Abstract
Predictions of one-dimensional protein structures such as secondary structures and contact numbers are useful for predicting three-dimensional structure and important for understanding the sequence-structure relationship. Here we present a new machine-learning method, critical random networks (CRNs), for predicting one-dimensional structures, and apply it, with position-specific scoring matrices, to the prediction of secondary structures (SS), contact numbers (CN), and residue-wise contact orders (RWCO). The present method achieves, on average, Q3 accuracy of 77.8% for SS, and correlation coefficients of 0.726 and 0.601 for CN and RWCO, respectively. The accuracy of the SS prediction is comparable to that obtained with other state-of-the-art methods, and accuracy of the CN prediction is a significant improvement over that with previous methods. We give a detailed formulation of the critical random networks-based prediction scheme, and examine the context-dependence of prediction accuracies. In order to study the nonlinear and multi-body effects, we compare the CRNs-based method with a purely linear method based on position-specific scoring matrices. Although not superior to the CRNs-based method, the surprisingly good accuracy achieved by the linear method highlights the difficulty in extracting structural features of higher order from an amino acid sequence beyond the information provided by the position-specific scoring matrices.
Keywords: protein structure prediction, one-dimensional structure, position-specific scoring matrix, critical random network
Predicting the three-dimensional structure of a protein from its amino acid sequence is an essential step toward achieving a thorough bottom-up understanding of complex biological phenomena. Recently, much progress has been made in developing so-called ab initio or de novo structure prediction methods1. In the standard approach to de novo structure predictions, a protein is represented as a physical object in three-dimensional (3D) space, and the global minimum of free energy surface is sought with a given force-field or a set of scoring functions. In the minimization process, structural features predicted from the amino acid sequence may be used as restraints to limit the conformational space to be sampled. Such structural features include so-called one-dimensional (1D) structures of proteins.
Protein 1D structures are 3D structural features projected onto strings of residue-wise structural assignments along the amino acid sequence2. For example, a string of secondary structures is a 1D structure. Other 1D structures include (solvent) accessibilities3, contact numbers4 and recently introduced residue-wise contact orders5. The contact number, also referred to as the coordination number or Ooi number6, of a residue is the number of contacts that the residue makes with other residues in the native 3D structure, while the residue-wise contact order of a residue is the sum of sequence separations between that residue and contacting residues. We have recently shown that it is possible to reconstruct the native 3D structure of a protein from a set of three types of native 1D structures, namely secondary structures (SS), contact numbers (CN), and residue-wise contact orders (RWCO)5. Therefore, these 1D structures contain rich information regarding the corresponding 3D structure, and their accurate prediction may be very helpful for predicting 3D structure.
Previously, we have developed a simple linear method of predicting contact numbers from the amino acid sequence4. Use of a multiple sequence alignment was shown to improve the accuracy of prediction, achieving an average correlation coefficient of 0.63 between predicted and observed contact numbers per protein. There, we used an amino acid frequency table obtained from the HSSP7 multiple sequence alignment.
In this paper, we build on the previous method by introducing a new framework called critical random networks (CRNs), and apply it to the prediction of secondary structures and residue-wise contact orders in addition to contact numbers. In this framework, a state vector of large dimension is associated with each site of a target sequence. The state vectors are connected via random nearest-neighbor interactions. The value of the state vector is determined by solving an equation of state. Then a 1D quantity of each site is predicted as a linear function of the state vector of the site as well as the corresponding local PSSM segment. This approach was inspired by the method of echo state networks (ESNs) which has been recently developed and successfully applied to complex time series analysis8,9. Unlike ESNs which treat an infinite series of input signals in one direction (from the past to the future), CRNs treat finite systems incorporating both up- and downstream information at the same time. Also, the so-called echo state property is not imposed on a network, but the system is instead set at a critical point of the network. As the input for the CRNs-based prediction, we employ position-specific scoring matrices (PSSMs) generated by PSI-BLAST10. With the combination of PSSMs and CRNs, accurate predictions of SS, CN and RWCO have been achieved.
Currently, almost all the accurate methods for one-dimensional structure predictions combine some kind of sophisticated machine-learning approach such as neural networks and support vector machines with PSSMs. The method presented here is no exception. This trend raises the question as to what extent the machine-learning approaches are effective. In this study, we address this issue by comparing the CRNs-based method with a purely linear method based on PSSMs. Although not as good as the CRNs-based method, the linear predictions are of surprisingly high quality. This result suggests that, although not insignificant, the effect of the machine-learning approaches is relatively of minor importance while the use of PSSMs is the most significant ingredient in one-dimensional structure prediction. The problem of how to effectively extract meaningful information from the amino acid sequence beyond that provided by PSSMs requires yet further study.
Materials and methods
Definition of 1D structures
Secondary structures (SS).
