Abstract
Membrane contact sites (MCSs) are regions of close apposition between different organelles that contribute to the functional integration of compartmentalized cellular processes. In recent years, we have gained insight into the molecular architecture of several contact sites, as well as into the regulatory mechanisms that underlie their roles in cell physiology. We provide an overview of two selected topics where lipid metabolism intersects with MCSs and organelle dynamics. First, the role of phosphatidic acid phosphatase, Pah1, the yeast homolog of metazoan lipin, toward the synthesis of triacylglycerol is outlined in connection with the seipin complex, Fld1/Ldb16, and lipid droplet formation. Second, we recapitulate the different contact sites connecting mitochondria and the endomembrane system and emphasize their contribution to phospholipid synthesis and their coordinated regulation. A comprehensive view is emerging where the multiplicity of contact sites connecting different cellular compartments together with lipid transfer proteins functioning at more than one MCS allow for functional redundancy and cross-regulation.
Keywords: endoplasmic reticulum, mitochondria, vacuole, organelle, phosphatidic acid, triacylglycerol
The pervasive importance of lipids in cell biology has become evident. From structural roles defining cellular compartments and surfaces with specific biological properties to functional roles in signal transduction processes and modulation of regulatory networks, the involvement of lipids in varied cellular functions is well-established. Concurrently, we have gained a broad understanding about the repertoire of proteins involved in synthesis, degradation, transport, and sensing of lipids, as well as the regulatory mechanisms that interconnect lipid metabolism with other cellular processes. The amenability of Saccharomyces cerevisiae to classical approaches of molecular genetics, and to high throughput analyses, makes this yeast a powerful organism to unveil the intricacies of this field. The yeast S. cerevisiae has played a foundational role in the field of molecular and cellular biology of lipids, including the identification of membrane contact sites (MCSs) and how they link lipid metabolism with organelle dynamics. We present advances in two topics where extensive and outstanding contributions have recently been made where their relevance transcends the yeast lipid field. We first review new information about the phosphatidic acid (PA) phosphatase, Pah1, the yeast homolog of human lipin, and its role in directing lipid flux into membrane synthesis or storage. We review the contribution of Pah1 to lipid droplet (LD) formation and its interaction with the seipin complex, Fld1/Ldb16, to regulate endoplasmic reticulum (ER)-LD contact site dynamics. Second, we recapitulate recent developments revealing MCS connections between the ER and mitochondria with other cellular compartments, and how these intersect with the maintenance of lipid homeostasis. Furthermore, we explore MCS redundancies that allow cells to cope with changing metabolic demand.
PA METABOLISM CO-COORDINATES MEMBRANE AND LD SYNTHESIS
PA is the common precursor for the synthesis of membrane phospholipids (PLs) and the major component of LDs, such as triacylglycerol (TAG) (Fig. 1). The de novo biosynthesis of PA, as well as its consumption for PL and TAG synthesis, takes place in the ER (1, 2). The size of the pool of PA localized at the ER impacts the expression of the genes involved in PL synthesis, as a transcriptional regulatory module directly connects PA availability at the ER with gene expression of PL biosynthetic genes in the nucleus (3, 4). In addition, protein synthesis dedicated to the secretory system proceeds through the ER and is coordinated through a regulatory network with the lipid biosynthetic capacity of the ER (5–8). The combined functional proficiency of these cellular processes is required for normal growth. The activity of Pah1, the yeast homolog of human lipin, has a prominent role, as this enzyme diverts the flux of PA from PL synthesis into TAG synthesis for accumulation into LDs based on the cellular need of membrane expansion and cell growth versus cessation of growth and lipid storage (9, 10). Consistent with such a central function, Pah1 is highly regulated posttranslationally. Pah1 is kept soluble and away from its membrane substrate, PA, by phosphorylation (11–13). Upon dephosphorylation by the conserved ER-localized protein phosphatase, Nem1/Spo7, Pah1 becomes membrane bound and active, generating diacylglycerol (DAG) for TAG synthesis (14, 15). Remarkably, the active form of Pah1 is unstable and rapidly degraded, resulting in a tightly controlled system for regulation of membrane versus storage lipid synthesis (16, 17). Interestingly, LDs are tightly associated with the ER at MCSs (18–21), and the activation and membrane recruitment of Pah1 takes place in the vicinity of LDs, suggesting a channeling mechanism of DAG delivery for TAG synthesis and sorting into LDs (22–25).
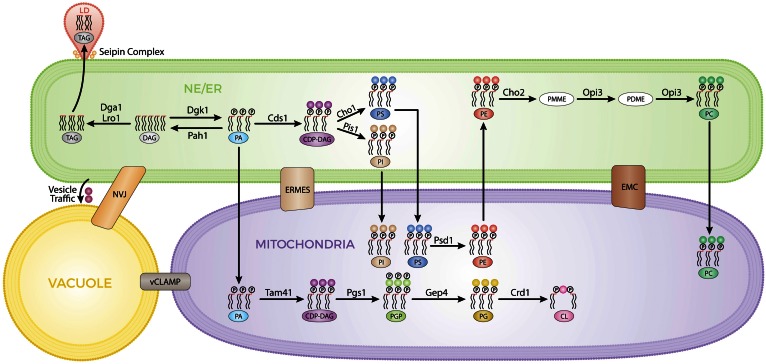
Fig. 1.
MCSs enable metabolic flux through different compartments for PL and TAG syntheses. This figure outlines several lipid biosynthetic pathways and their locations. The MCSs bridging the gaps between the different compartments are indicated. PA is the key lipid precursor for the synthesis of TAG and PLs taking place at the nuclear envelope (NE)/ER (green). TAG is sorted into LDs (pink) connected to the ER by the seipin complex. Most of the reactions for PL synthesis take place at the ER. PS decarboxylation mediated by Psd1 and generating PE occurs in the mitochondria (purple) with transport of PS and PE indicated by arrows across these compartments. Transports of PI, PC, and PA (for the synthesis of the specific mitochondrial lipids PG and CL) into mitochondria are also indicated by arrows from the ER and into mitochondria. ER-mitochondria encounter structure (ERMES) and ER membrane protein complex (EMC) are represented as boxes connecting both compartments. An alternate route through the vacuole (yellow) bridging ER and mitochondria is shown. Vacuole and NE/ER are connected indirectly by vesicle traffic through the endomembrane system, as well as by the NVJ contact site. The vacuolar and mitochondrial patch (vCLAMP) contact site is represented as a box connecting both compartments.
Below, we briefly describe the necessary conceptual framework regarding PA metabolism, which is reviewed in depth elsewhere (2, 10, 26, 27), to enable facile insight into the relevance of the research we review here. PA is synthesized in the ER where it is activated to CDP-DAG for subsequent synthesis of membrane lipids by the ER resident enzyme, Cds1 (Fig. 1). The high energy CDP-DAG is consumed at the ER for the synthesis of phosphatidylinositol (PI) catalyzed by the PI synthase, Pis1, and phosphatidylserine (PS) catalyzed by the PS synthase, Cho1. PS, in turn, generates phosphatidylethanolamine (PE) by decarboxylation catalyzed primarily by mitochondrially-localized Psd1, with a small contribution from Golgi/endosome localized Psd2. PE is subsequently converted into phosphatidylcholine (PC) by three successive methylations that take place in the ER. The first methylation is carried out by Cho2 and the second and third by Opi3. PA is also transported to the mitochondria where it is converted to CDP-DAG by the activity of the mitochondrial CDP-DAG synthase, Tam41 (28), for subsequent synthesis of the mitochondrial-specific lipids, phosphatidylglycerol (PG) by Pgs1/Gep4 and cardiolipin (CL) by Crd1 (Fig. 1) (29–31).
The PL, PA functions at a critical branching point in lipid metabolism where lipid biosynthetic flux can be directed into membrane PL synthesis concomitant with membrane proliferation, or into TAG synthesis and LD formation (Fig. 1). To generate TAG, PA is dephosphorylated at the ER to form DAG by the PA phosphatase, Pah1 (9). DAG is acylated by the acyltransferases, Dga1 and Lro1, to produce TAG for accumulation in LDs (32–35). DAG can also be phosphorylated back to PA by the ER-localized lipid kinase, Dgk1 (36, 37). There are three other PA phosphatases in yeast, Dpp1, Lpp1, and App1, which, together with Pah1, constitute the whole measurable PA phosphatase complement in yeast extracts (38). Dpp1, Lpp1, and App1 are not involved in diverting PA from PL synthesis to generate DAG for TAG synthesis, as the DAG, TAG, and PL relative composition of the triple dpp1Δ lpp1Δ app1Δ mutant do not differ appreciably from wild-type cells (38). Instead, Dpp1, Lpp1, and App1 are thought to be involved in lipid signaling or membrane structural/curvature changes associated with vesicular trafficking (10, 38).
The physiological relevance of Pah1 has been uncovered by studying pah1Δ yeast strains (9, 14, 22, 39–41). The absence of Pah1 leads to elevated levels of PA and other PLs, which is thought to cause the observed hyperproliferation of the nuclear ER (10, 11, 36). This metabolic block also leads to reduced amounts of DAG and TAG, a reduced number of LDs, and lipotoxicity, as the reduced level of DAG observed in pah1Δ cells hampers TAG synthesis and the ability to cope with excess exogenous fatty acids (9, 22, 39, 42). Additional phenotypes of pah1Δ cells are slow growth, thermosensitivity, and an impairment of pah1Δ cells to grow in nonfermentable carbon sources. The inability to grow on nonfermentable carbon sources is not believed to be due to a respiratory defect, instead a reduction of ATP level observed in pah1Δ cells as a consequence of deregulated PL synthesis (which consumes ATP) was proposed as the limiting factor for growth (41). In addition, pah1Δ cells exhibited an increased level of mitochondrial superoxides and a decreased tolerance to oxidative stress resulting in reduced chronological life span (41). Cells defective in Pah1 activity also displayed vacuolar fragmentation and decreased vacuolar fusion consistent with the observed reduction in vacuolar recruitment of several protein factors implicated in the vacuolar fusion process (40). Of interest, Pah1 has also been associated with the vacuolar membrane and the conversion from PA to DAG was proposed as a critical event leading to vacuole fusion (40).
PA can also be synthesized by Dgk1, an atypical DAG kinase that utilizes CTP for the synthesis of PA (37) (Fig. 1). Dgk1 is an integral membrane protein localized at the ER/nuclear membrane. Dgk1 antagonizes Pah1 activity, and they constitute a counteracting pair that controls the levels of PA and DAG at the nuclear ER. Indeed, Dgk1 was identified in two screens for multicopy suppressors of the lethality that arises from deregulated Pah1 activity (36). Dgk1 overexpression leads to an increased level of PA, derepression of UASINO-containing genes (see subsequent section), and nuclear membrane proliferation, phenotypes also observed in pah1Δ cells. Inactivation of the DGK1 gene suppresses phenotypes elicited by Pah1 deficiency, including elevated PA level, abnormal nuclear/ER morphology, and derepressed UASINO gene transcription (36). This interaction suggests that Dgk1 contributes to the PA pool that is a substrate for Pah1. In addition, the absence of Dgk1 restores LD formation to wild-type level in pah1Δ cells (22, 39). The susceptibility of pah1Δ cells to oxidative stress and their inability to grow in nonfermentable carbon sources were partially suppressed by loss of Dgk1 (41), while other phenotypes of pah1Δ cells, including decreased TAG synthesis and lipotoxicity, were not suppressed by Dgk1 deficiency (36, 39). This suggests that only a subset of the PA pool produced by Dgk1 overlaps with the function of that consumed by Pah1.
