ABSTRACT
N-myc down-regulated gene 1 (NDRG1) has been shown to regulate tumor growth and metastasis in various malignant tumors and also to be dysregulated in esophageal squamous cell carcinoma (ESCC). Here, we show that NDRG1 overexpression (91.9%, 79/86) in ESCC tumor tissues is associated with poor overall survival of esophageal cancer patients. When placed in stable transfectants of the KYSE 30 ESCC cell line generated by lentiviral transduction with the ectopic overexpression of NDRG1, the expression of transducin-like enhancer of Split 2 (TLE2) was decreased sharply, however β−catenin was increased. Mechanistically, NDRG1 physically associates with TLE2 and β−catenin to affect the Wnt pathway. RNA interference and TLE2 overexpression studies demonstrate that NDRG1 fails to active Wnt pathway compared with isogenic wild-type controls. Strikingly, NDRG1 overexpression induces the epithelial mesenchymal transition (EMT) through activating the Wnt signaling pathway in ESCC cells, decreased the expression of E-cadherin and enhanced the expression of Snail. Our study elucidates a mechanism of NDRG1-regulated Wnt pathway activation and EMT via affecting TLE2 and β-catenin expression in esophageal cancer cells. This indicates a pro-oncogenic role for NDRG1 in esophageal cancer cells whereby it modulates tumor progression.
KEYWORDS: β-catenin, EMT, esophageal cancer, NDRG1, TLE2, Wnt pathway
Abbreviations
- ANGPT1
Angiopoietin 1
- CDC25A
Cell division cycle 25A
- CDH1
Cadherin 1, type 1, E-cadherin (epithelial)
- CDH2
Cadherin 2, type 1, N-cadherin
- EMT
epithelial-mesenchymal transition
- ESCC
esophageal squamous cell carcinoma
- FGFBP1
Fibroblast growth factor binding protein 1
- FOXC2
Forkhead box C2
- FZD8
Frizzled class receptor 8
- IHC
immunohistochemistry
- KRT19
Keratin 19
- Lef
Lymphoid enhancer-binding factor
- MITF
Microphthalmia-associated transcription factor
- MMP1
Matrix metalloproteinase 1
- MSN
Moesin
- MTSS1
Metastasis suppressor 1
- MYC
V-myc avian myelocytomatosis viral oncogene homolog
- NDRG1
N-myc downstream-regulated gene 1
- NF-kB
Nuclear factor-kappa B
- NUDT13
Nudix (nucleoside diphosphate linked moiety X)-type motif 13
- OCLN
Occludin
- PLAUR
Plasminogen activator, urokinase receptor
- SERPINB5
Serpin peptidase inhibitor, clade B (ovalbumin), member 5
- SPP1
Secreted phosphoprotein 1
- SYK
Spleen tyrosine kinase
- TCF
Transcription factor
- TGFB1
Transforming growth factor-β 1
- TLE2
Transducin-like enhancer of split 2
- TMA
tissue microarray
- TSPAN13
Tetraspanin 13
- VIM
Vimentin
- WNT5A
Wingless-type MMTV integration site family, member 5A
- ZEB1
Zinc finger E-box binding homeobox 1
Introduction
Esophageal squamous cell carcinoma (ESCC) is the most popular pathological type of esophageal cancer. Mechanisms underlying ESCC development and progression are still unclear, and there's no early detection method with high sensitivity and specificity. Therefore, the 5-year survival rate of ESCC is no more than 10%. N-myc downregulated-gene 1 (NDRG1) is a member of the NDRG family, which consists of 4 members in mammals, NDRG1, NDRG2, NDRG3 and NDRG4.1 The NDRG1 gene is approximately 60 kb in length and is located on chromosome 8q24.3. Its protein is composed of 394 amino acids and is highly conserved, with a molecular weight of 43 kDa.2,3 The NDRG1 protein consists of an α/β hydrolase group enzyme, but it lacks a hydrolytic catalytic site and thus is deficient in enzymatic function. The C-terminus of NDRG1 encodes 3 tandem repeats that are involved in binding heavy metal ions.4 The N terminus of NDRG1 includes 2 myc boxes, which are called MBI and MBII. Both myc boxes are pivotal for the protein's function, which is implicated in many processes in cell biology, such as cell differentiation, stress, hormone response, and cell growth.5
NDRG1 has been shown to be a predominantly cytoplasmic protein, but its subcellular localization was predicted to include the cytoskeleton, microtubule organizing center, centrosome, nucleus and cell membrane, according to the UniProtKB/Swiss-Prot data for the NDRG1 gene (http://www.genecards.org/). NDRG1 cellular localization is dependent on and specific to the cell type. Although the amino acid sequence of the NDRG1 protein does not contain a predicted nuclear localization signal, NDRG1 appears to localize to the nucleus in some cell types.6,7 The expression of NDRG1 can be strongly induced by several stress stimuli, such as reducing agents, tunicamycin, Ni2+ compounds, calcium, and hypoxia.8-10 Various stimuli associated with carcinogenesis, including DNA damage, methylation and histone deacetylation-targeting drugs, oncogenes and tumor suppressor genes, alter the expression of NDRG1.3,11 NDRG1 expression during tumor progression is highly controversial, although it has been evaluated by immunohistochemical analysis in clinical cancer samples. Previous reports demonstrated that NDRG1 mRNA and protein levels were decreased in several malignancies, including gastric cancer, neuroblastoma, and colorectal cancer, compared with those in the corresponding normal tissues.12-16 NDRG1 was found to be a metastasis suppressor gene with potential roles in several functions, such as cell differentiation, cell cycle regulation and responses to hormones and stress.5,17,18 However, it was also reported that NDRG1 is upregulated in cervical adenocarcinoma, breast cancer, oral and oropharyngeal squamous cell carcinoma, and liver cancer.7,19-21 High NDRG1 expression is associated with angiogenesis and is a poor prognostic indicator because it is an oncogene.20
Previous results demonstrated that in ESCC specimens, NDRG1 mRNA expression was significantly reduced in tumors with more advanced pathology and local tumor invasion.22 Recently, mounting evidence has shown that the increased expression of NDRG1 protein is correlated with the malignant status of some cancers, including prostate cancer, lung cancer, and breast cancer, and it may therefore be a prognostic marker.7,23,24 There is no obvious correlation between protein and mRNA levels according to western blot assays and real-time RT-PCR, respectively, in esophageal cancer.25 It has also been reported that the ectopic overexpression of NDRG1 is highly associated with markers of metastasis, angiogenesis, apoptotic evasion, and enhanced NF-κB activity, suggesting that NDRG1 may play important roles in the progression of ESCC.1 Ureshino et al. demonstrated that higher expression of NDRG1 is closely correlated with poor prognosis in gastric cancer patients and promotes the metastasis of gastric cancer via the epithelial-mesenchymal transition (EMT).26 However, its function in malignant tumors is not fully clear. Previous data have shown that the overexpression of NDRG1 regulates NF-κB activation, which suggests it may also promote tumor progress and metastasis. To further investigate the role of NDRG1 in esophageal cancer, we explored the expression of NDRG1 in patients with ESCC and examined the mechanisms of NDRG1 ectopic overexpression and the relationship between NDRG1 and the Wnt signaling pathway in esophageal cancer.
Materials and methods
Cell culture and transfections
The esophageal cancer cell lines KYSE 30, KYSE 140, KYSE 150, KYSE 170, KYSE 180, KYSE 410 and KYSE 510 were obtained from Dr. Yutaka Shimada at Hyogo College of Medicine.27 EC 0156 was described previously.28 HEK293 cells were purchased from the Peking Union Medical College Cell Resource Center. All cell lines were grown in RPMI 1640 medium and supplemented with 10% FBS, 100 U/ml penicillin, and 100 µg/ml streptomycin at 37°C in 5% CO2.
For NDRG1 overexpression, the KYSE 30 cells were transfected with the pCMV6-entry-NDRG1 or pCMV6 empty vector (Origen, Rockville MD) by Lipofectamine 2000 (Invitrogen, USA).29 And the infection of KYSE 30 cells with NDRG1 RNAi-pLVTHM (shNDRG1) vector was used to stably knockdown the expression of NDRG1, in which pLVTHM bicistroniclentiviral vectors were purchased from Addgene (www.addgene.org). Within the pLVTHM vector, an additional H1 RNA polymerase III promoter permits the expression of a short hairpin RNA (shRNA) of NDRG1 for RNA interference (RNAi). The sense sequences for NDRG1 RNAi and scrambled control were 5′-GGA GTC CTT CAA CGA TTT G-3′ and 5′-GGG TCT TAG AAC TAG TTC C-3′ respectively.1 HEK293 cells were transiently transfected with pCMV6-entry-NDRG1, pCMV6-entry-TLE2, and pCMV6 empty vectors using the Lipofectamine 2000 reagent according to the manufacturer's protocol. All constructs were confirmed by sequencing.