Secondary structures were defined by the DSSP program11. For three-state SS prediction, the simple encoding scheme was employed. That is, α helices (H), β strands (E), and other structures (“coils”) defined by DSSP were encoded as H, E, and C, respectively. For SS prediction, we introduce feature variables (yiH, yiE, yiC) to represent each type of secondary structure at the i-th residue position, so that H is represented as (1, −1, −1), E as (−1, 1, −1), and C as (−1, −1, 1).
Contact numbers (CN).
Let Ci,j represent the contact map of a protein. Usually, the contact map is defined so that Ci,j= 1 if the i-th and j-th residues are in contact by some definition, or Ci,j= 0, otherwise. As in our previous study, we slightly modify the definition using a sigmoid function. That is,
| (1) |
where ri,j is the distance between Cβ (Cα for glycines) atoms of the i-th and j-th residues, d=12 Å is a cutoff distance, and w is a sharpness parameter of the sigmoid function which is set to 34,5. The rather generous cutoff length of 12 Å was shown to optimize the prediction accuracy4. The use of the sigmoid function enables us to use the contact numbers in molecular dynamic simulations5. Using the above definition of the contact map, the contact number of the i-th residue of a protein is defined as
| (2) |
The feature variable yi for CN is defined as yi= ni/log L where L is the sequence length of a target protein. The normalization factor log L is introduced because we have observed that the contact number averaged over a protein chain is roughly proportional to log L, and thus division by this value removes the size-dependence of predicted contact numbers.
Residue-wise contact orders (RWCO).
RWCOs were first introduced in Kinjo and Nishikawa5. Using the same notation as for contact numbers (see above), the RWCO of the i-th residue in a protein structure is defined by
| (3) |
The feature variable yi for RWCO is defined as yi= oi/L where L is the sequence length. Due to a similar reason as for CN, the normalization factor L was introduced to remove the size-dependence of the predicted RWCOs (the RWCO averaged over a protein chain is roughly proportional to the chain length).
Linear regression scheme
The input to the prediction scheme we develop in this paper is a position-specific scoring matrix (PSSM) of the amino acid sequence of a target protein. Let us denote the PSSM by U=(u1, ..., uL) where L is the sequence length of the target protein and ui is a 20-vector containing the scores of 20 types of amino acid residues at the i-th position: ui= (u1,i, ..., u20,i)t.
When predicting a type of 1D structure, we first predict the feature variable(s) for that type of 1D structure [i.e., yi= yiH, etc. for SS, ni/log L for CN, and oi/L for RWCO], and then the feature variable is converted to the target 1D structure. Prediction of the feature variable yi can be considered as a mapping from a given PSSM U to yi. More formally, we are going to establish the functional form of the mapping F in ŷi =F(U, i) where ŷi is the predicted value of the feature variable yi. In our previous paper, we showed that CN can be predicted to a moderate degree of accuracy by a simple linear regression scheme with a local sequence window4. Accordingly, we assume that the function F can be decomposed into linear (Fl) and nonlinear (Fn) parts: F = Fl+ Fn.
The linear part is expressed as
| (4) |
where M is the half window size of the local PSSM segment around the i-th residue, and {Dm,a} are the weights to be trained. To treat N- and C-termini separately, we introduced the “terminal residue” as the 21st kind of amino acid residue. The value of u21,i+m is set to unity if i+m<0 or i+m<L, or to zero otherwise. The “terminal residue” for the central residue (m=0) serves as a bias term and is always set to unity.
To establish the nonlinear part, we first introduce an N-dimensional “state vector” xi= (x1,i, ..., xN,i)t for the i-th sequence position where the dimension N is a free parameter. The value of xi is determined by solving the equation of state which is described in the next subsection. For the moment, let us assume that the equation of state has been solved, and denote the solution by . The state vector can be considered as a function of the whole PSSM U (i.e., ), and implicitly incorporates nonlinear and long-range effects. Now, the nonlinear part Fn is expressed as a linear projection of the state vector:
| (5) |
where {Ek} are the weights to be trained.
In summary, the prediction scheme is expressed as
| (6) |
Regarding ui–M, ..., ui+M and as independent variables, Eq. 6 reduces to a simple linear regression problem for which the optimal weights {Dm,a} and {Ek} are readily determined by using a least squares method. For CN or RWCO predictions, the predicted feature variable can be easily converted to the corresponding 1D quantities by multiplying by log L or L, respectively. For SS prediction, the secondary structure ŝi of the i-th residue is given by .
