Abstract
Objective
Increasing evidence indicates that the decreased expression of microRNA-133a (miR-133a) may be correlated with poor survival for cancer patients. Thus, we performed this meta-analysis to evaluate the prognostic value of decreased miR-133a in solid cancers.
Methods
Eligible studies were gathered by searching on PubMed, Web of Science, and Embase. Using the STATA 12.0 software, the pooled hazard ratios (HRs) and their corresponding 95% confidence intervals (CIs) for total and subgroup analyses were calculated to investigate the possible correlation between decreased miR-133a and overall survival (OS) of patients with cancer.
Results
Ten studies were enrolled in this meta-analysis. The pooled result showed that decreased expression of miR-133a predicted poor OS in solid cancer patients (HR =1.62, 95% CI: 1.16–2.24, P=0.004). Compared with the total pooled HR, further analyses indicated that the subgroups of digestive system neoplasms (HR =1.73, 95% CI: 1.20–2.51, P=0.003), frozen tissue preservation (HR =1.89, 95% CI: 1.41–2.53, P<0.001), and multivariate analysis (HR =2.07, 95% CI: 1.42–3.02, P<0.001) exhibited stronger connection between decreased miR-133a expression and OS outcome.
Conclusion
This meta-analysis suggested that decreased miR-133a was associated with poor OS in patients with solid cancer. Because of the data in our study are limited, additional studies are required to verify the poor prognosis of decreased miR-133a in solid tumors.
Keywords: prognosis, microRNA-133a, solid cancers, meta-analysis
Introduction
Cancer is one of the most important health problems of mankind. The 5-year relative survival rate for all cancers in Americans and Chinese is ~69% and 37%, respectively.1,2 Thus, the researches for prognostic indicators of cancer survival have important clinical values. Nowadays, numerous cancer prognostic factors have been discovered,3–5 and the prognostic values of microRNAs also have been proved by many studies.6–8
MicroRNAs are a kind of small noncoding RNA molecule in biology, containing ~22 nucleotides, that function in posttranscriptional regulation of gene expression and RNA silencing.9,10 MicroRNAs regulate tumorigenesis and cancer progression, also as gatekeepers of apoptosis for tumors.11,12 Through different mechanisms, microRNA can perform the role of a tumor promoter13,14 or cancer suppressor factor.15–17 Meanwhile, microRNA in cancers can also be up-18,19 or downregulated.20,21
MicroRNA-133a (miR-133a) plays an important role in cancer development and progression. Studies showed that miR-133a is a tumor suppressor.22,23 Consequently, decreased expression of miR-133a in cancer patients correlates with poor survival and prognosis.24–27 However, the results from some studies find that lower expression of miR-133a functioned as a favorable outcome factor.28,29 Thus, as these results so far seem to be controversial, it is still not enough to draw a conclusion for the prognostic value of decreased miR-133a.
As a fact, most researches assessing the implications of miR-133a expression in cancer were limited by the small sample sizes. Therefore, we performed this meta-analysis and expected to handle the inconsistencies of previous studies.
Materials and methods
Search strategy
The searches were performed by two reviewers (Jian Xiao and Yong Zou) independently. Articles were searched on PubMed, Web of Science, and Embase (updated to February 19, 2016) with language restriction of English. The search terms of keywords and their combination were as the following: “microRNA-133a OR mir-133a OR microRNA133a OR mir133a OR 133a” AND “survival OR prognosis OR prognostic” AND “cancer OR tumor OR tumour OR neoplasm OR neoplasma OR cancers OR tumors OR tumours OR neoplasms OR neoplasmas OR carcinoma OR carcinomas”. A manual search for the references of relevant articles was also performed to find out other potential studies. Any differences were resolved by discussion.
Inclusion and exclusion criteria
Eligible studies for this meta-analysis met the following inclusion criteria: 1) evaluated the correlation between miR-133a expression and overall survival (OS) of patients with any cancer types; 2) full-text was available; and 3) reported hazard ratios (HRs) with 95% confidence intervals (CIs) or sufficient data to evaluate the HRs and 95% CIs. The exclusion criteria were the following: 1) nonhuman researches, meta-analysis, and duplicated studies; 2) letters, case reports, comments, and meeting abstracts; and 3) neither reported HR and 95% CI nor parameters (such as survival curve) to indirectly obtain them.
Data extraction
Two investigators (Jian Xiao and Xiaoxiao Lu) reviewed the eligible articles independently. Any disagreements were brought to a conclusion by discussion. Information is collected from the following items: first author; year of publication, region, type of cancer, stage, cases, sex, median age, follow-up, test method, tissue preservation, cutoff value, patients with follow-up, high/low expression cases of miR-133a, outcome, analysis of variance, HR, and 95% CI as well as source of HR. No specific information represented unreported content of any items above. If both univariate and multivariate analyses of OS results were performed, HRs and 95% CIs were extracted preferentially from the multivariate analyses. When Kaplan–Meier curves were the only available information, OS data were gained from the previously stated method.30
Statistical analysis
STATA 12.0 software (StataCorp LP, College Station, TX, USA) was used to conduct all the statistical analyses. According to the cutoff values provided by the original published articles, miR-133a was defined as a high (or upregulated) and low (or downregulated) group. The HRs and their corresponding 95% CIs were performed to calculate the pooled HRs and 95% CIs. If HR >1 and its 95% CI did not overlap with 1, patients with decreased expression of miR-133a indicated a poor prognosis. The heterogeneity of combined HRs was figured out using Cochran’s Q test and Higgins’ I-squared statistic. If there was a result of P<0.05 or I2>50%, heterogeneity was defined, and then the random-effects model was applied. If not, a fixed-effects model was conducted. Subgroup analysis and meta-regression were further adopted to explore the possible explanations for heterogeneity. Sensitivity analysis, by successively omitting each study, was performed to assess the stability of the results. Publication bias was evaluated by Begg’s and Egger’s test. P<0.05 was considered statistically significant, and all the P-values were two-tailed.
