Abstract
Japanese encephalitis (JE) is one of the most severe viral infections of the central nervous system. No effective treatment for JE currently exists, because its pathogenesis remains largely unknown. The present study was designed to screen the potential microRNAs (miRNAs) involved in JE. Glioblastoma cells were collected, after being infected with the Japanese encephalitis virus (JEV). Total miRNAs were extracted and analyzed using an miRNA chip. One of the most severely affected miRNAs was selected, and the role of miR-370 in JEV infection was investigated. Cell viability and apoptosis of the host cells were evaluated. JEV replication was detected via analysis of gene E expression. Real-time polymerase chain reaction was used to determine the levels of endogenous miR-370 and expression of innate immunity-related genes. Following JEV infection, 114 miRNAs were affected, as evidenced by the miRNA chip. Among them, 30 miRNAs were upregulated and 84 were downregulated. The changes observed in five miRNAs were confirmed by real-time polymerase chain reaction. One of the significantly downregulated miRNAs was miR-370. Therefore, miR-370 mimic was transfected into the cells, following which the levels of endogenous miR-370 were significantly elevated. Concurrently, JEV replication was significantly reduced 24 hours after transfection of miR-370 mimic. Functionally, miR-370 mimic mitigated both JEV-induced apoptosis and the inhibition of host cell proliferation. Following JEV infection, interferon-β and nuclear factor-kappa B were upregulated, whereas miR-370 mimic prevented the upregulation of the genes induced by JEV infection. The present study demonstrated that miR-370 expression in host cells is downregulated following JEV infection, which further mediates innate immunity-related gene expression. Taken together, miR-370 mimic might be useful to prevent viral replication and infection-induced host cell injury.
Keywords: miR-370, Japanese encephalitis, glioblastoma cells, NF-κB
Introduction
Japanese encephalitis (JE) is caused by the Japanese encephalitis virus (JEV) and is one of the most severe viral infections of the central nervous system. One of the hallmarks of this infection is parenchymal inflammation of the brain. The mortality rate of JE is relatively high (35%–40%), and ~20%–70% of JE patients have permanent neurological sequelae.1,2 People’s Republic of China is one of the JE endemic countries and accounts for half of the total number of JE cases worldwide. Effective treatment for JE is not currently available. Supportive therapy and symptomatic treatment are implemented to alleviate suffering. Thus, understanding the mechanisms of JEV infection is particularly important.
The microRNAs (miRNAs) belong to a class of small noncoding RNA molecules with lengths of ~20–24 nucleotides. They function mainly through complementary binding to the target mRNA to facilitate its degradation or translation. A previous study reported that miRNAs perform critical roles in the regulation of cell proliferation, survival, and migration.3 Moreover, miRNAs have active roles in the modulation of innate immunity and viral replication.4 Hundreds of miRNAs have been discovered in the herpes simplex viruses (herpesviruses), polyomaviruses, adenoviruses, and retroviruses.5 Viral invasion affects host miRNA levels that can also inhibit target gene expression and lead to disease progression. Conversely, miRNAs from host cells could also target the corresponding RNA sequence and inhibit viral replication and proliferation, thereby alleviating the effects of viral infection. One miRNA has multiple target genes, and several miRNAs can work together to regulate the expression of a specific gene.6
The development of JE as it relates to innate immune mechanisms has been well defined.7 The expression of innate immunity-related genes was remarkably stimulated following JEV infection. The aim of the present study was to determine the miRNA expression profile following JE infection and investigate more specifically the function of miR-370 in JE and its effects on immunity-related gene expression.
Materials and methods
Cell culture and viral infection
This study was approved by the ethics committee of Jining Medical University. Cells of the U251 human glioblastoma cell line were obtained from the Cell Bank of Chinese Academy (Shanghai, People’s Republic of China). They were cultured in minimum essential medium (HyClone, South Logan, UT, USA), supplied with 15% fetal bovine serum (Lanzhou Minhai Biotech, Lanzhou, People’s Republic of China), at 37°C in an incubator (Thermo Fisher Scientific, Waltham, MA, USA) with 5% CO2. The JEV live vaccine strain SA14-14-2 was maintained in a laboratory at Jining Medical University. The cells were infected with JEV at a multiplicity of infection of 1. Twenty-four hours after infection, cells were collected for the miRNA chip assay. There were five repeats in each group, and differentially expressed miRNAs were further confirmed by real-time polymerase chain reaction (PCR).
