexa 647–conjugated 11A12 antibody (3 μg/ml) and Alexa 488–conjugated donkey anti-rabbit IgG (ThermoFisher Scientific; 1:200). After washing, the cells were fixed with 3% paraformaldehyde for 5 min and stained with DAPI to visualize DNA. Microscopy was performed as described earlier. Mice were fed a chow diet and housed in a barrier facility with a 12 h light-dark cycle. All studies were approved by UCLA’s Animal Research Committee.
Measurements of LPL activity
hLPL was prepared from a CHO cell line expressing V5-tagged hLPL (from Dr. Mark Doolittle, University of California, Los Angeles). LPL was purified by heparin–Sepharose chromatography on an ÄKTA pure HPLC (GE Healthcare) and eluted with a 0.1–2 M NaCl gradient in 20 mM NaPO4, pH 7.4. Taurodeoxycholate (final concentration, 5 mM) was added to the LPL (i.e., fractions eluting at ≥1 M NaCl) before storing at −80°C. The activity of purified hLPL was compared with that of a known quantity of bovine LPL. Lipase activity was measured with [3H]triolein that had been incorporated into Intralipid (0.5 μCi [3H]triolein/mg triglyceride). hLPL (6 μl, corresponding to the enzymatic activity of 5 ng of bovine LPL) was added to 200-μl incubation mixtures containing 5 mg of triglyceride/ml in 0.15 M Tris (pH 8.5) containing 0.1 M NaCl, 6% BSA (w/v), 16.7 units of heparin/ml, and 5% (v/v) heat-inactivated rat serum (as a source of apo-CII). mAbs (20 μg/ml, final concentration) were added to incubation mixtures 5 min before adding the LPL. Esterase activities were analyzed by adding 30 μl of the hLPL to 100-μl incubation mixtures of 50 mM Tris, 50 μM 1,2-di-O-lauryl-rac-glycero-3-glutaric acid 6′-methylresorufin ester (DGGR), 120 mM NaCl, 10 mg/ml BSA, and 0.5% Triton X-100 (pH 7.4). mAbs (200 μg/ml final concentration) were added to incubations 5 min before adding the LPL. Ester hydrolysis was determined by measuring the increase of resorufin fluorescence at λex 530 nm and λem 590 nm during the first 15 min of the incubation.
RESULTS
Testing the ability of LPL-specific mAbs to block the binding of LPL to GPIHBP1
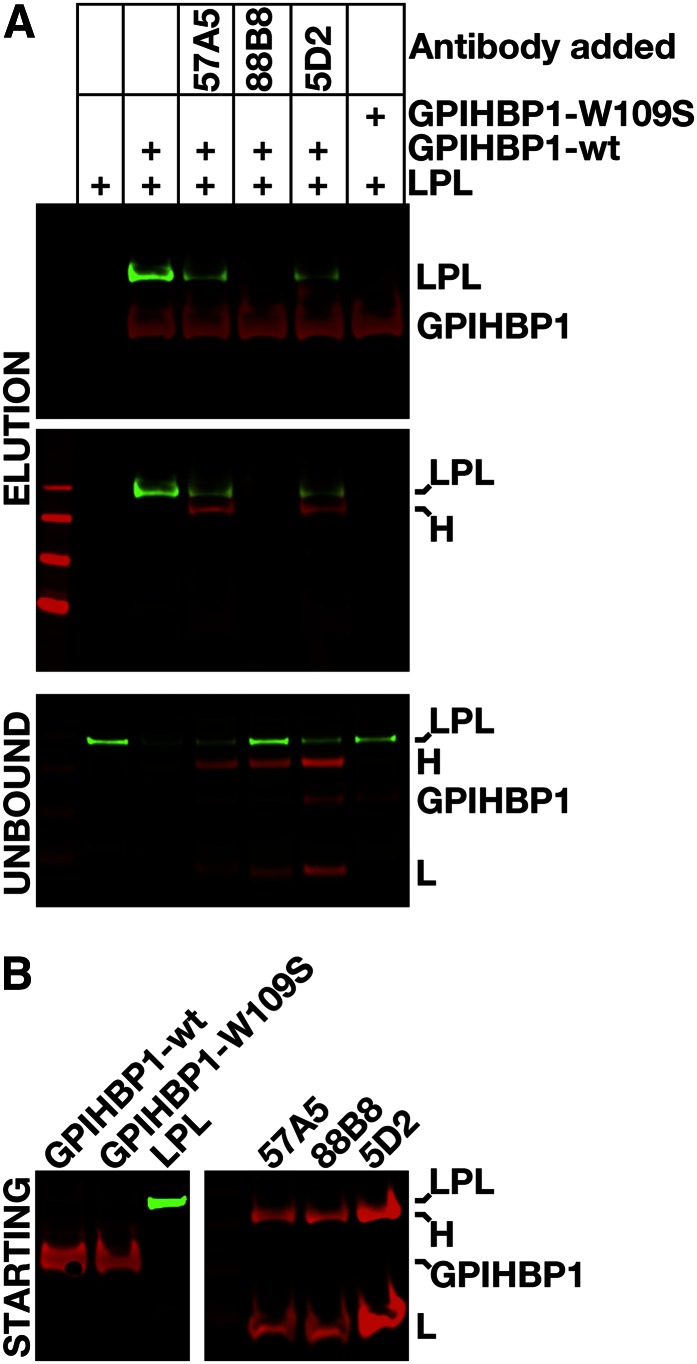
We used cell-free and cell-based LPL–GPIHBP1 binding assays (18, 23, 31, 33) to test the ability of three LPL-specific mAbs (5D2, 88B8, 57A5) (29, 30) to block the binding of hLPL to GPIHBP1. In the cell-free assay, we incubated GPIHBP1, LPL, and an LPL-specific mAb with agarose beads that had been coated with a GPIHBP1-specific mAb (11A12). After a 1-h incubation, the beads were washed, and the amounts of GPIHBP1 and GPIHBP1-bound LPL bound to the beads were assessed by Western blotting. mAb 88B8 abolished LPL binding to GPIHBP1; thus, no LPL could be eluted from the beads (Fig. 2). The ability of 88B8 (and 88B8 Fab′ fragments) to block the binding of LPL to GPIHBP1 was confirmed in additional independent experiments (supplemental Figs. S1, S2). mAbs 5D2 and 57A5 did not abolish the binding of LPL to GPIHBP1, but they did reduce LPL binding (Fig. 2).
