Abstract
A laboratory-built sheath liquid capillary electrophoresis-mass spectrometry interface was used to develop a qualitative method for fingerprinting analysis of 14 structurally similar flavones, flavonols, flavonones, and several representative glycosides in plant samples. The migration order of the flavonoids was dependent on a the number of hydroxyl groups present on the flavonoid B-ring, extent of conjugation, number of glycosidic functionalities, and ability of the flavonoid to form stable borate complexes with the background electrolyte. Parent ion scans of the flavonoids yielded [M-H]−, except for catechol containing flavonoids, which were detected as borate adducts. These adducts can be used diagnostically to determine the presence or absence of catechol groups on unknown polyphenolic compounds. Product ion scans of the flavonoid glycosides and borate adducts typically yielded the deprotonated aglycone fragment as the base peak, which could be used to confirm the base structure of the flavonoid. This method’s utility was demonstrated by analyzing flavonoids present in ethanolic extracts of Ginkgo biloba herbal supplements.
Introduction
Flavonoids are a class of polyphenolic compounds found in all plants, providing pigmentation and protecting the plants against pathogens and ultraviolet radiation. They are the most consumed polyphenolic compound in the human diet and are attributed with several therapeutic effects, including increased resistance to oxidants1, 2 and decreased occurrence of inflammation3, cardiovascular disease4, hypertension and cancer5, 6. Studies have shown that flavones and flavonols contain the highest antioxidant capacity5 and that their glycosidic forms retain some antioxidant activity7, 8.
Plant extracts have been used as medicinal treatments in most cultures for thousands of years. The popular Ginkgo biloba flavonoid extract EGb761, for example, has been reported to improve cognitive function by increasing dopamine levels in the brain, thereby improving memory9. In a population study, Gingko biloba flavonoid supplements were found to decrease the rate of cognitive decline in non-demented patients over 65 years of age when compared to patients not taking the supplement10. Additionally, these flavonoid supplements have also been shown to act as an agonist to 5-HT1A, resulting in relief to stress and depression11.
To identify potentially bioactive flavonoids from plant material, HPLC and GC are traditionally employed12-15. Capillary electrophoresis (CE) has potential advantages for flavonoid analysis, including faster analysis times, less consumption of precious sample, and high separation efficiencies for charged compounds. Conventional CE and micellar electrokinetic chromatography (MEKC)16 have been coupled to UV detection17-19, electrochemical20-22, and MS detection18, 23, 24 for the analysis of flavonoids. Of these detection systems, MS is capable of providing an additional dimension of separation based on mass, as well as pertinent structural information garnered from fragmentation studies. However, as noted by Rijke et al., little work has been conducted using CE-MS for flavonoid analysis25. This is because flavonoid separations by CE typically require selective background electrolyte components, such as borate, that can complex phenolic compounds, and micelles, that can act as a pseudo-stationary phase for flavonoids. These components are not volatile and are therefore rarely employed in CE-ESI-MS applications in favor of acetate and formate containing electrolytes18, 24. However, at low concentrations, additives can be used without significant spray degradation or instrument contamination due to the low mass loads of the electrophoresis capillary26-28.
Herein, we describe qualitative CE-ESI-MS method in negative ion mode for the detection of 14 common flavonoids utilizing an ammonium borate buffer as the BGE. The method is capable of separating and detecting five flavonoid glycones and nine aglycones in 13 minutes with separation efficiencies of up to 75,000 theoretical plates. Additionally, the catechol containing flavonoids were detected as borate adducts in the MS1 scans, adding an extra dimension of structural diagnostic information. Fragmentation data on these adducts is presented. Finally, the method was applied to the analysis of flavonoids in Ginkgo biloba herbal supplements.
Experimental
Reagents
Apigenin, chrysin, eriodoctyl, galangin, kaempferol, luteolin, naringenin, naringin, pinocembrin, quercetin, quercitrin, rutin, and ammonium biborate were purchased from Sigma (St. Louis, MO, USA). Apiin and apigetrin were purchased from Carl Roth (Karlsruhe, Germany). No further purification of the standards was conducted. Methanol, ethanol, isopropyl alcohol, acetic acid and ammonium hydroxide were purchased from Fisher Scientific (Pittsburg, PA, USA). Ultrapure water was obtained from a Milli-Q water purification system (Millipore, Bedford, MA, USA).
