Abstract
A method to search for local structural similarities in proteins at atomic resolution is presented. It is demonstrated that a huge amount of structural data can be handled within a reasonable CPU time by using a conventional relational database management system with appropriate indexing of geometric data. This method, which we call geometric indexing, can enumerate ligand binding sites that are structurally similar to sub-structures of a query protein among more than 160,000 possible candidates within a few hours of CPU time on an ordinary desktop computer. After detecting a set of high scoring ligand binding sites by the geometric indexing search, structural alignments at atomic resolution are constructed by iteratively applying the Hungarian algorithm, and the statistical significance of the final score is estimated from an empirical model based on a gamma distribution. Applications of this method to several protein structures clearly shows that significant similarities can be detected between local structures of non-homologous as well as homologous proteins.
Keywords: ligand binding sites, structural alignment, relational database, geometric indexing, Hungarian algorithm
According to the ‘sequence determines structure determines function’ paradigm, it should be possible to predict protein structure from its amino acid sequence, and in turn, to predict its function from the structure. It has been empirically proved, however, that ab initio approaches to the both of these problems are extremely difficult. Currently, the most practical and reliable methods for protein structure prediction are the ones based on sequence comparison. In such homology-based methods, sequence similarities imply structural similarities. It is tempting to assume that the same argument applies to the prediction of protein functions. That is, we expect that we can infer some functional information if there are some similarities between two protein structures. However, it has been demonstrated that the protein folds (approximate over-all structures) of proteins are not significantly correlated with their functions. Since many protein functions such as enzymatic catalysis and ligand binding are performed by a small subset of protein atoms or residues, it seems necessary to perform local structure comparison in addition to (or, instead of) fold comparison for inferring protein function by similarity.
A number of methods have been proposed for searching for local similarities in protein structures1. However, some of them limit the data size due to a prohibitive amount of CPU time and/or RAM space required2–4, while others sacrifice structural details or diversity for the efficiency of search5–7. The ever increasing structural data in the Protein Data Bank (PDB)8 include many proteins of unknown functions and hence making available efficient and thorough methods available for local structure comparison for inferring protein functions is a pressing matter. At the same time, however, such rapidly increasing data only make conventional methods more and more inefficient. It is required that methods for local structure comparison be able to follow the rapid increase of data with a reasonable scalability.
In this Note, we introduce techniques to construct a scalable method for similarity search for local protein structures. In this method, ligand binding sites consisting of protein atoms are first compiled as a table in a relational database management system (RDBMS)9. For a given protein structure as a query, the method searches for structurally equivalent atoms in the database that match the atoms in the query structure. This search process can be executed efficiently owing to the indexing mechanism of the RDBMS. We call this technique geometric indexing (GI). After identifying matching ligand binding sites, alignments at atomic resolution are obtained by using the Hungarian algorithm10,11. The present method is similar to the geometric hashing (GH) algorithm in spirit. However, since the total size of the structural data may well exceed several gigabytes, it is usually not possible to naively implement the GH method which must keep a huge hash table in RAM. On the other hand, an RDBMS stores all the data on a hard disk which is much cheaper and larger than RAM, and hence let us overcome the data size problem. In addition, almost any modern RDBMS provides an efficient indexing mechanism which allows us to retrieve data satisfying a given set of constraints rather quickly. By using the technique introduced here, it becomes possible to keep up with the rapidly increasing structural data without sacrificing the efficiency of searching or the details and diversity of structural information.
Materials and methods
Overview
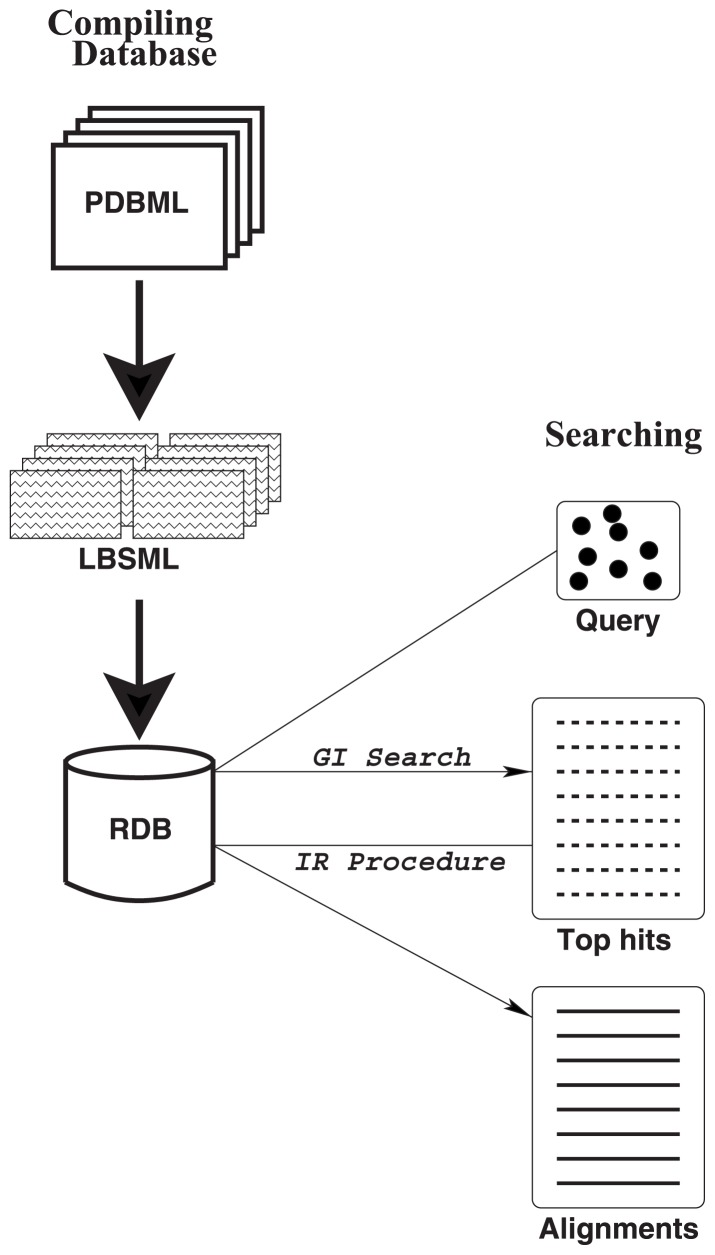
We first extract ligand binding sites (templates) from PDBML files12 and save them in XML files called LBSML (Ligand Binding Site Markup Language) files. An LBSML file contains information of atoms that are in contact with a ligand, along with reference sets (refsets) for local coordinate systems (see below). Then we compile refsets and atomic coordinates in local coordinate systems into a set of relational database (RDB) tables. This is a pre-processing stage and is carried out only once as long as we do not need to update the database (Fig. 1, left part).
