Abstract
A bioanalytical platform combining magnetic ligand fishing for α-glucosidase inhibition profiling and HPLC-HRMS-SPE-NMR for structural identification of α-glucosidase inhibitory ligands, both directly from crude plant extracts, is presented. Magnetic beads with N-terminus-coupled α-glucosidase were synthesized and characterized for their inherent catalytic activity. Ligand fishing with the immobilized enzyme was optimized using an artificial test mixture consisting of caffeine, ferulic acid, and luteolin before proof-of-concept with the crude extract of Eugenia catharinae. The combination of ligand fishing and HPLC-HRMS-SPE-NMR identified myricetin 3-O-α-l-rhamnopyranoside, myricetin, quercetin, and kaempferol as α-glucosidase inhibitory ligands in E. catharinae. Furthermore, HPLC-HRMS-SPE-NMR analysis led to identification of six new alkylresorcinol glycosides, i.e., 5-(2-oxopentyl)resorcinol 4-O-β-d-glucopyranoside, 5-propylresorcinol 4-O-β-d-glucopyranoside, 5-pentylresorcinol 4-O-[α-d-apiofuranosyl-(1→6)]-β-d-glucopyranoside, 5-pentylresorcinol 4-O-β-d-glucopyranoside, 4-hydroxy-3-O-methyl-5-pentylresorcinol 1-O-β-d-glucopyranoside, and 3-O-methyl-5-pentylresorcinol 1-O-[β-d-glucopyranosyl-(1→6)]-β-d-glucopyranoside.
The membrane-bound α-glucosidase enzyme in the brush border of the small intestine is one of the digestive enzymes responsible for release of absorbable glucose units from dietary carbohydrates. This enzyme plays a vital role in the management of postprandial hyperglycemia, i.e., the elevated and fluctuating after-meal blood glucose level associated with type 2 diabetes (T2D).1,2 Inhibition of α-glucosidase is one of the existing therapeutic strategies for managing blood glucose levels, with three clinically approved drugs: acarbose, miglitol, and voglibose.3 However, these drugs have been associated with various gastrointestinal side effects, such as abdominal pain, flatulence, and diarrhea,3,4 and new alternative drug leads and/or functional food constituents are needed. In recent years, considerable attention has been paid to natural products as inhibitory ligands of the α-glucosidase enzyme.5,6 However, the chemical complexity of, for example, plant extracts makes identification and characterization of inhibitory ligands from these complex mixtures a challenging and time-consuming task.
Several bioanalytical screening techniques have been proposed as advanced alternatives to the classical bioassay-guided fractionation. Among these is ligand fishing, a technique where a selected therapeutic target, e.g., a receptor or an enzyme, is immobilized on a solid support and used for fishing out ligands from complex mixtures. In natural product research, this technique has been used as a pharmacological profiling tool to discover biologically active metabolites in crude extracts.7–11 However, in most of the reported studies, ligand fishing has been followed by either preparative-scale isolation of the ligands10 (with chromatographic change from analytical-scale ligand fishing to targeted preparative-scale isolation being elusive and time-consuming) or analytical-scale LC-MS based identification11 (with limited structural information hampering structural elucidation or restricting identification to known compounds). The above challenges can be overcome by combining ligand fishing with a more powerful structural analysis tool, i.e., HPLC-HRMS-SPE-NMR, that already has proven to be a powerful technique for full structural identification of constituents directly from crude extracts at analytical-scale HPLC conditions.12,13
Magnetic beads (MBs) provide an excellent support for the immobilization of proteins.7,9,14,15 Their interaction with a magnetic force enables separation of MBs from a given aqueous matrix, and this is a major advantage in the ligand fishing procedure. In the current work, N-terminus-coupled α-glucosidase magnetic beads (AGN-TCMB) were synthesized, characterized, and used as a ligand fishing tool. The magnetic ligand fishing was used as a screening technology where a comparison of LC-MS profiles of the crude extract and the eluents from the fishing was used to pinpoint potential ligands. The m/z ratio and retention time of the pinpointed metabolites were subsequently used to target the HPLC-HRMS-SPE-NMR analysis toward ligands for the immobilized enzyme.
The combined ligand fishing/HPLC-HRMS-SPE-NMR platform was used to identify potential antihyperglycemic metabolites in the crude extract of Eugenia catharinae O. Berg. E. catharinae is a native Brazilian plant belonging to the Myrtaceae family, and a number of species from Eugenia are used in folk medicine for the treatment of diabetes.16–18 However, there has never been a chemical or pharmacological investigation of metabolites from E. catharinae. The combined magnetic ligand fishing/HPLC-HRMS-SPE-NMR analysis of this rarely investigated species permitted identification of four potential α-glucosidase inhibitory ligands, six new alkyresorcinol glycosides, and several other known metabolites.
RESULTS AND DISCUSSION
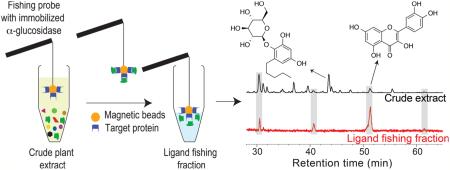
In the current work a new combination of magnetic ligand fishing with HPLC-HRMS-SPE-NMR was developed and applied for identification of α-glucosidase inhibitory ligands and new alkylresorcinol glycosides from E. catharinae. Magnetic beads with N-terminus-coupled α-glucosidase were synthesized and subsequently characterized for their ligand fishing properties. Comparison of LC-MS profiles of the crude mixture with the corresponding eluents after ligand fishing enabled unambiguous pinpointing of potential ligands in the complex extract. The m/z value and retention time (tR) of the pinpointed metabolites were subsequently used to target the HPLC-HRMS-SPE-NMR analysis toward the potential ligands for α-glucosidase (Figure 1).
Figure 1.
Schematic workflow for ligand fishing with N-terminus-coupled α-glucosidase magnetic beads combined with HPLC-HRMS-SPE-NMR analysis of E. catharinae extract. Path 1: Ligand fishing with α-glucosidase enzyme immobilized on magnetic beads followed by LC-HRMS analysis of both the crude extract and potential ligands fished out of the extract. Path 2: HPLC-HRMS-SPE-NMR analysis targeting potential α-glucosidase ligands pinpointed in the preceding ligand fishing experiment.
