Abstract
Flagellar motor proteins, MotA/B and PomA/B, are essential for the motility of Escherichia coli and Vibrio alginolyticus, respectively. Those complexes work as a H+ and a Na+ channel, respectively and play important roles in torque generation as the stators of the flagellar motors. Although Asp32 of MotB and Asp24 of PomB are believed to function as ion binding site(s), the ion flux pathway from the periplasm to the cytoplasm is still unclear. Conserved residues, Ala39 of MotB and Cys31 of PomB, are located on the same sides as Asp32 of MotB and Asp24 of PomB, respectively, in a helical wheel diagram. In this study, a series of mutations were introduced into the Ala39 residue of MotB and the Cys31 residue of PomB. The motility of mutant cells were markedly decreased as the volume of the side chain increased. The loss of function due to the MotB(A39V) and PomB(L28A/C31A) mutations was suppressed by mutations of MotA(M206S) and PomA(L183F), respectively, and the increase in the volume caused by the MotB(A39V) mutation was close to the decrease in the volume caused by the MotA(M206S) mutation. These results demonstrate that Ala39 of MotB and Cys31 of PomB form part of the ion flux pathway and pore with Met206 of MotA and Leu183 of PomA in the MotA/B and PomA/B stator units, respectively.
Keywords: membrane protein, ion flux, bacteria locomotion, motility, molecular motor, taxis
Flagellar motors are molecular machines powered by the electrochemical potential gradient of specific ions across the membrane1,2, and are classified by the respective coupling ion. For some bacteria, such as Escherichia coli and Salmonella enterica, the flagellar motor is driven by the H+ motive force, while the motors of alkalophilic Bacillus and marine Vibrio species are driven by the Na+ motive force1. Transmembrane proteins, MotA and MotB from E. coli3 and PomA and PomB from Vibrio alginolyticus, are essential for the motility of those bacteria1,4. In 2000, Sato and Homma purified the PomA/B complex and reconstituted it in liposomes5. Interestingly, those proteoliposomes were shown to have sodium-conducting activity in vitro5 and the complex was predicted to have a 4A:2B stoichiometry6. A cysteine-crosslinking study of MotB gave evidence that two MotB molecules are involved in each MotA/B complex7. Thus the complexes, MotA/B and PomA/B, work as H+ and Na+ channels, respectively, and play important roles in torque generation as the stators of the bacterial flagellar motors.
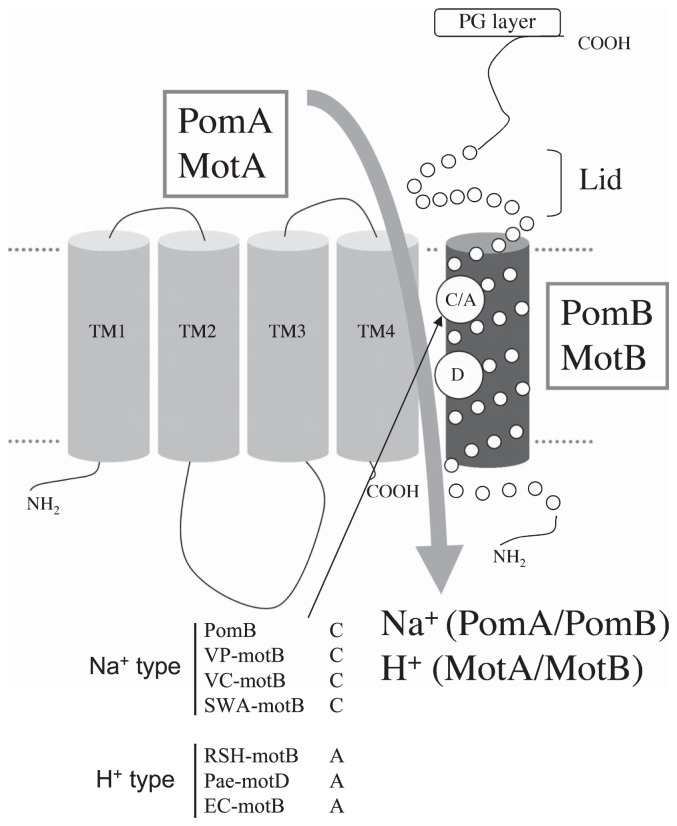
Blair and coworkers proposed an initial model of MotA4MotB2 (PomA4PomB2) organization8, in which a dimer of MotB transmembrane segments is located at the center of the complex. Transmembrane segments 3 (TM3) and 4 (TM4) of four MotA molecules are arranged in the inner layer around the MotB segments, and TM1 and TM2 are located on the outside. Thus TM3 and TM4 of MotA (PomA) are in close proximity to TM of MotB (PomB) (Figure 1). Only one carboxylic acid residue, Asp32 of MotB and Asp24 of PomB, exists in the predicted transmembrane regions of MotA/B and PomA/B, respectively (Figure 1). Because the D32N mutant of MotB9 and the D24C mutant of PomB10 do not mediate the flagellar motor rotation, those aspartic residues are believed to form a H+ binding site(s) of the MotA/B complex and a Na+ binding site(s) of the PomA/B complex. However, the ion flux pathways from the periplasm to the cytoplasm through these critical aspartate residues are still unclear. The ion flux mechanism of these protein complexes become a focus of great interest, in part because of its importance to the general understanding of the ion flux mechanism in membrane proteins, about which little is known.
Figure 1.