Figure 1.
Overview of the method. The left part (“Compiling database”) illustrates the pre-processing step. The right part (“Searching”) shows the search step for a given protein structure as a query.
Then a database search is carried out for a given protein structure as a query (Fig. 1, right part). A search is divided into two stages. In the first stage, called geometric indexing search (“GI Search” in Fig. 1), the database is scanned by exploiting the indexing mechanism of the RDBMS, and possible atomic correspondences are counted. In the second part (“IR Procedure” in Fig. 1), a predefined number of high-scoring templates are subject to iterative refinement of the alignment to the sub-structures of the query.
Data set
We downloaded all the PDBML12 files (43,755 entries) on June 6, 2007. From these PDB entries, those were discarded that do not contain a protein chain or that do not contain any hetero atoms other than water.
Definition of reference set (refset)
As in the geometric hashing algorithm, all atomic coordinates are expressed in various local coordinate systems defined by reference sets (refsets). To define refsets, we applied the Delaunay tessellation using the Qhull library13 to each PDB entry. This procedure yields a set of tetrahedra consisting of four atoms as the vertices that are closest to each other. Then we selected those tetrahedra whose volumes are between 2 and 10 Å3 and whose total accessible areas are greater than zero Å2. These tetrahedra serve as refsets. Although only three atoms are necessary to define a unique Cartesian coordinate system, we use four atoms of a tetrahedron to reduce the number of possible combinations for refsets in a later stage of similarity search.
We define atom types as follows. All the backbone atoms are treated uniquely so that backbone “N”, “Cα”, “C” and “O” are labeled as such and their types are denoted “BN”, “BA”, “BC”, and “BO”, respectively. The types of side chain atoms are assigned as the corresponding standard atom names (as annotated by the “type_symbol” tag of the PDBML file). We keep only those tetrahedra whose four vertices are of different atom types. Accordingly, we can lexicographically order the vertices of a tetrahedron unambiguously. We can also define the chirality of a tetrahedron (see below). Thus, the sequence of ordered atom types and chirality of a tetrahedron define the type of the tetrahedron. For example, a tetrahedron consisting of atoms of types “BN”, “BA”, “BC” and “S” with positive chirality is typed as “BA:BC:BC:S:+”.
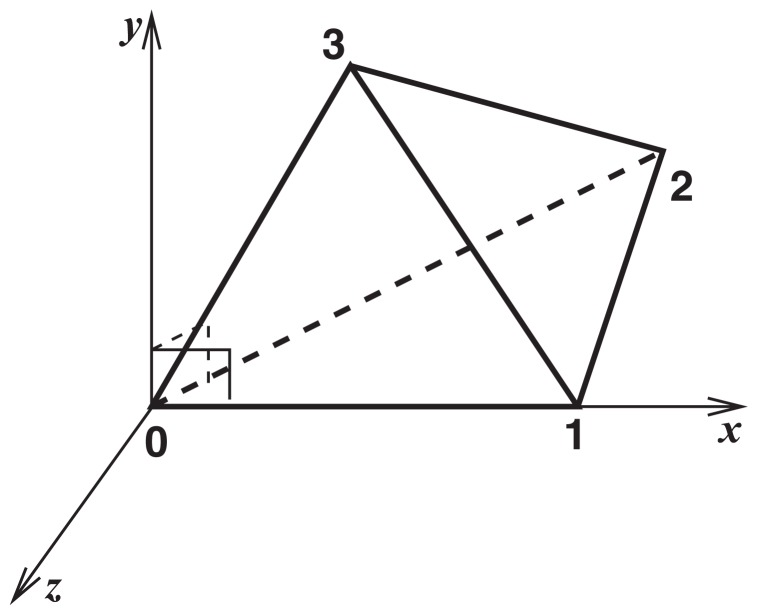
Let ri (i=0, ..., 3) be the coordinates of the four atoms of a refset (tetrahedron) in the original coordinate system (i.e., as in the PDB file). Here, the indices from 0 to 3 are so labeled in the lexicographical order of their atom types. When calculating the local coordinates of an atom in the refset, the origin is set to r0. The x-axis is defined by the unit vector parallel to r01 ≡r1–r0, that is, χ̄ ≡ (1/||r01||)r01. With r02≡r2–r0, the y-axis is defined by ŷ ≡ (1/||r02||) ○× r02. The z-axis is defined by ẑ≡○ × ŷ. Thus, for a given set of coordinates s in the original system, the local coordinates in the system spanned by the refsets {ri} are given as s′=[(s–r0) ·○, (s–r0) · ŷ, (s–r0) · ẑ]. This coordinate system spanned by a refset is illustrated in Figure 2. Using these notations, the definition of the chirality of a tetrahedron mentioned above is given as the sign of the dot product r03·ŷ. For example, the chirality of the tetrahedron in Figure 2 is positive. By explicitly including the chirality information, it is possible to discriminate the enantiomers for query and template structures. Therefore, for a given query structure, we can always find the templates of the correct chirality in the search stage described below.
Figure 2.
Local coordinate system defined by a refset (tetrahedron).
Extracting ligand binding sites
By using the annotations in PDBML files, we identified the so-called hetero atoms (ligand atoms), and all protein atoms that are in contact with any of the hetero atoms. Two atoms are defined to be in contact if their distance is less than or equal to 5 Å. For each ligand, we create an XML file containing a list of protein atoms that are in contact with it. We call this XML file an LBSML file. Atomic coordinates in an LBSML file are stored in the “extatom” style of the PDBML file12, so that the ligand binding site can be examined visually by using the PDBjViewer25. A set of protein atoms in contact with a ligand is called a ligand binding site. We also calculate refsets of the PDB entry. Along with the atomic coordinates of the ligand and the ligand binding site, the information of refsets and its type, volume, and lengths of edges of the tetrahedra defining the refsets is stored in an LBSML file. Refsets are saved in an LBSML file only if at least one of its vertex atoms is in contact with the ligand. The distance threshold for the contact between refset and ligand atoms was set to 5 Å. As a result, we constructed 162,626 LBSML files corresponding to the ligand binding sites. A set of atoms in a ligand binding site is also referred to as a template in the following.
Compilation of atomic coordinates and reference sets
We compile the information of LBSML files into tables of a relational database management system (RDBMS). The use of RDBMS allows us to handle a huge amount of structural data relatively efficiently. Basic information of LBSML files is saved in a table shown in Table 1.
Table 1.
Definition of the table for ligand binding sites
| CREATE TABLE lbsmldb ( | |||
| lbsml_id | INTEGER PRIMARY KEY, | ...... | (a) |
| lbsml | TEXT, | ...... | (b) |
| pdbx | TEXT, | ...... | (c) |
| ligand | TEXT, | ...... | (d) |
| natoms | INTEGER ); | ...... | (e) |
unique identifier;
file name;
PDB’s description of the protein;
PDB’s annotation of the ligand;
the number of protein atoms in contact with the ligand.
