Abstract
Elevated levels of DNA ligase IIIα (LigIIIα) have been identified as a biomarker of an alteration in DNA repair in cancer cells that confers hypersensitivity to a LigIIIα inhibitor, L67, in combination with a poly (ADP-ribose) polymerase inhibitor. Since LigIIIα functions in the nucleus and mitochondria, we examined the effect of L67 on these organelles. Here we show that, although the DNA ligase inhibitor selectively targets mitochondria, cancer and non-malignant cells respond differently to disruption of mitochondrial DNA metabolism. Inhibition of mitochondrial LigIIIα in cancer cells resulted in abnormal mitochondrial morphology, reduced levels of mitochondrial DNA and increased levels of mitochondrially-generated reactive oxygen species that caused nuclear DNA damage. In contrast, these effects did not occur in non-malignant cells. Furthermore, inhibition of mitochondrial LigIIIα activated a caspase 1-dependent apoptotic pathway that is known to be part of inflammatory responses induced by pathogenic microorganisms in cancer but not non-malignant cells. These results demonstrate that the disruption of mitochondrial DNA metabolism elicits different responses in non-malignant and cancer cells and suggests that the abnormal response in cancer cells may be exploited in the development of novel therapeutic strategies that selectively target cancer cells.
Keywords: DNA ligase, mitochondrial DNA, oxidative phosphorylation, reactive oxygen species, apoptosis
INTRODUCTION
Among the ATP-dependent DNA ligases encoded by the mammalian LIG genes, LIG1, LIG3, and LIG4 (1), DNA ligase I (LigI) is primarily responsible for joining Okazaki fragments during nuclear DNA replication. However, DNA ligase IIIα (LigIIIα) is essential for DNA replication in LigI-deficient cells (2–5). LigI and LigIIIα also appear to have overlapping functions in the repair of base damage and single-strand breaks (3–8). While DNA ligase IV is predominantly responsible for the repair of nuclear DNA double strand breaks (DSB)s by non-homologous end joining (NHEJ), LigI and Lig IIIα participate in alternative (alt) NHEJ pathways (9,10).
Unlike the nucleus, only one DNA ligase is present in mitochondria (3,4,11). Mitochondrial (mito) and nuclear (nuc) versions of LigIIIα are generated by alternative translation (11). Although mito LigIIIα is required to maintain mitochondrial DNA and is essential for cell viability under normal culture conditions, this lethality can be rescued by either addition of pyruvate and uridine to the culture media or expression of mitochondrially-targeted, heterologous DNA ligases, including the NAD-dependent E. coli LigA (3,4,12).
A subset of DNA ligase inhibitors preferentially sensitized cancer cells to DNA damaging agents (13). Subsequently, it was shown that BCR-ABL1-positive cell lines and samples from patients with chronic myeloid leukemia, in particular leukemia cells that had acquired resistance to imatinib, were hypersensitive to the LigI/III inhibitor L67 in combination with a poly (ADP-ribose) polymerase (PARP) inhibitor (14). A similar hypersensitivity was observed in breast cancer cell lines with either intrinsic or acquired resistance to anti-estrogens (15). Since LigIIIα knockdown had the same effect as L67 in combination with a PARP inhibitor, it appears that L67 exerts its cancer cell-specific effect by inhibition of LigIIIα (14,15). The hypersensitivity to the combination of L67 and a PARP inhibitor correlated with elevated expression of both LigIIIα and PARP1, and increased dependence on PARP1- and LigIIIα-dependent alt NHEJ (9,14–16).
Although the repair inhibitor combination does inhibit alt NHEJ (14,15), the observed synergy is unlikely to be due to the inhibition of two enzymes in the same pathway (9). Since LigIIIα has nuclear and mitochondrial functions, we examined the mechanism of L67-induced cytotoxicity. These studies revealed that L67 preferentially targets mito LigIIIα, resulting in mitochondrial dysfunction. Surprisingly, cancer cell mitochondria were more susceptible to L67 than mitochondria in non-malignant cells. The disruption of mitochondrial function in cancer cells resulted in elevated levels of mitochondrially-generated reactive oxygen species (ROS) and activation of a caspase 1-dependent apoptotic pathway that is involved in inflammatory responses induced by pathogenic microorganisms (17). In non-malignant cells, there was no increase in mitochondrially-generated ROS but oxidative phosphorylation (OXPHOS) was completely uncoupled and the cells became senescent.
MATERIALS AND METHODS
Cell lines
Human cervical (HeLa, 2012), colorectal (HCT116, 2006 and 2016) and breast (MDA-MB-231, 2008) cancers cell lines were purchased from ATCC and grown in the recommended media. A HeLa cell line that stably expresses mitochondrially-targeted E. coli LigA (mitoLigA) (4) after transfection with the plasmid pCAG-mitoLigAYFP-Neo that encodes E. coli LigA fused at its N terminus to the LigIIIα mitochondrial targeting sequence and at its C terminus to EYFP. The telomerase-immortalized human fibroblast cell line HCA-Ltrt from Dr. Murnane (2010), was grown in DMEM/F12 medium with 10% FBS. Normal breast epithelium MCF10A cells from Dr. Rassool (2012) were grown using recommended medium and mixture of additives (Lonza/Clonetics Corporation) with 5% horse serum and 100 ng/ml cholera toxin. Cell lines lacking mitochondria DNA (Rho minus cells) were established as described (18,19). The identity of commercially available cell lines was confirmed by STR profiling with the PowerPlex 1.2 System (Promega), most recently in 2016.
Colony Forming and Cell Growth Assays
To measure colony formation, cells were cultured in triplicate in 6-well plates overnight and then incubated with different concentrations of L67 for 14 days prior to staining with crystal violet. To measure cell growth, cells incubated with L67 for 48–72 hours. Genomic DNA was then stained with the CyQUANT NF reagent and quantitated according to the manufacturer’s protocol.
Ligation assay
Human Lig IIIα/His-taggedXRCC1 complex was expressed in and purified from insect cells (20). E. coli DNA ligase (LigA) was purchased from New England Biolabs. A labeled duplex oligonucleotide with a single ligatable nick was prepared as described previously (21). Lig IIIα/XRCC1 (0.05 pmol) and LigA (0.1 units) were incubated with L67 in the presence of either ATP (for Lig IIIα/XRCC1) or NAD (for LigA) at room temperature for 10 min prior to the addition of the DNA substrate (0.1 pmol) incubation in buffer containing 50 mM Tris-Cl pH 7.5, 12.5 mM NaCl, 6 mM MgCl2, 3% glycerol, 1 mM DTT, 0.25 mg/ml BSA, 1% DMSO at 37°C for 20 min in a final volume of 20 µl. After the addition of SDS to a final concetario of 0.5%, DNA was recovered with Streptavidin MagneSphere (Promega), suspended in formamide/dye solution and electrophoresed through a 12.5% denaturing polyacrylamide gel. Ligated and unligated DNAs were detected by autoradiography and quantitated by ImageJ (NIH).
Immunoblotting
Cells were lysed in 20 mM Tris-HCl, pH 7.5, 100 mM NaCl, 1 mM EDTA, 5% glycerol, 1% Triton X-100 containing a protease inhibitor cocktail (Sigma). Proteins were detected by immunoblotting using the following antibodies; actin, phospho-KAP1(serine 824), (Abcam); Chk2, KAP1 (GeneTex), phospho-Chk2, p53, phospho-p53(serine 15), PARP1, cleaved PARP1 (Cell Signaling); H2AX (R&D Systems) and γ-H2AX(serine 139) (Millipore). ATM (Cell Signalling) and phospho-ATM(serine 1981) (Abcam) were detected as described (22).
Measurement of γ-H2AX and mitochondrial superoxide (mSOX)
Overnight cultures were incubated in fresh medium containing DMSO with different concentrations of L67 for 24h in the absence or presence of 5 mM NAC. γ-H2AX and mSOX were detected by flow cytometry using an Alexa Fluor 647-conjugated antibody specific for γ-H2AX phosphorylated on serine139 and MitoSOX Red (Molecular Probes), respectively.