Synthesis and Characterization of N-Terminus-Coupled α-Glucosidase Immobilized Magnetic Beads
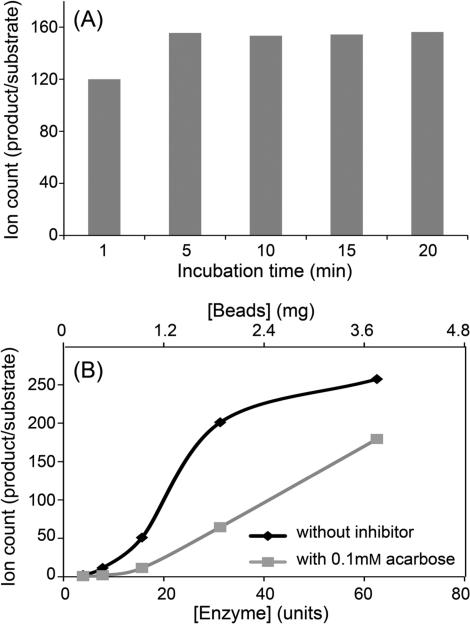
The N-terminus of the α-glucosidase enzyme was covalently bound to BcMag amine-terminated magnetic beads using gluteraldehyde as linker, and subsequently the imines were reduced in the presence of sodium cyanoborohydride (Borch reduction) (Figure 2). The unbound gluteraldehyde terminals were end-capped with hydroxylamine after immobilization of the α-glucosidase. In order to ensure that the immobilized enzyme retained its molecular integrity and inherent hydrolytic activity after immobilization, AGN-TCMB were incubated with p-nitrophenyl α-d-glucopyranoside (p-NPG), a standard substrate, for 1, 5, 10, 15, and 20 min at room temperature. The activity was assessed as the ratio of peak areas of the hydrolytic product p-nitrophenol (p-NP) and the nonhydrolyzed substrate (p-NPG) (Figure 3A). This shows that AGN-TCMB afforded approximately 150-fold more product than substrate from 5 to 20 min. In addition, the activities of different amounts of AGN-TCMB were tested with and without inhibitors (Figure 3B). It was observed that the presence of acarbose decreased the activity of the immobilized enzyme significantly. The results of these studies demonstrate that the enzymatic activity of α-glucosidase was maintained after immobilization, and incubation was performed at room temperature for 10 min for the remainder of the experiments.
Figure 2.
Synthesis of N-terminus-coupled α-glucosidase magnetic beads. The α-glucosidase enzyme is represented as H2N-AG.
Figure 3.
Assessment of enzymatic activity of the N-terminus-coupled α-glucosidase magnetic beads (AGN-TCMB) at different incubation times (A) and with different amounts of AGN-TCMB with and without the presence of inhibitor (B).
Ligand Fishing
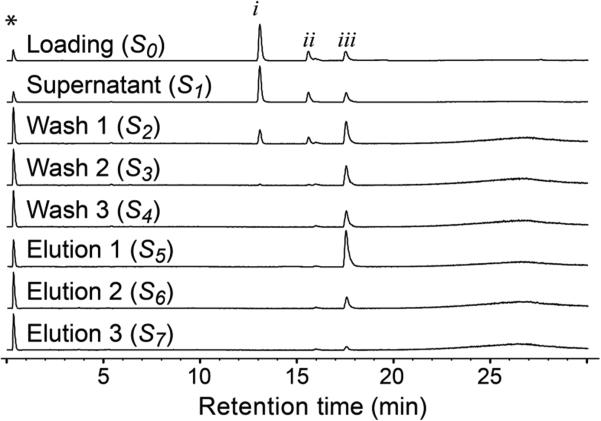
The ligand fishing experiment was developed and optimized using an equimolar mixture of caffeine, ferulic acid, and luteolin. As caffeine and ferulic acid do not bind to α-glucosidase, they were used as negative controls, while luteolin, a high affinity α-glucosidase binder (Ki value of 0.140 ± 0.002 mM),21 was used as a positive control. The ligand fishing procedure used in the current study consisted of three steps, i.e., loading, washing, and elution. In the loading step, i.e., adding dissolved sample to the AGN-TCMB, the amount of organic solvent in the loading solution was initially optimized. Previously, it has been shown that immobilized α-glucosidase can tolerate as high as 30% of MeOH in the aqueous buffer and still retain 12.5% of its activity compared to an experiment in aqueous buffer.19 However, in order to maintain molecular integrity of the enzyme and to prevent stripping of bound ligands from the immobilized enzyme, NH4OAc buffer (10 mM, pH 7.4) containing 5% MeOH was used. After incubation of AGN-TCMB with the loading solution (S0) for 10 min at room temperature, the supernatant (S1) was removed after magnetic separation of the beads. This was followed by three washing steps with NH4OAc buffer (S2–S4) to ensure removal of compounds with low or no affinity to the enzyme. In order to elute the bound ligands from the immobilized enzyme, the beads were incubated three times with 90% MeOH as elution buffer (S5–S7). At such high concentrations of organic solvent, it is anticipated that the immobilized enzyme will denature and thus release bound ligands.20 All solutions S0–S7 were subsequently analyzed by LC-MS. As seen in Figure 4, luteolin was successfully fished out of the artificial test mixture. Even though luteolin was present in all three washing steps, it was the only compound observed in all three elution steps (S4–S7), with the highest amount observed in elution fraction S5. The presence of luteolin in the washing steps S2–S4 is not surprising, because there will be a continuous ligand-dissociation/ligand-association from/with the ligand–protein complex at equilibrium. However, most importantly, caffeine and ferulic acid were not retained past the initial wash (S2), which demonstrated the specificity of the developed ligand fishing method with AGN-TCMB.
Figure 4.
Overlaid base peak chromatograms acquired from different solutions (S0–S7) during an AGN-TCMB-based ligand fishing procedure from an artificial test mixture containing caffeine (i), ferulic acid (ii), and luteolin (iii). Annotated peak (*) is from sodium formate internal standard.