Results
Eligible studies and characteristics
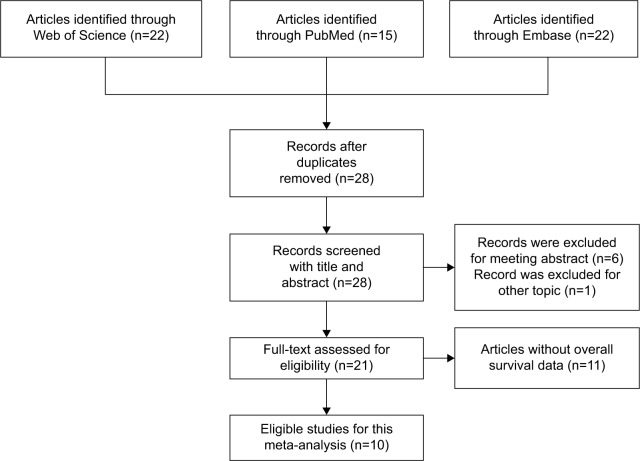
The initial search identified 59 potentially relevant records. After the duplicates were removed, 28 records were preserved. By further reviewing, 21 studies were determined to be of acceptable relevance and assessed for eligibility. However, eleven of them were excluded due to without OS data. Finally, ten studies met the eligible criteria and were included in this meta-analysis.24–29,31–34 The flow diagram on the selection process is shown in Figure 1.
Figure 1.
Flow diagram of the selection of eligible studies.
In total, 877 patients with follow-up data from four regions (People’s Republic of China, Iran, Japan, and Taiwan) were included in this study. Solid cancers included in our meta-analysis derived from five cancer types: osteosarcoma,24,28,33 non-small-cell lung cancer (NSCLC),26,34 esophageal cancer32 (or esophageal squamous cell carcinoma31), colorectal cancer,25,29 and pancreatic cancer.27 All of the miR-133a expression was tested by quantitative real-time polymerase chain reaction. The main characteristics of ten eligible studies are summarized in Tables 1 and 2.
Table 1.
Main characteristics of the eligible studies
| Study | Year | Region | Type of cancer | Stage | Cases | Sex (male/female) |
Median age (range) |
Follow-up (months) |
|---|---|---|---|---|---|---|---|---|
| Ji et al33 | 2013 | People’s Republic of China | Osteosarcoma | I–III | 92 | 64/28 | 20 (7–73) | 60 |
| Mirghasemi et al24 | 2015 | Iran | Osteosarcoma | I–IV | 35 | 18/17 | NSI | 78 |
| Lan et al34 | 2015 | People’s Republic of China | NSCLC | I–IV | 125 | 75/50 | 61.1 (23–90) | 51 |
| Fujiwara et al28 | 2014 | Japan | Osteosarcoma | NSI | 48 | 31/17 | NSI | 143 |
| Chen et al31 | 2014 | People’s Republic of China | ESCC | I–IV | 100 | 69/31 | 50 (40–80) | 50 |
| Akanuma et al32 | 2014 | Japan | Esophageal cancer | I–IV | 140 | 121/19 | NSI | 120 |
| Wan et al29 | 2014 | People’s Republic of China | Colorectal cancer | I–IV | 125 | 73/51 | 71.8 (29–95) | 65 |
| Wang et al26 | 2014 | Taiwan | NSCLC | I–III | 112 | 88/24 | NSI | 52 |
| Qin et al27 | 2013 | People’s Republic of China | Pancreatic cancer | I–II | 95 | 40/55 | NSI | 58 |
| Wang et al25 | 2014 | People’s Republic of China | Colorectal cancer | I–III | 169 | 96/73 | NSI | 77 |
Abbreviations: ESCC, esophageal squamous cell carcinoma; NSI, no specific information; NSCLC, non-small-cell lung cancer.
Table 2.