Fig. 2.
A cell-free LPL–GPIHBP1 binding assay to test the ability of LPL-specific mAbs 5D2, 57A5, and 88B8 to block the binding of V5-tagged hLPL to GPIHBP1. Secreted versions of wild-type (wt) GPIHBP1 and GPIHBP1-W109S were expressed in Drosophila S2 cells. Both GPIHBP1 proteins contain a uPAR epitope tag (23) as well as the epitope for mAb 11A12. The GPIHBP1 proteins (7.25 μg) were incubated with agarose beads coated with mAb 11A12, V5-tagged hLPL (825 ng), and either no antibody or mAbs 57A5, 88B8, or 5D2 (20 μg/ml final). We used a high molar concentration of mAbs so as to minimize the impact of differences in the affinity of the three different mAbs. After a 1-h incubation at 4°C, the beads were washed. GPIHBP1 and GPIHBP1-bound LPL were then released from the agarose beads by heating the beads in SDS-sample buffer. A: Western blots on the “unbound” fraction (proteins that did not bind to the agarose beads) and “elution” fractions with an IRdye800-V5 antibody (green), an IRdye680–antibody 11A12 (red), and an IRdye680-donkey anti–mouse IgG (red). LPL binding to GPIHBP1 was inhibited 53.8% with 57A5, 94.9% with 88B8, and 63.5% with 5D2, as judged by quantification with a Li-Cor scanner. H, heavy chain; L, light chain. B: Western blots performed on the “starting material” proteins that were added to the assay (mAbs, GPIHBP1, LPL).
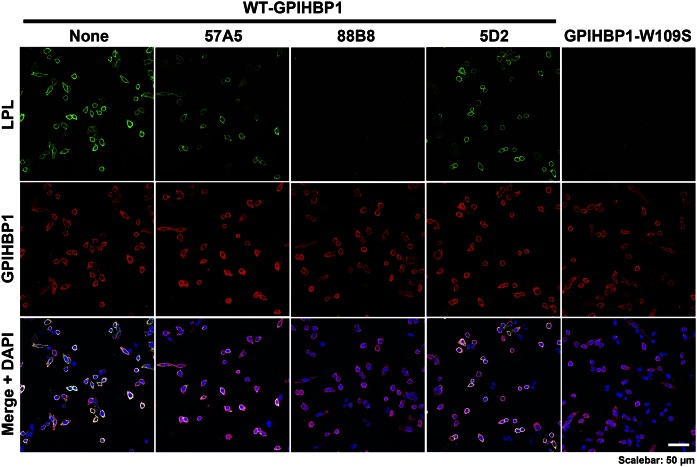
mAb 88B8 completely blocked binding of LPL to GPIHBP1 on the surface of GPIHBP1-transfected cells, as judged by immunofluorescence microscopy and Western blots of cell extracts (Fig. 3, supplemental Fig. S3). mAbs 57A5 and 5D2 caused partial inhibition of LPL binding to GPIHBP1 (Fig. 3, supplemental Fig. S3).
Fig. 3.
Immunofluorescence microscopy assay to evaluate the ability of mAbs 57A5, 88B8, and 5D2 to block the binding of LPL to GPIHBP1. CHO pgsA-745 cells were transiently transfected with S-protein–tagged wild-type human GPIHBP1 (or GPIHBP1-W109S, which lacks the ability to bind LPL). The cells were then incubated with V5-tagged hLPL and either no antibody or mAbs 57A5, 88B8, or 5D2 (20 mg/ml). After a 1-h incubation at 4°C, nonpermeabilized cells were stained for GPIHBP1 with an antibody against the S-protein tag (red) and LPL with a V5 antibody (green). Cell nuclei were stained with DAPI (blue).