PA DIRECTLY BINDS TRANSCRIPTIONAL REGULATORS OF LIPID SYNTHESIS
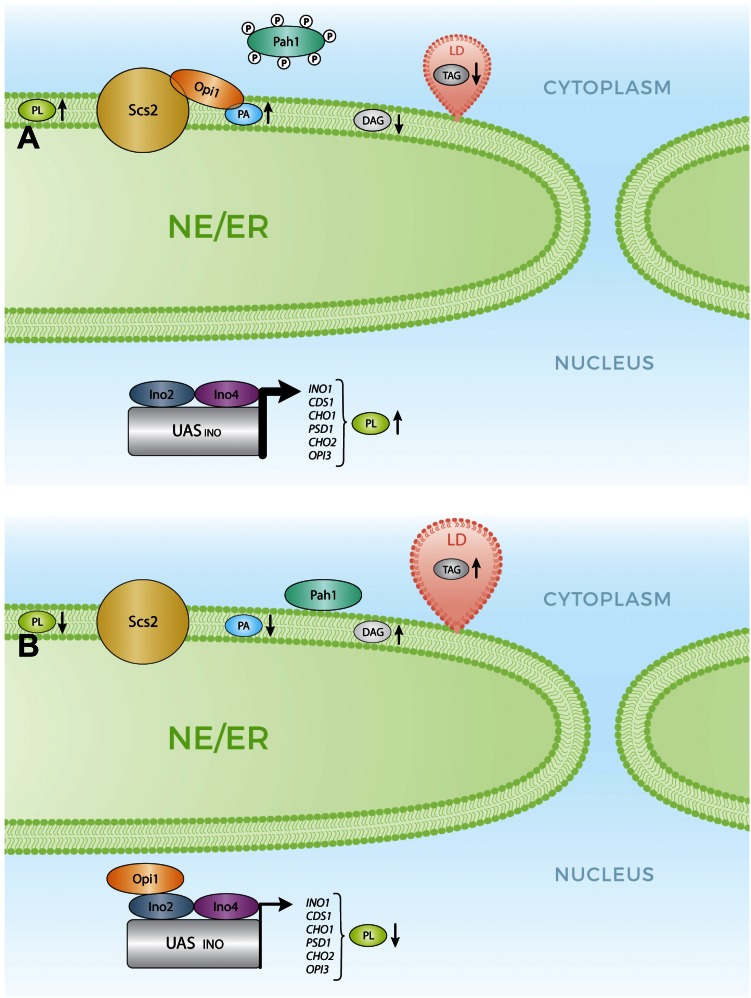
In addition to its function as a central precursor for PL and TAG syntheses, the pool of PA localized at the ER plays a regulatory role in modulating the expression of genes required for PL and fatty acid synthesis primarily through repressing transcription from genes containing an UASINO promoter element (3, 4, 43) (Fig. 2). Opi1 is a transcriptional repressor that senses the level of PA at the ER (43), and through the simultaneous interaction of PA and the integral ER protein, Scs2, Opi1 is bound to the ER and is inactive. A drop in PA level allows for Opi1 detachment from the ER membrane and translocation into the nucleus where it acts as a transcriptional repressor. The genes controlled by this PA-Opi1-regulated circuit contain the UASINO element in their promoters where the transcriptional activators, Ino2/Ino4, bind and promote transcription. Opi1 directly interacts with Ino2 and represses transcription. Thus, high levels of PA allow the transcription of PL synthesis genes, while low levels of PA do not prevent Opi1-mediated gene repression.
Fig. 2.
Activation status of PA phosphatase, Pah1, impacts on PL and NL homeostasis. A: Pah1 in its hyperphosphorylated form is kept off the ER and unable to reach its substrate, PA. An increased ER level of PA recruits the transcriptional repressor, Opi1, to the ER membrane through the simultaneous interaction with the integral protein, Scs2, allowing for the expression of genes involved in PL synthesis. A reduced level of DAG attenuates TAG synthesis and LD formation. B: Dephosphorylated Pah1 binds to ER membranes and hydrolyzes PA generating DAG leading to TAG synthesis and LD formation. Upon reduction of the PA level at the ER, Opi1 is released and translocates to the nucleus where it represses transcription and decreasing PL synthesis.
The PA-Opi1 regulatory circuit is highly responsive to inositol availability. INO1 encodes for inositol-3-phosphate synthase, which catalyzes the rate-limiting enzyme for the de novo synthesis of inositol. The amplitude of INO1 induction is the highest among the genes controlled by the PA-Opi1 regulatory circuit. Under inositol deprivation, INO1 is maximally expressed and PI synthesis depends on the rate of de novo inositol formation (Fig. 2). Under this growth condition, the total rate of CDP-DAG consumption does not exceed the rate of PA synthesis, leading to high levels of PA. Upon inositol supplementation, a dramatic drop of PA levels takes place as CDP-DAG is rapidly consumed by the high rate of PI synthesis resulting in Opi1 repressing INO1 transcription, along with other UASINO-containing genes (2, 4, 43). Other metabolic inputs, different from inositol, that impact on PA levels at the ER would affect Opi1-mediated PL synthesis gene expression. Indeed, the pah1Δ mutant exhibits elevated levels of PA and deregulated expression of UASINO-containing genes (9, 11, 14), while, conversely, the expression of a highly active hypophosphorylated Pah1 variant leads to a reduced level of PA and severe Opi1-mediated UASINO transcriptional repression (11).
PA PHOSPHATASE: REGULATING A MASTER REGULATOR
Pah1 is highly regulated through reversible phosphorylation, which in turn governs Pah1 membrane association, catalytic activity, and stability. Multiple phosphorylations take place on Pah1, catalyzed by direct phosphorylation by Cdc28-cyclinB kinase, Pho85-Pho80 kinase, and protein kinase A (10, 12, 13, 44). In its hyperphosphorylated form, Pah1 remains cytosolic and inactive (Fig. 3). The PA phosphatase activity of Pah1 is inhibited by direct phosphorylation of Pah1 by Pho85-Pho80 kinase or protein kinase A (13, 44). Dephosphorylation mediated by Nem1/Spo7 reactivates Pah1 (45). Pah1 is also phosphorylated by protein kinase C and protein kinase CKII (45, 46). Phosphorylation by protein kinase C only slightly increases Pah1 phosphatase activity, without noticeable effects on Pah1 localization or TAG synthesis (45). Phosphorylation by protein kinase CKII diminishes Pah1 phosphatase activity very modestly (46). Interestingly, the expression of a mutated variant of Pah1 carrying a phosphorylation-mimicking mutation at one of the CKII target sites combined with the seven nonphosphorylatable residues for Pho85-Pho80 kinase promotes a significant increase in TAG accumulation and LD number per cell (46). Of note, protein kinase CKII also targets Opi1, leading to its activation (47). Signaling through protein kinase CKII impinges on lipid metabolism, attenuating PL synthesis through the activation of Opi1 and by diverting PA into TAG synthesis.
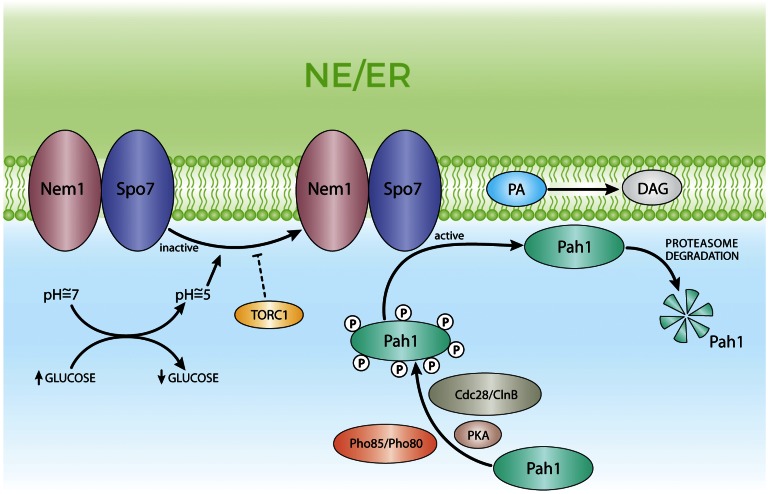
Fig. 3.
Mechanisms of regulation of PA phosphatase, Pah1. Pah1 is phosphorylated by protein kinase A (PKA), Pho80/Pho85, and Cdc28/ClnB. In its hyperphosphorylated status, Pah1 is inactive and kept away from the membranes. The ER resident phosphatase complex, Nem1/Spo7, dephosphorylates and activates Pah1. Pah1 translocates to the ER membrane reaching its substrate PA and generates DAG. The active form of Pah1 is a substrate of the proteasome, having a reduced half-life. In turn, the phosphatase complex, Nem1/Spo7, is activated by cytosol acidification upon glucose exhaustion through an uncharacterized mechanism. Activated TORC1 prevents Nem1/Spo7 activation promoting low levels of Pah1 activity.
The nuclear/ER resident phosphatase complex, Nem1/Spo7, is responsible for Pah1 dephosphorylation and activation (15). This process depends on the interaction of the acidic C-terminal tail of Pah1 with Nem1/Spo7 (23). Upon Nem1/Spo7-dependent dephosphorylation, Pah1 becomes membrane bound through its N-terminal amphipathic helix and can access its substrate, PA, generating DAG (48) (Fig. 3). TORC1 kinase participates in the regulation of Pah1 activity by modulating the activity of the Nem1/Spo7 phosphatase complex. Inhibition of TORC1 indirectly preserves the phosphorylation status of Nem1 and leads to the activation of Pah1 (49). Further evidence for the tight regulation of Pah1 activity by reversible phosphorylation arises from the observation that phenotypes provoked by the absence of Pah1 are similar to those elicited by Nem1 or Spo7 deficiency: PA builds up, there is upregulation of UASINO-containing genes, and there is aberrant nuclear/ER membrane expansion and reduced TAG content, which correlates with altered LD morphology (12, 14, 18, 22, 50, 51). The toxic effect of Nem1/Spo7 overexpression depends on the presence of Pah1 stressing the specific role of this phosphatase on the activation of Pah1 (14).
The steady state level of Pah1 substantially decreases as cells enter stationary phase (52, 53). Nonphosphorylatable variants of Pah1 also exhibit diminished steady state levels (12, 13). In support of the notion that the phosphorylation status of Pah1 modulates its abundance, it was shown that a defective Nem1/Spo7 phosphatase complex attenuated the diminution of Pah1 level for cells reaching late exponential phase (13), as did rapamycin-mediated inhibition of TORC1 (49). It was also noted that Pah1 level is higher in stationary phase cells treated with a proteasome inhibitor or defective in the main proteolytic activity of the proteasome (16). Using purified preparations of proteasome and a nonphosphorylated version of Pah1 purified from E. coli, in vitro experiments showed that Pah1 is degraded by the 20S proteasome in a ubiquitin-independent manner. Whereas phosphorylation by protein kinase CKII did not affect the susceptibility of Pah1 to 20S proteasome-mediated degradation (46), phosphorylation by protein kinase C slightly increased Pah1 instability and phosphorylation by protein kinase A or Pho85-Pho80 kinase attenuated Pah1 degradation (17). Consistent with this observation, in vivo analysis using nonphosphorylatable variants of Pah1 showed that abrogation of protein kinase C phosphorylation prevented the decrease in abundance of a Pah1 variant refractory to Pho85-Pho80 kinase phosphorylation (45). Using Pah1 purified from S. cerevisiae, which would be phosphorylated, it was observed that dephosphorylation by Nem1/Spo7 made Pah1 more susceptible to 20S proteasome degradation (17). The accumulated evidence suggests that changes in Pah1 phosphorylation are part of a sensitization mechanism whereby fully active dephosphorylated Pah1 becomes more susceptible to proteasome degradation. This mechanism allows for acute control of Pah1 to prevent the known toxic effects of unrestricted PA hydrolytic activity.
Pah1 is also regulated at transcriptional and posttranslational levels, coordinating lipid metabolism with other cellular processes. At the transcriptional level, Pah1 expression is regulated by growth phase. The expression of reporter genes driven from the PAH1 promoter increased as cells progressed into stationary phase, congruent with elevated PA phosphatase activity and TAG accumulation (53). However, this increment was not observed at the level of Pah1 abundance. Instead, as cells progressed into stationary phase and Pah1 became more active enzymatically, its steady state level decreased. As discussed above, proteasome-dependent degradation controls Pah1 abundance in stationary phase cells. Inositol supplementation did not affect the expression of a reporter gene driven by the PAH1 promoter for cells growing exponentially; whereas for cells in stationary phase, the presence of inositol in the growth media significantly increased the expression of the reporter gene. Although the canonical response through the Ino2/Ino4/Opi1 regulatory circuit to inositol availability is normally transcriptional repression, the fact that the single deletion of the INO2, INO4, or OPI1 gene erased the growth phase induction and the stimulatory effect of inositol on the expression of the PAH1 reporter gene supports the involvement of this regulatory circuit on the transcriptional control of PAH1 gene. Putative binding sites for the histone demethylases/transcription factors, Gis1 and Rph1, are present in the PAH1 promoter. In gis1Δ and rph1Δ mutants, PAH1 reporter gene expression showed loss of induction in stationary phase, as well as loss of inositol stimulation (53). Overall, these results reveal a complex network of transcriptional factors controlling the expression of PAH1 during growth, whose precise mechanism(s) has yet to be determined.