Refsets in each LBSML file were compiled in a table (Table 2) along with their features such as tetrahedron type, volume, and edge lengths as well as the reference to the LBSML file they are derived from, and their serial number (refset identifier) in the LBSML file (note there are usually multiple refsets in a single LBSML file). There were about 4.7 million refsets in total. The primary key of this table consists of a pair of the reference to LBSML file and the refset identifier. The types and local coordinates of atoms under each refset in an LBSML file are compiled into the same row as the refset.
Table 2.
Definition of the refset table
| CREATE TABLE refsetdb ( | |||
| lbsml_id | INTEGER, | ...... | (a) |
| irs | INTEGER, | ...... | (b) |
| PRIMARY KEY (lbsml_id, irs) | ...... | (c) | |
| tetra | TEXT, | ...... | (d) |
| tvol | DOUBLE PRECISION, | ...... | (e) |
| td01 | DOUBLE PRECISION, | ...... | (f) |
| td02 | DOUBLE PRECISION, | ...... | (f) |
| td03 | DOUBLE PRECISION, | ...... | (f) |
| td12 | DOUBLE PRECISION, | ...... | (f) |
| td23 | DOUBLE PRECISION, | ...... | (f) |
| td31 | DOUBLE PRECISION, | ...... | (f) |
| atype_id | INTEGER [ ], | ...... | (g) |
| xco | DOUBLE PRECISION [ ], | ...... | (h) |
| yco | DOUBLE PRECISION [ ], | ...... | (h) |
| zco | DOUBLE PRECISION [ ] | ...... | (h) |
| ); | |||
reference to “lbsmldb” (Table 1);
reference set identifier;
a pair of lbsml_id and irs makes the primary key of the refset.
tetrahedron type;
volume of tetrahedron;
“tdij” denotes the length of edge between vertices i and j of tetrahedron (A tetrahedron consists of four atoms denoted i, j=0, 1, 2, and 3).
types of the atoms spanned by the refset (encoded as integers).
local coordinates of the atoms spanned by the refset.
For any database systems, it is critical to create appropriate indexes for efficient information retrieval. According to Garcia-Molina et al.9, “an index is any data structure that takes as input a property of records — typically the value of one or more fields — and finds the records with that property ‘quickly’.” Here, we used an index based on the data structure called a B+ tree9. The refset table (Table 2) is indexed by the tetrahedron type, volume, and edge lengths with the SQL expression “CREATE INDEX tetraIdx ON refsetdb (tetra, tvol, td01, td02, td03, td12, td23, td31).”
The geometric indexing search method
Given a query protein structure, we search for ligand binding sites stored in the database that match a substructure of the query. To do so, we first define and select the refsets (tetrahedra) of the query structure by the same procedure as the templates except that contacts with hetero atoms are not taken into account (because they may not be present in the query structure). Then, for each refset of the query, we calculate the atomic coordinates of each atom under that refset. Next, we retrieve from the database those refsets whose tetrahedron types are the same as that of the query tetrahedron, and whose volume and edge lengths are close to the corresponding quantities of the tetrahedron of the query within predefined threshold. At the same time, those atomic coordinates which are based on the matching refsets are extracted from the database. This can be carried out with the SQL expression in Table 3. The retrieval of refsets and atomic coordinates are performed efficiently owing to the index constructed above. At this point, we have a list of tuples of atom type, coordinates, and LBSML file (lbsml_id) and refset identifiers (refset_id) returned by the SQL expression in Table 3. Then, for each local atomic coordinates of the query, we select from the tuple list those tuples whose atom type is the same as that of the query and coordinates close to those of the query. The query and template coordinates (xq, yq, zq) and (xt, yt, zt) are defined to be close if the distance between them is lower than a predefined constant Δc (Here we set Δc=2 Å). Finally, the LBSML file and refset identifiers, on which the retrieved atomic coordinates are based, are recorded, and the count of the triple (template LBSML file, and query and template refset identifier) is incremented.
Table 3.
Pseudo SQL expression for local structure search
| SELECT atype, xco, yco, zco, lbsml_id, irs FROM refsetdb | |
| WHERE tetra = ‘tq’ | |
| AND tvo1 BETWEEN | vq −Δv AND vq +Δv |
| AND td01 BETWEEN | d01−Δd AND d01+Δd |
| AND td02 BETWEEN | d02−Δd AND d02+Δd |
| AND td03 BETWEEN | d03−Δd AND d03+Δd |
| AND td12 BETWEEN | d12−Δd AND d12+Δd |
| AND td23 BETWEEN | d23−Δd AND d23+Δd |
| AND td31 BETWEEN | d31−Δd AND d31+Δd |
The table refsetdb is defined in Table 2. tq, vq, and dij are the type, volume, and edge length of a refset of the query. Δ’s are predefined constants for similarity thresholds. Expressions such as “vq−Δv” are given as constants in the actual code. We set Δv=1Å3 and Δd=2Å.
After all the query refsets are examined, we have a list of tuples of a LBSML file, a template refset identifier and a query refset identifier, as well as the count of each tuple. If the count is sufficiently large, the local structure in the LBSML file is likely to be present in the query structure. However, the count can be large just because there are a large number of atoms in certain templates. Therefore we use the score S( f, rt, rq) of the tuple of LBSML file f, template refset identifier rt and query refset identifier rq defined as
| (1) |
where cnt(f, rt, rq) is the count of the tuple (f, rt, rq) and Nf is the number of atoms in the template of the LBSML file f. We found that the best performance is attained with p=2, and this value is used throughout. We refer to this score as the “GI score” (after Geometric Indexing) in the following. The pairs of (f, rt, rq) are sorted in the decreasing order of SGI(f, rt, rq), and the top Ntop hits (say, Ntop = 10000) were saved for further refinement.
This search method, which we refer to as “GI search” in the following, is similar to the geometric hashing (GH) method14,15. However, it is not necessary to keep the database on memory, and atomic coordinates not matched directly by using a hash function. Instead, we use a conventional RDBMS for keeping the template information, and first select matching template refsets using an index of the database. In the present method, a matching refset serves not only as the basis of a local coordinate system but also as a seed alignment.
Iterative refinement of alignment (IR procedure)
By using the RDBMS-based search method, we can retrieve a set of ligand binding sites (and refsets) which are structurally similar to sub-structures of a query protein structure. At this point, however, the exact alignment of query and template atoms has not been obtained yet since all we have is the count of the tuple of LBSML files and template and query refset identifiers. As in the GH method, it is possible to obtain an alignment by using a strict definition of the neighbor of an atom in the RDBMS-based method. However, a small difference in the refsets could greatly perturb the quality of alignment. Therefore, it is desirable to employ a more robust method for refining the alignment at atomic resolution.