Stock solutions of the flavonoids were prepared by dissolving 1 mg of standard in 1 mL of methanol for aglycones and 70:30 methanol:water for glycones. The solutions were stored in the dark at 4 °C. Prior to dilution, the stock solutions were sonicated for 5 min to ensure flavonoid dissolution. The buffer pH was investigated in the range 8.7-10.0 and was adjusted using 0.1 M ammonium hydroxide. The BGE was degassed and replaced daily to avoid current drops due to bubbles in the CE capillary.
Instrumentation
The separation of 14 flavonoid standards was optimized using a P/ACE MDQ Capillary Electrophoresis System equipped with a diode array detector (Beckman Coulter, Palo Alto, CA, USA). A 65 cm × 50 μm i.d. × 360 μm o.d. capillary (Polymicro Technologies, Tucson, AZ, USA) was used with a 55 cm effective length for all CE-UV studies.
A sheath liquid CE-MS interface was fabricated on a Micromass Quattro Ultima triple quadrupole instrument (Waters, Millford, MA, USA) using a modified nanospray flange. The interface was constructed using a model MT1CS6 Valco mixing tee (VICI, Houston, TX, USA). The electrospray needle was made by cutting 30 gauge metal tubing (Component Supply Co., Fort Meade, FL, USA) into a 2 cm segment and sharpening the tip using a metal file in order to allow the use of lower electrospray voltages. The ESI needle was then secured into the front of the mixing tee using a 0.3 mm I.D Valco one piece fused silica adapter. Sheath liquid was supplied to the mixing tee via an infusion pump at 3 μL/min. The laboratory-built CE system was connected to the interface by threading a 65 cm length × 40 μm i.d. (105 μm o.d.) fused silica capillary through the back of the mixing tee using PEEK tubing and subsequently into the electrospray needle. The CE capillary was flush with the electrospray tip during analysis. Two separate Spellman power supplies provided separation and electrospray voltages. The CE power supply was operated in positive mode while the electrospray power supply was operated in negative mode. The mass spectrometer was operated at a cone voltage of −35 V and a cone temperature of 100 °C with 40 psi nitrogen bath gas flooding into the fish bowl flange. No drying gas was used.
Capillary preparation and infusion experiments
CE capillaries were sequentially rinsed with methanol, 0.1 M HCl, nanopure water, 0.1 M ammonium hydroxide, and BGE at 10 psi for 10 min. The capillary was also rinsed at 10 psi for 5 min with 0.1 M ammonium hydroxide and for 10 min with BGE between injections. Samples were injected onto the capillary hydrodynamically using 10 psi for 2 sec, which corresponds to 13.3 nL or 1.63% of the total capillary volume. Prior to daily CE-MS analysis, the sheath liquid line was rinsed with two tubing volumes of methanol and 80:20 IPA:water sheath liquid. The electrospray tip was rinsed with 50:50 water:methanol between runs to prevent the accumulation of salts. Infusion experiments to determine flavonoid fragment ions were conducted using 10 μm standards in BGE and infusing through the CE capillary using electroosmotic flow. The CID energy was increased in 5 eV steps until significant fragmentation of the parent was observed, which typically occurred at ~20 eV.
Preparation of Ginkgo biloba herbal supplement
Herbal supplements of 120 mg Ginkgo biloba were purchased at a local market. The plant material from the supplement was removed from a capsule and dissolved in 70:30 ethanol:water. The solution was sonicated for 1 hour and subsequently centrifuged (1000g) for 5 minutes. The supernatant was extracted and diluted 100-fold with BGE. The resultant solution was filtered through a 0.25 μm membrane filter twice and immediately injected into the CE system to minimize oxidation of the flavonoids.
Results and Discussion
Optimization of capillary electrophoresis conditions
The base structures of flavones, flavonols, and flavonones are shown in Figure 1, and the substitution patterns on the 14 flavonoids analyzed in this study are listed in Table 1. For simplicity, a Beckman CE-UV system equipped with an autosampler was used for the optimization of CE separation conditions. A borate based buffer was chosen for the separation because of its ability to form complexes with polyphenolic compounds and increase their effective negative charge and size[29, 30]. Migration in CE is based on the overall charge and hydrated radius of a molecule. Similar molecules possessing different borate complex formation constants can be resolved. However, borate buffer is normally considered incompatible with MS due to its low volatility. In our experiments, significant salt accumulation was not observed and the MS orifice was cleaned regularly to prevent buildup. Ammonium ion was chosen as the BGE cation due to its high volatility.
Fig. 1.
Basic structure of the flavonoids analyzed in this study. Underlined numbers represent common bond cleavages upon CID fragmentation.
Table 1.
