Abstract
Protein-protein interactions support most biological processes, and it is important to find specifically interacting partner proteins among homologous proteins in order to elucidate cellular functions such as signal transduction systems. Various high-throughput experimental methods for identifying these interactions have been invented, and used to generate a huge amount of data. Because these experiments have been applied to only a few organisms, and their accuracy is believed to be limited, it would be valuable to develop computational methods for predicting protein-protein interactions from their amino acid sequences or tertiary structural information. In this study, we describe a prediction method of interacting proteins based on homology-modeled complex structures. We employed the statistical residue-residue contact energy used in a previous study, and two types of new scores, simple electrostatic energy and sequence similarity between target sequences and template structures. The validity of each protein-protein complex model was measured using their single and combined scores. We applied our method to all the protein heterodimers of Saccharomyces cerevisiae. To evaluate the prediction performance of our method, we prepared two types of protein-protein interaction dataset: a complete dataset and high confidence dataset. The complete dataset (10,325 protein dimer models) contains all the yeast protein heterodimers whose complex structures can be modeled. Among them, pairs registered in the DIP database are defined as interacting pairs, and those not registered are defined as non-interacting protein pairs. The high confidence dataset (3,219 protein dimer models) is a more reliable subset of the complete dataset extracted using the criteria of the common subcellular localization. Both datasets show that sequence similarity has a much higher discrimination power than the other structure-based scores, but that the inclusion of contact energy results in significant improvement over predictions using sequence similarity alone. These results suggest that the sequence similarity is indispensable for the prediction, whereas structure scores can play supporting roles.
Keywords: protein-protein interaction, homology modeling, binding specificity, sequence similarity, contact energy
Protein-protein interactions support most important cellular functions, such as signal transduction, enzymatic activities, replication and translation. Recently, high-throughput screening methods, such as yeast-two-hybrid (Y2H) and tandem affinity purification (TAP), have generated large datasets of protein-protein interactions1–6. These interaction data are compiled in databases such as DIP, MIPS and BIND, which also contain data obtained by classical “low-throughput” methods7–9.
The high-throughput genome-wide screening experiments provide us with rich information about cellular processes. Because these techniques are costly and labor-intensive, however, these experiments have been performed only for a few organisms (e.g., Saccharomyces cerevisiae), even though complete genome sequences for more than two hundred organisms have been determined to date. To fill the gap between the vast amount of genome sequence data and the relatively smaller scope of interaction data, many researchers have worked to develop methods for computational prediction of protein-protein interactions from their amino acid sequences10,11.
Various approaches have been proposed to predict protein-protein interactions, such as gene fusion methods12,13, phylogenetic profiling methods14, co-evolution methods15,16, and homologous interaction methods17–21. Recently, several researchers proposed prediction methods based on 3D structures of protein-protein complexes22–26. These studies employed a common standard procedure. First, a structure of the two target proteins in complex is generated by comparative-modeling methods. For example, Alloy and Russell employed BLAST to find template structures for homology modeling; Lu et al. used a threading program developed by them for modeling multimers. In contrast to the residue-level coarse-grained models in these studies, Davis et al. used full-atomic models obtained from MODBASE27. Second, the validity of modeled structures is evaluated by interaction energies. Knowledge-based residue-residue contact energies were employed by each of these three studies cited above. Third, interaction energies are evaluated by applying various statistical scores. Alloy and Russell and Davis et al. employed the Z-score, using randomly shuffled sequences as the reference. Lu et al. also used the Z-score, but their reference state was a set of scores of all the template structures in a library. The prediction accuracies of all these studies were mainly confirmed by the overlaps with experimentally determined interactions. False predicted interactions have not been evaluated as extensively.
In this study, we also employed a structure-based approach, but we evaluated our predictions by discriminating between interacting and non-interacting protein pairs. In other words, we mainly focused on the interaction specificities among homologous protein pairs. We chose to do so because the specific interactions among similar homologous proteins are important for many cellular functions. There are many paralogous protein domains in eukaryotic genomes, and each has its own set of specific interacting partners. Proteins working in signal transduction pathways, especially protein kinases, G-proteins and transcription factors, have many similar homologues within genomes28. Binding specificities of these proteins are the basis of a complicated and robust signal transduction systems within the cell29.
One of the problems for evaluating reliability and coverage of predictions is that there is no gold standard for discriminating interacting and non-interacting protein pairs. This problem arises in part because high-throughput experiments of protein-protein interaction are believed to contain unreliable or inaccurate data30–32. Specifically, there is no gold standard for unambiguously defining non-interacting protein pairs. In this study, we prepared two types of dataset comprising interacting and non-interacting protein pairs: the “complete” dataset and the “high confidence” dataset. The complete dataset contains all the protein heterodimers whose complex structure can be modeled. Protein pairs registered in the DIP database are defined as interacting pairs, while those not registered are defined as non-interacting protein pairs. We expect these assumptions are safe for Saccharomyces cerevisiae, because the yeast is the most popular model organism for protein-protein interactions, and a huge amount of experimental data has been accumulated to date. However, the DIP database may contain both false positive data (i.e., protein pairs registered as interacting that do not, in fact, interact) and false negative data (unregistered protein pairs that actually interact in the cell). To evaluate our method more accurately, we therefore prepared the high confidence dataset, which is a more reliable subset of the complete dataset extracted using subcellular localization data. Recently, genome-wide analyses determining subcellular localization of yeast have been published33–35. We used data from these analyses to determine whether the proteins in each registered interacting pair share a common localization; if so, we regarded as interacting pair as reliable and included it in the high confidence dataset32,36,37. The performance of our method was evaluated by discriminating interacting and non-interacting protein pairs, using both the complete and high confidence dataset.
The outline of our prediction method is as follows. First, we predict the dimer structure of two target proteins by a homology modeling method. Sequence homology searches for the two target protein sequences are run against the sequence library of the component proteins of known dimer structures. If we find a dimer template structure that is composed of two proteins homologous to each target protein, a complex structure of the target proteins are modeled based on the template. To evaluate the validity of structure models, we employed three kinds of scores. First, we used knowledge-based residue-residue contact energy, which is used in each of the three previous studies discussed above. Second, because we expected long-range interaction between protein pairs and binding specificities to be provided by electrostatic interactions, we introduced a simple electrostatic energy. Third, we also employed a score based on sequence similarity between target and template proteins. The sequence similarity for interacting protein pairs has often been used in sequence-based predictions17–21; to date, however, it has not been used in combination with structural features. All three scores were transformed to a Z-score using randomized sequences as a reference. In contrast to previous studies, we analytically estimated the average and variance of energies. The performance using each of the three scores, both individually and in combination, was evaluated by recall-precision plots and maximum F-measure using the complete and high confidence datasets.