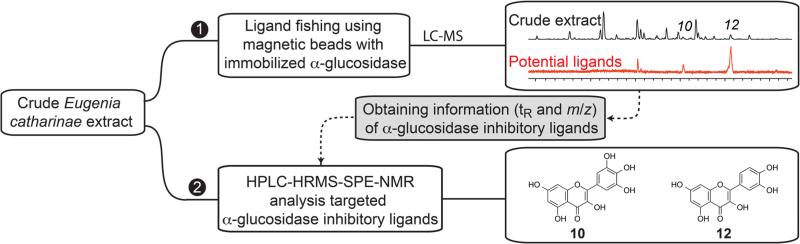
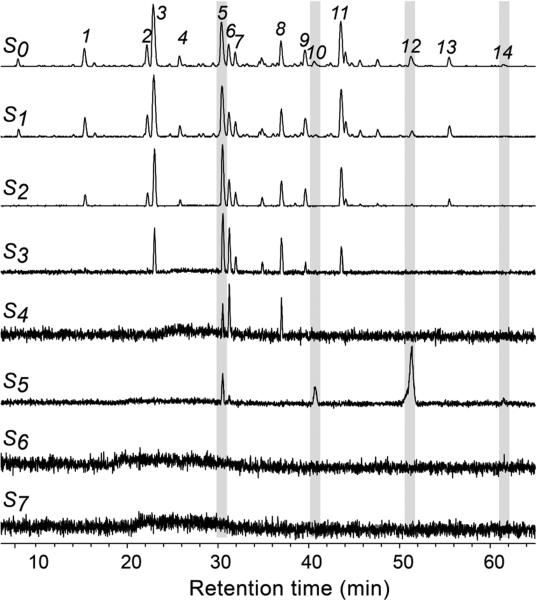
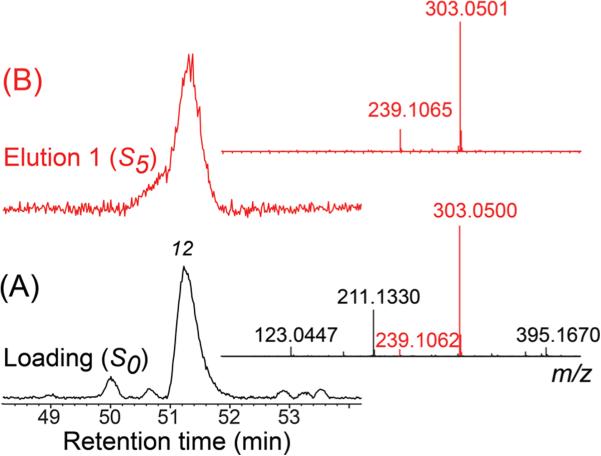
As a proof-of-concept of AGN-TCMB ligand fishing used as a targeting tool for HPLC-HRMS-SPE-NMR, a crude EtOAc extract of E. catharinae was analyzed using the optimized ligand fishing procedure outlined above. Comparison of the LC-HRMS base peak chromatograms of the loading solution (S0) and the first elution fraction (S5) readily identified peaks 5, 10, 12, and 14 as α-glucosidase ligands (Figure 5). However, further information is available from the full LC-HRMS data set acquired of all solutions S0–S7. A more detailed consideration of the HRMS data of the loading solvent S0 and the elution fraction S5 revealed some interesting information for peak 12. For the crude extract (loading (S0), Figure 6A), the HRMS data showed ions related to more than one metabolite. However, after the ligand fishing procedure (elution 1 (S5), Figure 6B), the HRMS data of peak 12 showed only characteristic ions related to the bound ligand, whereas ions related to the nonbinding and overlapping metabolite seen in Figure 6A were absent.
Figure 5.
Overlaid base peak chromatograms acquired of different solutions (S0–S7) obtained from AGN-TCMB-based ligand fishing from an EtOAc extract of E. catharinae.
Figure 6.
Expanded (50–54 min) base peak chromatogram of crude ethyl acetate extract of E. catharinae loaded on the magnetic beads for ligand fishing (A, black) and the eluents from the fishing experiment (B, red). Inserted to the right are the corresponding MS spectra of peak 12 from the two samples.
The LC-HRMS data sets obtained for all solutions can be regarded as one of the main advantages of ligand fishing compared to other bioactivity profiling tools, such as high-resolution α-glucosidase inhibition,22–24 high-resolution α-amylase inhibition,25 high-resolution monoamine oxidase-A inhibition,26 high-resolution radical scavenging,23,27 and high-resolution fungal plasma membrane H+-ATPase inhibition,28 which all have proven successful in combination with HPLC-HRMS-SPE-NMR. Thus, the structural information obtained with the LC-HRMS data sets from the ligand fishing experiments enables unambiguous assignment of overlapping HPLC peaks in the crude extract to the peak(s) representing the bioactive metabolite(s) obtained in the elution solvents S5–S7. Furthermore, the LC-HRMS method used for analysis of the samples from ligand fishing experiments provides a resolution of 60 data points per minute, which is superior to the typical resolution of 4.4–8.0 data points per minute obtained in the aforementioned high-resolution biochromatograms. Finally, the high-resolution biochromatograms suffer from background noise originating from overlapping constituents and residual chromatographic solvents and modifiers. In contrast, the background in the ligand fishing profiles is limited to the instrumental noise of the LC-MS system.
Identification of α-Glucosidase Inhibitory Ligands
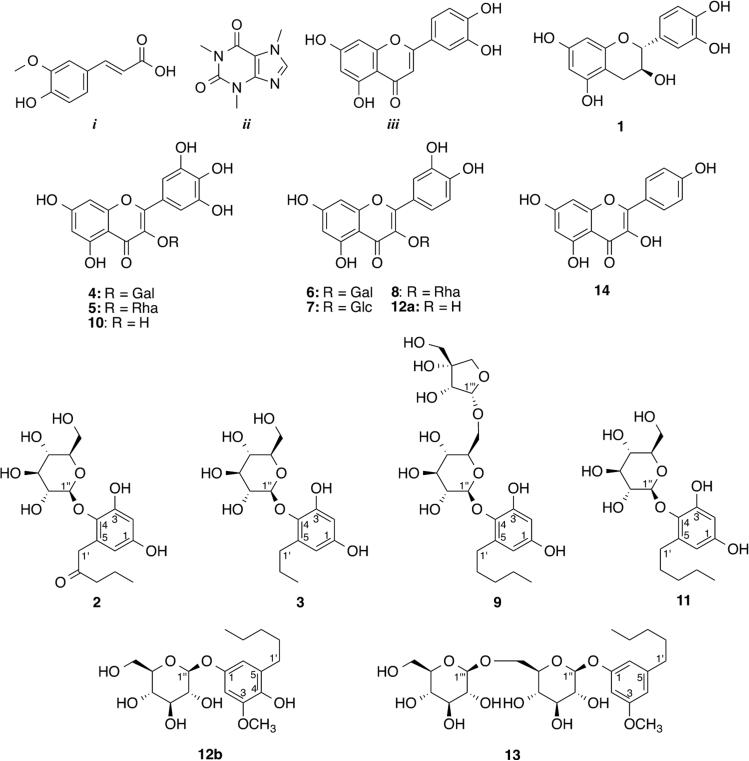
The α-glucosidase ligands identified from the ligand fishing experiments described above were targeted in the HPLC-HRMS-SPE-NMR analysis using the obtained tR and m/z information. Thus, peaks 5, 10, 12, and 14 from E. catharinae were trapped on SPE cartridges based on thresholds in the total ion current chromatogram and eluted to 1.7 mm NMR tubes (after drying SPE cartridges with N2 gas) for NMR analysis. Based on HRMS and 1H NMR data (Table S1, Supporting Information) and comparison with authentic reference samples, an in-house NMR database, and/or literature, peak 5 was identified as myricetin 3-O-α-l-rhamnopyranoside (myricitrin) (5),29 peak 10 as myricetin (10),30 peak 12 as quercetin (12a)30 coeluting with 12b (vide infra), and peak 14 as kaempferol (14).31 These four metabolites, identified as α-glucosidase ligands from the crude extract of E. catharinae, have previously been shown to inhibit α-glucosidase,30,32,33 which proves the effectiveness of using ligand fishing as a targeting tool for HPLC-HRMS-SPE-NMR analysis in the search for α-glucosidase inhibitors.