Representative flavonoids and the respective fragments used in this study. Bolded flavonoids contain catechol group.
| Compound Name | Monitored Mass |
R1 | R2 | R3 | R4 | R5 | Major Fragments |
|---|---|---|---|---|---|---|---|
| Flavones | |||||||
| 1) Chrysin | 253 | OH | OH | H | H | - | 209 (100), 181 (13) |
| 2) Apigenin | 269 | OH | OH | H | OH | - | 225 (100), 201 (25) |
| 3) Luteolin | 285 | OH | OH | OH | OH | - | 241 (100) |
| 4) Apigetrin | 431 | Glu | OH | H | OH | - | 269 (100) 311 (20) |
| 5) Apiin | 563 | Glu-Apio | OH | H | OH | - | 269 (100), 431(25) |
| Flavonones | |||||||
| 6) Pinocembrin | 255 | OH | OH | H | H | - | 213 (100), 211 (45) |
| 7) Naringenin | 271 | OH | OH | H | OH | - | 151 (100), 177 ( 23) |
| 8) Eriodictyol | 313 | OH | OH | OH | OH | - | 269 (100), 151 (10) |
| 9) Naringin | 580 | Glu-Rha | OH | H | OH | - | 271 (100), 459 (30) |
| Flavonols | |||||||
| 10) Galangin | 269 | OH | OH | OH | H | OH | 197 (100), 241 (98) |
| 11) Kaempferol | 285 | OH | OH | OH | H | OH | 257 (100), 213 (65) |
| 12) Quercetin | 345 | OH | OH | OH | OH | OH | 327 (100) 301 (35) |
| 13) Quercitrin | 491 | OH | OH | OH | OH | Rha | 473(100) |
| 14) Rutin | 609 | OH | OH | OH | OH | Glu-Rha | 301 (100), 591(20) |
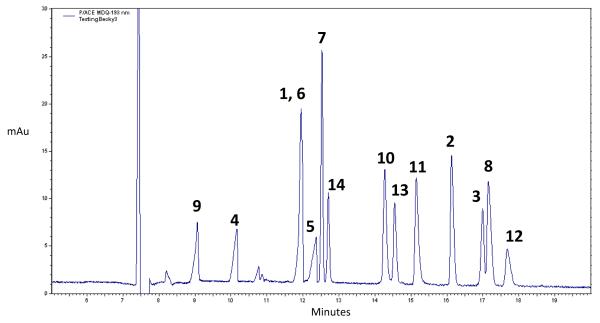
The optimized separation of the 14 flavonoids is shown in Figure 2. Biborate concentrations above 50 mM caused arcing between the ESI tip and the MS inlet, so only concentrations below this value were investigated. The effect of pH on the separation was investigated in the range of 8.7-10, where the flavonoids carry at least one negative charge. Additionally, the effect of methanol on the separation was investigated due to its ability to potentially increase separation efficiency and ESI spray volatility. Baseline resolution of the compounds was achieved in 18 min using a BGE chemistry of 25 mM ammonium biborate at pH = 9.3 with 10% methanol and a separation voltage of 25 kV. At pH values greater than 9.3, comigration of catechol containing flavonoids was observed, while methanol concentrations greater than 10% resulted in long analysis times (>20 min). Coelution of pinocembrin and chrysin, which differ by two mass units, was observed; however, if these two flavonoids are present in a single sample, they can be identified separately by mass spectrometric detection.
Fig. 2.
CE-UV separation of 14 flavonoid standards. Conditions: separation voltage: 25 kV, 65 cm × 50 μm i.d. × 360 μm o.d. capillary, 65 cm effective length, 25 mM ammonium biborate in 10% methanol, pH = 9.3. 100 μM standards dissolved in BGE.
The migration order of flavonoids under the optimal separations conditions can be used to identify the presence of specific structural features of unknown flavonoids. A primary factor that affected the migration properties of the flavonoids was their flavonoid subclass. For example, non-catechol flavonones were detected between 9-12.5 min, while flavonols were detected after 12.5 min. Another factor that determined the migration properties of the flavonoids was the extent of B-ring hydroxylation. A higher number of B-ring hydroxyl groups increased the migration time of the flavonoids. This effect is particularly true of quercetin, eriodictyol, and luteolin, all of which contain catechol functionalities. Catechols are known to form strong complexes with borate, resulting in an increased negative charge and longer migration times in normal mode CZE. This trend was observed for flavonoid glycones as well; however, because the flavonoid glycosides are much larger in size and lower in negative charge, they migrate more quickly toward the detector than even aglycones which contain one B-ring hydroxyl group.