Materials and methods
Datasets of heterodimer structures
Datasets of heterodimer structures are required for the library of template structures and for estimating values for statistical contact energies. We excluded the homodimers (pairs of identical proteins) because homodimeric crystal structures of single proteins are less reliable due to artificial crystal packing38. Heterodimers were defined as the proteins whose sequence identity is smaller than 50%. These sets comprise non-redundant representative tertiary structural data of heterodimers obtained from the PQS server39. The PQS server contains putative biological units of quarternary structures determined by X-ray crystallography, which are automatically chosen among the candidate complex structures generated by crystallographic symmetry operations of PDB data. The heterodimers datasets were generated by the following procedures: First, all the multimers included in the PQS server were separated into dimers. Dimers with fewer than five interacting residues (defined as a residue that has at least one Cβ atom located within 7 Å of Cβ atoms of another protein chain) were removed. Second, these dimers were clustered by single-linkage clustering algorithm40 according to similarities between dimers, defined as the lower sequence of the two sequence similarities between corresponding proteins. One representative dimer with the largest number of interacting residues was extracted from each cluster. We used the structural data from PQS (version of April 14, 2006). Two types of representative dataset were prepared using different threshold values of similarity of complexes. The former set comprises 1,687 heterodimers generated by the threshold of 40% similarity and is used as the dataset for calculation of contact energy; the latter comprises 2,635 heterodimers generated by the threshold of 95% similarity and used as the template structure library for the homology modeling.
Building complex structure models of yeast hetero protein pairs
From the UniProt ver. 49.4 database41, which is a curated protein sequence database with a high level of annotation, we extracted 5,314 Saccharomyces cerevisiae amino acid sequences. All the hetero pairs of the 5,314 yeast protein sequences were subjected to interaction prediction. To construct the sequence profile of each yeast amino acid sequence, PSI-BLAST was run against the nr database (version of September 22, 2006). The threshold for E-value (expected hits) was set to 0.001, and the number of iterations was set to three. Using the generated sequence profiles, we ran PSI-BLAST42 against the template structure library described above. For each target protein pair, we checked whether a dimer template structure consisting of two homologous proteins of each target protein exists in the database. If a dimer template structure was found for the target protein pairs, we required that the following conditions be met: (1) In the two alignments between target protein sequences and template complex, ratios of aligned interacting residues must not be smaller than 50%. (2) The numbers of aligned interacting residues must not be smaller than 10. If several template dimer structures were found, we selected the template whose lowest sequence identity is the highest among the template dimers. In this study, because a fast modeling method is necessary in order to allow us to deal with a large number of protein pairs, we use the conformation of aligned residues from the template structures, ignoring inserted residues, and did not build in side chain atoms for substituted residue.
Interacting and non-interacting protein pairs
The generated complex structure models were labeled either as “interacting” or “non-interacting” protein pairs. We prepared two types of the dataset using different criteria of interaction. In the “complete” dataset, if a protein pair of complex model is registered in protein-protein interaction databases, the pair is considered as an interacting pair. If it is not registered, it is considered as a non-interacting pair. Among many available protein-protein interaction databases, we chose the DIP database, because it contains data obtained via a wide range of experimental methods, such as yeast two hybrid, tandem affinity purification, affinity chromatography, in vitro binding, copurification, complex structures by X-ray crytallography. We used the dataset version of January 16, 2006. Although the DIP database contains a huge number of protein-protein interaction data, several latest experimental results are not yet registered. If we found complex template structures of almost identical (more than 95% sequence identity) proteins to target protein pairs, we relabeled these pairs as “interacting” pairs even if they are not registered in the DIP database, considering these experimentally determined complex structures as sufficiently well-supported to justify registration in the DIP database.
The complete dataset assumes all the interactions are already registered; however, high-throughput experiments of protein-protein interaction are believed to contain unreliable or inaccurate data, and protein pairs not registered in the DIP database may interact in the cell. To increase the reliability of the dataset, we prepared a “high confidence” dataset, a more reliable subset of the complete dataset extracted using subcellular localization information. Subcellular localization data was downloaded from the MIPS database (version of November 14, 2005), where one or more localized compartment types are assigned to each yeast protein. Localized compartment types consist of 19 types: extracellular, bud, cell wall, cell surface, plasma membrane, inner membrane, cytoplasm, cytoskeleton, endoplasmic reticulum, golgi body, transport vesicle, nuclear, mitochondria, peroxisome, endosome, vacuole, microsome, lipid particle, and other subcellular localization. The ratios of localized compartment types for the 5,211 registered yeast proteins are 34% for nuclear, 18% for cytoplasm, 18% for mitochondria, 5% for vacuole, 5% for endoplasmic reticulum, 4% for unknown, 2% for transport vesicle and 14% for other localized compartment types.
Protein pairs registered in the DIP database sharing at least one localizing compartment are selected for the high confident interacting protein pairs, those not registered in DIP not sharing any localizing compartments are selected for the high confident non-interacting protein pairs. The assumption of the high confidence dataset is that two proteins having different subcellular localizations do not interact each other, whereas reported pairs with similar localizations certainly interact in the cell.
Residue-residue statistical contact energy for protein-protein interaction
Residue-residue statistical contact energies were originally developed for coarse-grained models of protein folding and threading43–45. Recently, similar approaches were applied for evaluating protein-protein interaction46,47. In this study, we employed a typical log-odds formula for extracting the value of contact energies. A statistical contact energy econ(a, b) for contacting residues a and b in different polypeptide chains is estimated by the form of the log-odds score:
| (1) |
where P(a) and P(b) are the probabilities that amino acids a and b appear on the surface, Q(a, b) is the probability that amino acids a and b on the surface contact each other in the protein-protein interface. Surface residues of a protein are defined as those residues whose relative accessible surface areas are larger than 35%. Contacting residue pairs are defined as the residues in different chains, whose Cβ atoms are located within 7 Å of one another. Both probabilities are estimated using the dataset for calculation of contact energy (see Datasets of heterodimer structures). If the interface contacts between residues a and b are often found in the interface, the value of econ(a, b) is large and negative.
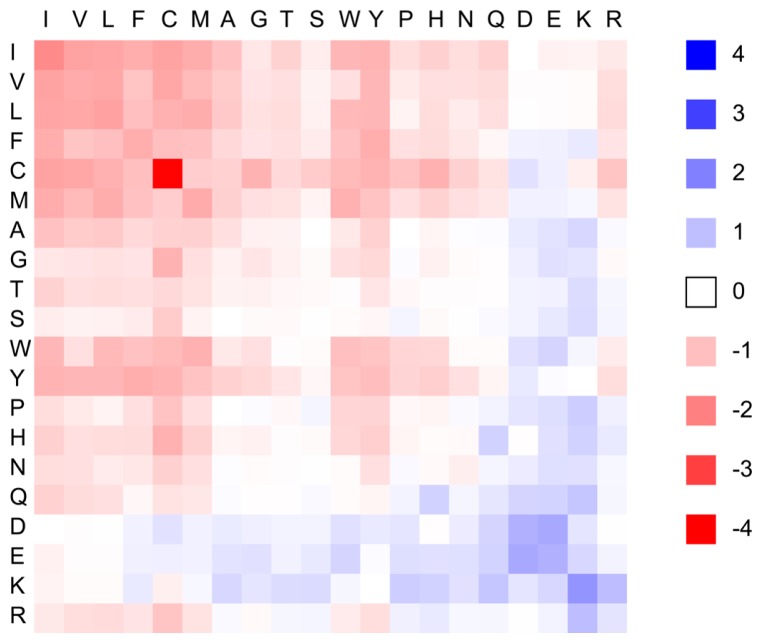
The estimated energy values are summarized in Figure 1. Hydrophobic residues are attractive to each other, especially in the case of the cysteine-cysteine pair. Hydrophilic residues, however, are generally repulsive even for differently charged residue pairs, such as the arginine-glutamic acid pair. These features are similar to those employed in previous studies46,47.