The following abbreviations are used throughout the paper: KYSE 30-NDRG1, a selected KYSE 30 clone that exogenously overexpresses NDRG1 via the pCMV6-entry vector, and KYS E30-Ctrl, KYSE 30 cells transfected with empty vector; KYSE 30-shNDRG1, a selected KYSE 30 clone with reduced NDRG1 expression compared with its scrambled control cells (KYSE 30-vec) generated by lentiviral transduction.
Sample collection and IHC staining
Surgical tissue specimens were collected after obtaining informed consent and all patients were diagnosed by 2 senior pathologists without chemo/radiotherapy before surgical operation. A total of 201 ESCC tumor and paired adjacent non-tumor tissue samples, including 29 fresh and 172 formalinfixed paraffin-embedded tissues, were collected from ESCC patients undergoing resection during the period from July 2006 to December 2008. The ESCC tissue microarrays (TMAs), contained primary tumor and adjacent esophageal epithelia tissues of 78 pairs and 8 tumor only ESCC patients, were purchased from the Outdo Biotechnology (Cat No. XT11-020, Shanghai, China). For immunohistochamical staining, the TMAs were incubated with the NDRG1 antibody or control IgG (1 μg/mL). After washing in PBS, they were incubated with a biotin-labeled secondary antibody. Signals were visualized using an ultrasensitive streptavidin-peroxidase system (Maxim Biotech, Fuzhou, China). After calculating the informative cases, the intensity of NDRG1 staining in individual case was quantified as described previously. A score of 4∼6 was defined as ‘weak expression’, 7∼9 as “moderate expression,” 10∼12 as “strong expression” and a score of 0∼3 as ‘negative’.30 The study has been approved by the Ethics Committee of Cancer Institute and Hospital, Chinese Academy of Medical Sciences (No. 12–130/664, Beijing, China).
Real-time PCR arrays
The Human Cancer Pathway Finder (PAHS-033A, Qiagen), Epithelial Mesenchymal Transition (PAHS-090A, Qiagen), and Wnt Signaling Pathway (PAHS-043A, Qiagen) PCR arrays were used to analyze the mRNA levels of 252 genes related to cell proliferation, apoptosis, the cell cycle, angiogenesis, invasion, metastasis, EMT, and Wnt signaling. The complete list of specific genes in this study is available in Supplementary Table 1, Table 2 and Table 3. Total RNA was isolated from the KYSE 30-NDRG1 and KYSE 30-vec cells using an RNeasy® mini kit (Qiagen, USA). The RNA yield and quality was assessed by UV absorbance and denaturing agarose gel electrophoresis. First-strand complementary synthesis reactions were performed using the SuperScript® III Reverse Transcriptase kit (Invitrogen, USA). The reactions were amplified using SsoFastTM EvaGreen® Supermix with a CFX 96TM real-time system (Chemoscience, USA). The complete list of gene-specific real-time primers used in this study is available in Supplementary Table 4. Data analysis of gene expression was performed using Excel-based PCR Array Data Analysis Software that was provided by the manufacturer (Qiagen, China). GAPDH served as an internal control to normalize the loading of the template cDNA. Each experiment was repeated at least twice, and the fold change in gene expression was assessed using the ΔCt method, according to a previous description.31
Immunoprecipitation and protein gel blot analysis
NDRG1 immunoprecipitation was performed according to a previously described protocol.32 Briefly, cells cultured in 10-cm dishes were lysed for 30 min on ice in lysis buffer containing 1% (w/v) Triton X-100, 0.15 M NaCl, 30 mM Tris-HCl (pH 7.5) and protease inhibitors (Roche, Germany). The lysates were sonicated and centrifuged at 10,000 g for 15 min at 4°C. A 1-mg sample of extracted protein in lysis buffer was incubated overnight with 5 μg of rabbit anti-NDRG1 antibody, which was followed by incubation with 20 μl of Dynabeads® Protein G (Invitrogen, USA) for 2 h at 4°C and 3 washes with lysis buffer. The beads were directly boiled in 1% SDS loading buffer for 5 min. Subcellular fractions, including nuclear and cytosolic fractions, were prepared using the proteoExtract sub-cellular proteome extraction kit (Calbiochem, USA) according to the manufacturer's instructions. The subcellular extraction of adherent tissue culture cells was performed as follows. Prior to extraction, the buffers were mixed well by vortexing. During the extraction procedure, the I-III and Benzonase® buffers were kept on ice, and Buffer IV and the Protease Inhibitor Cocktail were kept at room temperature. The different cellular fractions were extracted using the proteoExtract sub-cellular proteome extraction kit (Calbiochem, USA) according to the manufacturer's instructions.
Western blot analysis was performed as described previously 28 using the following antibodies: anti-NDRG1 and anti-Snail antibodies were purchased from Abcam Inc. (Abcam, MA, USA); anti-β-catenin from Santa Cruz Biotechnology (Santa Cruz, CA); anti-TLE2 from Bioworld (Bioworld, USA); anti-E-cadherin from Becton Dickinson and Company (BD, USA); and anti-β-actin from Sigma (Sigma, USA).
Immunofluorescent staining
KYSE 30-NDRG1 and KYSE 30-Ctrl cells were grown on glass coverslips and fixed in 4% paraformaldehyde for 30 min, rinsed 3 times with PBS, permeabilized with 0.1% Triton X-100, and blocked for 30 min with PBS containing 2% BSA. The cells on the coverslips were incubated for 1 h at room temperature with the indicated primary antibodies and then incubated at room temperature for 30 min with the appropriate fluorescent secondary antibodies. Fluorescence images were captured with a Nikon ECLIPSE 80i microscope.33
In vivo xenograft assay and animal ethics statement
Animal experiments were conducted as previously described.28 Briefly, 5 × 106 KYSE 30-NDRG1 or KYSE 30- Ctrl cells were suspended in 100 µl of PBS and injected subcutaneously into the right flanks of female nude mice (n = 3). To monitor the tumor growth, the tumor size was measured every 2–3 d using digital callipers. The tumor volume was calculated using the formula 0.5 × a × b2, where a is the length of the tumor, and b is the width. For immunohistochemical (IHC) staining, sections of paraffin-embedded xenograft tumor tissue were used for IHC as we have previously described.28 Briefly, sections were incubated with a primary antibody for NDRG1, Ki67, TLE2, MMP2, E-cadherin or β-catenin. After a PBS wash, they were incubated with a biotin-labeled secondary antibody. Signals were visualized using an ultrasensitive streptavidin-peroxidase system (Maxim Biotech, Fuzhou, China). All experimental procedures for the use of animals were previously reviewed and approved by the Animal Care and Use Committee of Cancer Institute and Hospital, Chinese Academy of Medical Sciences (Nos. NCC2013A012 and NCC2015A021, Beijing, China). All surgeries were performed under sodium pentobarbital anesthesia, and all efforts were made to minimize suffering.
Statistical analysis
Values are reported as the mean ± SD, and statistical significance was assessed with Student's t tests using Prism (v. 5, GraphPad). The relationship between the expression of NDRG1 protein and clinicopathologic characteristics was assessed by χ2 test. The xenograft tumor volume was compared between the KYSE 30-NDRG1 and KYSE-vec control groups by Student's t tests in the mouse xenograft assay. A P value of less than 0.05 was considered statistically significant.
Ethics statement
Surgical tissue specimens were collected after obtaining informed consent and approval from the Institutional Review Board of the Cancer Institute and Hospital of Chinese Academy of Medical Sciences (Beijing, China). All participants provided their written informed consent to participate in this study. The animal experiments protocol were passed the Animal Experimental Ethical Inspection of the Animal Care & Welfare Committee of Cancer Institute and Hospital, Chinese Academy of Medical Sciences & Peking Union Medical College (Beijing, China).