Identification of New Alkylresorcinol Glycosides
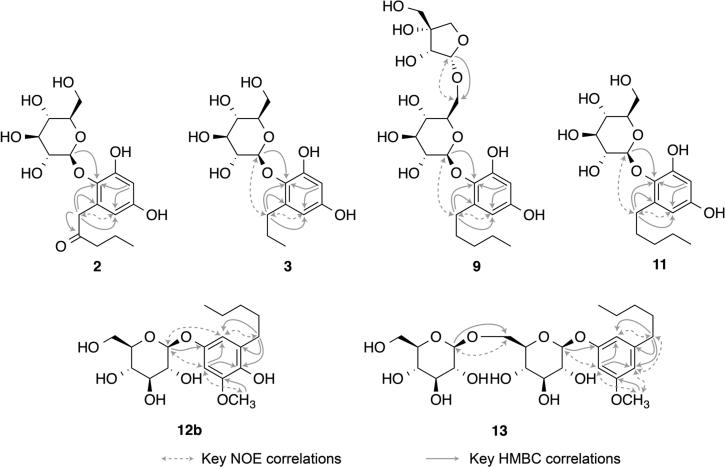
The 1H NMR spectrum of the content of peak 12 obtained in the HPLC-HRMS-SPE-NMR mode showed an additional set of resonances for a coeluting metabolite. This was consistent with our previous observation during comparison of HRMS spectra of peak 12 before (S0) and after (S5) the ligand fishing procedure (Figure 6A and B). The 1H NMR spectrum of this coeluting metabolite indicated that it could be a new alkylresorcinol glycoside, and it was therefore decided to perform a more thorough HPLC-HRMS-SPE-NMR analysis, including acquisition of 2D homo- (COSY and NOESY) and heteronuclear (HSQC and HMBC) experiments, of the remaining peaks 1–3, 5–9, and 11–13. Analysis of NMR data obtained in the HPLC-HRMS-SPE-NMR mode of the contents of peaks 2, 3, 9, and 11–13 revealed common structural features of mono- or disubstituted resorcinol aglycones, mono- or disaccharide glycans, and saturated alkyl substituents of different length and oxidation state.
The material eluted as peak 2 showed an [M + H]+ ion with m/z 373.1484, which in conjunction with the 13C NMR data corresponded to a molecular formula of C17H24O9. The 1H NMR spectrum showed an AB spin system [δ 6.26 (1H, d, J = 2.9, H-2) and 6.06 (1H, d, J = 2.9, H-6)] in agreement with two m-coupled aromatic proton resonances of a disubstituted resorcinol moiety. A 2-oxopentyl moiety at C-5 was identified based on HMBC correlations from the diastereotopic proton pair H-1′A/H-1′B [δ 3.73 d (16.3) and 4.01 d (16.3)] to C-5 at δ 130.5 and the carbonyl group at δ 211.8. This was further supported by the NOE correlation between H-1′A/H-1′B and H-6 (δ 6.06) of the resorcinol core. Furthermore, resonances for a β-glucopyranoside moiety were observed, and the site of glucosylation was established based on the 3J HMBC correlation from the anomeric proton resonance H-1″ (δ 4.45, d, J = 7.6 Hz) to C-4 (δ 138.3) of the resorcinol moiety. Finally, after analysis of all 2J and 3J HMBC correlations, the structure of the metabolite eluted as peak 2 was identified as the new 5-(2-oxopentyl)resorcinol 4-O-β-d-glucopyranoside (2). Fully assigned 1H and 13C NMR data are given in Table 1, selected HMBC and NOE correlations used for structure elucidation are shown in Figure 8, and 1H, COSY, NOESY, HSQC, and HMBC spectra obtained in the HPLC-HRMS-SPE-NMR mode are available in the Supporting Information.
Table 1.
NMR Spectroscopic Data of 2, 3, 9, 11, 12b, and 13 Obtained in the HPLC-HRMS-SPE-NMR Modea
| 2 |
3 |
9 |
11 |
12bc |
13 |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| pos. | δH (J in Hz) | δ C b | δH (J in Hz) | δ C b | δH (J in Hz) | δ C b | δH (J in Hz) | δ C b | δH (J in Hz) | δ C b | δH (J in Hz) | δ C b |
| 1 | n.d. | 155.2 | 155.5 | 155.1 | 151.6 | 159.6 | ||||||
| 2 | 6.26 d (2.9) | 103.5 | 6.16 d (2.9) | 101.9 | 6.17 d (2.8) | 101.9 | 6.16 d (2.9) | 101.8 | 6.66 d (2.5) | 100.6 | 6.54 t (2.1) | 101.1 |
| 3 | n.d. | 150.8 | 151.2 | 151.0 | 148.2 | 161.8 | ||||||
| 4 | 138.3 | 138.1 | 137.8 | 137.8 | 140.2 | 6.43 dd (2.1, 1.5) | 108.9 | |||||
| 5 | 130.5 | 138.1 | n. d. | n. d. | 130.1 | 146.2 | ||||||
| 6 | 6.06 d (2.9) | 109.1 | 6.12 d (2.9) | 107.9 | 6.12 d (2.8) | 107.8 | 6.12 d (2.9) | 107.7 | 6.50 d (2.5) | 111.0 | 6.53 dd (2.1, 1.5) | 110.1 |
| 1′ | 3.73 d (16.3) 4.01 d (16.3) |
45.5 | 2.73 ddd (13.5, 8.7, 6.8) 2.64 ddd (13.5, 9.1, 6.7) |
32.9 | 2.81 ddd (13.6, 8.3, 7.2) 2.60 ddd (13.6, 8.6, 7.3) |
30.7 | 2.77 ddd (13.6, 8.4, 7.4) 2.64 ddd (13.6, 8.4, 7.1) |
30.6 | 2.56 t (7.6) | 30.7 | 2.55 t (7.6) | 37.0 |
| 2′ | 211.8 | 1.59 m | 24.8 | 1.56 q (7.3) | 31.3 | 1.56 q (7.4) | 31.3 | 1.58 q (7.6) | 30.4 | 1.61 q (7.6) | 31.9 | |
| 3′ | 2.53 dt (17.2, 7.3) 2.49 dt (17.2, 7.4) |
44.7 | 0.95 t (7.3) | 14.3 | 1.35d m | 32.8 | 1.35f m | 32.8 | 1.33f m | 32.8 | 1.35 m | 32.5 |
| 4′ | 1.58 sext (7.3) | 17.8 | 1.35d m | 23.4 | 1.35f m | 23.5 | 1.33f m | 23.3 | 1.33 m | 23.3 | ||
| 5′ | 0.90 t (7.3) | 13.9 | 0.91 t (6.7) | 14.3 | 0.91 t (7.0) | 14.3 | 0.90 t (6.9) | 14.3 | 0.91 t (7.1) | 14.3 | ||
| 1″ | 4.45 d (7.6) | 107.6 | 4.44 d (7.9) | 107.7 | 4.41 d (7.8) | 107.6 | 4.44 d (7.9) | 107.7 | 4.73 d (7.5) | 103.5 | 4.89 d (7.3) | 101.9 |
| 2″ | 3.41d m | 75.1 | 3.47 dd (9.2, 7.9) | 75.2 | 3.45 dd (8.8, 8.0) | 75.2 | 3.47 dd (9.1, 7.9) | 75.2 | 3.41d m | 74.8 | 3.44d m | 74.7 |
| 3″ | 3.39d m | 77.8 | 3.40 dd (9.1, 9.1) | 77.9 | 3.40e m | 76.8g | 3.40 dd (9.1, 9.1) | 77.8 | 3.41d m | 77.8d | 3.45d m | 77.5 |
| 4″ | 3.39d m | 70.8 | 3.46 dd (9.6, 9.1) | 70.8 | 3.40e m | 70.9 | 3.46 dd (9.5, 9.1) | 70.8 | 3.35 dd (9.5, 8.8) | 71.4 | 3.43d m | 71.2 |
| 5″ | 3.24 m | 78.2 | 3.26 ddd (9.5, 4.7, 2.4) | 78.2 | 3.40e m | 77.8g | 3.26 ddd (9.5, 4.6, 2.3) | 78.0 | 3.41d m | 77.8d | 3.68 m | 76.9 |
| 6″ | 3.82 dd (12.1, 2.3) 3.72 dd (12.1, 5.1) |
62.1 | 3.84 dd (11.8, 2.4) 3.75 dd (11.8, 4.7) |
62.2 | 3.96 dd (11.3, 1.2) 3.64 dd (11.8, 4.6) |
68.1 | 3.84 dd (11.8, 2.3) 3.75 dd (11.8, 4.6) |
62.2 | 3.89 dd (11.9, 1.8) 3.75 dd (11.9, 5.8) |
62.6 | 4.15 dd (11.5, 2.0) 3.82 dd (11.5, 5.6) |
70.1 |