Figure 1.
Residue-residue statistical contact energy in protein-protein interfaces. In the horizontal and vertical axes, 20 amino acids are arranged in descending order of hydrophobicity. Energy values are represented from red (low energy) to blue (high energy).
The total contact energy Econ is the sum of the econ for all the contacted residue pairs including both surface and buried residues:
| (2) |
where N and M are the total number of the residues of proteins, and ai and aj are the amino acids of residues i and j.
Electrostatic energy for protein-protein interaction
Electrostatic interactions also play an important role in protein-protein interactions48. To validate our dimer models, we employed simplified electrostatic energies as proposed by Shaul and Schreiber49. An electrostatic energy eele between charges q1 and q2 is calculated by the following equation based on the Debye-Huckel theory:
| (3) |
where ɛr is the relative permittivity of water (=80). The variable r is a distance between the charges q1 and q2, and κ is Debye-Huckel screening parameter (=0.488 Å−1). The parameter a is set to 6 Å.
The total electrostatic energy Eele is the sum of the eele for all of the charged atom pairs:
| (4) |
where i and j are residues included in different proteins. The numbers N and M are the total number of residues, and Qi and Qj are the sets of charged atoms belonging to the residue i and j. The variable rst is a distance between atom s and t. The variable qs(ai) is the charge of the atom s of amino acid ai.
Formal charges are assigned to the atoms in the modeled complex structure: charge = −1 for aspartic acid and glutamic acid, and charge = +1 for lysine and arginine. To assign the charges for the model structures, we employed the charge rule proposed by Shaul and Schreiber49. For a substituted residue of the target sequence, total charges of the residue are equally assigned to the position of the selected atoms of the corresponding residue on the template structure. The location of the pseudo-charge on the amino acids is given in Table 1. For example, if the amino acid of the target protein is glutamic acid, and the corresponding amino acid of the template structure is threonine, a charge −0.5 is assigned to both OG1 and CG2 atoms of the threonine residue.
Table 1.
Atoms of amino acids where charges can be assigned
| Residue | Atom | Residue | Atom | Residue | Atom |
|---|---|---|---|---|---|
| GLU | OE1 | TRP | CE3 | SER | OG |
| GLU | OE2 | TYR | OH | ILE | CD1 |
| ASP | OD1 | PHE | CZ | MET | CE |
| ASP | OD2 | GLN | OE1 | LEU | CD1 |
| ARG | NH1 | GLN | NE2 | LEU | CD2 |
| ARG | NH2 | ASN | OD1 | VAL | CG1 |
| LYS | NZ | ASN | ND2 | VAL | CG2 |
| HIS | ND1 | CYS | SG | ALA | CB |
| HIS | NE2 | THR | OG1 | PRO | CG |
| TRP | CE2 | THR | CG2 | GLY | CA |
PDB atomic names are shown. These atoms are mainly taken from Shaul and Schreiber’s charge rules (Shaul and Schreiber, 2005). Atoms of proline and glycine have been added; OXT and the N-terminus atom have been removed.
Normalization of the energies
A Z-score is introduced to normalize the contact and electrostatic energies, and to remove biases of amino acid compositions of target proteins22–25. The Z-score for energy E is defined as follows:
| (5) |
where Mean[E] and Var[E] are the average and variance of E respectively for randomly shuffled amino acids sequences of the same composition. Z-score shows how many units of the standard deviation an energy of a protein pair is above or below the average by the random shuffling. Calculation of the averages and variances of the contact energy and electrostatic energy are described in the following sections. In contrast to studies by other groups, we analytically estimated the average and variance of energies without explicitly generating randomly shuffled sequences.
Mean and variance of contact energy for randomly shuffled sequences
We assume that random contacting amino acid pairs are generated by picking up two amino acids randomly from the surfaces of different proteins. For this random set of contacting amino acids, the average μcon and variance of the contact energy are calculated as follows:
| (6) |
| (7) |
where P(a) and P(b) are the proportions of amino acid a and b in surface residues for each protein, and A is the set of 20 genetically encoded amino acids. If we assume that the all the contacting protein pairs are independent in the shuffling process, the average and variance of the total contact energy Econ are calculated as follows:
| (8) |
| (9) |
where Ncontact is the total number of the contacting residues.
Mean and variance of electrostatic energy for randomly shuffled sequences
The average and variance of the electrostatic energy can be calculated in a similar way to that of the contact energy. We assume that random contacting amino acid pairs on the i-th and j-th positions of proteins are generated by picking up two amino acids randomly from the surfaces of different proteins. The average μele(i, j) and variance values of the electrostatic energy for the random sets are calculated as follows:
| (10) |
| (11) |
where the variable rst is the distance between atom s and t. The variables qs(a) and qt(b) are the charges of the atoms s and t of the i-th and j-th residues when they are replaced by amino acids a and b. P(a) and P(b) are the frequencies of amino acids a and b in surface residues for each protein. Qi and Qj are the set of charged atoms belonging to the residues i and j. If we assume that the all the protein pairs are independent in the shuffling process, the average and variance of the total electrostatic energy Eele are calculated as the sum of average and variance of each amino acids pairs:
| (12) |
| (13) |
where N and M are the total numbers of residues in each protein.
Sequence similarity between target and template
We employed sequence similarity between target protein and template protein as another feature for finding interacting proteins. We expected that two proteins will interact with each other if they have close homologues whose dimer structures have been experimentally determined. Here a Z-score is also introduced to measure sequence similarities. In this case, the number of identical residues Niden in the alignment is normalized by average and variance values for randomly shuffled sequences:
| (14) |
where Niden is the number of the identical residues, and Ncomp is the number of compared residues in the alignment with gaps removed. We assume that random shuffling is applied using the uniform distribution of amino acids (p is set to 1/20), and that the number of identical residue Niden obeys the binominal distribution. Because the other two Z-scores of energies have negative value for probable interfaces, the Z-score for sequence similarity was multiplied by minus one to facilitate comparison. Because we are modeling dimer structures, two different sequence similarities are obtained for one protein complex. We employed the higher score (in other words, the lower sequence similarity) for the purposes of discrimination.
The random shuffling process for sequence similarity is subtly different from that of contact and electrostatic energy. For contact and electrostatic energy, two amino acids on the surface are randomly chosen. In the case of the sequence similarity, the sequence of the template protein is fixed, and the sequence of the target protein is randomly generated using a uniform distribution of amino acids.