Borate complexation extended the migration times of catechol containing glycosylated flavonoids as well, as can be observed by the flavonoids rutin and quercitrin. Additionally, because rutin and quercitrin are 3-O-glycosides, the 7-position hydroxyl group is deprotonated at pH = 9.3, increasing their negative electrophoretic mobility relative to 7-O-glycosides. The 7-O-glycosides apigetrin and naringen are less negatively charged, resulting in fast migration times within 3 min of the neutral marker. Apiin, a 7-O-diglycoside would be expected to migrate more quickly toward the detector than apigetrin due to its size. However, it is detected 2 min after apigetrin, suggesting borate complexation could be occurring at its additional sugar substituent.
Flavonoid characterization by CE-MS/MS
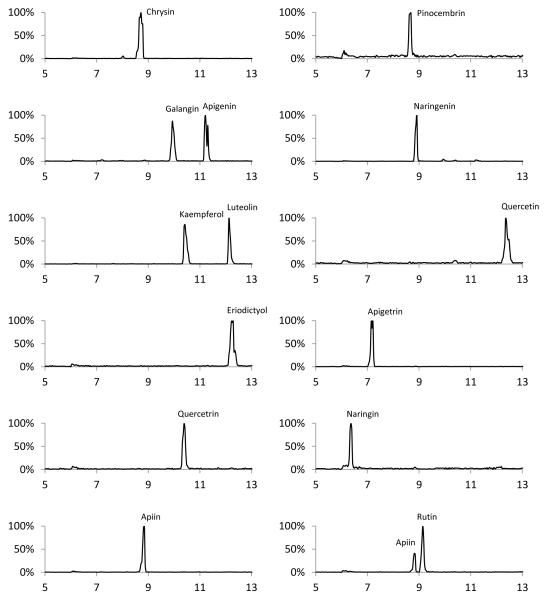
Sheath liquid composition was optimized by analyzing the signal stability of background ions present in the sheath solution. Without the assistance of drying gas, organic content of greater than 70% is required for spray formation at −3 kV spray voltage. Methanol based sheath liquids typically produced erratic signal fluctuations in negative mode electrospray. A fairly stable signal was observed by using 80:20 isopropyl alcohol (IPA):water. The utility of IPA sheath liquids has been reported31, 32, but methanol is still frequently used in CE-MS33, 34. The organic additive triethylamine was used to increase the signal of the 7-O-glycosides naringin, apigetrin, and apiin compared to ammonium acetate and acetic acid additives. These flavonoids have a higher pKa1, and require higher pH sheath liquids than that of the BGE to achieve a more negative charge. Sheath liquid flow rates from 1-10 μL/min were investigated, and 3 μL/min was chosen because it provided the best signal stability without significantly diluting the analyte. Using these sheath liquid conditions in tandem with the aforementioned optimized CE conditions, nine aglycones and five glycones were separated and detected in 13 min by CE-MS (Figure 3). The average number of theoretical plates for the flavonoid separation was 30,000, with as high as 75,000 theoretical plates for the flavonoid naringenin. As reported by Wang et. al., these CE peak efficiencies for flavonoids are significantly higher than those obtained using reverse phase LC35.
Fig. 3.
CE-MS separation of 14 flavonoid standards. Conditions: separation voltage: 25 kV, 65 cm × 40 μm i.d. × 105 μm o.d. capillary, 25 mM ammonium biborate in 10% methanol, pH = 9.3. Electrospray voltages = −3 kV. 25 μM standards dissolved in BGE.
Negative ionization mode was chosen for this study because the flavonoids were present as negative ions in the BGE. The retro-Diels Alder product ion of the A-ring at m/z 151 was observed during MS analysis of most of the flavonoid aglycones, and represents the 1,3 cleavage of the C-ring (Figure 1). The B-ring retro-Diels Alder fragment, which provides diagnostic information regarding B-ring hydroxylation and C-ring saturation, was not detected in these studies. Reports show that B-ring fragments can be detected in positive ion mode36, 37. However, because the flavonoids were separated as anions by CE, sensitive detection of them in positive ion mode was not successful. The difference between the parent ion mass and the [1,3 A]− ion mass can be used to infer the number of hydroxyl groups on the undetected fragment of the flavonoid; however, the location of the hydroxyl groups on the B- or C-ring cannot be determined. Losses of CO and CO2 were also common for the aglycones. Other ions characteristic of individual aglycones are in agreement with LC-MS/MS reports and possible mechanisms for these losses have been proposed elsewhere[36-39].