Since we assume that template and query atoms are approximately in the same refset, a reasonable set of possible alignments is obtained by the following procedure. First we regard the system of query and template atoms as a bipartite graph16 in which query atoms form one group and template atoms another, and edges are allowed only between the two groups. We assign an edge if the query atom i and template atom j are of the same atomic type and the distance dij between them is less than 2 Å. We assign a weight of wij=1–dij/2 to the edge. In an alignment, each query atom can match with at most one template atom. The best alignment is the one for which the sum of the matching edges is larger than or equal to any other alignments. This combinatorial optimization problem, called the maximum weight bipartite matching problem, can be readily solved by using the so-called Hungarian method10,11.
The refinement of alignment is performed iteratively as follows. First, by using the refset obtained by the RDBMS-based search, we construct a bipartite graph, and apply the Hungarian method to obtain the best matching (alignment). Second, we use the resulting alignment to rotate the template structure to optimally superpose onto the query structure. This can be carried out by a classical least squares technique such as the quaternion-based one of Diamond17. Third, based on the optimal superposition, we construct a new bipartite graph, and apply the Hungarian method. The second and third stages are iterated until convergence which is achieved after 4 or 5 iterations on average.
The score of an alignment based on the LBSML file f, template refset identifier rt and query refset identifier rq is calculated as
| (2) |
where the summation (Σ′) is over all the edges in the matching, Nali(f, rt, rq) is the number of aligned atom pairs and Nf is the number of atoms in the template of the LBSML file f. We refer to this score as the “IR score” (after Iterative Refinement) in the following.
Estimation of statistical significance
In order to estimate the statistical significance of the IR score defined above, we introduce a statistical model based on random sampling. After performing a GI search, we have a huge number of hits. Among those hits, we randomly select 2,000 of them for iterative refinement. As shown in the Results section, the distribution of the IR score of randomly selected alignments can be well approximated by a gamma distribution GAM(α, β) whose probability density function is given as
| (3) |
for x≥0 (note that the IR score is greater than or equal to 0 by definition). Let the mean and variance of the IR scores of the randomly selected alignments be m and v, respectively. Then the parameters α and β of the gamma distribution GAM(α, β) are given as α=m2/v and β=v/m, respectively. Then the P-value or the probability that the IR score T is greater than or equal to x is given as
| (4) |
which indicates that statistical significance of the IR score. That is, lower P-values indicate greater statistical significance.
Implementation
All the codes were written in the Objective Caml (OCaml) language (http://caml.inria.fr). The RDBMS employed was the PostgreSQL system (http://www.postgresql.org) which has been moderately optimized for the underlying hardware. All the computations were carried out on an Apple Power-Mac (dual 2.5 GHz PowerPC G5) with 8 gigabytes (GB) RAM.
Results
Execution time
We analyzed the execution time of a single search by using a mutant sperm whale myoglobin (PDB ID: 101m) as a query. The number of hits subject to the refinement was set to 50,000. The database consists of 162,626 ligand binding sites (LBSML files), 4,699,804 refsets (tetrahedra). In total, the hard disk space of 10 GB was consumed by the database.
The whole search process took 161 minutes of CPU time, in which 115 minutes were spent for the GI search, 45 minutes for the IR procedure. In the GI search, the SQL expressions for selecting compatible template refsets (Table 3) took 90 minutes, and other parts took 25 minutes. Thus, the execution of the SQL expression is the most time-consuming part of the whole process. This is because it involves access to the hard disk. In the PDB entry 101m, there were 376 refsets selected according to the criteria described above. The search time is roughly proportional to the number of refsets of the query. For each refset, an SQL expression for selecting compatible template refsets (see Table 3) was issued.
Effects of refinement
The scores used in the geometric indexing and iterative refinement stages are different (see Eqs. 1 and 2). Accordingly, the rank of high-scoring templates may change between before and after the refinement. To examine the effect of the refinement, we performed a search using the myoglobin (PDB ID: 101m) again. The top 50,000 hits of the GI search were used for the refinement.
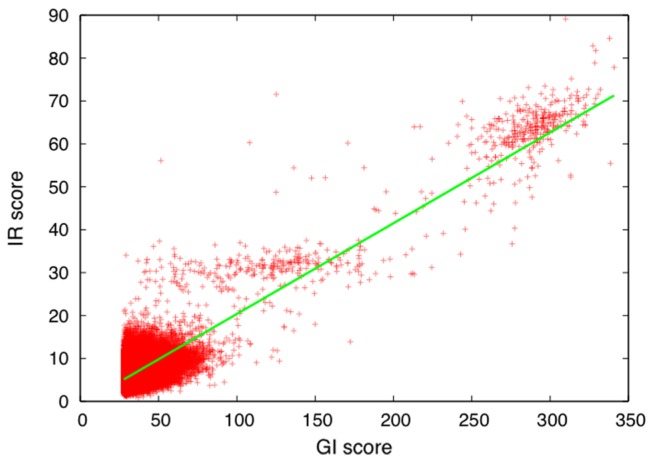
Figure 3 shows the two scores of each of the 50,000 templates. In general, the two scores correlate with each other very well, with a correlation coefficient of 0.87 in this case. But the rank of some templates may change dramatically upon refinement. The refinement greatly improved the scores of some templates of relatively low GI scores.
Figure 3.
Comparison of GI score and IR score. Each point represents a template included in the top 50,000 hits for the query (PDB ID: 101m). The regression line is also shown. The correlation coefficient between the scores is 0.87.
Modeling the distribution of IR scores
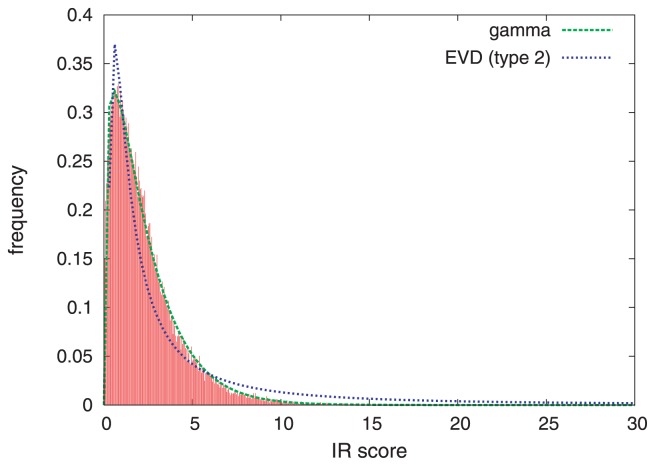
In order to estimate the statistical significance of IR score, we examined its distribution. We first performed a GI search, and then randomly selected 50,000 hits for iterative refinement. After the refinement, the histogram of the IR score was plotted. Fig. 4 is an example obtained for the query 101m. It is clearly seen that the distribution is well approximated by a gamma distribution (Fig. 4, green line). We also fitted the type-2 (Fréchet) extreme value distribution (since the IR score is non-negative), but the fit was not as good as the gamma distribution (Fig. 4, blue line). The same trend was observed for other proteins. Thus, we use the gamma distribution for calculating the statistical significance of the IR score. Since the parameters of the gamma distribution may be different depending on queries, they are calculated by random sampling each time a search is performed.