Seahorse Extracellular Flux analysis
In experiments performed on a Seahorse XF24 Flux analyzer, cells were maintained at 5% CO2 in DMEM media at 37°C. Cells (40,000) were plated on XF24 plates using Cell-Tak™ Cell and Tissue Adhesive (Corning) and growth medium was replaced with bicarbonate-free modified DMEM, the ‘assay medium’. In experiments performed on a Seahorse XF96e Flux analyzer, cells (20,000) were placed into each well using Cell-Tak™. After incubation for another 60 min in a 37°C incubator without CO2, oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) were measured essentially as previously described (18).
Mitochondrial morphology and DNA content
Cells were seeded on slides and grown overnight in phenol-free DMEM medium prior to incubation in fresh medium containing either DMSO or 30 µM L67 for 24h. Mitochondria were then stained by incubation with 250 nM Mitotracker Red CMXRos for 20 minutes at 37°C. After washing, cells were maintained in phenol free medium prior to imaging with a Zeiss LSM510 confocal system on an Axio Observer inverted microscope with a 63X 1.2 NA water immersion objective. Mitotracker Red was excited with a 543 nm HeNe laser and fluorescence emission collected with 560 nm LP filter.
Genomic DNA was isolated using the QIAamp DNA Kit (QIAGEN) and then quantitated with the Quant-iT PicoGreen dsDNA Assay Kit (Molecular Probes). Short (220 bp) and long (8.9 kb) fragments of mitochondrial DNA and a 4 kb fragment of the nuc gene encoding DNA polymerase δ were amplified using 3 ng of total genomic DNA using the primers (Table S2), amplification conditions (Table S3) and a KAPA LongRange HotStart PCR kit (KAPA BIOSYSTEMS). Mitochondrial DNA copy number was calculated as described (19).
Apoptosis Assay
Apoptotic cells were detected by flow cytometry using the PE Annexin V apoptosis detection Kit (BD Pharmingen) according to the manufacturer’s instructions. Where indicated, a caspase 1 inhibitor (50 µM Ac-YVAD-cmk, Sigma-Aldrich), a caspase 3 inhibitor (50 µM Z-DEVD-fmk, ApexBio) or a pan caspase inhibitor (50 µM Z-VAD-FMK, R&D Systems) was added to the media with L67 prior to incubation for 24 h.
β-Galactosidase assay
Expression of β-galactosidase was detected using the Senescence β-Galactosidase Staining Kit (Cell Signaling) according to the manufacturer’s instructions.
Caspase 1 Activity
Caspase 1 activity was measured by flow cytometry using a FLICA 660 in vitro caspase-1 detection Kit (Immunochemistry Technologies) according to the manufacturer’s instructions.
Statistical analysis
Data are expressed as mean ± SEM. For comparison of groups, we used the Student’s two-tailed T-test. A level of p<0.05 was regarded as statistically significant.
RESULTS
Cells without mitochondrial DNA are more resistant to the DNA ligase inhibitor L67
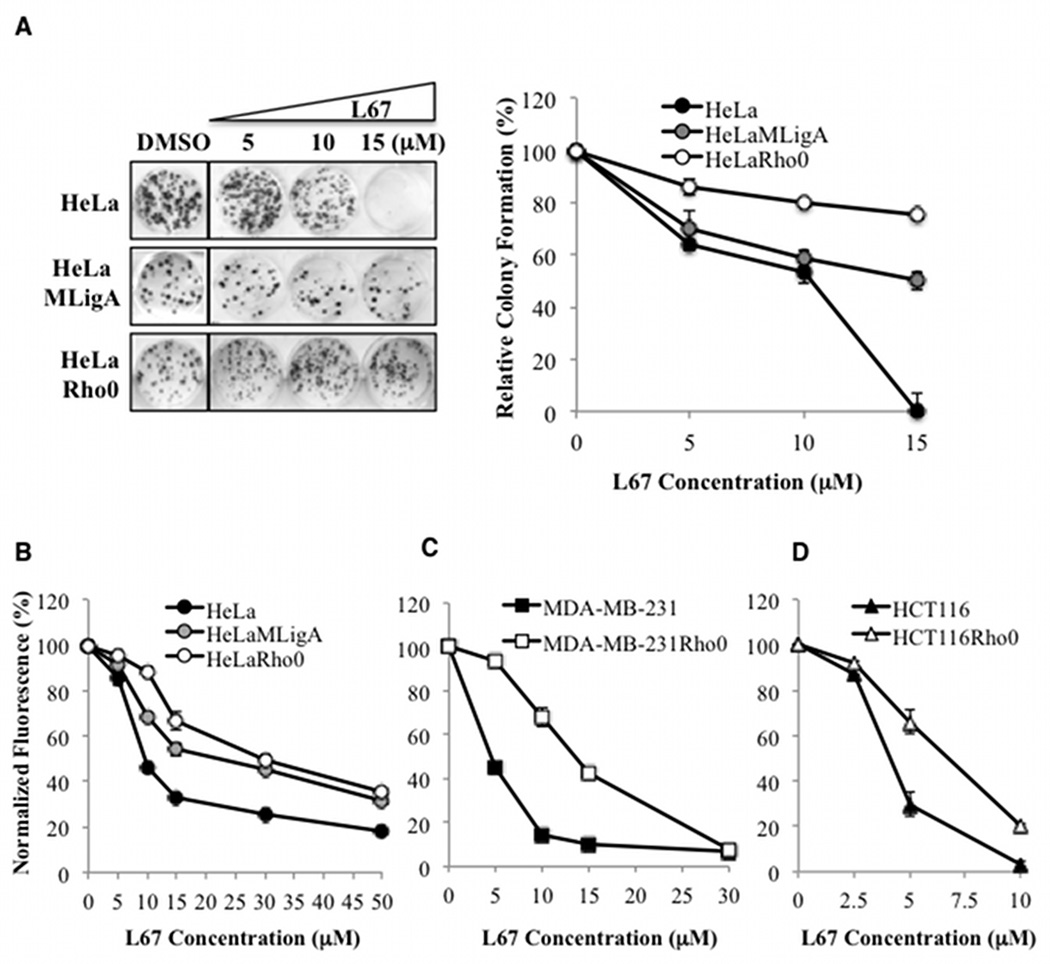
To determine the mechanisms underlying the cytotoxicity of the LigI/III inhibitor L67 (13), derivatives of cell lines established from cervical (HeLa, Supplementary Figure S1A), breast (MDA-MB-231) and colon (HCT116) cancers that lack mito DNA (Rho minus) were selected by growth in the presence of ethidium bromide, pyruvate and uridine (18). HeLa cells with abnormal mitochondria that lack mitochondrial DNA (Supplementary Figure S1A and B) were more resistant to L67 in colony forming (Fig. 1A) and cell proliferation assays (Fig. 1B) with the absence of mitochondrial DNA increasing the IC50 from 8.2 µM to 29.7 µM (Supplementary Table 1). The differential effect of L67 on HeLa and HeLa Rho minus cells was also observed when HeLa cells were grown in the media containing pyruvate and uridine (Supplementary Figure S1C). MDA-MB-231 and HCT 116 cells lacking mitochondrial DNA were also more resistant to L67 compared with their respective parental cells (Fig. 1C and D, and Supplementary Table 1), suggesting that, at lower concentrations, L67 causes cell death by targeting mitochondrial function.
Figure 1. Cancer cells lacking mitochondrial DNA are more resistant to L67.
A. Colony formation by HeLa, HeLaMLigA and HeLa Rho minus cells incubated with either vehicle control (DMSO) or the indicated concentration of L67. Left panel, the boxes indicate that the wells containing cells incubated with vehicle control were in the same plate but not contiguous with the wells containing cells incubated with L67. Right panel, cell survival results are shown graphically. Effect of L67 on the proliferation of; B. HeLa (filled circle), HeLaMLigA (gray circle) and HeLa Rho minus cells (empty circle); C. MDA-MB-231 (filled square), and MDA-MB-231 Rho minus (empty square); D. HCT116 (filled triangle), and HCT116 Rho minus cells (empty triangle), was determined as described in Materials and Methods. Data shown graphically are the mean ± SEM of three independent experiments and are expressed as a percentage of the values for untreated cells.