The miR-370 mimic and inhibitor were purchased from Thermo Fisher Scientific. Lipofectamine 3000 Transfection Reagent (Thermo Fisher Scientific) was used to promote transfection of miR-370 mimic and inhibitor. Following transfection of miR-370 mimic or inhibitor, endogenous miR-370 levels, cell proliferation (Cell Counting Kit-8 [CCK-8]; Beyotime, Ningbo, People’s Republic of China), apoptosis (TUNEL Apoptosis Detection Kit; Vazyme, Nanjing, People’s Republic of China), and JEV replication were all assessed.
Expression profile of miRNA
The Agilent miRNA chip (miRBase 21.0) was used to analyze the miRNA expression profile. Following extraction of miRNAs using specific isolation kits (mirVana; NY, USA), miRNAs were labeled with Hy3™, according to the protocol supplied with the MicroRNA Array kit. The GeneChip® Scanner 3000 7G 4C (Thermo Fisher Scientific) was used to scan the arrays, and data analyses were performed using the miRNA QC Tool software (Qiagen NV, Venlo, the Netherlands).
Cell proliferation and apoptosis
U251 cells were cultured in a 96-well plate (5×103 cells in each well). Following transfection of miR-370 mimic or inhibitor (50 nM), cells were infected with JEV. After treatment, cell proliferation was analyzed using the CCK-8 assay kit. CCK-8 solution (10 μL) was added to each well. An additional 4 hours of incubation facilitated CCK-8 staining of the cells. Absorbance was measured at 450 nm, using a microplate reader (SM600; Shanghai Yongchuan, Shanghai, People’s Republic of China). Apoptotic cells were detected using the TUNEL assay, according to the manufacturer’s instructions (Beyotime). Positive cells were counted using the Image-Pro Plus 6.0 image analysis software (National Institutes of Health, MA, USA).
Real-time PCR
Following treatment, the cells were collected and total miRNAs and mRNA were extracted. The concentration and purity of the total RNA were measured with the NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific). The mRNAs were then reverse transcribed. Real-time PCR combined with SYBR staining was used to amplify expression. The primers were designed via Primer Premier 6.0 software (Premier, Ottawa, Canada) and synthesized by Shanghai Biotech (Shanghai, People’s Republic of China). Expression of has-miR-370 (mature miRNA sequence: GCCUGCUGGGGUGGAACCUGGU) was normalized to that of U6 RNA (F-CTCGCTTCGGCAGCACA, R-ACGCTTCACGAATTTGCGT). TaqMan probes were designed based on the gene E (U47032) of JEV (5′-FAM-TTCTGAAGGCACCACCAAACAC-Eclipse-3′).8 The primers for tumor necrosis factor receptor-associated factor 3 (TRAF3), interferon regulatory factor-3 (IRF3), interferon-β (IFN-β), nuclear factor-kappa B (NF-κB), and glyceraldehyde 3-phosphate dehydrogenase are listed in Table 1.
Table 1.
Primers for real-time PCR
| Gene | GenBank accession no | Primer (5′-3′) | Length | Tm | Location |
|---|---|---|---|---|---|
| TRAF3 | NM_001139490 | For: TCACGGAGGTGATTAGAATGACT | 23 | 60.6 | 116-138 |
| Rev: ACAACCACGTCTATGGCCTTT | 21 | 61.7 | 242-222 | ||
| IRF3 | NM_001197128 | For: AGAGGCTCGTGATGGTCAAG | 20 | 61.3 | 260-279 |
| Rev: AGGTCCACAGTATTCTCCAGG | 21 | 60.3 | 359-339 | ||
| IFN-β | NM_002176 | For: ATGACCAACAAGTGTCTCCTCC | 22 | 61.6 | 1-22 |
| Rev: GGAATCCAAGCAAGTTGTAGCTC | 23 | 61.4 | 88-66 | ||
| NF-κB | NM_003998 | For: AACAGAGAGGATTTCGTTTCCG | 22 | 60.3 | 155-176 |
| Rev: TTTGACCTGAGGGTAAGACTTCT | 23 | 60.4 | 258-236 | ||
| GAPDH | NM_014364 | For: TGTGGGCATCAATGGATTTGG | 21 | 60.9 | 231-251 |
| Rev: ACACCATGTATTCCGGGTCAAT | 22 | 61.4 | 346-325 |
Note: “For” and “Rev” represent forward and reverse primers, respectively.