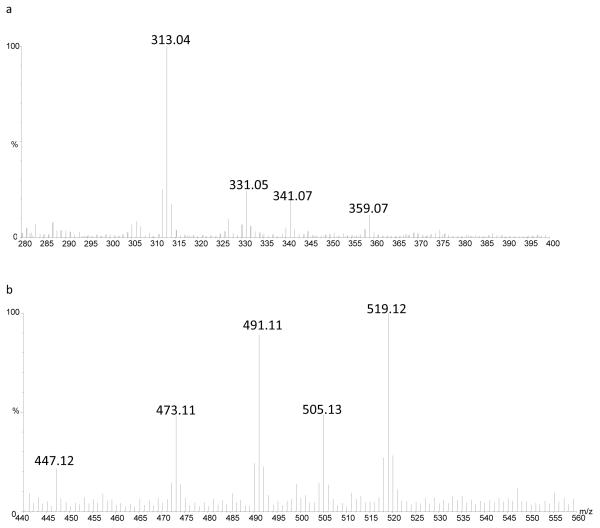
The fragmentation of catechol containing flavonoid aglycones is shown in Table 2. Borate adduct formation corresponding to [M + B(OH)3 − 2H2O − H]−, [M + B(OH)3 − H2O − H]−, [M + B(OH)3 + CH3OH − 2H2O − H]−, and [M + B(OH)3 + 2CH3OH – 3H2O –H]− were detected for all catechol containing flavonoids. For eriodictyol, [M + B(OH)3 - 2H2O - H] was its base peak at m/z 313 (Figure 4a). For quercetin and luteolin, the adducts were fairly evenly distributed in terms of abundance. Fragmentation of the borate adducts often resulted in detection of the free aglycone ion, except for the eriodictyol borate adduct, which underwent fragmentation similar to free aglycones. In MS1 scans, the borate adducts generate an increasing m/z pattern (M+26, 44, 58, and 72) that can be used to identify the presence of a catechol group on the aglycone or a cis-diol group on a sugar substituent. These diagnostic ions are particularly useful for flavonoids, where several isomers exist, and fragmentation pathways are fairly similar.
Table 2.
Fragmentation of catechol containing flavonoid aglycones.
| 3 | 3** | 3*** | 8* | 12 | 12** | |
|---|---|---|---|---|---|---|
| [M−H]− | 285 | 329 | 344 | 313 | 301 | 345 |
| [M−H2O−H]− | - | 311 | - | - | - | 327 |
| [M−CO−H]− | - | - | - | - | 273 | - |
| [M−CH3OH−H]− | - | - | 312 | - | - | - |
| [M−CO2−H]− | 241 | - | - | 269 | - | - |
| [Aglycone-H]− | - | 285 | 285 | - | - | 301 |
| [M−C2H4O2−H]− | - | - | - | 253 | - | 285 |
| [M−CO2−CO−H]− | - | - | - | - | 229 | - |
| [M−B-ring-H]− | - | - | - | - | 191 | - |
| [1,2 A]− | - | - | - | - | 179 | - |
| [1,3 A]− | 151 | - | - | 151 | 151 | - |
[M+B(OH)3−2H2O−H]−,
[M+B(OH)3−H2O−H]−,
[M+B(OH)3+CH3OH−2H2O−H]−
Fig. 4.
The formation of borate adducts for a) eriodictyol and b) quercitrin
The fragmentation of the flavonoid glycones and their most abundant borate adducts is shown in Table 3. Quercitrin and rutin were detected as [M-H]− as well as borate adducts (Figure 4b). Additionally, apiin was detected at [M+26-H]−, supporting borate complexation which would cause longer migration time relative to other non-catechol containing flavonoid glycosides. Interestingly, the base peak for rutin was [M+8-H]−, with the borate adduct [M+26-H]− being the second most abundant ion in its spectrum. The former of the two adducts was not expected, but it contained a similar isotopic distribution as the other borate adducts. Fragmentation of this adduct yielded the quercetin aglycone ion at m/z 301, which is expected from the quercetin derived glycoside. Upon CID fragmentation, the aglycone fragment was the base peak for all of the glycosides. For apigetrin and naringin, the [1,3 A]− fragment ion, with the sugar groups intact, was detected at approximately 20% abundance, that has been reported previously for flavonoid glycones40; however, in the case of naringin, this ion could also correspond to fragmentation along the glycoside directly connected to the aglycone38. The neutral loss of m/z 132 from apiin corresponds to the loss of its pentose substituent. Single sugar group neutral losses are possible for polyglycosides; however, the corresponding product ions were not observed for the diglycosides rutin or naringin.
Table 3.