Figure 4.
Distribution of IR scores of randomly selected templates. The red bars indicate the histogram of IR scores of randomly selected templates obtained for the query 101m. The green line is the probability density function (PDF) of the gamma distribution GAM(α, β) with the parameters α=1.32 and β=1.75 calculated from the mean and variance of the scores. The blue line is the PDF of the type 2 extreme value distribution with the parameters determined to best fit the histogram.
Examples of high-scoring alignments
We present in this section examples of high-scoring alignments for four query protein structures. These four proteins (myoglobin, subtilisin, cAMP-dependent protein kinase, and alcohol dehydrogenase) have well-characterized functions. Thus, it is relatively straightforward to discriminate true positives from false positives for these proteins, making them suitable for benchmarking. However, the precise definition of false positives is difficult as there is always a possibility that a query protein has a ligand-binding ability that has not been characterized as such. Therefore, we define false positives based on the physical plausibility (mainly, the presence of severe steric repulsions) as well as on our current biochemical knowledge of the query proteins.
Myoglobin
We first examine more closely the results obtained for the myoglobin (PDB ID: 101m) used above. We used the 50,000 hits by GI search for the further refinement. The heme binding site of myoglobins occupied the first 363 hits with IR scores (P-values) ranging from 89.1 (4.6×10−23) to 38.5 (3.9×10−10). Below the myoglobins were other globins such as hemoglobins and cytoglobins, all of which were identified by the heme binding sites. The first non-globin appeared at the 555th rank with IR score of 30.1 (P=5.3×10−8). This entry was an isopropanol binding site of single-strand selective monofunctional uracil DNA glycosylase (UDG; PDB ID: 1oe618). Visual inspection of the alignment suggests that this is likely to be a meaningless hit because the ligand is a constituent of the solvent and its binding site corresponds to an α helix (which is abundant in many proteins including globin). In fact, this isopropanol binding site of UDG (1oe6) shows a high score for any query that contains an α helix (data not shown), and is one of the frequently occurring false positives (see below). The next non-globin hit was the S-oxymethionine “binding” site of catalase (PDB ID: 2iuf19). S-oxymethionine here is actually a modified residue in the protein which happened to be annotated as HETATM in the PDBML file. This entry has a high score because the site is made of parts of α helices and α helices are common in globins. The next non-globin hit at the 489th rank with IR score of 26.1 (P=5.4×10−7) was a hypothetical protein from Pseudomonas aeruginosa (PDB ID: 1tu9). Although its function is not well known, the fold of this protein is globin-like (Y. Kim et al., unpublished) and the aligned atoms comprised the heme-binding site.
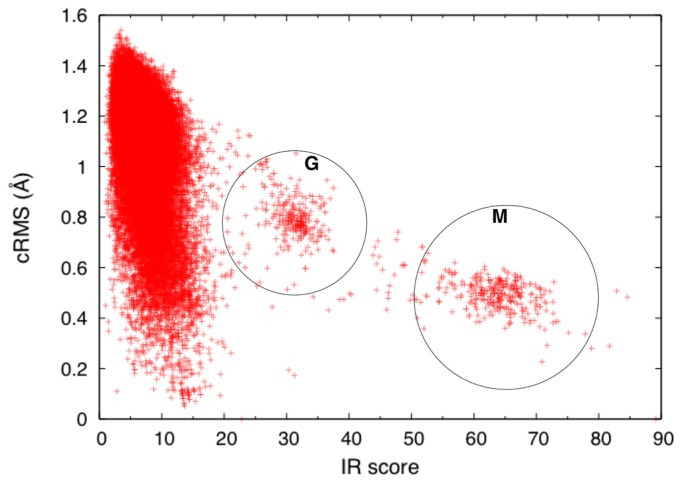
In general, good alignments should have high IR scores and low coordinate root mean square (cRMS) deviations. This trend is clearly observed in Figure 5. That is, good alignments should reside in the right bottom corner of the scatter plot of Figure 5. In this scatter plot, we can recognize two high-scoring clusters around IR score of 60–70 and 25–35, which correspond to closely related myoglobins and other globins, respectively. In the region of low IR scores, there are may templates with low cRMS values. A low IR score implies a small number of aligned atoms, hence the low cRMS values.
Figure 5.
Scatter plot of the IR scores and coordinate RMS deviations resulted from a search with the PDB entry 101m. The regions enclosed by the circles marked with M and G contain mostly myoglobins and other globins, respectively.
Subtilisin savinase
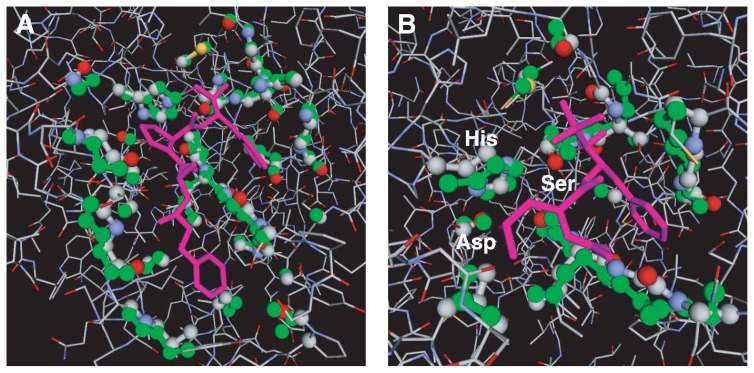
We next examine the result of a search with subtilisin savinase from Bacillus lentus (PDB ID: 1svn20) as a query. The top hit was the peptide binding site of subtilisin DY (PDB ID: 1bh621) with an IR score of 59.8 and P-value of 1.0×10−14 (Fig. 6A). Subsequent hits were subtilisins and related proteases. After these subtilisin-related templates (removing physically implausible templates), we found a Mn2+ binding site of Dicer from Giardia intestinalis (PDB ID: 2ffl22; P=1.5×10−5) and Mg2+ binding site of 30S ribosomal subunit S20 from Thermus thermophilus (PDB ID: 1i9423; P=1.8×10−5). But these ion binding sites reside within common loop structures, and hence they are likely to be biochemically/biologically insignificant. At the 255th rank, we found the active site of bovine γ-chymotrypsin (PDB ID: 7gch24) with an IR score of 20.9 (P-value 2.0×10−5). This protein has a different fold than subtilisins but shares the common catalytic triad consisting of three residues Ser, His, and Asp. The obtained atomic alignment indeed contains these catalytic residues. Namely, Asp32, His64, and Ser221 of subtilisin Savinase are aligned with Asp102, His57, and Ser195 of γ-trypsin (Fig. 6B).