The accumulated data clearly depict the importance of phosphorylation on the regulation of Pah1 function by affecting localization, catalytic activity, and stability. The involvement of direct phosphorylation of Pah1 by Cdc28-cyclin B kinase, Pho85-Pho80 kinase, protein kinase A, protein kinase C, and protein kinase CKII, as well as Pah1 dephosphorylation by the Nem1/Spo7 phosphatase, is well-established. Other protein kinases can phosphorylate Pah1 in vitro and many more phosphorylation sites have been identified on Pah1 that remain to be characterized (11, 54–57), underscoring the role of Pah1 as a signaling target for coordinating lipid metabolism with cellular processes.
PA PHOSPHATASE BALANCES MEMBRANE EXPANSION WITH LIPID STORAGE
Cell growth transition from exponential growth phase into stationary phase encompasses major metabolic changes. At the lipid level, this transition involves a switch from membrane synthesis to lipid storage, as revealed by the increase in cellular TAG and the accumulation of LDs. For cells growing logarithmically in glucose media, Pah1 was primarily cytoplasmic. Upon glucose exhaustion, Pah1 transiently localized to the nuclear membrane that contacts the vacuole [nuclear vacuolar junction (NVJ)] and then accumulated at two foci flanking the NVJ and in contact with LDs (58). However, the transient localization of Pah1 at the NVJ was not required for the lipid metabolic function of Pah1, as TAG and LD levels were not affected in nvj1Δ cells that had reached stationary phase. The localization of Pah1 with LDs and the increase in neutral lipid (NL) synthesis take place simultaneously. Analogously, acute glucose withdrawal triggered Pah1 localization into the NVJ, LDs, and other punctuate structures in cells growing exponentially (58). Cellular glucose depletion led to cytosol acidification followed by Nem1/Spo7 activation that, in turn, dephosphorylated Pah1. Dephosphorylated Pah1 binds to membranes and has access to its substrate, PA. The details of the mechanism leading to Nem1/Spo7 activation upon cytosol acidification have not been solved: mere acidification could activate the phosphatase or it could be mediated by TORC, as it is known that cytosol acidification upon glucose exhaustion activates TORC, and that Nem1/Spo7 is a downstream target of this kinase (49).
There is evidence that the Nem1/Spo7 complex and membrane bound Pah1 localize together at the vicinity of LDs, suggesting a channeling mechanism from PA hydrolysis to TAG synthesis and sorting into LDs. It was shown that Spo7 exhibited a typical ER distribution, whereas Pah1 exhibited a diffuse cytosolic localization for exponentially growing cells. Upon the overexpression of Dgk1 and consequent increase of PA at the ER, Spo7 concentrated to a crescent shape where Pah1 also colocalized (48), and this punctum was in close proximity to LDs (22). Upon Nem1 and Spo7 overexpression, Pah1 was recruited to the ER adjacent to LDs and led to an increased number of LDs per cell and formation of LD clusters (23).
In the absence of TAG and sterol ester synthesis, and consequent LD formation, Pah1 still targeted the nuclear envelope at stationary phase. It was observed that the recruitment of Pah1 to the nuclear membrane upon entry into stationary phase for cells unable to form LDs was accompanied by membrane proliferation, as revealed by altered nuclear morphology and organization of cortical ER. Similar defects were also observed when only TAG synthesis was prevented by inactivation of the DGA1 and LRO1 genes. At stationary phase, cells unable to form LDs exhibited increased amounts of DAG, ergosterol, fatty acids, and PLs compared with wild-type cells (58). The localization of Pah1 has clear effects on lipid metabolism and the subsequent formation of membranes versus LDs.
THE MAKING AND BREAKING OF LDs
LDs are the organelles responsible for the cellular storage of the NLs, TAG, and sterol esters. Stored NLs are a source of precursors for membrane synthesis, as well as substrates for energy production. Typically, a LD is a 0.3–0.4 μm sphere organized as a core of NLs wrapped in a PL monolayer. This PL monolayer allows for specific associations with numerous proteins, some of which are specific for LDs, whereas others are shared with the ER, the compartment from which LDs originate. Reflecting the most relevant function of LDs, the vast majority of proteins associated with LDs participate in NL synthesis and degradation (26). Consistent with a role for the ER in lipid homeostasis, and in particular as a site where the flux of lipid precursors is directed toward membrane synthesis or storage, LDs are not only formed from the ER, but most of them remain physically connected with the ER (19, 20).
Genome-wide screens for altered LD morphology and distribution identified genes involved in LD function. Mutants defective in PC synthesis through the PE methylation pathway due to mutation in the CHO2 or OPI3 genes, as well as mutants devoid of the PL synthesis transcriptional activator, Ino2 or Ino4, exhibited very similar LD morphology phenotypes. Loss of function mutations for each of these genes resulted in a few supersized LDs (SLDs) (diameter >1 μm) when cells were grown in minimal media (SC media). This phenotype is conditional because supplementation of the growth media with choline or inositol, which drive PL synthesis, or growth on YPD-rich media, which contain both choline and inositol, restored LD morphology to a wild-type appearance (51, 59). Under limiting PL synthesis, it is thought that SLD formation is favored, as the surface/volume ratio of LDs is minimized, while fusogenic PLs such as PE and PA accumulate simultaneously.
The genetic screens for cells with altered LD morphology also identified mutations in FLD1 (18, 60). FLD1 is the yeast homolog of the human gene, Berardinelli-Seip congenital lipodystrophy type 2 (BSCL2), mutations in which cause a severe form of congenital general lipodystrophy. BSCL2 encodes seipin, an integral protein of the ER that assembles into oligomeric complexes at ER-LD contact sites. It was noted that loss-of-function of Fld1 results in a LD phenotype very similar to that observed in lbd16Δ cells (59). Consistent with this, Fld1 and Lbd16 form a protein complex where Fld1 interacts with itself and with Ldb16 at ER-LD contact sites, referred to as the seipin complex (59, 61). In the absence of Fld1, Ldb16 becomes unstable and is targeted to the ER-associated degradation machinery. Both Fld1 and Lbd16 have a similar topologic array where their N and C termini, separated by two transmembrane domains and a luminal loop, face the cytosol. In spite of the similar domain architecture and the common function, there is no homology between these two proteins. Interestingly, the expression of human seipin complements the simultaneous absence of both proteins, as well as the absence of each one individually, suggesting that the function of the yeast seipin complex constituted by Fld1 and Ldb16 is performed by human seipin (59). The absence of Fld1 or Lbd16 elicits similar LD phenotypes, as well as similar perturbations on lipid metabolism. Typically, fld1Δ and ldb16Δ cells display a heterogeneous LD phenotype that depends on growth condition (18, 20, 25, 51, 59, 60). When grown in minimal media, most of the fld1Δ and ldb16Δ mutant cells present one or a few SLDs. The rest of the cell population develops one or two aggregates of small LDs entangled with ER membranes. Growth in YPD-rich media or in minimal media supplemented with inositol to drive PL synthesis results in the majority of cells containing aggregates of ER-entangled LDs and a small fraction presenting with a few SLDs. Under both media conditions, cells exhibited many very small LDs scattered throughout the cytoplasm. A possible contribution to the heterogeneity of LD morphology arises from the defect in LD inheritance observed for fld1Δ (20).
Deletion of the CHO2 gene, which decreases PC synthesis through the methylation pathway and whose inactivation also results in abnormal LD morphology, prevented the inositol-induced formation of ER-entangled LD aggregates (25, 51, 59), supporting the idea that augmented PL synthesis contributes to the change of LD morphology observed in seipin complex mutants upon inositol supplementation. This idea was further supported whereby overexpression of the CDP-DAG synthase encoded by CDS1, which catalyzes the conversion of PA into CDP-DAG that is the lipid precursor for the synthesis of most PLs, also promoted the LD morphology shift from SLD to LD aggregates (21).
THE CONTRIBUTION OF ER-LD CONTACT SITES TO LD FORMATION AND DYNAMICS
Several observations suggest that the Fld1/Ldb16 seipin complex could coordinate NL synthesis with LD biogenesis. The absence of the seipin complex delayed LD emergence leading to the accumulation of NLs in ER membranes (62) and the formation of LDs inside the nucleus (25, 62), revealing that the Fld1/Ldb16 complex directs LD emergence toward the cytosol. Interestingly, yeast cells devoid of the seipin complex also exhibit abnormal proliferation of the nuclear ER (21, 25, 59), a similar morphological feature also observed in cells defective in the PA phosphatase, Pah1. As discussed above, enrichment of PA at the ER upon PAH1 inactivation prevents Opi1 repression of UASINO-regulated genes. Derepressed levels of INO1 and OPI3, among other UASINO-regulated genes, were reported for both fld1Δ and ldb16Δ cells, suggesting that the absence of the seipin complex perturbs PA homeostasis and interferes with Opi1 function (51, 59, 63). However, unrestricted PL synthesis might not be the only cause for abnormal nuclear ER because opi1Δ cells exhibit wild-type nuclear ER (as well as normal LD morphology).
Alteration of PA homeostasis at the nuclear ER together with increased PL synthesis could trigger hyperproliferation of nuclear ER membranes. Indeed, three independent research groups reported that PA binding probes, such as Opi1, Pah1, and a reporter derived from Spo20 (Spo20 51-91), form puncta localized at, or very close to, LDs entangled with ER membranes in seipin complex-deficient cells (21, 24, 25). It has been reported that there is a slight increase in PA in the microsomal fraction isolated from fld1Δ cells (51). Coordination of LD formation with NL synthesis implies the localization of Pah1 (and other enzymes involved in NL synthesis) close to the seipin complex. Pah1 is recruited to nuclear membrane foci in contact with LDs (23, 58) and is required for LD biogenesis (22, 23, 62). Interestingly, in mammalian cells, it was observed that seipin interacts with lipin1, the human homolog of Pah1 (64). Thus, the seipin complex might function as a nucleation spot for NL biosynthetic enzymes.
Compatible with its proposed role facilitating LD biogenesis, the seipin complex could also work as a diffusion barrier delimiting LDs from the ER and thus contributing to LD identity. A recent proteome analysis of LDs isolated from fld1Δ and ldb16Δ cells showed that both mutants have a very similar LD proteome, but one that is very different to wild-type LDs. Many proteins residing in wild-type LDs were strongly reduced in LDs isolated from seipin complex-deficient cells (21). Conversely, several peripheral proteins carrying known or predicted lipid binding or membrane interacting domains were associated to LDs isolated from fld1Δ and ldb16Δ cells, but were not present in wild-type LDs (21). The absence of the seipin complex not only affected the partitioning of bona fide LD resident proteins, but also likely altered the LD PL monolayer, as revealed by the presence of ectopic peripheral proteins. LDs from seipin complex mutant cells are irregular in shape and their PL monolayer is wrinkled around the NL core (21). In addition, the ER-LD contact sites are expanded and bloated (21, 25). The assembly of purified Fld1 into a nonamer of toroid form (61), plus the evidence that Ldb16 interacts with Fld1 forming a macromolecular complex of still undefined structure (59), supports the idea of the seipin complex acting as a diffusion barrier at the ER-LD contacts sites.
Among the ectopic peripheral proteins bound to LDs in fld1Δ or ldb16Δ cells, the most abundant was Opi1 (21). Contemporarily with this report, two other groups showed the formation of Opi1 foci associated with LD in seipin complex-deficient cells (24, 25). In its membrane bound form, Opi1 interacts with both PA and the ER integral protein, Scs2. Remarkably, in seipin complex-deficient cells, Opi1 remained bound to LDs even if its Scs2-interacting motif was mutated or Scs2 was absent (21, 24). Analysis using the PA binding probe, Spo2051-91, consistently showed punctua associated with LDs from fld1Δ and ldb16Δ cells, further supporting the notion that the absence of seipin complex leads to PA accumulation in LDs (21, 24, 25). Based on the observation that the addition of inositol (and consequent decrease of total cellular PA) increased the number of mutant cells displaying GFP- Spo2051-91 foci, Grippa et al. (21) did not favor the notion that the recruitment of Opi1 and Spo2051-91 to LDs is due to binding to PA. Instead, they proposed that membrane-packing defects on the LD monolayer might drive the binding of these proteins to LDs. Supporting this idea, they showed that other ectopic peripheral proteins that bind membranes through an amphipathic helix were increased in LDs from seipin complex-deficient cells. Contrarily, Han et al. (24) proposed that the puncta represents clusters of PA molecules that are metabolically isolated and would define the neck between the ER and LD in the absence of of the seipin complex. The PA molecules were generated from multiple sources as the single ablation of genes involved in PA synthesis, such as GPT2, SLC1, ALE1, LOA1, DGK1, and SPO14, as well as increased expression of enzymes that consume PA, such as Pah1 or Cds1, did not hamper Opi1 recruitment to LDs.