Fragmentation of flavonoid glycosides and their borate adducts
| 4 | 5 | 5* | 9 | 13 | 13** | |
|---|---|---|---|---|---|---|
| [M−H]− | 431 | 563 | 589 | 579 | 447 | 491 |
| [M−H2O−H]− | - | - | 571 | - | - | 473 |
| [M-borate adduct-H]− | - | - | - | - | - | 447 |
| [M-apioside-H]− | - | 431 | - | - | - | - |
| [M-rhamnoside-H]− | - | - | - | - | - | 327 |
| [1,3 A]− | 311 | - | - | 459 | - | - |
| [Aglycone-H]− | 269 | 269 | 269 | 271 | 301 | 301 |
Flavonoid fingerprint analysis of Ginkgo biloba ethanolic extracts
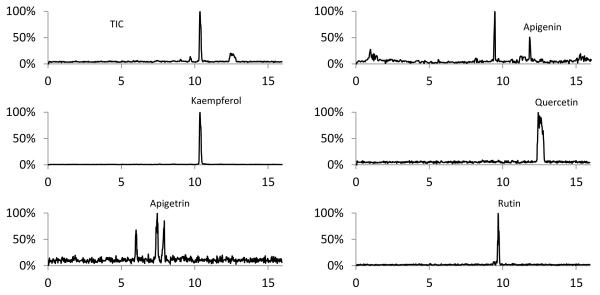
Ginkgo biloba flavonoids were extracted from herbal supplements by sonication in a 70:30 ethanol:water solution in order to extract both glycones and aglycones from the plant material. Hydrolysis of the plant material was not conducted because this results in the cleavage of O-glycosidic bonds, limiting the method to only the detection of aglycones41. The results for the CE-MS analysis of Ginkgo biloba flavonoids are shown in Figure 5. Based on CE-MS results, the most prominent flavonoids present in Gingko biloba are kaempferol and quercetin. Peak destacking of kaempferol and quercetin occurred in the 4-fold BGE diluted ethanolic extract of the Ginkgo biloba supplement, so a 100-fold dilution was conducted to minimize this effect. Apigenin, rutin, and apigetrin were also detected in the supplement, which is in agreement with other reports that show the presence of these flavonoids, as well as quercetin and kaempferol, in Ginkgo biloba samples42-44.
Fig. 5.
CE-MS of 100-fold dilution of the Ginkgo biloba supplement ethanol extract. *Apigenin (2) was not detected in the 100-fold dilution, but was detected in the 4-fold dilution of the extract.
Additional peaks with m/z 425, 431, 447, and 593 were detected. Fragmentation of the parent ion m/z 425 yielded no additional signature aglycone fragments, making its identification difficult. Fragmentation of the parent ions of m/z 431, 447, and 593 yielded the fragment ion m/z 285, suggesting these peaks could be kaempferol or luteolin glycosides, that have been reported in Ginkgo biloba extracts previously.45 While authentic standards would be required to determine these unknown compounds unequivocally, it can be presumed that they are kaempferol glycosides, as no borate adducts were detected that would signify the presence of the catechol moiety of luteolin glycosides.
Conclusion
A sheath liquid CE-MS method using a borate based buffer was developed for the detection and identification of the natural flavonoids apiin, apigenin, apigetrin, chrysin, eriodoctyl, galangin, kaempferol, luteolin, naringenin, naringin, pinocembrin, quercetin, quercitrin, and rutin in under 13 min. Flavonoid glycosides typically migrated faster than aglycones, followed by flavonones, flavonols, and flavones. The catechol containing compounds all migrated slowest due to borate complexation. MS studies showed that these borate complexes were detectable as m/z M+26, 44, 58, and 72, and these ions could be used diagnostically to differentiate catechol containing flavonoids from non-catechol containing flavonoids of the same m/z. MS/MS studies were conducted on the flavonoid standards, and the data was used to determine the presence of these compounds in Ginkgo biloba extract. The highest abundance flavonoids were quercetin and kaempferol, followed by apigetrin, rutin, and apigenin. The method described here shows the utility of CE to achieve high resolution separations. It also demonstrates the breadth of information that can be obtained from MS and MS/MS for the fingerprinting studies of flavonoids in a complex plant material, such as Ginkgo biloba.
Acknowledgements
We would like to thank Dr. Susan Lunte, Dr. John Stobaugh, and Dr. Heather Desaire for support in preparation of this article and the KU Mass Spectrometry Laboratory. This work was support by a grant from the National Institutes of Health R01-NS066466.