Critical random networks and the equation of state
We now describe the equation of state for the system of state vectors. We denote L state vectors along the amino acid sequence by X=(x1, ..., xL) ∈ RN×L, and define a nonlinear mapping gi: RN×L → RN for i=1, ..., L by
| (7) |
where β and α are positive constants, W is an N×N block-diagonal orthogonal random matrix, and V is an N×21 random matrix (a unit bias term is assumed in ui). The hyperbolic tangent function (tanh) is applied element-wise. We impose the boundary conditions as x0= xL+1= 0. In this equation, the term containing W represents nearest-neighbor interactions along the sequence. The amino acid sequence information is taken into account as an external field in the form of αVui. Next we define a mapping G: RN×L → RN×L by
| (8) |
Using this mapping G, the equation of state is defined as
| (9) |
That is, the state vectors are determined as a fixed point of the mapping G. More explicitly, Eq. 9 can be expressed as
| (10) |
for i=1, ..., L. That is, the state vector xi of the site i is determined by the interaction with the state vectors of the neighboring sites i-1 and i+1 as well as with the ‘external field’ ui of the site. The information on the external field at each site is propagated throughout the entire amino acid sequence via the nearest-neighbor interactions. Therefore, solving Eq. (10) means finding the state vectors that are consistent with the external field as well as the nearest-neighbor interactions, and each state vector in the obtained solution {xi} self-consistently embodies the information on the entire amino acid sequence in a mean-field sense.
For β<0.5, it can be shown that G is a contraction mapping in RN×L (with an appropriate norm defined therein). And hence, by the contraction mapping principle12, the mapping G has a unique fixed point independently of the strength α of the external field. When β is sufficiently smaller than 0.5, the correlation between two state vectors, say xi and xj, is expected to decay exponentially as a function of the sequential separation |i−j|. On the other hand, for β >0.5, the number of fixed points varies depending on the strength of the external field α. In this regime, we cannot reliably solve the equation of state (Eq. 9). In this sense, β=0.5 can be considered as a critical point of the system X. From an analogy with critical phenomena of physical systems13 (note the formal similarity of Eq. 10 with the mean field equation of the Ising model), the correlation length between state vectors is expected to diverge, or become long when the external field is finite but small. We call the system defined by Eq. 10 with β=0.5 a critical random network (CRN).
The equation of state (Eq. 10) is parameterized by two random matrices W and V, and consequently, so is the predicted feature variables ŷi. Following a standard technique of statistical learning such as neural networks14, we may improve the prediction accuracy by averaging ŷi obtained by multiple CRNs with different pairs of W and V. This averaging operation reduces the prediction errors due to the random fluctuations in the estimated parameters. We employ such an ensemble prediction with 10 sets of random matrices W and V in the following. The use of a larger number of random matrices for ensemble predictions improved the prediction accuracies slightly, but the difference was insignificant.
Numerics
Here we describe the value of the free parameters used, and a numerical procedure to solve the equation of state.
The half window size M in the linear part of Eq. 6 is set to 9 for SS and CN predictions, and to 26 for RWCO predictions. These values were found to be optimal in preliminary studies4,15. Regarding the dimension N of the state vector, we have found that N=2000 gives the best result after some experimentation, and this value is used throughout. Using a state vector with a dimension as large as 2000, it is expected that various properties of amino acid sequences can be extracted and memorized. If the dimension is too large, overfitting may occur, but we did not find such a case up to N=2000. Therefore, in principle, the state vector dimension could be even larger (but the computational cost becomes a problem).
Each element in the N×21 random matrix V in Eq. 10 is obtained from a uniform distribution in the range [−1, 1] and the strength parameter α is set to 0.01. Here and in the following, all random numbers were generated by the Mersenne twister algorithm16. The N×N random matrix W is obtained in the following manner. First we generate a random block diagonal matrix A whose block sizes are drawn from a uniform distribution of integers 2 to 20 (both inclusive), and the values of the block elements are drawn from the standard Gaussian distribution (zero mean and unit variance). By applying singular value decomposition, we have A=UΣVt where U and V are orthogonal matrices and Σ is a diagonal matrix of singular values. We set W=UVt which is orthogonal as well as block diagonal.
To solve the equation of state (Eq. 10), we use a simple functional iteration with a Gauss-Seidel-like updating scheme. Let ν denote the stage of iteration. We set the initial value of the state vectors (with ν =0) as
| (11) |
Then, for i=1, ..., L (in increasing order of i), we update the state vectors by
| (12) |
Next, we update them in the reverse order. That is, for i=L, ..., 1 (in decreasing order of i),
| (13) |
We then set ν← +1, and iterate Eqs. (12) and (13) until {xi} converges. The convergence criterion is
| (14) |
where ||·||RN denotes the Euclidean norm. Convergence is typically achieved within 100 to 200 iterations for one protein.
Preparation of training and test sets
We use the same set of proteins as used in our preliminary study15. In this set, there are 680 protein domains selected from the ASTRAL database17, each of which represents a superfamily from one of all-α, all-β, α/β, α+β or “multi-domain” classes of the SCOP database (release 1.65, December 2003)18. Conversely, each SCOP superfamily is represented by only one of the protein domains in the data set. Thus, no pair of protein domains in the data set are expected to be homologous to each other. For training the parameters and testing the prediction accuracy, 15-fold cross-validation is employed. The set of 680 proteins is randomly divided into two groups: one consisting of 630 proteins (training set), and the other consisting of 50 proteins (test set). For each training set, the regression parameters {Dm,a} and {Ei} are determined, and using these parameters, the prediction accuracy is evaluated for the corresponding test set. This procedure was repeated 15 times with different random divisions, leading to 15 pairs of training and test sets. In this way, there is some redundancy in the training and test sets although none of the pair of these sets share proteins in common. But this raises no problem since our objective is to estimate the average accuracy of the predictions. A similar validation procedure was also employed by Petersen et al.19 In total, 750 (=15×50) proteins were tested with which the averages of the measures of accuracy (see below) were calculated.