To confirm that the effect of L67 is dependent upon inhibition of mito LigIIIα, we stably expressed mitoLigA in HeLa cells (Supplementary Figure S2A). Although this enzyme can substitute for mitoLigIIIα (4), it is at least 6-fold more resistant to L67 than LigIIIα (Supplementary Figure S2B). Notably, expression of mitoLigA significantly increased the resistance of HeLa cells to L67 (Fig. 1A and B) albeit not to same level as the HeLa Rho minus cells (Supplementary Table 1). This may be due in part to a dominant negative effect of inhibiting endogenous LigIIIα.
Inhibition of mito LigIIIα results in increased mSOX and nuclear DNA damage in HeLa cells
The differential effect of L67 on the growth of parental and Rho minus cancer cells was most pronounced in a concentration range (5 to 15 µM, Fig. 1A–D), that encompasses the IC50 values for LigI (6 µM) and LigIII (8 µM) (13). Although LigI and LigIIIα participate in nuc DNA replication (2,5), incubation of synchronized cells with L67 at concentrations up to 15 µM for 24 h had only minor effects on cycle distribution (Supplementary Figure S3A). Furthermore, incubation with L67 concentrations up to 10 µM for 24 h only reduced bromodeoxyuridine (BrdU) incorporation by 10–15% (Supplementary Figure S3B). At higher concentrations of L67, there was an accumulation of HeLa cells, the derivative expressing mitoLigA and, to a lesser extent, Rho minus cells in S phase (Supplementary Figure S3A) that correlated with reduced BrdU incorporation (Supplementary Figure S3B).
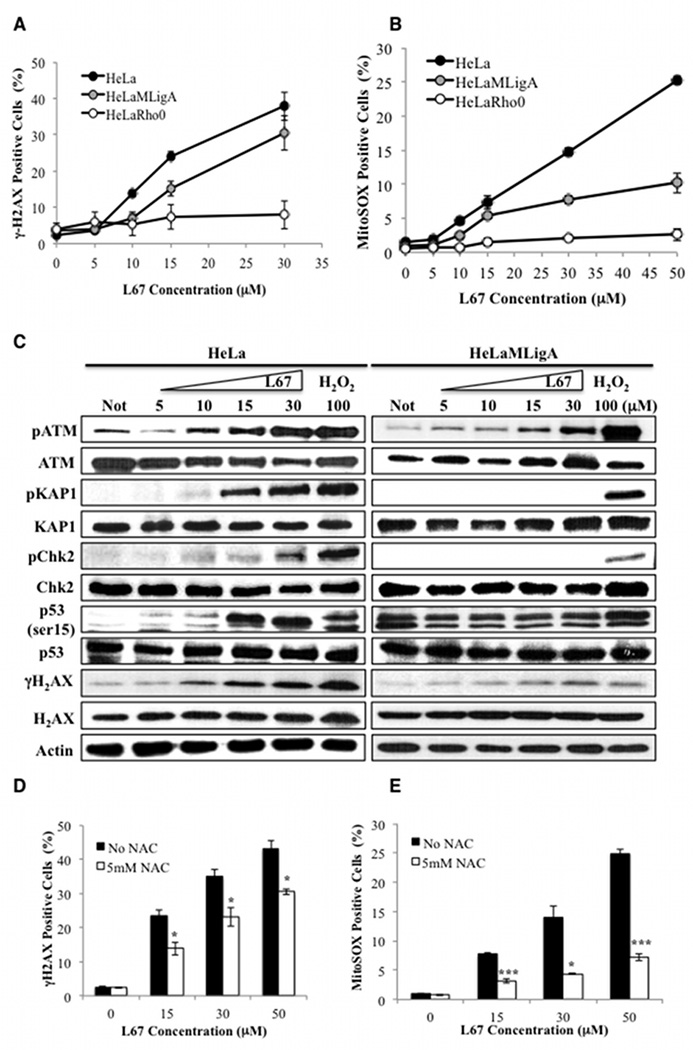
Interestingly, incubation of HeLa, but not HeLa Rho minus cells, with L67 at 10 and 15 µM for 24 h resulted in a dose-dependent increase in the formation of nuclear γH2AX foci (Supplementary Figure 4A) and steady state levels of γH2AX (Fig. 2A), consistent with replication stress and DSB induction in the nucleus (23). Expression of mitoLigA reduced γH2AX foci formation (Supplementary Fig. 4A) and the steady state levels (Fig. 2A) induced by L67 compared with the parental cells, but not to the extent observed in Rho minus cells (Fig. 2A and Supplementary Fig. 4A). Since mitochondria are the major cellular source of reactive oxygen species (ROS) and disruption of mitochondrial function can lead to elevated levels of ROS (24,25), we examined the effect of L67 on mSOX levels. Incubation with L67 for 24 h resulted in a concentration-dependent increase in mSOX levels in HeLa cells whereas expression of mitoLigA reduced the levels of mSOX induced by L67 and, as expected, there was no induction of mSOX in HeLa Rho minus cells (Fig. 2B).
Figure 2. Elevated levels of γH2AX, mSOX and ATM activation induced by L67 in HeLa cells: effect of L67 is attenuated by either the absence of mito DNA, expression of mitoLigA or co-incubation with an anti-oxidant.
γH2AX (A) and mSOX (B) in HeLa (filled circle), HeLaMLigA (gray circle) and HeLa Rho minus cells (empty circle) were detected by flow cytometry. C. The indicated proteins and their phosphorylated derivatives in extracts from HeLa (left panel) and HeLaMLigA (right panel) cells that had been incubated with or without (Not) L67 for 24 h were detected by immunoblotting. Where indicated, cells were incubated with 100 µM H202 for 1 h to induce the DNA damage response. Effect of the absence (black bars) or presence (white bars) of 5 mM n-acetyl cysteine on L67-induced γH2AX (D) and mSOX (E) in HeLa cells. Data shown graphically are the mean ± SEM of three independent experiments and are expressed as a percentage of the values for untreated cells. *p <0.05 and ***p <0.0001 using the unpaired two-tailed Student’s test.
Since γH2AX can be generated in response to other types of cellular stress in addition to DSBs (26), we examined the phosphorylation status of the ataxia telangiectasia mutated (ATM) kinase, which is recruited to and activated by DSBs, and ATM substrates (27). Expression of mitoLigA greatly attenuated L67-induced autophosphorylation of ATM and phosphorylation of γH2AX and several other ATM substrates but not p53, which was constitutively phosphorylated (Fig. 2C, compare left and right panels). While expression of mitoLigA attenuated the effects of L67 on cell survival and mSOX levels, mitochondrial function was abnormal (Fig. 4), suggesting that p53 may be activated in response to metabolic stress in these cells (28). Although ATM can be activated by DSB-independent mechanisms, including direct activation by ROS (27,29), incubation with L67 resulted in phosphorylation of KAP1, whose phosphorylation by ATM is DSB-dependent (30) (Fig. 2C). As expected, the free radical scavenger n-acetyl cysteine (NAC) reduced mSOX (Fig. 2E), ATM activation (Supplementary Fig. S4B) and γH2AX formation (Fig. 2D) induced by L67 in HeLa cells. Together, these results indicate that the nuclear DNA damage induced by L67 in HeLa cells is due to increased generation of ROS by mitochondria as result of inhibition of mito LigIIIα.