Results
NDRG1 overexpression in ESCC tumor tissues was associated with poor overall survival of esophageal cancer patients
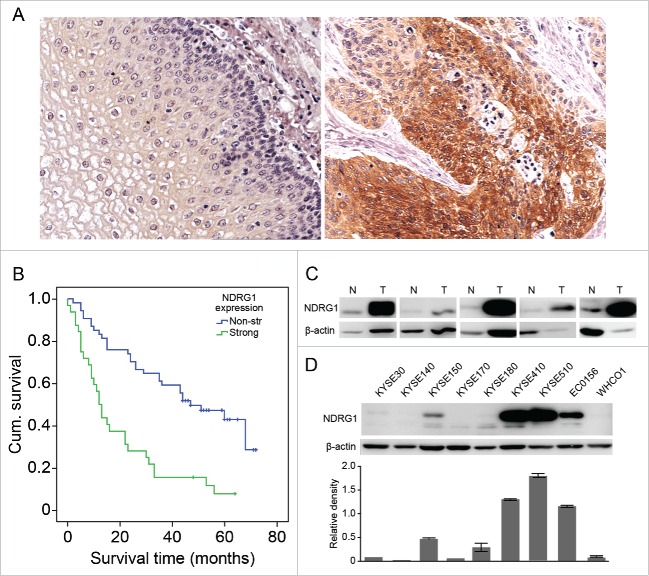
To assess the levels of NDRG1 expression in ESCC and adjacent normal tissues, we analyzed NDRG1 expression in TMAs of ESCC using IHC staining. Of the 86 ESCC cases with 78 paired adjacent tissues were analyzed, 86 tumor and 77 adjacent tissues were informative. In the tumor tissues, 18 (20.9%) had weak, 29 (33.7%) had moderate and 32 (37.2%) had strong staining of NDRG1, while 7 (8.1%) cases had negative expression. In contrast, the proportions of weak, moderate, strong and negative staining of NDRG1 in adjacent non-tumor tissues were 66.2% (51/77), 6.5% (5/77), 1.3% (1/77) and 26.0% (20/77), respectively. The upregulation of NDRG1 in ESCC tumor tissues compared with the levels in normal tissues was statistically significant (chi-square test, P < 0.001). NDRG1 protein was detected in the cytoplasm of ESCC and normal cells, with a definitive pattern in most tissues (Fig. 1A). We further analyzed the correlation between NDRG1 expression and patient survival. As shown in Fig. 1B, the estimated overall median survival time was 12 months (95% CI, 9–15) in the strongly expressing NDRG1 (strong) group and 47 months (95% CI, 29–65) in the weakly expressing NDRG1 (weak) group. The Kaplan-Meier analysis indicated that patients from the NDRG1-strong subset had significantly shorter overall median survival times than those in the NDRG1-week subset (P < 0.001). However, NDRG1 positivity did not have a significant association with other clinic pathological factors, including gender, age, pathological grade, tumor diameter, tumor position, lymphatic metastasis and AJCC stage (Table 1).
Figure 1.
NDRG1 overexpression was correlated with poor overall survival in esophageal cancer. (A) Expression of NDRG1 was analyzed by immunohistochemical analysis in TMAs containing 78 ESCC tumor and adjacent normal epithelial tissues, with duplicate cores used for each case. The majority of tumor areas strongly express NDRG1 in the cytoplasm. Magnification, 200×. (B) Kaplan-Meier curve combined with Log-rank analysis for patients with ESCC showing weak and strong NDRG1 expression. (C) NDRG1 expression in a subset of ESCC (T) and matched non-neoplastic surgical tissues (N) was analyzed by Western blot analysis. (D) Whole cell protein extracts from 9 esophageal cancer cell lines were subjected to protein gel blot analysis using antibodies against NDRG1. Quantitative values of relative NDRG1 levels were normalized to β-actin (mean ± SD).
Table 1.
Correlation between NDRG1 expression and clinical parameters.
| NDRG1 |
||||
|---|---|---|---|---|
| Variable |
Total (n=86) |
Negative (%) |
Positive (%) |
P value |
| Gender | ||||
| Male | 64 | 4 (57.1) | 60 (75.9) | 0.365 |
| Female | 22 | 3 (42.9) | 19 (24.1) | |
| Age | ||||
| ≤65 | 46 | 3 (42.9) | 43 (54.4) | 0.700 |
| >65 | 40 | 4 (57.1) | 36 (45.6) | |
| Pathological grad | ||||
| I, I-II | 28 | 4 (57.1) | 24 (41.8) | 0.440 |
| II, II-III, III | 58 | 3 (42.9) | 55 (58.2) | |
| Tumor diameter | ||||
| ≤4 .5 cm | 44 | 5 (71.4) | 39 (41.5) | 0.235 |
| >4.5 cm | 40 | 2 (28.6) | 38 (58.5) | |
| Position | ||||
| Up and middle | 31 | 3 (60.0) | 28 (60.9) | 1.000 |
| Low | 20 | 2 (40.0) | 18 (39.1) | |
| Lymphatic metastasis | ||||
| Negative | 50 | 5(83.3) | 45 (58.4) | 0.395 |
| Positive | 33 | 1(16.7) | 32 (41.6) | |
| AJCC stage | ||||
| 1B, 2 | 50 | 4 (66.7) | 46 (59.0) | 1.000 |
| 3A, 3B, 4 | 30 | 2 (33.3) | 28 (41.0) | |
| Tissue | ||||
| Normal | 77 | 20 (74.1) | 57 (47.2) | 0.002 |
| Cancer | 86 | 7 (25.9) | 79 (52.8) | |
To assess the expression of NDRG1, 5 ESCC cases with paired adjacent tissues and a panel of 9 additional ESCC cell lines were evaluated via western blot assay (Fig. 1C and 1D). The data showed that the adjacent tissues expressed low levels of NDRG1, whereas tumor tissue expressed much higher levels of NDRG1 protein, indicating that the level of NDRG1 protein expression was much stronger in ESCC than in paired adjacent tissues. The expression level of NDRG1 varied across the 9 cell lines, with KYSE 410, KYSE 510, and EC 0156 showing relatively high expression of NDRG1 and KYSKE 30, KYSE 140, KYSE 170, KYSE 180 and WHCO1 showing relatively low expression levels of NDRG1. Based on the cell panel data, the cell line KYSE 30 was chosen for further investigation of the effects of overexpressing NDRG1. Overall, these data further supported the idea that an increase in NDRG1 expression is a poor prognostic indicator in ESCC patients.
NDRG1 overexpression induces EMT in esophageal cancer cells
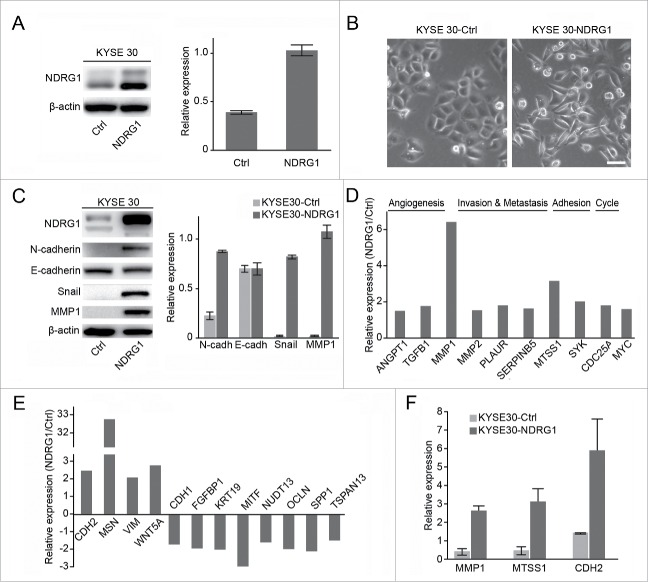
Our previous data showed that NDRG1 is a pro-oncogenic gene but that it performs a dispensable role in ESCC as a modulator of metastasis.1 NDRG1 overexpression significantly enhanced motility in wound-healing assays in KYSE 30 cells and also elevated MMP2 activity, a surrogate marker of metastasis.1 To functionally validate the role of NDRG1 in esophageal cancer, pCMV-NDRG1 transfection was performed in the KYS E30 esophageal cancer cell line. The expression of NDRG1 protein was analyzed in KYSE 30 cells with pCMV-NDRG1 (KYSE 30-NDRG1) by protein gel blot assay and was found to be significantly higher than in control cells (KYSE 30-Ctrl) (Fig. 2A). Interestingly, NDRG1 overexpression induced a change in KYSE 30 cells from an epithelial morphology to a spindle-like mesenchymal morphology (Fig. 2B). Consistent with the phenotypic changes associated with NDRG1 overexpression, we also observed an increase in the expression of mesenchymal markers, including N-cadherin, Snail and MMP1, but we did not observe decreased expression of the epithelial marker E-cadherin at the protein level (Fig. 2C). To provide new insights into the mechanisms by which NDRG1 might modulate metastasis, we performed an analysis of gene expression profiles in KYSE 30 cells overexpressing NDRG1 (KYS E30- NDRG1) versus mock cells (KYSE 30-Ctrl), focusing on genes in the angiogenesis, metastasis, adhesion, cycle and Wnt pathways using low-density real-time PCR arrays. The expression of 22 genes associated with angiogenesis, metastasis, adhesion, cycle and Wnt pathways, including ANGPT1, TGFB1, MMP1, MMP2, PLAUR, SERPINB5, MTSS1, SYK, CDC25A, MYC, CDH2, MSN, VIM, WNT5A, CDH1, FGFBP1, KRT19, MITF, NUDT13, OCLN, SPP1, and TSPAN13, were altered in response to NDRG1 overexpression. As shown in, Fig. 2D and 2E the expression levels of the angiogenesis-related genes ANGPT1 and TGFB1, invasion and metastasis-related genes MMP1, MMP2, PLAUR and SERPINB5, adhesion-related genes MTSS1 and SYK, cell cycle-related gene CDC25A, and oncogenic transcription factor MYC were significantly enhanced after NDRG1 overexpression. In contrast, the genes CDH1, FGFBP1, KRT19, MITF, NUDT13, OCLN, SPP1, and TSPAN13 were decreased. Thus, NDRG1 overexpression resulted in the induction of EMT in KYSE 30 cells. Quantitative RT-PCR analysis of MMP1, MTSS1 and CDH2 mRNA levels was validated additionally in KYSE 30-NDRG1 and mock cells (KYSE 30-Ctrl, Fig. 2F).