Preparation of a position-specific scoring matrix
To obtain the position-specific scoring matrix (PSSM) of a protein, we conducted ten iterations of a PSI-BLAST10 search of a customized sequence database with an E-value cutoff of 0.000520. The sequence database was compiled from the DAD database provided by the DNA Data Bank of Japan21, from which redundancy was removed by the program CD-HIT22 with a 95% identity cutoff. This database was subsequently filtered by the program PFILT used in the PSIPRED program23. We use the position-specific scoring matrices (PSSM) rather than the frequency tables for the prediction.
Measures of accuracy
For assessing the quality of SS predictions, we mainly use Q3 and SOV (the 1999 revision)24. The Q3 measure quantifies the percentage of correctly predicted residues, while the SOV measure evaluates the segment overlaps of secondary structural elements of predicted and native structures. Optionally, we use Qs and (with s being H, E, or C) and Matthews’ correlation coefficient MC. The Qs is defined by the percentage of correctly predicted SS type s out of the native SS type s, and is defined by the percentage of correctly predicted SS type s out of the predicted SS type s.
For CN and RWCO predictions, we use two measures for evaluating the prediction accuracy. The first one is the coefficient of the correlation (Cor) between the observed (ni) and predicted (n̂i) CN or RWCO4. The second is the RMS error normalized to the standard deviation of the native CN or RWCO (Dev A)4. While Cor measures the quality of relative values, Dev A measures that of absolute values of the predicted CN or RWCO.
Note that the measures Q3, SOV, Cor and Dev A are defined for a single protein chain. In practice, we average these quantities for all the proteins in the test sets to estimate the average accuracy of prediction. On the other hand, per-residue measures, Qs, and MC, were calculated using all the residues in the test data sets, rather than on a per-protein basis.
Results
We examine the prediction accuracies for SS, CN, and RWCO in turn. The main results are summarized in Table 1 and Figure 1. Finally, in order to examine the effect of nonlinear terms, we verify the results obtained using only linear terms (Eq. 4).
Table 1.
Summary of average prediction accuracies
| Struct. | Accuracy |
|---|---|
| SS | Q3= 77.8; SOV=77.3 |
| CN | Cor=0.726; Dev A=0.707 |
| RWCO | Cor=0.601; Dev A=0.881 |
Figure 1.
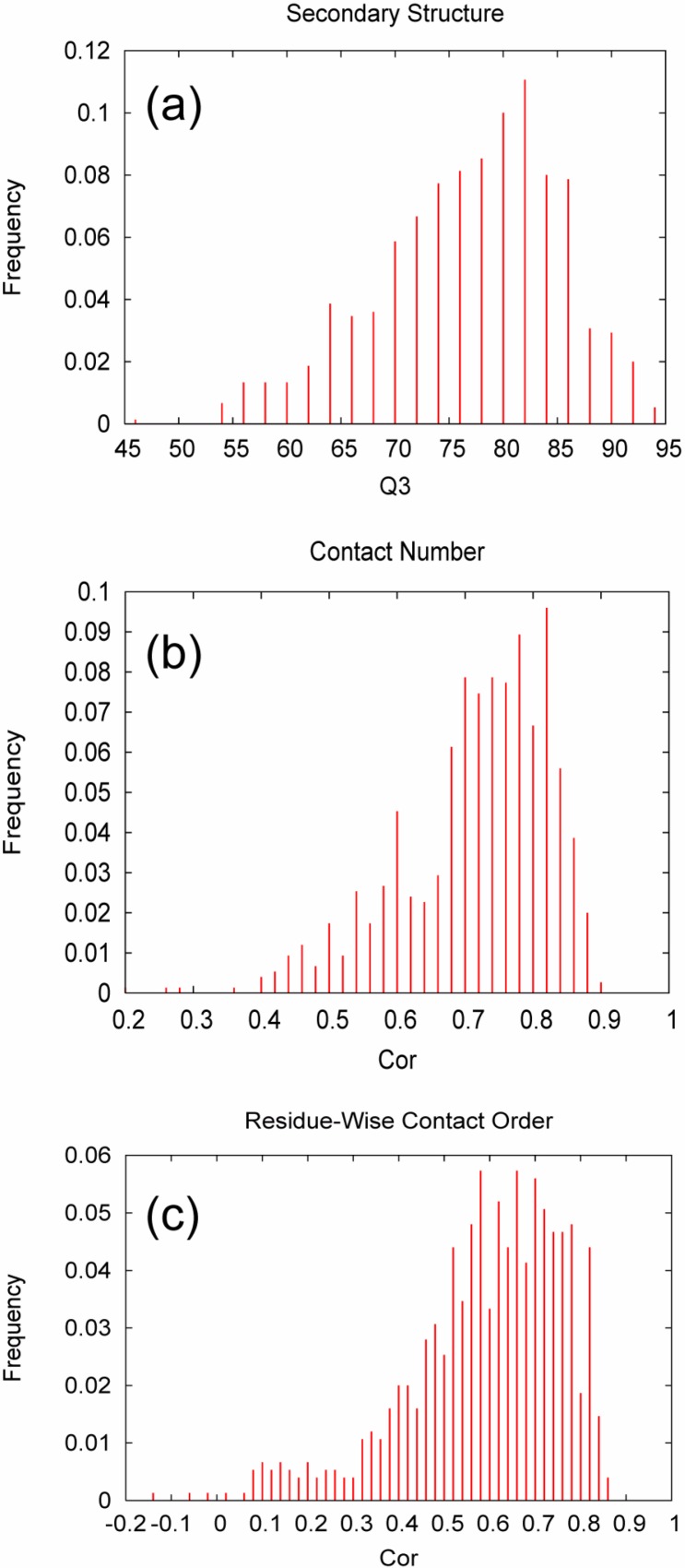
Histograms of accuracy measures obtained by ensemble predictions using 10 critical random networks. (a) Q3 for secondary structure prediction; (b) Cor for contact number prediction; (c) Cor for residue-wise contact order prediction.