Further relevant data of all identified studies
| Study | Year | Test method | Tissue preservation | Cutoff value | Patients with follow-up | High expression/low expression | Outcome | Analysis of variance | HR and 95% CI | Source of HR |
|---|---|---|---|---|---|---|---|---|---|---|
| Ji et al33 | 2013 | qRT-PCR | Frozen | Lower than the median value | 92 | 46/46 | OS | Univariate | 1.87 (1.12–3.31) | Survival curve |
| Mirghasemi et al24 | 2015 | qRT-PCR | Frozen | Lower than the median expression level | 35 | 16/19 | OS | Multivariate | 3.42 (1.33–9.37) | Reported |
| Lan et al34 | 2015 | qRT-PCR | FFPE | Lower than the median level of 1.60 | 57 | 30/27 | OS | Univariate | 0.74 (0.37–1.44) | Survival curve |
| Fujiwara et al28 | 2014 | qRT-PCR | FFPE | Lower than the optimal cutoff point | 48 | 8/40 | OS | Univariate | 0.25 (0.03–0.88) | Survival curve |
| Chen et al31 | 2014 | qRT-PCR | Frozen | Lower than the optimal cutoff value of 1.31 | 82 | 36/46 | OS | Univariate | 2.18 (1.33–3.73) | Survival curve |
| Akanuma et al32 | 2014 | qRT-PCR | FFPE | Lower than the set of 0.012 (normalized to RNU6B) | 84 | 62/22 | OS | Univariate | 1.98 (1.08–4.68) | Survival curve |
| Wan et al29 | 2014 | qRT-PCR | Frozen | Lower than the median value | 103 | 52/51 | OS | Univariate | 0.70 (0.33–1.42) | Survival curve |
| Wang et al26 | 2014 | qRT-PCR | Frozen | NSI | 112 | 68/44 | OS | Multivariate | 2.25 (1.13–4.29) | Reported |
| Qin et al27 | 2013 | qRT-PCR | Frozen | Lower than the median expression level | 95 | 47/48 | OS | Univariate | 2.33 (1.77–4.30) | Survival curve |
| Wang et al25 | 2014 | qRT-PCR | Frozen | Low than the median level | 169 | 84/85 | OS | Multivariate | 1.70 (1.01–2.86) | Reported |
Abbreviations: FFPE, formalin-fixed and paraffin-embedded; OS, overall survival; HR, hazard ratio; CI, confidence interval; NSI, no specific information; qRT-PCR, quantitative real-time polymerase chain reaction.
OS is associated with decreased miR-133a expression
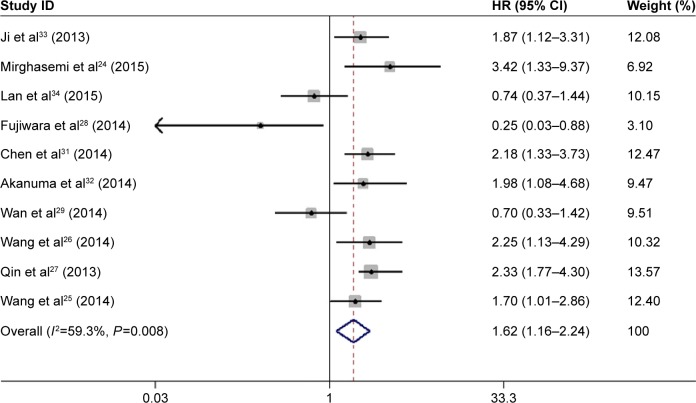
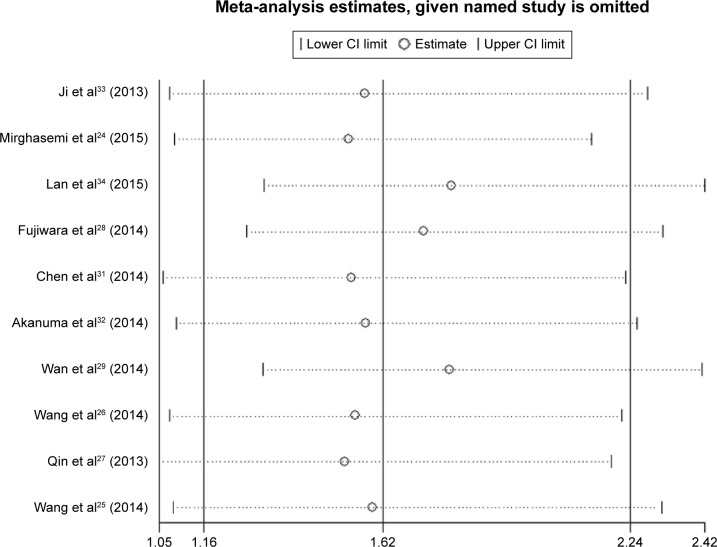
The pooled HR showed that decreased expression of miR-133a was significantly associated with unfavorable OS in patients with solid cancers (HR =1.62, 95% CI: 1.16–2.24, P=0.004) (Figure 2). However, obvious heterogeneity (I2=59.3%, P=0.008) was discovered by using a random-effects model. Therefore, sensitivity analysis was performed to assess the stability of the results by successively omitting each study. Results showed the pooled HRs did not vary substantially by excluding any individual study, indicating a better stability of this meta-analysis (Figure 3). Furthermore, a meta-regression was conducted to investigate the potential responsible factors for the heterogeneity. It found that none of these factors, including region, cancer type, cases (as well as the number of patients with follow-up data), maximum follow-up month, tissue preservation, cutoff value, and analysis of variance was contributing to the heterogeneity significance.
Figure 2.
Forest plot showing the association between decreased microRNA-133a and OS in patients with solid cancers (used a random-effects model).
Note: Weights are from random-effects analysis.
Abbreviations: CI, confidence interval; HR, hazard ratio; OS, overall survival.
Figure 3.
Sensitivity analysis for this meta-analysis.
Abbreviation: CI, confidence interval.