Putative membrane topology of stator proteins, PomA/B, MotA and MotB. Amino acid residues marked by a circle in the transmembrane helix of the B subunit are the sites of attention in this study. Abbreviations: VP, Vibrio parahaemolyticus; VC, Vibrio cholera; SWA, Shewanella oneidensis; RSH, Rhodobacter sphaeroides; Pae, Pseudomonas aeruginosa; EC, Escherichia coli.
To determine the residues which form the ion flux pathway( s) in the stator units of the H+ and Na+ driven flagellar motors, we have now analyzed the relationship between the motility of the cells and the volume of the side chains of amino acid residues that are predicted to be located inside the channel pore. Conserved residues, Cys31 in PomB and Ala39 in MotB10, are located on the same sides as Asp24 for PomB and Asp32 for MotB in a helical wheel diagram (Figure 1). In this study we introduced mutations at those sites, and measured the motility of cells containing the mutated proteins. We predicted that mutants having a bulky amino acid residue would hamper ion flux and disrupt the rotation of the flagellum depending on the volume of the amino acids introduced. In fact, it has been reported that the A39V mutant of MotB has a dominant and non-functional property11. In the present work, we compared the swimming speed against the volume of the amino acid side chain(s) between MotA/B and PomA/B. We conclude that Cys31 of PomB and Ala39 of MotB form ion flux pathways in the PomA/B and the MotA/B stator units of flagellar motors, respectively.
Materials and methods
Bacterial strains, plasmids, media and growth conditions
Escherichia coli strain DH5α or JM109 was used for DNA manipulations. The Vibrio alginolyticus strain NMB191 (ΔpomAB)12 and the E. coli strain RP6894 (ΔmotAB)13 were used in this study for the swimming assay. Mutant genes were constructed by PCR using the QuikChange site-directed mutagenesis method (Stratagene). All constructed plasmids were confirmed using an automated sequencer. V. alginolyticus cells were cultured at 30°C in VC medium (0.5% polypeptone, 0.5% yeast extract, 0.4% K2HPO4, 3% NaCl, 0.2% glucose) or in VPG500 medium (1% polypeptone, 0.4% K2HPO4, 500 mM NaCl, 0.5% glycerol). E. coli cells were cultured at 37°C in LB medium (1% Bacto tryptone, 0.5% yeast extract, and 0.5% NaCl) or at 30°C in TG medium (1% Bacto tryptone, 0.5% NaCl, and 0.5% glycerol). When necessary, kanamycin and ampicillin were added to final concentrations of 100 μg/ml and 50 μg/ml, respectively.
Swarming assay
Aliquots (1 μL) of overnight cultures in VC medium (for V. alginolyticus) or LB medium (for E. coli) were spotted onto VPG500 (for V. alginolyticus) or TG (for E. coli) plates containing 0.26% agar and 100 μg/ml kanamycin (for V. alginolyticus) or 50 μg/ml ampicillin (for E. coli) and were incubated at 30°C.
Measurements of swimming speed of V. alginolyticus and data processing
An overnight culture of V. alginolyticus in VC medium was incubated in VPG500 medium at a 100-fold dilution and cells were grown at 30°C to exponential growth phase. Cells were centrifuged at 3,500×g for 3 min, and the sedimented cells were resuspended in TMN50 (50 mM Tris-HCl, pH 7.5, 5 mM MgSO4, 5 mM glucose, 50 mM NaCl and 450 mM KCl). Cell suspensions were diluted 50-fold with TMN medium containing various concentrations of NaCl (ionic strength was kept constant to 500 mM by KCl), and the motility of cells were observed immediately using a dark-field microscope. Swimming speeds were determined from at least 200 individual cells as described previously14. Briefly, the speeds were calculated from individual cell tracks measured by a computer-assisted motion analysis program (MetaMorph). It should be noted that the swimming speeds and apparent Km values estimated here are 2~3 fold lower and 7-fold higher, respectively, than the values reported previously10. This resulted from differences in the data collection method. In this study, we employed software (CellTrack 1.2 beta) that could easily and rapidly analyze the motility of large numbers of cells14,15, and the error bars in Figures 3 and 4 are much smaller than those previously reported. Using this analysis, the data contains slow motile and stop-and-go-motion of the cells, and therefore the swimming speeds and apparent Km values were decreased and increased, respectively. It should also be noted that the speeds and Km values become almost identical when highly motile cells showing straight swimming behavior during the measurements were selected and analyzed (data not shown).
Figure 3.
(a) Relationship between the volume of the side chain of introduced amino acid residues and the relative swarming size as estimated from Figure 2. Circles colored blue or red show the WT MotA/B and its mutants or WT PomA/B and its mutants, respectively. Three independent experiments were averaged. (b) Relationship between the volume of the side chain of introduced amino acid residues and the relative swimming speed of cells in TG medium for MotA/B expressing E. coli, or in TMN50 medium for PomA/B expressing V. alginolyticus. Circles colored blue or red show the WT MotA/B and its mutants or the WT PomA/B and its mutants, respectively. Three to 5 independent experiments were averaged.
Figure 4.
Estimation of apparent Na+ binding affinity with WT PomA/B and PomB mutants. The swimming speeds of cells (WT; wild-type PomA/B, Ala; C31A, Thr; C31T, Gly; C31G, Met; C31M, Val; C31V, Ile; C31I, Ser; C31S) were plotted against NaCl concentrations. Data were fit by the Michaelis-Menten equation.