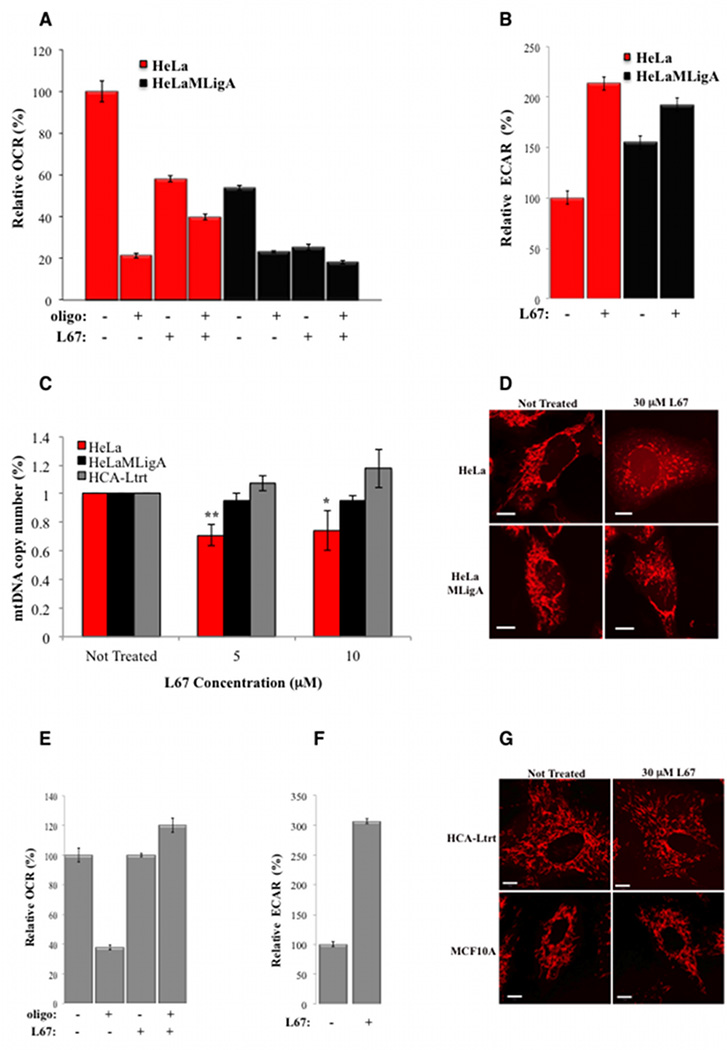
Figure 4. L67 causes a reduction in OCR and mitochondrial DNA, and abnormal mitochondrial morphology in HeLa but not HCA-Ltrt cells.
HeLa (red bars) and HeLaMLigA (black bars) cells were incubated with (+) or without (−) 10 µM L67 for 24 h. A. OCR measured in the presence (+) or absence of (−) 1 µM oligomycin. B. ECAR. OCR and ECAR were measured in a Seahorse Bioanalyzer as described in Materials and Methods. OCR and ECAR values for untreated HeLa and HCA-Ltrt cells values were set to 100%. Data shown graphically represent mean ± SEM of 4–6 replicates and 3–4 time points. For panels A and B, all treatment groups are significantly different than their untreated control with p values <0.001. C. Effect of L67 on mitochondrial DNA copy number was determined in HeLa (red bars), HeLaMLigA (black bars) and HCA-Ltrt (grey bars) cells as described in Materials and Methods after incubation in the presence or absence (Not Treated) of L67 for 24 h. Data shown graphically are the mean ± SEM of three independent experiments. *p <0.05, **p<0.001. D. Mitochondria in HeLa and HeLaMLigA were stained with Mitotracker Red and visualized by fluorescence microscopy after incubation in the presence or absence (Not Treated) of L67 for 24 h. Scale bars are 10 µm. OCR and ECAR were measured in HCA-Ltrt (grey bars) cells incubated with (+) or without (−) 10 µM L67 for 24 h as described above. E. OCR. With the exception of L67 treatment, all treatment groups are significantly different than the untreated control with a p value of <0.001 for oligo and a p value of <0.05 for oligo plus L67. F. ECAR. The treatment group is significantly different than the untreated control with a p value <0.001. G. Mitochondria in HCA-Ltrt and MCF10A cells were visualized as described above after incubation in the presence or absence (Not Treated) of L67 for 24 h.
L67 induces elevated mSOX in cancer but not non-malignant cell lines
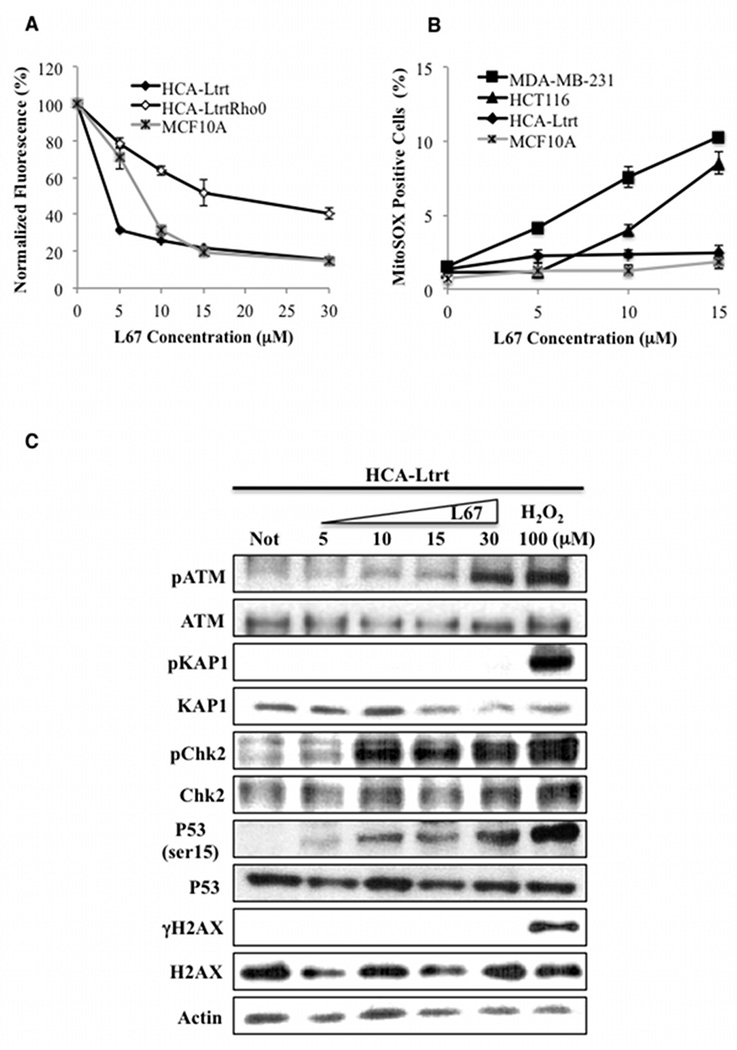
Similar to the cancer cell lines (Fig. 1), proliferation of a Rho minus derivative of a telomerase-immortalized fibroblast (HCA-Ltrt) was more resistant to L67 than the parental cell line (Fig. 3A). Surprisingly, while L67 had similar effects on cell cycle distribution (Supplementary Figures S3A and S5A) and the proliferation of cancer cell lines (Figs. 1A–D) and non-malignant cell lines established from normal tissues (Fig. 3A), elevated levels of mSOX were not detected in non-malignant cells, including HCA-Ltrt Rho minus cells (Fig. 3B and Supplementary Fig. S5B). Accordingly, L67 did not induce detectable phosphorylation of H2AX and KAP1 in HCA-Ltrt cells and ATM autophosphorylation was only observed with 30 µM L67 (Fig. 3C). Interestingly, phosphorylation of p53 and Chk2 occurred in the absence of detectable ATM activation (Fig. 3C), suggesting a response to a stress signal other than DNA damage (28).
Figure 3. Effect of L67 on the proliferation of non-maligant cells; no induction of mSOX and reduced ATM activation by L67 in non-malignant cell lines.
A. Proliferation of telomerase-immortalized fibroblasts HCA-Ltrt (filled diamond), HCA-Ltrt Rho minus cells (empty diamond) and MCF10A normal breast epithelial cells (crossed gray square). B. mSOX levels in HCA-Ltrt (filled diamond), MCF10A (crossed gray aquare), MDA-MB-231 (filled square) and HCT116 (filled triangle) cells. Cells were incubated in the presence or absence of L67 for 24. Data shown graphically are the mean ± SEM of three independent experiments and are expressed as a percentage of the values for untreated cells. C. The indicated proteins and their phosphorylated derivatives in extracts from HCA-Ltrt cells that had been incubated with or without (Not) L67 for 24 h were detected by immunoblotting. Where indicated, cells were incubated with 100 µM H202 for 1 h to induce the DNA damage response.