Defining the epitopes for LPL-specific mAbs
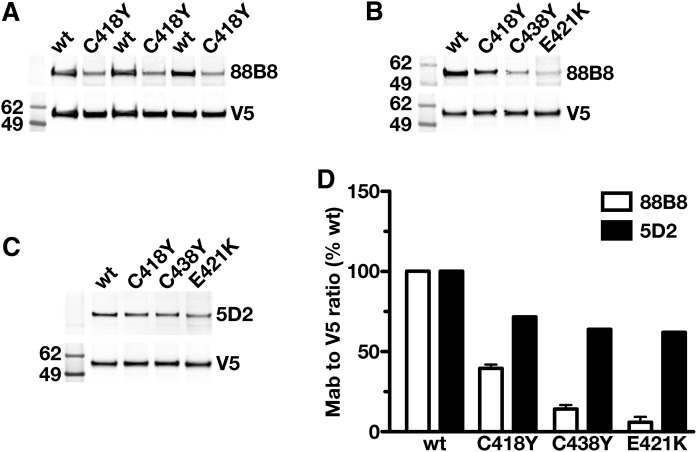
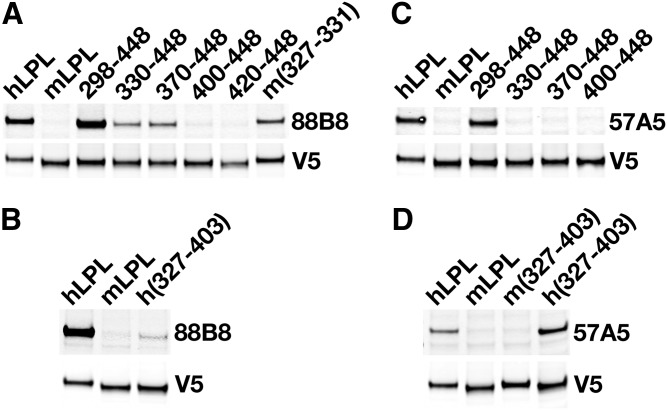
The 5D2 epitope is located within LPL residues 380–410 (29, 30), but data on the epitopes for mAbs 57A5 and 88B8 have not been reported. Preliminary Western blot studies indicated that both mAbs 57A5 and 88B8 bind to the carboxyl-terminal region of LPL (residues 298–448) (supplemental Fig. S4). Identical conclusions were reached by performing ELISAs on the medium of cells that had been transfected with Flag-tagged constructs encoding LPL’s amino- and carboxyl-terminal domains (K. Miyashita, unpublished observations). Because 88B8 and 88B8 Fab′ fragments block LPL binding to GPIHBP1 and also bound to LPL’s carboxyl-terminal domain, we suspected that the binding sites for mAb 88B8 and GPIHBP1 on LPL were similar and that LPL mutations known to interfere with GPIHBP1 binding (e.g., C418Y, E421K, C438Y) (27) would also interfere with 88B8 binding. Indeed, 88B8 bound more avidly to wild-type LPL than to mutant LPLs harboring C418Y, E421K, or C438Y mutations (Fig. 4A, B). The effect of those mutations on 5D2 binding was minimal (Fig. 4C, D). As expected, 88B8 bound avidly to hLPL but not mLPL; it bound avidly to a mLPL–hLPL chimera containing the entire carboxyl-terminal domain of hLPL (residues 298–448).
Fig. 4.
Testing the ability of mAbs 88B8 and 5D2 to bind to mutant forms of LPL with impaired ability to bind to GPIHBP1. CHO pgsA-745 cells were transfected with V5-tagged wild-type (wt) hLPL or mutant forms of hLPL (LPL-C418Y, LPL-C421K, LPL-C438Y) with either no ability (LPL-C418Y, LPL-C421K) or reduced ability (LPL-C438Y) to bind to GPIHBP1 (27). On the next day, the cells were washed, and cell lysates were prepared for Western blotting. Western blots were performed under nonreducing conditions. A: Western blot with IRdye800-labeled mAb 88B8 and an IRdye680-labeled V5 antibody. B: Western blot with mAb 88B8 (10 mg/ml) followed by an IRdye-800 donkey anti-mouse IgG, along with an IRdye680-V5 antibody. C: Western blot with IRdye800-labeled mAb 5D2 and an IRdye680-labeled V5 antibody. D: Quantification of mAbs 88B8 and 5D2 binding to LPL-C418Y, LPL-C438Y, and LPL-C421K (as determined by a Li-Cor scanner). Compared with wild-type LPL, the binding of the antibodies to LPL-C418Y was reduced by 70% for 88B8, 37% for 57A5, and 26% for 5D2.
Earlier studies implied LPL’s GPIHBP1-binding domain involved amino acids 403–438 (27), and the reduced binding of 88B8 to the LPL mutants suggested that the 88B8 epitope might be located in the same stretch of amino acids. To our surprise, the binding of 88B8 to LPL depended on upstream sequences within the primary sequence. mAb 88B8 bound weakly to mLPL–hLPL chimeras containing hLPL residues 330–448 or 370–448, and it failed to bind to chimeras containing hLPL residues 400–448 or 420–448 (Fig. 5A). The same pattern was observed for 88B8 Fab′ fragments (supplemental Fig. S5). These results suggested that LPL residues 298–330 are quite relevant to 88B8 binding. Within that region, the only amino acids that differ between the hLPL and mLPL sequences are residues 327–331. Those residues were important for the 88B8 epitope; when residues 327–331 in hLPL were replaced with the mLPL sequences, the binding of 88B8 was significantly reduced (Fig. 5A).
Fig. 5.
Western blot studies to assess epitopes for mAbs 88B8 and 57A5. CHO pgsA-745 cells were transfected with V5-tagged versions of mLPL, hLPL, or mLPL–hLPL chimeras (mLPL containing hLPL sequences 298–448, 330–448, 370–448, 400–448, 420–448, or 327–403; or hLPL containing mLPL sequences 327–331 or 327–403) (see Fig. 1 for description of the chimeras). After 24 h, the cells were washed and cell lysates were prepared for SDS-PAGE (nonreducing conditions) and Western blot studies. A, B: Binding of mAb 88B8 to different V5-tagged lipases. Blots were incubated with mAb 88B8 and an IRdye800-labeled donkey anti-mouse IgG, followed by an IRdye680-labeled V5 antibody. C, D: Binding of mAb 57A5 to different V5-tagged lipases. Blots were incubated with mAb 57A5 followed by an IRdye800-labeled donkey anti-mouse IgG and an IRdye680-V5 antibody.