Abbreviations: PCR, polymerase chain reaction; TRAF3, tumor necrosis factor receptor-associated factor 3; IRF3, interferon regulatory factor-3; IFN-β, interferon-β; NF-κB, nuclear factor-kappa B; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; Tm, melting temperature.
The Ct value for each gene was detected, and the expression of the target genes was calculated by the −ΔΔCT method.9,10 PCR was carried out using the real-time PCR LightCycler 96 (Hoffman-La Roche Ltd., Basel, Switzerland) as follows: initial denaturation at 95°C for 5 minutes, denaturation at 95°C for 30 seconds, annealing at 60°C for 13 seconds, and extension at 72°C for 30 seconds, for 40 cycles. The reaction system consisted of 10 μL 2× SYBR Fast qPCR Mix, 0.8 μL PCR forward/reverse primers (10 mM), 0.4 μL 50× ROX Reference Dye II, and 2 μL cDNA template.
Data analyses
Data were presented as mean ± standard deviation. All statistical analyses were performed using the SPSS 11.5 software (SPSS Inc., Chicago, IL, USA). Two-group comparison was conducted using the two-sample t-test. The comparisons among three groups or more were analyzed using one-way analysis of variance, followed by the Bonferroni post hoc test. A P-value <0.05 was considered statistically significant.
Results
Expression profile of miRNAs in U251 cells following JEV infection
Following JEV infection, 114 miRNAs were affected; 30 were upregulated and 84 were downregulated (Table 2). Five miRNAs with high fold changes were confirmed by real-time PCR (Figure 1). Among them, miR-342 and miR-335 were significantly upregulated. In contrast, miR-370, miR-114, and miR-410 were significantly downregulated (P<0.05).
Table 2.
Expression profile of miRNAs in U251 after JEV infection
| ProbeSet Name | Fold change (B1 A1) | Regulation (B1 A1) | Median CV (%) | Dyes |
|---|---|---|---|---|
| miR-21-5p | 4.2312 | Up | 3.69 | Cy3 |
| miR-150-5p | 3.1104 | Up | 6.79 | Cy3 |
| miR-342-3p | 2.6396 | Up | 7.15 | Cy3 |
| miR-663a | 3.2143 | Up | 6.18 | Cy3 |
| miR-3614-5p | 2.0065 | Up | 8.02 | Cy3 |
| miR-296-3p | 4.1056 | Up | 6.81 | Cy3 |
| miR-664b-5p | 3.3315 | Up | 7.03 | Cy3 |
| miR-335-3p | 3.4125 | Up | 6.76 | Cy3 |
| miR-155-5p | 2.6617 | Up | 6.88 | Cy3 |
| miR-887 | 3.9711 | Up | 6.18 | Cy3 |
| miR-3656 | 2.8991 | Up | 8.41 | Cy3 |
| miR-221-5p | 4.0061 | Up | 9.25 | Cy3 |
| miR-642a-3p | 0.4002 | Down | 6.87 | Cy3 |
| miR-370-5p | 0.2902 | Down | 8.04 | Cy3 |
| miR-191-3p | 0.3415 | Down | 7.07 | Cy3 |
| miR-145-3p | 0.3189 | Down | 5.78 | Cy3 |
| miR-1 | 0.2587 | Down | 6.81 | Cy3 |
| miR-let-7 | 0.3815 | Down | 5.79 | Cy3 |
| miR-270-3p | 0.1987 | Down | 5.97 | Cy3 |
| miR-114-3p | 0.3612 | Down | 5.74 | Cy3 |
| miR-199a-3p | 0.2986 | Down | 5.84 | Cy3 |
| miR-128 | 0.3476 | Down | 5.25 | Cy3 |
| miR-656 | 0.3628 | Down | 7.15 | Cy3 |
| miR-410 | 0.2284 | Down | 7.86 | Cy3 |
| miR-4485 | 0.3481 | Down | 5.84 | Cy3 |
| miR-217 | 0.2468 | Down | 6.83 | Cy3 |
| miR-484 | 0.1967 | Down | 6.01 | Cy3 |
| miR-25-3p | 0.3861 | Down | 4.91 | Cy3 |
| miR-127-3p | 0.2853 | Down | 2.05 | Cy3 |
Notes: A1, JEV group; B1, control group.
Abbreviations: miRNAs, microRNAs; JEV, Japanese encephalitis virus; CV, coefficient of variation.
Figure 1.