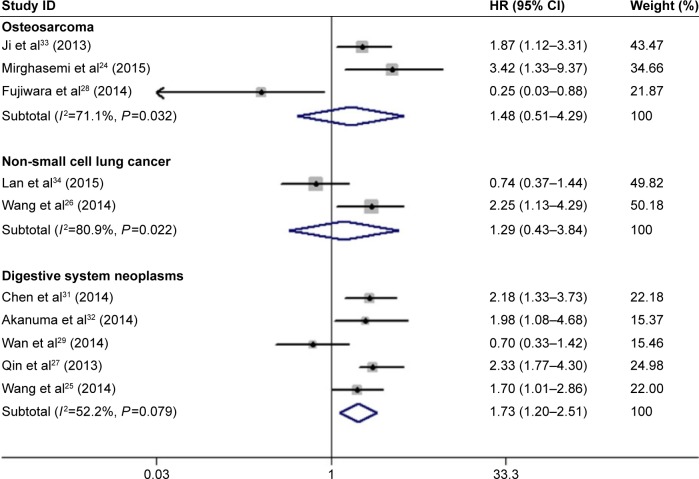
Subgroup analyses were performed according to the following categories: cancer type, region, tissue preservation, and analysis of variance. As the results shown in Figure 4 and Table 3, compared with the total pooled HR, decreased miR-133a exhibited a stronger correlation with poor OS in the subgroups of digestive system neoplasms (HR =1.73, 95% CI: 1.20–2.51, P=0.003), frozen tissue preservation (HR =1.89, 95% CI: 1.41–2.53, P<0.001), and multivariate analysis (HR =2.07, 95% CI: 1.42–3.02, P<0.001). In the meantime, these three subgroups also presented relatively low heterogeneity (I2=52.2%, P=0.079; I2=40.5%, P=0.121; and I2=0.0%, P=0.443, respectively) (Table 3). However, subgroups, such as osteosarcoma, NSCLC, formalin-fixed and paraffin-embedded (FFPE) tissue preservation, and univariate analysis, showed no statistically significant association between decreased miR-133a and OS outcome (Table 3).
Figure 4.
Forest plots showing the HRs and their corresponding CIs by cancer type subgroups (used a random-effects model).
Note: Weights are from random-effects analysis.
Abbreviations: CI, confidence interval; HR, hazard ratio.
Table 3.
Main results of subgroup analyses
| Categories | Subgroups | References | HR (95% CI) | P-value | Heterogeneity
|
|
|---|---|---|---|---|---|---|
| I2 (%) | P-value | |||||
| All | Total | 24–29, 31–34 | 1.62 (1.16–2.24) | 0.004 | 59.3 | 0.008 |
| Cancer type | Osteosarcoma | 24, 28, 33 | 1.48 (0.51–4.29) | 0.466 | 71.1 | 0.032 |
| Non-small-cell lung cancer | 26, 34 | 1.29 (0.43–3.84) | 0.644 | 80.9 | 0.022 | |
| Digestive system neoplasms | 25, 27, 29, 31, 32 | 1.73 (1.20–2.51) | 0.003 | 52.2 | 0.079 | |
| Region | East Asia | 25–29, 31–34 | 1.53 (1.09–2.14) | 0.013 | 60.3 | 0.010 |
| West Asia | 24 | 3.42 (1.29–9.08) | 0.014 | – | – | |
| Tissue | Frozen | 24–27, 29, 31, 33 | 1.89 (1.41–2.53) | <0.001 | 40.5 | 0.121 |
| preservation | FFPE | 28, 32, 34 | 0.88 (0.33–2.32) | 0.791 | 70.0 | 0.036 |
| Analysis of variance | Univariate | 27–29, 31–34 | 1.38 (0.88–2.15) | 0.156 | 68.9 | 0.004 |
| Multivariate | 24–26 | 2.07 (1.42–3.02) | <0.001 | 0.0 | 0.443 | |
Note: “–” indicates no data.
Abbreviations: HR, hazard ratio; CI, confidence interval; FFPE, formalin-fixed and paraffin-embedded.
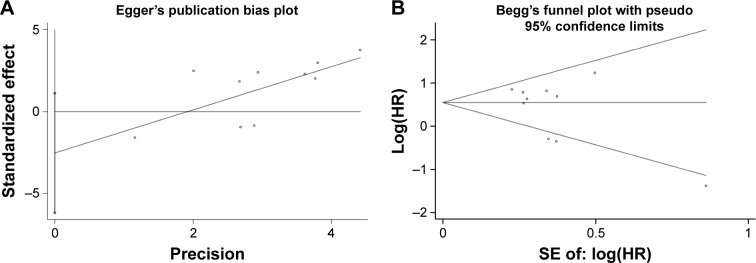
Publication bias assessment
As shown in Figure 5, we performed Begg’s funnel plot and Egger’s test to evaluate the publication bias for all articles in this meta-analysis. The P-values of Begg’s and Egger’s test were 0.37 and 0.15, respectively, suggesting no obvious risk of publication bias.
Figure 5.
Egger’s plot (A) and Begg’s funnel plot (B) for publication bias.
Abbreviation: SE, standard error.