Figure 2.
NDRG1 overexpression induces the epithelial mesenchymal transition in esophageal cancer cells. (A) The effect of NDRG1 exogenous expression was analyzed by western blot analysis. Quantitative values of relative NDRG1 levels were normalized to β-actin (mean ± SD). (B) The morphological changes of esophageal cancer cells overexpressed NDRG1. Image at 200x, 20 µm. (C) Whole cell protein extracts from KYSE 30-NDRG1 and KYSE 30-Ctrl cells were subjected to protein gel blot analysis using antibodies against NDRG1, E-cadherin, N-cadherin, Snail and MMP1. Quantitative values of relative protein levels were normalized to actin (mean ± SD). ((D)& E), Relative expression levels of genes associated with angiogenesis, metastasis, adhesion, the cell cycle and the Wnt pathway in KYSE 30 cells overexpressed NDRG1 (KYSE 30-NDRG1) compared with those of mock cells (KYS E30-Ctrl); results are based on analysis in real-time PCR arrays. (F) The mRNA levels of MMP1, MTSS1 and CDH2 were validated in both KYSE 30-NDRG1 and KYSE 30-Ctrl cells by RT-PCR. KYSE 30-NDRG1, NDRG1 overexpression KYSE 30 cells; KYSE 30-Ctrl, KYSE 30 cells transfected with vector control.
NDRG1 impacts the Wnt pathway via TLE2 and β-catenin in esophageal cancer cells
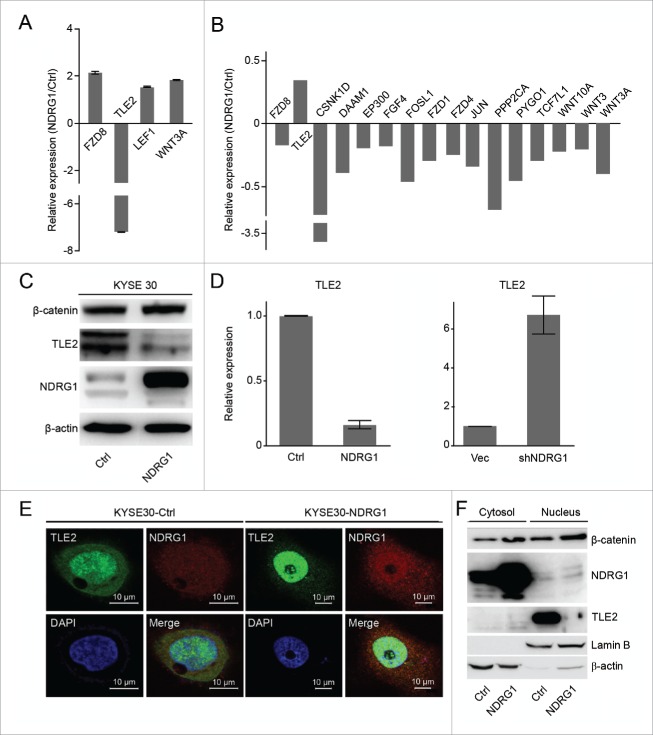
A key event following Wnt pathway activation is the molecular switch from transcriptional repression of TLE to the activation of TCF/Lef.34 Through quantitative gene expression analysis of key genes involved in metastasis, we found that Wnt pathway-associated genes, including Frizzled Class Receptor 8 (FZD8), Transducin-Like Enhancer of Split 2 (TLE2), Lymphoid Enhancer-Binding Factor 1 (LEF1) and Wingless-Type MMTV Integration Site Family, Member 3A (WNT3A), were changed; TLE2 expression was significantly decreased and other genes were greatly increased in NDRG1 overexpressing cells (KYSE 30-NDRG1) (Fig. 3A). The mRNA levels of Wnt pathway-associated genes in NDRG1 knock-down cells (KYSE 30-shNDRG1, shNDRG1) were assessed by quantitative RT-PCR and compared with mock cells (KYSE 30-vec, vec)(Fig. 3B). To identify functional links between NDRG1 and TLE2, we examined the effects of NDRG1 overexpression and knock-down on TLE expression. As shown in Fig. 3C and 3D, NDRG1 overexpression suppressed TLE2 expression at both the transcript and protein levels. Inversely, NDRG1 knock-down induced TLE2 upregulation at the transcript level in KYSE 30 cells. To further confirm that TLE expression is regulated by NDRG1, we assessed the co-localization of NDRG1 and TLE2 using immunofluorescence and western blot analysis following NDRG1 ectopic expression. As shown in Fig. 3E, we observed that NDRG1 overexpression slightly increased the nuclear localization of NDRG1 and TLE2 compared with that of the control group. Total cytosolic and nuclear fractions were isolated and tested for the expression of NDRG1, TLE2 and β-catenin by protein gel blot analysis. Significant accumulation of β-catenin and NDRG1 and a decrease in TLE protein levels were observed in the nuclear fractions of KYSE 30 cells (KYSE 30-NDRG1), indicating that NDRG1 regulates TLE2 transcript and protein expression (Fig. 3F).
Figure 3.
NDRG1 impacts the Wnt pathway via TLE2 and β-catenin in esophageal cancer cells. (A) Relative gene expression levels of FZD8, TLE2, LEF, and WNT3A were analyzed in KYSE 30 cells overexpressed NDRG1 (KYSE 30-NDRG1) compared with those in mock cells (KYSE 30-Ctrl) using real-time PCR arrays. (B) mRNA levels of Wnt pathway-associated genes in NDRG1 knock-down cells (KYSE 30-shNDRG1, shNDRG1) were assessed by quantitative RT-PCR and compared with mock cells (KYSE 30-vec, vec). (C) Protein levels of NDRG1, TLE2 and β-catenin from KYSE 30-NDRG1 and KYSE 30-Ctrl cells were measured by Western blot. (D) Quantitative RT-PCR results validated the mRNA levels of the TLE2 in KYSE 30-NDRG1 and NDRG1 knock-down (KYSE 30-shNDRG1) cells. (E) The colocalization of NDRG1 and TLE2 was analyzed by confocal microscopy in KYSE 30-NDRG1 and KYSE 30-Ctrl cells. TLE2 was mainly localized in the nuclei, while NDRG1 was expressed in both the nuclei and cytoplasma. NDRG1 and TLE2 colocalized in the nuclei.Green fluorescence, TLE2; red fluorescence, NDRG1; blue fluorescence, nuclei stained with DAPI. Scale bar, 10 μm. (F) Cytosolic and nuclear proteins were isolated from KYSE 30-NDRG1 and KYSE 30-Ctrl cells, and NDRG1, β-catenin and TLE2 expression was analyzed by western blot. Lamin B, nuclear marker; β-actin, cytoplasmic marker.