The finding that 88B8 bound (albeit weakly) to mLPL–hLPL chimeras containing hLPL residues 330–448 and 370–448 but failed to bind to a chimera with human residues 400–448 indicates that hLPL residues 330–400 are important for 88B8 binding. However, these results do not mean that residues 400–448 have no role in 88B8 binding, but rather that residues 400–448 are insufficient. First, the C418Y, E421K, and C438Y mutations clearly interfere with 88B8 binding (Fig. 4). Second, introducing human residues 327–403 into the mLPL expression vector resulted in only minimal restoration of 88B8 binding (Fig. 5B), implying that additional sequences at the carboxyl terminus of LPL (i.e., residues 403–448) were important for the 88B8 epitope.
mAb 57A5 bound avidly to hLPL and to a mLPL–hLPL chimera containing human residues 298–448 but not to chimeras containing human residues 330–448, 370–448, or 400–448 (Fig. 5C). In contrast to 88B8, 57A5 bound avidly to a mLPL–hLPL chimera containing human residues 327–403 (Fig. 5D).
Testing the impact of the LPL-specific mAbs on LPL activity
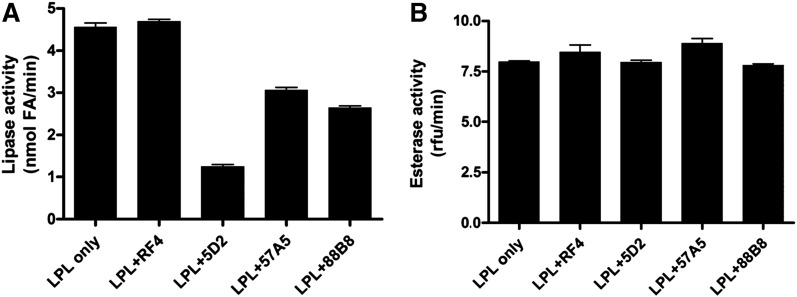
The tryptophan-rich motif within the 5D2 epitope (residues 380–410) is important for the ability of LPL to hydrolyze long-chain triacylglycerols but not short-chain water-soluble triacylglycerols (36). Thus, in our studies, antibody 5D2, which binds to the tryptophan-rich motif, inhibited LPL activity against triolein but not a soluble substrate (Fig. 6). mAbs 88B8 and 57A5 also reduced LPL activity against triolein but to a lesser degree (Fig. 6). None of the antibodies inhibited LPL catalytic activity with a water-soluble substrate (Fig. 6).
Fig. 6.
Testing the ability of mAbs 5D2, 57A5, and 88B8 to inhibit the catalytic activity of purified hLPL. A: LPL was added to incubation mixtures of lipid emulsion particles containing [3H]triolein in the presence of an irrelevant mAb (RF4) or each of the three LPL-specific mAbs (20 μg/ml). The activity of the hLPL in each assay corresponded to activity observed with 0.45 nM bovine LPL. B: hLPL, in an amount corresponding to the activity of 4.5 nM bovine LPL, was added to incubation mixtures containing the ester substrate DGGR in the presence of the mAbs (200 μg/ml). We deliberately used a high molar concentration of mAbs so as to minimize the impact of differences in the affinity of the three different mAbs. Data represent mean values of triplicate measurements ± SD.
Assessing LPL sequences relevant to binding GPIHBP1
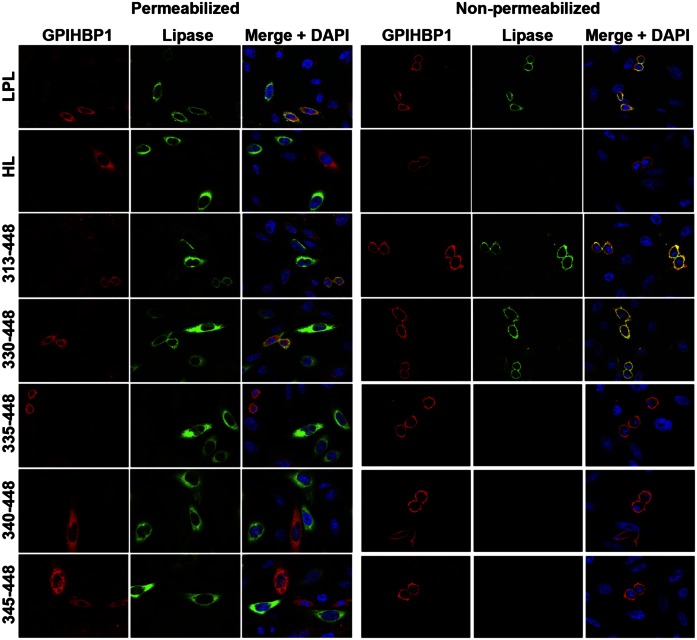
Earlier studies suggested that the binding of LPL to GPIHBP1 depended on LPL residues 403–438 (27). However, given that sequences throughout LPL’s carboxyl-terminal domain (residues 298–448) were important for 88B8 binding, we suspected that the same sequences might also play a role in GPIHBP1 binding. To explore this idea, we tested the ability of HL–LPL chimeras to bind to GPIHBP1 on the surface of CHO cells. Wong et al. (34) showed that it was possible to produce HL–LPL chimeras that are secreted and are catalytically active. We created HL–LPL chimeras containing hLPL residues 313–448, 330–448, 335–448, 340–448, and 345–448 (Fig. 1, supplemental Fig. S6). We then tested the ability of LPL, HL, and the HL–LPL chimeras to bind to GPIHBP1. We mixed CHO cells that had been transfected with a lipase construct with CHO cells that had been transfected with GPIHBP1. We then used immunofluorescence microscopy to assess the binding of the freshly secreted lipases to GPIHBP1 on GPIHBP1-transfected cells. As expected, full-length LPL bound avidly to GPIHBP1, but HL did not. HL–LPL chimeras containing LPL residues 313–448 and 330–448 bound to GPIHBP1, but chimeras containing LPL residues 335–448, 340–448, and 345–448 did not (Fig. 7). A Western blot experiment showed that cells expressing GPIHBP1 were capable of binding LPL as well as chimeras containing LPL residues 313–448 and 330–448, but not HL or a chimera containing LPL residues 335–448 (supplemental Fig. S7). Thus, a large segment of LPL’s carboxyl-terminal domain is required for GPIHBP1 binding—as was the case for 88B8 binding.