References
- 1.Yao LH, Jiang YM, Shi J, TomÁS-BarberÁN FA, Datta N, Singanusong R, Chen SS. Plant Foods Hum Nutr. 2004;59:113–122. doi: 10.1007/s11130-004-0049-7. [DOI] [PubMed] [Google Scholar]
- 2.Lee ML, R., Shen L, Yang L, Yen K, Hou W. Journal of Agricultural Food Chemistry. 2001;49:5551–5555. doi: 10.1021/jf010622j. [DOI] [PubMed] [Google Scholar]
- 3.Serafini M, Peluso I, Raguzzini A. Proceedings of the Nutrition Society. 2010;69:273–278. doi: 10.1017/S002966511000162X. [DOI] [PubMed] [Google Scholar]
- 4.Nandave MO, S. K., Arya DS. Natural Product Radiance. 2005;4:166–176. [Google Scholar]
- 5.Tanwar B, Modgil R. Spatula DD. 2012;2:59–68. [Google Scholar]
- 6.Tapas ARS, D. M., Kakde RB. Tropical Journal of Pharmaceutical Research. 2008;7:1089–1099. [Google Scholar]
- 7.Pietta P. Journal of Natural Products. 2000;63:1035–1042. doi: 10.1021/np9904509. [DOI] [PubMed] [Google Scholar]
- 8.Hopia AH, Marina Journal of American Oil Chemists Society. 1999;76:139–144. [Google Scholar]
- 9.Fehske CJ, Leuner K, Muller WE. Pharmacological research : the official journal of the Italian Pharmacological Society. 2009;60:68–73. doi: 10.1016/j.phrs.2009.02.012. [DOI] [PubMed] [Google Scholar]
- 10.Amieva H, Meillon C, Helmer C, Barberger-Gateau P, Dartigues JF. PloS one. 2013;8:e52755. doi: 10.1371/journal.pone.0052755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Winter JCT, D. Pharmacology Biochemistry and Behavior. 1999;62:543–547. doi: 10.1016/s0091-3057(98)00190-7. [DOI] [PubMed] [Google Scholar]
- 12.Ko Y-C, Lee R-J, Feng H-T, Lee M-R. Journal of the Chinese Chemical Society. 2013;60:1333–1338. [Google Scholar]
- 13.Deng F, Zito SW. Journal of chromatography. A. 2003;986:121–127. doi: 10.1016/s0021-9673(02)01921-0. [DOI] [PubMed] [Google Scholar]
- 14.Sutthanut K, Sripanidkulchai B, Yenjai C, Jay M. Journal of Chromatography A. 2007;1143:227–233. doi: 10.1016/j.chroma.2007.01.033. [DOI] [PubMed] [Google Scholar]
- 15.De Souza KCB, Schapoval EES, Bassani VL. Journal of Pharmaceutical and Biomedical Analysis. 2002;28:771–777. doi: 10.1016/s0731-7085(01)00693-8. [DOI] [PubMed] [Google Scholar]
- 16.Peres RG, Micke GA, Tavares MF, Rodriguez-Amaya DB. J Sep Sci. 2009;32:3822–3828. doi: 10.1002/jssc.200900312. [DOI] [PubMed] [Google Scholar]
- 17.Sanli S, Lunte CE. Analytical Methods. 2014;6:3858–3864. [Google Scholar]
- 18.Huck C, Stecher G, Ahrer W, Stogg WM, Buchberger W, Bonn GK. Journal of Separation Science. 2002;25:904–908. [Google Scholar]
- 19.Zhu J, Yu K, Chen X, Hu Z. Journal of Chromatography A. 2007;1166:191–200. doi: 10.1016/j.chroma.2007.08.018. [DOI] [PubMed] [Google Scholar]
- 20.Wang W, Lin P, Ma L, Xu K, Lin X. J Sep Sci. 2016 doi: 10.1002/jssc.201501287. [DOI] [PubMed] [Google Scholar]
- 21.Hendrickson HP, Kaufman AD, Lunte CE. Journal of Pharmaceutical and Biomedical Analysis. 1994;12:325–334. doi: 10.1016/0731-7085(94)90007-8. [DOI] [PubMed] [Google Scholar]
- 22.Wu T, Guan Y, Ye J. Food Chemistry. 2007;100:1573–1579. [Google Scholar]
- 23.Sawalha SMS, Arráez-Román D, Segura-Carretero A, Fernández-Gutiérrez A. Food Chemistry. 2009;116:567–574. [Google Scholar]
- 24.Carrasco-Pancorbo A, Neususs C, Pelzing M, Segura-Carretero A, Fernandez-Gutierrez A. Electrophoresis. 2007;28:806–821. doi: 10.1002/elps.200600382. [DOI] [PubMed] [Google Scholar]