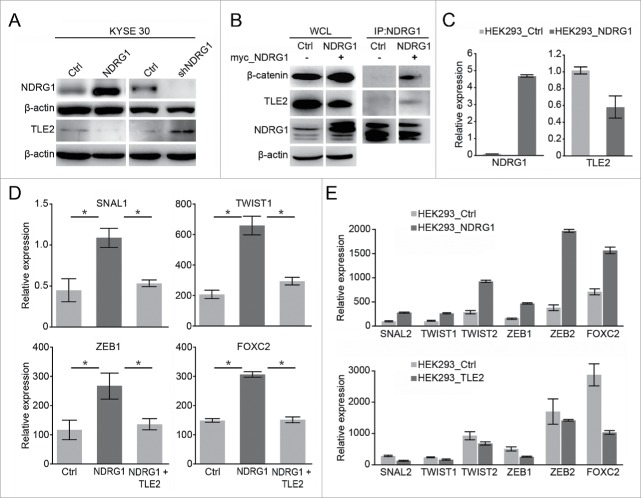
To determine whether NDRG1 overexpression or knock-down impacts TLE2 protein expression, we used western blot analysis to assess TLE2 expression. As shown in Fig. 4A, NDRG1 overexpression and knock-down altered TLE2 expression at the protein level in KYSE 30 cells. Because the overexpression of NDRG1 in KYSE 30 cells leads to transcriptional changes in several markers that are involved in EMT and because NDRG1 is primarily located in the cytoplasm and nucleus in KYSE 30 cells, we sought to determine whether NDRG1 interacts with TLE2 and β-catenin, which are upstream of EMT markers. To test this, anti-NDRG1 was used to immunoprecipitate endogenous NDRG1; we found that the TLE2 and β-catenin complex formed only in NDRG1-overexpressing KYSE 30 cells (KYSE30-NDRG1, Fig. 4B). It has been previously shown that Wnt signal activation is initiated through an interaction with Wnt growth factors and is transduced inside the cell by the protein β-catenin.34,35 To verify the subcellular localization of β-catenin and TLE, we analyzed the subcellular fractions of NDRG1-overexpressing and control KYSE 30 cells, including the cytosolic fraction and nuclear fractions. As shown in Fig. 3F, we found that β-catenin was enriched in both the cytosolic and nuclear fractions; in contrast, the inverse was found for TLE2, which was greatly decreased in the nuclear fraction after NDRG1 overexpression compared with the control group. To further confirm the epithelial and mesenchymal phenotypes, EMT-associated transcription factors, including Snail, Twist1, ZEB1, and FOXC2, were assessed by real-time PCR analysis. We showed that NDRG1 overexpression increased the levels of EMT-associated transcription factors (Fig. 4D). To confirm the role of NDRG1 in this process, 293T cells were transfected with pCMV-NDRG1. The expression of NDRG1 mRNA in these cells was significantly higher than that in control cells; conversely, TLE2 mRNA levels were decreased (Fig. 4C). EMT-associated transcription factors, including Snail, Twist1, ZEB1, and FOXC2, were upregulated in 293T cells that overexpressed NDRG1. These factors were relatively decreased in 293T cells that overexpressed TLE2 (Fig. 4E). Together, these data indicate that NDRG1 overexpression promotes the activation of the Wnt signaling pathway, through increased β-catenin and decreased TLE2.
Figure 4.
Alteration of NDRG1 expression affected TLE2 and EMT associated genes. (A) Comparison of protein expression levels of NDRG1 and TLE2 by protein gel blot analysis of total cell lysates in KYSE 30 overexpression (KYSE 30-NDRG1) and knock-down (KYSE 30-shNDRG1) esophageal cancer cells. (B) NDRG1 interacts with β-catenin and TLE2. NDRG1 was immunoprecipitated using an anti-NDKRG1 antibody. NDRG1, β-catenin and TLE2 were detected by western blot analysis. WCL, whole cell lysis; IP, immunoprecipitation; β-actin was a loading control. (C) The mRNA levels of the NDRG1 and TLE2 genes following transient transfection of HEK293 cells with NDRG1 were validated by quantitative real-time PCR. (D) Relative gene expression levels of SNAIL, TWIST1, ZEB1 and FOXC2 were analyzed by real-time PCR in transiently transfected mock (KYSE 30-Ctrl), NDRG1 (KYSE 30-NDRG1) and NDRG1 with TLE2 KYSE 30 cells. (E) The EMT-associated genes were measured by real-time PCR following the transient transfection of HEK293 cells with NDRG1 (HEK293-NDRG1), TLE2 (HEK293-TLE2) and vector alone (HEK293-Ctrl).
Ectopic overexpression of NDRG1 promotes the tumor progression in xenograft
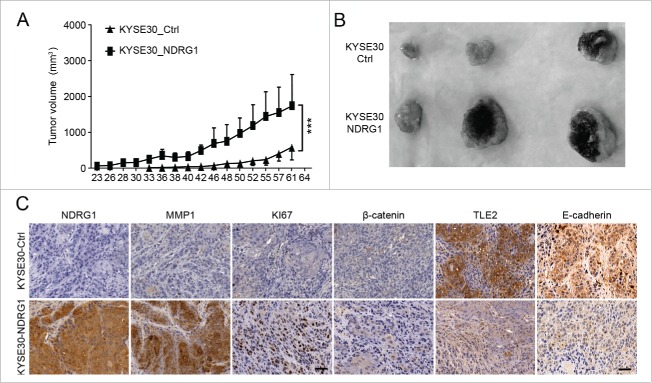
To examine whether NDRG1 ectopic overexpression could affect tumor growth in vivo, we performed in vivo experiments via subcutaneous transplantation of transduced cells into nude mice, whereby we compared tumor growth rates induced by KYSE 30-Vec and KYSE 30-NDRG1 cells in a xenograft model. As shown in Fig. 5A, the tumor growth rate of KYSE 30-NDRG1 cells was significantly faster than that of KYSE 30-Vec cells 20 d after inoculation by subcutaneous injection. We observed that the tumor size of NDRG1-overexpressing cells was much larger than that in the control group (Fig. 5B). Immunohistochemical analysis was investigated in xenograft sections. As shown in Fig. 5C, significantly higher NDRG1 levels were observed in the xenografts derived from NDRG1-overexpressing cells, indicating that NDRG1 overexpression was maintained during these in vivo experiments. MMP1 and Ki67 were highly upregulated in NDRG1-overexpressing xenografts compared with wild type xenografts. This result suggests that higher NDRG1 levels may induce Ki67 and MMP1 levels, which promote tumor growth in vivo, and is consistent with our in vitro data, in which NDRG1 overexpression significantly increased MMP1 at the transcript and protein expression levels.
Figure 5.
Ectopic overexpression of NDRG1 promotes tumor growth in vivo. (A) Tumor volume was measured every 2 to 3 d after subcutaneous inoculation with 5×106 cells/animal. KYSE 30 cells overexpressing NDRG1 (KYSE 30-NDRG1) enhanced the growth of the xenograft tumors compared with that of inoculations using parent cells (P < 0.001). (B) Photographs of the dissected xenografts using the same magnification scale. (C) Xenograft tissue was subjected to immunohistochemical analysis for NDRG1, MMP1, Ki67, β-catenin, TLE2 and E-cadherin, Scale bar, 100 μm.
Our previous data showed that NDRG1 overexpression reduced TLE2 and affected the Wnt signaling pathway. To further investigate whether NDRG1 overexpression could impact the Wnt signaling pathway, we analyzed the expression of TLE2, β-catenin and E-cadherin. NDRG1 overexpression significantly suppressed the expression of TLE2 and E-cadherin and slightly increased the levels of β-catenin protein. Together, the data from these in vivo experiments are consistent with the results we obtained from in vitro experiments. These results indicate that increased levels of NDRG1 affect the Wnt pathway by decreasing TLE2 and slightly increasing β-catenin levels, which promotes EMT in esophageal tumor cells.