Detection of PomA, PomB, MotA and MotB
Cells were cultured in VPG500 medium (for PomA and PomB) or in TG medium in the presence of 0.04% arabinose (for MotA and MotB) at the log phase of growth (optical density at 660 nm, ~0.5). Each cell suspension was dissolved in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer containing 5% 2-mercaptoethanol and was subjected to SDS-PAGE. Immunoblotting was performed using an anti-PomA antibody (PomA1312) and an anti-PomB antibody (PomB93) as described previously12. For immunoblotting, anti-E. coli MotA and anti-E. coli MotB antibodies, called MotA266 and MotB2, respectively, were raised against synthetic peptides (NH2-GCSERPSFIELEEHVRAVKN-OH for MotA266 and NH2-KNQAHPIIVVKRRKAKSHGC-OH for MotB2) which correspond to partial sequences of E. coli MotA (MotA Ser268 to Asn285) and MotB (MotB Lys2 to Gly20), and were affinity purified (Biologica Co.) prior to use.
Results
Effect of MotB A39X and PomB C31X mutations on the motility of E. coli and V. alginolyticus, respectively
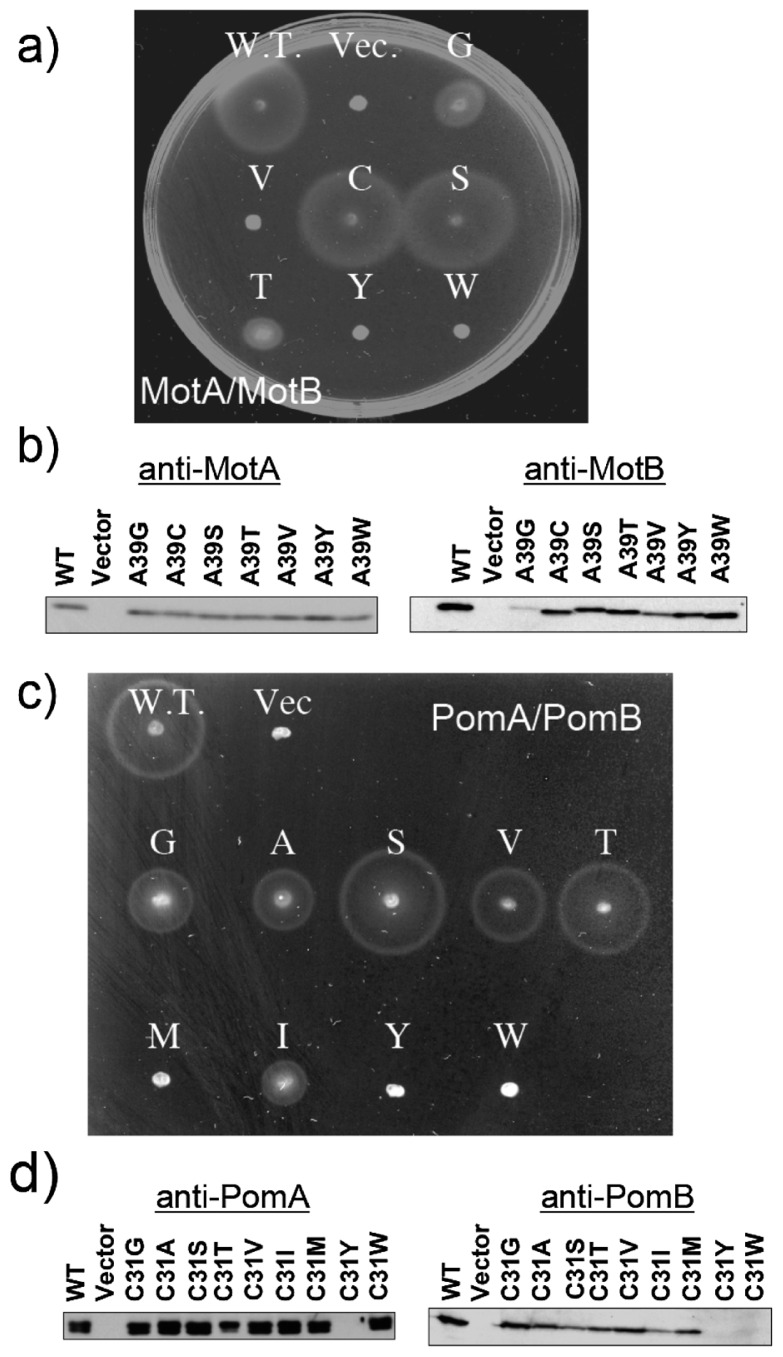
Some critical residues of MotA, MotB, PomA and PomB that are involved in torque generation have been reported2,3. In particular, Asp32 of MotB and Asp24 of PomB (Figure 1), which are unique acidic residues in the transmembrane segment, are expected to function in conveying H+ and Na+, respectively3. Two conserved residues, Cys31 in PomB and Ala39 in MotB, are located in the membrane-spanning region and are on the same sides as Asp24 for PomB and Asp32 for MotB in a helical wheel diagram (Figure 1). Figure 2a shows the swarming abilities of wild-type (WT) MotA/B, MotA/B(A39G), MotA/B(A39V), MotA/B(A39C), MotA/B(A39S), MotA/B(A39T), MotA/B(A39Y) and MotA/B(A39W). MotA/B(A39C) (C) and MotA/B(A39S) (S) showed no significant reduction in swarm size compared with WT MotA/B, whereas MotA/B(A39G) (G) and MotA/B(A39T) (T) reduced the swarming ability, and MotA/B(A39V) (V), MotA/B(A39Y) (Y) and MotA/B(A39W) (W) completely disrupted the swarming ability. Previously, Blair and coworkers demonstrated that the A39V mutant of MotB was dominant and non-functional11, which is consistent with our results. The expression level of the proteins was examined by Western blotting analysis. As shown in Figure 2b, protein expression levels of MotB mutants were almost the same as those of the WT, except for MotB(A39G) and MotB(A39V) mutants where amounts of MotB were reduced, while the amounts of MotA were not affected by any of the MotB mutations. Figure 2c shows the swarming abilities of the WT PomA/B, PomA/B(C31G), PomA/B(C31A), PomA/B(C31S), PomA/B(C31V), PomA/B(C31T), PomA/B(C31M), PomA/B(C31I), PomA/B(C31Y) and PomA/B(C31W). PomA/B(C31S) (S) and PomA/B(C31T) (T) showed no significant reduction in swarm size compared with WT PomA/B, whereas PomA/B(C31G) (G), PomA/B(C31A) (A) and PomA/B(C31V) (V) reduced the swarming ability, and PomA/B(C31M) (M), PomA/B(C31Y) (Y) and PomA/B(C31W) (W) completely disrupted the swarming ability. The reduction of the swarming size caused by C31A has been reported previously by Yorimitsu and coworkers16. The expression level of the proteins was examined by Western blotting analysis. As shown in Figure 2d, protein expression levels of PomB C31G, C31A, A31T, C31V and C31M mutants were almost the same as those of the WT PomB, whereas levels of PomB(C31S) and PomB(C31I) were reduced by the mutations, and PomB(C31Y) and PomB(C31W) did not show any PomB expression. As for C31Y, PomA was also not expressed. These results suggest that the introduction of a large residue (e.g. Tyr or Trp) into the Cys31 position disrupts protein folding or its insertion into the membrane. In any case, the side chain volumes at positions 39 of MotB and 31 of PomB seem to be important for function. To confirm this, we plotted the swarming rate against the volume of the introduced amino acid side chain (Figure 3a).