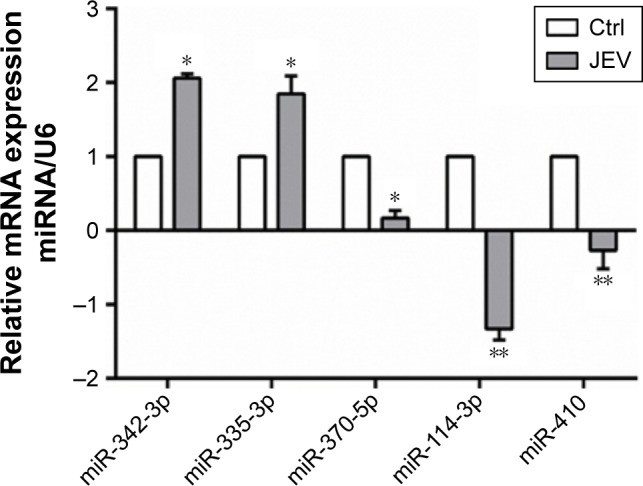
Effects of JEV on miRNA expressions in U251 cells.
Note: *P<0.05 and **P<0.01 compared with control.
Abbreviations: JEV, Japanese encephalitis virus; miRNA, microRNA; mRNA, messenger RNA; Ctrl, control.
Inhibition of JEV replication and JEV-induced cell injury by miR-370 mimic
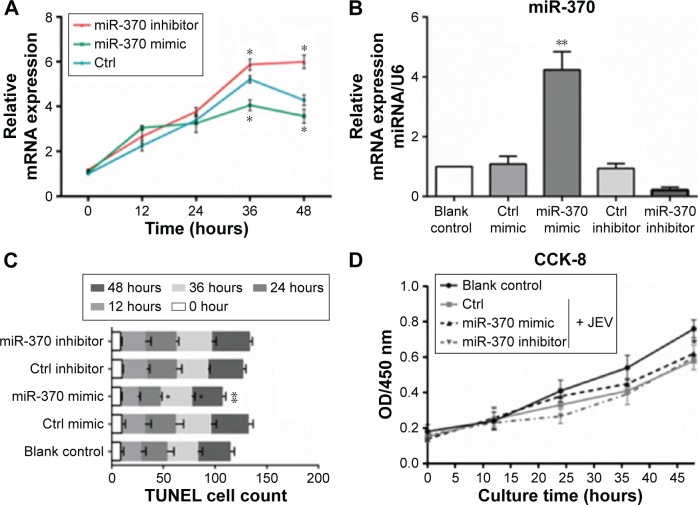
Endogenous miRNAs could be functionally mimicked or stimulated by exogenous miRNAs, whereas miRNA inhibitors specifically prevent the normal functioning of endogenous miRNAs. In the present study, miR-370 mimic and miR-370 inhibitor were selected to evaluate the role of miR-370 in JEV infection. As shown in Figure 2A, expression of gene E was significantly inhibited by miR-370 mimic at the 36-hour and 48-hour time points, whereas it was significantly enhanced by miR-370 inhibitor (versus control, P<0.05). These data suggest that viral replication was inhibited by miR-370 mimic and promoted by miR-370 inhibitor. In comparison with the control, miR-370 mimic significantly increased levels of endogenous miR-370 in U251 cells, 24 hours after transfection (Figure 2B). In successive experiments, cell proliferation and apoptosis were detected at 0, 12, 24, 36, and 48 hours following JEV infection. JEV infection significantly increased the number of apoptotic cells, 48 hours: F(3,15)=7.284, P<0.05 (Figure 2C) and inhibited cell proliferation, F(3,15)=4.21, P<0.05 (Figure 2D). In contrast, the effects of JEV on apoptosis were inhibited by miR-370 mimic, but not by miR-370 inhibitor (Figure 2C). JEV-induced inhibition of proliferation was slightly attenuated at the 48-hour time point by miR-370 mimic (P>0.05; Figure 2D).
Figure 2.
miR-370 mimic inhibition of JEV replication (A), endogenous miR-370 expression (B), JEV-induced apoptosis (C), and proliferation-inhibition (D).
Note: *P<0.05 and **P<0.01 compared with control inhibitor/mimic.
Abbreviations: JEV, Japanese encephalitis virus; mRNA, messenger RNA; miRNA, microRNA; Ctrl, control; OD, optical density; CCK-8, Cell Counting Kit-8; TUNEL, terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling.