Evaluation by recall-precision plots
To evaluate the discriminating powers between the interacting and non-interacting protein pairs, recall-precision plots were generated. Recall and precision are defined as follows:
| (15) |
| (16) |
where Ntp(S) is the number of interacting protein pairs with a score better than S, Nt is the number of interacting protein pairs and Np(S) is the number of pairs with a score better than S. Recall shows how many correct interactions are covered by the prediction, precision shows how reliable the prediction is. Recall and precision were calculated against all of the observed scores and plotted as a line on the plane. The line plotted more towards the upper right has larger Recall and Precision values than those toward the lower left. Generally speaking, predictions with high Recall value tend to have a low value of Precision. Thus, the maximum F-measure is introduced to find a good balance point between recall and precision. F-measure F(S) is defined as the harmonic mean of recall and precision, and the maximum F-measure Fmax is the largest F-measure among all of the observed scores:
| (17) |
| (18) |
Results and Discussion
Homology-modeled dimer structures of the interacting and non-interacting protein pairs
We modeled dimer structures of hetero protein pairs of Saccharomyces cerevisiae by the homology-modeling method. 10,325 models of protein pairs were generated; among them, 417 pairs were regarded as interacting, and 9,908 pairs were regarded as non-interacting. We call these pairs the complete dataset of protein-protein interaction. To select reliable data, the complete dataset is classified into three types of protein pairs: (i) Two proteins share at least one common localized compartment type. (ii) Subcellular localization of at least one protein is unknown. (iii) Two proteins do not share any localized compartment type. The classification is shown in Table 2. The interacting pairs in the complete dataset sharing at least one localized compartment are selected for the high confidence interacting pairs (380 pairs), and the non-interacting pairs not sharing any localized compartments are selected for the high confidence non-interacting pairs (2,839 pairs). Notably, the high confidence dataset contains only 37 fewer interacting pairs than the complete dataset, but 7,069 fewer non-interacting pairs. In other words, most of the protein pairs registered in DIP database have a similar localization, but there are many protein pairs that have a similar localization but nonetheless are not reported.
Table 2.
The classification of interacting and non-interacting protein pairs included in the complete dataset by subcellular localization
| Interacting pairs | Non-interacting pairs | ||
|---|---|---|---|
| (i) | Two proteins share at least one common localized compartment | 380 | 5,631 |
| (ii) | Subcellular localization of at least one protein is unknown | 10 | 1,438 |
| (iii) | Two proteins do not share any localized compartments | 27 | 2,839 |
|
| |||
| Total | 417 | 9,908 | |
The underlined numbers are for the complete dataset; bold numbers are for the high confidence dataset.
Network of the protein-protein interaction in the complete dataset
In order to have a full picture of these protein pairs, we drew a network of protein-protein interaction in the complete dataset (Fig. 2). In this network, nodes correspond to target proteins and edges correspond to target protein pairs whose dimer structure can be modeled. There are 1,036 nodes and 10,325 edges in the network. As there are approximately twenty-four times more non-interacting than interacting pairs, most of the edges are colored in blue. The network was separated into 64 clusters by single linkage clustering. Our network was more sparse than those appearing in previous experimental studies3,6, probably because we more stringently restricted the protein pairs that are able to be homology-modeled.
Figure 2.
The protein-protein interaction network of the interacting and non-interacting protein pairs included in the complete dataset. The graph was visualized by Cytoscape50. The nodes correspond to the target proteins; edges correspond to interactions. The interacting protein pairs are shown in red, the non-interacting ones in blue. The proteins including the domains of protein kinase catalytic subunit, WD40-repeat, G proteins, canonical RBD, ankyrin repeat, cyclin are colored green, cyan, red, yellow, gray and black, respectively. If the target protein includes more than two domains from the six types of domains, the node is colored according to the domain nearest to the N-terminus. The SCOP, which is the structural classification database of proteins, was used for identifying the domains51.
The largest cluster (Cluster A) has 573 proteins, and the second and third largest cluster (Cluster B and Cluster C) have 41 and 30 proteins, respectively. We focused on the target proteins included in Cluster A, and colored the nodes in the network according to the major domains included in Cluster A. Cluster A contains proteins involved in the signal transduction system. The numbers of the target proteins which include the domain of protein kinases catalytic subunit (green), WD40-repeat (cyan), G proteins (red), canonical RBD (yellow), ankyrin repeat (gray), cyclin (black) are 119, 97, 55, 50, 18 and 16, respectively. Cluster B contains proteins associated with ubiquitination, and consists of two major families: 17 domains of RING finger domain C3HC4 and 14 domains of ubiquitin conjugating enzyme UBC. Cluster C contains proteins involved in the DNA replication, and there were 23 domains of extended AAA ATPase, and 7 domains of DNA polymerase III clamp loader subunits C-terminal.
To show frequently appearing families in the network more precisely, we show statistics for the family pairs of the template complexes according to the interacting and non-interacting pairs included in the complete and high confidence dataset (Table 3). The family pairs of the non-interacting protein pairs are more biased than those of the interacting pairs, and the biases are mostly caused by the six major families colored in the network of Figure 2. For example, in case of the complete dataset, protein kinase catalytic subunit domains and ankyrin repeat domains form as many as 1,912 non-interacting protein pairs. Similar biases were also observed in the high confidence dataset, although its observed numbers of family pairs are smaller.
Table 3.