Discussion
NDRG1 is highly expressed in most epithelial cells and is associated with differentiation, the cell cycle and responses to stresses, such as hypoxia and CoCl2/Nickel.6,10,12,36 Previous studies have revealed that NDRG1 is a tumor suppressor, and the expression of NDRG1 has often been found to be downregulated in several malignancies.14,15 In fact, in esophageal cancer, NDRG1 levels appear to increase and are associated with their malignant status, indicating its usefulness as a prognostic marker.25 Although the multiple biological functions of NDRG1 have been investigated in several types of cancer, the role of NDRG1 in ESCC remains unclear. We therefore focused on studying NDRG1 in patients with esophageal cancer. Our results demonstrate that NDRG1 was expressed in 91.9% (79/86) of the ESCC samples but only in 74.0% (57/77) of matched adjacent normal tissues. The expression of NDRG1 was significantly upregulated in ESCC tumor tissues compared with that in normal tissues. In the ESCC tumor tissue analysis, we found that patients in the NDRG1-strong expression group had significantly shorter overall median survival times than patients in the NDRG1-weak group. Our data suggest that the upregulation of NDRG1 is closely correlated with poor survival in patients with esophageal cancer, indicating that NDRG1 may contribute to the malignant progression of esophageal carcinogenesis, consistent with the overexpression that has been described in prostate cancer, cervical adenocarcinoma, breast cancer, oral and oropharyngeal squamous cell carcinoma.7,19,23
The multiple mechanisms through which NDRG1 is involved in signaling pathways, including the NF-κB pathway, mammalian target of rapamycin (mTOR) signaling network, extracellular signal-regulated kinase (ERK) pathway, and Wnt pathway 11,37-39 have been demonstrated in several malignancies, but little is known about its biological function in esophageal cancer. Previous results showed that NDRG1 may have an oncogenic function in the progression of ESCC, through which it enhances metastasis, angiogenesis and apoptotic evasion in both in vitro and in vivo experimental studies.1,26 In the present study, we provide evidence showing that ectopic overexpression of NDRG1 induced a change from epithelial morphology in ESCC cells to a spindle-like mesenchymal morphology and induced higher expression of mesenchymal markers, including N-cadherin, Snail and MMP1. Using low-density real-time PCR arrays, we found that the 22 gene profiles we examined, which focused on genes in the angiogenesis, metastasis, adhesion, cell cycle and Wnt pathways, were both upregulated and downregulated in response to NDRG1 overexpression. The above-mentioned results indicate that NDRG1 overexpression may mediate the acquisition of metastatic potential that is associated with EMT, which eventually promotes the progression of tumor cells to malignancy in KYSE 30 cells.
The Wnt signaling pathway is widely known to be correlated with tumourigenesis.35,40 Previous studies have shown that activation of the Wnt pathway can induce EMT, which allows tumor cells to metastasize.14 In this study, using real-time PCR array analysis of the Wnt pathway, we found that Wnt pathway-associated genes, including FZD8, TLE2, LEF1 and WNT3A, were greatly increased, except for TLE2, which was significantly decreased, after NDRG1 overexpression. Cellular fraction assays showed that the accumulation of β-catenin and NDRG1 in the nuclear fraction of KYSE 30 cells was greatly increased, indicating that NDRG1 may impact the Wnt signaling pathway through effects on FZD8, TLE2, LEF1 and WNT3A gene transcription and β-catenin translocation. Previous investigations demonstrated that NDRG1 inhibited oncogenic Wnt signaling through FRAT1, PAK4, and GSK3β phosphorylation, which reduced total β-catenin levels.41,42 Other reports also indicated that NDRG1 suppressed the TGF-β-induced EMT phenotype via stabilizing E-cadherin and β-catenin at the cell membrane.37 Our present results strongly support the hypothesis that NDRG1-mediated promotion or suppression of tumor progression mainly depends on the tumor cell type and its differentiation status.3,43
In the canonical Wnt signaling pathway, a critical event is the switching of Wnt transcription factors from transcriptional repression of TLE2 to activation of TCF/Lef after β-catenin translocation and then binding to TCF/Lef, which is required to initiate the Wnt-specific transcriptional program.34,44 TLE2 belongs to the Groucho/TLE family, which interacts with a variety of DNA-binding proteins.44-46 TLE2 expression results in transcriptional repression of specific gene regulatory sequences, via the recruitment of Groucho. Buscarlet and colleagues demonstrated that active β-catenin binds to TCF/Lef at a site on Lef-1, where it displaces Groucho/TLE, which overlaps with the Lef-1-binding site.34,44,47 Other reports have shown that the modification of TLE by XIAP-mediated ubiquitylation decreases its affinity with β-catenin, thereby allowing the formation of the β-catenin/TCF complex and thus the switch to an activated Wnt signaling pathway.44 The molecular mechanism by which NDRG1 functions in ESCC remains unclear. Our experiments indicate that TLE2 is significantly decreased at both the transcript and the protein level after NDRG1 overexpression. On xenograft tissue, we validated that the expression level of TLE2 was significantly suppressed and that the level of β-catenin protein was slightly increased. These results indicate that the overexpression of NDRG1 affects the Wnt pathway by decreasing TLE2 and increasing β-catenin, which promotes EMT in esophageal tumor cells.
In conclusion, our results demonstrate the upregulation of NDRG1 in ESCC and its association with poor overall median survival. We also demonstrate a critical role for NDRG1 in inducing EMT in ESCC cells, upregulating mesenchymal markers, and activating the Wnt signaling pathway via decreased TLE2 and increased β-catenin in ESCC. These results suggest that NDRG1 mediates the regulation of TLE2 and β-catenin levels and thereby affects the Wnt pathway and promotes EMT in ESCC cells (Fig. 6). These results indicate a pivotal pro-oncogenic role for NDRG1 in ESCC through which it modulates tumor progression. Further studies of the molecular mechanisms involving NDRG1 may lead to the development of novel therapeutic targets for esophageal cancer.
Figure 6.
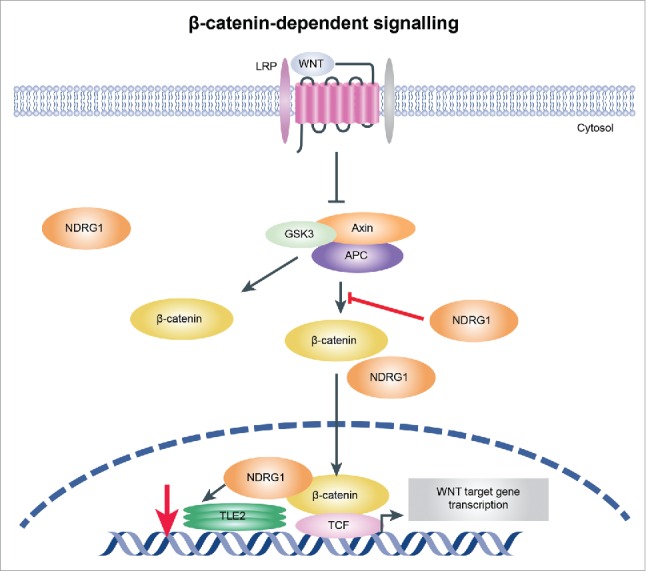
Illustration showing the mechanism by which NDRG1 mediates activation of the Wnt signaling pathway. Overexpression of NDRG1 results in decreased TLE2 expression and increased β-catenin levels, which may then switch to β-catenin/TCF complex assembly and initiation of Wnt signaling pathway activation, promoting EMT in esophageal tumor cells.
Supplementary Material
Disclosure of potential confllicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We would like to thank Dr. Yutaka Shimada at Hyogo College of Medicine for providing the KYSE 30, KYSE 140, KYSE 150, KYSE 170, KYSE 180 and KYSE 410 cell lines.
Author contributions
RA, YS, ZG, WW, LZ and FL conducted experiments and acquisition of data. YS, DTH, YX and XZ analyzed and interpretation of data, and wrote the manuscript. XZ designed the research.
Funding
This work was supported by grants from the NSFC (81572365, 81372591, 81321091 and 81572840) and the National High-Tech R & D Program (2012AA020206 and 2012AA02A503) and the State Key Projects for Basic Research (2014BAI09B07, 2014CBA02002, and 2014CBA02001) of China.