Effects of miR-370 mimic on the expression of innate immunity-related genes
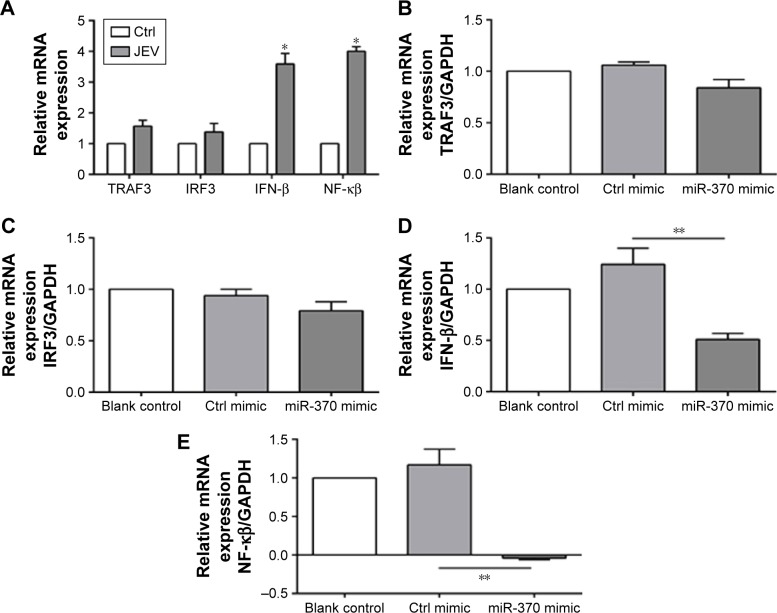
Glioblastoma cells were collected, and expression of innate immunity-related genes (TRAF3, IRF3, IFN-β, and NF-κB) was analyzed 24 hours after JEV infection. In comparison to the uninfected cells, those infected with JEV exhibited significant upregulation of IFN-β and NF-κB (Figure 3A). IRF3 and TRAF3 were evidently not affected following JEV infection or transfection of miR-370 mimic (Figure 3B and C). In contrast, miR-370 mimic significantly reduced JEV infection-induced upregulation of NF-κB expression and IFN-β expression (Figure 3D and E).
Figure 3.
Effects of miR-370 mimic on the expression of innate immunity-related genes.
Notes: (A) JEV increased IFN-β and NF-κB expression, (B) TRAF3, (C) IRF3, (D) IFN-β, and (E) NF-κB. *P<0.01 and **P<0.05 compared with control mimic/control.
Abbreviations: JEV, Japanese encephalitis virus; IFN-β, interferon-β; NF-κB, nuclear factor-kappa B; TRAF3, tumor necrosis factor receptor-associated factor 3; IRF3, interferon regulatory factor-3; mRNA, messenger RNA; Ctrl, control; GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
Discussion
The miRNAs are widely distributed in different types of cells and tissues. It is now clear that miRNAs are involved in several biological processes through complex regulatory mechanisms.11 The miRNAs and their target genes form a complex network to mediate cell proliferation, apoptosis, and biological functions.12 The present study demonstrated that JEV infection influences many miRNAs, including miR-370, that regulate the expression of innate immunity-related genes and induce lesions in host cells.
The functions of miRNAs in tumorigenesis and treatment have been extensively investigated.13,14 Recent studies also suggest that miRNAs perform critical roles in the mediation of innate immunity and virus replication.4,15 Viral nucleic acid can affect miRNA expression in the host cell, and this in turn can regulate gene expression and mediate viral replication. On the other hand, some reports indicate that miRNA could directly regulate the expression of viral genes, thus interfering with viral replication.4,16 Sonkoly et al17 found a close link between miRNAs and a chronic inflammatory skin disease that indicates the important immunomodulatory effect of miRNAs. These findings also reflect the mechanism by which miRNA regulates viral infection.
The first step to discover the immunoregulatory effects of miRNAs is to identify specific miRNAs. Direct cloning, the real-time fluorescence quantitative method, and northern blot have been commonly employed in the evaluation of differential miRNAs. However, these methods have obvious disadvantages, including low detection efficiency, poor reproducibility, and difficulty in high-throughput screening. Using miRNA microarray screening, O’Connell et al18 found that following poly(I:C) and IFN-β stimulation of mouse peritoneal macrophages, miR-155 was one of the significantly upregulated miRNAs. In the present study, we used an miRNA chip to detect the miRNA expression profile in U251 cells, following JEV infection. The Agilent miRNA chip was selected as well as the miRBase 21.0, as it is the latest available database. The fold changes detected in the real-time PCR were consistent with the results obtained from the miRNA chip. The miRNAs miR-342 and miR-335 were upregulated, whereas miR-370, miR-114, and miR-410 were downregulated after JEV infection. These results implicate that a series of miRNAs were affected after JEV infection. All these miRNAs could become the targets for further therapy.