L67 induces changes in mitochondrial function in both cancer and non-malignant cells but alterations in mitochondrial morphology and DNA copy number occurs only in cancer cells
As mentioned previously, expression of mitoLigA in HeLa cells resulted in altered mitochondrial function. The OCR was about 50% lower compared with the parental HeLa cells (Fig. 4A) whereas the proportion of oligomycin-insensitive oxygen consumption, indicative of uncoupled OXPHOS, was higher (~40%) compared with the parental HeLa cells (~20%). Incubation with L67 resulted in a similar reduction in OCR (40–50%) in both cell lines with about 80% of the remaining OCR due to uncoupled OXPHOS (Fig. 4A). The changes in OCR in the HeLa cells expressing mitoLigA resulted in a compensatory increase in ECAR, a measure of glycolysis. As expected, there was an increase in ECAR following incubation with L67 (Fig. 4B).
Since reduced mito DNA has been linked with elevated mSOX and increased sensitivity to DNA damaging agents (31), we examined the effect of L67 on mito DNA copy number. Incubation of HeLa cells with 5 and 10 µM L67 resulted in about a 25% reduction in mito DNA whereas there was no reduction in mito DNA in derivatives expressing mitoLigA (Fig. 4C). Furthermore, incubation with 30 µM L67 disrupted the filamentous network of mitochondria observed in untreated HeLa cells, resulting in a similar appearance to HeLa Rho minus cells with large irregular clumps of mitochondria and diffuse cell staining (Fig. 3D, and Supplementary Figure S1B). Thus, while expression of mitoLigA resulted in reduced OCR (Fig. 4A) and increased ECAR (Fig. 4B), it prevented the reduction of mito DNA and abnormal mitochondrial morphology induced by L67 (Fig. 4D). Notably, incubation of telomerase-immortalized fibroblasts with 10 µM L67 did not cause a reduction in OCR but OXPHOS was totally uncoupled (Fig. 4E) and there was a large compensatory increase in glycolysis (Fig. 4F). In addition, there were no reductions in mitochondrial DNA copy number (Fig. 4C) or alterations in mitochondrial morphology (Fig. 4G) in non-malignant cells.
Inhibition of mito LigIIIα induces apoptosis in cancer cells and senescence in non-malignant cells
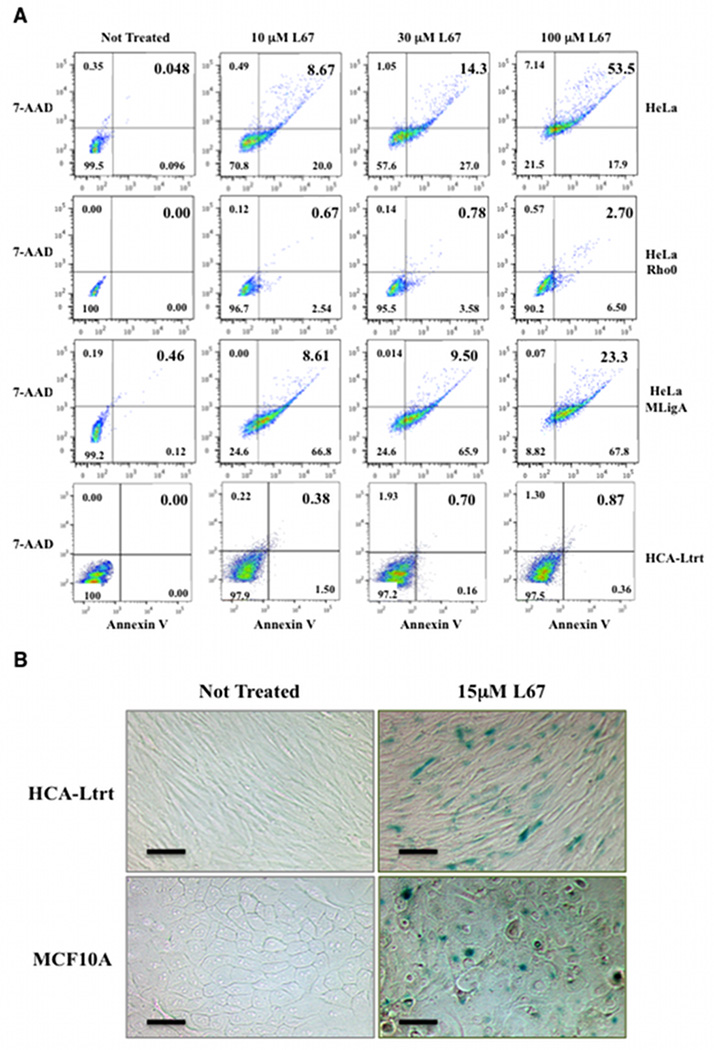
Following incubation with 10 µM L67 for 24 h, there were greater than 12-fold more apoptotic (7-AAD positive, PE Annexin V positive) HeLa and HeLa mitoLigA cells than apoptotic HeLa Rho minus cells (Fig. 5A), indicating that the majority of cell death induced by L67 is dependent upon mito DNA and presumably mitochondrial function. Although, at 10 µM L67, the extent of apoptosis was similar in the HeLa and HeLa mitoLigA cells (Fig. 5A), cell death was caspase-dependent in HeLa cells (Fig. 6) and caspase-independent in the HeLa mitoLigA cells (Fig. 6A and Supplementary Fig. 6). There was greater apoptosis in the HeLa cells compared with HeLa cells expressing mitoLigA at 100 µM L67 with apoptotic cells constituting about 50% of the HeLa cell population compared with 23% of the HeLa mitoLigA population (Fig. 5A), indicating that expression of mitoLigA reduces the cytotoxicity of L67. In contrast, very low levels of apoptosis were detected in HCA-Ltrt cells incubated with L67 (Fig. 5A). Since L67 reduced the proliferation of non-malignant cells (Fig. 3A), we asked whether L67 induced senescence. As shown in Figure 5B, incubation of HCA-Ltrt and MCF10A cells with L67 resulted in β-galactosidase expression, a marker of senescence. Thus, non-malignant and cancer cells respond differently to inhibition of mito LigIIIα with non-malignant cells becoming senescent whereas cancer cells undergo apoptosis.
Figure 5. L67 induces apoptosis in HeLa cells and senescence in non-malignant cells.
A. The fraction of late (upper right quadrant) and early (lower right) apoptotic cells in cultures of HeLa, HeLa Rho minus, HeLaMLigA and HCA-Ltrt cells after incubation in the presence or absence (Not Treated) of L67 for 24 h was determined by flow cytometry. B. β-galactosidase activity in HCA-Ltrt and MCF10A cells after incubation in the presence or absence (Not Treated) of L67 for 24 h was detected as described in Materials and Methods. Scale bars are 50 µm.