PERSPECTIVES ON Pah1 BIOLOGY: SIGNALING INPUTS AND COMPARTMENTALIZATION
The ability to divert the lipid biosynthetic flux into NLs and to segregate them into LDs has a major role in lipid homeostasis and contributes to normal cell physiology. In particular, TAG synthesis buffers the excess of lipid precursors not consumed for PL synthesis preventing lipotoxicity. In addition, the nutritional status impacts on the balance between PL synthesis and energy and membrane precursor storage. The PA phosphatase, Pah1, plays an essential role in this process. The several layers of regulation that modulate its function reveal the multiplicity of signals impinging on Pah1 activity. The activation of Pah1 mediated by the Nem1/Spo7 phosphatase complex is a critical step in this regulation. It is known that cytosolic acidification and the TORC pathway affect Nem1/Spo7 phosphatase activity. It would be important to identify what other signals might regulate the Nem1/Spo7 complex. Considering that the Nem1/Spo7 complex resides in the ER, it is plausible that this complex senses, directly or indirectly, membrane status and transduces this information modulating Pah1 activity. Another important aspect of this process, regarding the functional integrity of the ER, is the clustering of the factors required for TAG synthesis and LD formation. An orderly array of proteins and lipids facilitates substrate channeling and limits the distribution and concentration of DAG and TAG very near to or at LDs, preventing their potential disruptive effect on ER structure. Exploring the network of protein-protein and protein-lipid interactions among the factors involved in this process would contribute to an understanding of the mechanisms and regulations underlying the dynamics of the contact sites connecting the ER and LDs.
MAKING MITOCHONDRIAL MEMBRANES: THE CONTRIBUTION OF ER-MITOCHONDRIAL CONTACT SITES TO MITOCHONDRIAL MEMBRANE SYNTHESIS AND DYNAMICS
Different cellular compartments have characteristic lipid compositions (65, 66). Furthermore, lipids can be distributed asymmetrically across the leaflets of a membrane bilayer (67). Thus, the concerted synthesis, degradation, modification, and trafficking of lipids generate and sustain the highly heterogeneous lipid composition of organellar membranes (68–70). Mitochondria are organelles of endosymbiotic origin that, over evolution, have transferred most of their protein encoding genes into the cell host nucleus, with only a few genes still residing in the mitochondrial DNA (mtDNA) (71). Dedicated mitochondrial machineries allow for the import, folding, and assembly of the nuclear-encoded proteins required for proper mitochondrial physiology (72–75). Mitochondria are also equipped with a family of carriers that facilitate the transport of small soluble molecules like amino acids, nucleotides, and vitamins across the mitochondrial inner membrane, necessary for the interdependent metabolism at both sides of the mitochondrial double membrane (76, 77). Mitochondria also play an important role in lipid metabolism (28, 29, 31, 78): it is the site of synthesis of the mitochondrial-specific PLs, CL, and PG, generated from PA, which is imported from the ER (Fig. 1). In addition, mitochondria are responsible for the synthesis of ∼70% of cellular PE through the activity of Psd1. Psd1 resides on the mitochondrial inner membrane and its substrate, PS, is generated at the ER. PE has an essential role in mitochondrial physiology, but also contributes to the extramitochondrial pool of PE, as mitochondrial generated PE is transported back to the ER where it is used for the synthesis of PC. In addition, ER-synthesized PI and PC have to be imported into the mitochondria. The topologic array of enzymes involved in PL synthesis stresses the necessity of lipid transport systems connecting the mitochondria with other cellular compartments. Lipid trafficking to and from the mitochondria depends on efficient nonvesicular pathways (29–31, 78).
ERMES: A TETHERING COMPLEX WITH LIPID TRANSFER ACTIVITY
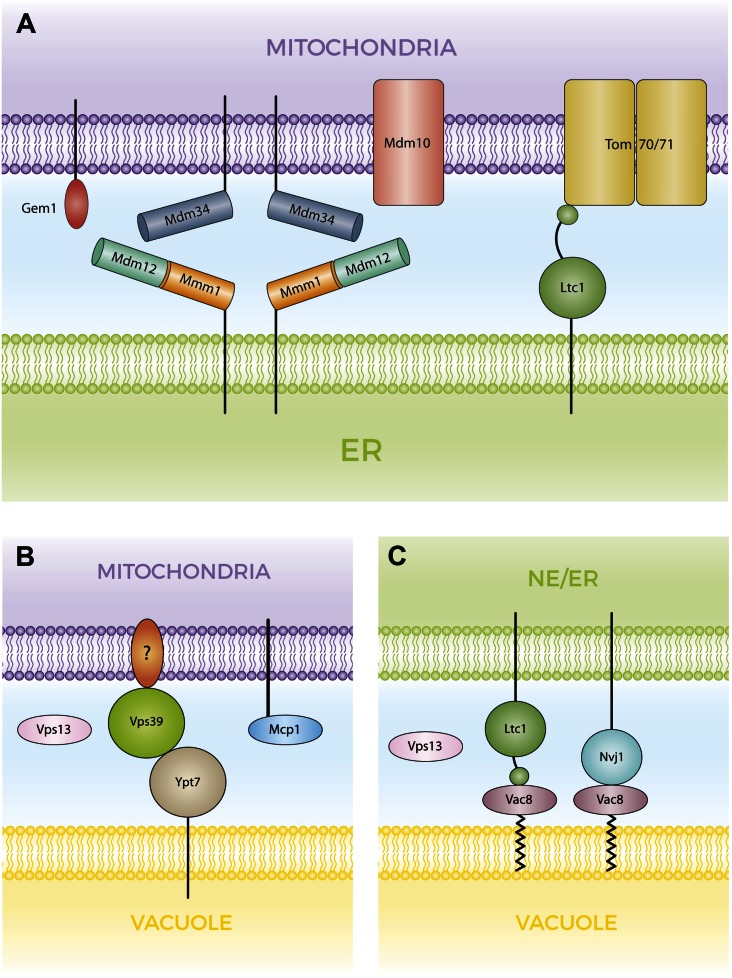
MCSs have been described, based on electron microscopy observations, as sites of closed apposition between different subcellular compartments. The biochemical characterization of a microsomal fraction that copurified associated with mitochondria as a domain of ER enriched in different lipid synthesis activities supported the notion that it represented ER-mitochondria contact sites where lipid synthesis and transport take place (79, 80). A protein complex named ER-mitochondria encounter structure (ERMES) was recently described as a tether between these compartments (81). Maintenance of mitochondrial morphology (Mmm)1, an ER integral protein, mitochondrial distribution and morphology (Mdm)12, a cytosolic protein, and two mitochondrial outer-membrane proteins, Mdm34 and Mdm10, are the core components of ERMES (Fig. 4). They interact with each other (82, 83) and are required for mitochondrial morphology, inheritance, and segregation of mtDNA (83–86). Their role in tethering ER and mitochondria was uncovered by the ability of a synthetic ER-mitochondria tether [construct helping in mitochondria-ER association (ChiMERA)] to overcome the respiratory deficiency of mdm12Δ cells (81). The four ERMES core proteins assemble together forming a few foci per cell at ER-mitochondria junctions. Null mutation for any of the genes encoding ERMES subunits leads to the remnant components losing their characteristic ER-mitochondria punctal distribution. Consistent with a model where the soluble Mdm12 bridges between the ER integral Mmm1 and mitochondrial Mdm34, Mdm12 localizes to mitochondria when Mmm1 is missing, and localizes to the ER when Mdm34 or Mdm10 is absent (81). Three ERMES subunits (Mmm1, Mdm12, and Mdm34) carry synaptotagmin-like mitochondrial and lipid binding protein (SMP) domains (87, 88). Structural analyses of the SMP domains of Mdm12 and Mmm1 revealed that they assemble into a heterotetramer forming an elongated crescent shape in which Mmm1 forms a central homodimer and a monomer of Mdm12 binds to each end of this dimer. A hydrophobic channel traverses this elongated heterotetramer (89). The SMP domains of Mdm12 and Mmm1 both exhibit PL binding activity with preference for PC (89). In addition, a ternary complex was described in which cytosolic Mdm12 bridges the integral ER Mmm1 to the mitochondrial Mdm34 (89). The structural and biochemical characterization of the three SMP domain-containing components of the ERMES complex provides evidence of the involvement of these proteins in the mechanical tethering of both compartments and supports their plausible role in mediating lipid transfer. The fourth subunit of the ERMES core complex, Mdm10, is a mitochondrial β-barrel protein. Mdm10 is also part of another complex, sorting and assembly machinery (SAM), required for the assembly of β-barrel proteins in the outer mitochondrial membrane (75, 90). Because Mdm10 seems to be present in limiting amount, the availability of Mdm10 for assembling into those different complexes might play a regulatory role.
Fig. 4.
Molecular organization of MCSs connecting the ER, mitochondria, and vacuole. A: ERMES and Ltc1-Tom70/71 tethering complexes at ER-mitochondria contact sites. The ERMES complex is composed of four subunits. The subunits Mmm1, Mdm12, and Mdm34 carry SMP domains, physically interact forming a ternary complex and are proposed to transport lipids between the ER and mitochondria. Mdm10 is a mitochondrial β-barrel protein shared with the SAM complex. Gem1 is a substoichiometric component of ERMES, but its role is poorly characterized. Ltc1 is an ER integral protein that contains a sterol binding domain and interacts with mitochondrial Tom70 and Tom71. B: vCLAMP complexes connect mitochondria and vacuole. The vacuolar Rab GTPases, Ypt7 and Vps39, are components of this complex, whereas the mitochondrial component is not known. Vps13 is observed associated with this MCS under fermenting growth conditions, whereas the involvement of Mcp1 in this complex was deduced by genetic analysis. C: The NVJ is characterized by the interaction of Vac8 and Nvj1. Ltc1 also localizes at this MCS through its interaction with Vac8 and independent of Nvj1. Under respiratory growth conditions, Vps13 localizes at the NVJ.
A fifth ERMES component, Gem1, was identified based on its copurification with the core ERMES complex (91, 92). Gem1 belongs to the mitochondrial Rho-like (Miro) class of the Ras GTPase superfamily. It is anchored to the outer mitochondrial membrane through a C-terminal transmembrane domain, with two GTPase domains flanking two calcium binding EF hands exposed to the cytoplasm (93, 94). Contrary to the absence of the ERMES core components, the absence of Gem1 does not prevent ERMES assembly, but, instead, fewer and larger foci are observed (91). Mutational analyses indicated that the first GTPase domain and the first EF hand are important for the localization of Gem1 to ERMES foci (91).
It was found that cells lacking MMM1, MDM12, MDM34, MDM10, or GEM1 have similar profiles of genetic interactions consistent with a common functional role. Interestingly, the similarity between the genetic interaction profiles is also shared by PSD1 (81), supporting the notion that the ERMES complex may be involved in mitochondrial lipid trafficking. Mitochondrial PL compositional analysis for mdm12Δ and mdm34Δ mutant cells showed a significant decrease in CL level (81), whereas PE and CL were both decreased in mmm1Δ and mdm10Δ cells (95). When the rate of conversion from PS to PE to PC was determined, ERMES mutants showed a 2- to 5-fold reduction compared with wild-type cells (81), implying that the ERMES complex may be directly involved in ER-mitochondria PL transport. However, the involvement of ERMES in PL transfer has been contested based on the marginal effect on mitochondrial lipid homeostasis observed upon inactivation of the complex (96). However, the identification of alternative MCSs involving mitochondria (see below) might explain the mild alteration of the mitochondrial PL profile observed for cells defective in ERMES.
In mdm12Δ and mdm34Δ cells, expression of the synthetic ER-mitochondria tether, ChiMERA, restored the ability to grow in nonfermentable carbon sources, restored mitochondrial morphology from giant globular into a tubular network, and partially restored the rate of PS to PC conversion; whereas, ChiMERA failed to alleviate these phenotypes for mmm1Δ and mdm10Δ cells (81). The inability of ChiMERA to compensate for the absence of Mmm1 and Mdm10 suggests that these two proteins perform other functions beyond ER-mitochondria tethering. Indeed, as was noted above, Mdm10 is also known to be part of the SAM complex (75, 90).