Fig. 7.
Immunofluorescence microscopy studies to assess the ability of LPL, HL, and LPL–HL chimeras to bind to GPIHBP1. CHO-K1 cells were electroporated with an S-protein–tagged human GPIHBP1 construct or a V5-tagged lipase expression vector (either LPL, HL, or HL–LPL chimeras containing LPL residues 313–448, 330–448, 335–448, 340–448, and 345–448) (see Fig. 1 for description of the constructs). The separately transfected cells were then mixed and plated on coverslips in 24-well plates. Twenty-four hours later, the cells were fixed in 3% paraformaldehyde, and blocked with 10% donkey serum in PBS/Mg/Ca. Some cells were permeabilized with 0.2% Triton X-100. Cells were then incubated for 1 h in blocking buffer with a goat polyclonal antibody against the S-protein tag (red) and a mouse mAb against the V5 tag (green), followed by a 30-min incubation with an Alexa 568–conjugated donkey anti-goat IgG (Invitrogen; 1:800) and an Alexa 488–conjugated donkey anti-mouse IgG (Invitrogen; 1:800). Cell nuclei were visualized with DAPI (blue). Cells expressing wild-type GPIHBP1 captured LPL secreted by neighboring LPL-expressing cells; hence, the GPIHBP1 and LPL signals colocalized on the merged image. HL–LPL (313–448) and HL–LPL (330–448) bound to GPIHBP1. HL and the remaining HL–LPL chimeras did not bind to GPIHBP1 (no colocalization of GPIHBP1 and the lipase on the merged image). HL–LPL chimeras containing LPL residues 370–448, 380–448, and 389–448 also failed to bind to GPIHBP1.
mAb 88B8 cannot bind to GPIHBP1-bound LPL but is still useful for immunohistochemistry studies
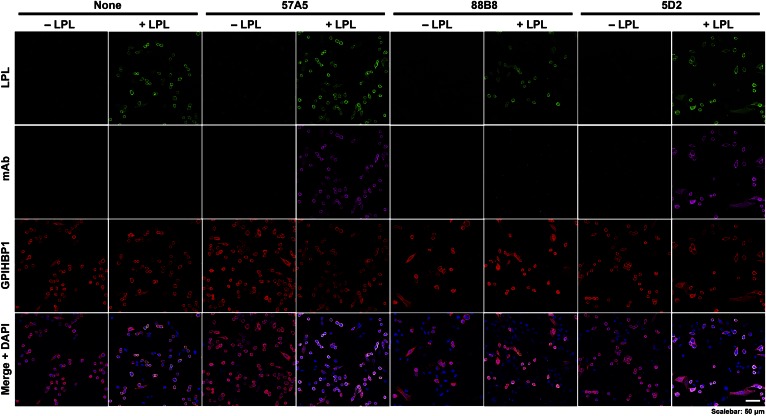
Because 88B8 and GPIHBP1 bind to similar sequences, we suspected that 88B8 would not bind to GPIHBP1-bound LPL on the surface of CHO cells. Indeed, this was the case (Fig. 8). In contrast, 5D2 and 57A5 did bind to GPIHBP1-bound LPL. Interestingly, the inability of 88B8 to bind to GPIHBP1-bound LPL did not interfere with its utility for immunohistochemistry studies. mAb 88B8 readily detected hLPL on capillaries after the LPL–GPIHBP1 complex was disrupted by methanol fixation (Fig. 9). mAb 5D2 also was useful for immunohistochemistry, but 57A5 was not (Fig. 9).
Fig. 8.
Assessing the ability of mAbs 57A5, 88B8, and 5D2 bind to GPIHBP1-bound LPL. CHO pgsA-745 cells were transiently transfected with S-protein–tagged human GPIHBP1. After 24 h, the cells were incubated with V5-tagged LPL for 1 h. The LPL was then removed and the cells were washed. mAbs 57A5, 88B8, and 5D2 (20 μg/ml) were then added to the cells and incubated for 1 h at 4°C. The cells were washed, fixed with paraformaldehyde, and then stained for GPIHBP1 with an antibody against the S-protein tag (red), and for LPL with a V5 antibody (green). mAbs 57A5, 88B8, and 5D2 were directly labeled (magenta).
Fig. 9.
Testing the capacity of mAbs 57A5, 88B8, and 5D2 to bind to hLPL in capillaries of the skeletal muscle of Lpl−/− mice carrying a hLPL transgene driven by the muscle creatine kinase promoter (Lpl−/−MCK-hLPL). For these studies, skeletal muscle from wild-type mice and Lpl−/−MCK-hLPL mice was harvested and embedded in OCT, and 7-mm-thick sections were cut, placed on slides, and fixed in methanol. Tissue sections were stained with a rabbit antibody against mouse CD31 (cyan), Alexa Fluor 568-labeled mAb (57A5, 88B8, or 5D2; green), and an Alexa Fluor 647-labeled antibody against GPIHBP1 (11A12, red).