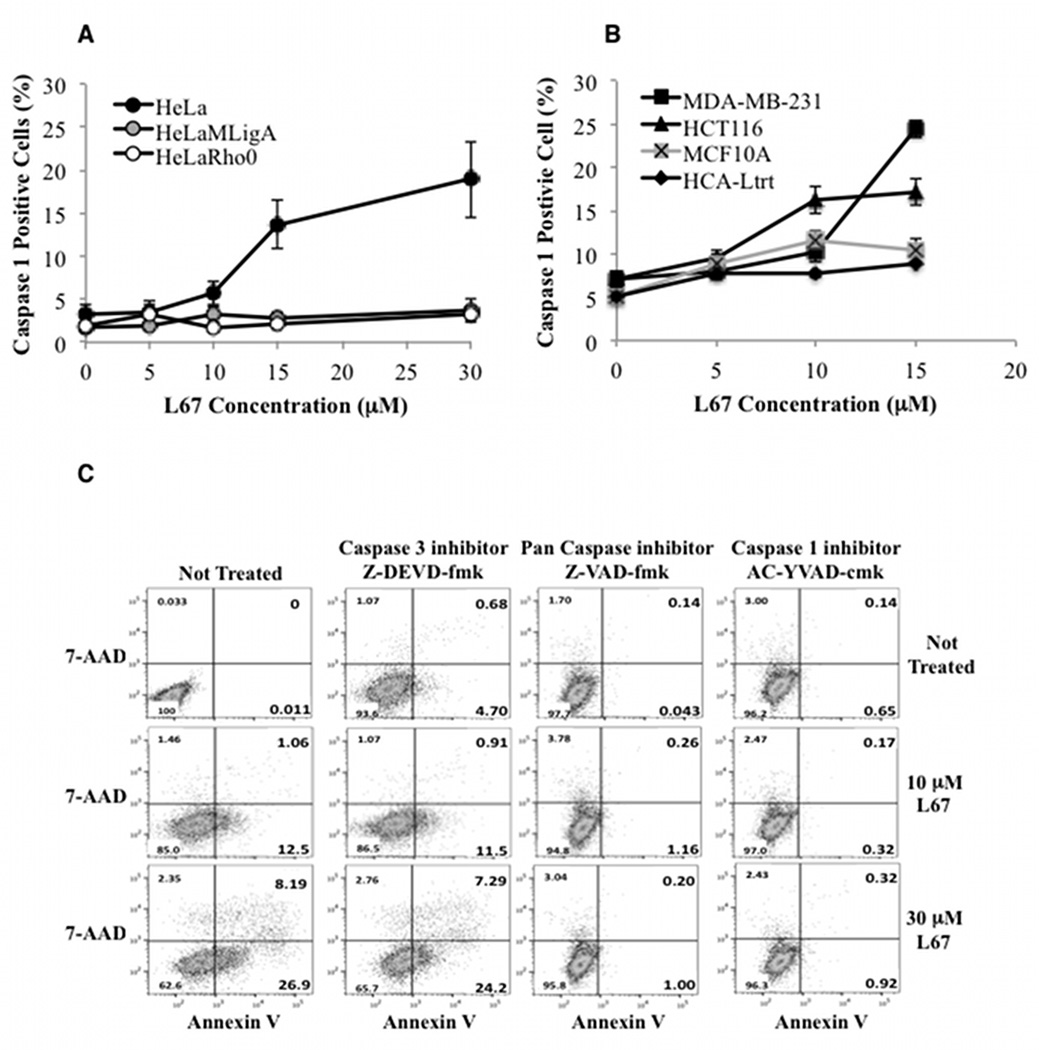
Figure 6. L67 induces caspase 1-dependent apoptosis in HeLa cells; attenuation of L67-induced apoptosis by loss of mitochondrial DNA, expression of mitoLigA or inhibition of caspase 1.
A. HeLa (filled circle), HeLaMLigA (gray circle) and HeLa Rho minus (empty circle); B. HCA-Ltrt (filled diamond), MCF10A (crossed gray aquare), MDA-MB-231 (filled square) and HCT116 (filled triangle) cells were incubated with or without L67 for 24 h. Caspase 1 activity was measured by flow cytometry. Data shown graphically are the mean ± SEM of three independent experiments. C. The effects of a caspase 3 inhibitor, a caspase 1 inhibitor and a pan caspase inhibitor on L67-induced apoptosis in HeLa cells.
Inhibition of mito LigIIIα activates a caspase 1-dependent cell death pathway in cancer but not non-malignant cells
No activation of caspases 7 and 9 that are involved in extrinsic and intrinsic apoptotic pathways was detected in HeLa cells incubated with L67 (Supplementary Figure S6). Instead, caspase 1, which is activated in the inflammatory response elicited by pathogens (17) and by the release of ROS and mito DNA into the cytoplasm (32–34), was activated in HeLa cells but not in either the Rho minus or mitoLigA expressing derivatives (Fig. 6A). Notably, incubation with L67 also activated caspase 1 in breast (MDA-MB-231) and colorectal (HCT116) cancer cell lines but not in non-malignant cell lines (Fig. 6B). To determine whether cell death is dependent upon caspase 1 activation, we examined the effect of the caspase inhibitors on L67-induced apoptosis in HeLa cells. As shown in Figure 6C, apoptosis was reduced by a caspase 1 inhibitor and a pan caspase inhibitor but not a caspase 3 inhibitor, confirming that cell death was mediated by caspase 1. Together these results demonstrate that inhibition of mito LigIIIα selectively induces cell death in cancer cells by activating a caspase 1-dependent apoptotic pathway.
DISCUSSION
Previous studies have shown that therapy-resistant forms of breast cancer and leukemia with elevated levels of LigIIIα and PARP-1 are hypersensitive to inhibition of LigIIIα in combination with a PARP inhibitor (14,15). Here we have shown that the LigIIIα inhibitor, L67, used in those studies preferentially targets mito LigIIIα and, at the concentrations used to preferentially kill therapy-resistant leukemia and breast cancer cells in combination with a PARP inhibitor (14,15), had no effect on nuclear DNA replication. Importantly, while L67 impacted mitochondrial function in both cancer and non-malignant cells, the effects and outcomes were very different. In cancer cell lines, incubation with L67 resulted in elevated levels of mitochondrially-generated ROS, abnormal mitochondrial morphology, reductions in OCR and mitochondrial DNA, and caspase 1-dependent cell death. Expression of L67-resistant mitoLigA in HeLa cells partially suppressed the increase in mSOX, and completely prevented changes in mitochondrial DNA copy number and morphology, and activation of caspase 1, demonstrating that the effects of L67 were dependent upon inhibition of mito LigIIIα. In contrast to cancer cells, L67 did not alter mitochondrial morphology, mitochondrial DNA copy number or mSOX levels and did not activate capsase 1 in non-malignant cells. Instead of apoptosis, L67 induced significant uncoupling of mitochondria with a compensatory increase in glycolysis and senescence in non-malignant cells. Thus, cancer cells are less able to mitigate the deleterious effects caused by disruption of mitochondrial DNA metabolism than non-malignant cells.
Since inhibition of mito LigIIIα did not cause an increase in steady state levels of mSOX in non-malignant cells even though there was no reduction in the OCR, we suggest that initial increases in the levels of mito DNA damage and/or mSOX activate anti-oxidant responses in non-malignant cells that prevent further increases in mSOX. Notably, we found that OXPHOS was totally uncoupled in the non-malignant cell lines incubated with L67. Since the uncoupling of oxygen consumption from electron transport reduces the generation of ROS by the electron transport chain (35), this alteration in non-malignant cells presumably attenuates increases in mSOX. In addition, non-malignant cells may be able to prevent the increase in mitochondrially-generated ROS by removing dysfunctional, damaged mitochondria via autophagy (32,34,36). Although mito dysfunction caused by inhibition of mito LigIIIα did not result in elevated levels of ROS, p53 was activated, presumably in response to the alterations in cellular energy metabolism, and may contribute to cellular senescence (28). The increase in glycolysis may either be an effort to compensate for the uncoupled OXPHOS or an indicator of senescence (37).
There are conflicting reports as to the effect of LigIIIα knockdown on mitochondrial DNA (12,38). In accord with the normal appearance of mitochondria in L67-treated non-malignant cells, these cells were able to maintain their mitochondrial DNA copy number. Recent studies have shown that there are at least two modes of mitochondrial DNA replication, one of which involves a conventional coupled leading and lagging strand whereas the other one involves RNA incorporation throughout the lagging strand (39). Since the latter mechanism does not require the joining of multiple Okazaki fragments (39), it is possible that non-malignant cells are able maintain mitochondrial DNA copy number by switching to this relatively DNA ligase-independent replication mechanism.
It is well established that cancers driven by oncogenes, such as Ras, c-Myc, BCR-ABL1 and FLT3-ITD, have elevated levels of ROS (40–42). While this increase in ROS activates signaling pathways that promote cell proliferation and metastasis and inhibit cell death pathways (43), redox homeostasis is presumably deregulated as part of the adaptation to constitutively higher ROS levels. Since treatment with an exogenous oxidizing agent induces elevated levels of mitochondrial superoxide and degradation of mitochondrial DNA (44–46) and reduced levels of LigIIIα increases the ROS-induced degradation of mitochondrial DNA (12), our results suggest that the elevated mSOX induced by L67 in cancer cells results from an inability to activate mechanisms, such as OXPHOS uncoupling and mitophagy, that reduce mitochondrially-generated ROS (35,36). Consequently, this initiates a vicious cycle in which elevated mSOX levels cause more DNA damage, resulting in reduced mitochondrial DNA copy number and more mito dysfunction that in turn generates more mSOX (31,45,47). Interestingly, expression of mitoLigA in HeLa cells did not totally prevent the increase in mSOX induced by L67, but fully maintained mitochondrial DNA copy number. Thus, our results support the idea that a combination of increased mSOX and inhibition of mito LigIIIα results in the degradation of mito DNA in HeLa cells.