Secondary structure prediction
The average accuracy of predicting secondary structures achieved by the ensemble CRNs-based approach is Q3= 77.8% and SOV=77.3 (Table 1). This is comparable to the current state-of-the-art predictors such as PSIPRED23. The results in terms of per-residue accuracies (Qs and ) are listed in Table 2. The values of Qs suggest that the present method underestimates α helices (H) and, especially, β strands (E) compared to coils C. However, when a residue is predicted as being H or E, the probability of the correct prediction is rather high, especially for E (
Table 2.
Summary of per-residue accuracies for SS predictions
| measure | H | E | C |
|---|---|---|---|
| Qs | 78.4 | 61.9 | 84.6 |
| 81.9 | 79.9 | 74.3 | |
| MC | 0.704 | 0.636 | 0.602 |
). The histogram of Q3 (Fig. 1a) shows that the peak of the histogram resides well beyond Q3= 80%, and that only 20% of the predictions exhibit a Q3 of less than 70%. These observations demonstrate the capability of the CRNs-based prediction schemes.
Contact number prediction
Using an ensemble of CRNs, a correlation coefficient (Cor) of 0.726 and normalized RMS error (Dev A) of 0.707 was achieved for CN predictions on average (Table 1). This result is a significant improvement over the previous method4 which yielded Cor=0.627 and Dev A=0.941. The median of the distribution of Cor (Fig. 1b) is 0.744, indicating that the majority of the predictions are of very high accuracy.
We have also examined the dependence of prediction accuracy on the structural class of target proteins (Table 3). Among all the structural classes, α/β proteins are predicted most accurately with Cor=0.757 and Dev A=0.668. The accuracy for other classes does not differ qualitatively although all-β proteins are predicted slightly less accurately.
Table 3.
Summary of CN predictions for each SCOP classa
| rangeb (Cor) | SCOP classc | ||||
|---|---|---|---|---|---|
|
| |||||
| a | b | c | d | e | |
| (−1, 0.5] | 8 | 6 | 3 | 14 | 1 |
| (0.5, 0.6] | 19 | 25 | 8 | 19 | 1 |
| (0.6, 0.7] | 29 | 29 | 22 | 54 | 3 |
| (0.7, 0.8] | 62 | 66 | 76 | 85 | 10 |
| (0.8, 0.9] | 43 | 38 | 57 | 67 | 3 |
| (0.9, 1.0] | 1 | 0 | 0 | 1 | 0 |
| total | 162 | 164 | 166 | 240 | 18 |
|
| |||||
| average Cor | 0.721 | 0.712 | 0.757 | 0.728 | 0.722 |
| average Dev A | 0.715 | 0.726 | 0.668 | 0.717 | 0.705 |
The number of occurrences of Cor for the proteins in the test sets, classified according to the SCOP database; average values of Cor and Dev A are also listed for each class.
The range “(x, y]” denotes x<Cor≤y.
a: all-α; b: all-β; c: α/β; d: α+β; e: multi-domain.