Figure 2.
Motility of MotB and PomB mutants on soft-agar plates (a)(c), and western blotting analysis of the proteins (b) (d). (a) Swarms of RP6894 (ΔmotAB) cells harboring a plasmid that express WT MotA/B, MotA/B(A39G) (G), MotA/B(A39V) (V), MotA/B(A39C) (C), MotA/B(A39S) (S), MotA/B(A39T) (T), MotA/B(A39Y) (Y) or MotA/B(A39W) (W). Transformants were cultured in liquid medium overnight, and aliquots were spotted onto a plate containing TG and 0.26% agar. Plates were incubated in the presence of 0.04% arabinose at 30°C for 10 hr. Vec. means a pBAD24 plasmid vector. (b) Western blotting analysis of MotA (left panel) and MotB (right panel) in whole cell extracts. Proteins were detected with an anti-MotA266 antibody and an anti-MotB2 antibody. (c) Swarms of NMB191 (ΔpomAB) cells harboring a plasmid that express WT PomA/B, PomA/B(C31G) (G), PomA/B(C31A) (A), PomA/B(C31S) (S), PomA/B(C31V) (V), PomA/B(C31T) (T), PomA/B(C31M) (M), PomA/B(C31I) (I), PomA/B(C31Y) (Y) or PomA/B(C31W) (W). Transformants were cultured in liquid medium overnight, and aliquots were spotted onto a plate containing VPG500 and 0.26% agar. Plates were incubated at 30°C for 6 hr. Vec. means a pSU41 plasmid vector. (b) Western blotting analysis of PomA (left panel) and PomB (right panel) in whole cell extracts. Proteins were detected with an anti-PomA1312 antibody and an anti-PomB93 antibody.
It is well-known that swarming size is affected not only by ion flux, but also by chemotactic responses of the cells18. To simply evaluate the motility, the swimming speeds of cells expressing the mutant proteins were measured by the software CellTrack beta 1.2 and were plotted against the volume of the introduced amino acid side chains (Figure 3b). The shapes of the graph were almost identical to that of Figure 3a, indicating that the mutations do not affect the chemotactic signal transduction pathway. These results suggest that Ala39 of MotB and Cys31 of PomB are located on the ion flux pathway of the stators of the H+ and the Na+ driven flagellar motors, respectively.
Apparent Km values for Na+ in the PomA/B complex and ion selectivity of MotA/B and PomA/B mutants
The Na+-driven motor has some advantages distinct from the H+-driven type. Namely, the apparent Km values for Na+ are easily estimated by measurements of swimming speeds at various NaCl concentrations10. Figure 4 shows the swimming speeds of cells containing WT or mutant PomA/B at various NaCl concentrations (Figure 4). The order the mutant stators in the Na+ affinity corresponds to the order of both the swarming rate and the swimming speed as shown in Figure 3, indicating that the differences in the swarming rate and the swimming speed originate from the Na+ affinity of PomA/B. The data fit well to the Michaelis-Menten equation (Figure 4), and the calculated kinetic parameters, Km and Vmax. The Km values were estimated as 33±4 mM for WT, 73±16 mM for C31S, 75±20 mM for C31T, 82±23 mM for C31V, 89±23 mM for C31A and 102±24 mM for C31G. The Vmax values were estimated as 28±1 μm/sec for WT, 24±2 μm/sec for C31S, 24±2 μm/sec for C31T, 18±2 μm/sec for C31V, 22±2 μm/sec for C31A and 8±1 μm/sec for C31G. The mutants having a bulky amino acid residue show small Km and Vmax values, suggesting that mutations hamper ion flux and disrupt the rotation of the flagellum depending on the volume of the amino acids introduced. These results support a model in which Cys31 faces toward the Na+ flux pathway in the PomA/B complex.