Discussion
In mammal cells, miR-133a performs many regulating functions. It regulates adipocyte browning in vivo35 and modulates osteogenic differentiation of the vascular smooth muscle cells.36 MiR-133a is also a biomarker for postmenopausal osteoporosis37 and can predict cardiac hypertrophy in chronic hemodialysis patients.38 In cancer research, miR-133a is regarded as a potential biomarker for breast cancer detection.39 It inhibits the growth of cervical cancer and gastric cancer by targeting epidermal growth factor receptor and insulin-like growth factor-1 receptor, respectively.40,41 Therefore, miR-133a is considered to be a tumor suppressor. Consequently, downregulated miR-133a induces cancer progression and predicts poor prognosis in cancer patients.24,25 However, no meta-analyses are performed so far to evaluate the prognostic value of decreased miR-133a in patients with solid cancers. So, we have done this meta-analysis.
In our meta-analysis, ten eligible articles met the inclusion criteria. The outcome of cancer patients was collected by OS data. By using a random-effects model to get the pooled HRs, the combined results indicated that decreased miR-133a expression was associated with an unfavorable prognosis in patients with solid cancers. Sensitivity analysis showed that no individual study was obviously affecting the overall result, indicating the pooled result of this meta-analysis is stable. However, due to evident heterogeneity between the included studies, we conducted further subgroup analyses regarding cancer type, region, tissue preservation, and analysis of variance. Despite osteosarcoma, NSCLC, FFPE tissue preservation, and univariate analysis subgroups showed no statistically significant correlation between decreased miR-133a and OS. While, compared with the total pooled HR, the subgroups of digestive system neoplasms, frozen tissue preservation, and multivariate analysis exhibited stronger connection between decreased miR-133a expression and OS outcome with relatively low heterogeneity. In addition, no publication bias was observed.
The results of this meta-analysis have several considerable implications. First, decreased miR-133a may be a common poor prognostic marker for solid cancers. The original researches of our meta-analysis were derived from five cancer types: osteosarcoma,24,28,33 NSCLC,26,34 esophageal cancer,31,32 colorectal cancer,25,29 and pancreatic cancer.27 As the pooled result from all these cancer types showed decreased miR-133a was correlated with a poor OS; thus, the conclusion can basically extend to all solid cancers.42,43 Second, decreased miR-133a probably is a reliable biomarker of poor survival for patients with digestive system neoplasms. Our analysis results showed that decreased miR-133a in patients with digestive system neoplasms exhibited poorer OS compared with osteosarcoma and NSCLC. However, the specific mechanism for this phenomenon needs to be explored in further research. Third, cancer tissues preserved by freezing may be more appropriate for microRNA detection than FFPE. Although studies showed consistent correlations between matched FFPE and frozen samples on the expression of microRNAs,44,45 the expression of a specific microRNA (eg, miR-133a) maybe is different. Unlike FFPF tissue preservation subgroup, our results indicated decreased miR-133a was obviously associated with poor prognosis in frozen tissues. It implicated that frozen tissues may reflect the expression of microRNA more actually than FFPE samples. Fourth, multivariate analysis maybe is more suitable for the researches on the correlation between microRNA expression and OS outcome in patients with cancer. The multivariate analysis subgroup in this meta-analysis exhibited stronger connection between decreased miR-133a expression and OS compared with the subgroup of univariate analysis. However, as the multivariate analysis ruled out the compounding effects from other factors, such as tumor size, stage, and nodal status,24–26 the pooled results from studies conducted by multivariate analysis may be considered as more reliable.
In cancer, microRNAs act as tumor suppressors when they downregulate different proteins with oncogenic activity.46 Similarly, microRNAs act as oncogenes if they downregulate genes involved in cell differentiation or as tumor suppressors.46 For the ten original studies included in our meta-analysis, most of them reported that miR-133a functions as a tumor suppressor and the decreased expression of miR-133a was correlated with poor prognostic in patients with solid cancer while a few other studies reported inconsistent results,28,29,34 indicating miR-133a maybe is a tumor promoter. We considered the reason for this paradox maybe is that miR-133a downregulated different target genes in different cancer types (Table 4).
Table 4.
The validated target genes of miR-133a in the eligible studies in this meta-analysis
| Study | Year | Type of cancer | Validated target genes |
|---|---|---|---|
| Ji et al33 | 2013 | Osteosarcoma | Bcl-xL and Mcl-1 |
| Fujiwara et al28 | 2014 | Osteosarcoma | SGMS2, UBA2, SNX30, and ANXA2 |
| Akanuma et al32 | 2014 | Esophageal cancer | FSCN1 and MMP14 |
| Wan et al29 | 2014 | Colorectal cancer | LASP1, CAV1, and FSCN1 |
| Wang et al26 | 2014 | Non-small-cell lung cancer | IGF-1R, TGFBR1, and EGFR |
| Qin et al27 | 2013 | Pancreatic cancer | FSCN1 |
Abbreviations: ANXA2, annexin A2; Bcl-xL, B-cell lymphoma-extra large; CAV1, caveolin-1; EGFR, epidermal growth factor receptor; FSCN1, Fascin-1; IGF-1R, insulin-like growth factor 1 receptor; LASP1, LIM and SH3 domain protein 1; MMP14, matrix metalloprotease 14; Mcl-1, myeloid cell leukemia-1; SGMS2, sphingomyelin synthase 2; SNX30, sorting nexin family member 30; TGFBR1, TGF-beta receptor type-1; UBA2, ubiquitin-like modifier activating enzyme 2.