Family pairs frequently appearing in template complexes
| Family pairs of the template structuresa | PDBb | Complete dataset | High confidence dataset | ||
|---|---|---|---|---|---|
|
|
|
||||
| Interc | Non-interd | Interc | Non-interd | ||
| Top 10 family pairs of the interacting protein pairs | |||||
|
| |||||
| 1. b.38.1.1/b.38.1.1 | 1b34AB | 33 | 24 | 33 | 0 |
| 2. d.153.1.4/d.153.1.4 | 1g65JK | 30 | 44 | 30 | 0 |
| 3. h.1.15.1/h.1.15.1 | 1gl2BC | 20 | 80 | 10 | 45 |
| 4. c.37.1.20-a.80.1.1/c.37.1.20-a.80.1.1 | 1sxjBC | 19 | 95 | 19 | 15 |
| 5. d.144.1.7/a.74.1.1-a.74.1.1 | 1finAB | 18 | 1662 | 14 | 559 |
| 6. c.3.1.3-d.16.1.6-c.3.1.3/c.37.1.8 | 1ukvGY | 13 | 61 | 12 | 13 |
| 7. d.144.1.7/d.211.1.1 | 1bi7AB | 10 | 1912 | 10 | 381 |
| 8. a.22.1.1/a.22.1.1 | 1id3AF | 9 | 12 | 9 | 0 |
| 9. i.1.1.1/i.1.1.1 | 1s1hJN | 8 | 16 | 8 | 9 |
| 10. a.116.1.1/c.37.1.8 | 1ow3AB | 6 | 342 | 5 | 99 |
|
| |||||
| Top 10 family pairs of the non-interacting protein pairs | |||||
|
| |||||
| 1. d.144.1.7/d.211.1.1 | 1g3nAB | 10 | 1912 | 10 | 381 |
| 2. a.74.1.1-a.74.1.1/d.144.1.7 | 1oiuBC | 18 | 1662 | 14 | 559 |
| 3. c.37.1.8-a.66.1.1-c.37.1.8/b.69.4.1 | 1gotAB | 1 | 530 | 1 | 321 |
| 4. a.116.1.1/c.37.1.8 | 1ow3AB | 6 | 342 | 5 | 99 |
| 5. j.66.1.1/d.144.1.7 | 1f3mAC | 1 | 319 | 1 | 112 |
| 6. c.10.2.4/d.58.7.1 | 1a9nAB | 2 | 257 | 1 | 109 |
| 7. c.37.1.8/c.10.1.2 | 1k5dAC | 4 | 239 | 3 | 87 |
| 8. c.45.1.1/d.144.1.7 | 1fq1AB | 0 | 204 | 0 | 59 |
| 9. a.48.1.1-a.39.1.7-d.93.1.1-g.44.1.1/d.20.1.1 | 1fbvAC | 3 | 189 | 3 | 38 |
| 10. a.118.1.1/c.37.1.8 | 1qbkBC | 4 | 184 | 4 | 44 |
SCOP ID included in the table are following; a.22.1.1:Nucleosome core histones, a.39.1.7:EF-hand modules in multidomain proteins, a.48.1.1:N-terminal domain of cb1 (N-cb1), a.66.1.1:Transducin (alpha subunit) insertion domain, a.74.1.1:Cyclin, a.80.1.1:DNA polymerase III clamp loader subunits C-terminal domain, a.116.1.1:BCR-homology GTPase activation domain (BH-domain), a.118.1.1:Armadillo repeat, b.38.1.1:Sm motif of small nuclear ribonucleoproteins SNRNP, b.69.4.1:WD40-repeat, c.3.1.3:GDI-like N domain, c.10.1.2:Rna1p (RanGAP1) N-terminal domain, c.10.2.4:U2A′-like, c.37.1.8:G proteins, c.37.1.20:Extended AAA-ATPase domain, c.45.1.1:Dual specificity phosphatase-like, d.16.1.6:GDI-like, d.20.1.1:Ubiquitin conjugating enzyme UBC, d.58.7.1:Canonical RBD, d.93.1.1:SH2 domain, d.144.1.7:Protein kinases catalytic subunit, d.153.1.4:Proteasome subunits, d.211.1.1:Ankyrin repeat, g.44.1.1:RING finger domain C3HC4, h.1.15.1:SNARE fusion complex, i.1.1.1:Ribosome complexes, j.66.1.1:pak1 autoregulatory domain.
PDB code of the template complexes.
Number of interacting protein pairs.
Number of non-interacting protein pairs.
Recently, researchers report that a protein-protein interaction network is a small world network, which is a network in which the length of the shortest path between any protein pairs tends to be small, but also has densely connected local neighborhood, and the number of interactions per proteins (degree) appears to follow a power law distribution52,53. Our non-interacting protein network was not a small world network, because its average length of the shortest path was not small (proteins are clustered into the 64 clusters), and number of interaction per proteins of our network did not follow a power law distribution (number of proteins with degree = 12 was larger than that of degree = 1). The deviation from the power law distribution was caused by the biased family distribution of non-interacting network.
Score distributions of the complete dataset for each feature
The Z-score distributions of three features (contact energy, electrostatic energy and sequence similarity between target and template) of the complete dataset are shown in Figure 3–5. As we assume that similar random surface amino acid pairs are generated in Z-score calculations of both contact and electrostatic energy, these Z-scores are comparable to each other. Z-scores for the contact energy ranged lower, and were distributed more widely, than Z-score for the electrostatic energy. The averages of Z-score of the contact energy for interacting and non-interacting protein pairs were −4.6 and −2.2, respectively, whereas those for the electrostatic energy were −0.77 and −0.15. The variances of the contact energies are 7.6 (interacting) and 4.6 (non-interacting) and those of the electrostatic energies are 0.99 (interacting) and 0.67 (non-interacting). As the differences of the averages between the interacting and non-interacting interacting protein pairs were 2.4 (contact energy) and 0.62 (electrostatic energy), the discrimination power of the contact energy seemed to be better than that of the electrostatic energy. The distribution of sequence similarities for the interacting protein pairs was not bell-shaped (as was the case for the contact and electrostatic energies), and was skewed toward the left. The distribution of the interacting pairs was broader than that of the non-interacting pairs; the variances of the Z-score distribution of sequence similarity are 394.2 (interacting) and 20.7 (non-interacting). The high confidence dataset also yields similar distributions (data not shown).
Figure 3.
Distributions of Z-scores of contact energy calculated for protein pairs included in the complete dataset. Black and gray bars correspond to interacting and non-interacting protein pairs respectively.
Figure 4.
Distributions of Z-score of electrostatic energy calculated for the protein pairs included in the complete dataset.
Figure 5.
Distributions of Z-score of sequence similarity calculated for the protein pairs included in the complete dataset.
Recall-precision plots
To evaluate the discrimination more strictly, we generated recall-precision plots for all three Z-scores, both individually and in combination. To generate combined scores, two or three Z-scores were added without any weights. Recall-precision plots are shown in Figure 6 (complete dataset) and Figure 7 (high confidence dataset); maximum F-measures of the recall-precision plot are summarized in Figure 8 (complete dataset) and Figure 9 (high confidence dataset). We also tested various weights such as Fischer’s discriminant method, but performance was not significantly improved. The basic characteristics of plots using the complete and high confidence dataset are similar, except that precision values and maximum F-measure of the high confidence dataset were generally higher than those of the complete dataset, probably because the number of non-interacting protein pairs (2,839 pairs) in the high confidence dataset was about one forth of that in the complete set (9,908 pairs). Similar biased results using co-localization datasets are reported in previous studies36,37.
Figure 6.
Recall-precision plots for discrimination between interacting and non-interacting protein pairs using single and combined scores in the complete dataset. “Con”: contact energy, “Ele”: electrostatic energy, “Seq”: sequence similarity. “Ele+Con”, “Seq+Con”, “Seq+Ele” and “Seq+Ele+Con” correspond to the plots using combined Z-scores. The purple triangle shows the performance of the method of Davis et al.25
Figure 7.
Recall-precision plots for discrimination between interacting and non-interacting protein pairs using single and combined scores in the high confidence dataset. Abbreviations as in Figure 6.
Figure 8.
The maximum F-measures with their recall and precision values for each recall-precision plot using single and combined Z-scores in the complete dataset. Abbreviations as in Figure 6. Dotted line: maximum F-measure of sequence similarity alone.
Figure 9.
The maximum F-measures with their recall and precision values for each recall-precision plot using single and combined Z-scores in case of the high confidence dataset. Abbreviations are the same as those used in Figure 6.
In both datasets, the discriminating power of sequence similarity alone was much higher than that of the contact and electrostatic energies. This high performance was consistent with other studies based on sequence similarities17–21. However, when the contact energy and the electrostatic energy were combined with the sequence similarity, the maximum F-measure was improved by 0.038 for the complete dataset. Similar improvements were observed for the high confidence dataset. This indicates that while sequence information is the most effective feature for detecting interacting protein pairs, structural information is able to improve prediction performance.