It has been reported that the V. alginolyticus polar flagellar motor can couple not only with Na+, but also with Li+, although the motility of cells in Li+ solutions as a cation is much lower than that in Na+ solutions19. Therefore, we investigated the effect of various monovalent cations on the motility of cells to test the ion selectively of the MotA/B and PomA/B mutants, and the results are listed in Table 1. The motility of the cells by the H+ flux was judged by inhibition of their swimming in buffer containing CCCP. However, the ion selectivity was not converted between H+ and Na+, as judged by their swarming and swimming speeds in buffers containing H+, Li+ or K+ ions, which implies that the ion selectivity is controlled by other site(s).
Table 1.
Ion dependence of the swimming motility of E. coli MotB mutants and V. alginolyticus PomB mutants
| Na+ | Li+ | K+ | H+ | |
|---|---|---|---|---|
| WT-MotA/B | − | − | − | ++ |
| MotB(A39G) | − | − | − | ++ |
| (A39T) | − | − | − | ++ |
| WT-PomA/B | ++ | + | − | − |
| PomB(C31G) | ++ | + | − | − |
| (C31V) | ++ | + | − | − |
| (C31L) | + | +/− | − | − |
The E. coli strain RP6894 (ΔmotAB) and the Vibrio alginolyticus strain NMB191 (ΔpomAB) were used as the host cell.
++, Vigorous swimming motility; +, slow swimming motility; +/−, very slow swimming motility; −, nonmotile.
Estimation of the counterpart of Ala39 (MotB) and Cys31 (PomB)
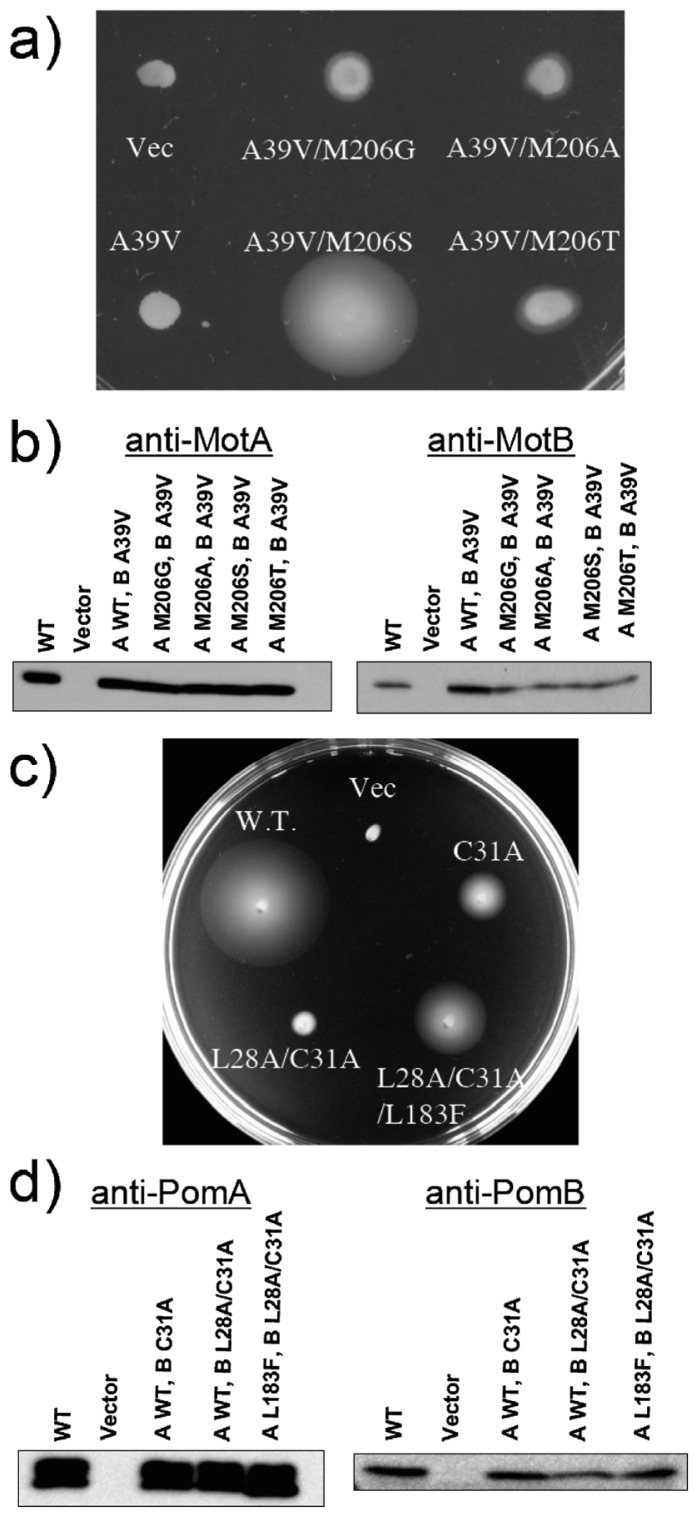
Blair and coworkers deduced the arrangement of core membrane segments of the MotA/B complex in E. coli using systematic cysteine-scanning mutagenesis and tryptophan replacement techniques7,8,20,21. On the basis of those reports, the side chain of Met206 in MotA (which corresponds to Leu183 in PomA) is thought to be located opposite to Ala39 in the channel. Here, randomized mutations were introduced in M206 of MotA in the A39V MotB mutant background. We chose A39V because valine is the smallest residue with non-motile behavior (Figures 2 and 3). As shown in Figure 5a, the loss of function caused by the A39V mutation was rescued by an additional mutation in MotA (M206S, M206G, M206A or M206T), and the increase of volume at position 39 by Val (50 Å3, compared to Ala) is close to the decrease of volume at position 206 by the substitution to Ser (72 Å3, compared to Met). Although volume difference between Met and Thr was much closer to the difference between Met and Ser, the recovery of the function by Met to Thr replacement is not so much. Therefore the orientation of the side chain of introduced amino acids may also be important for the ion flux. The protein expression level of MotA was not affected by any mutation at M206, although that of MotB was slightly affected by the mutations (Figure 5b). Similar experiments were carried out for PomA/B, and similar results were obtained. As shown in Figure 5c, the PomB(L28A/C31A) double mutant shows a slow-motile phenotype, whereas the function was rescued by an additional mutation of PomA(L183F). We confirmed that the protein expression level of PomA and PomB was not affected by any of the mutations (Figure 5d). Thus, we demonstrate here that Cys31 of PomB and Ala39 of MotB are located at the ion flux pathways, facing Leu183 of PomA (for PomB) and Met206 of MotA (for MotB) in the channel pores of the PomA/B and MotA/B stator units of the flagellar motors.