Residue-wise contact order prediction
For predicting RWCO, the average accuracy was such that Cor=0.601 and Dev A=0.881. Although these figures appear to be poor compared to those of the CN prediction described above, they are yet statistically significant. The distribution of Cor appears to be rather dispersed (Fig. 1c), indicating that the prediction accuracy strongly depends on the characteristics of each target protein. In a similar manner as for CN, we also examined the dependence of prediction accuracy on the structural class of target proteins (Table 4). In this case, we have found a notable dependence of accuracy on structural class. The best accuracy is obtained for α+β proteins with Cor=0.629 and Dev A=0.832. For these proteins, the distribution of Cor also shows a good tendency in that the fraction of poor predictions is relatively small (e.g., 14% for Cor<0.5). Interestingly, all-β proteins also show good accuracies but all-α proteins are particularly poorly predicted. These observations suggest that the correlation between the amino acid sequence and RWCO is strongly dependent on the structural class of the target protein. However, the rather dispersed distribution of Cor for each class (Table 4) also suggests that there are more detailed effects of the global context on the accuracy of RWCO prediction.
Table 4.
Summary of RWCO predictions for each SCOP classa
| range (Cor) | SCOP class | ||||
|---|---|---|---|---|---|
|
| |||||
| a | b | c | d | e | |
| (−1, 0.5] | 58 | 31 | 46 | 34 | 6 |
| (0.5, 0.6] | 29 | 37 | 31 | 56 | 4 |
| (0.6, 0.7] | 41 | 27 | 33 | 65 | 5 |
| (0.7, 0.8] | 24 | 47 | 40 | 72 | 3 |
| (0.8, 0.9] | 10 | 22 | 16 | 13 | 0 |
| total | 162 | 164 | 166 | 240 | 18 |
|
| |||||
| average Cor | 0.549 | 0.620 | 0.595 | 0.629 | 0.564 |
| average Dev A | 0.981 | 0.869 | 0.857 | 0.832 | 0.957 |
See Table 3 for notations.
Purely linear predictions with PSSMs
Almost all the modern methods for predicting 1D structures make use of PSSMs in combination with some kind of machine-learning technique such as feed-forward or recurrent neural networks or support vector machines. The present study is no exception. Curiously, machine-learning approaches have become so widespread that no attempt appears to have been made to test simplest linear predictors based on PSSMs. In this subsection, we present results of 1D predictions using only the linear terms (Eq. 4) but without CRNs. In this prediction scheme, the input is a local segment of a PSSM generated by PSI-BLAST, and a feature variable is predicted by a straight forward linear regression.
As can be clearly seen in Table 5, the results of the linear predictions are surprisingly good although not as good as with CRNs. For example, in SS prediction, the purely linear scheme achieved Q3 = 75.2% which is lower than that of the CRNs-based scheme by only 3.6 percentage points. Although this is of course a large difference in a statistical sense, there may not be a discernible difference as far as individual predictions are concerned. (However, the improvement in the SOV measure obtained by using CRNs is quite large, indicating that the nonlinear terms in CRNs are indeed able to extract cooperative features.) It is widely accepted that the upper limit of accuracy (Q3) of SS prediction based on a local window of a single sequence is less than 70%25. Therefore, more than 5 percentage points of the increase in Q3 is brought about simply by the use of PSSMs.
Table 5.
Summary of prediction accuracies using only linear terms
| Struct. | Accuracy |
|---|---|
| SS | Q3= 75.2; SOV=72.7 |
| CN | Cor=0.701; Dev A=0.735 |
| RWCO | Cor=0.584; Dev A=0.902 |
Similar observations also hold for CN and RWCO predictions (Table 5). In the case of CN prediction, we have previously obtained Cor=0.555 by a simple linear method with single sequences4. Therefore, the effect of PSSMs is even more dramatic than SS prediction. This may be due to the fact that the most conspicuous feature of amino acid sequences conserved among distant homologs (as detected by PSI-BLAST) is the hydrophobicity of amino acid residues26, which is closely related to contact numbers. Of course, the improvement with the use of PSSMs is largely made possible by the recent increase in amino acid sequence databases27.
The significance of criticality
The condition of criticality (β=0.5 in Eq. 10) is expected to enhance the extraction of the long-range correlations of an amino acid sequence, thus improving the prediction accuracy. To confirm this point, we tested the method by setting β=0.1 so that the network of state vectors is not at the critical point any more (otherwise the prediction and validation schemes were the same as above). The prediction accuracies obtained by these non-critical random networks were Q3= 76.7% and SOV=76.6 for SS, Cor=0.716 and Dev A=0.719 for CN, and Cor=0.589 and Dev A=0.897 for RWCO. These values are inferior to those obtained by the critical random networks (Table 1), although slightly better than the purely linear predictions (Table 5). Therefore, compared to the non-critical random networks, the critical random networks can indeed extract more information from the amino acid sequence and improve the prediction accuracies.