L67-induced cell death in cancer cells occurred via a caspase 1-dependent apoptotic pathway. Although caspase 1 can be activated by either inflammasomes or pyroptosomes (17), it seems likely that activation of caspase 1 by L67 is mediated by the inflammasome since reactive oxygen species and mitochondrial DNA have been shown to activate the NLRP3 inflammasome (32–34). Thus, we suggest that the L67-induced mitochondrial dysfunction triggers the mitochondrial permeability transition (48) in cancer cells with the subsequent release of superoxide and mito DNA into the cytoplasm, activating the inflammasome and caspase 1-dependent apoptosis (32–34).
Here we have shown that cancer cells are unable to effectively respond to the deleterious effects caused by the inhibition of LigIIIα, a key enzyme in mitochondrial DNA metabolism. Notably, the elevated levels of mitochondrially-generated ROS in cancer cells incubated with L67 result in increased nuclear DNA damage, suggesting that L67-induced mitochondria-dependent nuclear DNA damage underlies the synergistic activity of L67 with PARP inhibitors that reduce nuclear DNA repair (14,15). Furthermore, our studies showing that regulation of mitochondrial function is abnormal in cancer cells, not only provide further evidence that strategies targeting cancer cell mitochondria may have utility in cancer therapy (49,50), but may lead to the identification of novel therapeutic approaches that exploit these differences to selectively eliminate cancer cells by activating caspase 1-dependent apoptosis.
Supplementary Material
Acknowledgments
We thank Dr. Maria Jasin for the plasmid construct encoding mitoLigA and Dr. George Greco for the synthesis of L67.
Financial Support: This work was supported by US National Institute of Health Grants (R01 ES012512 and P01 CA92584 to AET). Flow cytometry and microscopy were carried out in University of New Mexico Cancer Center Shared Resources supported by NCI Cancer Center Support Grant P30 CA11800. Work was also support in part by PA CURE from the Department of Health, PA, (BVH).
Footnotes
Conflict of Interest: A.E Tomkinson is a co-inventor on US patents that cover the use of DNA ligase inhibitors as anti-cancer agents. The other authors declare that they have no conflicts of interest.
REFERENCES
- 1.Ellenberger T, Tomkinson AE. Eukaryotic DNA ligases: structural and functional insights. Annual review of biochemistry. 2008;77:313–338. doi: 10.1146/annurev.biochem.77.061306.123941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arakawa H, Bednar T, Wang M, Paul K, Mladenov E, Bencsik-Theilen AA, et al. Functional redundancy between DNA ligases I and III in DNA replication in vertebrate cells. Nucleic acids research. 2012;40(6):2599–2610. doi: 10.1093/nar/gkr1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gao Y, Katyal S, Lee Y, Zhao J, Rehg JE, Russell HR, et al. DNA ligase III is critical for mtDNA integrity but not Xrcc1-mediated nuclear DNA repair. Nature. 2011;471(7337):240–244. doi: 10.1038/nature09773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simsek D, Furda A, Gao Y, Artus J, Brunet E, Hadjantonakis AK, et al. Crucial role for DNA ligase III in mitochondria but not in Xrcc1-dependent repair. Nature. 2011;471(7337):245–248. doi: 10.1038/nature09794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Le Chalony C, Hoffschir F, Gauthier LR, Gross J, Biard DS, Boussin FD, et al. Partial complementation of a DNA ligase I deficiency by DNA ligase III and its impact on cell survival and telomere stability in mammalian cells. Cell Mol Life Sci. 2012;69(17):2933–2949. doi: 10.1007/s00018-012-0975-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barnes DE, Tomkinson AE, Lehmann AR, Webster AD, Lindahl T. Mutations in the DNA ligase I gene of an individual with immunodeficiencies and cellular hypersensitivity to DNA-damaging agents. Cell. 1992;69(3):495–503. doi: 10.1016/0092-8674(92)90450-q. [DOI] [PubMed] [Google Scholar]
- 7.Bentley DJ, Harrison C, Ketchen AM, Redhead NJ, Samuel K, Waterfall M, et al. DNA ligase I null mouse cells show normal DNA repair activity but altered DNA replication and reduced genome stability. Journal of cell science. 2002;115(Pt 7):1551–1561. doi: 10.1242/jcs.115.7.1551. [DOI] [PubMed] [Google Scholar]
- 8.Harrison C, Ketchen AM, Redhead NJ, O'Sullivan MJ, Melton DW. Replication failure, genome instability, and increased cancer susceptibility in mice with a point mutation in the DNA ligase I gene. Cancer research. 2002;62(14):4065–4074. [PubMed] [Google Scholar]
- 9.Audebert M, Salles B, Calsou P. Involvement of poly(ADP-ribose) polymerase-1 and XRCC1/DNA ligase III in an alternative route for DNA double-strand breaks rejoining. The Journal of biological chemistry. 2004;279(53):55117–55126. doi: 10.1074/jbc.M404524200. [DOI] [PubMed] [Google Scholar]
- 10.Simsek D, Brunet E, Wong SY, Katyal S, Gao Y, McKinnon PJ, et al. DNA ligase III promotes alternative nonhomologous end-joining during chromosomal translocation formation. PLoS Genet. 2011;7(6):e1002080. doi: 10.1371/journal.pgen.1002080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lakshmipathy U, Campbell C. The human DNA ligase III gene encodes nuclear and mitochondrial proteins. Molecular and cellular biology. 1999;19(5):3869–3876. doi: 10.1128/mcb.19.5.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shokolenko IN, Fayzulin RZ, Katyal S, McKinnon PJ, Wilson GL, Alexeyev MF. Mitochondrial DNA ligase is dispensable for the viability of cultured cells but essential for mtDNA maintenance. The Journal of biological chemistry. 2013;288(37):26594–26605. doi: 10.1074/jbc.M113.472977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen X, Zhong S, Zhu X, Dziegielewska B, Ellenberger T, Wilson GM, et al. Rational design of human DNA ligase inhibitors that target cellular DNA replication and repair. Cancer research. 2008;68(9):3169–3177. doi: 10.1158/0008-5472.CAN-07-6636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tobin LA, Robert C, Rapoport AP, Gojo I, Baer MR, Tomkinson AE, et al. Targeting abnormal DNA double strand break repair in tyrosine kinase inhibitor-resistant chronic meyloid leukemias. Oncogene. 2013;32:1784–1793. doi: 10.1038/onc.2012.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tobin LA, Robert C, Nagaria P, Chumsri S, Twaddell W, Ioffe OB, et al. Targeting abnormal DNA repair in therapy-resistant breast cancers. Molecular cancer research : MCR. 2012;10(1):96–107. doi: 10.1158/1541-7786.MCR-11-0255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sallmyr A, Tomkinson AE, Rassool F. Up-regulation of WRN and DNA ligase IIIα in Chromic myeloid leukemia: Consequences for the repair of DNA double strand breaks. Blood. 2008;112(4):1413–1423. doi: 10.1182/blood-2007-07-104257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Latz E, Xiao TS, Stutz A. Activation and regulation of the inflammasomes. Nat Rev Immunol. 2013;13(6):397–411. doi: 10.1038/nri3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qian W, Van Houten B. Alterations in bioenergetics due to changes in mitochondrial DNA copy number. Methods. 2010;51(4):452–457. doi: 10.1016/j.ymeth.2010.03.006. [DOI] [PubMed] [Google Scholar]
- 19.Santos JH, Meyer JN, Mandavilli BS, Van Houten B. Quantitative PCR-based measurement of nuclear and mitochondrial DNA damage and repair in mammalian cells. Methods Mol Biol. 2006;314:183–199. doi: 10.1385/1-59259-973-7:183. [DOI] [PubMed] [Google Scholar]