Figure 5.
(a) Swarms of RP6894 (ΔmotAB) cells harboring plasmids that express MotA/B(A39V) (A39V), MotA(M206G)/B(A39V) (A39V/M206G), MotA(M206A)/B(A39V) (A39V/M206A), MotA(M206S)/B(A39V) (A39V/M206S) or MotA(M206T)/B(A39V) (A39V/M206T). Transformants were cultured in liquid medium overnight, and aliquots were spotted onto a plate containing TG and 0.26% agar. Plates were incubated in the presence of 0.04% arabinose at 30°C for 20 hr. Vec. means a pBAD24 plasmid vector. (b) Western blotting analysis of MotA (left panel) and MotB (right panel) in whole cell extracts. Proteins were detected with an anti-MotA266 antibody and an anti-MotB2 antibody. (c) Swarms of NMB191 (ΔpomAB) cells harboring a plasmid that express PomA/B (WT), PomA/B(C31A) (C31A), PomA/B(L28A/C31A) (L28A/C31A) or PomA(L183F)/B(L28A/C31A) (L28A/C31A/L183F). Transformants were cultured in liquid medium overnight, and aliquots were spotted onto a plate containing VPG10 (1% polypeptone, 0.4% K2HPO4, 10 mM NaCl, 490 mM KCl, 0.5% glycerol) and 0.25% agar. Plates were incubated in the presence of 0.04% arabinose and 2.5 μg/ml chloramphenicol at 30°C for 12 hr. Vec. means a pBAD33 plasmid vector. (d) Western blotting analysis of PomA (left panel) and PomB (right panel) in whole cell extracts. Proteins were detected with an anti-PomA1312 antibody and an anti-PomB93 antibody.
Discussion
Flagellar motor proteins participate in the energy transduction from the electrochemical potential to a mechanical output, i.e. the rotation of the flagellum. The MotA/B complex and the PomA/B complex work as the stators and ion conductors in the H+-driven and Na+-driven flagellar motors, respectively. The interactions between MotA/B and H+, and between PomA/B and Na+ are critical for the flagellar motor rotation. Cation (H+, Na+, K+, Ca2+, etc.) channels work not only as energy converters like PomA/B, but also as systems to maintain ion concentrations inside cells22. Therefore, the ion flux mechanism has become a focus of much interest. In this study, we showed lines of evidence suggesting that Cys31 of PomB and Ala39 of MotB are located at the ion flux pathway with Leu183 of PomA and Met206 of MotA, respectively. Although, in general, systematic cysteine-scanning and tryptophan-scanning mutagenesis are highly effective approaches to investigate arrangements of membrane segments, the systematic point mutagenesis technique used here is also effective in analyzing the ion flux pathway within membrane protein complexes.
Where and how do the motor proteins recognize the coupling ion? Many mutations that impair motor function have been reported2,3. However, the roles of the residues are unclear. We expected that ion specificity might be affected by the mutations of Ala39 of MotB and Cys31 of PomB because these two residues construct ion channel wall. However, no change in ion specificity was observed in any of the mutants examined in this study (Table 1). This may be reasonable because neither of these two residues corresponds to any of the three residues within the membrane segment of B. clausii MotB that converted the H+-driven flagellar motor into the Na+-driven flagellar motor23. Interestingly, however, replacement of the corresponding three residues in PomA/B did not influence ion selectivity (unpublished data). It was also reported that replacement of the C-terminal region of Rhodobacter sphaeroides MotB with that of V. alginolyticus PomB converted the H+-driven flagellar motor into the Na+-driven one, suggesting that the peptidoglycan-binding domain is also a candidate region for determining ion specificity24. Thus, the ion selective mechanism remains unclear. Further studies are needed for the understanding of ion specificity determinants of the stator units.