Except for miR-133a, the miR-133 family also contains miR-133b. Studies found that miR-133b acts as a tumor suppressor and its expression was decreased in many types of solid cancers, such as NSCLC, bladder cancer, ovarian cancer, and gastric cancer.47–50 In addition, many other studies also reported that both miR-133a and miR-133b were decreased together in solid cancer tissues.31,50–52 Further considering the results of our current meta-analysis, all these evidences indicate that the decreased expression of miR-133 family maybe is the reliable predictor, suggesting patients with solid cancer will have a poor prognosis.
Nevertheless, when interpreting the results of this meta-analysis, several limitations should be noticed. One of the main limitations is the limited region for the included source of cancer patients. Patient populations in this study were limited to Asia, lacking data from other regions, such as Europe, America, Oceania, and Africa. Our results need to be proved by more studies from other regions. Another limitation is that some HR data were extracted from survival curves, which may introduce bias. Thus, the present statistics seem to be less reliable than those directly obtained from published studies. If possible, pooled HRs should be performed based on directly obtained data from the published studies. In addition, all of the included studies in our meta-analysis were designed for retrospective studies, which are more possible to be published when they report positive rather than negative results. Consequently, the association between decreased miR-133a and poor OS may have been overestimated. Finally, obvious heterogeneity showed in between studies, even though we used random-effects models to calculate the pooled HRs. The heterogeneity may be attributed to the inconformity in different population characteristics, types of cancer, treatment strategies, and so on, which will likely reduce the reliability of this study and those findings similar to ours.53,54
Conclusion
Our meta-analysis concluded that decreased miR-133a expression is significantly associated with poor OS in patients with solid cancer. However, our results also need to be cautiously considered because of the limitations described above. Further studies related to specific cancer types and large sample sizes are required to verify the prognostic value of decreased miR-133a in various cancers.
Acknowledgments
This work was funded by the National Natural Science Foundation of China (81572284).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Koutras A, Kotoula V, Fountzilas G. Prognostic and predictive role of vascular endothelial growth factor polymorphisms in breast cancer. Pharmacogenomics. 2015;16(1):79–94. doi: 10.2217/pgs.14.148. [DOI] [PubMed] [Google Scholar]
- 4.Aaltonen KE, Rosendahl AH, Olsson H, Malmstrom P, Hartman L, Ferno M. Association between insulin-like growth factor-1 receptor (IGF1R) negativity and poor prognosis in a cohort of women with primary breast cancer. BMC Cancer. 2014;14:794. doi: 10.1186/1471-2407-14-794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leclerc BG, Charlebois R, Chouinard G, et al. CD73 Expression is an independent prognostic factor in prostate cancer. Clin Cancer Res. 2016;22(1):158–166. doi: 10.1158/1078-0432.CCR-15-1181. [DOI] [PubMed] [Google Scholar]
- 6.Elhanati S, Ben-Hamo R, Kanfi Y, et al. Reciprocal Regulation between SIRT6 and miR-122 controls liver metabolism and predicts hepatocarcinoma prognosis. Cell Rep. 2016;14(2):234–242. doi: 10.1016/j.celrep.2015.12.023. [DOI] [PubMed] [Google Scholar]
- 7.Hur K, Toiyama Y, Okugawa Y, et al. Circulating microRNA-203 predicts prognosis and metastasis in human colorectal cancer. Gut. 2015 Dec 23; doi: 10.1136/gutjnl-2014-308737. Epub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang W, Wang X, Zhang Y, et al. Prognostic role of microRNA-150 in various carcinomas: a meta-analysis. Onco Targets Ther. 2016;9:1371–1379. doi: 10.2147/OTT.S97969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 10.Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 11.Nana-Sinkam SP, Croce CM. MicroRNA regulation of tumorigenesis, cancer progression and interpatient heterogeneity: towards clinical use. Genome Biol. 2014;15(9):445. doi: 10.1186/s13059-014-0445-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Subramanian S, Steer CJ. MicroRNAs as gatekeepers of apoptosis. J Cell Physiol. 2010;223(2):289–298. doi: 10.1002/jcp.22066. [DOI] [PubMed] [Google Scholar]