Figure 6.
Optimal superpositions of the query 1svn on templates. The wire-frame model in the CPK color scheme is the query protein 1svn. The template atoms are colored in green. Aligned atoms are in ball-and-stick model. The ligand of the template is the ball-and-stick model in magenta. A: Peptide-binding site of subtilisin DY (PDB ID: 1bh621). B: Peptide-binding site of γ-chymotrypsin (PDB ID: 7gch24); the labeled Ser, His, Asp are the aligned catalytic triad. The figures were created by using the PDBjViewer25.
cAMP-dependent protein kinase
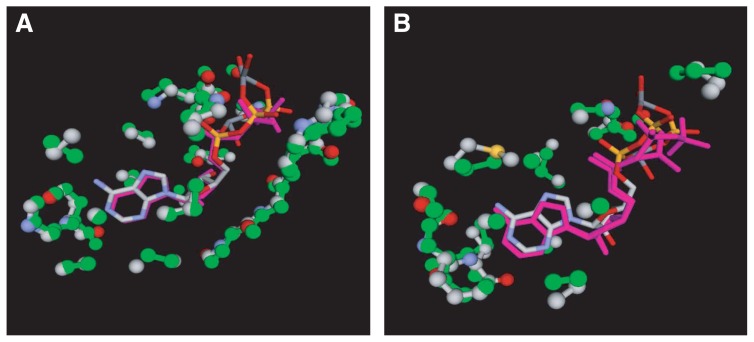
Our third example is the cAMP-dependent protein kinase, cAPK (PDB ID: 1atp26) from Mus musculus. This example is motivated by the work of Kobayashi and Go27 where they have found that the local structure of the nucleotide-binding site of cAPK is similar to those of other nucleotide-binding proteins with different folds. They listed five ATP-binding proteins that share similar local structures: glutaminyl-tRNA synthetase, D-Ala:D-Ala ligase (DD-ligase), casein kinase-1 (CK-1), seryl-tRNA synthetase, and glutamine synthetase27. According to the SCOP database28, CK-1 and cAPK belong to the same family, the protein kinase catalytic subunit family, although the sequence identity between them is as low as 19%. Among the five proteins listed by Kobayashi and Go, CK-1 exhibited a highly significant similarity with an IR score of 42.8 and P=8.9×10−11 (Fig. 7A). In contrast, we only found a weak similarity with glutathion synthetase, belonging to the same superfamily as DD-ligase, with a relatively low IR score of 12.5 (P=2.1×10−3; Fig. 7B). Most high-scoring templates were all kinases of the same fold. Other similarities listed by Kobayashi and Go were either not detected, or detected with wrong alignments. There are at least two possible explanations for this failure in detecting similar local structures. First, our criteria for selecting similar refsets may be too stringent so that possible hits are discarded during the GI search. Second, the number of aligned atoms as obtained by Kobayashi and Go is very small, ranging from 14 to 16, whereas some of obvious false hits contained more than 20 aligned atoms. The first point may be corrected by loosening the criteria at the cost of increased CPU time. The second point is more problematic, however. Kobayashi and Go used only ATP-binding proteins for their study while we used all the ligand-binding sites present in the current PDB. Accordingly, the signal-to-noise ratio is substantially lower in the present case. In order to overcome this problem, a more elaborate statistical method may be necessary.
Figure 7.
Optimal superpositions of the ATP-binding sites of the query cAMP-dependent protein kinase (cAPK; PDB ID: 1atp26) on templates. A: The template is the ATP-binding site of casein kinase-1 (PDB ID: 1csn29) from Schizosaccharomyces pombe. B: The template is the ATP-binding site of glutathion synthetase (PDB ID: 1m0w30) from Saccharomyces cerevisiae. The color scheme is the same as Fig. 6. The ligand of 1atp is also shown in the stick model with the CPK colors.
Alcohol dehydrogenase
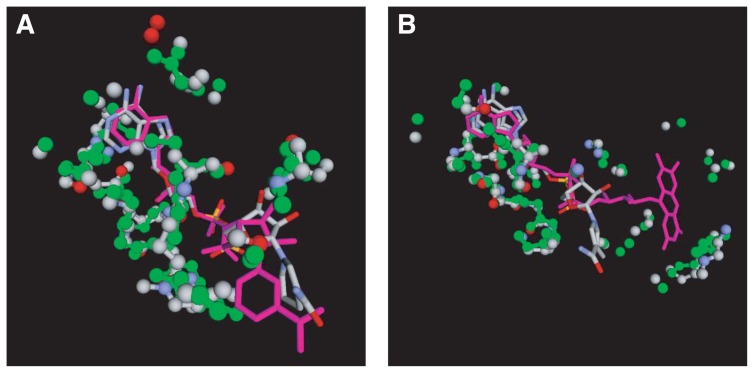
The fourth example is the alcohol dehydrogenase (ADH; PDB ID: 1het31) from Equus caballus (horse). The first 107 top hits are the nicotinamide-adenine-dinucleotide (NAD)-binding sites of ADHs from various species, which are followed by various kinds of other dehydrogenases such as formaldehyde dehydrogenase, sorbitol dehydrogenase, glucose dehydrogenase, and so on. We looked for structural similarities with proteins other than dehydrogenases, and have found a few such examples. One example is the NAD-binding site of the urocanase protein (PDB ID: 1x87; Tereshko et al., unpublished) with an IR score of 24.0 (P=2.7×10−6). According to the SCOP database, this protein belongs to the urocanase fold which is clearly different from the NAD(P)-binding Rossmann-fold domain of the ADH. The alignment (Fig. 8A) consists of 76 atom pairs yielding cRMS of 1.0 Å. Another example is the flavin-adenine dinucleotide (FAD)-binding site of p-hydroxybenzoate hydroxylase (PHBH; PDB ID: 1iuv32) which exhibited a significant IR score of 20.2 (P=2.3×10−5; Fig. 8B). PHBH belongs to the FAD/NAD(P)-binding domain fold which is different from the NAD(P)-binding Rossmann fold of ADH.
Figure 8.
Optimal superpositions of the NAD-binding sites of the query alcohol dehydrogenase (PDB ID: 1het)31 on templates. A: The template is the NAD-binding site of urocanase protein (PDB ID: 1x87; Tereshko et al., unpublished) from Bacillus stearothermophilus. B: The template is the FAD-binding site of p-hydroxybenzoate hydroxylase (PDB ID: 1iuv32) from Pseudomonas aeruginosa. The color scheme is the same as Fig. 6. The ligand of 1het is also shown in the stick model with the CPK colors.