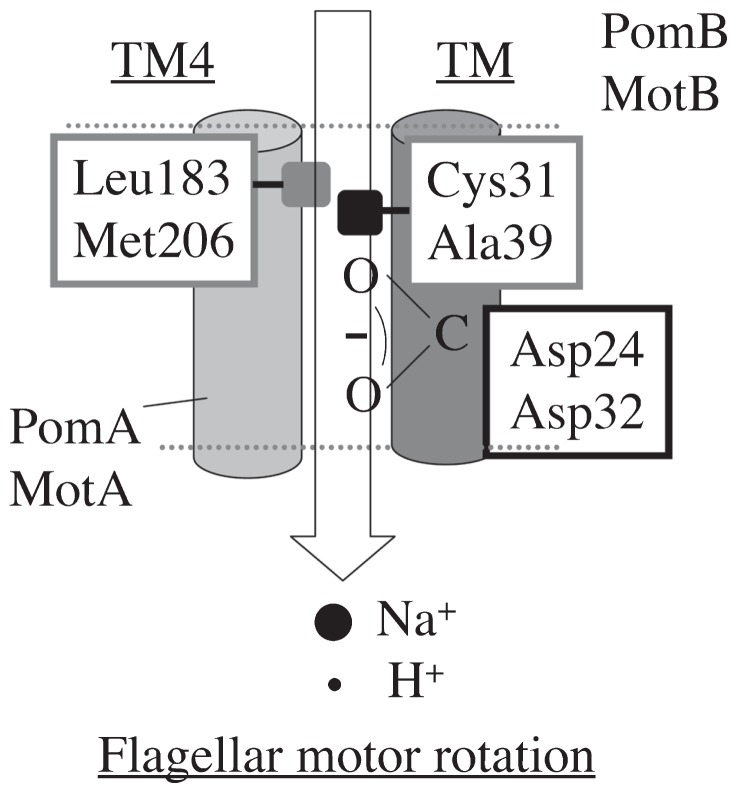
On the basis of our results combined with previous studies, we propose a model of the ion flux pathway through MotA/B and PomA/B ion channels (Figure 6). Asp24 of PomB and Asp32 of MotB form a Na+ and a H+ binding site, respectively. Residues conserved in the Na+-driven flagellar motors include Cys (Cys31 of PomB) and those in the H+-driven ones include Ala (Ala39 of MotB). Those residues are located at the periplasmic side of the membrane spanning domain of each subunit (Figure 1), and are important for ion translocation (Figures 2, 3 and 4). These results support a model in which Cys31 of PomB and Ala39 of MotB face toward the Na+ and H+ flux pathways in the complex, respectively. The loss of function caused by the MotB(A39V) and PomB(L28A/C31A) mutations was suppressed by the MotA(M206S) and PomA(L183F) mutations, respectively, and the increase in the side chain volume caused by MotB(A39V) (50 Å3) was close to the decrease caused by MotA-M206S (72 Å3) (Figure 5), which implies that Met206 of MotA and Leu183 of PomA form the ion channel pore with Ala39 of MotB and Cys31 of PomB, respectively (Figure 6).
Figure 6.
Model of the ion flux pathway. Asp24 of PomB and Asp32 of MotB form Na+ and H+ binding sites, respectively, Cys31(PomB)-Leu183(PomA) and Ala39(MotB)-Met206(MotA) serve as a ion channel wall and a regulator for H+ and Na+ flux, respectively. The ion flux leads to interaction changes and structural change(s) of FliG, and they mediate the flagellar motor rotation.
In 2000, Kojima et al. reported that the motility of the PomA(D31C) mutant showed a sharp dependence on the NaCl concentration, with a threshold at 38 mM25. We assume that a Na+ sensor exists at the periplasmic surface of PomA/B and propose a model in which PomB(Cys31)-PomA(Leu183) and MotB(Ala39)-MotA(Met206) regulate Na+ and H+ flux, respectively. Cys31 (Ala39) and Leu183 (Met206) are located in the transmembrane region of PomB (MotB) and the 4th transmembrane region of PomA (MotA), respectively. This is consistent with an earlier study by Blair and coworkers8, who reported that the transmembrane region of MotB is located near the third and fourth transmembrane region of MotA using a systematic cysteine-scanning mutagenesis approach, which is often used to study the structure and function of membrane proteins for which no three-dimensional structure is available. Using site-directed cysteine mutagenesis, Yakushi et al. reported that TM3 of PomA is in close proximity to the TM of PomB10.
It has been thought that ion flux drives the interactions and structural changes between MotA/B and FliG as well as PomA/B and FliG3. We believe that it would be structural changes of the rotor protein FliG that drive the flagellar motor rotation. The relationship between ion flux and structural changes of FliG is the next focus of our studies.
Acknowledgments
We would like to thank Drs. Kingo Takiguchi and Yohei Hizukuri for invaluable discussions.
This work was partially supported by grants from the Japanese Ministry of Education, Culture, Sports, Science, and Technology to YS (20050012), to SK (18770129) and to MH (18207011), and by Research Fellowships from the Japan Society for the Promotion of Science for Young Scientists to HT.