To validate the statistical significance of these improvements, we performed bootstrap sampling tests. The maximum F-measure was recalculated using protein pairs bootstrap-sampled from the all protein complex models. The sampling was repeated 1,000 times to generate 1,000 different maximum F-measures. In both datasets, among the 1,000 F-measures, all of the 1,000 F-measures of sequence similarity and contact energy (Seq+Con), and of all the three scores combined (Seq+Ele+Con) were larger than those of only sequence similarity (Seq). However, only 984 F-measures of sequence similarity and electrostatic energy (Seq+Ele) were larger than those of sequence similarity for the complete dataset. For the high confidence dataset, only 809 F-measures of Seq+Ele were larger than those of Seq. Thus, in both datasets, the improvement in discrimination after incorporation of contact energy was statistically significant (p<0.01), whereas, the improvement after incorporation of electrostatic energy was not. That is to say, sequence similarity has a much higher discriminating power than the other structure-based scores, but using contact energy results in significant improvement over predictions using sequence similarity alone.
The level of prediction accuracy practically required by users depends on their purposes. If a researcher needs to know interacting protein pairs without any confirming experiments, we would recommend the prediction with high Precision and low Recall. In contrast, if a researcher plans to perform a number of experiments to confirm protein-protein interactions, and needs candidates of interacting protein pairs, we would recommend the prediction with high Recall and low Precision. The improvement by our contact energy can contribute to the latter case, because Figure 6 indicates that the difference between the sequence similarity and the combined score is the largest in the region where Recall is high (0.4–0.5) and Precision is low (0.3–0.6).
Performance comparison with the previously published method
Generally speaking, it is difficult to quantitatively compare the protein-protein interaction prediction methods, because the criteria for interacting protein pairs and the libraries of complex structures can both differ. We compare the performance of our method with the latest related method proposed by Davis et al.25, by checking overlaps of their predictions with our complete dataset. Their method was based on the statistical contact energy in conjunction with functional annotation and subcellular localization data. The contact energy metric employed in their study was similar to ours, except that it was weighted by the ratio of contacting atoms to total atoms, and its contacting atomic types and threshold distance of contacts were deliberately chosen. Because their complex models were generated by structural alignments of monomer models to template complex structures, the number of model complex structure could be larger than ours if we employed the same structural library. Davis et al. applied their method to all the protein pairs of yeast, finally predicted 3,387 interacting protein pairs. Among the 3,387 predictions, 2,520 predictions are hetero (sequence identity is smaller than 50%) protein pairs, and only 300 pairs are included in our complete dataset; 84 pairs are interacting, and 216 are non-interacting pairs. The remaining 2,220 pairs are modeled by Davis et al., but not modeled by our method. This last difference was caused by the difference of the template structure library; we did not use homodimer templates to avoid artificial crystal packing, whereas they used all kinds of complex structures. We found that most of the remaining 2,220 hetero protein pairs were modeled using homodimer templates. Thus, by the equations (15) and (16), the values of recall and precision of the method of Davis et al. are,
Their values are plotted in Figure 6 (purple triangles). The performance of their method is better than that of our contact energy, and slightly better than that of our contact energy combined with electrostatic energy. This is probably due to their different estimation of contact energy and their filter by co-localization and co-functional annotation. However, the predictive performance of Davis et al.’s method is plotted under the line of sequence similarity (Seq in Fig. 6). Although the comparisons in the two studies were not performed on identical structural libraries and the assumption of our complete dataset is not absolutely correct, our results suggest that methods incorporating sequence similarity will yield more accurate predictions than methods incorporating only structure-based scores along with functional and localization data.
Conclusions
In this study, we developed a method for predicting protein-protein interaction based on dimer structure models, using two structural scores and sequence similarity. Because we restricted the protein pairs whose complex can be modeled by homology, the essence of our approach is the discrimination of specific interaction among similar homologous sequences. Previous structure-based prediction studies of protein-protein interaction have evaluated overlaps of predicted and experimentally observed interacting pairs, but have not checked as carefully the overlaps of non-interacting pairs. Because we believe that non-interacting protein pairs should be also evaluated, we prepared two kinds of datasets containing interacting and non-interacting protein pairs. The complete dataset contains all the hetero protein pairs whose complex structure can be modeled, and the high confidence dataset is the reliable subset using subcellular localization data. The two datasets have both assets and liabilities. On the one hand, reliability of interactions of the high confidence dataset should be higher than that of the complete dataset. On the other hand, precision values estimated from the high confidence dataset are biased to large values, because that set ignores co-localized protein pairs not registered in the DIP database.
Both datasets showed that the performance of a sequence similarity-based score was much greater than scores based on contact and electrostatic energies. Nonetheless, scores related to contact energy, as calculated from structural models, can contribute to improvements over the performance of sequence similarity alone. These results suggest that sequence similarity is indispensable for the prediction, whereas structure scores can play supporting roles. Our preliminary calculation showed that a score only using number of aligned interface residues had a high discrimination power, although it was smaller than that of contact energy. We suggest that the contact energy may indirectly check whether a modeled structure has a sufficient size of interface.
Electrostatic energy showed the worst performance, and did not significantly improve the performance of sequence similarity alone. There are several possible reasons for this poor performance. We employed the simplified electrostatic energy proposed by Shaul and Schreiber49. They reported that this energy successfully predicted the change of association rate kon, however, it may be insufficient to predict binding free energy. This energy ignores partial charges on polar atoms, it can not consider any polar interactions such as hydrogen bonds. Another reason is the inaccuracy of complex models of interacting protein pairs, which may more affect the performance of the electrostatic energy than that of the contact energy. It is because the electrostatic energy depends on sidechain conformations, whereas the contact energy does not. The omission of charges on binding ligands such as nucleotides and metal ions may be a serious problem. Many protein interactions of signal transduction systems are regulated by bindings of charged ligands, such as GTP and GDP.
Our results showed that combined score using sequence similarity and contact energy is the currently most accurately predictive score. Using the combined score, we now plan to apply our method to different organisms, and we hope to obtain new biological findings through our predicted interactions. We also plan to build a WWW server in order to make our prediction service freely available to other researchers.