Discussion
Comparison with other methods
Regarding the framework of 1D structure prediction, the critical random networks are most closely related to bidirectional recurrent neural networks (BRNNs)28, in that both can treat an entire amino acid sequence rather than only a local window segment. The main differences are the following. First, network weights between input and hidden layers as well as those between hidden units are trained in BRNNs, whereas the corresponding weights in CRNs (random matrices V and W, respectively, in Eq. 10) are fixed. Second, the output layer is nonlinear in BRNNs but linear in CRNs. Third, the network components that propagate sequence information from the N-terminus to C-terminus are decoupled from those in the opposite direction in BRNNs, but they are coupled in CRNs.
Regarding the accuracy of SS prediction, BRNNs29 and CRNs exhibit comparable results of Q3≈ 78%. However, a standard local window-based approach using feed-forward neural networks can also achieve this level of accuracy23. Thus, the CRNs-based method is not a single best predictor, but may serve as an addition to consensus predictions.
Although BRNNs have been also applied to CN prediction30, contact numbers are predicted as 2-state categorical data (buried or exposed) so the results cannot be directly compared. Nevertheless, we can convert CRNs-based real-value predictions into 2-state predictions. By using the same thresholds for the 2-state discretization as Pollastri et al.30 (i.e., the average CN for each residue type), we obtained Q2 = 75.6% per chain (75.1% per residue), and Matthews’ correlation coefficient MC=0.503 whereas those obtained by BRNNs are Q2= 73.9% (per residue) and MC= 0.478. Therefore, for 2-state CN prediction, the present method yields more accurate results.
Since the present study is the very first attempt to predict RWCOs, there are no alternative methods to compare with. However, the comparison of CRNs-based methods for SS and CN predictions with other methods suggests that the accuracy of the RWCO prediction presented here may be the best possible result using any of the statistical learning methods currently available for predicting 1D structures.
Possibilities for further improvements
In the present study, we employed the simplest possible architecture for CRNs in which different sites are connected via nearest-neighbor interactions. A number of possibilities exist for the elaboration of the architecture. For example, we may introduce short-cuts between distant sites to treat non-local interactions more directly. Since the prediction accuracies depend on the structural context of target proteins (Tables 3 and 4), it may be also useful to include more global features of amino acid sequences such as the bias of amino acid composition or the average of PSSM components. These possibilities are to be pursued in future studies.
Conclusion
We have developed a novel method, CRNs-based regression, for predicting 1D protein structures from the amino acid sequence. When combined with position-specific scoring matrices produced by PSI-BLAST, this method yields SS predictions as accurate as the best current predictors, CN predictions far better than previous methods, and RWCO predictions significantly correlated with observed values. We also examined the effect of PSSMs on prediction accuracy, and showed that most improvement is brought by the use of PSSMs although the further improvement due to the CRNs-based method is also significant. In order to achieve qualitatively better predictions, however, it seems necessary to take into account other, more global, information than is provided by PSSMs.
Acknowledgments
The authors thank Motonori Ota for critical comments on an early version of the manuscript, and Kentaro Tomii for advice on the use of PSI-BLAST. Most of the computations were carried out at the supercomputing facility of the National Institute of Genetics, Japan. This work was supported in part by a grant-in-aid from MEXT, Japan. The source code of the programs for the CRNs-based prediction as well as the lists of protein domains used in this study are available at http://maccl01.genes.nig.ac.jp/~akinjo/crnpred_suppl/.
References
- 1.Bonneau R, Baker D. Ab initio protein structure prediction: progress and prospects. Annu. Rev. Biophys. Biomol. Struct. 2001;30:173–189. doi: 10.1146/annurev.biophys.30.1.173. [DOI] [PubMed] [Google Scholar]
- 2.Rost B. Prediction in 1D: secondary structure, membrane helices, and accessibility. In: Bourne PE, Weissig H, editors. Structural Bioinformatics. Wiley-Liss, Inc.; Hoboken, U.S.A.: 2003. pp. 559–587. chapter 28. [PubMed] [Google Scholar]
- 3.Lee B, Richards FM. The interpretation of protein structures: Estimation of static accessibility. J. Mol. Biol. 1971;55:379–400. doi: 10.1016/0022-2836(71)90324-x. [DOI] [PubMed] [Google Scholar]
- 4.Kinjo AR, Horimoto K, Nishikawa K. Predicting absolute contact numbers of native protein structure from amino acid sequence. Proteins. 2005;58:158–165. doi: 10.1002/prot.20300. [DOI] [PubMed] [Google Scholar]
- 5.Kinjo AR, Nishikawa K. Recoverable one-dimensional encoding of protein three-dimensional structures. Bioinformatics. 2005;21:2167–2170. doi: 10.1093/bioinformatics/bti330. [DOI] [PubMed] [Google Scholar]
- 6.Nishikawa K, Ooi T. Prediction of the surface-interior diagram of globular proteins by an empirical method. Int. J. Peptide Protein Res. 1980;16:19–32. doi: 10.1111/j.1399-3011.1980.tb02931.x. [DOI] [PubMed] [Google Scholar]
- 7.Sander C, Schneider R. Database of homology-derived protein structures. Proteins. 1991;9:56–68. doi: 10.1002/prot.340090107. [DOI] [PubMed] [Google Scholar]
- 8.Jaeger H. The “echo state” approach to analysing and training recurrent neural networks. 2001. Technical Report 148, GMD — German National Research Institute for Computer Science.