References
- 1.Yorimitsu T, Homma M. Na(+)-driven flagellar motor of Vibrio. Biochim Biophys Acta. 2001;1505:82–93. doi: 10.1016/s0005-2728(00)00279-6. [DOI] [PubMed] [Google Scholar]
- 2.Berg HC. The rotary motor of bacterial flagella. Annu Rev Biochem. 2003;72:19–54. doi: 10.1146/annurev.biochem.72.121801.161737. [DOI] [PubMed] [Google Scholar]
- 3.Blair DF. Flagellar movement driven by proton translocation. FEBS Lett. 2003;545:86–95. doi: 10.1016/s0014-5793(03)00397-1. [DOI] [PubMed] [Google Scholar]
- 4.McCarter LL. Polar flagellar motility of the Vibrionaceae. Microbiol Mol Biol Rev. 2001;65:445–462. doi: 10.1128/MMBR.65.3.445-462.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sato K, Homma M. Functional reconstitution of the Na(+)-driven polar flagellar motor component of vibrio alginolyticus. J Biol Chem. 2000;275:5718–5722. doi: 10.1074/jbc.275.8.5718. [DOI] [PubMed] [Google Scholar]
- 6.Sato K, Homma M. Multimeric structure of PomA, a component of the Na+-driven polar flagellar motor of vibrio alginolyticus. J Biol Chem. 2000;275:20223–20228. doi: 10.1074/jbc.M002236200. [DOI] [PubMed] [Google Scholar]
- 7.Braun TF, Blair DF. Targeted disulfide cross-linking of the MotB protein of Escherichia coli: evidence for two H(+) channels in the stator Complex. Biochemistry. 2001;40:13051–13059. doi: 10.1021/bi011264g. [DOI] [PubMed] [Google Scholar]
- 8.Braun TF, Al-Mawsawi LQ, Kojima S, Blair DF. Arrangement of core membrane segments in the MotA/MotB proton-channel complex of Escherichia coli. Biochemistry. 2004;43:35–45. doi: 10.1021/bi035406d. [DOI] [PubMed] [Google Scholar]
- 9.Zhou J, Sharp LL, Tang HL, Lloyd SA, Billings S, Braun TF, Blair DF. Function of protonatable residues in the flagellar motor of Escherichia coli: a critical role for Asp 32 of MotB. J Bacteriol. 1998;180:2729–2735. doi: 10.1128/jb.180.10.2729-2735.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yakushi T, Maki S, Homma M. Interaction of PomB with the third transmembrane segment of PomA in the Na+-driven polar flagellum of Vibrio alginolyticus. J Bacteriol. 2004;186:5281–5291. doi: 10.1128/JB.186.16.5281-5291.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blair DF, Kim DY, Berg HC. Mutant MotB proteins in Escherichia coli. J Bacteriol. 1991;173:4049–4055. doi: 10.1128/jb.173.13.4049-4055.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yorimitsu T, Sato K, Asai Y, Kawagishi I, Homma M. Functional interaction between PomA and PomB, the Na(+)-driven flagellar motor components of Vibrio alginolyticus. J Bacteriol. 1999;181:5103–5106. doi: 10.1128/jb.181.16.5103-5106.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang H, Blair DF. Regulated underexpression of the FliM protein of Escherichia coli and evidence for a location in the flagellar motor distinct from the MotA/MotB torque generators. J Bacteriol. 1995;177:3485–3495. doi: 10.1128/jb.177.12.3485-3495.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sudo Y, Furutani Y, Kandori H, Spudich JL. Functional importance of the interhelical hydrogen bond between Thr204 and Tyr174 of sensory rhodopsin II and its alteration during the signaling process. J Biol Chem. 2006;281:34239–34245. doi: 10.1074/jbc.M605907200. [DOI] [PubMed] [Google Scholar]
- 15.Sudo Y, Spudich JL. Three strategically placed hydrogen-bonding residues convert a proton pump into a sensory receptor. Proc Natl Acad Sci USA. 2006;103:16129–16134. doi: 10.1073/pnas.0607467103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yorimitsu T, Kojima M, Yakushi T, Homma M. Multimeric structure of the PomA/PomB channel complex in the Na+-driven flagellar motor of Vibrio alginolyticus. J Biochem (Tokyo) 2004;135:43–51. doi: 10.1093/jb/mvh005. [DOI] [PubMed] [Google Scholar]
- 17.Chothia C. Principles that determine the structure of proteins. Annu Rev Biochem. 1984;53:537–572. doi: 10.1146/annurev.bi.53.070184.002541. [DOI] [PubMed] [Google Scholar]
- 18.Falke JJ, Bass RB, Butler SL, Chervitz SA, Danielson MA. The two-component signaling pathway of bacterial chemotaxis: a molecular view of signal transduction by receptors, kinases, and adaptation enzymes. Annu Rev Cell Dev Biol. 1997;13:457–512. doi: 10.1146/annurev.cellbio.13.1.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu JZ, Dapice M, Khan S. Ion selectivity of the Vibrio alginolyticus flagellar motor. J Bacteriol. 1990;172:55236–5244. doi: 10.1128/jb.172.9.5236-5244.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sharp LL, Zhou J, Blair DF. Features of MotA proton channel structure revealed by tryptophan-scanning mutagenesis. Proc Natl Acad Sci USA. 1995;92:7946–7950. doi: 10.1073/pnas.92.17.7946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim EA, Price-Carter M, Carlquist WC, Blair DF. Membrane segment organization in the stator complex of the flagellar motor: implications for proton flow and proton-induced conformational change. Biochemistry. 2008;47:11332–11339. doi: 10.1021/bi801347a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fraser SP, Pardo LA. Ion channels: functional expression and therapeutic potential in cancer. Colloquium on Ion Channels and Cancer. EMBO Rep. 2008;9:512–515. doi: 10.1038/embor.2008.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Terahara N, Krulwich TA, Ito M. Mutations alter the sodium versus proton use of a Bacillus clausii flagellar motor and confer dual ion use on Bacillus subtilis motors. Proc Natl Acad Sci USA. 2008;105:14359–14364. doi: 10.1073/pnas.0802106105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Asai Y, Kawagishi I, Sockett RE, Homma M. Coupling ion specificity of chimeras between H(+)- and Na(+)-driven motor proteins, MotB and PomB, in Vibrio polar flagella. EMBO J. 2000;19:3639–3648. doi: 10.1093/emboj/19.14.3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kojima S, Shoji T, Asai Y, Kawagishi I, Homma M. A slow-motility phenotype caused by substitutions at residue Asp31 in the PomA channel component of a sodium-driven flagellar motor. J Bacteriol. 2000;182:3314–3318. doi: 10.1128/jb.182.11.3314-3318.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]