- 25.de Rijke E, Out P, Niessen WM, Ariese F, Gooijer C, Brinkman UA. Journal of chromatography. A. 2006;1112:31–63. doi: 10.1016/j.chroma.2006.01.019. [DOI] [PubMed] [Google Scholar]
- 26.Vanhoenacker G, De Villers A, Lazou K, De Keukeleire D, Sandra P. Chromatographia. 2001;54:309–315. [Google Scholar]
- 27.Bednár̆ P, Papoušková B, Müller L, Barták P, Stávek J, Pavloušek P, Lemr K. Journal of Separation Science. 2005;28:1291–1299. doi: 10.1002/jssc.200500071. [DOI] [PubMed] [Google Scholar]
- 28.Brensinger K, Rollman C, Copper C, Genzman A, Rine J, Lurie I, Moini M. Forensic science international. 2016;258:74–79. doi: 10.1016/j.forsciint.2015.11.007. [DOI] [PubMed] [Google Scholar]
- 29.Vaher M, Koel M. Journal of Chromatography A. 2003;990:225–230. doi: 10.1016/s0021-9673(02)02013-7. [DOI] [PubMed] [Google Scholar]
- 30.Ballus CA, Meinhart AD, de Oliveira RG, Godoy HT. Food Research International. 2012;45:136–144. [Google Scholar]
- 31.Tong P, Zhang L, He Y, Tang S, Cheng J, Chen G. Talanta. 2010;82:1101–1106. doi: 10.1016/j.talanta.2010.05.045. [DOI] [PubMed] [Google Scholar]
- 32.Xu XB, Liu DB, Guo XM, Yu SJ, Yu P. Journal of chromatography. A. 2014;1366:65–72. doi: 10.1016/j.chroma.2014.09.019. [DOI] [PubMed] [Google Scholar]
- 33.Schiavone NM, Sarver SA, Sun L, Wojcik R, Dovichi NJ. Journal of chromatography. B, Analytical technologies in the biomedical and life sciences. 2015;991:53–58. doi: 10.1016/j.jchromb.2015.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Banks JF., Jr. Journal of Chromatography A. 1995;712:245–252. [Google Scholar]
- 35.Wang S-P, Huang K-J. Journal of Chromatography A. 2004;1032:273–279. doi: 10.1016/j.chroma.2003.11.099. [DOI] [PubMed] [Google Scholar]
- 36.Cuyckens F, Ma YL, Pocsfalvi G, Claeysi M. Analusis. 2000;28:888–895. [Google Scholar]
- 37.Justino GC, Borges CM, Florencio MH. Rapid communications in mass spectrometry : RCM. 2009;23:237–248. doi: 10.1002/rcm.3869. [DOI] [PubMed] [Google Scholar]
- 38.Cuyckens F, Rozenberg R, de Hoffmann E, Claeys M. Journal of mass spectrometry : JMS. 2001;36:1203–1210. doi: 10.1002/jms.224. [DOI] [PubMed] [Google Scholar]
- 39.Fabre N, Rustan I, de Hoffmann E, Quetin-Leclercq J. Journal of the American Society for Mass Spectrometry. 2001;12:707–715. doi: 10.1016/S1044-0305(01)00226-4. [DOI] [PubMed] [Google Scholar]
- 40.Yan C, Liu S, Zhou Y, Song F, Cui M, Liu Z. Journal of the American Society for Mass Spectrometry. 2007;18:2127–2136. doi: 10.1016/j.jasms.2007.09.012. [DOI] [PubMed] [Google Scholar]
- 41.van Beek TA, Montoro P. Journal of chromatography. A. 2009;1216:2002–2032. doi: 10.1016/j.chroma.2009.01.013. [DOI] [PubMed] [Google Scholar]
- 42.Wang J, Han S. Chromatographia. 2013;76:715–718. [Google Scholar]
- 43.Guo RZ, Liu XG, Gao W, Dong X, Fanali S, Li P, Yang H. Journal of chromatography. A. 2015;1422:147–154. doi: 10.1016/j.chroma.2015.10.008. [DOI] [PubMed] [Google Scholar]
- 44.Cao Y, Chu Q, Fang Y, Ye J. Analytical and bioanalytical chemistry. 2002;374:294–299. doi: 10.1007/s00216-002-1436-2. [DOI] [PubMed] [Google Scholar]
- 45.Luo JL, F., Liu Y, Shih Y, Lo C. Journal of Food and Drug Analysis. 2013;21:27–39. [Google Scholar]