- 9.Jaeger H, Haas H. Harnessing nonlinearity: predicting chaotic systems and saving energy in wireless communication. Science. 2004;304:78–80. doi: 10.1126/science.1091277. [DOI] [PubMed] [Google Scholar]
- 10.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DL. Gapped blast and PSI-blast: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kabsch W, Sander C. Dictionary of protein secondary structure: Pattern recognition of hydrogen bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 12.Takahashi W. Nonlinear Functional Analysis: Fixed Point Theorems and Related Topics. Kindai Kagaku Sha; Tokyo: 1988. (in Japanese). [Google Scholar]
- 13.Goldenfeld N. Lectures on phase transitions and the renormalization group, volume 85 of Frontiers in Physics. Addison-Wesley, Reading; Massachusetts: 1992. [Google Scholar]
- 14.Haykin S. Neural Networks: A Comprehensive Foundation. 2nd ed. Prentice-Hall, Upper Saddle River; New Jersey: 1999. [Google Scholar]
- 15.Kinjo AR, Nishikawa K. Predicting residue-wise contact orders of native protein structure from amino acid sequence. 2005. arXiv.org:q-bio.BM/0501015. [DOI] [PMC free article] [PubMed]
- 16.Matsumoto M, Nishimura T. Mersenne twister: a 623-dimensionally equidistributed uniform pseudorandom number generator. ACM Trans. Model. Comput. Simul. 1998;8:3–30. [Google Scholar]
- 17.Chandonia JM, Hon G, Walker NS, Lo Conte L, Koehl P, Levitt M, Brenner SE. The astral compendium in 2004. Nucleic Acids Res. 2004;32:D189–D192. doi: 10.1093/nar/gkh034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: A structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 19.Petersen TN, Lundegaard C, Nielsen M, Bohr H, Bohr J, Brunak S, Gippert GP, Lund O. Prediction of protein secondary structure at 80% accuracy. Proteins. 2000;41:17–20. [PubMed] [Google Scholar]
- 20.Tomii K, Akiyama Y. FORTE: a profile-profile comparison tool for protein fold recognition. Bioinformatics. 2004;20:594–595. doi: 10.1093/bioinformatics/btg474. [DOI] [PubMed] [Google Scholar]
- 21.Tateno Y, Saitou N, Okubo K, Sugawara H, Gojobori T. DDBJ in collaboration with mass-sequencing teams on annotation. Nucleic Acids Res. 2005;33:D25–D28. doi: 10.1093/nar/gki020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li W, Jaroszewski L, Godzik A. Tolerating some redundancy significantly speeds up clustering of large protein databases. Bioinformatics. 2002;18:77–82. doi: 10.1093/bioinformatics/18.1.77. [DOI] [PubMed] [Google Scholar]
- 23.Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 24.Zemla A, Venclovas C, Fidelis K, Rost B. A modified definition of sov, a segment-based measure for protein secondary structure prediction assessment. Proteins. 1999;34:220–223. doi: 10.1002/(sici)1097-0134(19990201)34:2<220::aid-prot7>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 25.Crooks GE, Brenner SE. Protein secondary structure: entropy, correlations and prediction. Bioinformatics. 2004;20:1603–1611. doi: 10.1093/bioinformatics/bth132. [DOI] [PubMed] [Google Scholar]
- 26.Kinjo AR, Nishikawa K. Eigenvalue analysis of amino acid substitution matrices reveals a sharp transition of the mode of sequence conservation in proteins. Bioinformatics. 2004;20:2504–2508. doi: 10.1093/bioinformatics/bth297. [DOI] [PubMed] [Google Scholar]
- 27.Przybylski D, Rost B. Alignments grow, secondary structure prediction improves. Proteins. 2002;46:197–205. doi: 10.1002/prot.10029. [DOI] [PubMed] [Google Scholar]
- 28.Baldi P, Brunak S, Frasconi P, Soda G, Pollastri G. Exploiting the past and the future in protein secondary structure prediction. Bioinformatics. 1999;15:937–946. doi: 10.1093/bioinformatics/15.11.937. [DOI] [PubMed] [Google Scholar]
- 29.Pollastri G, Przybylski D, Rost B, Baldi P. Improving the prediction of protein secondary structure in three and eight classes using recurrent neural networks and profiles. Proteins. 2002;47:228–235. doi: 10.1002/prot.10082. [DOI] [PubMed] [Google Scholar]
- 30.Pollastri G, Baldi P, Fariselli P, Casadio R. Prediction of coordination number and relative solvent accessibility in proteins. Proteins. 2002;47:142–153. doi: 10.1002/prot.10069. [DOI] [PubMed] [Google Scholar]