| 1‴ | 4.96 d (2.1) | 110.7 | 4.34 d (7.8) | 104.7 | ||||||||
| 2‴ | 3.92 d (2.1) | 77.9 | 3.22 dd (9.1, 7.8) | 74.9 | ||||||||
| 3‴ | 79.2 | 3.34 t (8.9) | 77.7e | |||||||||
| 4‴ | 3.92 d (9.6) 3.75 d (9.6) |
74.9 | 3.29 t (8.9) | 71.3 | ||||||||
| 5‴ | 3.59 s | 65.6 | 3.24 m | 77.7e | ||||||||
| 6‴ | 3.86 dd (11.7, 2.1) 3.66 dd (11.7, 5.7) |
62.7 | ||||||||||
| 3-OCH3 | 3.83 (s) | 56.4 | 3.77 s | 55.6 | ||||||||
Spectra acquired at 300 K in mefhanol-d4. 1H resonance frequency 600 MHz and 13C resonance frequency ISO MHz.
Chemical shift values obtained from 2D HSQC and HMBC experiments.
NMR analysis of compound 12a was performed in a nonseparable mixture containing 12a and 12b. n.d.: not detected.
Overlapping signals within the column, multiplicities expressed as m.
Overlapping signals within the column, multiplicities expressed as m.
Overlapping signals within the column, multiplicities expressed as m.
Assignments may be interchanged within the column.
Figure 8.
Selected NOE correlations (double-headed stippled arrows) and HMBC correlations (single-headed arrow pointing from H to C) used for structure elucidation of 2, 3, 9, 11, 12b, and 13.
The material eluted as peaks 3 and 11 showed [M + H]+ ions with m/z 331.1382 and 359.1695, corresponding to molecular formulas of C15H22O8 and C17H26O8, respectively. They were both identified as analogues closely related to 2, with similar AB spin systems for a 4,5-disubstituted resorcinol moiety and a β-glucopyranosyl unit at C-4 based on a 3J HMBC correlation from H-1″ to C-4 (see Figure 8 and Table 1). The main difference between the material eluted as peak 3 and 11 was the presence of a propyl and a pentyl group, respectively. For both compounds, the position of alkylation was identified as C-5 of the resorcinol moiety based on 3J HMBC correlations from H-1′A/H-1′B to C-5 and NOEs between H-1′A/H-1′B and H-6 as well as H-1″. Thus, the materials eluted as peak 3 and 11 were identified as 5-propylresorcinol 4-O-β-d-glucopyranoside (3) and 5-pentylresorcinol 4-O-β-d-glucopyranoside (11), respectively. Both compounds are new, and fully assigned 1H and 13C NMR data are given in Table 1 and 1H, COSY, NOESY, HSQC and HMBC spectra obtained in the HPLC-HRMS-SPE-NMR mode are available in the Supporting Information.
After identification of the potential α-glucosidase inhibitory ligand coeluted under peak 12 as quercetin (12a), the nonbinding minor coeluent was also analyzed using extensive 1D and 2D NMR experiments obtained in the HPLC-HRMS-SPE-NMR mode. The minor metabolite showed an [M + H]+ ion at m/z 373.1842, corresponding to a molecular formula of C18H28O8. The 1H NMR and HSQC spectrum showed the overall patterns of a disubstituted resorcinol moiety, a β-glucopyranoside unit, and a pentyl group, but with additional 1H (δ 3.83 s) and 13C (δ 56.4) NMR resonances for a methoxy group. The glucosylation was at C-1 based on a 3J HMBC correlation between the anomeric proton of the β-glucopyranoside [δ 4.73 d (7.5)] and C-1 (δ 151.6) and NOEs between H-1″ and H-2 as well as H-6 (Figure 8). Methoxylation was at C-3 based on 3J HMBC correlation from the methoxy singlet to C-3 and an NOE correlation between 3-OCH3 and H-2. Finally, the alkylation was at C-5 based on 2J and 3J HMBC correlations from H-1′ [δ 2.56 t (7.6)] to C-5 (δ 130.1), C-4 (δ 140.2), and C-6 (δ 111.0) as well as an NOE correlation between H-1′ and H-6. After full assignment of all 1H and 13C NMR resonances (Table 1), the structure of the minor coeluent under peak 12 was identified as 4-hydroxy-3-O-methyl-5-pentylresorcinol 1-O-β-d-glucopyranoside (12b), which is a new compound. 1H, COSY, NOESY, HSQC, and HMBC spectra obtained in the HPLC-HRMS-SPE-NMR mode are available in the Supporting Information.
The material eluted as peaks 9 and 13 showed [M + H]+ ions at m/z 491.2113 and 519.2425, corresponding to molecular formulas of C22H34O12 and C24H38O12, respectively, which indicated they were diglycosylated analogues of the above-described monoglycosylated alkylresorcinols. Thus, the material eluted as peak 9 was identified as 5-pentylresorcinol 4-O-[α-d-apiofuranosyl-(1→6)]-β-d-glucopyranoside (9), the 6-apiofuranosyl-substituted analogue of 11, based on selected HMBC and NOE correlations (Figure 8) used for structure elucidation. The material eluted as peak 13 was identified as 3-O-methyl-5-pentylresorcinol 1-O-[β-d-glucopyranosyl-(1→6)]-β-d-glucopyranoside (13), i.e., the only monosubstituted resorcinol identified in E. catharinae. The positions of alkylation, methoxylation, and glucosylation of the resorcinol moiety were readily identified based on HMBC and NOE correlations as shown in Figure 8. The same applies to the additional β-glucopyranoside moiety attached to C-6″ as seen from 3J HMBC correlation from H-1‴ [δ 4.34 d (7.8)] to C-6″ (δ 70.1) as well as an NOE correlation between H-1‴ and H-6″A/H-6″B (Figure 8). Compounds 9 and 13 are new, and fully assigned 1H and 13C NMR data are given in Table 1 and 1H, COSY, NOESY, HSQC, and HMBC spectra obtained in the HPLC-HRMS-SPE-NMR mode are available in the Supporting Information.