- 20.Della-Maria J, Zhou Y, Tsai MS, Kuhnlein J, Carney JP, Paull TT, et al. Human Mre11/Human Rad50/Nbs1 and DNA Ligase IIIα/XRCC1 Protein Complexes Act Together in an Alternative Nonhomologous End Joining Pathway. The Journal of biological chemistry. 2011;286(39):33845–33853. doi: 10.1074/jbc.M111.274159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Song W, Levin DS, Varkey J, Post S, Bermudez VP, Hurwitz J, et al. A conserved physical and functional interaction between the cell cycle checkpoint clamp loader and DNA ligase I of eukaryotes. The Journal of biological chemistry. 2007 doi: 10.1074/jbc.M703774200. [DOI] [PubMed] [Google Scholar]
- 22.Gamper AM, Rofougaran R, Watkins SC, Greenberger JS, Beumer JH, Bakkenist CJ. ATR kinase activation in G1 phase facilitates the repair of ionizing radiation-induced DNA damage. Nucleic acids research. 2013;41(22):10334–10344. doi: 10.1093/nar/gkt833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bonner WM, Redon CE, Dickey JS, Nakamura AJ, Sedelnikova OA, Solier S, et al. GammaH2AX and cancer. Nat Rev Cancer. 2008;8(12):957–967. doi: 10.1038/nrc2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tann AW, Boldogh I, Meiss G, Qian W, Van Houten B, Mitra S, et al. Apoptosis induced by persistent single-strand breaks in mitochondrial genome: critical role of EXOG (5'-EXO/endonuclease) in their repair. The Journal of biological chemistry. 2011;286(37):31975–31983. doi: 10.1074/jbc.M110.215715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Turrens JF. Mitochondrial formation of reactive oxygen species. J Physiol. 2003;552(Pt 2):335–344. doi: 10.1113/jphysiol.2003.049478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cleaver JE, Feeney L, Revet I. Phosphorylated H2Ax is not an unambiguous marker for DNA double-strand breaks. Cell Cycle. 2011;10(19):3223–3224. doi: 10.4161/cc.10.19.17448. [DOI] [PubMed] [Google Scholar]
- 27.Shiloh Y, Ziv Y. The ATM protein kinase: regulating the cellular response to genotoxic stress, and more. Nat Rev Mol Cell Biol. 2013;14(4):197–210. [PubMed] [Google Scholar]
- 28.Jones RG, Plas DR, Kubek S, Buzzai M, Mu J, Xu Y, et al. AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Molecular cell. 2005;18(3):283–293. doi: 10.1016/j.molcel.2005.03.027. [DOI] [PubMed] [Google Scholar]
- 29.Guo Z, Kozlov S, Lavin MF, Person MD, Paull TT. ATM activation by oxidative stress. Science. 2010;330(6003):517–521. doi: 10.1126/science.1192912. [DOI] [PubMed] [Google Scholar]
- 30.Goodarzi AA, Noon AT, Deckbar D, Ziv Y, Shiloh Y, Lobrich M, et al. ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Molecular cell. 2008;31(2):167–177. doi: 10.1016/j.molcel.2008.05.017. [DOI] [PubMed] [Google Scholar]
- 31.Mei H, Sun S, Bai Y, Chen Y, Chai R, Li H. Reduced mtDNA copy number increases the sensitivity of tumor cells to chemotherapeutic drugs. Cell death & disease. 2015;6:e1710. doi: 10.1038/cddis.2015.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakahira K, Haspel JA, Rathinam VA, Lee SJ, Dolinay T, Lam HC, et al. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat Immunol. 2011;12(3):222–230. doi: 10.1038/ni.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shimada K, Crother TR, Karlin J, Dagvadorj J, Chiba N, Chen S, et al. Oxidized mitochondrial DNA activates the NLRP3 inflammasome during apoptosis. Immunity. 2012;36(3):401–414. doi: 10.1016/j.immuni.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou R, Yazdi AS, Menu P, Tschopp J. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2011;469(7329):221–225. doi: 10.1038/nature09663. [DOI] [PubMed] [Google Scholar]
- 35.Echtay KS, Roussel D, St-Pierre J, Jekabsons MB, Cadenas S, Stuart JA, et al. Superoxide activates mitochondrial uncoupling proteins. Nature. 2002;415(6867):96–99. doi: 10.1038/415096a. [DOI] [PubMed] [Google Scholar]
- 36.Zhong Z, Umemura A, Sanchez-Lopez E, Liang S, Shalapour S, Wong J, et al. NF-kappaB Restricts Inflammasome Activation via Elimination of Damaged Mitochondria. Cell. 2016;164(5):896–910. doi: 10.1016/j.cell.2015.12.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.James EL, Michalek RD, Pitiyage GN, de Castro AM, Vignola KS, Jones J, et al. Senescent human fibroblasts show increased glycolysis and redox homeostasis with extracellular metabolomes that overlap with those of irreparable DNA damage, aging, and disease. Journal of proteome research. 2015;14(4):1854–1871. doi: 10.1021/pr501221g. [DOI] [PubMed] [Google Scholar]
- 38.Lakshmipathy U, Campbell C. Antisense-mediated decrease in DNA ligase III expression results in reduced mitochondrial DNA integrity. Nucleic acids research. 2001;29(3):668–676. doi: 10.1093/nar/29.3.668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Holt IJ, Jacobs HT. Unique features of DNA replication in mitochondria: a functional and evolutionary perspective. Bioessays. 2014;36(11):1024–1031. doi: 10.1002/bies.201400052. [DOI] [PubMed] [Google Scholar]
- 40.Sallmyr A, Fan J, Datta K, Kim KT, Grosu D, Shapiro P, et al. Internal tandem duplication of FLT3 (FLT3/ITD) induces increased ROS production, DNA damage, and misrepair: implications for poor prognosis in AML. Blood. 2008;111(6):3173–3182. doi: 10.1182/blood-2007-05-092510. [DOI] [PubMed] [Google Scholar]
- 41.Hlavata L, Aguilaniu H, Pichova A, Nystrom T. The oncogenic RAS2(val19) mutation locks respiration, independently of PKA, in a mode prone to generate ROS. The EMBO journal. 2003;22(13):3337–3345. doi: 10.1093/emboj/cdg314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vafa O, Wade M, Kern S, Beeche M, Pandita TK, Hampton GM, et al. c-Myc can induce DNA damage, increase reactive oxygen species, and mitigate p53 function: a mechanism for oncogene-induced genetic instability. Molecular cell. 2002;9(5):1031–1044. doi: 10.1016/s1097-2765(02)00520-8. [DOI] [PubMed] [Google Scholar]
- 43.Sabharwal SS, Schumacker PT. Mitochondrial ROS in cancer: initiators, amplifiers or an Achilles' heel? Nat Rev Cancer. 2014;14(11):709–721. doi: 10.1038/nrc3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Furda AM, Marrangoni AM, Lokshin A, Van Houten B. Oxidants and not alkylating agents induce rapid mtDNA loss and mitochondrial dysfunction. DNA repair. 2012;11(8):684–692. doi: 10.1016/j.dnarep.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yakes FM, Van Houten B. Mitochondrial DNA damage is more extensive and persists longer than nuclear DNA damage in human cells following oxidative stress. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(2):514–519. doi: 10.1073/pnas.94.2.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shokolenko I, Venediktova N, Bochkareva A, Wilson GL, Alexeyev MF. Oxidative stress induces degradation of mitochondrial DNA. Nucleic acids research. 2009;37(8):2539–2548. doi: 10.1093/nar/gkp100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Santos JH, Hunakova L, Chen Y, Bortner C, Van Houten B. Cell sorting experiments link persistent mitochondrial DNA damage with loss of mitochondrial membrane potential and apoptotic cell death. The Journal of biological chemistry. 2003;278(3):1728–1734. doi: 10.1074/jbc.M208752200. [DOI] [PubMed] [Google Scholar]
- 48.Green DR, Van Houten B. SnapShot: Mitochondrial quality control. Cell. 2011;147(4):950, 50 e1. doi: 10.1016/j.cell.2011.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Viale A, Pettazzoni P, Lyssiotis CA, Ying H, Sanchez N, Marchesini M, et al. Oncogene ablation-resistant pancreatic cancer cells depend on mitochondrial function. Nature. 2014;514(7524):628–632. doi: 10.1038/nature13611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang X, Fryknas M, Hernlund E, Fayad W, De Milito A, Olofsson MH, et al. Induction of mitochondrial dysfunction as a strategy for targeting tumour cells in metabolically compromised microenvironments. Nat Commun. 2014;5:3295. doi: 10.1038/ncomms4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.