DISCUSSION
In the current studies, we identified a hLPL–specific mAb, 88B8, that abolishes the binding of hLPL to GPIHBP1 in both cell-free and cell-based LPL–GPIHBP1 binding assays. mAb 88B8 binding to LPL was impaired by the very same LPL missense mutations (C418Y, E421K, C438Y) that are known to interfere with the binding of LPL to GPIHBP1 (27), suggesting that 88B8 and GPIHBP1 binding sites are very similar. We suspected initially that the LPL sequences required for 88B8 binding would be confined to residues ∼403–438, but this was not the case. Additional upstream sequences (residues 298–400) proved to be important for 88B8 binding. mAb 88B8 bound avidly to a mLPL–hLPL chimera containing hLPL residues 298–448 and weakly to chimeras containing residues 330–448 and 370–448. mAb 88B8 did not bind to a chimera containing hLPL residues 400–448. The fact that extensive sequences within LPL’s carboxyl-terminal domain are required for 88B8 binding led us to suspect that the same sequences would be required for GPIHBP1 binding. Indeed, studies with HL–LPL chimeras showed that a large portion of LPL’s carboxyl-terminal domain was required for GPIHBP1 binding. HL–LPL chimeras containing hLPL residues 313–448 and 330–448 bound to GPIHBP1, but chimeras containing LPL residues 335–448 or 340–448 did not. Thus, the ability of GPIHBP1 to bind to LPL depends on residues 330–448 and not simply residues 403–438 (as we had originally suspected). These results add considerably to our understanding of LPL sequences required for LPL–GPIHBP1 interactions.
To fully understand 88B8–LPL interactions or LPL–GPIHBP1 interactions, cocrystal structures are required. However, with the mutagenesis-based binding data that are in hand, we believe that the simplest interpretation is that both 88B8 and GPIHBP1 interact with a complex epitope that depends on the proper folding of a large portion of LPL’s carboxyl-terminal domain (residues 298–448). It is equally possible that LPL residues 298–400 are simply required for the proper conformation of a more compact binding site (residues ∼400–448). We do not believe that the absence of GPIHBP1 binding to the HL–LPL chimera containing residues 389–448 means that residues 389–448 are unimportant for GPIHBP1 binding. First, the C418Y and E421K mutations abolished GPIHBP1 binding and do so without affecting LPL catalytic activity—implying that those mutations did not cause global changes in LPL structure. Second, and perhaps more importantly, Mysling et al. (11) recently reported, using hydrogen–deuterium exchange/mass spectrometry studies, that the amide hydrogens in LPL residues 419–425 were protected from deuterium exchange by GPIHBP1 binding (i.e., that the binding of GPIHBP1 to those LPL sequences limited their accessibility to solvent). In a similar fashion, we believe that residues 400–448 are relevant to 88B8 binding, despite the fact that 88B8 did not bind to the mLPL–hLPL chimera containing hLPL residues 400–448. First, 88B8 binding was disrupted by C418Y, E421K, and C438Y mutations. Second, 88B8 could not bind to the mLPL–hLPL chimera containing hLPL residues 298–403.
The epitope for 57A5 was simpler: it bound to hLPL residues 298–448 but not to residues 330–448, implying that residues 298 to 330 were crucial for the epitope. Unlike 88B8, 57A5 and 5D2 did not abolish LPL binding to GPIHBP1, but partial inhibition was clearly evident. We suspect that the binding of 5D2 and 57A5 locks LPL into a conformation with reduced affinity for GPIHBP1—or alternatively that these antibodies create a steric hindrance to GPIHBP1 binding. In earlier studies, 5D2 inhibited LPL activity against triolein but not a soluble short-chain triacylglycerol (37). We confirmed those findings and found that the same property applies, at least to some degree, to 88B8 and 57A5. We suspect that the binding of all three antibodies creates a steric hindrance or locks the carboxyl-terminal domain into a suboptimal conformation for triolein hydrolysis.
mAbs 5D2 and 57A5 had no difficulty binding to GPIHBP1-bound LPL on the surface of cultured cells, whereas 88B8 was unable to bind, reflecting the fact that GPIHBP1 and 88B8 have overlapping binding sites. Remarkably, 88B8 was still useful for immunohistochemistry. Once the LPL–GPIHBP1 complex had been disrupted by methanol fixation, mAb 88B8 readily bound to the LPL on capillaries, colocalizing with GPIHBP1 and the endothelial cell marker CD31. mAb 5D2, but not 57A5, also detected LPL in capillaries.