The significantly upregulated miRNAs included miR-370, a finding that is consistent with those of previous publications.19,20 The DLK1/DIO3 region of chromosome 14, in which miR-370 is located, is believed to regulate tumorigenesis. A previous study reported that miR-146a and miR-370 coordinate EV71-induced apoptosis.21 Although there are some critical differences between the procedures of this study and those of the present study, including host cell type (rhabdomyosarcoma cells) and viral type (EV71), downregulation of miR-370 was a similar outcome following viral infection. Furthermore, miR-370 has been widely reported to regulate cell viability.22 Based on these findings, miR-370 was selected as the target miRNA for the present study. It should be noted that the present results showed that endogenous miR-370 expression was significantly downregulated in U251 cells following JEV infection. Apoptosis and inhibition of cell proliferation were significantly enhanced, 48 hours after JEV infection. It is interesting to note that miRNA-370 mimic reversed the effects of infection. These results suggest that miR-370 could inhibit JEV-induced injury to a certain extent.
Real-time PCR was used to detect expression of gene E (U47032) and evaluate the effects of miR-370 on JEV replication. Replication of JEV was significantly inhibited by miR-370 mimic. Furthermore, expression of the genes involved in the innate immune response pathway was compared. Compared with the control, IFN-β and NF-κB were significantly upregulated following JEV infection. Expression of NF-κB and IFN-β was significantly reduced by miR-370 mimic. On the one hand, miR-370 mimic could inhibit the action of NF-κB and ultimately downregulate the expression of IFN-β. Another report has shown that miR-370 inhibits its target gene, lin28a, which subsequently regulates RelA/p65 expression and activates the NF-κB pathway.23 On the other hand, miR-370 mimic possibly inhibited gene expression by preventing viral replication.
Conclusion
Although the roles of other miRNAs were also assessed, the results suggest that miR-370 mimic inhibits JEV replication and attenuates JEV-induced injury of host cells. Moreover, innate immunity-related gene expression was attenuated, following miR-370 treatment. These data provide evidence that miR-370 could serve as a therapeutic target for JE.
Acknowledgments
This study was supported by the Natural Science Foundation of Shandong Province, People’s Republic of China (ZR2015PH026 and ZR2015YL023).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Zheng Y, Li M, Wang H, Liang G. Japanese encephalitis and Japanese encephalitis virus in mainland China. Rev Med Virol. 2012;22(5):301–322. doi: 10.1002/rmv.1710. [DOI] [PubMed] [Google Scholar]
- 2.Wang H, Liang G. Epidemiology of Japanese encephalitis: past, present, and future prospects. Ther Clin Risk Manag. 2015;11:435–448. doi: 10.2147/TCRM.S51168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu L, Liu J, Cheng G. Role of microRNAs in schistosomes and schistosomiasis. Front Cell Infect Microbiol. 2014;4:165. doi: 10.3389/fcimb.2014.00165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ren Z, Ambros VR. Caenorhabditis elegans microRNAs of the let-7 family act in innate immune response circuits and confer robust developmental timing against pathogen stress. Proc Natl Acad Sci U S A. 2015;112(18):E2366–E2375. doi: 10.1073/pnas.1422858112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tang Q, Wu YQ, Chen DS, Zhou Q, Chen HC, Liu ZF. Bovine herpesvirus 5 encodes a unique pattern of microRNAs compared with bovine herpesvirus 1. J Gen Virol. 2014;95(pt 3):671–678. doi: 10.1099/vir.0.061093-0. [DOI] [PubMed] [Google Scholar]