- 13.Zhang L, Sun ZJ, Bian Y, Kulkarni AB. MicroRNA-135b acts as a tumor promoter by targeting the hypoxia-inducible factor pathway in genetically defined mouse model of head and neck squamous cell carcinoma. Cancer Lett. 2013;331(2):230–238. doi: 10.1016/j.canlet.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zou CD, Zhao WM, Wang XN, et al. MicroRNA-107: a novel promoter of tumor progression that targets the CPEB3/EGFR axis in human hepatocellular carcinoma. Oncotarget. 2016;7(1):266–278. doi: 10.18632/oncotarget.5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen S, Sun KX, Liu BL, Zong ZH, Zhao Y. MicroRNA-505 functions as a tumor suppressor in endometrial cancer by targeting TGF-alpha. Mol Cancer. 2016;15(1):11. doi: 10.1186/s12943-016-0496-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi ZC, Chu XR, Wu YG, et al. MicroRNA-375 functions as a tumor suppressor in osteosarcoma by targeting PIK3CA. Tumour Biol. 2015;36(11):8579–8584. doi: 10.1007/s13277-015-3614-9. [DOI] [PubMed] [Google Scholar]
- 17.Jin Y, Zhao M, Xie Q, Zhang H, Wang Q, Ma Q. MicroRNA-338-3p functions as tumor suppressor in breast cancer by targeting SOX4. Int J Oncol. 2015;47(4):1594–1602. doi: 10.3892/ijo.2015.3114. [DOI] [PubMed] [Google Scholar]
- 18.Knudsen KN, Nielsen BS, Lindebjerg J, Hansen TF, Holst R, Sorensen FB. microRNA-17 Is the Most Up-Regulated Member of the miR-17-92 Cluster during Early Colon Cancer Evolution. PLoS One. 2015;10(10):e0140503. doi: 10.1371/journal.pone.0140503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He XJ, Ma YY, Yu S, et al. Up-regulated miR-199a-5p in gastric cancer functions as an oncogene and targets klotho. BMC Cancer. 2014;14:218. doi: 10.1186/1471-2407-14-218. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Zhu ED, Li N, Li BS, et al. miR-30b, down-regulated in gastric cancer, promotes apoptosis and suppresses tumor growth by targeting plasminogen activator inhibitor-1. PLoS One. 2014;9(8):e106049. doi: 10.1371/journal.pone.0106049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang X, Huang H, Li Z, et al. MiR-495 is a tumor-suppressor microRNA down-regulated in MLL-rearranged leukemia. Proc Natl Acad Sci U S A. 2012;109(47):19397–19402. doi: 10.1073/pnas.1217519109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kojima S, Chiyomaru T, Kawakami K, et al. Tumour suppressors miR-1 and miR-133a target the oncogenic function of purine nucleoside phosphorylase (PNP) in prostate cancer. Br J Cancer. 2012;106(2):405–413. doi: 10.1038/bjc.2011.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang W, Liu K, Liu S, Ji B, Wang Y, Liu Y. MicroRNA-133a functions as a tumor suppressor by targeting IGF-1R in hepatocellular carcinoma. Tumour Biol. 2015;36(12):9779–9788. doi: 10.1007/s13277-015-3749-8. [DOI] [PubMed] [Google Scholar]
- 24.Mirghasemi A, Taheriazam A, Karbasy SH, et al. Down-regulation of miR-133a and miR-539 are associated with unfavorable prognosis in patients suffering from osteosarcoma. Cancer Cell Int. 2015;15:86. doi: 10.1186/s12935-015-0237-6. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25.Wang LL, Du LT, Li J, et al. Decreased expression of miR-133a correlates with poor prognosis in colorectal cancer patients. World J Gastroenterol. 2014;20(32):11340–11346. doi: 10.3748/wjg.v20.i32.11340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang LK, Hsiao TH, Hong TM, et al. MicroRNA-133a suppresses multiple oncogenic membrane receptors and cell invasion in non-small cell lung carcinoma. PLoS One. 2014;9(5):e96765. doi: 10.1371/journal.pone.0096765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qin Y, Dang X, Li W, Ma Q. miR-133a functions as a tumor suppressor and directly targets FSCN1 in pancreatic cancer. Oncol Res. 2013;21(6):353–363. doi: 10.3727/096504014X14024160459122. [DOI] [PubMed] [Google Scholar]
- 28.Fujiwara T, Katsuda T, Hagiwara K, et al. Clinical relevance and therapeutic significance of microRNA-133a expression profiles and functions in malignant osteosarcoma-initiating cells. Stem Cells. 2014;32(4):959–973. doi: 10.1002/stem.1618. [DOI] [PubMed] [Google Scholar]
- 29.Wan TM, Lam CS, Ng L, et al. The clinicopathological significance of miR-133a in colorectal cancer. Dis Markers. 2014;2014:919283. doi: 10.1155/2014/919283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krieg A, Riemer JC, Telan LA, Gabbert HE, Knoefel WT. CXCR4 – A prognostic and clinicopathological biomarker for pancreatic ductal adenocarcinoma: a meta-analysis. PLoS One. 2015;10(6):e0130192. doi: 10.1371/journal.pone.0130192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen G, Peng J, Zhu W, et al. Combined downregulation of microRNA-133a and microRNA-133b predicts chemosensitivity of patients with esophageal squamous cell carcinoma undergoing paclitaxel-based chemotherapy. Med Oncol. 2014;31(11):263. doi: 10.1007/s12032-014-0263-6. [DOI] [PubMed] [Google Scholar]
- 32.Akanuma N, Hoshino I, Akutsu Y, et al. MicroRNA-133a regulates the mRNAs of two invadopodia-related proteins, FSCN1 and MMP14, in esophageal cancer. Br J Cancer. 2014;110(1):189–198. doi: 10.1038/bjc.2013.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ji F, Zhang H, Wang Y, et al. MicroRNA-133a, downregulated in osteosarcoma, suppresses proliferation and promotes apoptosis by targeting Bcl-xL and Mcl-1. Bone. 2013;56(1):220–226. doi: 10.1016/j.bone.2013.05.020. [DOI] [PubMed] [Google Scholar]