Acknowledgments
We are grateful to Drs. Kei Yura, Hidetoshi Kono, Kensuke Nakamura, and Gautam Basu for stimulating discussions and encouragements. We thank Drs. Toshio Hakoshima and Naotake Ogasawara for supports and suggestions throughout the work. We also thank Drs. Andrej Sali and Fred P. Davis for providing us the list of their predicted interacting protein pairs to validate our prediction performance. This work was supported by the Special Coordination Funds Promoting Science and Technology and a Grant-in-Aid for Scientific Research on Priority Area (C), Genome Information Science, from MEXT (the Ministry of Education, Culture, Sports, Science and Technology of Japan). N. Fukuhara was supported by a Grand-in-Aid for the 21st Century COE Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
References
- 1.Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, Qureshi-Emili A, Li Y, Godwin B, Conover D, Kalbfleisch T, Vijayadamodar G, Yang M, Johnston M, Fields S, Rothberg JM. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 2.Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci USA. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gavin AC, Bosche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon AM, Cruciat CM, Remor M, Hofert C, Schelder M, Brajenovic M, Ruffner H, Merino A, Klein K, Hudak M, Dickson D, Rudi T, Gnau V, Bauch A, Bastuck S, Huhse B, Leutwein C, Heurtier MA, Copley RR, Edelmann A, Querfurth E, Rybin V, Drewes G, Raida M, Bouwmeester T, Bork P, Seraphin B, Kuster B, Neubauer G, Superti-Furga G. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 4.Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, Millar A, Taylor P, Bennett K, Boutilier K, Yang L, Wolting C, Donaldson I, Schandorff S, Shewnarane J, Vo M, Taggart J, Goudreault M, Muskat B, Alfarano C, Dewar D, Lin Z, Michalickova K, Willems AR, Sassi H, Nielsen PA, Rasmussen KJ, Andersen JR, Johansen LE, Hansen LH, Jespersen H, Podtelejnikov A, Nielsen E, Crawford J, Poulsen V, Sorensen BD, Matthiesen J, Hendrickson RC, Gleeson F, Pawson T, Moran MF, Durocher D, Mann M, Hogue CWV, Figeys D, Tyers M. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- 5.Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, Rau C, Jensen LJ, Bastuck S, Dumpelfeld B, Edelmann A, Heurtier MA, Hoffman V, Hoefert C, Klein K, Hudak M, Michon AM, Schelder M, Schirle M, Remor M, Rudi T, Hooper S, Bauer A, Bouwmeester T, Casari G, Drewes G, Neubauer G, Rick JM, Kuster B, Bork P, Russell RB, Superti-Furga G. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- 6.Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, Punna T, Peregrin-Alvarez JM, Shales M, Zhang X, Davey M, Robinson MD, Paccanaro A, Bray JE, Sheung A, Beattie B, Richards DP, Canadien V, Lalev A, Mena F, Wong P, Starostine A, Canete MM, Vlasblom J, Wu S, Orsi C, Collins SR, Chandran S, Haw R, Rilstone JJ, Gandi K, Thompson NJ, Musso G, Onge PS, Ghanny S, Lam MHY, Butland G, Altaf-Ul AM, Kanaya S, Shilatifard A, O’Shea E, Weissman JS, Ingles CJ, Hughes TR, Parkinson J, Gerstein M, Wodak SJ, Emili A, Greenblatt JF. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 7.Bader GD, Betel D, Hogue CWV. BIND: the Biomolecular interaction network database. Nucleic Acids Res. 2003;31:248–250. doi: 10.1093/nar/gkg056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salwinski L, Miller CS, Smith AJ, Pettit FK, Bowie JU, Eisenberg D. The database of interacting proteins: 2004 update. Nucleic Acids Res. 2004;32:D449–D451. doi: 10.1093/nar/gkh086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guldener U, Munsterkotter M, Oesterheld M, Pagel P, Ruepp A, Mewes HW, Stumpflen V. MPact: the MIPS protein interaction resource on yeast. Nucleic Acids Res. 2006;34:D436–D441. doi: 10.1093/nar/gkj003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salwinski L, Eisenberg D. Computational methods of analysis of protein-protein interactions. Curr Opin Struct Biol. 2003;13:377–382. doi: 10.1016/s0959-440x(03)00070-8. [DOI] [PubMed] [Google Scholar]
- 11.Bork P, Jensen LJ, von Mering C, Ramani AK, Lee I, Marcotte EM. Protein interaction networks from yeast to human. Curr Opin Struct Biol. 2004;14:292–299. doi: 10.1016/j.sbi.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 12.Enright AJ, Iliopoulos I, Kyrpides NC, Ouzounis CA. Protein interaction maps for complete genomes based on gene fusion events. Nature. 1999;402:86–90. doi: 10.1038/47056. [DOI] [PubMed] [Google Scholar]
- 13.Marcotte EM, Pellegrini M, Ng HL, Rice DW, Yeates TO, Eisenberg D. Detecting protein function and protein-protein interactions from genome sequences. Science. 1999;285:751–753. doi: 10.1126/science.285.5428.751. [DOI] [PubMed] [Google Scholar]
- 14.Pellegrini M, Marcotte EM, Thompson MJ, Eisenberg D, Yeates TO. Assigning protein functions by comparative genome analysis: Protein phylogenetic profiles. Proc Natl Acad Sci USA. 1999;96:4285–4288. doi: 10.1073/pnas.96.8.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pazos F, Valencia A. Similarity of phylogenetic trees as indicator of protein-protein interaction. Protein Eng. 2001;14:609–614. doi: 10.1093/protein/14.9.609. [DOI] [PubMed] [Google Scholar]
- 16.Sato T, Yamanishi Y, Kanehisa M, Toh H. The inference of protein-protein interactions by co-evolutionary analysis is improved by excluding the information about the phylogenetic relationships. Bioinformatics. 2005;21:3482–3489. doi: 10.1093/bioinformatics/bti564. [DOI] [PubMed] [Google Scholar]
- 17.Matthews LR, Vaglio P, Reboul J, Ge H, Davis BP, Garrels J, Vincent S, Vidal M. Identification of potential interaction networks using sequence-based searches for conserved protein-protein interactions of “Interologs”. Genome Res. 2001;11:2120–2126. doi: 10.1101/gr.205301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wojcik J, Schachter V. Protein-protein interaction map inference using interacting domain profile pairs. Bioinformatics. 2001;17:S296–S305. doi: 10.1093/bioinformatics/17.suppl_1.s296. [DOI] [PubMed] [Google Scholar]
- 19.Wojcik J, Boneca IG, Legrain P. Prediction, assessment and validation of protein interaction maps in bacteria. J Mol Biol. 2002;323:763–770. doi: 10.1016/s0022-2836(02)01009-4. [DOI] [PubMed] [Google Scholar]
- 20.McDermott J, Samudrala R. Enhanced functional information from predicted protein networks. Trends Biotechnol. 2004;22:60–62. doi: 10.1016/j.tibtech.2003.11.010. [DOI] [PubMed] [Google Scholar]
- 21.Patil A, Nakamura H. HINT: a database of annotated protein-protein interactions and their homologs. Biophysics. 2005;1:21–24. doi: 10.2142/biophysics.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aloy P, Russell RB. Interrogating protein interaction networks through structural biology. Proc Natl Acad Sci USA. 2002;99:5896–5901. doi: 10.1073/pnas.092147999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu L, Lu H, Skolnick J. MULTIPROSPECTOR: An algorithm for the prediction of protein-protein interactions by multimeric threading. Proteins. 2002;49:350–364. doi: 10.1002/prot.10222. [DOI] [PubMed] [Google Scholar]