Discussion
We have demonstrated that the present method can detect non-trivial similarities in protein local structures at atomic resolution in a reasonable CPU time. Here we discuss a few remaining issues to be solved and possibilities for further improvements.
Recurring false positives
It was often observed that certain ligand binding sites exhibited high scores regardless of query structures. Such examples include the isopropanol binding site of UDG and the S-oxymethionine binding site of catalase as mentioned above in the example of myoglobin. These and other recurring false hits are almost always part of super-secondary structures which consist of α-helices and β-strands which are highly regular and abundant. Another source of error is the ambiguous definition of “ligands”. For example, the ligand in the S-oxymethionine binding site of catalase (2iuf19) described above is actually a modified residue in the protein, not another molecule than the protein itself. In this case, most part of the ligand (S-oxymethionine) should be treated as a part of the protein. Many of the ligands treated in this study are biologically irrelevant but are present as a part of the solvent. Such examples include the isopropanol in the PDB entry 1oe618 described above. Therefore, it would be helpful to include only biologically relevant ligands in the database although this may require a great deal of effort in the absence of proper annotations.
Increasing sensitivity
In the proposed method, we first select candidates based on the attributes of refsets, such as the volume and edge length of tetrahedra. In the current implementation, the criteria for refsets are relatively stringent so that it is not guaranteed that all the possibly important refsets are stored in the database (e.g., tetrahedra containing multiple atoms of the same type). This may be a reason why the present method failed to detect some of the known similarities between cAPK and other proteins of different folds. In order not to miss such important refsets, it may be possible to use backbone-based refsets33. However, the naive definition of backbone-based refsets (defined by three atoms N, Cα, C) is extremely inefficient because all such refsets are essentially identical and we have to retrieve all such refsets every time we issue an SQL query similar to that of Table 3. Therefore, we need to add some extra attributes to efficiently select relevant candidates for retaining efficiency. For example, we may use similarity between amino acid residues or backbone dihedral angles for restricting possible candidates.
A better statistical model may also improve the sensitivity. Currently we employ a simple gamma distribution that depend only on the IR score. However, we observed that the IR score depends on cRMS in a systematic manner so that some false hits with relatively high IR scores with large cRMS values may be eliminated. Therefore, it may be helpful to estimate the cRMS-dependent parameters for the gamma distribution.
Improving efficiency
The method presented here can be relatively efficiently executed on a small desktop computer. The key idea is to use a conventional RDBMS to handle the large amount of structural data. The most time-consuming part is the access to data stored on a hard disk. Conventional RDBMS implements a cache mechanism so that frequently accessed data are stored in memory when possible. Using this mechanism, it is possible speed up the similarity search by simply implementing the GI method in a computer with a large memory. This will automatically lead to the efficiency comparable to the GH method. However, unlike naive implementations of the GH method, the present GI method does not break even when the data size grows to such an extent that it does not fit into the memory.
Another possible improvement may be made by reducing the number of query refsets to be examined. The current implementation requires a CPU time proportional to the number of refsets of the query, which ranges from ~100 to 2,000 or more in typical proteins. In the examples given above, a search with myoglobin (PDB ID: 101m) with 376 refsets took approximately 160 minutes while a search with alcohol dehydrogenase (PDB ID: 1het) with 1654 refsets took 730 minutes (~12 hours). If we can eliminate many of the query refsets which are unlikely to be ligand binding sites, the computational time may be greatly reduced.
Conclusion
We have developed a method for searching for local atomic structures of proteins in database that are structurally similar to sub-structures of a given query protein structure. In particular, we presented techniques based on a conventional relational database management system to practically deal with the huge amount of structural data currently available in the Protein Data Bank. In spite of the facts that the size of the database is massive and that the resolution of the alignments obtained by the method is of the atomic level, the present method can yield search results typically within a few hours using an ordinary desktop computer. With further improvements discussed above, the present method seems to be a promising approach to routinely searching for local structural similarity at atomic resolution, and to functional annotation of newly determined protein structures. Finally it is noted that the core idea of the present method is a very general one, and is obviously applicable to other similar problems such as, for example, the similarity search of molecular surfaces3,34 where the geometric hashing tech nique is applicable in principle, but prohibitive in practice due to a huge data size.
Acknowledgments
The authors thank Drs. Daron Standley and Kengo Kinoshita for critical comments on an early version of the manuscript. This work was supported by a grant-in-aid from the Institute for Bioinformatics Research and Developmenet, the Japan Science and Technology Agency.
References
- 1.Jones S, Thornton JM. Searching for functional sites in protein structures. Curr Opin Struct Biol. 2004;8:3–7. doi: 10.1016/j.cbpa.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 2.Kinoshita K, Sadanami K, Kidera A, Go N. Structural motif of phosphate-binding site common to various protein superfamilies: all-against-all structural comparison of protein-mononucleotide complexes. Protein Eng. 1999;12:11–14. doi: 10.1093/protein/12.1.11. [DOI] [PubMed] [Google Scholar]
- 3.Kinoshita K, Nakamura H. Identification of protein biochemical functions by similarity search using the molecular surface database eF-site. Protein Sci. 2003;12:1589–1595. doi: 10.1110/ps.0368703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brakoulias A, Jackson RM. Towards a structural classification of phosphate binding sites in protein-nucleotide complexes: an automated all-against-all structural comparison using geometric matching. Proteins. 2004;56:250–260. doi: 10.1002/prot.20123. [DOI] [PubMed] [Google Scholar]