- 34.Lan D, Zhang X, He R, et al. MiR-133a is downregulated in non-small cell lung cancer: a study of clinical significance. Eur J Med Res. 2015;20:50. doi: 10.1186/s40001-015-0139-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu W, Bi P, Shan T, et al. miR-133a regulates adipocyte browning in vivo. PLoS Genet. 2013;9(7):e1003626. doi: 10.1371/journal.pgen.1003626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liao XB, Zhang ZY, Yuan K, et al. MiR-133a modulates osteogenic differentiation of vascular smooth muscle cells. Endocrinology. 2013;154(9):3344–3352. doi: 10.1210/en.2012-2236. [DOI] [PubMed] [Google Scholar]
- 37.Wang Y, Li L, Moore BT, et al. MiR-133a in human circulating monocytes: a potential biomarker associated with postmenopausal osteoporosis. PLoS One. 2012;7(4):e34641. doi: 10.1371/journal.pone.0034641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wen P, Song D, Ye H, et al. Circulating MiR-133a as a biomarker predicts cardiac hypertrophy in chronic hemodialysis patients. PLoS One. 2014;9(10):e103079. doi: 10.1371/journal.pone.0103079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shen J, Hu Q, Schrauder M, et al. Circulating miR-148b and miR-133a as biomarkers for breast cancer detection. Oncotarget. 2014;5(14):5284–5294. doi: 10.18632/oncotarget.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Song X, Shi B, Huang K, Zhang W. miR-133a inhibits cervical cancer growth by targeting EGFR. Oncol Rep. 2015;34(3):1573–1580. doi: 10.3892/or.2015.4101. [DOI] [PubMed] [Google Scholar]
- 41.Gong Y, Ren J, Liu K, Tang LM. Tumor suppressor role of miR-133a in gastric cancer by repressing IGF1R. World J Gastroenterol. 2015;21(10):2949–2958. doi: 10.3748/wjg.v21.i10.2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dai SL, Zhou J, Pan C, et al. Prognostic value of microRNA-145 in patients with various cancers: a meta-analysis. Cancer Biomark. 2015;15(4):507–513. doi: 10.3233/CBM-150475. [DOI] [PubMed] [Google Scholar]
- 43.Wang F, Zhong S, Zhang H, et al. Prognostic value of microRNA-182 in cancers: a meta-analysis. Dis Markers. 2015;2015:482146. doi: 10.1155/2015/482146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang X, Chen J, Radcliffe T, Lebrun DP, Tron VA, Feilotter H. An array-based analysis of microRNA expression comparing matched frozen and formalin-fixed paraffin-embedded human tissue samples. J Mol Diagn. 2008;10(6):513–519. doi: 10.2353/jmoldx.2008.080077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meng W, McElroy JP, Volinia S, et al. Comparison of microRNA deep sequencing of matched formalin-fixed paraffin-embedded and fresh frozen cancer tissues. PLoS One. 2013;8(5):e64393. doi: 10.1371/journal.pone.0064393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shenouda SK, Alahari SK. MicroRNA function in cancer: oncogene or a tumor suppressor? Cancer Metastasis Rev. 2009;28(3–4):369–378. doi: 10.1007/s10555-009-9188-5. [DOI] [PubMed] [Google Scholar]
- 47.Chen SW, Wang TB, Tian YH, Zheng YG. Down-regulation of microRNA-126 and microRNA-133b acts as novel predictor biomarkers in progression and metastasis of non small cell lung cancer. Int J Clin Exp Pathol. 2015;8(11):14983–14988. [PMC free article] [PubMed] [Google Scholar]
- 48.Chen XN, Wang KF, Xu ZQ, et al. MiR-133b regulates bladder cancer cell proliferation and apoptosis by targeting Bcl-w and Akt1. Cancer Cell Int. 2014;14:70. doi: 10.1186/s12935-014-0070-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu X, Li G. MicroRNA-133b inhibits proliferation and invasion of ovarian cancer cells through Akt and Erk1/2 inactivation by targeting epidermal growth factor receptor. Int J Clin Exp Pathol. 2015;8(9):10605–10614. [PMC free article] [PubMed] [Google Scholar]
- 50.Liu Y, Zhang X, Zhang Y, et al. Identification of miRNomes in human stomach and gastric carcinoma reveals miR-133b/a-3p as therapeutic target for gastric cancer. Cancer Lett. 2015;369(1):58–66. doi: 10.1016/j.canlet.2015.06.028. [DOI] [PubMed] [Google Scholar]
- 51.Kano M, Seki N, Kikkawa N, et al. miR-145, miR-133a and miR-133b: Tumor-suppressive miRNAs target FSCN1 in esophageal squamous cell carcinoma. Int J Cancer. 2010;127(12):2804–2814. doi: 10.1002/ijc.25284. [DOI] [PubMed] [Google Scholar]
- 52.Qiu T, Zhou X, Wang J, et al. MiR-145, miR-133a and miR-133b inhibit proliferation, migration, invasion and cell cycle progression via targeting transcription factor Sp1 in gastric cancer. FEBS Lett. 2014;588(7):1168–1177. doi: 10.1016/j.febslet.2014.02.054. [DOI] [PubMed] [Google Scholar]
- 53.Qu J, Li M, Zhong W, Hu C. Prognostic role of microRNA-25 in cancers: evidence from a meta-analysis. Int J Clin Exp Med. 2015;8(8):12921–12927. [PMC free article] [PubMed] [Google Scholar]
- 54.Wang S, Hu J, Zhang D, Li J, Fei Q, Sun Y. Prognostic role of microRNA-31 in various cancers: a meta-analysis. Tumour Biol. 2014;35(11):11639–11645. doi: 10.1007/s13277-014-2492-x. [DOI] [PubMed] [Google Scholar]