- 6.Thum T. MicroRNA therapeutics in cardiovascular medicine. EMBO Mol Med. 2012;4(1):3–14. doi: 10.1002/emmm.201100191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fadnis PR, Ravi V, Desai A, Turtle L, Solomon T. Innate immune mechanisms in Japanese encephalitis virus infection: effect on transcription of pattern recognition receptors in mouse neuronal cells and brain tissue. Viral Immunol. 2013;26(6):366–377. doi: 10.1089/vim.2013.0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Uchil PD, Satchidanandam V. Phylogenetic analysis of Japanese encephalitis virus: envelope gene based analysis reveals a fifth genotype, geographic clustering, and multiple introductions of the virus into the Indian subcontinent. Am J Trop Med Hyg. 2001;65(3):242–251. doi: 10.4269/ajtmh.2001.65.242. [DOI] [PubMed] [Google Scholar]
- 9.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 10.Zhu G, Li J, He L, Wang X, Hong X. MPTP-induced changes in hippocampal synaptic plasticity and memory are prevented by memantine through the BDNF-TrkB pathway. Br J Pharmacol. 2015;172(9):2354–2368. doi: 10.1111/bph.13061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pogribny IP, Beland FA, Rusyn I. The role of microRNAs in the development and progression of chemical-associated cancers. Toxicol Appl Pharmacol. 2015 Nov 24; doi: 10.1016/j.taap.2015.11.013. Epub. [DOI] [PubMed] [Google Scholar]
- 12.Takasaki S. Roles of microRNAs in cancers and development. Methods Mol Biol. 2015;1218:375–413. doi: 10.1007/978-1-4939-1538-5_24. [DOI] [PubMed] [Google Scholar]
- 13.Li C, Gao Y, Zhang K, et al. Multiple roles of microRNA-100 in human cancer and its therapeutic potential. Cell Physiol Biochem. 2015;37(6):2143–2159. doi: 10.1159/000438572. [DOI] [PubMed] [Google Scholar]
- 14.Lv LY, Wang YZ, Zhang Q, Zang HR, Wang XJ. miR-539 induces cell cycle arrest in nasopharyngeal carcinoma by targeting cyclin-dependent kinase 4. Cell Biochem Funct. 2015;33(8):534–540. doi: 10.1002/cbf.3152. [DOI] [PubMed] [Google Scholar]
- 15.Bidarimath M, Khalaj K, Wessels JM, Tayade C. MicroRNAs, immune cells and pregnancy. Cell Mol Immunol. 2014;11(6):538–547. doi: 10.1038/cmi.2014.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaul V, Krams S. MicroRNAs as master regulators of immune responses in transplant recipients. Curr Opin Organ Transplant. 2015;20(1):29–36. doi: 10.1097/MOT.0000000000000148. [DOI] [PubMed] [Google Scholar]
- 17.Sonkoly E, Wei T, Janson PC, et al. MicroRNAs: novel regulators involved in the pathogenesis of psoriasis? PLoS One. 2007;2(7):e610. doi: 10.1371/journal.pone.0000610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O’Connell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc Natl Acad Sci U S A. 2007;104(5):1604–1609. doi: 10.1073/pnas.0610731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goswami S, Banerjee A, Kumari B, et al. Differential expression and significance of circulating microRNAs in cerebrospinal fluid of acute encephalitis patients infected with Japanese encephalitis virus. Mol Neurobiol. 2016 Feb 9; doi: 10.1007/s12035-016-9764-y. Epub. [DOI] [PubMed] [Google Scholar]
- 20.Thounaojam MC, Kundu K, Kaushik DK, et al. MicroRNA 155 regulates Japanese encephalitis virus-induced inflammatory response by targeting Src homology 2-containing inositol phosphatase 1. J Virol. 2014;88(9):4798–4810. doi: 10.1128/JVI.02979-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang YL, Ho BC, Sher S, Yu SL, Yang PC. miR-146a and miR-370 coordinate enterovirus 71-induced cell apoptosis through targeting SOS1 and GADD45beta. Cell Microbiol. 2015;17(6):802–818. doi: 10.1111/cmi.12401. [DOI] [PubMed] [Google Scholar]
- 22.Wu Z, Sun H, Zeng W, He J, Mao X. Upregulation of MircoRNA-370 induces proliferation in human prostate cancer cells by downregulating the transcription factor FOXO1. PLoS One. 2012;7(9):e45825. doi: 10.1371/journal.pone.0045825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu WP, Yi M, Li QQ, et al. Perturbation of microRNA-370/Lin-28 homolog A/nuclear factor kappa B regulatory circuit contributes to the development of hepatocellular carcinoma. Hepatology. 2013;58(6):1977–1991. doi: 10.1002/hep.26541. [DOI] [PubMed] [Google Scholar]