References
- 1.Wei W, Bracher-Manecke JC, Zhao X, Davies NH, Zhou L, Ai R, Oliver L, Vallette F, Hendricks DT. Oncogenic but non-essential role of N-myc downstream regulated gene 1 in the progression of esophageal squamous cell carcinoma. Cancer Biol Ther 2013; 14:164-74; PMID:23192272; http://dx.doi.org/ 10.4161/cbt.22956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Botelho NK, Schneiders FI, Lord SJ, Freeman AK, Tyagi S, Nancarrow DJ, Hayward NK, Whiteman DC, Lord RV. Gene expression alterations in formalin-fixed, paraffin-embedded Barrett esophagus and esophageal adenocarcinoma tissues. Cancer Biol Ther 2010; 10:172-9; PMID:20543560; http://dx.doi.org/ 10.4161/cbt.10.2.12166 [DOI] [PubMed] [Google Scholar]
- 3.Li J, Ying J, Fan Y, Wu L, Ying Y, Chan AT, Srivastava G, Tao Q. WNT5A antagonizes WNT/β-catenin signaling and is frequently silenced by promoter CpG methylation in esophageal squamous cell carcinoma. Cancer Biol Ther 2010; 10:617-24; PMID:20603606; http://dx.doi.org/ 10.4161/cbt.10.6.12609 [DOI] [PubMed] [Google Scholar]
- 4.Jain M, Kumar S, Upadhyay R, Lal P, Tiwari A, Ghoshal UC, Mittal B. Influence of apoptosis (BCL2, FAS), cell cycle (CCND1) and growth factor (EGF, EGFR) genetic polymorphisms on survival outcome: an exploratory study in squamous cell esophageal cancer. Cancer Biol Ther 2007; 6:1553-8; PMID:17912028; http://dx.doi.org/ 10.4161/cbt.6.10.4709 [DOI] [PubMed] [Google Scholar]
- 5.Sun J, Zhang D, Bae DH, Sahni S, Jansson P, Zheng Y, Zhao Q, Yue F, Zheng M, Kovacevic Z, et al.. Metastasis suppressor, NDRG1, mediates its activity through signaling pathways and molecular motors. Carcinogenesis 2013; 34:1943-54; PMID:23671130; http://dx.doi.org/ 10.1093/carcin/bgt163 [DOI] [PubMed] [Google Scholar]
- 6.Lachat P, Shaw P, Gebhard S, van Belzen N, Chaubert P, Bosman FT. Expression of NDRG1, a differentiation-related gene, in human tissues. Histochem Cell Biol 2002; 118:399-408; PMID:12432451; http://dx.doi.org/ 10.1007/s00418-002-0460-9 [DOI] [PubMed] [Google Scholar]
- 7.Wang H, Qian H, Yu J, Zhang X, Zhang L, Fu M, Liang X, Zhan Q, Lin C. Administration of PUMA adenovirus increases the sensitivity of esophageal cancer cells to anticancer drugs. Cancer Biol Ther 2006; 5:380-5; PMID:16481741; http://dx.doi.org/ 10.4161/cbt.5.4.2477 [DOI] [PubMed] [Google Scholar]
- 8.Takaoka M, Kim SH, Okawa T, Michaylira CZ, Stairs DB, Johnstone CN, Andl CD, Rhoades B, Lee JJ, Klein-Szanto AJ, et al.. IGFBP-3 regulates esophageal tumor growth through IGF-dependent and independent mechanisms. Cancer Biol Ther 2007; 6:534-40; PMID:17457048; http://dx.doi.org/ 10.4161/cbt.6.4.3832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Y, Zhu H, Quan L, Zhou C, Bai J, Zhang G, Zhan Q, Xu N. Downregulation of survivin by RNAi inhibits the growth of esophageal carcinoma cells. Cancer Biol Ther 2005; 4:974-8; PMID:16082195; http://dx.doi.org/ 10.4161/cbt.4.9.1914 [DOI] [PubMed] [Google Scholar]
- 10.Yang Y, Goldstein BG, Chao HH, Katz JP. KLF4 and KLF5 regulate proliferation, apoptosis and invasion in esophageal cancer cells. Cancer Biol Ther 2005; 4:1216-21; PMID:16357509; http://dx.doi.org/ 10.4161/cbt.4.11.2090 [DOI] [PubMed] [Google Scholar]
- 11.Fang BA, Kovacevic Z, Park KC, Kalinowski DS, Jansson PJ, Lane DJ, Sahni S, Richardson DR. Molecular functions of the iron-regulated metastasis suppressor, NDRG1, and its potential as a molecular target for cancer therapy. Biochim Biophys Acta 2014; 1845:1-19; PMID:24269900; http://dx.doi.org/ 10.1016/j.bbagrm.2013.11.001 [DOI] [PubMed] [Google Scholar]
- 12.Chang X, Xu X, Ma J, Xue X, Li Z, Deng P, Zhang S, Zhi Y, Chen J, Dai D. NDRG1 expression is related to the progression and prognosis of gastric cancer patients through modulating proliferation, invasion and cell cycle of gastric cancer cells. Mol Biol Rep 2014; 41:6215-23; PMID:24985974; http://dx.doi.org/ 10.1007/s11033-014-3501-2 [DOI] [PubMed] [Google Scholar]
- 13.Chen J, Li S, Yang Z, Lu G, Hu H. Correlation between NDRG1 and PTEN expression in endometrial carcinoma. Cancer Sci 2008; 99:706-10; PMID:18377423; http://dx.doi.org/ 10.1111/j.1349-7006.2008.00749.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mao Z, Sun J, Feng B, Ma J, Zang L, Dong F, Zhang D, Zheng M. The metastasis suppressor, N-myc downregulated gene 1 (NDRG1), is a prognostic biomarker for human colorectal cancer. PLoS One 2013; 8:e68206; PMID:23874544; http://dx.doi.org/ 10.1371/journal.pone.0068206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matsuzaki E, Hiratsuka S, Hamachi T, Takahashi-Yanaga F, Hashimoto Y, Higashi K, Kobayashi M, Hirofuji T, Hirata M, Maeda K. Sphingosine-1-phosphate promotes the nuclear translocation of β-catenin and thereby induces osteoprotegerin gene expression in osteoblast-like cell lines. Bone 2013; 55:315-24; PMID:23612487; http://dx.doi.org/ 10.1016/j.bone.2013.04.008 [DOI] [PubMed] [Google Scholar]
- 16.Strzelczyk B, Szulc A, Rzepko R, Kitowska A, Skokowski J, Szutowicz A, Pawelczyk T. Identification of high-risk stage II colorectal tumors by combined analysis of the NDRG1 gene expression and the depth of tumor invasion. Ann Surg Oncol 2009; 16:1287-94; PMID:19259744; http://dx.doi.org/ 10.1245/s10434-009-0381-0 [DOI] [PubMed] [Google Scholar]
- 17.Lu WJ, Chua MS, So SK. Suppressing N-Myc downstream regulated gene 1 reactivates senescence signaling and inhibits tumor growth in hepatocellular carcinoma. Carcinogenesis 2014; 35:915-22; PMID:24302615; http://dx.doi.org/ 10.1093/carcin/bgt401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee JC, Chung LC, Chen YJ, Feng TH, Juang HH. N-myc downstream-regulated gene 1 downregulates cell proliferation, invasiveness, and tumorigenesis in human oral squamous cell carcinoma. Cancer Lett 2014; 355:242-52; PMID:25218595; http://dx.doi.org/ 10.1016/j.canlet.2014.08.035 [DOI] [PubMed] [Google Scholar]
- 19.Dos Santos M, da Cunha Mercante AM, Nunes FD, Leopoldino AM, de Carvalho MB, Gazito D, Lopez RV, Chiappini PB, de Carvalho Neto PB, Fukuyama EE, et al.. Prognostic significance of NDRG1 expression in oral and oropharyngeal squamous cell carcinoma. Mol Biol Rep 2012; 39:10157-65; PMID:22972152; http://dx.doi.org/ 10.1007/s11033-012-1889-0 [DOI] [PubMed] [Google Scholar]
- 20.Nishio S, Ushijima K, Tsuda N, Takemoto S, Kawano K, Yamaguchi T, Nishida N, Kakuma T, Tsuda H, Kasamatsu T, et al.. Cap43/NDRG1/Drg-1 is a molecular target for angiogenesis and a prognostic indicator in cervical adenocarcinoma. Cancer Lett 2008; 264:36-43; PMID:18281151; http://dx.doi.org/ 10.1016/j.canlet.2008.01.020 [DOI] [PubMed] [Google Scholar]
- 21.Sibold S, Roh V, Keogh A, Studer P, Tiffon C, Angst E, Vorburger SA, Weimann R, Candinas D, Stroka D. Hypoxia increases cytoplasmic expression of NDRG1, but is insufficient for its membrane localization in human hepatocellular carcinoma. FEBS Lett 2007; 581:989-94; PMID:17316623; http://dx.doi.org/ 10.1016/j.febslet.2007.01.080 [DOI] [PubMed] [Google Scholar]
- 22.Ando T, Ishiguro H, Kimura M, Mitsui A, Kurehara H, Sugito N, Tomoda K, Mori R, Takashima N, Ogawa R, et al.. Decreased expression of NDRG1 is correlated with tumor progression and poor prognosis in patients with esophageal squamous cell carcinoma. Dis Esophagus 2006; 19:454-8; PMID:17069588; http://dx.doi.org/ 10.1111/j.1442-2050.2006.00618.x [DOI] [PubMed] [Google Scholar]