- 5.Wallace AC, Borkakoti N, Thornton JM. TESS: A geometric hashing algorithm for deriving 3D coordinate templates for searching structural databases. application to enzyme active sites. Protein Sci. 1997;6:2308–2323. doi: 10.1002/pro.5560061104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stark A, Sunyaev S, Russell RB. A model for statistical significance of local similarities in structure. J Mol Biol. 2003;326:1307–1316. doi: 10.1016/s0022-2836(03)00045-7. [DOI] [PubMed] [Google Scholar]
- 7.Jambon M, Imberty A, Deléage G, Geourjon C. A new bioinformatic approach to detect common 3D sites in protein structures. Proteins. 2003;52:137–145. doi: 10.1002/prot.10339. [DOI] [PubMed] [Google Scholar]
- 8.Berman H, Henrick K, Nakamura H. Announcing the worldwide protein data bank. Nature Struct Biol. 2003;10:980. doi: 10.1038/nsb1203-980. [DOI] [PubMed] [Google Scholar]
- 9.Garcia-Molina H, Ullman JD, Widom J. Database Systems: The Complete Book. Prentice Hall; Upper Saddle River, NJ, U.S.A: 2002. [Google Scholar]
- 10.Lawler E. Combinatorial Optimization: Networks and Matroids. Dover; New York, U.S.A: 2001. [Google Scholar]
- 11.Gupta A, Ying L. Technical Report RC 21576 (97320) IBM T. J. Watson Research Center; 1999. On algorithms for finding maximum matchings in bipartite graphs. [Google Scholar]
- 12.Wesbrook J, Ito N, Nakamura H, Henrick K, Berman HM. PDBML: the representation of archival macromolecular structure data in XML. Bioinformatics. 2005;21:988–992. doi: 10.1093/bioinformatics/bti082. [DOI] [PubMed] [Google Scholar]
- 13.Barber C, Dobkin D, Huhdanpaa H.The Quickhull algorithm for convex hulls ACM Trans on Math Software 22469–483.1996http://www.qhull.org. [Google Scholar]
- 14.Wolfson HJ, Rigoutsos I. Geometric hashing: An overview. IEEE Comput Sci Eng. 1997;4:10–21. [Google Scholar]
- 15.Nussinov R, Wolfson HJ. Efficient detection of three-dimensional structural motifs in biological macromolecules by computer vision techniques. Proc Natl Acad Sci USA. 1991;88:10495–10499. doi: 10.1073/pnas.88.23.10495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bollobás B. Modern Graph Theory. Springer-Verlag; New York, U.S.A: 1998. [Google Scholar]
- 17.Diamond R. A note on the rotational superposition problem. Acta Cryst. 1988;A44:211–216. [Google Scholar]
- 18.Wibley JEA, Waters TR, Haushalter K, Verdine GL, Pearl LH. Structure and specificity of the vertebrate anti-mutator uracil-dna glycosylase smug1. Mol Cell. 2003;6:1647–1659. doi: 10.1016/s1097-2765(03)00235-1. [DOI] [PubMed] [Google Scholar]
- 19.Alfonso-Prieto M, Borovik A, Carpena X, Murshudov G, Melik-Adamyan W, Fita I, Rovira C, Loewen PC. The structures and electronic configuration of compound I intermediates of helicobacter pylori and penicillium vitale catalases determined by X-ray crystallography and QM/MM density functional theory calculations. J Am Chem Soc. 2007;129:4193–4205. doi: 10.1021/ja063660y. [DOI] [PubMed] [Google Scholar]
- 20.Betzel C, Klupsch S, Papendorf G, Hastrup S, Branner S, Wilson K. Crystal structure of the alkaline proteinase savinase from bacillus lentus at 1.4 å resolution. J Mol Biol. 1992;223:427–445. doi: 10.1016/0022-2836(92)90662-4. [DOI] [PubMed] [Google Scholar]
- 21.Eschenburg S, Genov N, Peters K, Fittkau S, Stoeva S, Wilson K, Betzel C. Crystal structure of subtilisin DY, a random mutant of subtilisin carlsberg. Eur J Biochem. 1998;257:309–318. doi: 10.1046/j.1432-1327.1998.2570309.x. [DOI] [PubMed] [Google Scholar]
- 22.Macrae IJ, Zhou K, Li F, Repic A, Brooks AN, Cande WZ, Adams PD, Doudna JA. Structural basis for double-stranded rna processing by dicer. Science. 2006;311:195–198. doi: 10.1126/science.1121638. [DOI] [PubMed] [Google Scholar]
- 23.Pioletti M, Schlunzen F, Harms J, Zarivach R, Gluhmann M, Avila H, Bashan A, Bartels H, Auerbach T, Jacobi C, Hartsch T, Yonath A, Franceschi F. Crystal structures of complexes of the small ribosomal subunit with tetracycline, edeine and IF3. EMBO J. 2001;20:1829–1839. doi: 10.1093/emboj/20.8.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brady K, Wei A, Ringe D, Abeles R. Structure of chymotrypsin-trifluoromethyl ketone inhibitor complexes: comparison of slowly and rapidly equilibrating inhibitors. Biochemistry. 1990;29:7600–7607. doi: 10.1021/bi00485a009. [DOI] [PubMed] [Google Scholar]
- 25.Kinoshita K, Nakamura H. eF-site and PDBjViewer: database and viewer for protein functional sites. Bioinformatics. 2004;20:1329–1330. doi: 10.1093/bioinformatics/bth073. [DOI] [PubMed] [Google Scholar]
- 26.Zheng JH, Trafny EA, Knighton DR, Xuong NH, Taylor SS, Ten Eyck LF, Sowadski JM. 2.2-angstrom refined crystal-structure of the catalytic subunit of cAMP-dependent protein-kinase complexed with MnATP and a peptide inhibitor. Acta Crystallogr. 1993;D49:362–365. doi: 10.1107/S0907444993000423. [DOI] [PubMed] [Google Scholar]
- 27.Kobayashi N, Go N. A method to search for similar protein local structures at ligand-binding sites and its application to adenine recognition. Eur Biophys J. 1997;26:135–144. doi: 10.1007/s002490050065. [DOI] [PubMed] [Google Scholar]
- 28.Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: A structural classification of proteins database for the investigation of sequences and structures. J Mol Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 29.Xu R, Carmel G, Sweet R, Kuret J, Cheng X. Crystal structure of casein kinase-1, a phosphate-directed protein kinase. EMBO J. 1995;14:1015–1023. doi: 10.1002/j.1460-2075.1995.tb07082.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gogos A, Shapiro L. Large conformational changes in the catalytic cycle of glutathione synthase. Structure. 2002;10:1669–1676. doi: 10.1016/s0969-2126(02)00906-1. [DOI] [PubMed] [Google Scholar]
- 31.Meijers R, Morris RJ, Adolph HW, Merli A, Lamzin VS, Cedergren-Zeppezauer ES. On the enzymatic activation of NADH. J Biol Chem. 2001;276:9316–9321. doi: 10.1074/jbc.M010870200. [DOI] [PubMed] [Google Scholar]
- 32.Gatti DL, Entsch B, Ballou DP, Ludwig ML. pH-dependent structural changes in the active site of p-hydroxybenzoate hydroxylase point to the importance of proton and water movements during catalysis. Biochemistry. 1996;35:567–578. doi: 10.1021/bi951344i. [DOI] [PubMed] [Google Scholar]
- 33.Pennec X, Ayache N. A geometric algorithm to find small but highly similar 3D substructures in proteins. Bioinformatics. 1998;14:516–522. doi: 10.1093/bioinformatics/14.6.516. [DOI] [PubMed] [Google Scholar]
- 34.Shulman-Peleg A, Nussinov R, Wolfson HJ. Recognition of functional sites in protein structures. J Mol Biol. 2004;339:607–633. doi: 10.1016/j.jmb.2004.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]