Identification of Additional Compounds
Additionally, a known flavan-3-ol (catechin, 134) and four flavonol glycosides (myricetin 3-O-β-d-galactopyranoside, 4;30 quercetin 3-O-β-d-galactopyranoside, 6;35 quercetin 3-O-β-d-glucopyranoside 7,36 and quercetin 3-O-α-l-rhamnopyranoside, 830) were identified by analysis of HRMS data and comparing 1H NMR chemical shifts with literature values. Retention times, HRMS, and assigned 1H NMR data of these compounds are provided in Table S1, Supporting Information.
In summary, a bioanalytical platform combining magnetic ligand fishing and HPLC-HRMS-SPE-NMR was developed for analysis of α-glucosidase inhibitory ligands direct from crude plant extracts. A proof-of-concept study was performed using a native Brazilian plant, E. catharinae. This has resulted in identification of four α-glucosidase inhibitory ligands (5, 10, 12a, and 14) from this rarely investigated species. HPLC-HRMS-SPE-NMR analysis of other metabolites in this plant resulted in identification of six new alkylresorcinol glycosides (2, 3, 9, 11, 12b, and 13) and five known flavonoids (1, 4, and 6–8). It was demonstrated that the presented bioanalytical platform is a powerful tool for screening crude plant extracts for ligands of selected therapeutic targets.
EXPERIMENTAL SECTION
Materials
Pyridine (99.8%), glutaraldehyde, hydroxylamine hydrochloride, sodium cyanoborohydride, ammonium acetate, methanol, acetonitrile, quercetin, kaempferol, myricetin, luteolin, caffeine, ferulic acid, acarbose, p-nitrophenyl α-d-glucopyranoside (p-NPG), and α-glucosidase type I (EC 3.2.20, from Saccharomyces cerevisiae, lyophilized powder) were obtained from Sigma-Aldrich (St. Louis, MO, USA). BcMag amine-terminated magnetic beads (30 mg/mL, 1 μm) were purchased from Bioclone (San Diego, CA, USA). Methanol-d4 was purchased from Eurisotop (Gif-Sur-Yvette Cedex, France). Water was purified by deionization and 0.22 μm membrane filtration using a Millipore system (Billerica, MA, USA), and other solvents were all analytical grade.
Plant Material and Extraction
The leaves of Eugenia catharinae O. Berg were collected in Florianópolis-SC, Brazil, in July 2010 from a healthy and agrotoxic-free plant. A voucher specimen has been deposited in the Herbarium FLOR, Universidade Federal de Santa Catarina, Santa Catarina, Brazil, and was catalogued as FLOR 27820. The air-dried and powdered leaves (500.0 g) were extracted with 96% EtOH at room temperature for 5 days. The solvent was removed by rotatory evaporation below 55 °C, and the crude extract was resuspended in EtOH/H2O (20:80 v/v). The resulting suspension was partitioned with n-hexane and EtOAc. The EtOAc fraction was dried and used for analysis.
Preparation of N-Terminus-Coupled α-Glucosidase Magnetic Beads
Immobilization of N-terminus-coupled α-glucosidase magnetic beads was performed using a previously published protocol with slight modifications.9 Briefly, a suspension of 1 mL (30 mg) of BcMag amine-terminated magnetic beads was magnetically separated from the supernatant using a magnetic separator (Dynal MPC-S) and washed with 3 × 1 mL of coupling buffer (10 mM aqueous pyridine, pH 6.0). The beads were then resuspended in 1 mL of 5% gluteraldehyde solution in coupling buffer and rotated in an orbital rotator for 1 day. After magnetic separation, the beads were washed with 3 × 1 mL of coupling buffer. A 4.5 mg/mL solution of α-glucosidase (86 units/mg) was prepared in the coupling buffer, and 1 mL of this solution was added to the activated beads. The mixture was rotated at room temperature for 4 days. After removing the supernatant, the beads were resuspended in 1 mL of an end-capping solution (100 mM hydroxylamine in the coupling buffer containing 2.5% of sodium cyanoborohydride), and the rotation continued for 1 day. Finally, the supernatant was discarded and N-terminus-coupled α-glucosidase magnetic beads (AGN-TCMB) were washed with 3 × 1 mL of assay buffer (10 mM ammonium acetate, pH 7.4) and stored in 1 mL of the same buffer at 4 °C.
Immobilized Enzyme Activity Studies
A 5 mg amount of AGN-TCMB was added to each of five 1.5 mL Eppendorf tubes, followed by 20 μL of 0.4 mM p-NPG. The samples were incubated for 5, 10, 15, 20, and 25 min, respectively, before magnetic separation, and the supernatant was collected and submitted for LC-MS analysis. From similar AGN-TCMB, two sets of samples were aliquoted in the following amounts: 3.75, 1.875, 0.938, 0.469, and 0.234 mg. One set was dissolved in 500 μL of assay buffer, whereas the other set was dissolved in 500 μL of assay buffer containing 10 mM acarbose. To all solutions was added 20 μL of 0.4 mM p-NPG, and the resulting mixture was incubated for 10 min. After magnetic separation, the supernatant was collected and subjected to LC-MS analysis on an Agilent 1100 system (Agilent Technologies, Palo Alto, CA, USA) equipped with a vacuum degasser (G1322A), a binary pump (1312A), an autosampler (G1313A), and a mass selective detector (G1946B) with atmospheric pressure ionization. All LC-MS experiments were performed using a single injection loading of 10 μL on a Phenomenex C18(2) Luna column (100 mm × 4.6 mm i.d., 3 μm particle size, 100 Å pore size) (Phenomenex, Inc., Torrance, CA, USA). Separations were performed at 25 °C with a flow rate of 0.5 mL/min using the following binary mobile phase of solvent A (H2O acidified with 0.5% formic acid) and solvent B (MeCN acidified with 0.5% formic acid). The following gradient elution profile was used for the separation: 0 min, 0% B; 20 min, 100% B; 25 min, 100% B; 26 min, 0% B.