MULTIPLE ROLES FOR ERMES IN MITOCHONDRIAL BIOLOGY
In addition to its proposed role in lipid transport, ERMES has other roles in mitochondrial biology. Consistent with the observation that ER tubules wrap around mitochondria marking division sites (97) and the defects in mitochondrial inheritance in ERMES mutants (84), ERMES is localized at sites of mitochondrial constriction. Gem1 facilitates the resolution of ERMES-associated constrictions into newly generated mitochondrial tips (98), and as ERMES components colocalize with replicating mitochondrial nucleoids (83, 99), ERMES and its regulator, Gem1, have been determined to contribute to the distribution of mitochondrial nucleoids. ERMES is also involved in mitophagy, as ERMES mutants have a defect in mitophagy, whereas other autophagy pathways are not impaired, and ERMES colocalizes with sites of mitophagosome formation. The expression of the artificial ER mitochondria tether, ChiMERA, restored mitophagy. It was proposed that ERMES aids in the synthesis of PLs for expansion of the mitophagosome (100). Recently, it was also reported that mitochondria-peroxisome interactions take place through ERMES. Mdm10, Mdm12, and Mdm34 were required for proper localization of Pex11, whereas the absence of the ER-resident ERMES component, Mmm1, did not affect Pex11 localization. Pex11 was shown to physically interact with Mdm34 and was required for formation of contact sites between peroxisomes and mitochondria (101).
The numerous phenotypes associated to ERMES mutants and the many functional roles assigned to this complex are remarkable (102). Its role as a physical tether between the ER and mitochondria is superimposed with its probable lipid transfer activity. Do those several mutant phenotypes rise from the lack of a single function? The ability of the artificial tether, ChiMERA, to alleviate some of the phenotypes of ERMES mutants stresses the importance of ERMES as a tether and suggests the existence of lipid transfer proteins (LTPs) that resume their activities when MCS are restored. Even though the tether function is disengaged when an ERMES subunit is missing, could the remnant SMP-containing subunits of ERMES transfer lipids if the tether is artificially restored? The precise molecular role of this complex is also veiled by the existence of redundant MCSs connecting the mitochondria with other compartments that act homeostatically to sustain mitochondria functionality upon ERMES failure. Despite the fact that ERMES is not a conserved eukaryotic complex and is missing in metazoans and plants (103), the increasing understanding of its functional interactions sheds light on uncovered aspects of mitochondrial biology.
MITOCHONDRIAL CONNECTIONS BEYOND ERMES
Multicopy extragenic suppressors of phenotypes associated with loss of ERMES components have recently been identified (104). Specifically, a multicopy suppressor screen for genes whose increased dose could restore growth of mdm10Δ cells on a nonfermentable carbon source at 37°C led to the isolation of two mitochondrial proteins, Mdm10 complementing protein (Mcp)1 and Mcp2 (104). The overexpression of these proteins reverted the globular mitochondrial morphology of mdm10Δ cells into a normal tubular shape. The altered mitochondrial PL profile of mdm10Δ cells was also restored by increased expression of these proteins. Mcp1 and Mcp2 overexpression alleviated the growth defect on nonfermentable carbon source for loss-of-function mutants of the core ERMES complex (mdm12Δ, mdm34Δ, and mmm1Δ). In addition, the combined absence of either Mcp1 or Mcp2 with any of the ERMES components resulted in severe growth defects (104). Although the precise molecular function of these two proteins is not known, evidence suggests that they are involved in alternative redundant function with ERMES.
Another ER-mitochondria tethering complex has recently been identified (105). ER membrane protein complex (EMC) is a conserved multisubunit complex (106) that interacts with Tom5, a nonessential subunit of the mitochondrial protein translocase of the outer membrane complex (Fig. 1). The absence of five of the six subunits of the EMC (Emc1, Emc2, Emc3, Emc5, and Emc6) lead to a reduction of PS to PE conversion similar to that observed for psd1Δ cells. In support of a role for EMC in facilitating transfer of PS to the mitochondria for its conversion to PE, this quintuple mutant had a 50% reduction of PS and PE mitochondrial levels. In addition, this mutant had reduced contact between ER and mitochondria and did not proliferate on nonfermentable carbon sources. Importantly, this quintuple mutant carrying a thermo-sensitive allele of MMM1, encoding the ER-anchored component of ERMES, was not viable at the restrictive temperature and had a severe defect in PS transfer to mitochondria. The expression of the synthetic ER-mitochondria tether, ChiMERA, rescued both the growth and the PL metabolic defects of the EMC quintuple mmm1Δ mutant (105). In contrast to ERMES, which is not present in metazoans and plants, EMC is widely conserved among eukaryotes (103, 106). No component of EMC or its mitochondrial interactor, Tom5, has been characterized as a LTP. How lipid transfer occurs at EMC tethers remains unknown.
vCLAMPING THE MITOCHONDRIA TO THE VACUOLE
Two different approaches described the same MCS, vacuole and mitochondria patch (vCLAMP), connecting mitochondria to vacuoles (Fig. 4). Both approaches led to the identification of Vps39, a component of the homotypic fusion and vacuole protein sorting (HOPS) complex as a critical factor for the formation of vCLAMP (107, 108). In one approach, based on the consistent observation of contact sites between vacuoles and mitochondria, Hönscher et al. (108) overexpressed components of the HOPS tethering complex and determined the effect on vacuole-mitochondria MCSs under the assumption that the HOPS complex might be involved, and that the high levels of tethering machinery would expand these MCSs. Indeed, the overexpression of Vps39 enlarged the apposition regions between mitochondria and vacuoles. Furthermore, GFP-Vps39 was detected at the enlarged vCLAMP sites. Whereas the HOPS components, Vps41 and Vps11, were not required for Vps39-dependent expansion of vCLAMP, the absence of the Rab GTPase Ypt7 led to detachment of GFP-Vps39 from membranes, with the phosphorylation status of Vps39 affecting its ability to assemble into vCLAMP. vCLAMP expansion mediated by increased expression of Vps39 ameliorated the growth defect of the ERMES mutants, mdm10Δ and mdm12Δ, on fermentable and nonfermentable carbon sources, supporting the idea that ERMES and vCLAMP perform some redundant functions aimed to maintain proper mitochondrial physiology. The number of ERMES contact sites increased when cells were respiring, whereas vCLAMP sites decreased, suggesting that these two complexes are reciprocally regulated (108).
In the approach to explore mitochondrial connectivity, Elbaz-Alon et al. (107) performed a genome-wide genetic screen based on the observation that a dysfunctional ERMES complex does not elicit a lethal phenotype. This implies redundancy of mitochondrial connections with the endomembrane system, and if such a redundant pathway fails an expansion of the ERMES contact sites was anticipated as a compensatory homeostatic response. Using a Mdm34-GFP fusion as a reporter for ERMES foci, Elbaz-Alon et al. (107) screened a collection of nonessential single gene deletions and hypomorphic alleles of essential genes for altered Mdm34-GFP pattern. They found that the absence of the HOPS subunits, Vps39 and Vam7, promoted an increased number of ERMES foci per cell. Light and electron microscopy showed that GFP-Vps39 was present at sites of close apposition between mitochondria and vacuoles. Importantly, the absence of the ERMES complex resulted in mitochondria that were almost entirely surrounded by vCLAMP, revealing a reciprocal interplay between them. Consistent with the notion that these redundant pathways contribute to an essential function, the combination of impaired ERMES and vCLAMP functions was lethal (107).
To explore the impact that these two MCSs had on the mitochondrial PL metabolism, a strain defective in VPS39 and expressing MDM34 under the repressible GalS promoter was generated. After 24 h of growth on glucose, the mutant strain exhibited a reduction of 40% of PE and CL when mitochondrial lipid fractions were compared against wild-type. There was also a nearly 2-fold accumulation of newly synthesized PS, and although the rate of PE synthesis was not affected, the synthesis of its product, PC, was also reduced more than 2-fold. The depletion of the ERMES complex in the absence of vCLAMP led to a severe alteration of mitochondrial PL homeostasis, further supporting the idea that both MCSs facilitate, either directly or indirectly, mitochondrial lipid transport (107).
ERMES mutants acquire suppressors easily (86, 109), and the characterization of suppressed mmm1Δ mutant strains able to grow on a nonfermentable carbon source led to the identification of two dominant mutations in Vps13 as causative of the suppressed phenotype (109). VPS13 has been identified in several screens for trafficking mutants (110–112) and encodes a cytosolic protein that is peripherally associated with endosomes (113) and involved in prospore membrane biogenesis (114). One dominant allele of VPS13 was further studied and was shown to restore mitochondria morphology from the giant globular form characteristic of ERMES mutants to a normal tubular network, and also restored mtDNA stability. However, ERMES complexes were not reassembled. Congruent with the idea that Vps13 performed a redundant function with the ERMES complex, vps13Δ cells, also deficient in ERMES, were not viable. Localization of Vps13 using a functional GFP variant showed that, in addition to being found in scattered puncta, presumably endosomes, it also resided at vacuole-mitochondria MCSs. It is known that vCLAMP formation is repressed under the respiring condition, and in support of the idea that Vps13 integrates vCLAMP, Vps13 relocalized to NVJs upon switching cells from a fermentable to nonfermentable carbon source, whereas dominant variants of Vps13 did not relocalize to NVJ contact sites. Importantly, genetic interaction analyses suggest that Vps13, Vps39, and Mcp1 function in the same pathway, whereas Mcp2 does not (109). Consistent with the dynamic nature of ERMES and vCLAMP, it was observed that these two complexes were reversibly lost under nutrient stress. The vCLAMP marker, mCherry-Vps39, shifted reversibly from vacuole membrane and vCLAMP contact sites into multiple cytosolic puncta upon glucose withdrawal. The ERMES subunit, Mdm34-GFP, fluctuated from few foci congruent with few ERMES complexes per cell, to a diffuse cytosolic distribution depending on glucose availability (115). The observed diffuse cytosolic distribution of Mdm34 under glucose deprivation is interesting because Mdm34 is thought to be an integral protein of the external mitochondrial membrane.
Characterization of vCLAMP reveals not only the redundancy of contact sites bridging the mitochondria with the endomembrane system through different tethering complexes but also the subjacent regulatory cross-talk that controls the formation and extension of MCSs upon different metabolic inputs. A further molecular characterization of this new MCS and the elucidation of the reciprocal regulation of ERMES and vCLAMP formation are very much awaited.
LIPID BINDING PROTEINS REGULATE MCS CROSS-TALK
A new family of LTPs was recently described, based on bioinformatic analysis (116), that is characterized by the presence of a domain distantly related to the sterol binding steroidogenic acute regulatory transfer (StART) domain. Members of this lipid binding family contain additional domains involved in membrane interactions and a transmembrane domain at their C termini. Whereas most fungi contain three members of this protein family, there are six proteins carrying this StART-like domain in S. cerevisiae due to genome duplication. Ysp1 and its paralog Ysp2 and Sip3 and its paralog LTP anchored at a MCS (Lam)4 are ER proteins present at ER-plasma membrane (PM) MCSs, while Lam5 and its paralog Lam6 [also named lipid transfer at contact site (Ltc)1] are localized at the NVJ and ER-mitochondria contact sites. The combined genetic and biochemical evidence suggest that all members of this family bind sterol (116, 117).
Consistent with the localization of Ltc1 at ER-mitochondria contact sites, two other research groups identified Ltc1 as an interactor with the ERMES complex based on proteomic analyses (118, 119). The absence of Ltc1 alone did not impair growth on fermentable or nonfermentable carbon sources or affect mitochondrial morphology or assembly of the ERMES complex; however, it did aggravate the growth defect of ERMES mutants, suggesting that Ltc1 and ERMES fulfill different roles toward mitochondrial functionality (118, 119). Murley et al. (118) showed that the majority of Ltc1 localized at ER-mitochondria MCSs with about half colocalizing with ERMES foci. A minor fraction of Ltc1 was distributed between the NVJ and non-NVJ ER-vacuole MCSs. Complementary proteomic analyses using Ltc1 as bait identified, in addition to ERMES subunits, the vacuolar protein, Vac8, which is involved in NVJ formation, and two subunits of the translocase of the mitochondrial outer membrane, Tom70 and Tom71. In the absence of Tom70/Tom71, most of Ltc1 was found at NVJ MCSs and, reciprocally, when Vac8 was missing, Ltc1 localized to ER-mitochondria MCSs. Consistent with its possession of a StART-like domain, the soluble region of Ltc1 was demonstrated to transfer sterol in vitro. ERMES deficiency led to altered mitochondrial ergosterol levels (104), and Ltc1 was required for the partitioning of ergosterol-enriched areas in vacuolar membranes (118). Considering that sterols are synthesized in the ER, that Ltc1 transfers sterols between membranes in vitro, and that Ltc1 is found at ER-mitochondria/ERMES and NVJ MCSs, Ltc1 is a candidate sterol transfer protein that enables mitochondrial and vacuolar ergosterol homeostasis through inter-organelle sterol transfer at MCSs.