Supplementary Material
Footnotes
Abbreviations:
- DAPI
- 4′,6-diamidino-2-phenylindole
- GPIHBP1
- glycosylphosphatidylinositol-anchored high density lipoprotein binding protein 1
- hLPL
- human LPL
- mAb
- monoclonal antibody
- mLPL
- mouse LPL
- TRL
- triglyceride-rich lipoprotein
This work was supported by grants from the National Heart, Lung, and Blood Institute (HL090553, HL087228, and HL125335) and a Transatlantic Network Grant from the Fondation Leducq (12CVD04). C.M.A. was supported by a Ruth L. Kirschstein National Research Service Award (T32HL69766). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors have no financial interests to declare.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Korn E. D. 1955. Clearing factor, a heparin-activated lipoprotein lipase. II. Substrate specificity and activation of coconut oil. J. Biol. Chem. 215: 15–26. [PubMed] [Google Scholar]
- 2.Korn E. D. 1955. Clearing factor, a heparin-activated lipoprotein lipase. I. Isolation and characterization of the enzyme from normal rat heart. J. Biol. Chem. 215: 1–14. [PubMed] [Google Scholar]
- 3.Havel R. J., and Gordon R. S. Jr. 1960. Idiopathic hyperlipemia: metabolic studies in an affected family. J. Clin. Invest. 39: 1777–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Merkel M., Eckel R. H., and Goldberg I. J.. 2002. Lipoprotein lipase: genetics, lipid uptake, and regulation. J. Lipid Res. 43: 1997–2006. [DOI] [PubMed] [Google Scholar]
- 5.Young S. G., and Zechner R.. 2013. Biochemistry and pathophysiology of intravascular and intracellular lipolysis. Genes Dev. 27: 459–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fong L. G., Young S. G., Beigneux A. P., Bensadoun A., Oberer M., Jiang H., and Ploug M.. 2016. GPIHBP1 and plasma triglyceride metabolism. Trends Endocrinol. Metab. 27: 455–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beigneux A. P., Davies B., Gin P., Weinstein M. M., Farber E., Qiao X., Peale P., Bunting S., Walzem R. L., Wong J. S., et al. . 2007. Glycosylphosphatidylinositol-anchored high density lipoprotein–binding protein 1 plays a critical role in the lipolytic processing of chylomicrons. Cell Metab. 5: 279–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davies B. S. J., Beigneux A. P., Barnes R. H. II, Tu Y., Gin P., Weinstein M. M., Nobumori C., Nyrén R., Goldberg I. J., Olivecrona G., et al. . 2010. GPIHBP1 is responsible for the entry of lipoprotein lipase into capillaries. Cell Metab. 12: 42–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goulbourne C. N., Gin P., Tatar A., Nobumori C., Hoenger A., Jiang H., Grovenor C. R., Adeyo O., Esko J. D., Goldberg I. J., et al. . 2014. The GPIHBP1-LPL complex is responsible for the margination of triglyceride-rich lipoproteins in capillaries. Cell Metab. 19: 849–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ioka R. X., Kang M-J., Kamiyama S., Kim D-H., Magoori K., Kamataki A., Ito Y., Takei Y. A., Sasaki M., Suzuki T., et al. . 2003. Expression cloning and characterization of a novel glycosylphosphatidylinositol-anchored high density lipoprotein-binding protein, GPI-HBP1. J. Biol. Chem. 278: 7344–7349. [DOI] [PubMed] [Google Scholar]
- 11.Mysling S., Kristensen K. K., Larsson M., Beigneux A. P., Gardsvoll H., Fong L. G., Bensadouen A., Jorgensen T. J., Young S. G., and Ploug M.. 2016. The acidic domain of the endothelial membrane protein GPIHBP1 stabilizes lipoprotein lipase activity by preventing unfolding of its catalytic domain. eLife. 5: e12095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Franssen R., Young S. G., Peelman F., Hertecant J., Sierts J. A., Schimmel A. W. M., Bensadoun A., Kastelein J. J. P., Fong L. G., Dallinga-Thie G. M., et al. . 2010. Chylomicronemia with low postheparin lipoprotein lipase levels in the setting of GPIHBP1 defects. Circ Cardiovasc Genet. 3: 169–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Olivecrona G., Ehrenborg E., Semb H., Makoveichuk E., Lindberg A., Hayden M. R., Gin P., Davies B. S., Weinstein M. M., Fong L. G., et al. . 2010. Mutation of conserved cysteines in the Ly6 domain of GPIHBP1 in familial chylomicronemia. J. Lipid Res. 51: 1535–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Charrière S., Peretti N., Bernard S., Di Filippo M., Sassolas A., Merlin M., Delay M., Debard C., Lefai E., Lachaux A., et al. . 2011. GPIHBP1 C89F neomutation and hydrophobic C-terminal domain G175R mutation in two pedigrees with severe hyperchylomicronemia. J. Clin. Endocrinol. Metab. 96: E1675–E1679. [DOI] [PubMed] [Google Scholar]
- 15.Yamamoto H., Onishi M., Miyamoto N., Oki R., Ueda H., Ishigami M., Hiraoka H., Matsuzawa Y., and Kihara S.. 2013. Novel combined GPIHBP1 mutations in a patient with hypertriglyceridemia associated with CAD. J. Atheroscler. Thromb. 20: 777–784. [DOI] [PubMed] [Google Scholar]
- 16.Rios J. J., Shastry S., Jasso J., Hauser N., Garg A., Bensadoun A., Cohen J. C., and Hobbs H. H.. 2012. Deletion of GPIHBP1 causing severe chylomicronemia. J. Inherit. Metab. Dis. 35: 531–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coca-Prieto I., Kroupa O., Gonzalez-Santos P., Magne J., Olivecrona G., Ehrenborg E., and Valdivielso P.. 2011. Childhood-onset chylomicronaemia with reduced plasma lipoprotein lipase activity and mass: identification of a novel GPIHBP1 mutation. J. Intern. Med. 270: 224–228. [DOI] [PubMed] [Google Scholar]