Ligand Fishing Analysis of Model Mixture
A model mixture consisting of ferulic acid, caffeine, and luteolin (0.17 mM each, equimolar) was prepared in the assay buffer (S0) and subjected to ligand fishing as follows. To 10 mg of AGN-TCMB was added 600 μL of the above mixture, and the resulting suspension was incubated for 10 min at room temperature. The supernatant (S1) was collected following a magnetic separation, and the beads were washed with 3 × 400 μL of NH4OAc buffer (S2–S4). Finally, the beads were eluted with 3 × 400 μL of 90% MeOH (S5–S7). S1–S7 were subjected to LC-MS analysis using similar system and chromatographic conditions (column oven temperature, flow rate, and mobile phase composition) as described in the HPLC-HRMS-SPE-NMR analysis section. The separation was performed using the following gradient profile: 0 min, 0% B; 20 min, 100% B; 30 min, 100% B.
Ligand Fishing Analysis of E. catharinae
A solution of the EtOAc extract of E. catharinae (3.5 mg/mL; S0) was prepared in NH4OAc buffer containing 5% MeOH, and 600 μL of this was added to 10 mg of AGN-TCMB. After incubation for 10 min at room temperature followed by magnetic separation, the supernatant (S1) was removed, and the beads were washed with 3 × 400 μL of assay buffer to yield S2–S4, and 3 × 400 μL of 90% MeOH to yield eluent solutions S5–S7. S1–S7 were subjected to LC-HRMS analysis using the HPLC-HRMS-SPE-NMR system and chromatographic conditions (column oven temperature, flow rate, and mobile phase composition) described in the next section. The separation was performed using the following gradient profile: 0 min, 0% B; 20 min, 100% B; 30 min, 100% B.
HPLC-HRMS-SPE-NMR
HPLC separations were performed with an Agilent 1260 system consisting of a degasser, a quaternary pump, an autosampler, a column oven, a diode array detector, and a Phenomenex C18(2) Luna column (150 mm × 4.6 mm i.d., 3 μm particle size, 100 Å pore size) (Phenomenex, Inc.). The column was operated at 25 °C, and the flow rate was maintained at 0.5 mL/min. The aqueous eluent (A) consisted of H2O/MeCN (95:5, v/v), and the organic eluent (B) consisted of MeCN/H2O (95:5, v/v), both acidified with 0.1% formic acid. Separations were performed using the following gradient elution profile: 0 min, 5% B; 58 min, 35% B; 60 min, 100% B; 75 min, 100% B. The column eluate was connected to a T-piece splitter directing ca. 1% of the flow to a Bruker micrOTOF-Q mass spectrometer equipped with an electrospray ionization (ESI) interface (Bruker Daltonik, Bremen, Germany). Mass spectra were acquired in both positive and negative ion mode, using a drying temperature of 200 °C, a nebulizer pressure of 2.0 bar, and a drying gas flow of 7 L/min. The remaining ca. 99% of the HPLC eluate was directed to the photodiode-array detector and then, after diluting with 1 mL/min of H2O by means of a Knauer Smartline pump 120 (Knauer, Berlin, Germany), to a Prospect 2 SPE-unit (Spark Holland, Emmen, The Netherlands). Selected chromatographic peaks were trapped on SPE cartridges (Hysphere GP-phase, 10 × 2 mm i.d., from Spark Holland), preconditioned with 500 μL of MeCN and subsequently equilibrated with 500 μL of H2O. A total of 10 cumulative trappings were performed for peaks 1–14 after 10 repeated separations using injections corresponding to 2.25 mg of extract (15 μL injection volumes of 150 mg/mL samples). After trapping, the cartridges were dried with a stream of nitrogen gas for 1 h and subsequently eluted with methanol-d4 into 1.7 mm o.d. NMR tubes (Bruker Biospin, Karlsruhe, Germany) by means of a Gilson 215 liquid handler (Gilson, Middleton, WI, USA). The tubes were filled with 30 μL of eluted sample and sealed with plastic balls. Chromatographic separation, mass spectrometry, and analyte trapping on SPE cartridges were controlled using Hystar ver. 3.2 software (Bruker Daltonik), whereas the elution process was mediated by Prep Gilson ST ver. 1.2 software (Bruker Biospin).
NMR Experiments
NMR experiments were performed with a Bruker Avance III NMR system (operating at a 1H frequency of 600.13 MHz) equipped with a Bruker SampleJet sample changer and a cryogenically cooled gradient inverse triple-resonance 1.7 mm TCI probehead (Bruker Biospin, Karlsruhe, Germany) optimized for 1H and 13C observation. Bruker standard pulse sequences were used throughout this study. Topspin ver. 3.2 (Bruker Biospin) was used for acquisition and processing of NMR data, whereas IconNMR ver.4.2 (Bruker Biospin) was used for controlling automated sample change and acquisition. 1D 1H NMR spectra were acquired in automation (temperature equilibration to 300 K, optimization of lock parameters, gradient shimming, and setting of receiver gain) with 30-degree pulses, 3.66 s interpulse intervals, and HOD signal presaturation during relaxation delay (4.0 s). A total of 64k data points were collected and multiplied with an exponential function corresponding to line-broadening of 0.3 Hz prior to Fourier transform. 2D homo- and heteronuclear experiments were acquired with 2048 data points in the direct dimension and 128 (DQF-COSY and HMBC) or 256 (multiplicity-edited HSQC and phase-sensitive NOESY) data points in the indirect dimension. The HMBC and HSQC experiments were optimized for nJH,C = 8 Hz and 1JH,C = 145 Hz, respectively. HMBC and HSQC spectra were processed to 2k × 1k data matrices, after linear prediction (32 coefficients) in F1 and application of a sine-bell window function in F1 and F2.
Supplementary Material
Figure 7.
Structures of compounds used for the model mixture (i–iii) and compounds 1–14 identified in E. catharinae.
ACKNOWLEDGMENTS
HPLC equipment used for high-resolution bioassay profiles was obtained via a grant from The Carlsberg Foundation. The 600 MHz HPLC-HRMS-SPE-NMR system used in this work was acquired through a grant from Apotekerfonden af 1991, The Carlsberg Foundation, and the Danish Agency for Science, Technology and Innovation via the National Research Infrastructure funds. This research was supported in part by the Intramural Research Program of the NIA (R.M.) and by a grant from The Danish Research Council for Independent Research | Technology and Production (SGW: DFF - 4005-00387).
Footnotes
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jnat-prod.5b00603.
Table S1 with HRMS and 1H NMR data and Figures S1–S30 with 1D and 2D NMR spectra of 2, 3, 9, 11, 12b, and 13 obtained in the HPLC-HRMS-SPE-NMR mode (PDF)
The authors declare no competing financial interest.