Interestingly, a minor fraction of Ltc1 also localized to vCLAMP (119), in addition to ERMES and NVJ, and overexpression of Ltc1 led to the expansion of all of these MCSs. Almost all the mitochondria were in contact with the ER with 60% of their circumference covered with ER when Ltc1 was overexpressed. NVJs were also expanded and pieces of the nucleus were engulfed by the vacuole. vCLAMPs were easily detected when Ltc1 was overexpressed, whereas they were hardly visible in wild-type cells. In addition, vCLAMPs were invaded by tubules of ER when Ltc1 was overexpressed. It was also reported that the reciprocal interplay between the extent of ERMES and vCLAMP sites was dependent on the level of Ltc1 at each site. These observations suggest that the level of Ltc1 at each MCS determines their extent. Variation of Ltc1 expression level combined with changes in its relative affinity and recruitment for each site can result in cross-talk regulation for these MCSs. The role of the sterol binding/transfer activity of Ltc1 in maintaining MCS cross-talk would be interesting to explore.
FINAL REMARKS: MULTIPLICITY OF MCS AND TETHERING FACTORS IS A RECURRENT THEME
It is clear from the examples illustrated in this review that there are numerous redundancies in aspects of inter-organelle MCSs. Beyond those discussed in depth here, other examples have been described. For example, six ER integral proteins have been identified as tethering factors required for the formation of cortical ER (cER), Ist2, Scs2, Scs22, and the three tricalbins, Tcb1/2/3. In the absence of these six proteins, there is a 90% reduction of cER and a consequent accumulation of cytoplasmic ER (120). In spite of the dramatic collapse of the cER, the StART-like domain containing proteins Ysp1, Ysp2, Sip3, and Lam4 persisted at ER-PM junctions in this sextuple mutant, revealing the existence of redundant factors involved in the formation of ER-PM MCSs (116). Analogously, the localization of Ltc1 at ER-vacuole contact sites was dependent on Vac8. However, in the absence of Nvj1, the perinuclear binding partner of Vac8 (121), the localization of Ltc1 was not disturbed, indicating that other nuclear-vacuole contact sites are formed through Vac8, but independent of Nvj1 (118). These examples further illustrate the multiplicity of tethering complexes occurring between subcellular compartments.
Another relevant observation obtained through these studies is the occurrence of proteins like Ltc1 and Vps13 at more than one MCS. The accumulated evidence suggests that they contribute to MCS dynamics under different physiological conditions by facilitating cross-talk between different contact sites.
Overall, the recent identification and characterization of several tethering complexes, LTPs, and other protein factors involved in the assembly and regulation of MCSs connecting different cell compartments has opened avenues for new and exciting insights in this field: from a more thorough molecular description of the MCS to the deciphering of the signaling networks that modulate the dynamics of MCSs to adjust subcellular connectivity to changing metabolic conditions.
Footnotes
Abbreviations:
- cER
- cortical endoplasmic reticulum
- ChiMERA
- construct helping in mitochondria-endoplasmic reticulum association
- CL
- cardiolipin
- DAG
- diacylglycerol
- EMC
- endoplasmic reticulum membrane protein complex
- ER
- endoplasmic reticulum
- ERMES
- endoplasmic reticulum-mitochondria encounter structure
- HOPS
- homotypic fusion and vacuole protein sorting
- Lam
- lipid transfer protein anchored at a membrane contact site
- LD
- lipid droplet
- Ltc
- lipid transfer at contact site
- LTP
- lipid transfer protein
- Mcp
- maintenance of mitochondrial morphology 10 complementing protein
- MCS
- membrane contact site
- Mdm
- mitochondrial distribution and morphology
- Mmm
- maintenance of mitochondrial morphology
- mtDNA
- mitochondrial DNA
- NL
- neutral lipid
- NVJ
- nuclear vacuolar junction
- PA
- phosphatidic acid
- PC
- phosphatidylcholine
- PE
- phosphatidylethanolamine
- PG
- phosphatidylglycerol
- PI
- phosphatidylinositol
- PL
- phospholipid
- PM
- plasma membrane
- PS
- phosphatidylserine
- SAM
- sorting and assembly machinery
- SLD
- supersized LD
- SMP
- synaptotagmin-like mitochondrial and lipid binding protein
- StART
- steroidogenic acute regulatory transfer
- TAG
- triacylglycerol
- vCLAMP
- vacuole and mitochondria patch
The work on the science described in this review was supported by a Discovery Grant from the Canadian Network for Research and Innovation in Machining Technology, Natural Sciences and Engineering Research Council of Canada to C.R.M.
REFERENCES
- 1.Athenstaedt K., and Daum G.. 1999. Phosphatidic acid, a key intermediate in lipid metabolism. Eur. J. Biochem. 266: 1–16. [DOI] [PubMed] [Google Scholar]
- 2.Henry S. A., Kohlwein S. D., and Carman G. M.. 2012. Metabolism and regulation of glycerolipids in the yeast Saccharomyces cerevisiae. Genetics. 190: 317–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Henry S. A., and Patton-Vogt J. L.. 1998. Genetic regulation of phospholipid metabolism: yeast as a model eukaryote. Prog. Nucleic Acid Res. Mol. Biol. 61: 133–179. [DOI] [PubMed] [Google Scholar]
- 4.Carman G. M., and Henry S. A.. 2007. Phosphatidic acid plays a central role in the transcriptional regulation of glycerophospholipid synthesis in Saccharomyces cerevisiae. J. Biol. Chem. 282: 37293–37297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Travers K. J., Patil C. K., Wodicka L., Lockhart D. J., Weissman J. S., and Walter P.. 2000. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 101: 249–258. [DOI] [PubMed] [Google Scholar]
- 6.Garbarino J., Padamsee M., Wilcox L., Oelkers P. M., D’Ambrosio D., Ruggles K. V., Ramsey N., Jabado O., Turkish A., and Sturley S. L.. 2009. Sterol and diacylglycerol acyltransferase deficiency triggers fatty acid-mediated cell death. J. Biol. Chem. 284: 30994–31005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jonikas M. C., Collins S. R., Denic V., Oh E., Quan E. M., Schmid V., Weibezahn J., Schwappach B., Walter P., Weissman J. S., et al. 2009. Comprehensive characterization of genes required for protein folding in the endoplasmic reticulum. Science. 323: 1693–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schuck S., Prinz W. A., Thorn K. S., Voss C., and Walter P.. 2009. Membrane expansion alleviates endoplasmic reticulum stress independently of the unfolded protein response. J. Cell Biol. 187: 525–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han G-S., Wu W-I., and Carman G. M.. 2006. The Saccharomyces cerevisiae Lipin homolog is a Mg2+-dependent phosphatidate phosphatase enzyme. J. Biol. Chem. 281: 9210–9218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pascual F., and Carman G. M.. 2013. Phosphatidate phosphatase, a key regulator of lipid homeostasis. Biochim. Biophys. Acta. 1831: 514–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O’Hara L., Han G-S., Peak-Chew S., Grimsey N., Carman G. M., and Siniossoglou S.. 2006. Control of phospholipid synthesis by phosphorylation of the yeast lipin Pah1p/Smp2p Mg2+-dependent phosphatidate phosphatase. J. Biol. Chem. 281: 34537–34548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choi H-S., Su W-M., Morgan J. M., Han G-S., Xu Z., Karanasios E., Siniossoglou S., and Carman G. M.. 2011. Phosphorylation of phosphatidate phosphatase regulates its membrane association and physiological functions in Saccharomyces cerevisiae: identification of SER(602), THR(723), and SER(744) as the sites phosphorylated by CDC28 (CDK1)-encoded cyclin-dependen. J. Biol. Chem. 286: 1486–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choi H-S., Su W-M., Han G-S., Plote D., Xu Z., and Carman G. M.. 2012. Pho85p-Pho80p phosphorylation of yeast Pah1p phosphatidate phosphatase regulates its activity, location, abundance, and function in lipid metabolism. J. Biol. Chem. 287: 11290–11301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Santos-Rosa H., Leung J., Grimsey N., Peak-Chew S., and Siniossoglou S.. 2005. The yeast lipin Smp2 couples phospholipid biosynthesis to nuclear membrane growth. EMBO J. 24: 1931–1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Su W-M., Han G-S., and Carman G. M.. 2014. Yeast Nem1-Spo7 protein phosphatase activity on Pah1 phosphatidate phosphatase is specific for the Pho85-Pho80 protein kinase phosphorylation sites. J. Biol. Chem. 289: 34699–34708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pascual F., Hsieh L-S., Soto-Cardalda A. I., and Carman G. M.. 2014. Yeast Pah1p phosphatidate phosphatase is regulated by proteasome-mediated degradation. J. Biol. Chem. 289: 9811–9822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hsieh L-S., Su W-M., Han G-S., and Carman G. M.. 2015. Phosphorylation regulates the ubiquitin-independent degradation of yeast Pah1 phosphatidate phosphatase by the 20S proteasome. J. Biol. Chem. 290: 11467–11478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Szymanski K. M., Binns D., Bartz R., Grishin N. V., Li W-P., Agarwal A. K., Garg A., Anderson R. G. W., and Goodman J. M.. 2007. The lipodystrophy protein seipin is found at endoplasmic reticulum lipid droplet junctions and is important for droplet morphology. Proc. Natl. Acad. Sci. USA. 104: 20890–20895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jacquier N., Choudhary V., Mari M., Toulmay A., Reggiori F., and Schneiter R.. 2011. Lipid droplets are functionally connected to the endoplasmic reticulum in Saccharomyces cerevisiae. J. Cell Sci. 124: 2424–2437. [DOI] [PubMed] [Google Scholar]
- 20.Wolinski H., Kolb D., Hermann S., Koning R. I., and Kohlwein S. D.. 2011. A role for seipin in lipid droplet dynamics and inheritance in yeast. J. Cell Sci. 124: 3894–3904. [DOI] [PubMed] [Google Scholar]
- 21.Grippa A., Buxó L., Mora G., Funaya C., Idrissi F-Z., Mancuso F., Gomez R., Muntanyà J., Sabidó E., and Carvalho P.. 2015. The seipin complex Fld1/Ldb16 stabilizes ER-lipid droplet contact sites. J. Cell Biol. 211: 829–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adeyo O., Horn P. J., Lee S., Binns D. D., Chandrahas A., Chapman K. D., and Goodman J. M.. 2011. The yeast lipin orthologue Pah1p is important for biogenesis of lipid droplets. J. Cell Biol. 192: 1043–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karanasios E., Barbosa A. D., Sembongi H., Mari M., Han G-S., Reggiori F., Carman G. M., and Siniossoglou S.. 2013. Regulation of lipid droplet and membrane biogenesis by the acidic tail of the phosphatidate phosphatase Pah1p. Mol. Biol. Cell. 24: 2124–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Han S., Binns D. D., Chang Y-F., and Goodman J. M.. 2015. Dissecting seipin function: the localized accumulation of phosphatidic acid at ER/LD junctions in the absence of seipin is suppressed by Sei1pΔNterm only in combination with Ldb16p. BMC Cell Biol. 16: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wolinski H., Hofbauer H. F., Hellauer K., Cristobal-Sarramian A., Kolb D., Radulovic M., Knittelfelder O. L., Rechberger G. N., and Kohlwein S. D.. 2015. Seipin is involved in the regulation of phosphatidic acid metabolism at a subdomain of the nuclear envelope in yeast. Biochim. Biophys. Acta. 1851: 1450–1464. [DOI] [PubMed] [Google Scholar]