- 24.Lu L, Arakaki AK, Lu H, Skolnick J. Multimeric threading-based prediction of protein-protein interactions on a genomic scale: application to the Saccharomyces cerevisiae proteome. Genome Res. 2003;13:1146–1154. doi: 10.1101/gr.1145203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davis FP, Braberg H, Shen MY, Pieper U, Sali A, Madhusudhan MS. Protein complex compositions predicted by structural similarity. Nucleic Acids Res. 2006;34:2943–2952. doi: 10.1093/nar/gkl353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grigoryan G, Keating AE. Structure-based prediction of bZIP partnering specificity. J Mol Biol. 2006;355:1125–1142. doi: 10.1016/j.jmb.2005.11.036. [DOI] [PubMed] [Google Scholar]
- 27.Pieper U, Eswar N, Davis FP, Braberg H, Madhusudhan MS, Rossi A, Marti-Renom M, Karchin R, Webb BM, Eramian D, Shen MY, Kelly L, Melo F, Sali A. MODBASE: a database of annotated comparative protein structure models and associated resources. Nucleic Acids Res. 2006;34:D291–D295. doi: 10.1093/nar/gkj059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rubin GM, Yandell MD, Wortman JR, Gabor Miklos GL, Nelson CR, Hariharan IK, Fortini ME, Li PW, Apweiler R, Fleischmann W, Cherry JM, Henikoff S, Skupski MP, Misra S, Ashburner M, Birney E, Boguski MS, Brody T, Brokstein P, Celniker SE, Chervitz SA, Coates D, Cravchik A, Gabrielian A, Galle RF, Gelbart WM, George RA, Goldstein LS, Gong F, Guan P, Harris NL, Hay BA, Hoskins RA, Li J, Li Z, Hynes RO, Jones SJM, Kuehl PM, Lemaitre B, Littleton JT, Morrison DK, Mungall C, O’Farrell PH, Pickeral OK, Shue C, Vosshall LB, Zhang J, Zhao Q, Zheng XH, Zhong F, Zhong W, Gibbs R, Venter JC, Adams MD, Lewis S. Comparative genomics of the eukaryotes. Science. 2000;287:2204–2215. doi: 10.1126/science.287.5461.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gomperts BD, Kramer IM, Tatham PER. Signal Transduction. Academic Press; San Diego: 2002. Protein domains and signal transduction; pp. 393–410. [Google Scholar]
- 30.Deane CM, Salwinski L, Xenarios I, Eisenberg D. Protein Interactions: two methods for assessment of the reliability of high throughput observations. Mol Cell Prot. 2002;1:349–356. doi: 10.1074/mcp.m100037-mcp200. [DOI] [PubMed] [Google Scholar]
- 31.von Mering C, Krause R, Snel B, Cornell M, Oliver SG, Fields S, Bork P. Comparative assessment of large-scale data sets of protein-protein interactions. Nature. 2002;417:399–403. doi: 10.1038/nature750. [DOI] [PubMed] [Google Scholar]
- 32.Sprinzak E, Sattath S, Margalit H. How reliable are experimental protein-protein interaction data? J Mol Biol. 2003;327:919–923. doi: 10.1016/s0022-2836(03)00239-0. [DOI] [PubMed] [Google Scholar]
- 33.Kumar A, Agarwal S, Heyman JA, Matson S, Heidtman M, Piccirillo S, Umansky L, Drawid A, Jansen R, Liu Y, Cheung KH, Miller P, Gerstein M, Roeder GS, Snyder M. Subcellular localization of the yeast proteome. Genes & Dev. 2002;16:707–719. doi: 10.1101/gad.970902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O’Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 35.Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O’Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 36.Ben-Hur A, Noble WS. Choosing negative examples for the prediction of protein-protein interactions. BMC Bioinformatics. 2006;7:S2. doi: 10.1186/1471-2105-7-S1-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li MH, Wang XL, Lin L, Liu T. Effect of example weights on prediction of protein-protein interactions. Computat Biol Chem. 2006;30:386–392. doi: 10.1016/j.compbiolchem.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 38.Aloy P, Ceulemans H, Stark A, Russell RB. The relationship between sequence and interaction divergence in proteins. J Mol Biol. 2003;332:989–998. doi: 10.1016/j.jmb.2003.07.006. [DOI] [PubMed] [Google Scholar]
- 39.Henrick K, Thornton JM. PQS: a protein quaternary structure file server. Trends Biochem Sci. 1998;23:358–361. doi: 10.1016/s0968-0004(98)01253-5. [DOI] [PubMed] [Google Scholar]
- 40.Johnson RA, Wichern DW. Applied multivariate statistical analysis. Prentice-Hall; London: 1998. p. 740. [Google Scholar]
- 41.Wu CH, Apweiler R, Bairoch A, Natale DA, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M, Martin MJ, Mazumder R, O’Donovan C, Redaschi N, Suzek B. The universal protein resource (UniProt): an expanding universe of protein information. Nucleic Acids Res. 2006;34:D187–D191. doi: 10.1093/nar/gkj161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miyazawa S, Jernigan RL. Estimation of effective interresidue contact energies from protein crystal structures: Quasi-chemical approximation. Macromolecules. 1985;18:534–552. [Google Scholar]
- 44.Sippl MJ. Calculation of conformational ensembles from potentials of mean force: An approach to the knowledge-based prediction of local structures in globular proteins. J Mol Biol. 1990;213:859–883. doi: 10.1016/s0022-2836(05)80269-4. [DOI] [PubMed] [Google Scholar]
- 45.Jones DT, Taylor WR, Thornton JM. A new approach to protein fold recognition. Nature. 1992;358:86–89. doi: 10.1038/358086a0. [DOI] [PubMed] [Google Scholar]
- 46.Moont G, Gabb HA, Sternberg MJE. Use of pair potentials across protein interfaces in screening predicted docked complexes. Proteins. 1999;35:364–373. [PubMed] [Google Scholar]
- 47.Lu H, Lu L, Skolnick J. Development of unified statistical potentials describing protein-protein interactions. Biophys J. 2003;84:1895–1901. doi: 10.1016/S0006-3495(03)74997-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sheinerman FB, Norel R, Honig B. Electrostatic aspects of protein-protein interactions. Curr Opin Struct Biol. 2000;10:153–159. doi: 10.1016/s0959-440x(00)00065-8. [DOI] [PubMed] [Google Scholar]
- 49.Shaul Y, Schreiber G. Exploring the charge space of protein-protein association: A proteomic study. Proteins. 2005;60:341–352. doi: 10.1002/prot.20489. [DOI] [PubMed] [Google Scholar]
- 50.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Andreeva A, Howorth D, Brenner SE, Hubbard TJP, Chothia C, Murzin AG. SCOP database in 2004: refinements integrate structure and sequence family data. Nucleic Acids Res. 2004;32:D226–D229. doi: 10.1093/nar/gkh039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jeong H, Mason SP, Barabasi AL, Oltvai ZN. Lethality and centrality in protein networks. Nature. 2001;411:41–42. doi: 10.1038/35075138. [DOI] [PubMed] [Google Scholar]
- 53.Wagner A. The yeast protein interaction network evolves rapidly and contains few redundant duplicate genes. Mol Biol Evol. 2001;18:1283–1292. doi: 10.1093/oxfordjournals.molbev.a003913. [DOI] [PubMed] [Google Scholar]