REFERENCES
- 1.Bischoff H. Clin. Invest. Med. 1995;18:303–311. [PubMed] [Google Scholar]
- 2.van de Laar FA. Vasc. Health Risk. Manag. 2008;4:1189–1195. doi: 10.2147/vhrm.s3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mooradian A, Thurman J. Drugs. 1999;57:19–29. doi: 10.2165/00003495-199957010-00003. [DOI] [PubMed] [Google Scholar]
- 4.van de Laar FA, Lucassen PL, Akkermans RP, van de Lisdonk EH, Rutten GE, van Weel C. Diabetes Care. 2005;28:154–163. doi: 10.2337/diacare.28.1.154. [DOI] [PubMed] [Google Scholar]
- 5.Mata R, Cristians S, Escandón-Rivera S, Juárez-Reyes K, Rivero-Cruz I. J. Nat. Prod. 2013;76:468–483. doi: 10.1021/np300869g. [DOI] [PubMed] [Google Scholar]
- 6.Yin Z, Zhang W, Feng F, Zhang Y, Kang W. Food Science and Human Wellness. 2014;3:136–174. [Google Scholar]
- 7.Lourenco VK, Jiang Z, Zhang X, Vieira LC, Corrêa AG, Cardoso CL, Cass QB, Moaddel R. Talanta. 2013;116:647–652. doi: 10.1016/j.talanta.2013.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qing L-S, Xue Y, Zheng Y, Xiong J, Liao X, Ding L-S, Li B-G, Liu Y-MJ. Chromatogr. A. 2010;1217:4663–4668. doi: 10.1016/j.chroma.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yasuda M, Wilson DR, Fugmann SD, Moaddel R. Anal. Chem. 2011;83:7400–7407. doi: 10.1021/ac201403y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu Y-T, Jia Y-W, Liu Y-M, Liang J, Ding L-S, Liao XJ. Agric. Food Chem. 2014;62:10679–10686. doi: 10.1021/jf503687e. [DOI] [PubMed] [Google Scholar]
- 11.Qing L-S, Xue Y, Deng W-L, Liao X, Xu X-M, Li B-G, Liu Y-M. Anal. Bioanal. Chem. 2011;399:1223–1231. doi: 10.1007/s00216-010-4399-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Staerk D, Kesting JR, Sairafianpour M, Witt M, Asili J, Emami SA, Jaroszewski JW. Phytochemistry. 2009;70:1055–1061. doi: 10.1016/j.phytochem.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 13.Schmidt B, Jaroszewski JW, Bro R, Witt M, Stærk D. Anal. Chem. 2008;80:1978–1987. doi: 10.1021/ac702064p. [DOI] [PubMed] [Google Scholar]
- 14.Marszałł M. Pharm. Res. 2011;28:480–483. doi: 10.1007/s11095-010-0284-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sanghvi M, Moaddel R, Wainer IW. J. Chromatogr. A. 2011;1218:8791–8798. doi: 10.1016/j.chroma.2011.05.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bokesch HR, Wamiru A, Le Grice SFJ, Beutler JA, McKee TC, McMahon JB. J. Nat. Prod. 2008;71:1634–1636. doi: 10.1021/np8002518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grangeiro MS, Calheiros-Lima AP, Martins MF, Arruda LF, Garcez-do-Carmo L, Santos WC. J. Ethnopharmacol. 2006;108:26–30. doi: 10.1016/j.jep.2006.04.021. [DOI] [PubMed] [Google Scholar]
- 18.Omar R, Li L, Yuan T, Seeram NP. J. Nat. Prod. 2012;75:1505–1509. doi: 10.1021/np300417q. [DOI] [PubMed] [Google Scholar]
- 19.Prodanović R, Milosavić N, Jovanović S, Prodanović O, Ćirković Velicković T, Vujcić Z, Jankov R. Biocatal. Biotransform. 2006;24:195–200. [Google Scholar]
- 20.Fernández A, Sinanoglu O. Biophys. Chem. 1985;21:163–166. doi: 10.1016/0301-4622(85)80002-8. [DOI] [PubMed] [Google Scholar]
- 21.Yan J, Zhang G, Pan J, Wang Y. Int. J. Biol. Macromol. 2014;64:213–223. doi: 10.1016/j.ijbiomac.2013.12.007. [DOI] [PubMed] [Google Scholar]
- 22.Schmidt JS, Nyberg NT, Staerk D. Food Chem. 2014;161:192–198. doi: 10.1016/j.foodchem.2014.03.062. [DOI] [PubMed] [Google Scholar]
- 23.Wubshet SG, Schmidt JS, Wiese S, Staerk D. J. Agric. Food Chem. 2013;61:8616–8623. doi: 10.1021/jf402949y. [DOI] [PubMed] [Google Scholar]
- 24.Liu B, Kongstad KT, Qinglei S, Nyberg NT, Jager AK, Staerk D. J. Nat. Prod. 2015;78:294–300. doi: 10.1021/np5009416. [DOI] [PubMed] [Google Scholar]
- 25.Okutan L, Kongstad KT, Jager AK, Staerk D. J. Agric. Food Chem. 2014;62:11465–11471. doi: 10.1021/jf5047283. [DOI] [PubMed] [Google Scholar]
- 26.Grosso C, Jager AK, Staerk D. Phytochem. Anal. 2013;24:141–147. doi: 10.1002/pca.2393. [DOI] [PubMed] [Google Scholar]
- 27.Wiese S, Wubshet SG, Nielsen J, Staerk D. Food Chem. 2013;141:4010–4018. doi: 10.1016/j.foodchem.2013.06.115. [DOI] [PubMed] [Google Scholar]
- 28.Kongstad KT, Wubshet SG, Johannesen A, Kjellerup L, Winther A-ML, Jager AK, Staerk D. J. Agric. Food Chem. 2014;62:5595–5602. doi: 10.1021/jf501605z. [DOI] [PubMed] [Google Scholar]
- 29.Chung S-K, Kim Y-C, Takaya Y, Terashima K, Niwa M. J. Agric. Food Chem. 2004;52:4664–4668. doi: 10.1021/jf049526j. [DOI] [PubMed] [Google Scholar]
- 30.Wubshet SG, Moresco HH, Tahtah Y, Brighente IMC, Staerk D. Phytochemistry. 2015;116:246–252. doi: 10.1016/j.phytochem.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 31.Liao C-R, Kuo Y-H, Ho Y-L, Wang C, Yang C, Lin C-W, Chang Y-S. Molecules. 2014;19:9515–9534. doi: 10.3390/molecules19079515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moradi-Afrapoli F, Asghari B, Saeidnia S, Ajani Y, Mirjani M, Malmir M, Dolatabadi Bazaz R, Hadjiakhoondi A, Salehi P, Hamburger M, Yassa N. Daru, J. Pharm. Sci. 2012;20:37. doi: 10.1186/2008-2231-20-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang H, Du Y-J, Song H-C. Food Chem. 2010;123:6–13. [Google Scholar]
- 34.Benavides A, Montoro P, Bassarello C, Piacente S, Pizza C. J. Pharm. Biomed. Anal. 2006;40:639–647. doi: 10.1016/j.jpba.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 35.Ek S, Kartimo H, Mattila S, Tolonen A. J. Agric. Food Chem. 2006;54:9834–9842. doi: 10.1021/jf0623687. [DOI] [PubMed] [Google Scholar]
- 36.Han J-T, Bang M-H, Chun O-K, Kim D-O, Lee C-Y, Baek N-I. Arch. Pharmacal Res. 2004;27:390–395. doi: 10.1007/BF02980079. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.