- 26.Kohlwein S. D., Veenhuis M., and van der Klei I. J.. 2013. Lipid droplets and peroxisomes: key players in cellular lipid homeostasis or a matter of fat–store ’em up or burn ’em down. Genetics. 193: 1–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang C-W. 2015. Lipid droplet dynamics in budding yeast. Cell. Mol. Life Sci. 72: 2677–2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tamura Y., Harada Y., Nishikawa S., Yamano K., Kamiya M., Shiota T., Kuroda T., Kuge O., Sesaki H., Imai K., et al. 2013. Tam41 is a CDP-diacylglycerol synthase required for cardiolipin biosynthesis in mitochondria. Cell Metab. 17: 709–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Horvath S. E., and Daum G.. 2013. Lipids of mitochondria. Prog. Lipid Res. 52: 590–614. [DOI] [PubMed] [Google Scholar]
- 30.Scharwey M., Tatsuta T., and Langer T.. 2013. Mitochondrial lipid transport at a glance. J. Cell Sci. 126: 5317–5323. [DOI] [PubMed] [Google Scholar]
- 31.Tamura Y., Sesaki H., and Endo T.. 2014. Phospholipid transport via mitochondria. Traffic. 15: 933–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oelkers P., Tinkelenberg A., Erdeniz N., Cromley D., Billheimer J. T., and Sturley S. L.. 2000. A lecithin cholesterol acyltransferase-like gene mediates diacylglycerol esterification in yeast. J. Biol. Chem. 275: 15609–15612. [DOI] [PubMed] [Google Scholar]
- 33.Oelkers P., Cromley D., Padamsee M., Billheimer J. T., and Sturley S. L.. 2002. The DGA1 gene determines a second triglyceride synthetic pathway in yeast. J. Biol. Chem. 277: 8877–8881. [DOI] [PubMed] [Google Scholar]
- 34.Sorger D., and Daum G.. 2002. Synthesis of triacylglycerols by the acyl-coenzyme A:diacyl-glycerol acyltransferase Dga1p in lipid particles of the yeast Saccharomyces cerevisiae. J. Bacteriol. 184: 519–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sorger D., and Daum G.. 2003. Triacylglycerol biosynthesis in yeast. Appl. Microbiol. Biotechnol. 61: 289–299. [DOI] [PubMed] [Google Scholar]
- 36.Han G-S., O’Hara L., Carman G. M., and Siniossoglou S.. 2008. An unconventional diacylglycerol kinase that regulates phospholipid synthesis and nuclear membrane growth. J. Biol. Chem. 283: 20433–20442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Han G-S., O’Hara L., Siniossoglou S., and Carman G. M.. 2008. Characterization of the yeast DGK1-encoded CTP-dependent diacylglycerol kinase. J. Biol. Chem. 283: 20443–20453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chae M., Han G-S., and Carman G. M.. 2012. The Saccharomyces cerevisiae actin patch protein App1p is a phosphatidate phosphatase enzyme. J. Biol. Chem. 287: 40186–40196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fakas S., Qiu Y., Dixon J. L., Han G. S., Ruggles K. V., Garbarino J., Sturley S. L., and Carman G. M.. 2011. Phosphatidate phosphatase activity plays key role in protection against fatty acid-induced toxicity in yeast. J. Biol. Chem. 286: 29074–29085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sasser T., Qiu Q. S., Karunakaran S., Padolina M., Reyes A., Flood B., Smith S., Gonzales C., and Fratti R. a.. 2012. Yeast lipin 1 orthologue Pah1p regulates vacuole homeostasis and membrane fusion. J. Biol. Chem. 287: 2221–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Park Y., Han G-S., Mileykovskaya E., Garrett T. A., and Carman G. M.. 2015. Altered lipid synthesis by lack of yeast Pah1 phosphatidate phosphatase reduces chronological life span. J. Biol. Chem. 290: 25382–25394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Han G-S., Siniossoglou S., and Carman G. M.. 2007. The cellular functions of the yeast lipin homolog PAH1p are dependent on its phosphatidate phosphatase activity. J. Biol. Chem. 282: 37026–37035. [DOI] [PubMed] [Google Scholar]
- 43.Loewen C. J. R., Gaspar M. L., Jesch S. A., Delon C., Ktistakis N. T., Henry S. A., and Levine T. P.. 2004. Phospholipid metabolism regulated by a transcription factor sensing phosphatidic acid. Science. 304: 1644–1647. [DOI] [PubMed] [Google Scholar]
- 44.Su W. M., Han G. S., Casciano J., and Carman G. M.. 2012. Protein kinase A-mediated phosphorylation of Pah1p phosphatidate phosphatase functions in conjunction with the Pho85p-Pho80p and Cdc28p-Cyclin B kinases to regulate lipid synthesis in yeast. J. Biol. Chem. 287: 33364–33376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Su W-M., Han G-S., and Carman G. M.. 2014. Cross-talk phosphorylations by protein kinase C and Pho85p-Pho80p protein kinase regulate Pah1p phosphatidate phosphatase abundance in Saccharomyces cerevisiae. J. Biol. Chem. 289: 18818–18830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hsieh L-S., Su W-M., Han G-S., and Carman G. M.. 2016. Phosphorylation of yeast Pah1 phosphatidate phosphatase by casein kinase II regulates its function in lipid metabolism. J. Biol. Chem. 291: 9974–9990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chang Y-F., and Carman G. M.. 2006. Casein kinase II phosphorylation of the yeast phospholipid synthesis transcription factor Opi1p. J. Biol. Chem. 281: 4754–4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Karanasios E., Han G-S., Xu Z., Carman G. M., and Siniossoglou S.. 2010. A phosphorylation-regulated amphipathic helix controls the membrane translocation and function of the yeast phosphatidate phosphatase. Proc. Natl. Acad. Sci. USA. 107: 17539–17544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dubots E., Cottier S., Péli-Gulli M-P., Jaquenoud M., Bontron S., Schneiter R., and De Virgilio C.. 2014. TORC1 regulates Pah1 phosphatidate phosphatase activity via the Nem1/Spo7 protein phosphatase complex. PLoS One. 9: e104194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Siniossoglou S., Santos-Rosa H., Rappsilber J., Mann M., and Hurt E.. 1998. A novel complex of membrane proteins required for formation of a spherical nucleus. EMBO J. 17: 6449–6464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fei W., Shui G., Zhang Y., Krahmer N., Ferguson C., Kapterian T. S., Lin R. C., Dawes I. W., Brown A. J., Li P., et al. 2011. A role for phosphatidic acid in the formation of “supersized” lipid droplets. PLoS Genet. 7: e1002201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Soto-Cardalda A., Fakas S., Pascual F., Choi H-S., and Carman G. M.. 2012. Phosphatidate phosphatase plays role in zinc-mediated regulation of phospholipid synthesis in yeast. J. Biol. Chem. 287: 968–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pascual F., Soto-Cardalda A., and Carman G. M.. 2013. PAH1-encoded phosphatidate phosphatase plays a role in the growth phase- and inositol-mediated regulation of lipid synthesis in Saccharomyces cerevisiae. J. Biol. Chem. 288: 35781–35792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ptacek J., Devgan G., Michaud G., Zhu H., Zhu X., Fasolo J., Guo H., Jona G., Breitkreutz A., Sopko R., et al. 2005. Global analysis of protein phosphorylation in yeast. Nature. 438: 679–684. [DOI] [PubMed] [Google Scholar]
- 55.Dephoure N., Howson R. W., Blethrow J. D., Shokat K. M., and O’Shea E. K.. 2005. Combining chemical genetics and proteomics to identify protein kinase substrates. Proc. Natl. Acad. Sci. USA. 102: 17940–17945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mah A. S., Elia A. E. H., Devgan G., Ptacek J., Schutkowski M., Snyder M., Yaffe M. B., and Deshaies R. J.. 2005. Substrate specificity analysis of protein kinase complex Dbf2-Mob1 by peptide library and proteome array screening. BMC Biochem. 6: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chi A., Huttenhower C., Geer L. Y., Coon J. J., Syka J. E. P., Bai D. L., Shabanowitz J., Burke D. J., Troyanskaya O. G., and Hunt D. F.. 2007. Analysis of phosphorylation sites on proteins from Saccharomyces cerevisiae by electron transfer dissociation (ETD) mass spectrometry. Proc. Natl. Acad. Sci. USA. 104: 2193–2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Barbosa A. D., Sembongi H., Su W-M., Abreu S., Reggiori F., Carman G. M., and Siniossoglou S.. 2015. Lipid partitioning at the nuclear envelope controls membrane biogenesis. Mol. Biol. Cell. 26: 3641–3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang C-W., Miao Y-H., and Chang Y-S.. 2014. Control of lipid droplet size in budding yeast requires the collaboration between Fld1 and Ldb16. J. Cell Sci. 127: 1214–1228. [DOI] [PubMed] [Google Scholar]
- 60.Fei W., Shui G., Gaeta B., Du X., Kuerschner L., Li P., Brown A. J., Wenk M. R., Parton R. G., and Yang H.. 2008. Fld1p, a functional homologue of human seipin, regulates the size of lipid droplets in yeast. J. Cell Biol. 180: 473–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Binns D., Lee S., Hilton C. L., Jiang Q. X., and Goodman J. M.. 2010. Seipin is a discrete homooligomer. Biochemistry. 49: 10747–10755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cartwright B. R., Binns D. D., Hilton C. L., Han S., Gao Q., and Goodman J. M.. 2015. Seipin performs dissectible functions in promoting lipid droplet biogenesis and regulating droplet morphology. Mol. Biol. Cell. 26: 726–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hancock L. C., Behta R. P., and Lopes J. M.. 2006. Genomic analysis of the Opi- phenotype. Genetics. 173: 621–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sim M. F. M., Dennis R. J., Aubry E. M., Ramanathan N., Sembongi H., Saudek V., Ito D., O’Rahilly S., Siniossoglou S., and Rochford J. J.. 2012. The human lipodystrophy protein seipin is an ER membrane adaptor for the adipogenic PA phosphatase lipin 1. Mol. Metab. 2: 38–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schneiter R., Brügger B., Sandhoff R., Zellnig G., Leber A., Lampl M., Athenstaedt K., Hrastnik C., Eder S., Daum G., et al. 1999. Electrospray ionization tandem mass spectrometry (ESI-MS/MS) analysis of the lipid molecular species composition of yeast subcellular membranes reveals acyl chain-based sorting/remodeling of distinct molecular species en route to the plasma membrane. J. Cell Biol. 146: 741–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ejsing C. S., Sampaio J. L., Surendranath V., Duchoslav E., Ekroos K., Klemm R. W., Simons K., and Shevchenko A.. 2009. Global analysis of the yeast lipidome by quantitative shotgun mass spectrometry. Proc. Natl. Acad. Sci. USA. 106: 2136–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Panatala R., Hennrich H., and Holthuis J. C. M.. 2015. Inner workings and biological impact of phospholipid flippases. J. Cell Sci. 128: 2021–2032. [DOI] [PubMed] [Google Scholar]
- 68.Kohlwein S. D., Daum G., Schneiter R., and Paltauf F.. 1996. Phospholipids: synthesis, sorting, subcellular traffic - the yeast approach. Trends Cell Biol. 6: 260–266. [DOI] [PubMed] [Google Scholar]
- 69.Dickson R. C. 2010. Roles for sphingolipids in Saccharomyces cerevisiae. Adv. Exp. Med. Biol. 688: 217–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jacquier N., and Schneiter R.. 2012. Mechanisms of sterol uptake and transport in yeast. J. Steroid Biochem. Mol. Biol. 129: 70–78. [DOI] [PubMed] [Google Scholar]
- 71.Kurland C. G., and Andersson S. G.. 2000. Origin and evolution of the mitochondrial proteome. Microbiol. Mol. Biol. Rev. 64: 786–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Truscott K. N., Brandner K., and Pfanner N.. 2003. Mechanisms of protein import into mitochondria. Curr. Biol. 13: R326–R337. [DOI] [PubMed] [Google Scholar]
- 73.Lithgow T., and Schneider A.. 2010. Evolution of macromolecular import pathways in mitochondria, hydrogenosomes and mitosomes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365: 799–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Opalińska M., and Meisinger C.. 2015. Metabolic control via the mitochondrial protein import machinery. Curr. Opin. Cell Biol. 33: 42–48. [DOI] [PubMed] [Google Scholar]
- 75.Höhr A. I. C., Straub S. P., Warscheid B., Becker T., and Wiedemann N.. 2015. Assembly of β-barrel proteins in the mitochondrial outer membrane. Biochim. Biophys. Acta. 1853: 74–88. [DOI] [PubMed] [Google Scholar]