- 18.Plengpanich W., Young S. G., Khovidhunkit W., Bensadoun A., Karnman H., Ploug M., Gardsvoll H., Leung C. S., Adeyo O., Larsson M., et al. . 2014. Multimerization of glycosylphosphatidylinositol-anchored high density lipoprotein-binding protein 1 (GPIHBP1) and familial chylomicronemia from a serine-to-cysteine substitution in GPIHBP1’s Ly6 domain. J. Biol. Chem. 289: 19491–19499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beigneux A. P., Franssen R., Bensadoun A., Gin P., Melford K., Peter J., Walzem R. L., Weinstein M. M., Davies B. S., Kuivenhoven J. A., et al. . 2009. Chylomicronemia with a mutant GPIHBP1 (Q115P) that cannot bind lipoprotein lipase. Arterioscler. Thromb. Vasc. Biol. 29: 956–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gonzaga-Jauregui C., Mir S., Penney S., Jhangiani S., Midgen C., Finegold M., Muzny D. M., Wang M., Bacino C. A., Gibbs R. A., et al. . 2014. Whole-exome sequencing reveals GPIHBP1 mutations in infantile colitis with severe hypertriglyceridemia. J. Pediatr. Gastroenterol. Nutr. 59: 17–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rabacchi C., D’Addato S., Palmisano S., Lucchi T., Bertolini S., Calandra S., and Tarugi P.. 2016. Clinical and genetic features of three patients with familial chylomicronemia due to mutations in GPIHBP1 gene. J. Clin. Lipidol. In press . [DOI] [PubMed] [Google Scholar]
- 22.Ariza M. J., Martinez-Hernandez P. L., Ibarretxe D., Rabacchi C., Rioja J., Grande-Aragon C., Plana N., Tarugi P., Olivecrona G., Calandra S., and Valdivielso P.. 2016. Novel mutations in the GPIHBP1 gene identified in 2 patients with recurrent acute pancreatitis. J. Clin. Lipidol. 10: 92–100.e1. [DOI] [PubMed] [Google Scholar]
- 23.Beigneux A. P., Fong L. G., Bensadoun A., Davies B. S., Oberer M., Gardsvoll H., Ploug M., and Young S. G.. 2015. GPIHBP1 missense mutations often cause multimerization of GPIHBP1 and thereby prevent lipoprotein lipase binding. Circ. Res. 116: 624–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beigneux A. P., Davies B. S., Tat S., Chen J., Gin P., Voss C. V., Weinstein M. M., Bensadoun A., Pullinger C. R., Fong L. G., et al. . 2011. Assessing the role of the glycosylphosphatidylinositol-anchored high density lipoprotein-binding protein 1 (GPIHBP1) three-finger domain in binding lipoprotein lipase. J. Biol. Chem. 286: 19735–19743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Henderson H. E., Hassan F., Marais D., and Hayden M. R.. 1996. A new mutation destroying disulphide bridging in the C-terminal domain of lipoprotein lipase. Biochem. Biophys. Res. Commun. 227: 189–194. [DOI] [PubMed] [Google Scholar]
- 26.Henderson H., Leisegang F., Hassan F., Hayden M., and Marais D.. 1998. A novel Glu421Lys substitution in the lipoprotein lipase gene in pregnancy-induced hypertriglyceridemic pancreatitis. Clin. Chim. Acta. 269: 1–12. [DOI] [PubMed] [Google Scholar]
- 27.Voss C. V., Davies B. S., Tat S., Gin P., Fong L. G., Pelletier C., Mottler C. D., Bensadoun A., Beigneux A. P., and Young S. G.. 2011. Mutations in lipoprotein lipase that block binding to the endothelial cell transporter GPIHBP1. Proc. Natl. Acad. Sci. USA. 108: 7980–7984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Machida T., Miyashita K., Sone T., Tanaka S., Nakajima K., Saito M., Stanhope K., Havel P., Sumino H., and Murakami M.. 2015. Determination of serum lipoprotein lipase using a latex particle-enhanced turbidimetric immunoassay with an automated analyzer. Clin. Chim. Acta. 442: 130–135. [DOI] [PubMed] [Google Scholar]
- 29.Chang S-F., Reich B., Brunzell J. D., and Will H.. 1998. Detailed characterization of the binding site of the lipoprotein lipase-specific monoclonal antibody 5D2. J. Lipid Res. 39: 2350–2359. [PubMed] [Google Scholar]
- 30.Liu M. S., Ma Y., Hayden M. R., and Brunzell J. D.. 1992. Mapping of the epitope on lipoprotein lipase recognized by a monoclonal antibody (5D2) which inhibits lipase activity. Biochim. Biophys. Acta. 1128: 113–115. [DOI] [PubMed] [Google Scholar]
- 31.Gin P., Beigneux A. P., Voss C., Davies B. S., Beckstead J. A., Ryan R. O., Bensadoun A., Fong L. G., and Young S. G.. 2011. Binding preferences for GPIHBP1, a glycosylphosphatidylinositol-anchored protein of capillary endothelial cells. Arterioscler. Thromb. Vasc. Biol. 31: 176–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ben-Zeev O., Mao H. Z., and Doolittle M. H.. 2002. Maturation of lipoprotein lipase in the endoplasmic reticulum. Concurrent formation of functional dimers and inactive aggregates. J. Biol. Chem. 277: 10727–10738. [DOI] [PubMed] [Google Scholar]
- 33.Beigneux A. P., Gin P., Davies B. S. J., Weinstein M. M., Bensadoun A., Fong L. G., and Young S. G.. 2009. Highly conserved cysteines within the Ly6 domain of GPIHBP1 are crucial for the binding of lipoprotein lipase. J. Biol. Chem. 284: 30240–30247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wong H., Davis R. C., Nikazy J., Seebart K. E., and Schotz M. C.. 1991. Domain exchange: characterization of a chimeric lipase of hepatic lipase and lipoprotein lipase. Proc. Natl. Acad. Sci. USA. 88: 11290–11294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sattler W., Levak-Frank S., Radner H., Kostner G. M., and Zechner R.. 1996. Muscle-specific overexpression of lipoprotein lipase in transgenic mice results in increased alpha-tocopherol levels in skeletal muscle. Biochem. J. 318: 15–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lookene A., Groot N. B., Kastelein J. J., Olivecrona G., and Bruin T.. 1997. Mutation of tryptophan residues in lipoprotein lipase. Effects on stability, immunoreactivity, and catalytic properties. J. Biol. Chem. 272: 766–772. [DOI] [PubMed] [Google Scholar]
- 37.Wong H., Davis R. C., Thuren T., Goers J. W., Nikazy J., Waite M., and Schotz M. C.. 1994. Lipoprotein lipase domain function. J. Biol. Chem. 269: 10319–10323. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.