- 23.Symes AJ, Eilertsen M, Millar M, Nariculam J, Freeman A, Notara M, Feneley MR, Patel HR, Masters JR, Ahmed A. Quantitative analysis of BTF3, HINT1, NDRG1 and ODC1 protein over-expression in human prostate cancer tissue. PLoS One 2013; 8:e84295; PMID:24386364; http://dx.doi.org/ 10.1371/journal.pone.0084295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang D, Tian X, Jiang Y. NDRG1/Cap43 overexpression in tumor tissues and serum from lung cancer patients. J Cancer Res Clin Oncol 2012; 138:1813-20; PMID:22722710; http://dx.doi.org/ 10.1007/s00432-012-1236-9 [DOI] [PubMed] [Google Scholar]
- 25.Sohda M, Mochida Y, Kato H, Miyazaki T, Nakajima M, Fukuchi M, Manda R, Fukai Y, Masuda N, Ono M, et al.. Overexpression of Cap43 is associated with malignant status of esophageal cancer. Anticancer Res 2009; 29:965-70; PMID:19414333 [PubMed] [Google Scholar]
- 26.Ureshino H, Murakami Y, Watari K, Izumi H, Kawahara A, Kage M, Arao T, Nishio K, Yanagihara K, Kinoshita H, et al.. N-myc downstream regulated gene 1 (NDRG1) promotes metastasis of human scirrhous gastric cancer cells through epithelial mesenchymal transition. PLoS One 2012; 7:e41312; PMID:22844455; http://dx.doi.org/1728357 10.1371/journal.pone.0041312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shimada Y, Imamura M, Wagata T, Yamaguchi N, Tobe T. Characterization of 21 newly established esophageal cancer cell lines. Cancer 1992; 69:277-84; PMID:1728357; http://dx.doi.org/ 10.1002/1097-0142(19920115)69:2%3c277::AID-CNCR2820690202%3e3.0.CO;2-C [DOI] [PubMed] [Google Scholar]
- 28.Xu Y, Zhou L, Huang J, Liu F, Yu J, Zhan Q, Zhang L, Zhao X. Role of Smac in determining the chemotherapeutic response of esophageal squamous cell carcinoma. Clin Cancer Res 2011; 17:5412-22; PMID:21676925; http://dx.doi.org/ 10.1158/1078-0432.CCR-11-0426 [DOI] [PubMed] [Google Scholar]
- 29.He S, Wang L, Miao L, Wang T, Du F, Zhao L, Wang X. Receptor interacting protein kinase-3 determines cellular necrotic response to TNF-α. Cell 2009; 137:1100-11; PMID:19524512; http://dx.doi.org/ 10.1016/j.cell.2009.05.021 [DOI] [PubMed] [Google Scholar]
- 30.Hao XP, Pretlow TG, Rao JS, Pretlow TP. Beta-catenin expression is altered in human colonic aberrant crypt foci. Cancer Res 2001; 61:8085-8; PMID:11719432 [PubMed] [Google Scholar]
- 31.Roy S, Chakravarty D, Cortez V, De Mukhopadhyay K, Bandyopadhyay A, Ahn JM, Raj GV, Tekmal RR, Sun L, Vadlamudi RK. Significance of PELP1 in ER-negative breast cancer metastasis. Mol Cancer Res 2012; 10:25-33; PMID:22086908; http://dx.doi.org/ 10.1158/1541-7786.MCR-11-0456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu Y, Lin Z, Zhao N, Zhou L, Liu F, Cichacz Z, Zhang L, Zhan Q, Zhao X. Receptor interactive protein kinase 3 promotes Cisplatin-triggered necrosis in apoptosis-resistant esophageal squamous cell carcinoma cells. PLoS One 2014. June 24; 9(6):e100127; PMID:24959694; http://dx.doi.org/ 10.1371/journal.pone.0100127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Han B, Li W, Sun Y, Zhou L, Xu Y, Zhao X. A prolyl-hydroxylase inhibitor, ethyl-3,4-dihydroxybenzoate, induces cell autophagy and apoptosis in esophageal squamous cell carcinoma cells via up-regulation of BNIP3 and N-myc downstream-regulated gene-1. PLoS One 2014; 9:e107204; PMID:25232961; http://dx.doi.org/ 10.1371/journal.pone.0107204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wissing MD, Mendonca J, Kim E, Shim JS, Kaelber NS, Kant H, Hammers H, Commes T, Van Diest PJ, Liu JO, et al.. Identification of cetrimonium bromide and irinotecan as compounds with synthetic lethality against NDRG1 deficient prostate cancer cells. Cancer Biol Ther 2013; 14:401-10; PMID:23377825; http://dx.doi.org/ 10.4161/cbt.23759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clevers H, Nusse R. Wnt/β-catenin signaling and disease. Cell 2012; 149:1192-205; PMID:22682243; http://dx.doi.org/ 10.1016/j.cell.2012.05.012 [DOI] [PubMed] [Google Scholar]
- 36.Chen B, Zaveri PG, Longtine MS, Nelson DM. N-myc downstream-regulated gene 1 (NDRG1) mediates pomegranate juice protection from apoptosis in hypoxic BeWo cells but not in primary human trophoblasts. Placenta 2015; 36:847-53; PMID:26028238; http://dx.doi.org/ 10.1016/j.placenta.2015.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen Z, Zhang D, Yue F, Zheng M, Kovacevic Z, Richardson DR. The iron chelators Dp44mT and DFO inhibit TGF-β-induced epithelial-mesenchymal transition via up-regulation of N-Myc downstream-regulated gene 1 (NDRG1). J Biol Chem 2012; 287:17016-28; PMID:22453918; http://dx.doi.org/ 10.1074/jbc.M112.350470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu W, Iiizumi-Gairani M, Okuda H, Kobayashi A, Watabe M, Pai SK, Pandey PR, Xing F, Fukuda K, Modur V, et al.. KAI1 gene is engaged in NDRG1 gene-mediated metastasis suppression through the ATF3-NFkappaB complex in human prostate cancer. J Biol Chem 2011; 286:18949-59; PMID:21454613; http://dx.doi.org/ 10.1074/jbc.M111.232637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pearce LR, Sommer EM, Sakamoto K, Wullschleger S, Alessi DR. Protor-1 is required for efficient mTORC2-mediated activation of SGK1 in the kidney. Biochem J 2011; 436:169-79; PMID:21413931; http://dx.doi.org/ 10.1042/BJ20102103 [DOI] [PubMed] [Google Scholar]
- 40.MacDonald BT, Tamai K, He X. Wnt/β-catenin signaling: components, mechanisms, and diseases. Dev Cell 2009; 17:9-26; PMID:19619488; http://dx.doi.org/ 10.1016/j.devcel.2009.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu W, Xing F, Iiizumi-Gairani M, Okuda H, Watabe M, Pai SK, Pandey PR, Hirota S, Kobayashi A, Mo YY, et al.. N-myc downstream regulated gene 1 modulates Wnt-β-catenin signalling and pleiotropically suppresses metastasis. EMBO Mol Med 2012; 4:93-108; PMID:22246988; http://dx.doi.org/ 10.1002/emmm.201100190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jin R, Liu W, Menezes S, Yue F, Zheng M, Kovacevic Z, Richardson DR. The metastasis suppressor NDRG1 modulates the phosphorylation and nuclear translocation of β-catenin through mechanisms involving FRAT1 and PAK4. J Cell Sci 2014; 127:3116-30; PMID:24829151; http://dx.doi.org/ 10.1242/jcs.147835 [DOI] [PubMed] [Google Scholar]
- 43.Melotte V, Qu X, Ongenaert M, van Criekinge W, de Bruine AP, Baldwin HS, van Engeland M. The N-myc downstream regulated gene (NDRG) family: diverse functions, multiple applications. FASEB J 2010; 24:4153-66; PMID:20667976; http://dx.doi.org/ 10.1096/fj.09-151464 [DOI] [PubMed] [Google Scholar]
- 44.Hanson AJ, Wallace HA, Freeman TJ, Beauchamp RD, Lee LA, Lee E. XIAP monoubiquitylates Groucho/TLE to promote canonical Wnt signaling. Mol Cell 2012; 45:619-28; PMID:22304967; http://dx.doi.org/ 10.1016/j.molcel.2011.12.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Buscarlet M, Perin A, Laing A, Brickman JM, Stifani S. Inhibition of cortical neuron differentiation by Groucho/TLE1 requires interaction with WRPW, but not Eh1, repressor peptides. J Biol Chem 2008; 283:24881-8; PMID:18611861; http://dx.doi.org/ 10.1074/jbc.M800722200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roth M, Bonev B, Lindsay J, Lea R, Panagiotaki N, Houart C, Papalopulu N. FoxG1 and TLE2 act cooperatively to regulate ventral telencephalon formation. Development 2010; 137:1553-62; PMID:20356955; http://dx.doi.org/ 10.1242/dev.044909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.He Z, Liu Y, Liang D, Wang Z, Robertson ES, Lan K. Cellular corepressor TLE2 inhibits replication-and-transcription- activator-mediated transactivation and lytic reactivation of Kaposi's sarcoma-associated herpesvirus. J Virol 2010; 84:2047-62; PMID:19939918; http://dx.doi.org/ 10.1128/JVI.01984-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.