- 76.Palmieri F. 2014. Mitochondrial transporters of the SLC25 family and associated diseases: a review. J. Inherit. Metab. Dis. 37: 565–575. [DOI] [PubMed] [Google Scholar]
- 77.Schaedler T. A., Faust B., Shintre C. A., Carpenter E. P., Srinivasan V., van Veen H. W., and Balk J.. 2015. Structures and functions of mitochondrial ABC transporters. Biochem. Soc. Trans. 43: 943–951. [DOI] [PubMed] [Google Scholar]
- 78.Osman C., Voelker D. R., and Langer T.. 2011. Making heads or tails of phospholipids in mitochondria. J. Cell Biol. 192: 7–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gaigg B., Simbeni R., Hrastnik C., Paltauf F., and Daum G.. 1995. Characterization of a microsomal subfraction associated with mitochondria of the yeast, Saccharomyces cerevisiae. Involvement in synthesis and import of phospholipids into mitochondria. Biochim. Biophys. Acta. 1234: 214–220. [DOI] [PubMed] [Google Scholar]
- 80.Achleitner G., Gaigg B., Krasser A., Kainersdorfer E., Kohlwein S. D., Perktold A., Zellnig G., and Daum G.. 1999. Association between the endoplasmic reticulum and mitochondria of yeast facilitates interorganelle transport of phospholipids through membrane contact. Eur. J. Biochem. 264: 545–553. [DOI] [PubMed] [Google Scholar]
- 81.Kornmann B., Currie E., Collins S. R., Schuldiner M., Nunnari J., Weissman J. S., and Walter P.. 2009. An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science. 325: 477–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Boldogh I. R., Nowakowski D. W., Yang H-C., Chung H., Karmon S., Royes P., and Pon L. A.. 2003. A protein complex containing Mdm10p, Mdm12p, and Mmm1p links mitochondrial membranes and DNA to the cytoskeleton-based segregation machinery. Mol. Biol. Cell. 14: 4618–4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Youngman M. J., Hobbs A. E. A., Burgess S. M., Srinivasan M., and Jensen R. E.. 2004. Mmm2p, a mitochondrial outer membrane protein required for yeast mitochondrial shape and maintenance of mtDNA nucleoids. J. Cell Biol. 164: 677–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Burgess S. M., Delannoy M., and Jensen R. E.. 1994. MMM1 encodes a mitochondrial outer membrane protein essential for establishing and maintaining the structure of yeast mitochondria. J. Cell Biol. 126: 1375–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sogo L. F., and Yaffe M. P.. 1994. Regulation of mitochondrial morphology and inheritance by Mdm10p, a protein of the mitochondrial outer membrane. J. Cell Biol. 126: 1361–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Berger K. H., Sogo L. F., and Yaffe M. P.. 1997. Mdm12p, a component required for mitochondrial inheritance that is conserved between budding and fission yeast. J. Cell Biol. 136: 545–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kopec K. O., Alva V., and Lupas A. N.. 2010. Homology of SMP domains to the TULIP superfamily of lipid-binding proteins provides a structural basis for lipid exchange between ER and mitochondria. Bioinformatics. 26: 1927–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kopec K. O., Alva V., and Lupas A. N.. 2011. Bioinformatics of the TULIP domain superfamily. Biochem. Soc. Trans. 39: 1033–1038. [DOI] [PubMed] [Google Scholar]
- 89.AhYoung A. P., Jiang J., Zhang J., Khoi Dang X., Loo J. A., Zhou Z. H., and Egea P. F.. 2015. Conserved SMP domains of the ERMES complex bind phospholipids and mediate tether assembly. Proc. Natl. Acad. Sci. USA. 112: E3179–E3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Meisinger C., Pfannschmidt S., Rissler M., Milenkovic D., Becker T., Stojanovski D., Youngman M. J., Jensen R. E., Chacinska A., Guiard B., et al. 2007. The morphology proteins Mdm12/Mmm1 function in the major beta-barrel assembly pathway of mitochondria. EMBO J. 26: 2229–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kornmann B., Osman C., and Walter P.. 2011. The conserved GTPase Gem1 regulates endoplasmic reticulum-mitochondria connections. Proc. Natl. Acad. Sci. USA. 108: 14151–14156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Stroud D. A., Oeljeklaus S., Wiese S., Bohnert M., Lewandrowski U., Sickmann A., Guiard B., van der Laan M., Warscheid B., and Wiedemann N.. 2011. Composition and topology of the endoplasmic reticulum-mitochondria encounter structure. J. Mol. Biol. 413: 743–750. [DOI] [PubMed] [Google Scholar]
- 93.Frederick R. L., McCaffery J. M., Cunningham K. W., Okamoto K., and Shaw J. M.. 2004. Yeast Miro GTPase, Gem1p, regulates mitochondrial morphology via a novel pathway. J. Cell Biol. 167: 87–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Koshiba T., Holman H. A., Kubara K., Yasukawa K., Kawabata S., Okamoto K., MacFarlane J., and Shaw J. M.. 2011. Structure-function analysis of the yeast mitochondrial Rho GTPase, Gem1p: implications for mitochondrial inheritance. J. Biol. Chem. 286: 354–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Osman C., Haag M., Potting C., Rodenfels J., Dip P. V., Wieland F. T., Brügger B., Westermann B., and Langer T.. 2009. The genetic interactome of prohibitins: coordinated control of cardiolipin and phosphatidylethanolamine by conserved regulators in mitochondria. J. Cell Biol. 184: 583–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nguyen T. T., Lewandowska A., Choi J-Y., Markgraf D. F., Junker M., Bilgin M., Ejsing C. S., Voelker D. R., Rapoport T. A., and Shaw J. M.. 2012. Gem1 and ERMES do not directly affect phosphatidylserine transport from ER to mitochondria or mitochondrial inheritance. Traffic. 13: 880–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Friedman J. R., Lackner L. L., West M., DiBenedetto J. R., Nunnari J., and Voeltz G. K.. 2011. ER tubules mark sites of mitochondrial division. Science. 334: 358–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Murley A., Lackner L. L., Osman C., West M., Voeltz G. K., Walter P., and Nunnari J.. 2013. ER-associated mitochondrial division links the distribution of mitochondria and mitochondrial DNA in yeast. eLife. 2: e00422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hobbs A. E., Srinivasan M., McCaffery J. M., and Jensen R. E.. 2001. Mmm1p, a mitochondrial outer membrane protein, is connected to mitochondrial DNA (mtDNA) nucleoids and required for mtDNA stability. J. Cell Biol. 152: 401–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Böckler S., and Westermann B.. 2014. Mitochondrial ER contacts are crucial for mitophagy in yeast. Dev. Cell. 28: 450–458. [DOI] [PubMed] [Google Scholar]
- 101.Mattiazzi Ušaj M., Brložnik M., Kaferle P., Žitnik M., Wolinski H., Leitner F., Kohlwein S. D., Zupan B., and Petrovič U.. 2015. Genome-wide localization study of yeast Pex11 identifies peroxisome-mitochondria interactions through the ERMES complex. J. Mol. Biol. 427: 2072–2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lang A., John Peter A. T., and Kornmann B.. 2015. ER-mitochondria contact sites in yeast: beyond the myths of ERMES. Curr. Opin. Cell Biol. 35: 7–12. [DOI] [PubMed] [Google Scholar]
- 103.Wideman J. G., Gawryluk R. M. R., Gray M. W., and Dacks J. B.. 2013. The ancient and widespread nature of the ER-mitochondria encounter structure. Mol. Biol. Evol. 30: 2044–2049. [DOI] [PubMed] [Google Scholar]
- 104.Tan T., Ozbalci C., Brügger B., Rapaport D., and Dimmer K. S.. 2013. Mcp1 and Mcp2, two novel proteins involved in mitochondrial lipid homeostasis. J. Cell Sci. 126: 3563–3574. [DOI] [PubMed] [Google Scholar]
- 105.Lahiri S., Chao J. T., Tavassoli S., Wong A. K. O., Choudhary V., Young B. P., Loewen C. J. R., and Prinz W. A.. 2014. A conserved endoplasmic reticulum membrane protein complex (EMC) facilitates phospholipid transfer from the ER to mitochondria. PLoS Biol. 12: e1001969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wideman J. G. 2015. The ubiquitous and ancient ER membrane protein complex (EMC): tether or not? F1000Res. 4: 624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Elbaz-Alon Y., Rosenfeld-Gur E., Shinder V., Futerman A. H., Geiger T., and Schuldiner M.. 2014. A dynamic interface between vacuoles and mitochondria in yeast. Dev. Cell. 30: 95–102. [DOI] [PubMed] [Google Scholar]
- 108.Hönscher C., Mari M., Auffarth K., Bohnert M., Griffith J., Geerts W., van der Laan M., Cabrera M., Reggiori F., and Ungermann C.. 2014. Cellular metabolism regulates contact sites between vacuoles and mitochondria. Dev. Cell. 30: 86–94. [DOI] [PubMed] [Google Scholar]
- 109.Lang A. B., Peter A. T. J., Walter P., and Kornmann B.. 2015. ER-mitochondrial junctions can be bypassed by dominant mutations in the endosomal protein Vps13. J. Cell Biol. 210: 883–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bankaitis V. A., Johnson L. M., and Emr S. D.. 1986. Isolation of yeast mutants defective in protein targeting to the vacuole. Proc. Natl. Acad. Sci. USA. 83: 9075–9079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Robinson J. S., Klionsky D. J., Banta L. M., and Emr S. D.. 1988. Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol. Cell. Biol. 8: 4936–4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Brickner J. H., and Fuller R. S.. 1997. SOI1 encodes a novel, conserved protein that promotes TGN-endosomal cycling of Kex2p and other membrane proteins by modulating the function of two TGN localization signals. J. Cell Biol. 139: 23–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Huh W-K., Falvo J. V., Gerke L. C., Carroll A. S., Howson R. W., Weissman J. S., and O’Shea E. K.. 2003. Global analysis of protein localization in budding yeast. Nature. 425: 686–691. [DOI] [PubMed] [Google Scholar]
- 114.Park J-S., and Neiman A. M.. 2012. VPS13 regulates membrane morphogenesis during sporulation in Saccharomyces cerevisiae. J. Cell Sci. 125: 3004–3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Suresh H. G., da Silveira Dos Santos A. X., Kukulski W., Tyedmers J., Riezman H., Bukau B., and Mogk A.. 2015. Prolonged starvation drives reversible sequestration of lipid biosynthetic enzymes and organelle reorganization in Saccharomyces cerevisiae. Mol. Biol. Cell. 26: 1601–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gatta A. T., Wong L. H., Sere Y. Y., Calderón-Noreña D. M., Cockcroft S., Menon A. K., and Levine T. P.. 2015. A new family of StART domain proteins at membrane contact sites has a role in ER-PM sterol transport. eLife. 4: 07253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.González Montoro A., and Ungermann C.. 2015. StARTing to understand membrane contact sites. Trends Cell Biol. 25: 497–498. [DOI] [PubMed] [Google Scholar]
- 118.Murley A., Sarsam R. D., Toulmay A., Yamada J., Prinz W. A., and Nunnari J.. 2015. Ltc1 is an ER-localized sterol transporter and a component of ER-mitochondria and ER-vacuole contacts. J. Cell Biol. 209: 539–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Elbaz-Alon Y., Eisenberg-Bord M., Shinder V., Stiller S. B., Shimoni E., Wiedemann N., Geiger T., and Schuldiner M.. 2015. Lam6 regulates the extent of contacts between organelles. Cell Reports. 12: 7–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Manford A. G., Stefan C. J., Yuan H. L., Macgurn J. A., and Emr S. D.. 2012. ER-to-plasma membrane tethering proteins regulate cell signaling and ER morphology. Dev. Cell. 23: 1129–1140. [DOI] [PubMed] [Google Scholar]
- 121.Pan X., Roberts P., Chen Y., Kvam E., Shulga N., Huang K., Lemmon S., and Goldfarb D. S.. 2000. Nucleus-vacuole junctions in Saccharomyces cerevisiae are formed through the direct interaction of Vac8p with Nvj1p. Mol. Biol. Cell. 11: 2445–2457. [DOI] [PMC free article] [PubMed] [Google Scholar]